Whether multidrug resistance (MDR) is associated with mortality in patients with Pseudomonas aeruginosa bloodstream infections (BSI) remains controversial. Here, we explored the prognostic factors of P. aeruginosa BSI with emphasis on antimicrobial resistance and virulence. All P. aeruginosa BSI episodes in a 5-year period were retrospectively analyzed.
KEYWORDS: Pseudomonas aeruginosa, bloodstream infections, antimicrobial resistance, virulence, mortality
ABSTRACT
Whether multidrug resistance (MDR) is associated with mortality in patients with Pseudomonas aeruginosa bloodstream infections (BSI) remains controversial. Here, we explored the prognostic factors of P. aeruginosa BSI with emphasis on antimicrobial resistance and virulence. All P. aeruginosa BSI episodes in a 5-year period were retrospectively analyzed. The impact in early (5-day) and late (30-day) crude mortality of host, antibiotic treatment, and pathogen factors was assessed by multivariate logistic regression analysis. Of 243 episodes, 93 (38.3%) were caused by MDR-PA. Crude 5-day (20%) and 30-day (33%) mortality was more frequent in patients with MDR-PA (34.4% versus 11.3%, P < 0.001 and 52.7% versus 21.3%, P < 0.001, respectively). Early mortality was associated with neutropenia (adjusted odds ratio [aOR], 9.21; 95% confidence interval [CI], 3.40 to 24.9; P < 0.001), increased Pitt score (aOR, 2.42; 95% CI, 1.34 to 4.36; P = 0.003), respiratory source (aOR, 3.23; 95% CI,2.01 to 5.16; P < 0.001), inadequate empirical therapy (aOR, 4.57; 95% CI, 1.59 to 13.1; P = 0.005), shorter time to positivity of blood culture (aOR, 0.88; 95% CI, 0.80 to 0.97; P = 0.010), an exoU-positive genotype (aOR, 3.58; 95% CI, 1.31 to 9.79; P = 0.013), and the O11 serotype (aOR, 3.64; 95% CI, 1.20 to 11.1; P = 0.022). These risk factors were similarly identified for late mortality, along with an MDR phenotype (aOR, 2.18; 95% CI, 1.04 to 4.58; P = 0.040). Moreover, the O11 serotype (15.2%, 37/243) was common among MDR (78.4%, 29/37) and exoU-positive (89.2%, 33/37) strains. Besides relevant clinical variables and inadequate empirical therapy, pathogen-related factors such as an MDR phenotype, an exoU-positive genotype, and the O11 serotype adversely affect the outcome of P. aeruginosa BSI.
INTRODUCTION
Pseudomonas aeruginosa is a severe cause of bloodstream infections (BSI), with mortality rates above 30%, despite advances in medical care (1, 2). The presence of underlying diseases, the source of infection, and the severity of acute presentation are key host factors modulating prognosis (3, 4). Delayed adequate antimicrobial therapy is also independently associated with increased mortality (5, 6). In addition, pathogen-related factors, such as antimicrobial resistance and virulence traits are crucial elements which may affect the clinical outcomes of P. aeruginosa infections (7).
In this regard, a concern for P. aeruginosa infections is in the global emergence of multidrug resistant (MDR) and extensively drug resistant (XDR) strains, which limit the selection of effective antimicrobial therapies (8, 9). Successful selection of chromosomal mutations and the growing acquisition of transferable resistance determinants, particularly those encoding carbapenemases (e.g., GES, VIM, or IMP), are responsible for this increasing threat (10). Of note, some MDR/XDR strains, denominated high-risk clones, have a clonal epidemic population structure with limited sequence types (ST111, ST175, ST235) and a well-described ability to disseminate and cause severe infections (11, 12). However, despite the global spread of these high-risk clones, the real impact of multidrug resistance is still controversial. In many cases, MDR P. aeruginosa strains incur biological costs that compromise their pathogenic potential (13). However, this effect may vary significantly depending on the specific genetic context of the strains (14).
P. aeruginosa employs the toxins of the type III secretion system (TTSS) to interact with specific host targets and establish infection (15). Of the four TTSS effector proteins (ExoS, ExoT, ExoU, and ExoY), ExoU has been associated with poor outcomes in both clinical and experimental studies (16–19). In addition, it has been reported that some P. aeruginosa strains expressing lipopolysaccharide O-antigen serotypes, such as O1 and O11, may induce a worse prognosis in respiratory tract infections (20, 21). However, the correlation between resistance phenotype, TTSS genotype, and O serotype, and how these impact P. aeruginosa BSI, has not been consistently explored in clinical studies.
The assessment of host, pathogen, and treatment factors, which may account for the severity and mortality of P. aeruginosa BSI, may be of help in improving patient outcomes. Thus, the main objective of this study was to explore the prognostic factors affecting mortality in a large cohort of patients with P. aeruginosa BSI, with an emphasis on antimicrobial resistance and virulence.
(The preliminary results of this study were presented as a poster presentation at the XXIII Congress of the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC), Madrid, Spain, 23 to 25 May 2019.)
RESULTS
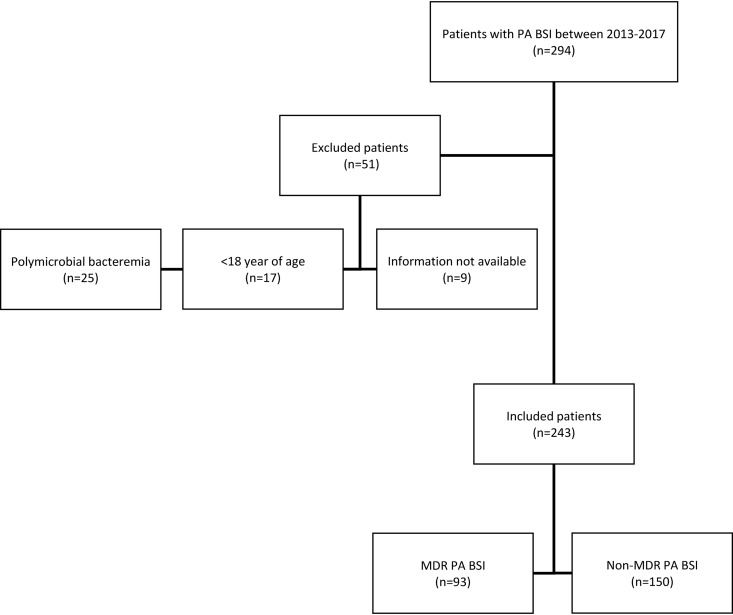
Among 294 patients with P. aeruginosa BSI, 51 were excluded from the study owing to polymicrobial bacteremia (n = 25), an age of less than 18 years (n = 17), or incomplete clinical or microbiological information (n = 9). Finally, 243 patients with laboratory-confirmed P. aeruginosa BSI were included in this study (Fig. 1).
FIG 1.
Study flow diagram. Non-duplicated clinical isolates from P. aeruginosa bloodstream infection patients between 2013 and 2017. Only the first episode of bacteremia recorded for each individual patient was included. MDR, multidrug resistant; PA, P. aeruginosa; BSI, bloodstream infections.
MDR versus non-MDR phenotype.
