Abstract
Bidirectional selection for either high or low responsiveness to endurance running has created divergent rat phenotypes of high-response trainers (HRT) and low-response trainers (LRT). We conducted proteome profiling of HRT and LRT gastrocnemius of 10 female rats (body weight 279 ± 35 g; n = 5 LRT and n = 5 HRT) from generation 8 of selection. Differential analysis of soluble proteins from gastrocnemius was conducted by label-free quantitation. Genetic association studies were conducted in 384 Russian international-level athletes (age 23.8 ± 3.4 yr; 202 men and 182 women) stratified to endurance or power disciplines. Proteomic analysis encompassed 1,024 proteins, 76 of which exhibited statistically significant (P < 0.05, false discovery rate <1%) differences between HRT and LRT muscle. There was significant enrichment of enzymes involved in glycolysis/gluconeogenesis in LRT muscle but no enrichment of gene ontology phrases in HRT muscle. Striated muscle-specific serine/threonine-protein kinase-beta (SPEG-β) exhibited the greatest difference in abundance and was 2.64-fold greater (P = 0.0014) in HRT muscle. Coimmunoprecipitation identified 24 potential binding partners of SPEG-β in HRT muscle. The frequency of the G variant of the rs7564856 polymorphism that increases SPEG gene expression was significantly greater (32.9 vs. 23.8%; OR = 1.6, P = 0.009) in international-level endurance athletes (n = 258) compared with power athletes (n = 126) and was significantly associated (β = 8.345, P = 0.0048) with a greater proportion of slow-twitch fibers in vastus lateralis of female endurance athletes. Coimmunoprecipitation of SPEG-β in HRT muscle discovered putative interacting proteins that link with previously reported differences in transforming growth factor-β signaling in exercised muscle.
Keywords: artificial selection model, coimmunoprecipitation, endurance training, exercise capacity, label-free quantitation, liquid chromatography mass spectrometry, responsiveness to exercise, skeletal muscle
INTRODUCTION
Exercise training has a positive impact on health and is broadly considered to be effective in the prevention of chronic diseases, including Type 2 diabetes mellitus and cardiovascular disease (56). In humans, maximum aerobic capacity (V̇o2max) is a strong and independent indicator of mortality (40) and also modifies risk associated with factors collectively termed the metabolic syndrome, including insulin resistance, abdominal obesity, atherogenic dyslipidemia, hypertension, and a proinflammatory and prothrombotic state (16). An individual’s V̇o2max is a product of their genetic heritage as well as their recent history of habitual activity or exercise training. However, there is broad interindividual variation in the response of humans to exercise training (5, 22). The Health, Risk Factors, Exercise Training and Genetics (HERITAGE) family study reported 47% of the variance in responsiveness (improvement in V̇o2max) to training may be attributable to genetic and other familial factors (4). In Caucasian men and women, the average increase in V̇o2max after 20 wk supervised training was 400 ml/min, but the magnitude of improvement in V̇o2max was broadly distributed, for example, the V̇o2max of some individuals increased by more than 1,000 ml/min (relative increase of ~50%). In contrast, other individuals failed to show a measurable change in V̇o2max or responded negatively and exhibited a decrease in V̇o2max in response to the standardized exercise stimulus. Moreover, changes in insulin sensitivity varied widely, and 42% of the 596 HERITAGE participants that underwent an intravenous glucose tolerance test exhibited no change or a decrease in insulin sensitivity after exercise training (7). Such an inability to respond to aerobic exercise may have serious health consequences, and investigation of the mechanisms underpinning this phenomenon is needed to identify targets and candidate biomarkers for more personalized therapies.
In humans a mixture of genetic and environmental factors contributes to the broad range of responsiveness to exercise training, and this presents challenges to identifying the underlying mechanistic links between exercise and improvements in health outcomes. The genetic factors that contribute to V̇o2max (and therefore disease risk) interact with environmental factors and can be divided into intrinsic and acquired components. The intrinsic component governs an individual’s baseline aerobic capacity and disease risk-profile in the nontrained sedentary state, whereas the acquired component governs the individual’s responsiveness to an environment of high physical activity such as regular endurance training. The intrinsic and acquired components of exercise capacity are each selectable traits, and we have used bidirectional artificial selection in rats to develop models of either high versus low intrinsic running capacity or high versus low responsiveness to endurance training. Selection on intrinsic running capacity has generated high-capacity runners (HCR) that resemble endurance-trained individuals and low-capacity runners (LCR) that have a significantly heightened disease risk profile, including significantly poorer cardiovascular function (e.g., (57)), peripheral insulin sensitivity [e.g., (47)], and life expectancy [e.g., (30)]. Selection on the acquired component of exercise capacity has generated high-response-to-training (HRT) and low-response-to-training (LRT) rats (31, 48) that do not differ in their intrinsic running capacity but have significantly different responses to a standardized regimen of endurance training. After 8 wk of moderate endurance exercise, the maximal running capacity of HRT rats increases by on average 50%, whereas LRT rats either fail to respond positively or decrease in maximal running capacity on average by 50%. Lessard et al. (33) reports LRT exhibit primary metabolic defects including poor glucose tolerance and elevated plasma triacylglyceride levels in the untrained sedentary state, and when exposed to endurance training LRT fail to promote skeletal muscle angiogenesis and exhibit an altered inflammatory response to acute exercise. It is not yet clear whether common molecular targets from hypothesis-led literature on muscle responses to exercise training are differentially regulated in HRT/LRT muscle. For example, Lessard et al. (33) reports no difference in the response of signaling proteins (e.g., AMPK and Akt) or mitochondrial capacity between HRT/LRT rats exposed to exercise training. In contrast, Marton et al. (37) conclude there are significant differences in markers of mitochondrial biogenesis in the muscles HRT and LRT rats exposed to controlled exercise training.
Wider analysis of molecular differences that regress with low versus high responsiveness to endurance training may provide further mechanistic insight to the regulators of the training response and the elevated disease risk that is associated with a lack of adaptation to endurance training. Nevertheless, finding the common denominators that underpin differences in the change in exercise capacity in response to endurance training is challenging because the genetic underpinning of training responsiveness is highly convoluted. Previous attempts to find predictors of exercise responsiveness have been performed at the transcript level, which offers powerful bioinformatic analysis through reverse engineering of nucleotide data and has highlighted complex relationships such as the repression of negative regulators (e.g., miRNA) that target selective transcription factors (28). Proteomic analysis offers an alternative and pragmatic approach to discovering new information that may be complementary to other -omic approaches (59), and we (9, 10) have previously used proteomics to highlight differences in HCR/LCR muscle that segregate with intrinsic running capacity. The proteome/protein complement of a tissue is what defines that tissue and is the net result of complex upstream events involving genetic and environmental interactions. The proteome is often regarded as a product downstream of transcriptional processing, translational regulation and protein degradative processes. However, proteins that already exist within the cell are what “sense” and transduce environmental stimuli and so also reside upstream of gene transcription and other regulatory processes. To gain new insight to muscle proteome differences associated with the responsiveness to endurance training, we conducted high-definition mass spectrometry [HDMSE (8)] profiling of proteins in HRT and LRT gastrocnemius. We report comprehensive differences between HRT and LRT muscle, including a greater abundance of straited muscle-specific serine/threonine protein kinase-beta (SPEG-β) in HRT gastrocnemius. We identify putative binding partners of SPEG-β in rat muscle that link with previously reported differences in transforming growth factor-β (TGF-β) signaling. Moreover, genetic analysis in international-level Russian athletes found the G variant of the rs7564856 polymorphism that increases SPEG gene expression was significantly greater in endurance than power athletes, which offers some external validation of the potential role of SPEG-β in modulating muscle responsiveness to endurance training.
METHODS
Rat model of artificial selection on responsiveness to endurance exercise training.
HRT and LRT rats were generated from a large-scale bidirectional selection program on the response to endurance exercise described in Koch et al. (31). In brief, genetically heterogeneous rats (n = 152) from the N:NIH outcrossed stock were used to develop the HRT and LRT strains by selective mating of males and females that exhibited either the greatest (HRT) or least (LRT) response (improvement in exercise capacity) to a standardized and progressive regimen of endurance training. The maximal running capacity (maximum distance run; DIST) of each animal was measured by a speed-ramped treadmill test to exhaustion that was performed before (DIST1) and after (DIST2) an 8 wk program of 24 exercise sessions beginning when the animals were ~11–13 wk of age. The volume of endurance training progressed each session by increments of 1 m/min running speed and 0.5 min duration, beginning at 10 m/min for 20 min in the 1st week and finishing at 21 m/min for 31.5 min in the 8th week. All exercise tests and training sessions were performed on a motorized treadmill at a 15° incline, and the response to training was calculated as the change in maximal running distance (ΔDIST = DIST2 − DIST1). At each generation, 10 male and 10 female rats that represented the extremes of training response were bred to develop HRT and LRT strains. Rats used in the current work were females from generation 8 of selection and were shipped from the US to the University of Jyväskylä (Jyväskylä, Finland) at 10 mo of age. The animals were housed in standard conditions (temperature 22°C, humidity 50 ± 10%, light from 8:00 AM to 8:00 PM) and had free access to tap water and food pellets (R36; Labfor, Stockholm, Sweden). The study was approved by the National Animal Experiment Board of Finland (permit number ESLH-2007-06894/Ym-23), and at the age of 17 mo the maximal running capacity of the rats was tested according to the speed-ramped protocol used previously at the University of Michigan. Two weeks after the maximal running test five LRT and five HRT rats (body mass 290 ± 30 g and 268 ± 40 g, respectively) were euthanized, and gastrocnemius muscles were excised, snap-frozen in liquid nitrogen, and stored at −80°C before being shipped to Liverpool John Moores University (Liverpool, UK) for proteomic analysis.
