Abstract
Retinal disease represents a growing global problem, both in terms of quality of life and economic impact, yet new therapies are not being developed at a sufficient rate to meet this mounting need. In this context, retinal organoids derived from human induced pluripotent stem cells hold significant promise for improving upon the current drug development process, increasing the speed and efficiency of moving potential therapeutic agents from bench to bedside. These organoid systems display the cell–cell and cell–matrix interactions, cellular heterogeneity, and physiological responses reflective of human biology and, thus, have the ability to replicate retinal disease pathology in a way that 2-dimensional cell cultures and animal models have been heretofore unable to achieve. However, organoid technology is not yet mature enough to meet the high-throughput demands of the first stages of drug screening. Hence, the augmentation of the existing drug development pipeline with retinal organoids, rather than the replacement of existing pathway components, may provide a way to harness the benefits of this improved pathological modeling. In this study, we outline the possible benefits of such a symbiosis, discuss other potential uses, and highlight barriers that remain to be overcome.
Keywords: retina, drug screening, hiPSC, organoids, cell culture models
Introduction
The retina, the means through which most people primarily perceive the world, is a complex and delicate organ. Part of the central nervous system, the human retina lacks regenerative capacity, and thus, its damage results in permanent vision impairment or total blindness.
Retinal degenerative diseases are debilitating conditions that affect millions of people worldwide. Globally it is estimated that 8.4 million people suffer from age related macular degeneration (AMD), 4 million from glaucoma, 2.6 million from diabetic retinopathy,1 and over 1 million from retinitis pigmentosa.2 What's more, the number of people affected appears to be increasing with global population age.1 This raises significant concerns, both for individual quality of life and for economic impact, as the annual financial burden of adult visual disorders surpasses $35 billion in the United States alone.3
Despite this growing problem, the treatment and management of retinal diseases remain challenging. While novel regenerative strategies are being devised in research laboratories, most of these potential therapies have not yet made their way to the clinic, leaving current patients to rely on traditional approaches focused on disease prevention or progress retardation that are often insufficient.
The clear need for new therapeutic drugs for retinal diseases has been met with a relative lack of progress. As of 2015, only 13.8% of drugs across all indications were estimated to progress from phase I clinical trials to approval.2 Drugs that do reach the market will have accrued an average development cost of five billion dollars of R&D.3 We argue that this is due, in part, to the current inefficiencies of the drug development pipeline.
Traditionally, drug screening begins in 2-dimensional (2D), dissociated cell cultures, often based on immortalized cell lines. However, 2D cultures carry inherent inaccuracies as they lack the cellular diversity, organization, and mechanical and chemical signaling dynamics inherent to the 3-dimensional (3D) biology found in vivo. Thus, intricacies inherent to cellular physiology and, importantly, pathophysiology can be absent. This can mask unanticipated blunted, toxic, or even exacerbated effects when the same therapies are trialed in humans, leading to wasted resources while potentially overlooking other more effective therapies. Moreover, the use of animal models in preclinical studies, although valuable and necessary, cannot completely overcome these inadequacies, as they are oftentimes unable to fully recapitulate human pathophysiology, resulting in a leaky screening pipeline.
Thus, a tremendous effort is being undertaken by researchers around the world to generate innovative, physiologically relevant models that can overcome these obstacles, maximizing the efficiency of the drug development pipeline. This is in line with the National Eye Institute's Audacious Goals Initiative statement which, while specifically discussing neural regeneration, called for improved and increased translational research around vision restoration.4
At the forefront of this effort is the utilization of retinal organoids derived from human stem cells to model diseases and test novel therapeutics. In this scenario, 3D retinal organoids are in a unique position to enhance or replace several stages of the existing drug development pipeline. In this article, we present an overview of some of the latest innovations in organoid technology and retinal disease modeling and suggest ways in which they could be incorporated into the existing drug development pathway.
From Dissociated Cells to Retinas in a Dish
Dissociated, 2D cell cultures have been traditionally used for drug screening due to their relative simplicity, ease of use, and scalability, as well as the availability of numerous existing quantitative and automatable technologies and assays. These models have been invaluable and will likely continue to have their place in the drug discovery pipeline.