An MDR phenotype was documented in 93 (38.3%) isolates (87 [93.5%] were XDR and 6 [6.5%] were MDR non-XDR), while a non-MDR phenotype was observed in 150 (61.7%) isolates (127 [84.7%] were moderately resistant and 23 [15.3%] were multidrug susceptible). The main variables related to the MDR phenotype are detailed in Table 1. Patients with an MDR phenotype had a greater proportion of respiratory infections (35.5% versus 14.7%, P < 0.001) with a higher Pitt score (2 [1 to 3] versus 2 [0 to 3], P = 0.069) and septic shock (34.4% versus 22.7%, P = 0.064), compared to those with a non-MDR phenotype. Combined empirical antimicrobial therapy was used in 47.7% of cases and showed higher odds of being adequate in comparison with monotherapy (79.3% versus 49.6%, P < 0.001). Of note, inadequate empirical antimicrobial therapy was higher in patients with an MDR phenotype (59.1% versus 20.0%, P < 0.001). Moreover, an MDR phenotype determined significant differences in both early (34.4% versus 11.3%, P < 0.001) and late crude mortality (52.7% versus 21.3%, P < 0.001) (Fig. S1A and S1D in the supplemental material).
TABLE 1.
Comparative analysis of cases with MDR and non-MDR bloodstream infections caused by P. aeruginosa
| Variablee | Totala,d | MDRb | Non-MDRc | P |
|---|---|---|---|---|
| Median age (IQR) | 66.0 (55.0–77.0) | 63.0 (55.0–73.0) | 67.0 (55.0–78.0) | 0.079 |
| No. (%) of males | 161 (66.3) | 56 (60.2) | 105 (70.0) | 0.153 |
| Comorbidity | ||||
| Charlson comorbidity index | 2 (2–3) | 2 (2–4) | 2 (1–3) | 0.513 |
| No. diabetes mellitus (%) | 63 (25.9) | 31 (33.3) | 32 (21.3) | 0.054 |
| No. end-stage renal disease (%) | 41 (16.9) | 12 (12.9) | 29 (19.3) | 0.261 |
| No. solid malignancy (%) | 50 (20.6) | 19 (20.4) | 31 (20.7) | 0.965 |
| No. hematological malignancy (%) | 62 (25.5) | 31 (33.3) | 31 (20.7) | 0.040 |
| No. severe neutropenia (%) | 58 (23.9) | 30 (32.3) | 28 (18.7) | 0.024 |
| No. transplant (%) | 52 (21.4) | 19 (20.4) | 33 (22.0) | 0.897 |
| No. prior MDR colonization (%) | 34 (13.9) | 31 (33.3) | 3 (2.0) | <0.001 |
| No. ICU admission in previous 3 mo. (%) | 44 (18.1) | 17 (18.3) | 27 (18.0) | 0.956 |
| Prior invasive procedures | ||||
| No. venous catheter (%) | 146 (60.1) | 70 (75.3) | 76 (50.7) | <0.001 |
| No. urinary catheter (%) | 107 (44.0) | 41 (44.1) | 66 (44.0) | 0.989 |
| No. mechanical ventilation (%) | 36 (14.8) | 17 (18.3) | 19 (12.7) | 0.312 |
| No. surgery in previous mo. (%) | 81 (33.3) | 37 (39.8) | 44 (29.3) | 0.124 |
| No. antimicrobial therapy in previous mo. (%) | 170 (69.9) | 82 (88.2) | 88 (58.7) | <0.001 |
| No. carbapenems (%) | 54 (22.2) | 36 (38.7) | 18 (12.0) | <0.001 |
| No. fluoroquinolones (%) | 52 (21.4) | 29 (31.2) | 23 (15.3) | 0.006 |
| Acquisition type | ||||
| No. nosocomial (%) | 137 (56.4) | 71 (76.3) | 66 (44.0) | <0.001 |
| No. healthcare-associated (%) | 82 (33.7) | 22 (23.7) | 60 (40.0) | 0.013 |
| No. community (%) | 24 (9.9) | 0 (0) | 24 (16.0) | <0.001 |
| Ward of admission | ||||
| No. medical (%) | 120 (49.4) | 29 (31.2) | 91 (60.7) | <0.001 |
| No. onco-hematological (%) | 51 (21.0) | 28 (30.1) | 23 (15.3) | 0.009 |
| No. surgical (%) | 29 (11.9) | 14 (15.1) | 15 (10.0) | 0.328 |
| No. critical care (%) | 43 (17.7) | 22 (23.7) | 21 (14.0) | 0.081 |
| Primary source of infection | ||||
| No. high-risk source (%) | 150 (61.7) | 67 (72.0) | 83 (55.3) | 0.014 |
| No. unknown (%) | 26 (10.7) | 8 (8.6) | 18 (12.0) | 0.536 |
| No. respiratory (%) | 55 (22.6) | 33 (35.5) | 22 (14.7) | <0.001 |
| No. abdominal (%) | 50 (20.6) | 21 (22.6) | 29 (19.3) | 0.656 |
| No. soft tissue (%) | 19 (7.8) | 5 (5.4) | 14 (9.3) | 0.330 |
| Low-risk source | 93 (38.3) | 26 (27.9) | 67 (44.7) | 0.014 |
| No. urinary (%) | 67 (27.6) | 18 (19.4) | 49 (32.7) | 0.035 |
| No. vascular catheter (%) | 23 (9.5) | 7 (7.5) | 16 (10.7) | 0.557 |
| No. other (%) | 3 (1.2) | 1 (1.1) | 2 (1.3) | 1.000 |
| Clinical presentation | ||||
| No. Pitt bacteremia score ≥2 (%) | 135 (55.6) | 60 (64.5) | 75 (50.0) | 0.038 |
| No. septic shock (%) | 66 (27.2) | 32 (34.4) | 34 (22.7) | 0.064 |
| No. inadequate empiric antibiotic (%) | 85 (34.9) | 55 (59.1) | 30 (20.0) | <0.001 |
| Carbapenemase type/ST | ||||
| No. VIM-2/ST175 (%) | 43 (17.7) | 43 (46.2) | 0 (0) | <0.001 |
| No. GES-5/ST235 (%) | 33 (13.6) | 33 (35.5) | 0 (0) | <0.001 |
| TTSS genotype | ||||
| No. exoU+/exoS− (%) | 50 (20.6) | 33 (35.5) | 17 (11.3) | <0.001 |
| No. exoU−/exoS+ (%) | 185 (76.1) | 59 (63.4) | 126 (84.0) | <0.001 |
| No. exoU−/exoS− (%) | 8 (3.3) | 1 (1.1) | 7 (4.7) | 0.001 |
| O-antigen serotype | ||||
| No. O1 (%) | 38 (15.6) | 8 (8.6) | 30 (20.0) | 0.028 |
| No. O4 (%) | 50 (20.6) | 44 (47.3) | 6 (4.0) | <0.001 |
| No. O6 (%) | 38 (15.6) | 6 (6.5) | 32 (21.3) | <0.001 |
| No. O11 (%) | 37 (15.2) | 29 (31.2) | 8 (5.3) | <0.001 |
| No. other O-types (%) | 80 (32.9) | 6 (6.5) | 74 (49.3) | <0.001 |
| Median TTP (hours) of blood culture (IQR)f | 16.0 (12.0–19.0) | 16.0 (12.0–19.0) | 15.0 (12.0–18.0) | 0.553 |
| Outcome | ||||
| No. 5-day mortality (early mortality) (%) | 49 (20.2) | 32 (34.4) | 17 (11.3) | <0.001 |
| No. 30-day mortality (late mortality) (%) | 81 (33.3) | 49 (52.7) | 32 (21.3) | <0.001 |
Total cases; n = 243, 100%.
