The worldwide spread of multidrug-resistant Enterobacterales is a serious threat to public health. Here, we compared the MICs of plazomicin, amikacin, gentamicin, and tobramycin against 303 multinational multidrug-resistant Gram-negative bacilli. We followed Clinical and Laboratory Standards Institute (CLSI) guidelines and applied CLSI breakpoints as well as those of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) for amikacin, gentamicin, and tobramycin and of the U.S.
KEYWORDS: plazomicin, antimicrobial resistance, Enterobacterales
ABSTRACT
The worldwide spread of multidrug-resistant Enterobacterales is a serious threat to public health. Here, we compared the MICs of plazomicin, amikacin, gentamicin, and tobramycin against 303 multinational multidrug-resistant Gram-negative bacilli. We followed Clinical and Laboratory Standards Institute (CLSI) guidelines and applied CLSI breakpoints as well as those of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) for amikacin, gentamicin, and tobramycin and of the U.S. Food and Drug Administration for plazomicin. Overall, the highest percentage of susceptible isolates (80.2%) was demonstrated for plazomicin, which had the lowest MIC50 (1 μg/ml) of the aminoglycosides studied. Of the 42 isolates resistant to plazomicin, 34 had MICs of ≥128 μg/ml, with 33 of the 34 having MICs of >128 μg/ml for amikacin, gentamicin, and tobramycin. Among the 42 blaNDM-positive isolates, 35.7% were plazomicin susceptible, with the percentage of isolates susceptible to amikacin being 38.1% or 35.7% when applying the CLSI or EUCAST breakpoint, respectively. The 20 blaOXA-48-like-positive isolates showed 50.0% susceptibility to plazomicin. Among 35 isolates with blaCTX-M as their only characterized resistance mechanism, 68.6% were plazomicin susceptible, while the percentage susceptible to amikacin was 74.3% or 62.9% when applying the CLSI or EUCAST breakpoint, respectively. Among the 117 blaKPC-positive isolates, 94.9% were susceptible to plazomicin, whereas when the CLSI and EUCAST breakpoints were applied, 43.6% and 25.6%, respectively, were susceptible to amikacin; 56.4% and 44.4%, respectively, were susceptible to gentamicin; and 5.1% and 4.3%, respectively, were susceptible to tobramycin.
INTRODUCTION
The U.S. Centers for Disease Control and Prevention classifies carbapenem-resistant Enterobacteriaceae (CRE) to be an urgent threat to human health (1). Likewise, the World Health Organization’s Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics lists CRE, alongside 3rd-generation-cephalosporin-resistant Enterobacteriaceae, as one of the three Priority 1: Critical pathogens, together with carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa (2). Aminoglycosides are a consideration for the therapy of infections caused by multidrug-resistant aerobic Gram-negative bacilli. Discovered in the 1940s, they bind bacterial ribosomes and interfere with protein synthesis (3). Unfortunately, resistance to most commonly used aminoglycosides is increasing worldwide (4). There are four main mechanisms of aminoglycoside resistance. The most common involves aminoglycoside-modifying enzymes (AMEs), which are grouped into acetyltransferases, nucleotidyltransferases, and phosphotransferases (5). Another mechanism, 16S rRNA methyltransferases (16S-RMTases), confers high-level resistance to all available aminoglycosides but is not as common as AMEs (6). Finally, efflux, as well as the decreased permeability of aminoglycosides, has been described (7). Aminoglycosides that are less affected by these resistance mechanisms are needed.
Plazomicin (PLZ), formerly ACHN-490, is a semisynthetic derivate of sisomicin (8). Its structure avoids modification by most AMEs (9). AAC(2′)-Ia, carried by Providencia stuartii, confers plazomicin resistance, as does the AME APH (2ʺ)-Iva (8, 10, 11). We compared the activities of plazomicin, amikacin (AMK), gentamicin (GEN), and tobramycin (TOB) against 303 clinical isolates of Enterobacterales. Ceftolozane-tazobactam, imipenem-relebactam, imipenem, ertapenem, meropenem, ceftriaxone, and cefepime susceptibility was previously reported for 277 isolates (12).
RESULTS
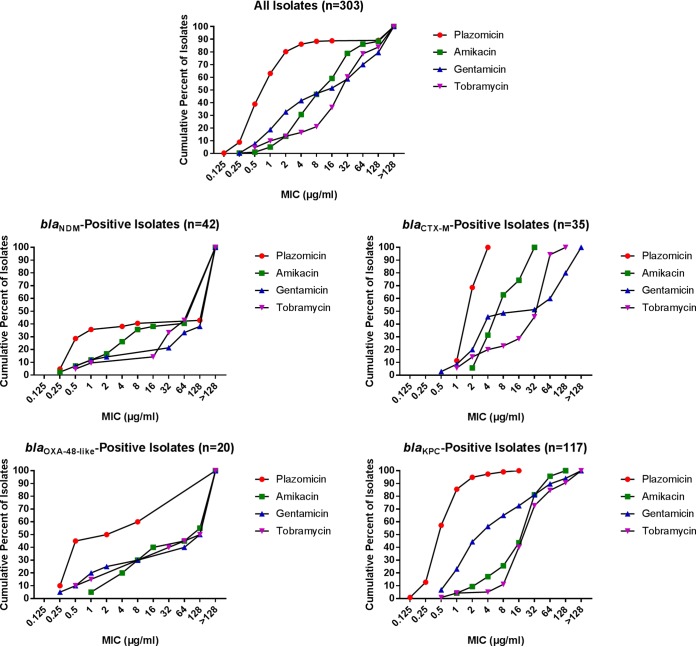
The cumulative numbers of isolates inhibited at specific concentrations are shown in Fig. 1, with the full data set being shown in Table S1 in the supplemental material. Table S2 shows the MIC50 and MIC90, as well as the percentage of isolates susceptible (S), intermediate (I; or susceptible increased exposure [IE]), and resistant (R). Overall, plazomicin was the aminoglycoside with the highest percentage of susceptible isolates. Using FDA breakpoints, 80.2% of the isolates were plazomicin susceptible, with 5.9% being intermediate and 13.9% being resistant (Table S2). Of the 42 isolates resistant to plazomicin, 33 had a MIC of >128 μg/ml, with one having a MIC of 128 μg/ml; the 34 isolates with plazomicin MICs of ≥128 μg/ml also had MICs of >128 μg/ml to amikacin, gentamicin, and tobramycin, except for 1 isolate, which had a tobramycin MIC of 16 μg/ml. Of the 42 isolates resistant to plazomicin, 26 were blaNDM positive, 10 were blaOXA-48-like positive, and 3 were blaKPC positive.