Processing of rat muscle samples.
Muscles were pulverized in liquid nitrogen then homogenized on ice in eight volumes of 1% NP-40, 50 mM Tris pH 7.4 containing Complete protease inhibitor (Roche Diagnostics, Lewes, UK). Samples were incubated on ice for 15 min then centrifuged at 1,000 rpm, 4°C for 5 min. Supernatants were cleared by centrifugation (12,000 g, 4°C for 45 min), and protein concentrations were measured with the Bradford assay (Sigma, Poole, Dorset, UK). Each sample was adjusted to 5 μg/μl, and in-solution tryptic digestion was performed in preparation for label-free quantitation (LFQ). Aliquots containing 100 μg protein were precipitated in five volumes of acetone for 1 h at −20°C. Pellets were resuspended in 0.1% (wt/vol) Rapigest SF (Waters, Milford, MA) in 50 mM ammonium bicarbonate and incubated at 80°C for 15 min. DTT was added (final concentration 1 mM) and incubated at 60°C for 15 min followed by incubation while protected from light in the presence of 5 mM iodoacetamide at 4°C. Sequencing-grade trypsin (Promega, Madison, WI) was added at an enzyme-to-protein ratio of 1:50, and digestion allowed to proceed at 37°C overnight. Digestion was terminated by the addition of 2 μl concentrated trifluoroacetic acid, and peptide solutions were cleared by centrifugation at 13 000 g for 15 min. Samples were diluted 1:1 with a tryptic digest of yeast alcohol dehydrogenase 1 (100 fmol/μl) to enable the amount of each identified protein to be quantified, as described previously (51).
LFQ by HDMSE.
Peptide mixtures were analyzed by nanoscale reverse-phase ultraperformance liquid chromatography (UPLC) (nanoACQUITY; Waters, Milford, MA) and online ion-mobility mass spectrometry (IMS) (SYNAPT G2-S; Waters, Manchester, UK). Samples (200 ng tryptic peptides) were loaded in aqueous 0.1% (vol/vol) formic acid via a Symmetry C18 5 μm, 2 cm × 180 μm trap column (Waters, Milford, MA). Separation was conducted at 35°C through an HSS T3 C18 1.8 μm, 25 cm × 75 μm analytical column (Waters, Milford, MA). Peptides were eluted with a gradient rising to 40% acetonitrile 0.1% (vol/vol) formic acid over 90 min at a flow rate of 300 nl/min. Additionally, a Lockmass reference (100 fmol/μl Glu-1-fibrinopeptide B) was delivered to the NanoLockSpray source of the mass spectrometer at a flow rate of 500 nl/min and was sampled at 60 s intervals. For all measurements, the mass spectrometer was operated in a positive electrospray ionization mode at a resolution of >25,000 FWHM. Prior to analysis, the time-of-flight (TOF) analyzer was calibrated with a NaCsI mixture from m/z 50 to 1,990. HDMSE analyses were conducted within the Tri-wave ion guide. Accumulated ions were separated according to their drift time characteristics in the N2 gas-filled mobility cell before collision-induced dissociation alternating between low (4 eV) and elevated (14–40 eV) collision energies at a scan speed of 0.9 s per function over 50–2,000 m/z. Analytical data were LockMass corrected postacquisition using the doubly charged monoisotopic ion of the Glu-1-fibrinopeptide B. Charge reduction and deconvolution of potential parent-fragment correlation were achieved in the first instance by means of retention and drift time alignment, as described previously (35).
HDMSE spectra were aligned with Progenesis QI for Proteomics (QI-P; Nonlinear Dynamics, Newcastle, UK). Prominent ion features (~1,200 per chromatogram) were used as vectors to match each data set to a common reference chromatogram. An analysis window of 10–100 min and 50–1,650 m/z was selected, which encompassed a total of 47,109 features (charge states of +2, +3, or +4) and 3,924 of these features were separated by IMS. Protein identifications and quantitative information were extracted by the dedicated algorithms in ProteinLynx GlobalSERVER (PLGS) v3.0 (Waters, Milford, MA). Peak lists were searched against the UniProt database restricted to “Rattus” (8,071 entries). The initial ion-matching requirements were ≧1 fragment per peptide, ≧3 fragments per protein, and ≧1 peptide per protein. The enzyme specificity was trypsin allowing 1 missed cleavage, carbamidomethyl of cysteine (fixed), and oxidation of methionine (variable). Parent- and fragment-ion ppm errors were calculated empirically, and decoy databases were used to calculate the identification error rate. Scoring of the database searches was refined by correlation of physicochemical properties of fragmented peptides from theoretical and experimental data. Peptide identifications were imported to QI-P and filtered to exclude peptides with scores <5.5 (34). In total, 16,749 peptides were identified, and 1,018 had been resolved by IMS.
Coimmunoprecipitation and gel electrophoresis-liquid chromatography-tandem mass spectrometry.
Coimmunoprecipitation (Co-IP) experiments were performed using a rabbit anti-SPEG polyclonal Ab (HPA018904; Sigma-Aldrich, Poole, Dorset, UK). Negative control Co-IP experiments were conducted using rabbit anti-NDRG2 monoclonal Ab (Ab174850; Abcam plc) or by incubating samples with Protein-A Dynabeads (Thermoscientific, Runcorn, UK) only. Protein A Dynabeads were suspended in phosphate-buffered saline with 0.05% Tween-20 (PBS-T) and rotated for 30 min at room temperature (RT) with 1 μg of polyclonal antibody in 50 μl of PBS-T. The bead-antibody complex was washed five times in 50 μl of PBS-T and incubated with 500 μg of muscle protein for 3 h at 4°C on a sample mixer. The bead-Ab sample complexes were washed three times in PBS-T, and proteins were extracted from the beads by two sequential incubations in 5 μl of LDS sample buffer (NuPAGE; Thermo Scientific, Runcorn, UK) for 4 min each at 95°C. Samples were electrophoresed through 7% Tris-Acetate precast gels (NuPAGE; Thermo Scientific, Runcorn, UK) and stained for 1 h with colloidal Coomassie blue (Bio-Rad, Deeside, UK). Each gel lane was cut into 7 × 5 mm segments, and each segment was diced in to 1 mm3 pieces, and tryptic in-gel digestion was performed as described previously (20). Each segment was processed separately in preparation for nanoscale reverse-phase UPLC (NanoAcquity; Waters, Milford, MA) and online ESI QTOF MS/MS (Q-TOF Premier; Waters, Manchester, UK). Peptides were desalted using C18 ZipTips (Millipore, Billerica, MA) and loaded by partial-loop injection on to a 180 μm ID × 2 cm long 100 Å, 5 µm BEH C18 Symmetry trap column (Waters, Milford, MA) at flow rate of 5 μl/min for 3 min in 2.5% (vol/vol) ACN, 0.1% (vol/vol) FA. Separation was conducted at 35°C via a 75 μm ID × 25 cm long 130 Å, 1.7 µm BEH C18 analytical reverse-phase column (Waters). Peptides were eluted with a linear gradient that rose to 37.5% ACN 0.1% (vol/vol) FA over 90 min at a flow rate of 300 nl/min. Eluted peptides were sprayed directly in to the MS via a NanoLock Spray source and Picotip emitter (New Objective, Woburn, MA). Additionally, a LockMass reference (100 fmol/μl Glu-1-fibrinopeptide B) was delivered to the NanoLock Spray source of the mass spectrometry (MS) and was sampled at 240 s intervals. For all measurements, the MS was operated in positive ESI mode at a resolution of 10,000 FWHM. Before analysis, the TOF analyzer was calibrated with fragment ions of [Glu-1]-fibrinopeptide B from m/z 50 to 1,990. Peptide MS were recorded between 350 and 1,600 m/z. Data-dependent tandem mass spectrometry (MS/MS) spectra were collected over the range 50–2,000 m/z. The five most abundant precursor ions of charge 2+, 3+, or 4+ were selected for fragmentation using an elevated (20–40 eV) collision energy. A 30 s dynamic exclusion window was used to avoid repeated selection of peptides for MS/MS.
MS/MS spectra were searched against the UniProt database restricted to Rattus (8,071 sequences) with Mascot Distiller (www.matrixscience.com) and a locally implemented Mascot server (v.2.2.03; www.matrixscience.com). Enzyme specificity was trypsin (allowing 1 missed cleavage), carbamidomethyl modification of cysteine (fixed), deamidation of asparagine and glutamine (variable), oxidation of methionine (variable), and m/z errors of 0.3 Da.