However, it is becoming increasingly evident that they are not sufficient to ensure the discovery of successful drug candidates. This includes human induced pluripotent stem cell (hiPSC)-derived 2D cultures, which incorporate important advantages over immortalized cell lines and primary cell cultures, yet have so far only led to the identification of a small number of candidate therapeutic compounds through high throughput screening (HTS).5,6 This scenario is likely due to the fact that dissociated cell cultures fail to account for the effects of cell morphology and chemistry due to cell–cell interactions in 3D native environments and, thus, fail to recapitulate important aspects of disease pathophysiology. These models also lack additional immediate support cell types, as well as distant organ cell types, each of which may affect drug potency or represent sources of toxicity.
One promising strategy to overcome these problems is the recent development of zebrafish-based whole organism screening platforms.7–9 Zebrafish (Danio rerio) constitutes the only vertebrate model to date that can truly be used for HTS in whole organisms, allowing the possibility to evaluate drug toxicity and in vivo effects on all organs and systems simultaneously. Their small size, transparency, and permeability to small molecules facilitate their use in chemical screening platforms.9 Moreover, a versatile and readily accessible quantitative screening method has been developed for whole organism zebrafish screening that is capable of achieving true HTS capacities.10
However, despite their advantages, zebrafish systems pose similar drawbacks as other animal models in that their ability to recapitulate human pathophysiology may be limited. Thus, the development of improved human-based in vitro systems is still a highly desirable component that can contribute significant value and efficiencies to the drug development pipeline.
In this context, 3D stem cell technologies stand to improve upon many of the shortfalls of 2D cultures. Human stem cells can be induced to differentiate in vitro into highly organized 3D retinal tissue or “retinal organoids.” These organoids are capable of faithfully recapitulating the retina's native histoarchitecture and cellular composition,11–13 providing a far more realistic snapshot of in vivo biology. Moreover, these organoids have been shown to achieve advanced levels of cellular maturation, including the ability to respond to light.12,14–16
Of particular interest is the use of hiPSC for these applications. These cells have the advantage of being readily available, as they can be produced from a variety of sources, such as fibroblasts, blood, or urine, and offer the unprecedented opportunity of generating retinal tissues that carry a patient's individual genetic makeup.5,6,15 This technology thus has the potential to realize the promise of personalized medicine and decrease the likelihood of rejection in transplantation approaches.17 Moreover, patient-derived 3D retinas can be applied to the generation of improved disease models through disease-causing mutations; can be used to screen for drugs that are active in a patient-specific genetic background; and can be applied to the design of “clinical trials in a dish” by facilitating the sampling of a diverse population in drug screening assays.
Finally, retinal organoids can be applied not only to the screening of compounds and small molecules, but also to the testing of gene therapy strategies and certain drug delivery approaches, including the use of nanoparticles.
Retinal Organoid-Based Disease Models
To develop improved drug screening assays based on organoid technologies, it is imperative to generate adequate and robust disease models that can be used to evaluate drug effects on disease pathophysiological hallmarks through phenotypic screening. Several studies have already displayed some success in recreating retinal pathology from patient-derived cells.
X-linked retinitis pigmentosa has been recreated in 3D retinal organoids to investigate the disease mechanisms associated with retinitis pigmentosa GTPase regulator (RPGR) mutations. This study implicated the loss of RPGR-driven Gelsolin activation and consequent dysregulation of actin disassembly in the disease pathogenesis.18 Furthermore, a later study using organoids generated from 3 retinitis pigmentosa patients harboring RPGR mutations demonstrated that CRISPR-Cas9-mediated correction of these mutations rescued photoreceptor structure and electrophysiological properties and reversed the observed ciliopathy.19 Retinal organoids were also used to help elucidate the localization and role of receptor expression enhancing protein 6 (REEP6; the mutation of which is known to cause autosomal recessive retinitis pigmentosa), in retinal homeostasis, providing insights into disease pathophysiology.20
In a different study, the role of the small GTPase ARL3 and its activating protein retinitis pigmentosa 2 (RP2) was studied using retinal organoids derived from a patient carrying an RP2 nonsense mutation. The findings of this study suggest a role for these proteins in the trafficking of specific kinesins to cilia tips and raise the possibility that translational read-through inducing drugs, such as PTC124, could be beneficial for patients with this type of retinal dystrophy.21 Another ciliopathy, Leber Congenital Amaurosis, has been replicated in organoids generated from patient cells in a study that investigated the pathogenic mechanisms caused by a common mutation in the cilia-related gene CEP290. This study demonstrated the restoration of normal ciliary trafficking after treatment with antisense morpholinos as a potential treatment strategy.22
Finally, a retinal organoid model of an inherited form of glaucoma has been developed using hiPSC from a patient with an optineurin mutation (OPTN E50K). This study showed that patient-derived retinal ganglion cells exhibited a dramatic increase in apoptosis, which could be rescued by the addition of neuroprotective factors.23
Altogether, these studies demonstrate the feasibility of modeling patient specific disease in retinal organoids and set a precedent for the use of these models in the identification of therapeutic strategies. In addition, CRISPR/Cas9 technology can be used to introduce specific disease-causing mutations in hiPSC lines for organoid generation, expanding the repertoire of disease models with the valuable advantage of allowing the testing of these models vis-a-vis their isogenic controls.