FIG 1.
Finlandograms showing the cumulative percentage of isolates inhibited at the specified concentrations (in micrograms per milliliter) for all isolates and specified groups. The blaCTX-M and blaOXA-48-like groups included isolates with those mechanisms as their only defined β-lactam resistance mechanism.
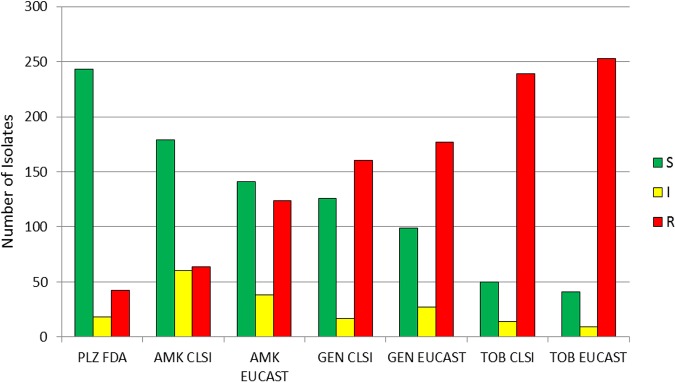
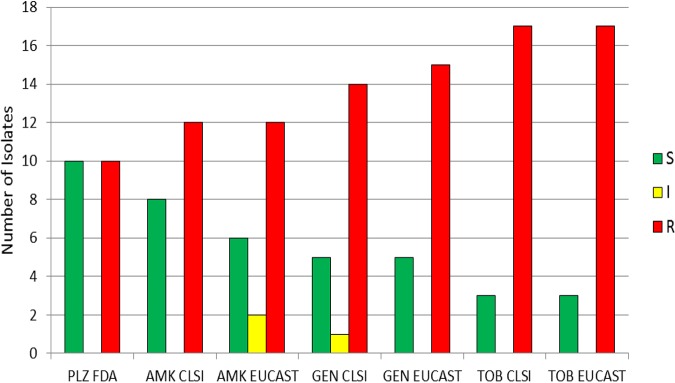
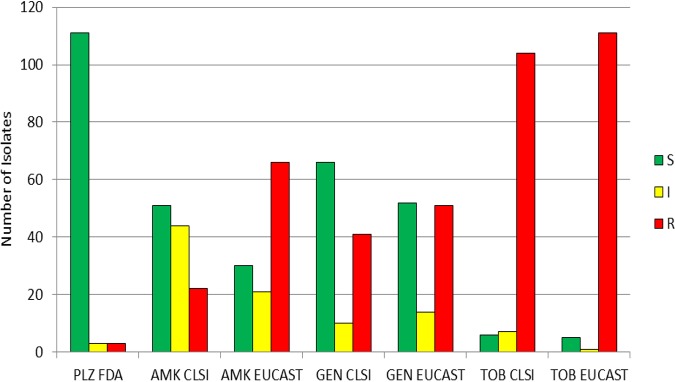
Applying European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria to the aminoglycosides tested generally led to lower rates of susceptibility and higher rates of resistance compared to what was found using Clinical and Laboratory Standards Institute (CLSI) criteria (Fig. 2). For example, the percentage of isolates susceptible to amikacin, gentamicin, and tobramycin was 59.1%, 41.6%, and 16.5%, respectively, using CLSI criteria, and 46.5%, 32.7%, and 13.5%, respectively, using EUCAST criteria. The percentage of isolates resistant to amikacin, gentamicin, and tobramycin was 21.1%, 52.8%, and 78.9%, respectively, using CLSI criteria and 40.9%, 58.4%, and 83.5%, respectively, using EUCAST criteria (Table S2).
FIG 2.
Number of isolates susceptible, intermediate, and resistant to aminoglycosides, determined using CLSI, EUCAST, and FDA breakpoints, including all isolates studied. S, susceptible; I, CLSI intermediate and EUCAST susceptible increased exposure categories; R, resistant; PLZ, plazomicin; AMK, amikacin; GEN, gentamicin; TOB, tobramycin.
blaNDM-positive isolates.