Cases with MDR infections; n = 93, 38.3%.
Cases with non-MDR infections; n = 150, 61.7%.
Data are expressed as n (%) or median (IQR).
MDR, multidrug-resistant; ICU, intensive care unit; ST, sequence type; TTSS, type III secretion system; TTP, time to positivity.
Time from the start of incubation to the alert signal in the blood culture system.
Antimicrobial susceptibility testing results for P. aeruginosa isolates are displayed in Table 2. Most XDR isolates were susceptible only to colistin (100%), amikacin (56.3%), and ceftazidime-avibactam (49.4%). For XDR strains, the most commonly identified carbapenemase was VIM-2 (43 strains [18.0%]), followed by GES-5 (33 strains [13.6%]). Pulsed-field gel electrophoresis showed two major epidemic clonal lineages within XDR strains (A = 33 and B = 43). Multilocus sequence typing analysis revealed that these XDR strains were frequently linked to high-risk clones, including ST175 (43 strains [18.0%]) and ST235 (33 strains [13.6%]). Table S1 shows a comparative analysis between MDR/XDR ST175 and ST235 high-risk clones. Patients with BSI caused by strains belonging to the ST235 clone presented a more severe clinical presentation and a poorer prognosis. Ceftolozane-tazobactam was only active against the noncarbapenemase-producing strains. Also, ceftazidime-avibactam was only active against GES-5 carbapenemase-producing isolates.
TABLE 2.
Antimicrobial susceptibility data for the 243 isolates of P. aeruginosa causing bloodstream infections
| No. of isolates susceptible to the indicated antibiotic (MIC [mg/liter]) (%)a: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate type (no.)b | PIP/TZ (≤16) | CAZ (≤8) | FEP (≤8) | ATM (≤16) | IMP (≤4) | MER (≤2) | GEN (≤4) | TOB (≤4) | AMI (≤8) | CIP (≤0.5) | COL (≤2) | TOL/TZ (≤4) | CAZ/AVI (≤8) |
| All (243) | 154 (63.4) | 152 (62.6) | 152 (62.6) | 26 (10.7) | 144 (59.3) | 144 (59.3) | 157 (64.6) | 163 (67.1) | 205 (84.4) | 133 (54.7) | 243 (100) | 10 (11.2) | 43 (49.4) |
| MDR (93) | |||||||||||||
| Non-XDR (6) | 2 (33.3) | 2 (33.3) | 2 (33.3) | 0 (0) | 4 (66.7) | 5 (91.7) | 5 (91.7) | 5 (91.7) | 6 (100) | 4 (66.7) | 6 (100) | ND | ND |
| XDR (87) | 2 (2.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (4.5) | 9 (10.1) | 49 (56.3) | 0 (0) | 87 (100) | 10 (11.2) | 43 (49.4) |
| VIM-2/ST175 (43) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 43 (100) | 0 (0) | 43 (100) | 0 (0) | 0 (0) |
| GES-5/ST235 (33) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 33 (100) | 0 (0) | 33 (100) |
| Othersc (11) | 2 (18.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (36.4) | 9 (81.8) | 6 (54.5) | 0 (0) | 11 (100) | 10 (90.9) | 10 (90.9) |
| Non-MDR (150) | |||||||||||||
| ModR (127) | 127 (100) | 127 (100) | 127 (100) | 3 (2.3) | 117 (92.2) | 116 (91.3) | 125 (98.4) | 126 (99.2) | 127 (100) | 106 (83.4) | 127 (100) | ND | ND |
| MultiS (23) | 23 (100) | 23 (100) | 23 (100) | 23 (100) | 23 (100) | 23 (100) | 23 (100) | 23 (100) | 23 (100) | 23 (100) | 23 (100) | ND | ND |
PIP/TZ, piperacillin-tazobactam; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; IMP, imipenem; MER, meropenem; GEN, gentamicin; TOB, tobramycin; AMI, amikacin; CIP, ciprofloxacin; COL, colistin; TOL/TZ, ceftolozane-tazobactam; CAZ/AVI, ceftazidime-avibactam; ND, not done.
MDR, multidrug resistant; XDR, extensively drug resistant; ModR, moderately resistant; MultiS, multidrug susceptible; ST, sequence type.
Eleven extensively drug-resistant P. aeruginosa isolates had no carbapenemase genes (blaGES, blaVIM, or blaIMP) identified.
exoU-positive versus exoU-negative genotype.
The presence of exoT and exoY genes was documented in most strains (235 [96.7%] and 227 [93.4%], respectively). Concomitantly, all strains were positive for either exoU or exoS genes (50 [20.6%] and 185 [76.1%], respectively), except for 8 (3.3%) strains that were negative for both genes. The main variables related to the exoU genotype are shown in Table S2. Patients with an exoU-positive genotype had a greater proportion of respiratory infections (36.0% versus 19.2%, P = 0.019) with a higher Pitt score (3 [1 to 4] versus 2 [0 to 3], P = 0.035) and septic shock (52.0% versus 20.7%, P < 0.001), compared to the exoU-negative genotype. Likewise, an exoU-positive genotype determined significant differences in both early (40.0% versus 15.0%, P < 0.001) and late (60.0% versus 26.4%, P < 0.001) crude mortality (Fig. S1B and S1E). Because exoT and exoY genotypes were mostly positive, these genes were not included in the analysis.
O11 versus non-O11 serotypes.
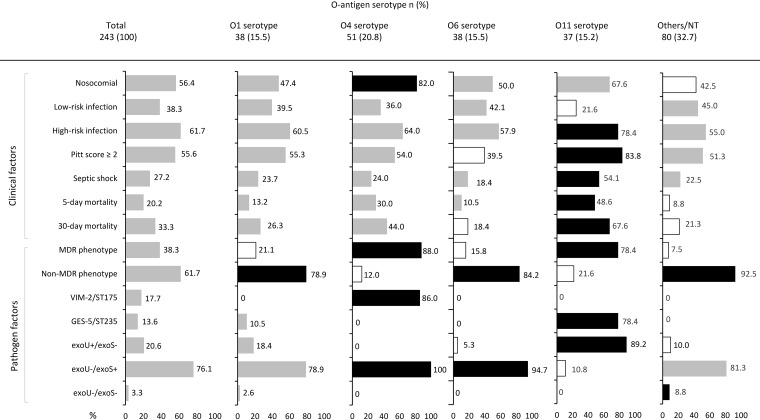
The O-antigen serotype was documented in most strains (213 [87.7%]), while 30 (12.3%) isolates were nontypeable. The most prevalent serotype was O4 (50 [20.6%]) followed by O1 (38 [15.6%]), O6 (38 [15.6%]), and O11 (37 [15.2%]) serotypes. The main characteristics according to O-antigen serotype are shown in Fig. 2. Patients infected with O11 serotype strains had a greater proportion of high-risk sources (78.4% versus 58.7%, P = 0.027), chiefly respiratory source, with a higher Pitt score (3 [2 to 3] versus 2 [0 to 3], P = 0.012) and septic shock (54.1% versus 22.3%, P < 0.001) compared to other O serotypes. In addition, there was a significant increase in 5-day and 30-day crude mortality in patients with an O11 serotype BSI compared to other O serotypes (48.6% versus 15.0%, P < 0.001 and 67.6% versus 27.2%, P < 0.001, respectively) (Fig. S1C and S1F).