While retinal organoid technologies are ideally suited for modeling monogenic diseases of the neural retina, their potential for modeling more complex diseases for drug screening applications should not be overlooked. For example, known environmental stressors can be added to the culture media to mimic at least some aspects of retinal disease, a practice that has been widely used in other culture systems.24 Finally, the promise hiPSC-derived models hold for personalized medicine includes the investigation of polygenic diseases or those with unknown etiologies and even the optimization of patient-specific therapeutic targets. However, important limitations of time and cost would need to be overcome to realize that promise.
Screening Technologies for Retinal Organoids
The application of retinal organoids in phenotypic drug screening has been hampered by a dearth of quantitative assays suitable for 3D models. Traditional 2D cell-based screening platforms utilizing fluorescence, imaging, and biochemical assays take advantage of their 2D arrangement and often homogenous cellular composition to have instant access to the entire test population for the measurement of experimental variables. Three-dimensional cultures lack this inherent geometric advantage, and their analysis is further complicated by the presence of multiple cell types organized in specific and biologically meaningful arrangements. Thus, measurement of experimental variables is more challenging in these systems because of issues like the need for spatial focus/resolution, penetration of dyes/lasers for imaging, drug diffusion kinetics, and difficulty parsing measurements from the various cell types present.
To overcome these limitations, we recently developed a fluorescent reporter-based quantitative screening platform for 3D retinal organoids. This platform, termed 3D automated reporter quantification, is based on fluorescence intensity measurements in multiwell plates and features speed, sensitivity, and reproducibility parameters compatible with high-throughput applications.25 As proof of concept of the utility of the platform in drug testing, we used JC-1, a fluorescence-based mitochondrial depolarization marker, to evaluate mitochondrial health in retinal organoid photoreceptors in longitudinal studies.25
Alternatively, technologies that have been successfully used for drug screening in other 3D systems could also be adapted to retinal organoids, thus expanding the breath of output measurements and screening platforms for these models. For example, luminescence-based assays have been used to assess the effect of drug candidates on cell viability in “tumorspheres,” cancer-like organoids derived from hiPSCs.26 This type of assay could be seamlessly adapted to retinal organoids for the development of a screening platform, yet its reliance on cell lysis means that outcome measurements necessarily represent the response of the tissue as a whole rather than of a specific cell type.
Other potentially useful assays for the assessment of global effects of drug treatments on retinal organoids include fluorescence-based dihydroethidium staining to evaluate oxidative stress in the retina25 or colorimetric assays for cell viability such as WST-8 or MTS, where a substrate is reduced by enzymatic cellular activity resulting in the formation of a yellow-color formazan dye that is proportional to the number of live cells.27,28 However, these assays should be optimized for retinal organoid models to ensure proper tissue penetration and linear correlation with cellular status in a 3D context.