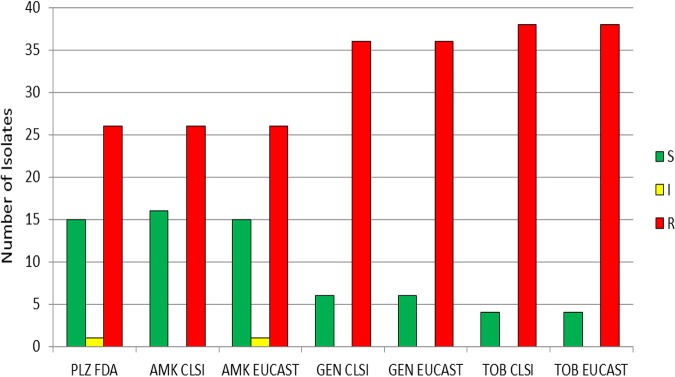
MICs were separately analyzed for the 42 blaNDM (New Dehli metallo-β-lactamase)-harboring isolates (Fig. 3). Using FDA breakpoints for plazomicin, 35.7% were susceptible, 2.4% were intermediate, and 61.9% were resistant (Table S2). A total of 38.1%, 14.3%, and 9.5% were susceptible to amikacin, gentamicin, and tobramycin, respectively, when applying CLSI breakpoints. A total of 61.9%, 85.7%, and 90.5% were resistant to amikacin, gentamicin, and tobramycin, respectively, when using CLSI breakpoints. Applying EUCAST breakpoints yielded similar results, with 35.7%, 14.3%, and 9.5% of the isolates being susceptible to amikacin, gentamicin, and tobramycin, respectively. A total of 61.9%, 85.7%, and 90.5% were resistant to amikacin, gentamicin, and tobramycin, respectively, using EUCAST breakpoints. Among the blaNDM-positive isolates, the percentage susceptible to amikacin and plazomicin was similar and higher than that the percentage susceptible to gentamicin or tobramycin. More than half of the blaNDM-positive isolates (57.1% for plazomicin and tobramycin, 59.5% for amikacin, and 61.9% for gentamicin) had MIC values >128 μg/ml for the studied aminoglycosides. The MIC90 and MIC50 for all aminoglycosides studied were >128 μg/ml for isolates harboring blaNDM (Table S2).
FIG 3.
Aminoglycoside susceptibility of isolates with the NDM (New Delhi metallo-β-lactamase) resistance mechanism (n = 42), determined using CLSI, EUCAST, and FDA breakpoints. S, susceptible; I, CLSI intermediate and EUCAST susceptible increased exposure categories; R, resistant; PLZ, plazomicin; AMK, amikacin; GEN, gentamicin; TOB, tobramycin.
blaCTX-M-positive isolates.
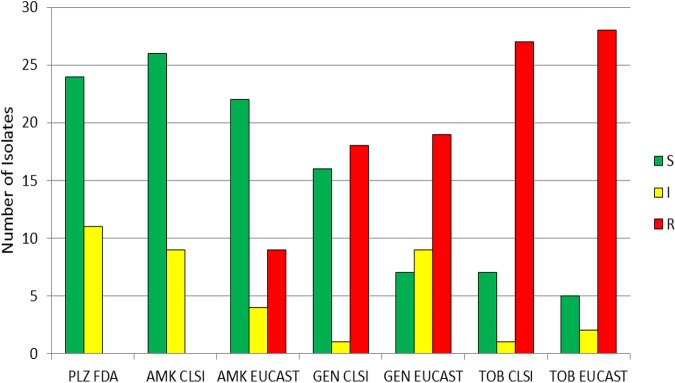
There were 35 isolates that possesed blaCTX-M as their only defined β-lactam resistance mechanism (Fig. 4). A total of 68.6% were susceptible, 31.4% were intermediate, and none were resistant using FDA breakpoints for plazomicin (Table S2). Using CLSI criteria, 74.3% were susceptible, 25.7% were intermediate, and none were resistant to amikacin; using EUCAST breakpoints, 62.9% were susceptible, 11.4% were IE, and 25.7% were resistant to amikacin. Applying CLSI breakpoints for gentamicin, 45.7% were susceptible, 2.9% were intermediate, and 51.4% were resistant, while 20.0% were susceptible, 25.7% were IE, and 54.3% were resistant using EUCAST criteria. Applying CLSI breakpoints for tobramycin, 20.0% were susceptible, 2.9% were intermediate, and 77.1% were resistant; 14.3% were susceptible, 5.7% were IE, and 80.0% were resistant using EUCAST breakpoints. Plazomicin had a MIC50 of 2 μg/ml and a MIC90 of 4 μg/ml for isolates harboring blaCTX-M as their only defined β-lactam resistance mechanism (Table S2). Amikacin’s MIC50 was 8 μg/ml and its MIC90 was 32 μg/ml for these isolates (Table S2). For these isolates, the gentamicin and tobramycin MIC50 was 32 and 64 μg/ml, respectively, and the MIC90 was >128 and 64 μg/ml, respectively. Analogous to the findings for the blaNDM-positive isolates, the percentage of isolates susceptible to plazomicin was similar to the percentage susceptible to amikacin, while fewer isolates were susceptible to gentamicin and tobramycin. Using CLSI criteria, the percentage of isolates susceptible to amikacin was higher than the percentage susceptible to plazomicin, while the percentage susceptible to amikacin was lower than the percentage susceptible to plazomicin using EUCAST breakpoints. For gentamicin, a difference was observed when comparing EUCAST and CLSI criteria, as nearly half (45.7%) of the isolates were gentamicin susceptible when applying CLSI criteria but only 20.0% were gentamicin susceptible when applying EUCAST criteria. Using EUCAST criteria, 9 of the 35 isolates were redefined from CLSI susceptible to the EUCAST IE category and 1 was redefined from CLSI intermediate to EUCAST resistant.