FIG 2.
Clinical and pathogen factors in P. aeruginosa isolates from bloodstream infection patients according to O-antigen serotype. The proportion of isolates for each variable is indicated in the bar chart. Statistical significance (P ≤ 0.05) by χ2 or Fisher test is represented by colored bars (black, more prevalent; white, less prevalent). MDR, multidrug resistant; NT, nontypeable.
Association between O-antigen serotype, resistance phenotype, and TTSS genotype.
As shown in Fig. 2, serotypes O4 and O11 were more frequently associated with an MDR phenotype than were other O serotypes (88.0% versus 25.4%, P < 0.001 and 78.4% versus 31.1%, P < 0.001, respectively). In fact, within the MDR phenotype, a significant association was observed between O4 and O11 serotypes and the XDR phenotype (86.0% versus 22.8%, P < 0.001 and 78.4% versus 28.2%, P < 0.001, respectively). Likewise, within the XDR phenotype, the O4 serotype was identified in all the VIM-2/ST175 (43 [100%]) strains and the O11 serotype in most of GES-5/ST235 (29 [87.9%]) strains. Concomitantly, the O11 serotype was more frequently associated with the exoU-positive genotype than other O serotypes (89.2% versus 8.3%, P < 0.001). In contrast, O4 and O6 serotypes were more frequently associated with the exoU-negative genotype than were other O serotypes (100% versus 70.0%, P < 0.001 and 94.7% versus 72.5%, P = 0.002, respectively). In summary, the O4 serotype was strongly associated with the XDR phenotype (VIM-2/ST175 clone) and the exoU-negative genotype, whereas the O11 serotype was positively linked to the XDR phenotype (GES-5/ST235 clonal complex) and the exoU-positive genotype.
5-day and 30-day crude mortality.
Outcome was able to evaluated in 240 patients (98.8%). Among them, 46 (19.2%) died within 5 days. Univariate and multivariate analyses are shown in Table 3 and Table 4. A statistically significant inverse “dose-response” effect was observed between the time to positivity of blood culture and the mortality rate (Fig. S2). Because an interconnected nature has been observed of pathogen-related factors, which can result in collinearity, we performed a multivariate analysis using three separate models, including either the MDR phenotype or adequate empirical therapy, and the exoU genotype or O11 serotype. After adjustment for significant variables, host factors, including neutropenia, primary infection of respiratory tract, increased Pitt score, and a shorter time to positivity of blood culture, showed a significant association with increased 5-day mortality in all three models. In addition, inadequate empirical antimicrobial treatment (aOR, 4.57; 95% CI,1.59 to 13.1; P = 0.005), along with infection by an exoU-positive strain (aOR, 3.58; 95% CI, 1.31 to 9.79; P = 0.013), or by an O11 serotype strain (aOR, 3.64; 95% CI, 1.20 to 11.1; P = 0.022), proved to be independent predictors of 5-day mortality in each model. Bacteremia by an MDR isolate showed a trend toward higher mortality when adjusted by other parameters (aOR, 2.39; 95% CI, 0.97 to 5.87; P = 0.057).
TABLE 3.
Univariate analysis of predictors factors for 5-day and 30-day crude mortality of patients with P. aeruginosa bloodstream infections.
| Variablee | Early mortality (5-day)g |
Late mortality (30-day)g
|
||||
|---|---|---|---|---|---|---|
| Nonsurvivorsa | Survivorsb | P | Nonsurvivorsc | Survivorsd | P | |
| Median age (IQR) | 66.0 (57.0–77.0) | 58.0 (53.0–74.0) | 0.059 | 66.0 (57.0–77.0) | 62.0 (53.0–76.0) | 0.157 |
| No. (%) of males | 28 (60.9) | 131 (67.5) | 0.493 | 44 (56.4) | 115 (71.0) | 0.037 |
| Charlson comorbidity index | 2 (2–4) | 2 (2–3) | 0.767 | 2 (2–3) | 2 (1–3) | 0.776 |
| No. hematological malignancy (%) | 24 (52.2) | 37 (19.1) | <0.001 | 29 (37.2) | 32 (19.8) | 0.006 |
| No. severe neutropenia (%) | 26 (56.5) | 32 (16.5) | <0.001 | 33 (42.3) | 25 (15.4) | <0.001 |
| No. nosocomial acquisition (%) | 34 (73.9) | 101 (52.1) | 0.012 | 59 (75.6) | 76 (46.9) | <0.001 |
| No. high-risk source (%) | 44 (95.7) | 103 (53.1) | <0.001 | 66 (84.6) | 81 (50.0) | <0.001 |
| No. respiratory (%) | 29 (63.0) | 24 (12.4) | <0.001 | 35 (44.9) | 18 (11.1) | <0.001 |
| No. Pitt bacteremia score ≥2 (%) | 40 (86.9) | 92 (47.4) | <0.001 | 62 (79.5) | 70 (43.2) | <0.001 |
| No. septic shock (%) | 27 (58.7) | 37 (19.1) | <0.001 | 35 (44.9) | 29 (17.9) | <0.001 |
| No. inadequate empiric antibiotic (%) | 22 (47.8) | 63 (32.5) | 0.074 | 30 (38.5) | 55 (33.9) | 0.589 |
| No. MDR phenotype (%) | 31 (67.4) | 61 (31.4) | <0.001 | 48 (61.5) | 44 (27.2) | <0.001 |
| No. VIM-2/ST175 (%) | 13 (28.3) | 29 (14.9) | 0.055 | 20 (25.6) | 22 (13.6) | 0.034 |
| No. GES-5/ST235 (%) | 15 (32.6) | 18 (9.3) | <0.001 | 23 (29.5) | 10 (6.2) | <0.001 |
| No. exoU+/exoS− genotype (%) | 19 (41.3) | 30 (15.5) | <0.001 | 29 (37.2) | 20 (12.3) | <0.001 |
| No. O4 serotype (%) | 14 (30.4) | 35 (18.0) | 0.095 | 21 (26.9) | 28 (17.3) | 0.118 |
| No. O11 serotype (%) | 18 (39.1) | 19 (9.8) | <0.001 | 25 (32.1) | 12 (7.4) | <0.001 |
| Median TTP (hours) of blood culture (IQR)f | 14.0 (11.0–16.0) | 16.0 (12.0–19.0) | 0.005 | 14.0 (11.0–17.0) | 17.0 (13.0–19.0) | 0.001 |
Early nonsurvivors; n = 46, 19.2%.
Early survivors; n = 194, 80.8%.
Late nonsurvivors; n = 78, 32.5%.
Late survivors; n = 162, 67.5%.
MDR, multidrug-resistant; ST, sequence type; TTP, time to positivity.
Time from the start of incubation to the alert signal in the blood culture system.
Three patients were excluded from analysis due to palliative treatment, thus the total was n = 240.
TABLE 4.