Image-based technologies for high content screening are also being adapted to 3D systems and, thus, hold promise for retinal organoid-based screening platforms (for a thorough review on this topic, see Li et al.).29 Finally, the development of multielectrode arrays now allows for the electrophysiological recording of hundreds of cells in a tissue simultaneously30–33 and has already been applied to retinal organoid systems.16
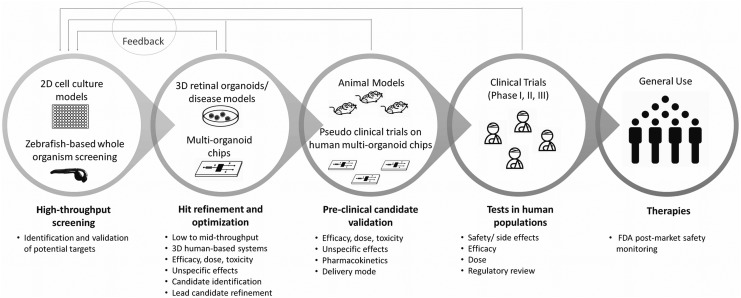
Overall, however, the implementation of these organoid technologies at a scale necessary to meet the demands of HTS, which is commonly thought of as >10,000 screens per day, with ultra-HTS defined as >100,000 screens per day,34 is not currently possible due to technical limitations discussed below, as well as the potential high cost of such an endeavor. Thus, we propose a new and more realistic drug development pipeline that could capitalize on the advantages of these powerful technologies for drug development applications (Fig. 1). In this optimized model, 3D organoid systems stand as an intermediate step between 2D cultures and animal models, in a secondary screening and validation phase, reducing unnecessary and costly animal experimentation without the need to meet initial screening quantities necessary for the first steps of drug discovery.
FIG. 1.
Proposed drug development pipeline utilizing retinal organoids both as an interim step and as an augmentation to preclinical trials.
Still, even at this smaller scale, careful consideration should be given to certain aspects of the study design, namely: (1) The use of multiple independent clones per phenotype (at least 3 biological replicates), although not always possible due to accessibility and costs, is an ideal scenario to increase reproducibility and disease-specific efficacy. In addition, cell lines derived from different sexes should be included in validation assays in cases where sex is a disease-relevant variable; (2) Technical replicates and multiple dose concentrations are critical for added rigor in these secondary screening phases and for dose–response assessment in validation steps; (3) Positive and negative controls should be included in every assay (these are assay-dependent, but could include, for example, wild-type organoids and vehicle treated disease models); and (4) Screening performance can be determined using Z′ factor or SSMD* statistical parameters.5,6,25
Barriers and Solutions
Despite the significant advances in stem cell-derived organoid technologies and their numerous advantages over other systems, there still exist several challenges to overcome before the potential of 3D retinal organoids for drug development can be fully realized. These include:
Time
The time and attention required for the culture of complex 3D structures are inherently greater than their 2D counterparts. Particularly, the generation of retinal organoids mimics native retinal development both spatially and temporally.12 This is a remarkable feature, but it implies a time frame of several months to almost a year, analogous to embryologic timelines, for the generation of laminated, relatively mature retinas from hiPSC, which can pose a significant challenge, both regarding time and costs, for industrial applications
Thus, efforts are being undertaken to overcome this issue. For example, the use of rotating bioreactors for the growth of 3D retinal tissue from mouse stem cells has been shown to improve the growth and slightly accelerate the differentiation rate of organoids compared to static protocols.35 Yet no technology currently exists for the generation of relatively mature retinal organoids at a significantly faster rate. Thus, until accelerated organoid generation and differentiation can be achieved, this issue can be bypassed by maintaining banks of retinal organoid tissue through a continuous pipeline of organoid generation, allowing for rapid, on-demand experimentation.
Maturation
Another concern for the application of retinal organoids in drug development is the fact that they constitute developing systems. Thus, while congenital retinal dystrophies may be more easily modeled by these organoids, some prevalent retinal diseases don't present until late in life, in mature retinas. Moreover, the pathogenesis of these diseases may be influenced by a lifetime of epigenetic modifications and environmental insults, posing additional challenges for their modeling with organoid systems.36 In addition, as retinal organoids age and photoreceptors acquire more mature characteristics, the cells in the inner layers of the retina start to degenerate, beginning with retinal ganglion cells. This could constitute a limitation for investigations requiring complete retinal structures with fully mature and differentiated cellular components.
Therefore, strategies to aid in the aging of retinal organoids or to promote the survival of all retinal cell types for long periods of time constitute critical and currently unmet needs in this field.