Genetic association studies in human athletes.
Genetic association studies were conducted in 384 Russian international-level athletes that have been reported in previous studies [e.g., (2, 42)]. Participants (age 23.8 ± 3.4 yr; 202 men and 182 women) included in the current cohort were of Caucasian Eastern European descent and were stratified into two groups. Group 1 (n = 258) included endurance athletes (3–10 km runners, biathletes, 5–10 km skaters, cross-country skiers, marathon runners, 0.8–25 km swimmers, rowers/kayakers, race walkers, 1.5–10 km speed skaters, and triathletes). Of those, 35 female endurance athletes were also involved in the muscle biopsy study. Group 2 (n = 126) comprised power athletes (50–100 m swimmers, sprint cyclers, 100–400 m runners, 500–1,000 m speed skaters, and short-trackers, track and field jumpers, heptathletes/decathletes, and throwers). The study was approved by the Ethics Committee of the Physiological Section of the Russian National Committee for Biological Ethics and Ethics Committee of the Federal Research and Clinical Center of Physical-chemical Medicine of the Federal Medical and Biological Agency of Russia. Written informed consent was obtained from each participant. The study complied with the guidelines set out in the Declaration of Helsinki and ethical standards in sport and exercise science research.
Venous blood samples (4 ml) were collected in EDTA-coated tubes (Vacuette EDTA, Greiner Bio-One) and were transported to the laboratory at 4°C. DNA was extracted from leukocytes on the same day with a commercial kit according to the manufacturer's instructions (Technoclon). DNA quality was assessed by agarose gel electrophoresis and HumanOmni1-Quad BeadChips (Illumina) were used for genotyping of 1,140,419 single nucleotide polymorphisms (SNPs). In addition, Human OmniExpress BeadChips (Illumina) were used for genotyping of >700,000 SNPs in the 35 female athletes that also gave muscle samples for fiber-type analysis. The assay required 200 ng of DNA sample as input with a concentration of at least 50 ng/µl. Exact concentrations of DNA in each sample were measured using a Qubit Fluorometer (Invitrogen). All further procedures were performed according to the instructions of Infinium HD Assay.
Evaluation of muscle fiber composition in human athletes.
Samples of the vastus lateralis muscle of 35 female athletes were obtained with the Bergström needle biopsy procedure under local anesthesia with 1% lidocaine solution. Serial cross sections (7 μm thick) were cut from frozen muscle samples with a microtome (Leica Microsystems, Wetzlar, Germany) and were thaw-mounted on Polysine glass slides. Sections were air-dried for 15 min at RT and then washed in 3 × 5 min incubations in PBS before being incubated (RT for 1 h) in PBS containing primary Ab against slow or fast isoforms of myosin heavy chain (M8421, 1:5,000; M4276; 1:600, respectively; Sigma-Aldrich). Muscle sections were washed for 3 × 5 min in PBS and then incubated (RT for 1 h) in PBS containing secondary Ab conjugated with FITC (F0257; 1:100; Sigma-Aldrich). Sections were washed in PBS (3 × 5 min), placed in mounting media, and covered with a coverslip before imaging with a fluorescent microscope (Eclipse Ti-U; Nikon). All analyzed images contained >100 fibers, and the ratio of the number of stained fibers to the total fiber number was calculated. Fibers stained in serial sections with antibodies against slow and fast isoforms were considered as hybrid fibers.
Statistical analysis.
Data are presented as means and standard deviation unless otherwise stated. Differences in protein abundance measured by LFQ of LRT and HRT samples (n = 5 in each group) were investigated by one-way analysis of variance, corrected using q-values (53) to a false discovery rate of 1%. Functional enrichment testing was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov).
Genetic variations in or near the human SPEG gene were investigated using the Genotype Tissue Expression (GTEx) database (17). Statistical analysis of genotype data was conducted using PLINK v1.90, R (version 3.4.3), and GraphPad InStat (GraphPad Software) software. Genotype distribution and allele frequencies between athletes in group 1 or group 2 were compared by χ2 tests. Quantitative trait (proportion of slow-twitch muscle fibers) SNP association was tested in a linear additive model. P values < 0.05 were considered statistically significant.
RESULTS
LRT and HRT rats had a maximum running capacity (DIST1) of 852 ± 176 m and 642 ± 98 m, respectively, at ~11 wk of age. After 8 wk endurance training, the maximum running capacity (DIST2) of LRT (539 ± 107 m) had decreased by 49% (ΔDIST −313 ± 144 m), whereas the maximum running capacity of HRT rats had increased by 44% (ΔDIST +376 ± 111 m). The running capacity of the animals was retested at 17 mo of age, and there was no difference between LRT (114 ± 122 m) and HRT (173 ± 102 m) groups.
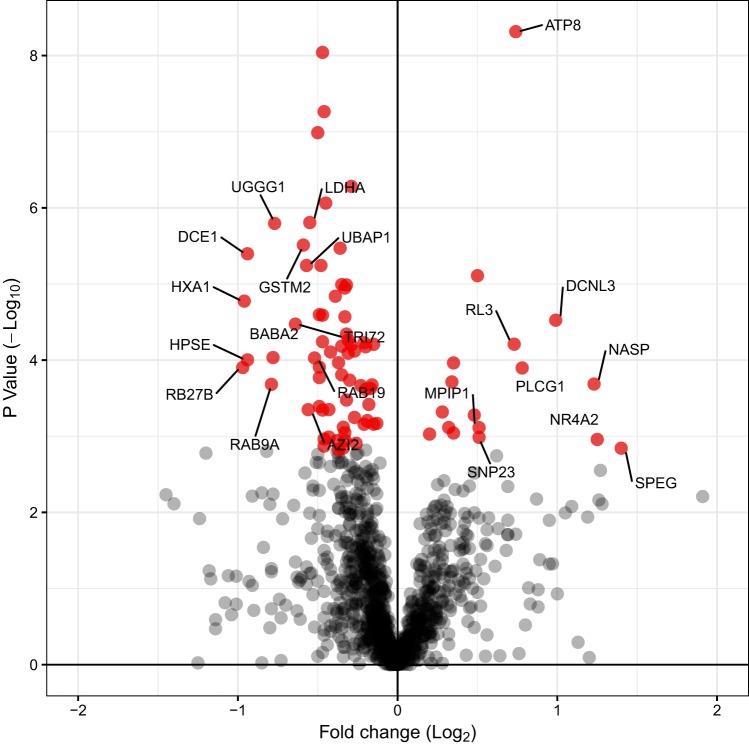
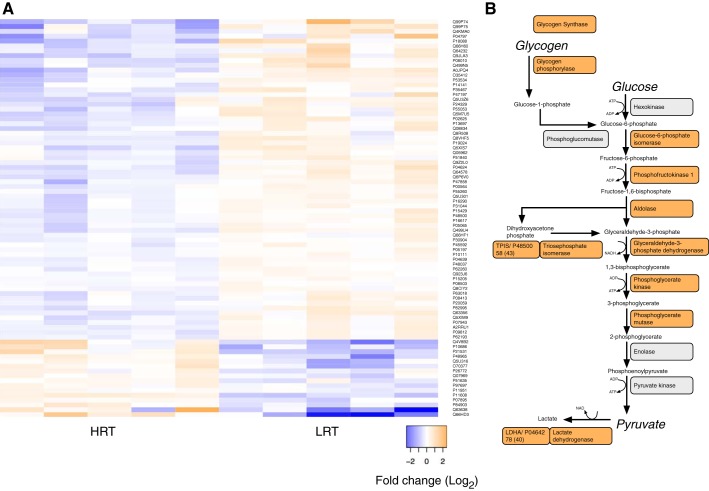
LFQ encompassed 1,024 proteins that were confidently (FDR <1%) identified in each of the HRT and LRT samples (n = 5, per group). Protein identifications and normalized abundance data are available at https://doi.org/10.6084/m9.figshare.9995087. Differential analysis of proteins quantified using three or more peptides revealed the relative abundance of 76 proteins differed significantly (P < 0.05, q < 0.01) between HRT and LRT groups (Fig. 1). Thirteen proteins were more abundant in HRT muscle (Table 1), whereas 63 proteins were more abundant in LRT (Table 2 and Fig. 2A). There was significant enrichment of proteins associated with the KEGG metabolic pathways glycolysis and gluconeogenesis in LRT muscle, and 12 enzymes involved in muscle glycogen/glucose metabolism (Fig. 2B) were more abundant compared with HRT muscle. In contrast, there was no significant enrichment of gene ontology phrases or KEGG metabolic pathways among the 13 proteins that were more abundant in HRT muscle. The protein most enriched in gastrocnemius of HRT rats was striated muscle-specific serine/threonine-protein kinase beta (SPEG-β; also known as striated muscle preferentially expressed gene). Co-IP gel electrophoresis-liquid chromatography-tandem mass spectrometry identified 24 potential binding partners of SPEG-β in HRT muscle (Table 3). There was no significant enrichment of gene ontology among the potential SPEG-β binding partners, and we were unable to identify binding partners that were specific to either HRT or LRT.