FIG 4.
Aminoglycoside susceptibility of isolates with the CTX-M (cefotaxime-hydrolyzing β-lactamase) resistance mechanism (n = 35) as their only defined β-lactam resistance mechanism, determined using CLSI, EUCAST, and FDA breakpoints. S, susceptible; I, CLSI intermediate and EUCAST susceptible increased exposure categories; R, resistant; PLZ, plazomicin; AMK, amikacin; GEN, gentamicin; TOB, tobramycin.
blaOXA-48-like-positive isolates.
We separately analyzed the 20 isolates carrying only blaOXA-48-like (Fig. 5). Using FDA breakpoints for plazomicin, 50.0% were susceptible and 50.0% were resistant to plazomicin (Table S2). Applying CLSI breakpoints, 40.0% were susceptible and 60.0% were resistant to amikacin. Thirty percent were susceptible, 10.0% were IE, and 60.0% were resistant using EUCAST breakpoints. By the CLSI criteria for gentamicin, 25.0% were susceptible, 5.0% were intermediate, and 70.0% were resistant; and by the EUCAST criteria, 25.0% were susceptible and 75.0% were resistant. Applying both CLSI and EUCAST criteria for tobramycin, 15.0% were susceptible and 85.0% were resistant. The MIC50 for plazomicin was 2 μg/ml and the MIC90 for plazomicin was >128 μg/ml for blaOXA-48-like-positive isolates (Table S2). Of the 10 isolates resistant to plazomicin, 8 had MIC values of >128 μg/ml. For amikacin, gentamicin, and tobramycin, the MIC50 was 128 μg/ml and the MIC90 was >128 μg/ml. Applying CLSI and EUCAST criteria, a higher percentage of isolates were susceptible to plazomicin than to amikacin; as many isolates were susceptible to plazomicin as were resistant to plazomicin. With both CLSI and EUCAST criteria, lower percentages of isolates were susceptible to gentamicin and tobramycin than to plazomicin and amikacin.
FIG 5.
Aminoglycoside susceptibility of isolates with only an OXA-48-like (oxacillin-hydrolyzing β-lactamase) resistance mechanism (n = 20), determined using CLSI, EUCAST, and FDA breakpoints. S, susceptible; I, CLSI intermediate and EUCAST susceptible increased exposure categories; R, resistant; PLZ, plazomicin; AMK, amikacin; GEN, gentamicin; TOB, tobramycin.
blaKPC-positive isolates.
In total, 117 isolates were blaKPC positive (Fig. 6). With FDA breakpoints, 94.9% were susceptible to plazomicin, while 2.6% were intermediate and 2.6% were resistant to plazomicin (Table S2). Using CLSI breakpoints for amikacin, 43.6% were susceptible, while 37.6% were intermediate and 18.8% were resistant; applying EUCAST breakpoints, 25.6% were susceptible, 17.9% were IE, and 56.4% were resistant to amikacin. Using CLSI criteria for gentamicin, 56.4% were susceptible, 8.5% were intermediate, and 35.0% were resistant. Applying EUCAST breakpoints for gentamicin, 44.4% were susceptible, 12.0% were IE, and 43.6% were resistant. Overall, more blaKPC-positive isolates were susceptible to gentamicin than to amikacin using CLSI and EUCAST breakpoints. Fewer isolates were susceptible to gentamicin than to plazomicin with either the CLSI or the EUCAST criteria. Applying CLSI breakpoints, 5.1% of isolates were susceptible to tobramycin, while 6.0% were intermediate and 88.9% were resistant; following EUCAST criteria, 4.3% were susceptible, 0.9% were IE, and 94.9% were resistant. The MIC50 for plazomicin was 0.5 μg/ml and the MIC90 was 2 μg/ml for blaKPC-positive isolates (Table S2). The MIC50 and MIC90 for amikacin were 32 and 64 μg/ml, respectively. For gentamicin, the MIC50 and MIC90 were 4 and 128 μg/ml, respectively, and for tobramycin, the MIC50 and the MIC90 were 32 and 128 μg/ml, respectively.
FIG 6.
Aminoglycoside susceptibility of isolates with the KPC (Klebsiella pneumoniae carbapenemase) resistance mechanism (n = 117), determined using CLSI, EUCAST, and FDA breakpoints. S, susceptible; I, CLSI intermediate and EUCAST susceptible increased exposure categories; R, resistant; PLZ, plazomicin; AMK, amikacin; GEN, gentamicin; TOB, tobramycin.
Amikacin-, gentamicin-, or tobramycin-resistant isolates.