Lack of extra-retinal structures
Another limitation of current retinal organoid technologies for drug development is their lack of vascularization and absence of inflammatory cell types, features that limit the kinds of diseases that can be replicated in these models. For example, pathologies such as diabetic retinopathy, in which a vascular insult is central to the pathophysiology, cannot be faithfully recapitulated. Thus, screening for this type of diseases would be limited to the addition of specific inflammatory factors or other known cellular stressors to the culture media for neuroprotective drug development.
Similarly, the lack of juxtaposition of a polarized retinal pigmented epithelium (RPE) to the retinal structure limits their application to diseases in which this interaction plays an important role in disease pathophysiology such as AMD. For this reason, research is underway by multiple groups to try to generate novel organoid-based models that recapitulate the retina-RPE interaction. Even then, such models will be more likely to be applied to candidate drug validation rather than to larger scale screening.
Genetic variation
Much has been said about the potential of iPSC-derived models to replicate retinal diseases, but additional challenges surface when considering the generation of data that are generalizable to the public. Variability between iPSC lines have been reported, particularly with regards to their differentiation efficiency.5 However, with current protocols, once these cells are differentiated into retinal organoids they follow an endogenous developmental program of retinal cell differentiation, organization, and maturation that is remarkably consistent in organoids from different sources at the same time of differentiation.12
Interestingly, there is evidence that most of the genetic37 and transcriptional38 diversity between iPSC clones can be attributed to interdonor variability, rather than to differences between lines generated from different cell sources derived from the same donor.37 That is, this variability seems to stem from a true representation of the genetic diversity in a population.
Therefore, efforts are being undertaken to overcome this barrier. Namely, the hiPSC Initiative provides a large database of iPSCs for researchers to consult when selecting their clones and evaluating their findings.39 What's more, statistical analyses and tools have been developed to appropriately adjust sample sizes and ensure adequate power despite these variations.40 In contrast, this characteristic can be seen as an opportunity, as iPSC technologies can be applied to the development of a “clinical trial in a dish” where a candidate drug could be validated for its effect on a broader sample of the population.
Scalability and automation
Current protocols for the generation of retinal organoids can be laborious and, in most cases, still require manual isolation or selection of tissues, increasing costs and hampering automation. However, the use of bioreactors and robotic technologies for media exchange, reagent addition, and organoid sorting and plating can provide some level of automation, as well as improve scalability. In addition, retinal organoid cultures have recently been adapted to a 96-well plate format, presenting important advantages for large-scale applications.16
Cost
Each of the aforementioned barriers contributes to the overall cost of retinal organoid production and utilization. The extensive time required for the development of mature organoids necessitates a steady flow of organoid generation or otherwise lengthens the time to experimentation, increasing capital consumption. The high cost of reagents for stem cell-based cultures and the intensive manual labor involved in certain stages of the differentiation protocol may make the cost of large-scale drug screening in organoids prohibitive except perhaps for major drug companies. Scalability and automation are key to decreasing time and labor requirements and, thus, cost.
As these barriers are addressed, the overall obstacle of cost will decrease as well. Until then, utilization of organoid technology as an intermediate between HTS and clinical trials is a more realistic proposition and an investment that may be justified by the reduction in costs from having fewer unsuccessful drugs advancing to the significantly more expensive clinical trial stages.
Mimicking Complexity
Evaluating target tissues for drug effects on pathologic mechanisms is at the heart of drug screening. iPSCs have the ability to generate multiple tissue types, providing the opportunity to pursue additional levels of complexity to better mimic human conditions. Indeed, the last few years have seen a booming of organoid technologies with the derivation of brain,41,42 gut,43–45 lung,46 kidney,47 and liver48–50 organoids, among others, from human stem cells. Thus, the screening of drugs on microfluidic chips containing patient-derived tissues representing multiple organs to evaluate not only efficacy but also potential toxicity and side effects represents an ambitious prospective avenue for development.51 This concept has the immediate value of more accurately simulating the clinical context and, while it will not replace clinical trials, it has the potential to reduce the numbers of fruitless trials with their associated costs and ethical quandaries, by providing for a more efficient drug development pipeline.
