LETTER
Recently, two novel plasmid-mediated tigecycline resistance genes, tet(X3) and tet(X4), were identified in Acinetobacter baumannii and Escherichia coli, respectively, both of which can significantly compromise the efficacy of tigecycline (1–3), which is one of the few available drugs that can be used to treat infections caused by extensively drug-resistant pathogens (4). The presence of tet(X) variant genes on conjugative plasmids significantly increases the speed of dissemination of tigecycline resistance among these pathogens (1). Furthermore, several studies indicated that ISCR2 may mediate the transposition process of tet(X4) (5, 6). Despite these initial reports, the existence of tet(X) genes and the mechanisms of their transfer need further exploration.
During our annual surveillance of antibiotic resistance among bacteria of food-producing animal origin in Guangdong Province, China, in 2018, a tet(X4)-positive strain 16EC was identified. The strain was recovered from a fecal swab sample from an apparently healthy pig and was identified as E. coli using both matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonik GmbH, Bremen, Germany) and 16S rRNA gene sequencing. Antimicrobial susceptibility testing was performed using the broth microdilution method according to the Clinical and Laboratory Standards Institute document M100S-S25 (7, 8) and with the EUCAST breakpoint (http://www.eucast.org/clinical_breakpoints/) for tigecycline. Strain 16EC was resistant to all tested tetracycline antibiotics, including tigecycline (MIC = 32 mg/liter), but susceptible to colistin, meropenem, ciprofloxacin, ceftazidime, and cefepime (see Table S1 in the supplemental material). S1-pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting were performed to determine the location of tet(X4) in the genome of 16EC, the result of which located tet(X4) on the bands with sizes of ∼125 kb, ∼30 kb, and ∼9 kb (Fig. S1).
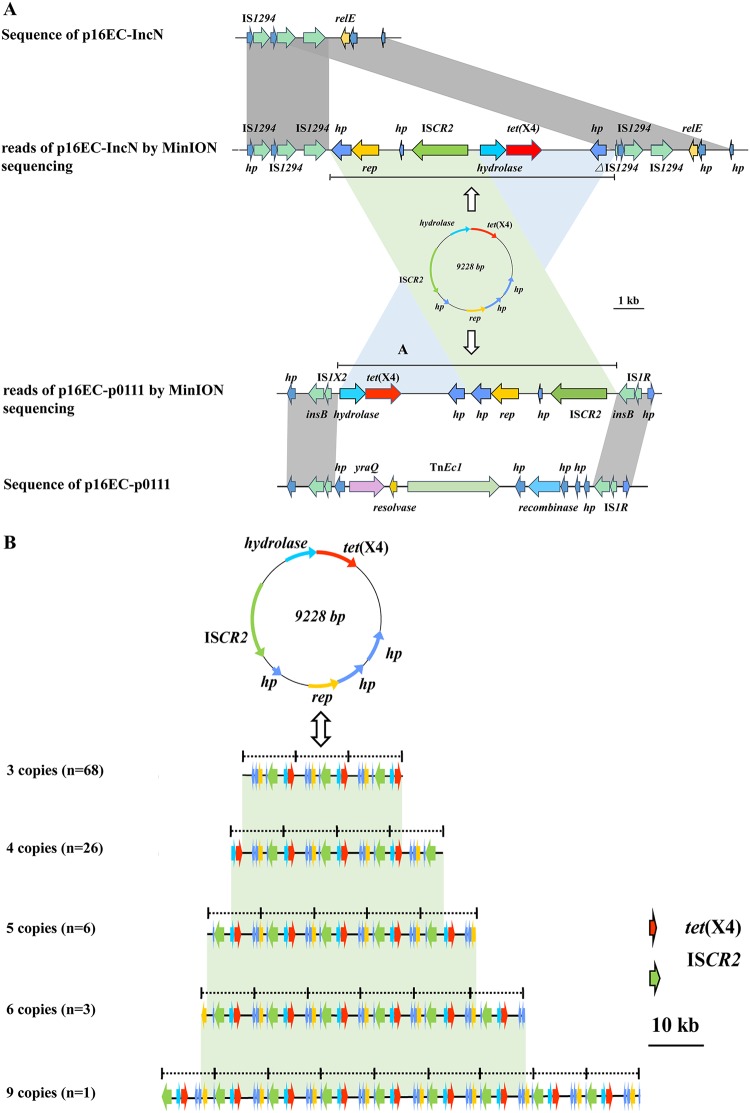
The genomic DNA of 16EC was then subjected to 300-bp paired-end whole-genome sequencing using the Illumina HiSeq 2500 system (Annoroad, Beijing, China), while the plasmid was sequenced using the MinION system (Oxford Nanopore Technologies, Oxford, UK); then, the high-quality de novo assembly was performed with Unicycler v0.4.7, which simultaneously combines short-read (Illumina reads) and long-read (MinION reads) data (9–11). A total of 0.5 Gb and 1.08 Gb of clean data were obtained from the Illumina sequencing and MinION sequencing platforms, respectively. The N50 value of the MinION sequencing was 18,245 bp. Sequencing results revealed the presence of three plasmids in 16EC, the 126,656-bp p0111-type plasmid p16EC-p0111, the 124,991-bp IncN-type plasmid p16EC-IncN, and a smaller 9,228-bp plasmid, p16EC-9K. p16EC-9K contained one intact copy of ISCR2 along with six other open reading frames (ORFs), encoding a Tet(X4), a replicon protein, a hydrolase, and three hypothetical proteins (Fig. 1). The rep gene showed 84% nucleotide sequence identity and 86% amino acid identity to the rep region of the ColE2-type plasmid pETEC58 (GenBank accession number FN649416.1) from E. coli (12). To further confirm the function of ColE2-like rep and to exclude the interference of ISCR2 in the transfer of p16EC-9K, we ligated a partial sequence of p16EC-9K without the ISCR2 element with an ampicillin resistance gene, ampR, and transformed the recombinant plasmid into DH5α. The tet(X4)-positive and ISCR2-negative transformants exhibited resistance to tigecycline (MIC, 16 mg/liter). S1-PFGE and Southern blotting results verified the existence of this recombinant plasmid in transformant (Fig. S2). These results confirmed the function of the ColE2-like replicon of p16EC-9K in E. coli.