Sixty-four of the 303 tested isolates were resistant to amikacin by applying CLSI criteria (Fig. 2). Of those 64, 39.1% were susceptible, 3.1% were intermediate, and 57.8% were resistant to plazomicin using FDA criteria. One hundred twenty-four isolates were resistant to amikacin according to EUCAST criteria, of which 64.5% were susceptible, 4.8% were intermediate, and 30.6% were resistant to plazomicin using the FDA criteria. Applying CLSI breakpoints, 160 of the tested isolates were resistant to gentamicin (Fig. 2); of the 160 gentamicin-resistant isolates, 68.1% were susceptible to plazomicin, 6.9% were intermediate, and 25.0% were resistant with FDA breakpoints. Using EUCAST breakpoints, 177 isolates were resistant to gentamicin; of those, 70.6% were susceptible, 6.2% were intermediate, and 23.2% were resistant to plazomicin with the FDA breakpoints. According to the CLSI criteria, 239 of the 303 tested isolates were resistant to tobramycin (Fig. 2), and of these, 77.4% were susceptible, 5.0% were intermediate, and 17.6% were resistant to plazomicin when applying the FDA breakpoints. Using the EUCAST criteria, 253 isolates were resistant to tobramycin (Fig. 2); of those 253 isolates, 77.5% were susceptible, 5.9% were intermediate, and 16.6% were resistant to plazomicin using the FDA breakpoints.
DISCUSSION
In our set of 303 multidrug-resistant aerobic Gram-negative bacilli with a diversity of underlying resistance mechanisms, the highest overall percent susceptibility was found for plazomicin, and among the commonly used aminoglycosides, plazomicin had the lowest MIC50, findings generally in accordance with those of other studies (13–17).
In our set of blaKPC-positive isolates, the highest percent susceptibility was found for plazomicin, at 94.9%, whereas when the CLSI and EUCAST breakpoints were applied, 43.6% and 25.6%, respectively, were susceptible to amikacin; 56.4% and 44.4%, respectively, were susceptible to gentamicin; and 5.1% and 4.3%, respectively, were susceptible to tobramycin. Notably, there was a difference in the characterization of amikacin resistance when using the CLSI and EUCAST breakpoints. Applying the CLSI breakpoint for amikacin, 18.8% of the blaKPC-positive isolates were resistant, whereas 56.4% were resistant using the EUCAST breakpoint. Among the blaCTX-M-positive isolates (which had no other defined resistance mechanism), 45.7% were susceptible to gentamicin using the CLSI breakpoint, while when applying the EUCAST breakpoint, only 20.0% were gentamicin susceptible. Looking at these blaCTX-M-positive isolates and using CLSI criteria, the highest percent susceptibility among the aminoglycosides tested was found for amikacin; however, applying EUCAST criteria, the percentage of isolates susceptible to plazomicin was higher than that to the other aminoglycosides tested. Differences between CLSI and EUCAST breakpoints, combined with the application of FDA breakpoints for plazomicin, impact interpretation of the activity of aminoglycosides. The reason(s) for the lack of susceptibility to plazomicin of some of the blaCTX-M-positive isolates is unknown and needs further study.
The findings for the blaNDM-positive isolates are notable. blaNDM was described in 2009 in a Swedish patient of Indian origin who had been hospitalized in New Delhi, India—hence the name New Delhi metallo-β-lactamase—before that patient was referred to Örebro in Sweden (18). In 2013, Hidalgo et al. described an association of the NDM carbapenemase with an aminoglycoside 16S-RMTase, RmtF (19). 16S-RMTases confer high-level pan-aminoglycoside resistance (HL-PAR); Taylor et al. described in 2018 that 94.5% of 806 pan-aminoglycoside-resistant Enterobacteriaceae from the United Kingdom and Ireland were positive for at least one 16S-RMTase (20). In 2019, Galani et al. reported on 300 Greek carbapenem-nonsusceptible isolates collected between November 2014 and April 2016, of which 50 were blaNDM positive. Twenty-three (7.7%) of the 300 isolates harbored 16S-RMTases, which led to high rates of aminoglycoside resistance, but none were NDM producing. All of their blaNDM-positive isolates were susceptible to plazomicin (14). Similarly, in a Brazilian study published in 2018 by Martins et al., the 69 blaNDM-positive isolates reported on had plazomicin MICs of ≤4 μg/ml (17). In our set of 42 blaNDM-positive isolates, 35.7% were susceptible to plazomicin, although the MIC50 and MIC90 were >128 μg/ml. Of the 26 plazomicin-resistant blaNDM-positive isolates, 1 had a MIC of 8 μg/ml, while the other 25 had MICs of ≥128 μg/ml. Overall, there appear to be geographically varying associations between the NDM carbapenemase and 16S-RMTases (21). Using FDA and EUCAST breakpoints for plazomicin and amikacin, respectively, in our set of blaNDM-positive isolates, an equal percentage of isolates would be considered susceptible; applying FDA and CLSI criteria, respectively, more isolates would be considered susceptible to amikacin than to plazomicin. Twenty-five blaNDM-positive isolates, one blaCTX-M-1-LIKE-positive isolate, one blaTEM-1-positive isolate, one blaOXA-48-like-positive isolate, one blaOXA-48-like-positive isolate, four blaOXA-181-positive isolates, and two blaOXA-232-positive isolates showed HL-PAR. These 34 (11.2%) isolates may harbor 16S-RMTases, increased efflux, multiple AMEs, or combinations thereof. It would be interesting to further evaluate the genetic profile of this group of 34 isolates.
There are limitations to this study, in that resistance mechanisms were selectively defined; we did not evaluate for efflux pumps or porin mutations and evaluated only a subset of β-lactamases. We did not assess for AMEs, 16S-RMTases, or other possible aminoglycoside resistance mechanisms. Further, the number of isolates harboring specific resistance mechanisms evaluated was limited, so we could not determine MIC50 and MIC90 values for isolates harboring every resistance mechanism specifically.