Multivariate analysis of predictor factors for 5-day and 30-day crude mortality of patients with P. aeruginosa bloodstream infections.
| Variableb | 5-day mortality (early mortality)a,c |
30-day mortality (late mortality)c
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 |
Model 2 |
Model 3 |
Model 1 |
Model 2 |
||||||
| aOR (CI 95%) | P | aOR (CI 95%) | P | aOR (CI 95%) | P | aOR (CI 95%) | P | aOR (CI 95%) | P | |
| Severe neutropenia | 9.47 (3.52–25.5) | <0.001 | 6.57 (2.62–16.5) | <0.001 | 9.21 (3.40–24.9) | <0.001 | 2.97 (1.38–6.35) | 0.005 | 2.80 (1.32–5.92) | 0.007 |
| Respiratory infection | 3.22 (2.02–5.14) | <0.001 | 2.94 (1.87–4.62) | <0.001 | 3.23 (2.01–5.16) | <0.001 | 1.93 (1.30–2.86) | 0.001 | 1.95 (1.31–2.90) | 0.001 |
| Nosocomial acquisition | __ | __ | __ | __ | __ | __ | 1.62 (1.10–2.37) | 0.012 | 1.56 (1.07–2.27) | 0.019 |
| Pitt score ≥2 | 2.49 (1.39–4.44) | 0.002 | 2.14 (1.23–3.71) | 0.007 | 2.42 (1.34–4.36) | 0.003 | 1.86 (1.28–2.70) | 0.001 | 1.81 (1.25–2.63) | 0.002 |
| exoU+/exoS− genotype | 2.99 (1.06–8.42) | 0.038 | 3.58 (1.31–9.79) | 0.013 | 3.89 (1.65–9.19) | 0.002 | ||||
| O11 serotype | 3.64 (1.20–11.1) | 0.022 | 3.63 (1.42–9.31) | 0.007 | ||||||
| MDR phenotype | 2.39 (0.97–5.87) | 0.057 | 2.18 (1.04–4.58) | 0.04 | 2.17 (1.03–4.58) | 0.042 | ||||
| Inadequate empiric antibiotic | 4.57 (1.59–13.1) | 0.005 | 4.17 (1.42–12.2) | 0.009 | __ | __ | __ | __ | ||
| Median TTP (hours) of blood culture (IQR)d | 0.88 (0.80–0.97) | 0.009 | 0.90 (0.82–0.98) | 0.014 | 0.88 (0.81–0.97) | 0.01 | 0.91 (0.86–0.97) | 0.005 | 0.92 (0.86–0.97) | 0.008 |
aOR, adjusted odds ratio; CI, confidence interval.
MDR, multidrug-resistant; TTP, time to positivity; —, variables included in the initial model of multivariate analysis, then discarded in a stepwise backward selection process.
Three patients were excluded from analysis due to palliative treatment, thus the total was n = 240.
Time from the start of incubation to the alert signal in the blood culture system.
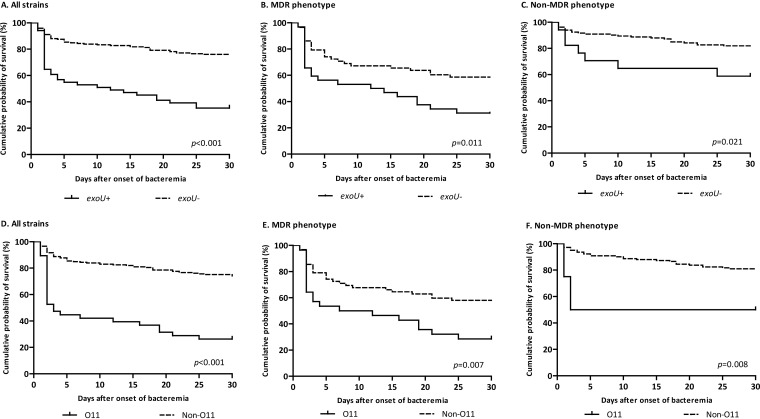
Seventy-eight (32.5%) patients died within 30 days. Similar risk factors for 30-day mortality were observed in the univariate and multivariate analyses (Table 3 and Table 4). The MDR phenotype proved to be an independent risk factor for mortality (aOR, 2.18; 95% CI, 1.04 to 4.58; P = 0.040). Again, a shorter time to positivity of blood culture was associated with higher mortality. Of interest, the nosocomial acquisition of bacteremia proved to be an independent predictor of 30-day mortality (aOR, 1.62; 95% CI, 1.10 to 2.37; P = 0.012). A stratified analysis according to MDR phenotype revealed that an exoU-positive genotype and O11 serotype were associated with increased 30-day mortality both in patients with an MDR phenotype (log-rank test, P = 0.011 and P = 0.007) and a non-MDR phenotype (log-rank test, P = 0.021 and P = 0.008) (Fig. 3).
FIG 3.
Kaplan-Meier curves showing the crude impact of exoU genotype (A, B, and C) and O11 serotype (D, E, and F) on 30-day mortality in patients with P. aeruginosa bloodstream infections according to the resistance phenotype. Statistical significance was determined by the log-rank test.
DISCUSSION
In this study, we have performed a detailed clinical and microbiological investigation to explore the risk factors affecting the prognosis of P. aeruginosa BSI. Our results confirm the high mortality associated with this infection, with 20% and 33% of patients dying within the first 5 and 30 days, respectively. This finding is consistent with recent data showing the high lethality of this condition compared with bacteremia caused by other microorganisms (22). We have shown that the poor observed outcome is the result of dynamic factors operating at the level of the host, the microorganism, and the antimicrobial therapy (14). With respect to the pathogen, both the development of antimicrobial resistance and its virulence must be addressed in order to have a complete perspective on the clinical problem.
After adjustment for potential confounders, our results suggest that patients with an MDR P. aeruginosa BSI have about roughly twice the odds of dying compared to patients infected with non-MDR isolates. Several studies have identified an association between antimicrobial resistance and an adverse clinical prognosis (1, 3, 5). However, the real impact of multidrug resistance is not so well established. In many cases, acquisition of antimicrobial resistance may be accompanied by a fitness cost and decreased virulence, therefore reducing disease severity and consequently mortality (7, 13, 14). However, this effect may vary significantly depending on the specific genetic context of the involved strains (14). Data presented here suggest that P. aeruginosa pathogenicity not only depends on the fitness cost of antimicrobial resistance, but also on the presence of some virulence determinants such as the TTSS genotype and the lipopolysaccharide O-antigen serotype.
Accordingly, we found that the exoU-positive genotype and the O11 serotype were risk factors for mortality, independently of other variables, including multidrug resistance (Fig. 3). The TTSS genotype is considered one of the most important virulence determinants of P. aeruginosa (15, 23). Of the four TTSS effector proteins (ExoS, ExoT, ExoU, and ExoY), ExoU has been associated with poor outcomes in both clinical and experimental research (16–19). A recent experimental study showed that the exoU gene was expressed and ExoU was produced early during acute pneumonia in a mouse model. This exotoxin possesses phospholipase A2 activity that causes rapid plasma membrane disruption and necrotic cell death. Therefore, it promotes bacterial transmigration by killing epithelial cells (17). In a Spanish multicenter study, Peña and colleagues elegantly showed the exoU-positive genotype to be an independent predictor of early mortality in P. aeruginosa BSI (18). Similarly, the O-antigen serotype has been used for the classification of P. aeruginosa isolates and plays an important immunogenic and structural role (20, 21, 24–26). Among P. aeruginosa isolates, O11 is one of the most prevalent serotypes worldwide and has been correlated with poorer prognosis in nosocomial pneumonia (20). In an experimental model of acute lung infection, serotype O11 was found to be associated with increased lung injury, probably related to the presence of ExoU (21). Unluckily, in our series most O11 serotype strains also carried the exoU-positive genotype, so it was difficult to distinguish whether this specific serotype behaved as a direct virulence factor or was only a surrogate marker of a hazardous isolate of P. aeruginosa. Therefore, definitive conclusions regarding the involvement of the O11 serotype in a poorer outcome should be taken with caution.