Fig. 1.
Differential analysis of high-definition mass spectrometry (HDMSE) data. Volcano plot presenting the log2 fold-difference in abundance between high-response trainer (HRT) versus low-response trainer (LRT) gastrocnemius and the statistical significance determined by one-way ANOVA (n = 5, per group). Proteins that were statistically different (P < 0.05) and had a false discovery rate <1% are highlighted in red. Proteins that exhibit a >1-fold difference in abundance are annotated by UniProt protein identifiers.
Table 1.
Proteins more abundant in HRT gastrocnemius
| Description | Accession | Score | Peptides | Delta | P Value |
|---|---|---|---|---|---|
| Striated muscle-specific serine/threonine-protein kinase | Q63638 | 109 | 18 (4) | 2.64 | 1.44E-03 |
| Nuclear autoantigenic sperm protein | Q66HD3 | 44 | 7 (3) | 2.35 | 2.1E-04 |
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1 | P10686 | 57 | 9 (4) | 1.72 | 1.3E-04 |
| ATP synthase protein 8 | P11608 | 24 | 4 (3) | 1.67 | 4.85E-09 |
| cGMP-gated cation channel alpha-1 | Q62927 | 83 | 11 (3) | 1.53 | 1.81E-03 |
| Stromal interaction molecule 1 | P84903 | 38 | 6 (4) | 1.42 | 7.78E-06 |
| Ras-related protein Rab-35 | Q5U316 | 106 | 14 (4) | 1.4 | 5.3E-04 |
| Alcohol dehydrogenase [NADP+] | P51635 | 80 | 13 (6) | 1.28 | 1.1E-04 |
| 10 kDa heat shock protein, mitochondrial | P26772 | 65 | 10 (7) | 1.27 | 9.1E-04 |
| Inositol monophosphatase | P97697 | 123 | 18 (10) | 1.26 | 1.9E-04 |
| Platelet glycoprotein 4 | Q07969 | 28 | 5 (4) | 1.25 | 7.7E-04 |
| Superoxide dismutase [Mn], mitochondrial | P07895 | 51 | 10 (6) | 1.21 | 4.8E-04 |
| Cytochrome c oxidase subunit 6C-2 | P11951 | 69 | 11 (5) | 1.15 | 9.4E-04 |
Protein description and Accession relate to the Swiss-Prot database entry identified from MSe searches performed in Progenesis QI-P via GLPS. Delta is the fold difference relative to low-response trainers (LRT). Values are reported for proteins quantified using 3 or more peptides and exhibiting significant (P < 0.05) differences in abundance at a false discovery rate of <1%. HRT, high-response trainer.
Table 2.
Proteins more abundant in LRT gastrocnemius
| Description | Accession | Score | Peptides | Delta | P Value |
|---|---|---|---|---|---|
| Membrane/cytoskeletal/vesicle/microtubule | |||||
| Ras-related protein Rab-27B | Q99P74 | 38 | 6 (3) | 1.96 | 1.25E-04 |
| Ras-related protein Rab-9A | Q99P75 | 43 | 6 (6) | 1.72 | 2.08E-04 |
| Tripartite motif-containing protein 72 | A0JPQ4 | 261 | 28 (22) | 1.56 | 3.37E-05 |
| Signal-induced proliferation-associated 1-like protein 1 | O35412 | 327 | 52 (21) | 1.41 | 1.24E-04 |
| Growth arrest-specific protein 8 | Q499U4 | 51 | 8 (6) | 1.38 | 9.09E-09 |
| Myosin-Ie | Q63356 | 96 | 16 (6) | 1.37 | 8.65E-07 |
| Potassium voltage-gated channel subfamily A member 5 | P19024 | 65 | 12 (7) | 1.25 | 1.03E-05 |
| Protocadherin Fat 3 | Q8R508 | 279 | 52 (17) | 1.23 | 1.84E-04 |
| Annexin A4 | P55260 | 169 | 25 (14) | 1.16 | 5.92E-05 |
| Cofilin-1 | P45592 | 104 | 15 (8) | 1.15 | 6.96E-04 |
| Annexin A6 | P48037 | 523 | 67 (44) | 1.12 | 2.36E-04 |
| Dynein heavy chain 12, axonemal | Q923J6 | 797 | 132 (57) | 1.11 | 6.2E-05 |
| Microtubule-associated protein 1B | P15205 | 188 | 30 (15) | 1.11 | 6.94E-04 |
| Glucose metabolic processes | |||||
| Glyceraldehyde-3-phosphate dehydrogenase | P04797 | 473 | 62 (34) | 1.77 | 1.58E-03 |
| L-lactate dehydrogenase A chain | P04642 | 450 | 78 (40) | 1.47 | 1.56E-06 |
| Fatty acid-binding protein, epidermal | P55053 | 34 | 4 (3) | 1.38 | 1.09E-03 |
| Glucose-6-phosphate isomerase | Q6P6V0 | 485 | 65 (46) | 1.37 | 5.45E-08 |
| Glycogen phosphorylase, brain form (fragment) | P53534 | 631 | 83 (23) | 1.34 | 7.83E-05 |
| 6-phosphofructokinase, muscle type | P47858 | 477 | 63 (35) | 1.31 | 1.14E-03 |
| Glycogen phosphorylase, muscle form | P09812 | 1275 | 188 (104) | 1.28 | 3.39E-06 |
| Fructose-bisphosphate aldolase A | P05065 | 554 | 76 (43) | 1.28 | 1.02E-05 |
| Beta-enolase | P15429 | 687 | 101 (26) | 1.27 | 7.6E-04 |
| Glycogen [starch] synthase, muscle | A2RRU1 | 284 | 41 (28) | 1.26 | 2.7E-05 |
| Triosephosphate isomerase | P48500 | 368 | 58 (43) | 1.25 | 1.13E-05 |
| Phosphoglycerate kinase 1 | P16617 | 512 | 79 (54) | 1.25 | 4.55E-05 |
| Aldose reductase | P07943 | 209 | 30 (22) | 1.24 | 5.43E-05 |
| Phosphoglycerate mutase 2 | P16290 | 290 | 40 (15) | 1.24 | 8.11E-05 |
| Mitochondrion | |||||
| Acyl-CoA synthetase family member 2, mitochondrial | Q499N5 | 36 | 7 (5) | 1.41 | 1.69E-04 |
| Trans-2,3-enoyl-CoA reductase | Q64232 | 41 | 5 (3) | 1.37 | 1.35E-03 |
| NADP-dependent malic enzyme | P13697 | 70 | 11 (7) | 1.3 | 1.59E-03 |
| Citrate synthase, mitochondrial | Q8VHF5 | 213 | 25 (19) | 1.3 | 1.49E-03 |
| Voltage-dependent anion-selective channel protein 1 | Q9Z2L0 | 293 | 35 (28) | 1.27 | 6.57E-05 |
| ADP/ATP translocase 1 | Q05962 | 275 | 35 (8) | 1.21 | 7.6E-05 |
| Phosphatidylethanolamine-binding protein 1 | P31044 | 106 | 11 (9) | 1.2 | 1.23E-03 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Q66HF1 | 378 | 47 (28) | 1.15 | 1.52E-03 |
| Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | P08503 | 153 | 23 (14) | 1.1 | 6.77E-04 |
| Calcium handling | |||||
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | Q64578 | 1060 | 176 (64) | 1.41 | 1.03E-07 |
| Protein S100-A1 | P35467 | 37 | 4 (3) | 1.4 | 2.53E-05 |
| Parvalbumin alpha | P02625 | 272 | 45 (31) | 1.39 | 5.71E-06 |
| Chaperones/protein folding | |||||
| UDP-glucose:glycoprotein glucosyltransferase 1 | Q9JLA3 | 138 | 24 (7) | 1.7 | 1.6E-06 |
| Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A | Q08834 | 52 | 9 (3) | 1.35 | 4.46E-04 |
| Heat shock protein HSP 90-alpha | P82995 | 296 | 45 (16 | 1.31 | 1.45E-05 |
| Heat shock cognate 71 kDa protein | P63018 | 486 | 52 (17) | 1.28 | 1.56E-04 |
| T-complex protein 1 subunit beta | Q5XIM9 | 64 | 9 (7) | 1.26 | 1.13E-03 |
| Peptidyl-prolyl cis-trans isomerase A | P10111 | 123 | 13 (10) | 1.12 | 2.12E-04 |
| Signal transduction | |||||
| RAC-beta serine/threonine-protein kinase | P47197 | 110 | 20 (9) | 1.39 | 4.53E-04 |
| Calcium/calmodulin-dependent protein kinase type II beta chain | P08413 | 63 | 10 (4) | 1.29 | 1.08E-04 |
| 14-3-3 protein epsilon | P62260 | 202 | 25 (15) | 1.14 | 2.42E-04 |
| Cell stress | |||||
| Glutathione S-transferase Mu 2 | P08010 | 337 | 37 (12) | 1.5 | 3.08E-06 |
| Macrophage migration inhibitory factor | P30904 | 36 | 7 (7) | 1.17 | 2.16E-04 |
| Dual oxidase 1 | Q8CIY2 | 72 | 14 (4) | 1.15 | 6.68E-05 |
| Skeletal muscle specific | |||||
| Carbonic anhydrase 3 | P14141 | 249 | 43 (31) | 1.26 | 1.43E-03 |
| Creatine kinase M-type | P00564 | 549 | 94 (58) | 1.21 | 5.66E-04 |
| Four and a half LIM domains protein 1 | Q9WUH4 | 229 | 30 (23) | 1.19 | 1.68E-03 |
| Posttranscriptional processing | |||||
| Protein mago nashi homolog | Q27W02 | 67 | 9 (3) | 1.42 | 1.73E-03 |
| Serine/threonine-protein kinase PRP4 homolog | Q5RKH1 | 64 | 12 (6) | 1.29 | 1.87E-03 |
| Protein turnover (ribosome/proteasome) | |||||
| 26S protease regulatory subunit 4 | P62193 | 98 | 15 (6) | 1.22 | 6.32E-05 |
| NSFL1 cofactor p47 | O35987 | 126 | 19 (12) | 1.2 | 1.72E-03 |
| Elongation factor 2 | P05197 | 376 | 52 (34) | 1.13 | 3.82E-04 |
| Lipoprotein | |||||
| Apolipoprotein A-I | P04639 | 82 | 12 (7) | 1.22 | 5.22E-07 |
| Hemopexin | P20059 | 262 | 35 (27) | 1.14 | 6.22E-04 |
| Uncharacterized | |||||
| Coiled-coil domain-containing protein 146 | Q66H60 | 110 | 19 (7) | 1.39 | 5.73E-05 |
| Coiled-coil domain-containing protein 67 | Q5U3Z6 | 132 | 26 (10) | 1.35 | 1.03E-03 |
Protein description and Accession relate to the Swiss-Prot database entry identified from MSe searches performed in Progenesis QI-P via GLPS. Fold difference relative to high-response trainer (HRT). Values are reported for protein quantified using 3 or more peptides and exhibiting significant differences in abundance at a false discovery rate of <1%. LRT, low-response trainer.