We compared the aminoglycoside susceptibility of the 277 previously evaluated isolates (12) with their susceptibility to imipenem-relebactam, imipenem, meropenem, ertapenem, ceftolozane-tazobactam, ceftriaxone and cefepime, applying CLSI breakpoints for all except imipenem-relebactam, for which FDA breakpoints were applied. Of the previously evaluated 277 isolates, 24 were resistant to all 11 antimicrobials (8.7%). Nineteen of those 24 isolates were blaNDM positive and 5 were blaOXA-48-like positive. Using EUCAST and FDA criteria, 23 isolates (8.3%) were resistant to all tested antimicrobials; of those, 19 were positive for blaNDM and 4 were positive for blaOXA-48-like. Of the 31 previously evaluated blaNDM-positive isolates, all were resistant to imipenem-relebactam, imipenem, ertapenem, meropenem, ceftolozane-tazobactam, cefepime, and ceftriaxone, using CLSI breakpoints; in this set of 31 isolates, 64.5% were resistant, 3.2% were intermediate, and 32.3% were susceptible to plazomicin, when applying FDA breakpoints. All 48 blaCTX-M-positive isolates were susceptible to imipenem-relebactam and imipenem, while 95.8% were susceptible to ertapenem, 100.0% were susceptible to meropenem, 83.3% were susceptible to ceftolozane-tazobactam, 12.5% were susceptible to cefepime, and 0.0% were susceptible to ceftriaxone. Using FDA breakpoints for plazomicin, 75.0% of the same 48 blaCTX-M-positive isolates were susceptible. One hundred eight blaKPC-positive isolates had been previously tested; of those, 92.6% were susceptible, 3.7% were intermediate, and 3.7% were resistant to imipenem-relebactam. The percentages of resistance to imipenem, ertapenem, meropenem, ceftolozane-tazobactam, cefepime, and ceftriaxone were between 88.0 and 97.2%. Of the blaKPC-positive isolates previously evaluated (12), 94.4% were susceptible, 2.8% were intermediate, and 2.8% were resistant to plazomicin, using FDA breakpoints.
In conclusion, plazomicin showed in vitro activity superior to that of amikacin, gentamicin, and tobramycin against this collection of clinically relevant multidrug-resistant aerobic Gram-negative bacilli.
MATERIALS AND METHODS
A total of 277 aerobic multidrug-resistant Gram-negative bacilli previously collected from the United States, Canada, and Singapore and genotypically characterized by select β-lactamase gene-specific PCR, with or without amplified product sequencing, were studied (22–32). Additionally, 26 carbapenemase-producing Enterobacterales were collected from the Mayo Clinic, Rochester, MN, and characterized by select β-lactamase gene PCR assays. All isolates carried one or more of blaCMY, blaCTX-M, blaFOX, blaIMI, blaIMP, blaKPC, blaNDM, blaOXA-48-like, blaSHV, blaSME, and blaTEM, with 198 harboring a carbapenemase gene. The isolates, which included 12 species/species complexes, including 157 Klebsiella pneumoniae complex isolates, 98 Escherichia coli isolates, 21 Enterobacter cloacae complex isolates, 8 Citrobacter freundii complex isolates, 5 Serratia marcescens isolates, 4 Citrobacter koseri isolates, 3 Klebsiella aerogenes isolates, 2 P. stuartii isolates, 2 Morganella morganii isolates, 1 Citrobacter sedlakii isolate, 1 Klebsiella oxytoca isolate, and 1 Proteus mirabilis isolate, were stored at −80°C in MicroBank vials (Pro-Lab Diagnostics, Round Rock, TX).
F1 and subsequent F2 generations of the study isolates were grown on BBL Trypticase soy agar with 5% sheep blood (Becton, Dickinson and Company, Sparks, MD). MICs were determined using broth microdilution following CLSI guidelines (33). E. coli ATCC 25922 was used as a quality control strain. Preparations of plazomicin (PLZ; Achaogen, South San Francisco, CA), amikacin (AMK; MilliporeSigma, St. Louis, MO), gentamicin (GEN; MilliporeSigma), and tobramycin (TOB) (MilliporeSigma) were made according to CLSI guidelines (33). The antibiotic concentrations tested ranged from 0.125 to 256 μg/ml. The MIC50 and MIC90, as well as the percentage and cumulative percentage S, percentage I (including the percentage in the IE category), and percentage R, were determined using CLSI (33, 34) and EUCAST (35) breakpoints. CLSI breakpoints for gentamicin and tobramycin of ≤4 μg/ml for S, 8 μg/ml for I, and ≥16 μg/ml for R and for amikacin of ≤16 μg/ml for S, 32 μg/ml for I, and ≥64 μg/ml for R were applied. EUCAST breakpoints for gentamicin and tobramycin (standard dosing regimen) of ≤2 μg/ml for S, 4 μg/ml for IE, and >4 μg/ml for R and for amikacin (standard dosing regimen) of <8 μg/ml for S, 16 μg/ml for IE, and >16 μg/ml for R were also applied. For plazomicin, FDA breakpoints of ≤2 μg/ml for S, 4 μg/ml for I, and ≥8 μg/ml for R (36) were applied.