Advances in microfluidic technologies, biomaterials, and microfabrication are already bringing this prospect closer to realization. However, multi-organoid-on-a-chip technologies still need to confront several challenges. Many of the limitations mentioned above for retinal organoids are common to other organoid systems as well. Yet organoid-on-a-chip technologies present further challenges related to the optimization of media conditions that can be applied to all organoids being tested simultaneously, the optimization of the level of maturation required for each component, the mass ratios of different organoids, and lack of vascularization, among others. And this is in addition to matters related to chip materials, extracellular matrices, microfluidics optimization, etc.51,52
Despite these difficulties, several groups have engineered initial prototypes experimenting with multiorgan physiology on a chip. Encouraging results have been obtained using 2D human cell-based chips to evaluate multitissue toxicity with representation of cardiac, muscle, neuronal, and liver cell types.53 This study tested the response of the system to five different drugs with known side effects and obtained results that were in agreement with published human and animal toxicity data, demonstrating the usefulness of these multitissue chip approaches to recapitulate aspects of human physiology. Moreover, important advances have been made in the last few years for the adaptation of this chip technology to 3D human cultures, with representation of heart, lung and liver tissues,54 and intestine, liver, skin, and kidney.55
While many barriers need to be overcome as described above, the further development of these systems using patient iPSC-derived organoids could provide the opportunity for personalized pharmacokinetic studies.54
Future drug screening paradigms utilizing retinal organoids incorporated in these chips could evaluate off-target toxicity and toxicity of possible metabolites from hepatic processing (Fig. 1). In addition, retinal organoids could be considered as standard additions to other multiorgan chips to assess retinal toxicity of possible nonretinal therapies, as vision changes are considered serious side effects for any drug.
Conclusions
The current drug development pipeline does not meet the needs of the growing population of people with retinal disease. R&D investigators should consider adopting novel screening methods, specifically the use of hiPSC-derived organoid models, to enhance existing protocols. With recent advances in technology, the integration of organoids, whether individually or in conjugation with chips, represents not only an opportunity to lower costs and improve clinical trial success rates but also to change the shape of drug development entirely through the personalization of drug identification. Today's drug development pathways are not sufficient, and organoids developed from hiPSCs just might be tomorrow's answer.
Acknowledgment
This work was supported, in part, by a Challenge Grant to the Department of Ophthalmology at the University of Colorado from Research to Prevent Blindness.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Flaxman S.R., Bourne R.R.A., Resnikoff S., et al. Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis. Lancet Glob. Health. 5:e1221–e1234, 2017 [DOI] [PubMed] [Google Scholar]
- 2. Wong C.H., Siah K.W., and Lo A.W.. Estimation of clinical trial success rates and related parameters. Biostatistics. 20:273–286, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herper M. The Cost Of Creating A New Drug Now $5 Billion, Pushing Big Pharma To Change, 2013: https://www.forbes.com/sites/matthewherper/2013/08/11/how-the-staggering-cost-of-inventing-new-drugs-is-shaping-the-future-of-medicine/#2d55d28f13c3 Last accessed April25, 2019
- 4. Goldberg J.L., and Guido W.. Report on the national eye institute audacious goals initiative: regenerating the optic nerve. Invest. Ophthalmol. Vis. Sci. 57:1271–1275, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Engle S.J., and Vincent F.. Small molecule screening in human induced pluripotent stem cell-derived terminal cell types. J. Biol. Chem. 289:4562–4570, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elitt M.S., Barbar L., and Tesar P.J.. Drug screening for human genetic diseases using iPSC models. Hum. Mol. Genet. 27:R89–R98, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kithcart A., and MacRae C.A.. Using zebrafish for high-throughput screening of novel cardiovascular drugs. JACC Basic Transl. Sci. 2:1–12, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang G., Rajpurohit S.K., Delaspre F., et al. First quantitative high-throughput screen in zebrafish identifies novel pathways for increasing pancreatic beta-cell mass. eLife. 4, 2015; DOI: 10.7554/eLife.08261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mathias J.R., Saxena M.T., and Mumm J.S.. Advances in zebrafish chemical screening technologies. Future Med. Chem. 4:1811–1822, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walker S.L., Ariga J., Mathias J.R., et al. Automated reporter quantification in vivo: high-throughput screening method for reporter-based assays in zebrafish. PLoS One. 7:e29916, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eiraku M., Takata N., Ishibashi H., et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 472:51, 2011 [DOI] [PubMed] [Google Scholar]