Fig. 2.
Enzymes of the glycolytic pathway are more abundant in LRT gastrocnemius. A: heat map of 76 proteins that differed significantly (P < 0.05, q < 0.01) in abundance between high-response trainer (HRT) and low-response trainer (LRT) gastrocnemius (n = 5, per group). LRT muscle exhibited significant enrichment of proteins associated with the KEGG metabolic pathways glycolysis and gluconeogenesis. B: 12 enzymes involved in glycogen/glucose metabolism were more abundant in LRT muscle.
Table 3.
Putative protein-interaction partners of SPEG-β in skeletal muscle
| Accession | Description; Protein Identifier | MOWSE Score | Sequence Coverage, % |
|---|---|---|---|
| C0HL12 | Adhesion G protein-coupled receptor B1; AGRB1 | 29 | 3.1 |
| Q5U2S6 | Ankyrin repeat and SOCS box protein 2; ASB2 | 35 | 3.8 |
| P70673 | ATP-sensitive inward rectifier potassium channel 11; KCJ11 | 68 | 5.9 |
| Q3KR97 | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1; BI2L1 | 31 | 4.5 |
| P41350 | Caveolin-1; CAV1 | 64 | 32.6 |
| P01026 | Complement C3; CO3 | 35 | 4.4 |
| Q01205 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial; ODO2 | 51 | 7.3 |
| P39052 | Dynamin-2; DYN2 | 36 | 4.5 |
| M0R8U1 | Dynein heavy chain 5, axonemal; DYH5 | 30 | 2.5 |
| Q5U4E6 | Golgin subfamily A member 4; GOGA4 | 40 | 4.6 |
| P97636 | Interleukin-18; IL18 | 29 | 13.4 |
| O35790 | N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase; PIGL | 34 | 5.2 |
| P51839 | Olfactory guanylyl cyclase GC-D; GUC2D | 31 | 0.8 |
| Q8K4M9 | Oxysterol-binding protein-related protein 1; OSBL1 | 30 | 3.5 |
| Q505J8 | Phenylalanine–tRNA ligase alpha subunit; SYFA | 29 | 2.2 |
| P33568 | Retinoblastoma-associated protein; RB | 34 | 1.4 |
| Q53UA7 | Serine/threonine-protein kinase TAO3; TAOK3 | 137 | 8.1 |
| P02770 | Serum albumin; ALBU | 46 | 5.6 |
| A4ZYQ5 | Solute carrier family 2, facilitated glucose transporter member 7; GTR7 | 28 | 2.1 |
| Q63638* | Striated muscle-specific serine/threonine-protein kinase; SPEG | 68 | 1.1 |
| P46462 | Transitional endoplasmic reticulum ATPase; TERA | 116 | 18.7 |
| Q6AY56 | Tubulin alpha-8 chain; TBA8 | 31 | 11.1 |
| Q5PQS3 | Ventricular zone-expressed PH domain-containing protein homolog 1; MELT | 32 | 3.1 |
| Q8K3Y6 | Zinc finger CCCH-type antiviral protein 1; ZCCHV | 30 | 4 |
MOWSE, molecular weight search.
Six SNPs (rs13386459, rs907683, rs72965313, rs4674396, rs745027, rs7564856) located in close proximity (i.e., in high linkage disequilibrium) to the SPEG gene were significantly (P < 5*10−8–10−13) associated with SPEG gene expression in human skeletal muscle. Of those, the rs7564856 was available for genotyping in the athletic cohorts by microarray analysis. According to GTEx data, the G allele of the rs7564856 SNP was reported to be associated (P = 2.7*10−8) with increased expression of the SPEG gene and may be favorable for endurance sports. The G allele of rs7564856 was significantly greater (32.9 vs. 23.8%; OR = 1.6, P = 0.009) in endurance athletes compared with power athletes (Fig. 3). We also found the G allele was significantly (β = 8.345, P = 0.0048) associated with increased proportion of slow-twitch muscle fibers in female endurance athletes.
Fig. 3.
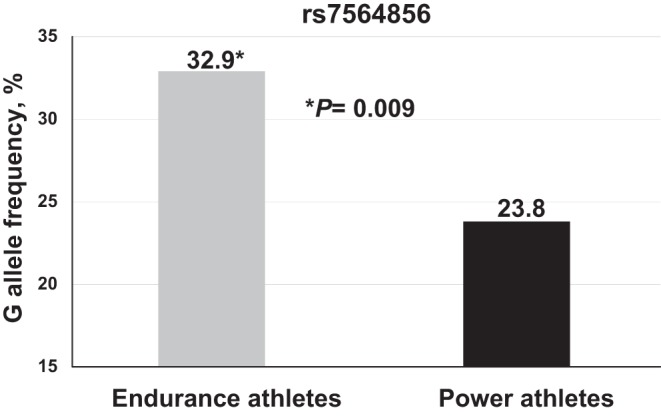
Striated muscle-specific serine/threonine-protein kinase (SPEG) rs7564856 G allele frequency is greater in international-level endurance athletes. Genetic association studies in Russian international-level athletes of Caucasian Eastern European descent, stratified into endurance athletes (n = 258) and power athletes (n = 126). The frequency of the G allele of rs7564856 reported to be associated with increased expression of the SPEG gene was significantly (P = 0.009) greater in endurance compared with power athletes.
DISCUSSION
We have used robust HDMSE profiling to compare the abundance of more than 1,000 proteins in gastrocnemius of rats artificially selected (31) as HRT or LRT. Stringent differential analysis (P values filtered to 1% FDR) identified widespread differences that cosegregate with exercise responsiveness and highlighted potential targets for future mechanistic research. SPEG-β exhibited the greatest difference in abundance and was 2.64-fold greater in HRT muscle. SPEG-β has been highlighted in at least two (18, 43) earlier nontargeted proteomic analyses of acute muscle responses to exercise. In the current work, proteins that coimmunoprecipitated with SPEG-β in HRT muscle include novel targets involved in c-Jun NH2-terminal kinase (JNK) signaling, which has recently emerged as a regulator of the adaptive response of muscle to exercise training (32). In humans, the G allele of SNP rs7564856 near the SPEG-β locus was associated with differences in endurance performance and muscle fiber type. These findings provide new insight to the role of SPEG-β in muscle and heighten interest in SPEG-β as a target for mechanistic research in molecular exercise physiology.