Thus, we were particularly interested in assessing the association between O serotypes, TTSS genotypes, and resistance phenotypes. In agreement with previous studies, we observed that O4 and O11 isolates exhibited mostly an MDR phenotype and an exoS-positive or exoU-positive genotype, respectively (24–26). Given this connection, the isolation of an O4 and/or O11 serotype P. aeruginosa is meaningful for the patient’s prognosis and for the empirical antibiotic choice. In our setting, O-antigen serotyping could be a simple, useful procedure for the rapid presumptive identification of MDR/XDR isolates susceptible only to colistin, amikacin, and some of them to ceftazidime-avibactam (8, 9). On the other hand, the identification of serotypes other than O4 or O11 would significantly decrease the chances of an MDR isolate. While this association has been shown in our hospital, the coincidence of the O4 and O11 serotypes with ST175 and ST235 international high-risk clones could make these observations of interest in many other locations (19, 27, 28). In fact, in a recent Spanish nationwide study, including 1445 P. aeruginosa isolates, Del Barrio-Tofiño and colleagues found that O4 and O11 serotypes are linked to the MDR/XDR profile of widespread ST175 and ST235 clones, respectively (29).
The strength of our multivariate analysis is reinforced by its adjustment with other relevant variables also influencing a patient’s prognosis. Severe neutropenia is a serious condition usually concurrent with underlying hematological diseases, which easily gives way to uncontrolled infection and death (30). The severity of the clinical presentation, especially in the setting of respiratory infection, has a well-documented impact on mortality (1, 2, 4, 5, 19). BSI nosocomially acquired was also an independent predictor of late mortality, and probably stands as a surrogate marker of a clinical patient’s complexity. In addition, inadequate initial empirical therapy has also been associated with poor prognosis (2, 5, 6), likely reflecting the low number of valid options in the setting of multidrug resistance.
The odds-ratio coefficients were also adjusted by the time to positivity of blood cultures. Although there’s potential for this to be influenced by the volume of blood inoculated in the bottles, or by an over-long delay in the sampling processing, the performance of blood cultures has become a standard, easy, and automatized procedure. From an overall perspective, the time to positivity stands as a surrogate marker of the inoculum, meaning that cases with a high bacterial burden will result in positive blood cultures sooner than infections of low inoculum. While our area under the receiver operating characteristic (ROC) curves were not good-enough to identify a precise time cutoff (not shown), Fig. S2 illustrates well the association of this variable with the likelihood of death, which has also been shown in other BSI (31).
Our study has the inherent limitations of a retrospective analysis. Although the confounding variables have been controlled for, they may be subject to the usual biases. Also, our study reflects the experience of just a single medical center and the results may not be applicable to other locations with a different epidemiology. In addition, we did not consider in our analysis the doses or optimized administration (e.g., extended-infusion) of antipseudomonal antibiotics. Despite this, our study provides some insights about the association between antimicrobial resistance and virulence traits, as well as the implications of host, pathogen, and antimicrobial treatment on patient´s outcomes.
In conclusion, MDR P. aeruginosa BSI represents a serious infection, associated with significant crude mortality. Overall, many of our MDR cases illustrate that the coexistence of specific virulence traits along with the acquisition of resistance determinants implies a “perfect storm” infection and a poorer prognosis for the patient. In this context, O-antigen serotyping is a tool potentially capable of rapid identification of MDR/XDR and virulent strains, thus guiding the choices of antimicrobial therapy, including novel β-lactam- β-lactamase inhibitor combinations, and supporting the close monitoring of patients.
MATERIALS AND METHODS
Study design.
This retrospective observational cohort study was conducted at the Hospital Universitario 12 de Octubre, a 1300-bed tertiary-care teaching hospital, in Madrid, Spain. The study included all patients with laboratory-confirmed P. aeruginosa BSI from January 2013 to December 2017. Only the first episode of bacteremia recorded for each individual patient was included. Nonduplicated clinical isolates from P. aeruginosa BSI patients were collected. Patients less than 18 years of age, with polymicrobial bacteremia, or those with incomplete medical records were excluded.
Ethical approval.
This study was approved by the Research Ethics Committee of our institution (Health Research Institute, Hospital Universitario 12 de Octubre, Madrid, Spain) (reference number TP17/0041), which exempted the need to seek written informed consent due to the observational nature of the study. All the data collected were anonymized.
Clinical variables and definitions.
Patient data were collected via chart review and included the following factors: (i) age; (ii) sex; (iii) comorbidities; (iv) severity of underlying diseases measured by the Charlson comorbidity index (32); (v) presence of severe neutropenia; (vi) antimicrobial treatment received in the previous month; (vii) prior known MDR P. aeruginosa colonization; (viii) intensive care admission in the previous 3 months; (ix) invasive procedures performed prior to the diagnosis of BSI (i.e., need for mechanical ventilation, use of venous catheter or urinary catheter); (x) surgery in the previous month; (xi) acquisition type (community, health care-associated, and nosocomial); (xii) ward of admission at the time of BSI (medical, onco-hematological, surgical, or intensive care); (xiii) source of bacteremia; (xiv) presentation with septic shock (33); (xv) Pitt bacteremia score (34); and (xvi) adequate empirical therapy. The main outcome variables were crude mortality at 5 days (early mortality) and 30 days (late mortality) after the onset of BSI.
Nosocomial bacteremia was defined as infection occurring more than 48 h after hospital admission. Healthcare-associated bacteremia was defined according to criteria previously described by Friedman et al. (35). Source of bacteremia was divided into 2 categories: (i) high-risk sources, which included the respiratory tract, intraabdominal, skin and soft tissues and those of unknown origin; and (ii) low-risk sources, which included urinary tract and vascular catheter (6). Septic shock was defined as sepsis associated with evidence of organ hypoperfusion and systolic blood pressure of <90 mm Hg or the need for vasopressors to maintain blood pressure (32). Severe neutropenia was defined as an absolute neutrophil count of <500 neutrophils/mm3. Adequate empirical antibiotic therapy was considered when at least 1 antipseudomonal antibiotic with in vitro activity was administered during the first 24 h after taking the blood sample. Crude mortality was defined as death by any cause.
Microbiological variables and molecular studies.
Blood cultures were processed using the BacT/Alert 3D blood culture system (bioMérieux, Marcy l´Etoile, France). Time to positivity, defined as the period between the start of incubation in the blood culture instrument and the automated growth signal, was documented by the system software and recorded automatically for each positive blood culture. Identification was carried out using MALDI-TOF mass spectrometry (Bruker Daltonics Inc., Bremen, Germany). Antimicrobial susceptibility testing was performed using a semiautomated microdilution system (MicroScan, Beckman Coulter diagnostics, Indianapolis, US), including the following antimicrobial agents: ceftazidime, cefepime, aztreonam, piperacillin-tazobactam, imipenem, meropenem, gentamicin, tobramycin, amikacin, ciprofloxacin, and colistin. MICs of ceftolozane-tazobactam and ceftazidime-avibactam were also determined by Etest (bioMérieux, Marcy l´Etoile, France). Breakpoints were in accordance with the European Committee on Antimicrobial Susceptibility Testing (EUCAST) v.8.1 (www.eucast.org).