Supplementary Material
ACKNOWLEDGMENTS
The plazomicin used in this study was generously provided by Achaogen. We acknowledge and thank the Clinical Bacteriology Laboratory staff at Mayo Clinic, especially Audrey N. Schuetz, Peggy C. Kohner, and Nicolynn C. Cole, and the Infectious Diseases Research Laboratory staff at Mayo Clinic, especially Suzannah M. Schmidt-Malan. We thank the following individuals and groups for providing isolates included in this study: D. Jane Hata from Mayo Clinic in Jacksonville, FL; Mary K. Hayden and Karen Lolans from Rush University Medical Center, Chicago, IL; Paul C. Schreckenberger from Loyola, Chicago, IL; James R. Johnson from the VA Medical Center in Minneapolis, MN, and the Hennepin County Medical Center in Minneapolis, MN; Patricia J. Simner from the Canadian Antimicrobial Resistance Alliance and the CANWARD study; George G. Zhanel and Daryl J. Hoban from the University of Manitoba; Koh Tse Hsien from the Singapore General Hospital; and Partha Pratim De, Sanjay Ryan Menon, and Shawn Vasoo from Tan Tock Seng Hospital in Singapore.
Wim A. Fleischmann reports that he is chief executive officer and a shareholder at SelfCare UG (Haftungsbeschränkt), registered in Germany as HRB 760532. Robin Patel reports grants from CD Diagnostics, Merck, Hutchison Biofilm Medical Solutions, Accelerate Diagnostics, Contrafect, TenNor Therapeutics Limited, and Shionogi. Robin Patel is a consultant to Curetis, Specific Technologies, Next Gen Diagnostics, PathoQuest, Selux Dx, and Qvella; monies are paid to Mayo Clinic. In addition, Robin Patel has a patent on Bordetella pertussis/B. parapertussis PCR issued, a patent on a device/method for sonication with royalties paid by Samsung to Mayo Clinic, and a patent on an antibiofilm substance issued. Robin Patel receives travel reimbursement from ASM and IDSA, an editor’s stipend from IDSA, and honoraria from the NBME, Up-to-Date, and the Infectious Diseases Board Review Course.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf. Accessed 21 April 2019.
- 2.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/. Accessed 21 April 2019.
- 3.Kotra LP, Haddad J, Mobashery S. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother 44:3249–3256. doi: 10.1128/aac.44.12.3249-3256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross AS, Opal S, Kopecko DJ. 1983. Progressive increase in antibiotic resistance of Gram-negative bacterial isolates. Walter Reed Hospital, 1976 to 1980: specific analysis of gentamicin, tobramycin, and amikacin resistance. Arch Intern Med 143:2075–2080. doi: 10.1001/archinte.1983.00350110053015. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballaben AS, Andrade LN, Galetti R, Ferreira JC, McElheny CL, Mettus RT, da Silva P, de Oliveira Garcia D, Darini ALC, Doi Y. 2018. Diversity of high-level aminoglycoside resistance mechanisms among Gram-negative nosocomial pathogens in Brazil. Antimicrob Agents Chemother 62:e01550-18. doi: 10.1128/AAC.01550-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garneau-Tsodikova S, Labby KJ. 2016. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm 7:11–27. doi: 10.1039/C5MD00344J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhanel GG, Lawson CD, Zelenitsky S, Findlay B, Schweizer F, Adam H, Walkty A, Rubinstein E, Gin AS, Hoban DJ, Lynch JP, Karlowsky JA. 2012. Comparison of the next-generation aminoglycoside plazomicin to gentamicin, tobramycin and amikacin. Expert Rev Anti Infect Ther 10:459–473. doi: 10.1586/eri.12.25. [DOI] [PubMed] [Google Scholar]
- 9.Aggen JB, Armstrong ES, Goldblum AA, Dozzo P, Linsell MS, Gliedt MJ, Hildebrandt DJ, Feeney LA, Kubo A, Matias RD, Lopez S, Gomez M, Wlasichuk KB, Diokno R, Miller GH, Moser HE. 2010. Synthesis and spectrum of the neoglycoside ACHN-490. Antimicrob Agents Chemother 54:4636–4642. doi: 10.1128/AAC.00572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin K, Clarke AJ. 2001. Overexpression and characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase of Providencia stuartii. Antimicrob Agents Chemother 45:2238–2244. doi: 10.1128/AAC.45.8.2238-2244.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox G, Ejim L, Stogios PJ, Koteva K, Bordeleau E, Evdokimova E, Sieron AO, Savchenko A, Serio AW, Krause KM, Wright GD. 2018. Plazomicin retains antibiotic activity against most aminoglycoside modifying enzymes. ACS Infect Dis 4:980–987. doi: 10.1021/acsinfecdis.8b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt-Malan SM, Mishra AJ, Mushtaq A, Brinkman CL, Patel R. 2018. In vitro activity of imipenem-relebactam and ceftolozane-tazobactam against resistant Gram-negative bacilli. Antimicrob Agents Chemother 62:e00533-18. doi: 10.1128/AAC.00533-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castanheira M, Davis AP, Mendes RE, Serio AW, Krause KM, Flamm RK. 2018. In vitro activity of plazomicin against Gram-negative and Gram-positive isolates collected from U.S. hospitals and comparative activities of aminoglycosides against carbapenem-resistant Enterobacteriaceae and isolates carrying carbapenemase genes. Antimicrob Agents Chemother 62:e00313-18. doi: 10.1128/AAC.00313-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galani I, Nafplioti K, Adamou P, Karaiskos I, Giamarellou H, Souli M, Study Collaborators. 