Lessard et al. (33) reports hyperactivation of JNK in sedentary LRT muscle and alterations to normal TGF-β signaling in response to endurance training. Follow-up analyses (32) found JNK phosphorylates the linker region of similar to mothers against decapentaplegic homolog 2 (SMAD2) and, thereby, inhibits canonical myostatin-TGF-β signaling. In human muscle, JNK phosphorylation of the SMAD2 linker region is particularly robust after resistance rather than endurance exercise (32). Phosphorylation of the SMAD2 linker region potentiates muscle adaptation to resistance exercise, but activation of JNK in response to endurance exercise (i.e., as in LRT muscle) appears to be an inappropriate response and is associated with blunted adaptation to endurance training (33). The events upstream of exercise-induced phosphorylation of SMAD2 by JNK have not yet been elucidated. Interestingly, several of the proteins that coimmunoprecipitated with SPEG-β in HRT muscle (Table 3) are implicated in JNK and TGF-β signaling and could represent viable candidates to investigate differences in TGF-β-mediated responses to endurance exercise. Caveolin-1 (CAV1) coimmunoprecipitated with SPEG-β, and a greater expression of CAV1 mRNA was previously highlighted by transcriptome profiling of HRT/LRT muscle (28). CAV1 inhibits TGF-β-mediated activation of SMAD2 by promoting degradation of TGF-β receptors (32). Similarly, ventricular zone-expressed PH domain-containing protein homolog 1 (VEPH1) may interfere with TGF-β-targeted signaling by impeding nuclear accumulation of SMAD2 (49). Serine/threonine-protein kinase (TAOK3), which was initially named JNK inhibitory kinase (55), downregulates the activity of JNK (26) and, therefore, may be the most likely of the putative SPEG-β interacting partners to play a mechanistic role in modulating muscle adaption to endurance training.
Targeted investigations of SPEG-β binding partners in skeletal muscle report SPEG-β interacts with myotubularin and colocalizes with other proteins of the junctional sarcoplasmic reticulum (SR), including the dihydropyridine receptor, sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (SERCA), and the Z-band protein, desmin (1). Accordingly, skeletal muscles of SPEG knockout mice exhibit centronuclear myopathy (1), and deficits in force that are associated with impaired RyR1-meditated Ca2+ release from the SR (23). Similar protein-protein interactions also occur in cardiac muscle (46), where SPEG-β phosphorylates SERCA2a and promotes SERCA2a oligomerization, which is associated with enhanced Ca2+ transport activity (45). In the current analysis, SERCA2 coimmunoprecipitated in control experiments as well as with SPEG-β and so was removed from the list (Table 3) of putative SPEG-β interacting partners. However, dynamin 2 (DNM2) was among the putative interaction partners of SPEG-β. Overexpression of DNM2 increases resting intracellular calcium concentration ([Ca2+]i) and is associated with impaired contractile properties and centronuclear myopathy (14). Valosin-containing protein (VSP, also known as transitional endoplasmic ATPase) belongs to the AAA+ ATPase family and immunoprecipitated with SPEG-β. VSP is required for lysosomal network dynamics, and upon its inhibition lysosomal network and autophagy are impaired (25).
Pathway analysis of proteins enriched in HRT muscle did not find significant enrichment of functional groups, but manual interrogation of Table 1 reveals the protein list is punctuated by features that are common to exercise-trained muscle. For example, HRT muscle has a greater abundance of the fatty acid translocase (CD36), mitochondrial superoxide dismutase (Mn SOD), and 10 kDa heat shock protein (CH10) that are also more abundant in diaphragm of exercise-trained rats (52). Consistent with these findings, gene expression of CD36 is also greater in HRT muscle (28). However, SPEG was not among the genes annotated on the array used in the previous (28) transcriptomic analysis. Notably, there was no overlap between proteins enriched in HRT muscle and those previously reported (9) to be enriched in the soleus proteome of HCR rats selected for high intrinsic running capacity. This is consistent with the difference in genetic heritability of training responsiveness vs. intrinsic capacity in rats (31) and humans (6).
HRT muscle had greater abundance of proteins associated with store-operated Ca2+ entry [SOCE (44)], including cGMP-gated cation channel alpha-1 (CNGA1), inositol monophosphatase (IMP), stromal interaction molecule 1 (STIM1), and 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1 (PLCG1). SOCE may assist in maintaining the myoplasmic pool of Ca2+ during repeated contractions and in doing so may modulate fatigue (41). STIM1 localizes to the longitudinal SR and enables rapid activation (13) of SOCE. Muscles of STIM1-haploinsufficent animals have impaired refilling of internal Ca2+ stores when subjected to repeated stimulation, fatigue sooner, and achieve smaller tetanic forces. In hepatocytes, PLCG1 can activate SOCE independently of STIM1 (36), but the role of PLCG1 in skeletal muscle has not yet been reported. CNGA1 is activated by SOCE (58) at physiological extracellular concentrations, which enables propagation of Ca2+ currents through CNGA1 (15). Maintenance of phosphatidylinositol 4,5-bisphosphate (PIP2) levels by IMP supports both SOCE and Ca2+ oscillating signals (3). Ca2+ signaling is indeed diversely involved in exercise-related adaptations. Oscillations in, and moderate rises to, [Ca2+]i upregulate mitochondrial biogenesis (24) and ATP production (19), whereas a prolonged elevation in [Ca2+]i is associated with muscle weakness and atrophy (38). Although speculative, exercise-related perturbations in [Ca2+]i in LCR muscle may result in prolonged increase in [Ca2+]i culminating in negative effects that eventually lead to a low-responsiveness phenotype. Increased [Ca2+]i results in oligomerization and overexpression of VDAC1, which leads to the formation of apoptosomes and cell death (27). VDAC1 is 1.27-fold more abundant in LRT, and the S100a1 Ca2+-binding protein, which is enriched in slow-twitch fibers (12) was 1.4-fold more abundant in LRT. Accounting for a lesser percentage of slow-twitch fibers in LRT gastrocnemius makes the relative difference per fiber more pronounced.
LRT gastrocnemius exhibited pronounced enrichment of enzymes involved in glucose metabolic processes (Fig. 2B), which may link with the previously (33) reported difference in myofiber profile between HRT and LRT. In the plantaris of LRT rats, the proportion of type I slow-twitch fibers (7%) is similar to sedentary Wistar rats (10a) but significantly less than in HRT muscle (20%), whereas no difference in myosin heavy chain (MyHC) type IIa myofiber abundance is observed (33). The current proteomic analysis focused on soluble muscle proteins, and it is not possible to report the myofiber profile of HRT/LRT gastrocnemius muscle from the current data. The 1.4-fold greater abundance of sarcoplasmic/endoplasmic reticulum ATPase-1 and parvalbumin in LRT muscle alongside the conspicuous enrichment of glycolytic enzymes may suggest a faster-twitch myofiber profile in LRT compared with HRT gastrocnemius. However, some mitochondrial proteins were also more abundant in LRT compared with HRT muscle (Table 2).
A potential difference in myofiber profile between HRT/LRT gastrocnemius raises the question of whether differences in SPEG-β protein abundance may also be a consequence of a difference in MyHC profile. Drexler et al. (12) reports comparative proteome analysis of mouse fast-twitch extensor digitorum longus (EDL) and slow-twitch soleus and reports SPEG-β among those proteins enriched in EDL. Murgia et al. (39) reports deep proteome analysis of individual myofibers extracted from vastus lateralis of younger and older human adults. We extracted SPEG-β abundance data from Murgia et al. (39) and investigated differences due to either MyHC fiber type or age by two-way analysis of variance. The abundance of SPEG-β was not different (P = 0.8144) between muscles of younger and older adults, but SPEG-β abundance was significantly (P = 0.0288) different across different myofiber types. SPEG-β abundance was 65% greater (P = 0.0129) in MyHC IIx than type I fibers, while fibers containing MyHC IIa were intermediate, and SPEG-β abundance was 21% greater (P = 0.7463) in MyHc IIa containing fibers compared with MyHC type I (Tukey HSD post-hoc analysis). Therefore, potential differences in myofiber profile between HRT and LRT gastrocnemius seem to be an unlikely explanation for the greater abundance of SPEG-β in HRT muscle.
SPEG-β is serine/threonine protein kinase homologous to proteins of the myosin light chain kinase family (54). In C2C12 myoblasts, expression of the alpha isoform of SPEG co-occurs with myoblast differentiation and the emergence of myosin heavy chain expression (21), whereas SPEG-β is solely detected in adult muscle in vivo (21), suggesting expression of the beta isoform may be instigated during postnatal maturation in response to contractile activity. SPEG-β is a phosphoprotein, and we (18) reported greater phosphorylation of SPEG-β in the rat heart 0–3 h after an incremental exercise test to V̇o2max. Similarly, Potts et al. (43) report phosphorylation of SPEG-β occurs in mouse skeletal muscle 1 h after a protocol of maximal-intensity contractions. In the current work, the abundance of SPEG-β is greater (2.64-fold, P = 0.0014) in gastrocnemius of rats that exhibit high responsiveness to endurance training. Although we did not investigate the phosphorylation status of SPEG-β in HRT/LRT gastrocnemius, the evidence to date suggests SPEG-β functions as a mechanosensitive kinase in striated muscle. In the future, it would be interesting to investigate whether either SPEG-β abundance or phosphorylation status changes in response to long-term endurance training. We were unable to find data reporting SPEG-β abundance in trained versus untrained muscle, but we did identify that the frequency of the G variant of SNP rs7564856 (that increases the expression of the SPEG gene) is greater in endurance compared with power athletes. The G allele of SNP rs7564856 was also significantly associated with increased proportion of slow-twitch muscle fibers in female endurance athletes. In addition, in 130,000 UK Biobank participants, rs7564856 is associated (P = 1.63*10−11) with the pulmonary ratio of forced expiratory volume in the first one second/forced vital capacity (29), which is a surrogate indicator of V̇o2max (11).