MDR P. aeruginosa isolates were defined as strains nonsusceptible to at least 1 agent in 3 or more antipseudomonal antimicrobial categories. XDR isolates were defined as nonsusceptible to at least 1 agent in all but 2 or fewer antipseudomonal antimicrobial categories; thus, an XDR isolate was also included as MDR (36). All other P. aeruginosa isolates were considered non-MDR strains, and this category included moderately resistant (nonsusceptible to ≥1 agent in <3 antimicrobial categories) and multidrug-susceptible (susceptible to all antimicrobial agents) strains (18).
The detection of carbapenemase genes (blaGES, blaKPC, blaVIM, and blaIMP) was investigated by PCR and sequencing. Clonal relatedness among XDR isolates was first evaluated by pulsed-field gel electrophoresis (37). One representative XDR isolate was further analyzed by multilocus sequence typing (38) using an available database (https://pubmlst.org/paeruginosa/). The detection of exoS, exoT, exoU, and exoY genes was performed by PCR and sequencing as described previously (23). The O serotype was determined by agglutination using monovalent antiserum (Bio-Rad, Marnes-la-Coquette, France) to 16 somatic O-antigens as described previously (20). Nontypeable strains did not agglutinate with any antisera.
Statistical analysis.
The results were expressed as medians and interquartile ranges (IQR) for continuous variables or as absolute and relative frequencies for categorical variables. Continuous and categorical parameters were compared using the Student’s t test or the Mann-Whitney U test, and the χ2 test or Fisher’s exact test, respectively. Linear trends were assessed by the Mantle-Haenszel test. Independent risk factors of mortality were identified using a logistic regression model, including variables with P values of ≤0.1 in the univariate analysis. Given the high number of potential predictors of mortality, a backward stepwise algorithm was used to identify the best-fitting subset of variables for use in the final multivariate regression model. The likelihood ratio criteria were used for choosing the best model. Absence of collinearity among variables included in the initial model was verified. Kaplan-Meier survival curves were used for survival analysis, and the log-rank test was used to compare differences between groups. Patients who were discarded for active antimicrobial therapy due to end-stage disease were considered not evaluable for the analysis of crude mortality. All statistical tests were two-tailed and a P value of ≤0.05 was considered statistically significant. Analyses were performed using the SPSS statistical package v.20.0 (SPSS Inc., Chicago, IL, USA) and graphics were generated with Prism software v.5.0 (GraphPad Inc., La Jolla, CA, USA).
Supplementary Material
ACKNOWLEDGMENTS
We thank Mar Aguilera, Antonia Martín, and Esther Zabala (Department of Clinical Microbiology, Hospital Universitario 12 de Octubre, Madrid, Spain) for technical assistance.
This study was supported by Plan Nacional de I+D+ i 2013 to 2016, Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016), and cofinanced by the European Development Regional Fund via “A way to achieve Europe.” E. V. was also supported by a “Juan Rodés” fellowship grant (Instituto de Salud Carlos III).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Peña C, Suarez C, Gozalo M, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez F, Tubau F, Martínez-Martínez L, Oliver A. 2012. Prospective multicenter study of the impact of carbapenem resistance on mortality in Pseudomonas aeruginosa bloodstream infections. Antimicrob Agents Chemother 56:1265–1272. doi: 10.1128/AAC.05991-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peña C, Suarez C, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez F, Tubau F, Oliver A, Martínez-Martínez L. 2013. Effect of adequate single-drug vs combination antimicrobial therapy on mortality in Pseudomonas aeruginosa bloodstream infections: a post hoc analysis of a prospective cohort. Clin Infect Dis 57:208–216. doi: 10.1093/cid/cit223. [DOI] [PubMed] [Google Scholar]
- 3.Tam VH, Rogers CA, Chang KT, Weston JS, Caeiro JP, Garey KW. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa bacteremia on patient outcomes. Antimicrob Agents Chemother 54:3717–3722. doi: 10.1128/AAC.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joo E-J, Kang C-I, Ha YE, Kang S-J, Park SY, Chung DR, Peck KR, Lee NY, Song J-H. 2011. Risk factors for mortality in patients with Pseudomonas aeruginosa bacteremia: clinical impact of antimicrobial resistance on outcome. Microb Drug Resist 17:305–312. doi: 10.1089/mdr.2010.0170. [DOI] [PubMed] [Google Scholar]
- 5.Kang C-I, Kim S-H, Kim H-B, Park S-W, Choe Y-J, Oh M-D, Kim E-C, Choe K-W. 2003. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis 37:745–751. doi: 10.1086/377200. [DOI] [PubMed] [Google Scholar]
- 6.Kang C, Kim S, Park WB, Kim H, Kim E, Oh M, Choe K, Lee K. 2005. Bloodstream infections caused by antibiotic-resistant Gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother 49:760–766. doi: 10.1128/AAC.49.2.760-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beceiro A, Tomas M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev 26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Barrio-Tofiño E, López-Causapé C, Cabot G, Rivera A, Benito N, Segura C, Montero MM, Sorlí L, Tubau F, Gómez-Zorrilla S, Tormo N, Durá-Navarro R, Viedma E, Resino-Foz E, Fernández-Martínez M, González-Rico C, Alejo-Cancho I, Martínez JA, Labayru-Echverria C, Dueñas C, Ayestarán I, Zamorano L, Martinez-Martinez L, Horcajada JP, Oliver A. 2017. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother 61:e01589-17. doi: 10.1128/AAC.01589-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.del Barrio-Tofiño E, Zamorano L, Cortes-Lara S, López-Causapé C, Sánchez-Diener I, Cabot G, Bou G, Martínez-Martínez L, Oliver A, Galán F, Gracia I, Rodríguez MA, Martín L, Sánchez JM, Viñuela L, García MV, Lepe JA, Aznar J, López-Hernández I, Seral C, Javier Castillo-García F, López-Calleja AI, Aspiroz C, de la Iglesia P, Ramón S, Riera E, Cruz Pérez M, Gallegos C, Calvo J, Dolores Quesada M, Marco F, Hoyos Y, Pablo Horcajada J, Larrosa N, González JJ, Tubau F, Capilla S, Pérez-Moreno MO, Centelles MJ, Padilla E, Rivera A, Mirelis B, Elisa Rodríguez-Tarazona R, Arenal-Andrés N, del Pilar Ortega M, Megías G, García I, Colmenarejo C, González JC, Martínez NM, Gomila B, Giner S, Tormo N, Garduño E, Agulla JA, Seoane A, Pita J, Vidal IP, Guzmán DM, García M, Pérez del Molino ML, Barbeito G, Artiles F, Azcona-Gutiérrez JM, Sáenz Y, Antonio Oteo J, González A, Villa J, Chaves F, Cercenado E, Alarcón T, Zurita ND, Merino I, Morosini MI, Cantón R, Isabel Sánchez M, Moreno L, Yagüe G, Leiva J, Luis Barrios J, Canut A, Oteo J. 2019. Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J Antimicrob Chemother 74:1825–1835. doi: 10.1093/jac/dkz147. [DOI] [PubMed] [Google Scholar]