2019. Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect Dis 19:167. doi: 10.1186/s12879-019-3801-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Diaz MD, Culebras E, Rodriguez-Avial I, Rios E, Vinuela-Prieto JM, Picazo JJ, Rodriguez-Avial C. 2017. Plazomicin activity against 346 extended-spectrum-beta-lactamase/AmpC-producing Escherichia coli urinary isolates in relation to aminoglycoside-modifying enzymes. Antimicrob Agents Chemother 61:e02454-16. doi: 10.1128/AAC.02454-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore DM, Mushtaq S, Warner M, Zhang JC, Maharjan S, Doumith M, Woodford N. 2011. Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J Antimicrob Chemother 66:48–53. doi: 10.1093/jac/dkq408. [DOI] [PubMed] [Google Scholar]
- 17.Martins AF, Bail L, Ito CAS, da Silva Nogueira K, Dalmolin TV, Martins AS, Rocha JLL, Serio AW, Tuon FF. 2018. Antimicrobial activity of plazomicin against Enterobacteriaceae-producing carbapenemases from 50 Brazilian medical centers. Diagn Microbiol Infect Dis 90:228–232. doi: 10.1016/j.diagmicrobio.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hidalgo L, Hopkins KL, Gutierrez B, Ovejero CM, Shukla S, Douthwaite S, Prasad KN, Woodford N, Gonzalez-Zorn B. 2013. Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in Enterobacteriaceae isolated in India and the UK. J Antimicrob Chemother 68:1543–1550. doi: 10.1093/jac/dkt078. [DOI] [PubMed] [Google Scholar]
- 20.Taylor E, Sriskandan S, Woodford N, Hopkins KL. 2018. High prevalence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacteriaceae in the UK and Ireland. Int J Antimicrob Agents 52:278–282. doi: 10.1016/j.ijantimicag.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Serio AW, Keepers T, Krause KM. 2019. Plazomicin is active against metallo-beta-lactamase-producing Enterobacteriaceae. Open Forum Infect Dis 6:ofz123. doi: 10.1093/ofid/ofz123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohner PC, Robberts FJ, Cockerill FR III, Patel R. 2009. Cephalosporin MIC distribution of extended-spectrum-β-lactamase- and pAmpC-producing Escherichia coli and Klebsiella species. J Clin Microbiol 47:2419–2425. doi: 10.1128/JCM.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasoo S, Cunningham SA, Cole NC, Kohner PC, Menon SR, Krause KM, Harris KA, De PP, Koh TH, Patel R. 2015. In vitro activities of ceftazidime-avibactam, aztreonam-avibactam, and a panel of older and contemporary antimicrobial agents against carbapenemase-producing Gram-negative bacilli. Antimicrob Agents Chemother 59:7842–7846. doi: 10.1128/AAC.02019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balm MN, La MV, Krishnan P, Jureen R, Lin RT, Teo JW. 2013. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect 19:E421–E423. doi: 10.1111/1469-0691.12247. [DOI] [PubMed] [Google Scholar]
- 25.Cunningham SA, Noorie T, Meunier D, Woodford N, Patel R. 2013. Rapid and simultaneous detection of genes encoding Klebsiella pneumoniae carbapenemase (blaKPC) and New Delhi metallo-beta-lactamase (blaNDM) in Gram-negative bacilli. J Clin Microbiol 51:1269–1271. doi: 10.1128/JCM.03062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh TH, Cao D, Shan QY, Bacon A, Hsu LY, Ooi EE. 2013. Acquired carbapenemases in Enterobacteriaceae in Singapore, 1996–2012. Pathology 45:600–603. doi: 10.1097/PAT.0b013e3283650b1e. [DOI] [PubMed] [Google Scholar]
- 27.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/aac.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. 2012. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac Surveill Response J 3:19–24. doi: 10.5365/WPSAR.2011.2.4.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teo JW, Kurup A, Lin RT, Hsien KT. 2013. Emergence of clinical Klebsiella pneumoniae producing OXA-232 carbapenemase in Singapore. New Microbes New Infect 1:13–15. doi: 10.1002/2052-2975.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teo JW, La MV, Krishnan P, Ang B, Jureen R, Lin RT. 2013. Enterobacter cloacae producing an uncommon class A carbapenemase, IMI-1, from Singapore. J Med Microbiol 62:1086–1088. doi: 10.1099/jmm.0.053363-0. [DOI] [PubMed] [Google Scholar]
- 31.Robberts FJ, Kohner PC, Patel R. 2009. Unreliable extended-spectrum beta-lactamase detection in the presence of plasmid-mediated AmpC in Escherichia coli clinical isolates. J Clin Microbiol 47:358–361. doi: 10.1128/JCM.01687-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham SA, Vasoo S, Patel R. 2016. Evaluation of the Check-Points Check MDR CT103 and CT103 XL microarray kits by use of preparatory rapid cell lysis. J Clin Microbiol 54:1368–1371. doi: 10.1128/JCM.03302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed CLSI document M07 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0 http://www.eucast.org. Accessed 15 May 2019.
- 36.U.S. Food and Drug Administration. Plazomicin—injection. FDA-identified interpretive criteria. https://www.fda.gov/drugs/development-resources/plazomicin-injection. Accessed 19 May 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.