In conclusion, artificial selection for high responsiveness to endurance training is associated with a greater abundance of SPEG-β in rat gastrocnemius. Co-IP of SPEG-β in HRT muscle discovered putative interacting proteins that may link with previously reported differences in TGF-β signaling in exercised muscle. In humans, genetic polymorphisms near the SPEG gene locus are associated with higher expression of SPEG-β, which may favor endurance phenotypes. These findings alongside recent reports of acute phosphorylation of SPEG-β in cardiac (18) and skeletal muscle (43) in response to acute exercise support the hypothesis that SPEG-β is an important component in the adaptational response of muscle to exercise and warrants further mechanistic study.
GRANTS
The exercise rat models are funded by the National Institutes of Health’s Office of Research Infrastructure Programs grant P40OD-021331. The rat models for low and high exercise response to training are maintained as an international resource with support from the Department of Physiology & Pharmacology, University of Toledo College of Medicine, Toledo, OH. The Russian study was supported in part by Russian Science Foundation Grant 17-15-01436 “Comprehensive analysis of the contribution of genetic, epigenetic and environmental factors in the individual variability of the composition of human muscle fibers”; DNA sample collection, genotyping and determination of muscle fiber composition of Russian subjects.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.L.B., L.G.K., and J.G.B. conceived and designed research; D.K., J.C., H.K., E.A.S., O.V.B., A.K.L., D.V.P., E.V.G., I.I.A., and J.G.B. performed experiments; D.K., J.C., H.K., E.A.S., O.V.B., A.K.L., D.V.P., E.V.G., I.I.A., and J.G.B. analyzed data; D.K., H.K., E.A.S., O.V.B., A.K.L., D.V.P., E.V.G., I.I.A., and J.G.B. interpreted results of experiments; J.G.B. prepared figures; D.K. and J.G.B. drafted manuscript; D.K., J.C., H.K., S.L.B., L.G.K., and J.G.B. edited and revised manuscript; D.K., J.C., H.K., E.A.S., O.V.B., A.K.L., D.V.P., E.V.G., I.I.A., S.L.B., L.G.K., and J.G.B. approved final version of manuscript.
ACKNOWLEDGMENTS
Contact L. G. Koch (Lauren.Koch2@UToledo.Edu) or S. L. Britton (brittons@umich.edu) for information on the exercise rat models.
REFERENCES
- 1.Agrawal PB, Pierson CR, Joshi M, Liu X, Ravenscroft G, Moghadaszadeh B, Talabere T, Viola M, Swanson LC, Haliloğlu G, Talim B, Yau KS, Allcock RJ, Laing NG, Perrella MA, Beggs AH. SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy. Am J Hum Genet 95: 218–226, 2014. doi: 10.1016/j.ajhg.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmetov I, Kulemin N, Popov D, Naumov V, Akimov E, Bravy Y, Egorova E, Galeeva A, Generozov E, Kostryukova E, Larin A, Mustafina L, Ospanova E, Pavlenko A, Starnes L, Żmijewski P, Alexeev D, Vinogradova O, Govorun V. Genome-wide association study identifies three novel genetic markers associated with elite endurance performance. Biol Sport 32: 3–9, 2015. doi: 10.5604/20831862.1124568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alswied A, Parekh AB. Ca2+ Influx through Store-operated Calcium Channels Replenishes the Functional Phosphatidylinositol 4,5-Bisphosphate Pool Used by Cysteinyl Leukotriene Type I Receptors. J Biol Chem 290: 29555–29566, 2015. doi: 10.1074/jbc.M115.678292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard C, An P, Rice T, Skinner JS, Wilmore JH, Gagnon J, Pérusse L, Leon AS, Rao DC. Familial aggregation of VO(2max) response to exercise training: results from the HERITAGE Family Study. J Appl Physiol (1985) 87: 1003–1008, 1999. doi: 10.1152/jappl.1999.87.3.1003. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard C, Rankinen T. Individual differences in response to regular physical activity. Med Sci Sports Exerc 33 Suppl: S446–S451, 2001. doi: 10.1097/00005768-200106001-00013. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard C, Rankinen T, Chagnon YC, Rice T, Pérusse L, Gagnon J, Borecki I, An P, Leon AS, Skinner JS, Wilmore JH, Province M, Rao DC. Genomic scan for maximal oxygen uptake and its response to training in the HERITAGE Family Study. J Appl Physiol (1985) 88: 551–559, 2000. doi: 10.1152/jappl.2000.88.2.551. [DOI] [PubMed] [Google Scholar]
- 7.Boulé NG, Weisnagel SJ, Lakka TA, Tremblay A, Bergman RN, Rankinen T, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C; HERITAGE Family Study . Effects of exercise training on glucose homeostasis: the HERITAGE Family Study. Diabetes Care 28: 108–114, 2005. doi: 10.2337/diacare.28.1.108. [DOI] [PubMed] [Google Scholar]
- 8.Burniston JG, Connolly J, Kainulainen H, Britton SL, Koch LG. Label-free profiling of skeletal muscle using high-definition mass spectrometry. Proteomics 14: 2339–2344, 2014. doi: 10.1002/pmic.201400118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burniston JG, Kenyani J, Gray D, Guadagnin E, Jarman IH, Cobley JN, Cuthbertson DJ, Chen Y-W, Wastling JM, Lisboa PJ, Koch LG, Britton SL. Conditional independence mapping of DIGE data reveals PDIA3 protein species as key nodes associated with muscle aerobic capacity. J Proteomics 106: 230–245, 2014. doi: 10.1016/j.jprot.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burniston JG, Kenyani J, Wastling JM, Burant CF, Qi NR, Koch LG, Britton SL. Proteomic analysis reveals perturbed energy metabolism and elevated oxidative stress in hearts of rats with inborn low aerobic capacity. Proteomics 11: 3369–3379, 2011. doi: 10.1002/pmic.201000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Cornachione AS, Benedini-Elias PC, Polizello JC, Carvalho LC, Mattiello-Sverzut AC. Characterization of fiber types in different muscles of the hindlimb in female weanling and adult Wistar rats. Acta Histochem Cytochem 44: 43–50, 2011. doi: 10.1267/ahc.10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimopoulou I, Tsintzas OK, Daganou M, Cokkinos DV, Tzelepis GE. Contribution of lung function to exercise capacity in patients with chronic heart failure. Respiration 66: 144–149, 1999. doi: 10.1159/000029356. [DOI] [PubMed] [Google Scholar]
- 12.Drexler HC, Ruhs A, Konzer A, Mendler L, Bruckskotten M, Looso M, Günther S, Boettger T, Krüger M, Braun T. On marathons and Sprints: an integrated quantitative proteomics and transcriptomics analysis of differences between slow and fast muscle fibers. Mol Cell Proteomics 11: M111.010801, 2012. doi: 10.1074/mcp.M111.010801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards JN, Friedrich O, Cully TR, von Wegner F, Murphy RM, Launikonis BS. Upregulation of store-operated Ca2+ entry in dystrophic mdx mouse muscle. Am J Physiol Cell Physiol 299: C42–C50, 2010. doi: 10.1152/ajpcell.00524.2009. [DOI] [PubMed] [Google Scholar]
- 14.Fraysse B, Guicheney P, Bitoun M. Calcium homeostasis alterations in a mouse model of the Dynamin 2-related centronuclear myopathy. Biol Open 5: 1691–1696, 2016. doi: 10.1242/bio.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frings S, Seifert R, Godde M, Kaupp UB. Profoundly different calcium permeation and blockage determine the specific function of distinct cyclic nucleotide-gated channels. Neuron 15: 169–179, 1995. doi: 10.1016/0896-6273(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 16.Grundy SM, Brewer HB Jr, Cleeman JI, Smith SC Jr, Lenfant C; American Heart Association; National Heart, Lung, and Blood Institute . Definition of metabolic syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation 109: 433–438, 2004. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 17.GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration & Visualization—EBI; Genome Browser Data Integration & Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis & Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group; Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature 550: 204–213, 2017. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, Isserlin R, Emili A, Burniston JG. Exercise-responsive phosphoproteins in the heart. J Mol Cell Cardiol 111: 61–68, 2017. doi: 10.1016/j.yjmcc.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Hajnóczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell 82: 415–424, 1995. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- 20.Holloway KV, O’Gorman M, Woods P, Morton JP, Evans L, Cable NT, Goldspink DF, Burniston JG. Proteomic investigation of changes in human vastus lateralis muscle in response to interval-exercise training. Proteomics 9: 5155–5174, 2009. doi: 10.1002/pmic.200900068. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh CM, Fukumoto S, Layne MD, Maemura K, Charles H, Patel A, Perrella MA, Lee ME. Striated muscle preferentially expressed genes α and β are two serine/threonine protein kinases derived from the same gene as the aortic preferentially expressed gene-1. J Biol Chem 275: 36966–36973, 2000. doi: 10.1074/jbc.M006028200. [DOI] [PubMed] [Google Scholar]