- 12. Zhong X., Gutierrez C., Xue T., et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 5:4047, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Phillips M.J., Wallace K.A., Dickerson S.J., et al. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest. Ophthalmol. Vis. Sci. 53:2007–2019, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fligor C.M., Langer K.B., Sridhar A., et al. Three-dimensional retinal organoids facilitate the investigation of retinal ganglion cell development, organization and neurite outgrowth from human pluripotent stem cells. Sci. Rep. 8:14520, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li G., Xie B., He L., et al. Generation of retinal organoids with mature rods and cones from urine-derived human induced pluripotent stem cells. Stem Cells Int. 2018:4968658, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hallam D., Hilgen G., Dorgau B., et al. Human-induced pluripotent stem cells generate light responsive retinal organoids with variable and nutrient-dependent efficiency. Stem Cells. 36:1535–1551, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Canto-Soler V., Flores-Bellver M., and Vergara M.N.. Stem cell sources and their potential for the treatment of retinal degenerations. Invest. Ophthalmol. Vis. Sci. 2016;57:ORSFd1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Megaw R., Abu-Arafeh H., Jungnickel M., et al. Gelsolin dysfunction causes photoreceptor loss in induced pluripotent cell and animal retinitis pigmentosa models. Nat. Commun. 8:271, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng W.L., Gao M.L., Lei X.L., et al. Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 10:1267–1281, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arno G., Agrawal S.A., Eblimit A., et al. Mutations in REEP6 cause autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 99:1305–1315, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwarz N., Lane A., Jovanovic K., et al. Arl3 and RP2 regulate the trafficking of ciliary tip kinesins. Hum. Mol. Genet. 26:3451, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parfitt D.A., Lane A., Ramsden C.M., et al. Identification and correction of mechanisms underlying inherited blindness in human iPSC-derived optic cups. Cell Stem Cell. 18:769–781, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohlemacher S.K., Sridhar A., Xiao Y., et al. Stepwise differentiation of retinal ganglion cells from human pluripotent stem cells enables analysis of glaucomatous neurodegeneration. Stem Cells. 34:1553–1562, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beharry K.D., Cai C.L., Valencia G.B., et al. Human retinal endothelial cells and astrocytes cultured on 3-D scaffolds for ocular drug discovery and development. Prostaglandins Other Lipid Mediat. 134:93–107, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vergara M.N., Flores-Bellver M., Aparicio-Domingo S., et al. Three-dimensional automated reporter quantification (3D-ARQ) technology enables quantitative screening in retinal organoids. Development. 144:3698–3705, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Subedi A., Shimizu T., Ryo A., Sanada E., Watanabe N., and Osada H.. Discovery of novel selenium derivatives as Pin1 inhibitors by high-throughput screening. Biochem. Biophys. Res. Commun. 474:528–533, 2016 [DOI] [PubMed] [Google Scholar]
- 27. Subedi A., Futamura Y., Nishi M., Ryo A., Watanabe N., and Osada H.. High-throughput screening identifies artesunate as selective inhibitor of cancer stemness: involvement of mitochondrial metabolism. Biochem. Biophys. Res. Commun. 477:737–742, 2016 [DOI] [PubMed] [Google Scholar]
- 28. Choucha Snouber L., Bunescu A., Naudot M., et al. Metabolomics-on-a-chip of hepatotoxicity induced by anticancer drug flutamide and its active metabolite hydroxyflutamide using HepG2/C3a microfluidic biochips. Toxicol. Sci. 132:8–20, 2013 [DOI] [PubMed] [Google Scholar]
- 29. Li L., Zhou Q., Voss T.C., Quick K.L., and LaBarbera D.V.. High-throughput imaging: focusing in on drug discovery in 3D. Methods. 96:97–102, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berdondini L., Imfeld K., Maccione A., et al. Active pixel sensor array for high spatio-temporal resolution electrophysiological recordings from single cell to large scale neuronal networks. Lab. Chip. 9:2644–2651, 2009 [DOI] [PubMed] [Google Scholar]