- 10.Potron A, Poirel L, Nordmann P. 2015. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 12.Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Geisinger E, Isberg RR. 2017. Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. J Infect Dis 215:S9–17. doi: 10.1093/infdis/jiw402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juan C, Peña C, Oliver A. 2017. Host and pathogen biomarkers for severe Pseudomonas aeruginosa infections. J Infect Dis 215:S44–51. doi: 10.1093/infdis/jiw299. [DOI] [PubMed] [Google Scholar]
- 15.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Solh AA, Hattemer A, Hauser AR, Alhajhusain A, Vora H. 2012. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Med 40:1157–1163. doi: 10.1097/CCM.0b013e3182377906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell HA, Logan LK, Hauser AR. 2013. Type III secretion of ExoU is critical during early Pseudomonas aeruginosa pneumonia. mBio 4:e00032. doi: 10.1128/mBio.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peña C, Cabot G, Gómez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez-López F, Tubau F, Martínez-Martínez L, Oliver A. 2015. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis 60:539–548. doi: 10.1093/cid/ciu866. [DOI] [PubMed] [Google Scholar]
- 19.Recio R, Villa J, Viedma E, Orellana MÁ, Lora-Tamayo J, Chaves F. 2018. Bacteraemia due to extensively drug-resistant Pseudomonas aeruginosa sequence type 235 high-risk clone: facing the perfect storm. Int J Antimicrob Agents 52:172–179. doi: 10.1016/j.ijantimicag.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Lu Q, Eggimann P, Luyt C-E, Wolff M, Tamm M, François B, Mercier E, Garbino J, Laterre P-F, Koch H, Gafner V, Rudolf MP, Mus E, Perez A, Lazar H, Chastre J, Rouby J-J. 2014. Pseudomonas aeruginosa serotypes in nosocomial pneumonia: prevalence and clinical outcomes. Crit Care 18:R17. doi: 10.1186/cc13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Berre R, Nguyen S, Nowak E, Kipnis E, Pierre M, Quenee L, Ader F, Lancel S, Courcol R, Guery BP, Faure K. 2011. Relative contribution of three main virulence factors in Pseudomonas aeruginosa pneumonia. Crit Care Med 39:2113–2120. doi: 10.1097/CCM.0b013e31821e899f. [DOI] [PubMed] [Google Scholar]
- 22.Thaden JT, Park LP, Maskarinec SA, Ruffin F, Fowler VG, van Duin D. 2017. Results from a 13-year prospective cohort study show increased mortality caused by Pseudomonas aeruginosa compared to other bacteria. Antimicrob Agents Chemother 61:e02671-16. doi: 10.1128/AAC.02671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feltman H, Jain M, Peterson L, Schulert G, Khan S, Hauser AR. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659–2669. doi: 10.1099/00221287-147-10-2659. [DOI] [PubMed] [Google Scholar]
- 24.Faure K, Shimabukuro D, Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. 2003. O-antigen serotypes and type III secretory toxins in clinical isolates of Pseudomonas aeruginosa. J Clin Microbiol 41:2158–2160. doi: 10.1128/jcm.41.5.2158-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamasbi RJ, Proudfoot EM. 2008. Phenotypic and genotypic characteristics of clinical isolates of Pseudomonas aeruginosa: rate of occurrence and distribution of different serotypes, antimicrobial susceptibility profiles, and molecular typing. Lab Med 39:155–161. doi: 10.1309/1BAWW0951N7V71CE. [DOI] [Google Scholar]
- 26.Berthelot P, Attree I, Plésiat P, Chabert J, de Bentzmann S, Pozzetto B, Grattard F. 2003. Genotypic and phenotypic analysis of type III secretion system in a cohort of Pseudomonas aeruginosa bacteremia isolates: evidence for a possible association between O serotypes and exo genes. J Infect Dis 188:512–518. doi: 10.1086/377000. [DOI] [PubMed] [Google Scholar]
- 27.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother 53:4930–4933. doi: 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viedma E, Juan C, Villa J, Barrado L, Ángeles Orellana M, Sanz F, Otero JR, Oliver A, Chaves F. 2012. VIM-2-producing multidrug-resistant Pseudomonas aeruginosa ST175 clone, Spain. Emerg Infect Dis 18:1235–1241. doi: 10.3201/eid1808.111234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Barrio-Tofiño E, Sánchez-Diener I, Zamorano L, Cortes-Lara S, López-Causapé C, Cabot G, Bou G, Martínez-Martínez L, Oliver A, Galán F, Gracia I, Rodríguez MA, Martín L, Sánchez JM, Viñuela L, García MV, Lepe JA, Aznar J, López-Hernández I, Seral C, Castillo-García FJ, López-Calleja AI, Aspiroz C, de la Iglesia P, Ramón S, Riera E, Pérez MC, Gallegos C, Calvo J, Quesada MD, Marco F, Hoyos Y, Horcajada JP, Larrosa N, González JJ, Tubau F, Capilla S, Pérez-Moreno MO, Centelles MJ, Padilla E, Rivera A, Mirelis B, Rodríguez-Tarazona RE, Arenal-Andrés N, del Pilar Ortega M, Megías G, García I, Colmenarejo C, González JC, Martínez NM, Gomila B, Giner S, Tormo N, Garduño E, Agulla JA, Seoane A, Pita J, Vidal IP, Guzmán DM, García M, Pérez del Molino ML, Barbeito G, Artiles F, Azcona-Gutiérrez JM, Sáenz Y, Oteo JA, González A, Villa J, Chaves F, Cercenado E, Alarcón T, Zurita ND, Merino I, Morosini MI, Cantón R, Sánchez MI, Moreno L, Yagüe G, Leiva J, Barrios JL, Canut A, Oteo J. 2019. Association between Pseudomonas aeruginosa O-antigen serotypes, resistance profiles and high-risk clones: results from a Spanish nationwide survey. J Antimicrob Chemother 74:3217–3220. doi: 10.1093/jac/dkz346. [DOI] [PubMed] [Google Scholar]
- 30.Marin M, Gudiol C, Ardanuy C, Garcia-Vidal C, Calvo M, Arnan M, Carratalà J. 2014. Bloodstream infections in neutropenic patients with cancer: differences between patients with haematological malignancies and solid tumours. J Infect 69:417–423. doi: 10.1016/j.jinf.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 31.Liao CH, Lai CC, Hsu MS, Huang YT, Chu FY, Hsu HS, Hsueh PR. 2009. Correlation between time to positivity of blood cultures with clinical presentation and outcomes in patients with Klebsiella pneumoniae bacteraemia: prospective cohort study. Clin Microbiol Infect 15:1119–1125. doi: 10.1111/j.1469-0691.2009.02720.x. [DOI] [PubMed] [Google Scholar]
- 32.Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 33.Singer M, Deutschman CS, Seymour C, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, Poll T, Der, Vincent JL, Angus DC. 2016. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow JW, Fine MJ, Shlaes DM, Quinn JP, Hooper DC, Johnson MP, Ramphal R, Wagener MM, Miyashiro DK, Yu VL. 1991. Enterobacter bacteremia: clinical features and emergence of antibiotic resistance during therapy. Ann Intern Med 115:585. doi: 10.7326/0003-4819-115-8-585. [DOI] [PubMed] [Google Scholar]
- 35.Friedman ND, Kaye KS, Stout JE, Mcgarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, Macfarquhar J, Walton AL, Reller LB, Sexton DJ. 2002. Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med:791–798. doi: 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]
- 36.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 37.Tenover FC, Arbeit RD, Goering RV, Mickelsen Pa, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis : criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.