FIG 1.
Characteristics of the novel ColE2-like plasmid p16EC-9K bearing tet(X4) and the tandem structure of multicopy of ISCR2-tet(X4). Arrows indicate genes and their directions. Regions in gray represent two linked areas with high sequence similarity. (A) Linear alignment of the original plasmid reads generated by MinION sequencing and the final polished sequences generated by HiSeq sequencing. A schematic of p16EC-9K is shown between the linear sequences. (B) Structure of the tandem tet(X4)-ISCR2 segment and diagram showing the various copy number arrangements detected in the sequencing reads.
Although sequencing data showed that tet(X4) was only located on the p16EC-9K plasmid, the ∼125-kb tet(X4)-carrying band confirmed by Southern blotting was missing from the two assembled plasmids, p16EC-p0111 and p16EC-IncN, of 16EC. To probe the reason, original reads of ≥20 kb were subjected to BLASTn comparison against the tet(X4), with strict cutoffs for alignment length and identity (coverage, ≥90%; identity, ≥85%). A total of 111 tet(X4)-positive reads of >20 kb in size were selected following screening of 47,151 original MinION sequencing reads and compared against the sequences of the two 16EC plasmids, p16EC-p0111 and p16EC-IncN, using BLASTn analysis. Six (37 kb to 90 kb) and one (34 kb) of the 111 reads showed high similarity to regions within p16EC-p0111 and p16EC-IncN, respectively (Fig. S3). The tet(X4)-carrying fragments in these seven reads showed high nucleotide sequence identity (>85%) to that of p16EC-9K (coverage, >99%). However, the arrangement of the p16EC-9K-associated ORFs differed among the long reads; ISCR2 was located upstream of tet(X4) in the p16EC-IncN reads but was located downstream of tet(X4) in the p16EC-p0111 reads (Fig. 1A). These results confirmed that tet(X4) was also located on the two ∼125-kb plasmids (p16EC-p0111 and p16EC-IncN), which is consistent with the result of Southern blotting. However, it remains largely unclear why the direction of p16EC-9K insertion into the two plasmids differed and how these processes occurred. We propose two possible formation processes. (i) In plasmid fusion, p16EC-9K was fused into p16EC-p0111 and p16EC-IncN mediated by IS1 and IS1294, respectively, through recombination (Fig. 1A). (ii) In transposition, p16EC-9K was transited into these two large plasmids by ISCR2, which differed from other insertion sequences in that it lacked terminal inverted repeats and is thought to transpose via a mechanism termed rolling-circle transposition (13, 14). As such, tet(X4) could theoretically be transposed using a single intact copy of ISCR2. Furthermore, the low number of tet(X4)-positive p16EC-p0111 (n = 6) and p16EC-IncN (n = 1) reads might account for why the tet(X4) gene was absent on these plasmids following de novo assembly of the MinION and Illumina HiSeq sequencing reads.