- 31. Portelli G., Barrett J.M., Hilgen G., et al. Rank order coding: a retinal information decoding strategy revealed by large-scale multielectrode array retinal recordings. eNeuro. 3, 2016; DOI: 10.1523/ENEURO.0134-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maccione A., Hennig M.H., Gandolfo M., et al. Following the ontogeny of retinal waves: pan-retinal recordings of population dynamics in the neonatal mouse. J. Physiol. 592:1545–1563, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abbott J., Ye T., Qin L., et al. CMOS nanoelectrode array for all-electrical intracellular electrophysiological imaging. Nat. Nanotechnol. 12:460–466, 2017 [DOI] [PubMed] [Google Scholar]
- 34. Szymanski P., Markowicz M., and Mikiciuk-Olasik E.. Adaptation of high-throughput screening in drug discovery-toxicological screening tests. Int. J. Mol. Sci. 13:427–452, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DiStefano T., Chen H.Y., Panebianco C., et al. Accelerated and improved differentiation of retinal organoids from pluripotent stem cells in rotating-wall vessel bioreactors. Stem Cell Rep. 10:300–313, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Artero Castro A., Lukovic D., Jendelova P., and Erceg S.. Concise review: human induced pluripotent stem cell models of retinitis pigmentosa. Stem Cells. 36:474–481, 2018 [DOI] [PubMed] [Google Scholar]
- 37. Burrows C.K., Banovich N.E., Pavlovic B.J., et al. Genetic variation, not cell type of origin, underlies the majority of identifiable regulatory differences in iPSCs. PLoS Genet. 12:e1005793, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schuster J., Halvardson J., Pilar Lorenzo L., et al. Transcriptome profiling reveals degree of variability in induced pluripotent stem cell lines: impact for human disease modeling. Cell. Reprogram. 17:327–337, 2015 [DOI] [PubMed] [Google Scholar]
- 39. Streeter I., Harrison P.W., Faulconbridge A., Flicek P., Parkinson H., and Clarke L.. The human-induced pluripotent stem cell initiative-data resources for cellular genetics. Nucleic Acids Res. 45:D691–D697, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Germain P.L., and Testa G.. Taming human genetic variability: transcriptomic meta-analysis guides the experimental design and interpretation of iPSC-based disease modeling. Stem Cell Rep. 8:1784–1796, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qian X., Jacob F., Song M.M., Nguyen H.N., Song H., and Ming G.L.. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 13:565–580, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lancaster M.A., Renner M., Martin C.A., et al. Cerebral organoids model human brain development and microcephaly. Nature. 501:373–379, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spence J.R., Mayhew C.N., Rankin S.A., et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 470:105–109, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Forster R., Chiba K., Schaeffer L., et al. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Rep. 2:838–852, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nadkarni R.R., Abed S., Cox B.J., et al. Functional enterospheres derived in vitro from human pluripotent stem cells. Stem Cell Rep. 9:897–912, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dye B.R., Dedhia P.H., Miller A.J., et al. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife. 5, 2016; DOI: 10.7554/eLife.19732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takasato M., Er P.X., Chiu H.S., et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 526:564–568, 2015 [DOI] [PubMed] [Google Scholar]
- 48. Ang L.T., Tan AKY, Autio M.I., et al. A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep. 22:2190–2205, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takebe T., Sekine K., Enomura M., et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 499:481–484, 2013 [DOI] [PubMed] [Google Scholar]
- 50. Takebe T., Sekine K., Kimura M., et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 21:2661–2670, 2017 [DOI] [PubMed] [Google Scholar]
- 51. Miranda C.C., Fernandes T.G., Diogo M.M., and Cabral J.M.S.. Towards multi-organoid systems for drug screening applications. Bioengineering (Basel). 5, 2018; DOI: 10.3390/bioengineering5030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wikswo J.P., Curtis E.L., Eagleton Z.E., et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab. Chip. 13:3496–3511, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oleaga C., Bernabini C., Smith A.S., et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 6:20030, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Skardal A., Murphy S.V., Devarasetty M., et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 7:8837, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maschmeyer I., Lorenz A.K., Schimek K., et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab. Chip. 15:2688–2699, 2015 [DOI] [PubMed] [Google Scholar]