Unexpectedly, 104 of 111 original reads with various copy numbers of a tandem structure consisting of ISCR2 and tet(X4) [ISCR2-tet(X4)] were observed (Fig. 1B). Considering the size and the composition of these repeats, it seems likely that they were formed by the fusion of multiple p16EC-9K plasmids, which may be generated by rolling-circle replication through ISCR2. The results of Southern blotting, as the ∼30-kb tet(X4)-positive band, which may contain 3 copies of tet(X4)-ISCR2, further confirmed the existence of these structures. However, the specific mechanism remains unclear.
In conclusion, we characterized the polymorphism existence of the mobile tigecycline-resistant gene tet(X4) in E. coli 16EC. tet(X4) was located on three different Inc plasmids, p16EC-p0111, p16EC-IncN, and a novel ColE2-like plasmid, p16EC-9K, the latter of which may play an important role in the mobilization of tet(X4) through either recombination or rolling-circle transposition via insertion sequence (IS) elements. Further studies are needed to fully understand the mechanism of multicopy variant formation and the transfer mode of the tet(X4) gene.
Data availability.
The complete sequences of the plasmids p16EC-9K, p16EC-p0111, and p16EC-IncN have been deposited in NCBI under the GenBank accession numbers MN381965, MN086777, and MN086778, respectively. The original reads of p16EC-p0111 and p16EC-IncN containing tet(X4) have been deposited in the Figshare database (https://figshare.com/s/0b1391fe51e66212c58c).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Key R&D Program of China (2018YFD0500300) and the National Natural Science Foundation of China (81861138051 and 81661138002).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 2.Sun J, Chen C, Cui CY, Zhang Y, Liu X, Cui ZH, Ma XY, Feng Y, Fang LX, Lian XL, Zhang RM, Tang YZ, Zhang KX, Liu HM, Zhuang ZH, Zhou SD, Lv JN, Du H, Huang B, Yu FY, Mathema B, Kreiswirth BN, Liao XP, Chen L, Liu YH. 2019. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol 4:1457–1464. doi: 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai L, Du P, Du Y, Sun H, Zhang P, Wan Y, Lin Q, Fanning S, Cui S, Wu Y. 2019. Detection of plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from pork, Sichuan and Shandong Provinces, China, February 2019. Eurosurveillance 24:1900340 10.2807/1560-7917.ES.2019.24.25.1900340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brust K, Evans A, Plemmons R. 2014. Favourable outcome in the treatment of carbapenem-resistant Enterobacteriaceae urinary tract infection with high-dose tigecycline. J Antimicrob Chemother 69:2875–2876. doi: 10.1093/jac/dku185. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Chen L, Zhang Y, Cui CY, Wu XT, He Q, Liao XP, Liu YH, Sun J. 2019. Detection of chromosome-mediated tet(X4)-carrying Aeromonas caviae in a sewage sample from a chicken farm. J Antimicrob Chemother 74:3628–3630. doi: 10.1093/jac/dkz387. [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Cui CY, Zhang Y, He Q, Wu XT, Li G, Liao XP, Kreiswirth BN, Liu YH, Chen L, Sun J. 2019. Emergence of mobile tigecycline resistance mechanism in Escherichia coli strains from migratory birds in China. Emerg Microbes Infect 8:1219–1222. doi: 10.1080/22221751.2019.1653795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CLSI. 2018. Performance standards for antimicrobial susceptibility testing; 25th informational supplement M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.CLSI. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 4th ed, VET01-A4 and supplement VET01-S2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, Xie M, Dong N, Lin D, Yang X, Wong MHY, Chan EW, Chen S. 2018. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7:1–9. doi: 10.1093/gigascience/gix132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R, Chen K, Chan EW, Chen S. 2019. Characterization of the stability and dynamics of Tn6330 in an Escherichia coli strain by nanopore long reads. J Antimicrob Chemother 74:1807–1811. doi: 10.1093/jac/dkz117. [DOI] [PubMed] [Google Scholar]
- 12.Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, Savarino SJ, Turner SM, Pallen MJ, Penn CW, Parkhill J, Turner AK, Johnson TJ, Thomson NR, Smith SG, Henderson IR. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J Bacteriol 192:5822–5831. doi: 10.1128/JB.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacques M, Michael C. 2005. Insertion sequences. Microbiol Mol Biol R 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete sequences of the plasmids p16EC-9K, p16EC-p0111, and p16EC-IncN have been deposited in NCBI under the GenBank accession numbers MN381965, MN086777, and MN086778, respectively. The original reads of p16EC-p0111 and p16EC-IncN containing tet(X4) have been deposited in the Figshare database (https://figshare.com/s/0b1391fe51e66212c58c).