- 22.Hubal MJ, Gordish-Dressman H, Thompson PD, Price TB, Hoffman EP, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Seip RL, Clarkson PM. Variability in muscle size and strength gain after unilateral resistance training. Med Sci Sports Exerc 37: 964–972, 2005. [PubMed] [Google Scholar]
- 23.Huntoon V, Widrick JJ, Sanchez C, Rosen SM, Kutchukian C, Cao S, Pierson CR, Liu X, Perrella MA, Beggs AH, Jacquemond V, Agrawal PB. SPEG-deficient skeletal muscles exhibit abnormal triad and defective calcium handling. Hum Mol Genet 27: 1608–1617, 2018. doi: 10.1093/hmg/ddy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivarsson N, Mattsson CM, Cheng AJ, Bruton JD, Ekblom B, Lanner JT, Westerblad H. SR Ca2+ leak in skeletal muscle fibers acts as an intracellular signal to increase fatigue resistance. J Gen Physiol 151: 567–577, 2019. doi: 10.1085/jgp.201812152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson ML, Irving BA, Lanza IR, Vendelbo MH, Konopka AR, Robinson MM, Henderson GC, Klaus KA, Morse DM, Heppelmann C, Bergen HR III, Dasari S, Schimke JM, Jakaitis DR, Nair KS. Differential Effect of Endurance Training on Mitochondrial Protein Damage, Degradation, and Acetylation in the Context of Aging. J Gerontol A Biol Sci Med Sci 70: 1386–1393, 2015. doi: 10.1093/gerona/glu221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapfhamer D, King I, Zou ME, Lim JP, Heberlein U, Wolf FW. JNK pathway activation is controlled by Tao/TAOK3 to modulate ethanol sensitivity. PLoS One 7: e50594, 2012. doi: 10.1371/journal.pone.0050594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keinan N, Pahima H, Ben-Hail D, Shoshan-Barmatz V. The role of calcium in VDAC1 oligomerization and mitochondria-mediated apoptosis. Biochim Biophys Acta 1833: 1745–1754, 2013. doi: 10.1016/j.bbamcr.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Keller P, Vollaard NBJ, Gustafsson T, Gallagher IJ, Sundberg CJ, Rankinen T, Britton SL, Bouchard C, Koch LG, Timmons JA. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol (1985) 110: 46–59, 2011. doi: 10.1152/japplphysiol.00634.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kichaev G, Bhatia G, Loh P-R, Gazal S, Burch K, Freund MK, Schoech A, Pasaniuc B, Price AL. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am J Hum Genet 104: 65–75, 2019. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch LG, Kemi OJ, Qi N, Leng SX, Bijma P, Gilligan LJ, Wilkinson JE, Wisløff H, Høydal MA, Rolim N, Abadir PM, van Grevenhof EM, Smith GL, Burant CF, Ellingsen O, Britton SL, Wisløff U. Intrinsic aerobic capacity sets a divide for aging and longevity. Circ Res 109: 1162–1172, 2011. doi: 10.1161/CIRCRESAHA.111.253807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch LG, Pollott GE, Britton SL. Selectively bred rat model system for low and high response to exercise training. Physiol Genomics 45: 606–614, 2013. doi: 10.1152/physiolgenomics.00021.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessard SJ, MacDonald TL, Pathak P, Han MS, Coffey VG, Edge J, Rivas DA, Hirshman MF, Davis RJ, Goodyear LJ. JNK regulates muscle remodeling via myostatin/SMAD inhibition. Nat Commun 9: 3030, 2018. doi: 10.1038/s41467-018-05439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lessard SJ, Rivas DA, Alves-Wagner AB, Hirshman MF, Gallagher IJ, Constantin-Teodosiu D, Atkins R, Greenhaff PL, Qi NR, Gustafsson T, Fielding RA, Timmons JA, Britton SL, Koch LG, Goodyear LJ. Resistance to aerobic exercise training causes metabolic dysfunction and reveals novel exercise-regulated signaling networks. Diabetes 62: 2717–2727, 2013. doi: 10.2337/db13-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin Y, Hradetzky E, Bahn S. Quantification of proteins using data-independent analysis (MSE) in simple andcomplex samples: a systematic evaluation. Proteomics 11: 3273–3287, 2011. doi: 10.1002/pmic.201000661. [DOI] [PubMed] [Google Scholar]
- 35.Li GZ, Vissers JPC, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9: 1696–1719, 2009. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 36.Litjens T, Nguyen T, Castro J, Aromataris EC, Jones L, Barritt GJ, Rychkov GY. Phospholipase C-γ1 is required for the activation of store-operated Ca2+ channels in liver cells. Biochem J 405: 269–276, 2007. doi: 10.1042/BJ20061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marton O, Koltai E, Takeda M, Koch LG, Britton SL, Davies KJA, Boldogh I, Radak Z. Mitochondrial biogenesis-associated factors underlie the magnitude of response to aerobic endurance training in rats. Pflugers Arch 467: 779–788, 2015. doi: 10.1007/s00424-014-1554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matecki S, Dridi H, Jung B, Saint N, Reiken SR, Scheuermann V, Mrozek S, Santulli G, Umanskaya A, Petrof BJ, Jaber S, Marks AR, Lacampagne A. Leaky ryanodine receptors contribute to diaphragmatic weakness during mechanical ventilation. Proc Natl Acad Sci USA 113: 9069–9074, 2016. doi: 10.1073/pnas.1609707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murgia M, Toniolo L, Nagaraj N, Ciciliot S, Vindigni V, Schiaffino S, Reggiani C, Mann M. Single Muscle Fiber Proteomics Reveals Fiber-Type-Specific Features of Human Muscle Aging. Cell Reports 19: 2396–2409, 2017. doi: 10.1016/j.celrep.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 40.Myers J, Kaykha A, George S, Abella J, Zaheer N, Lear S, Yamazaki T, Froelicher V. Fitness versus physical activity patterns in predicting mortality in men. Am J Med 117: 912–918, 2004. doi: 10.1016/j.amjmed.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 41.Pan Z, Brotto M, Ma J. Store-operated Ca2+ entry in muscle physiology and diseases. BMB Rep 47: 69–79, 2014. doi: 10.5483/BMBRep.2014.47.2.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickering C, Suraci B, Semenova EA, Boulygina EA, Kostryukova ES, Kulemin NA, Borisov OV, Khabibova SA, Larin AK, Pavlenko AV, Lyubaeva EV, Popov DV, Lysenko EA, Vepkhvadze TF, Lednev EM, Leońska-Duniec A, Pająk B, Chycki J, Moska W, Lulińska-Kuklik E, Dornowski M, Maszczyk A, Bradley B, Kana-Ah A, Cięszczyk P, Generozov EV, Ahmetov II. A Genome-Wide Association Study of Sprint Performance in Elite Youth Football Players. J Strength Cond Res 33: 2344–2351, 2019. doi: 10.1519/JSC.0000000000003259. [DOI] [PubMed] [Google Scholar]
- 43.Potts GK, McNally RM, Blanco R, You J-S, Hebert AS, Westphall MS, Coon JJ, Hornberger TA. A map of the phosphoproteomic alterations that occur after a bout of maximal-intensity contractions. J Physiol 595: 5209–5226, 2017. [Erratum in J Physiol 597: 967–968, 2019.] doi: 10.1113/JP273904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prakriya M, Lewis RS. Store-Operated Calcium Channels. Physiol Rev 95: 1383–1436, 2015. doi: 10.1152/physrev.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quan C, Li M, Du Q, Chen Q, Wang H, Campbell D, Fang L, Xue B, MacKintosh C, Gao X, Ouyang K, Wang HY, Chen S. SPEG Controls Calcium Reuptake Into the Sarcoplasmic Reticulum Through Regulating SERCA2a by Its Second Kinase-Domain. Circ Res 124: 712–726, 2019. doi: 10.1161/CIRCRESAHA.118.313916. [DOI] [PubMed] [Google Scholar]
- 46.Quick AP, Wang Q, Philippen LE, Barreto-Torres G, Chiang DY, Beavers D, Wang G, Khalid M, Reynolds JO, Campbell HM, Showell J, McCauley MD, Scholten A, Wehrens XHT. SPEG (Striated Muscle Preferentially Expressed Protein Kinase) Is Essential for Cardiac Function by Regulating Junctional Membrane Complex Activity. Circ Res 120: 110–119, 2017. doi: 10.1161/CIRCRESAHA.116.309977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivas DA, Lessard SJ, Saito M, Friedhuber AM, Koch LG, Britton SL, Yaspelkis BB III, Hawley JA. Low intrinsic running capacity is associated with reduced skeletal muscle substrate oxidation and lower mitochondrial content in white skeletal muscle. Am J Physiol Regul Integr Comp Physiol 300: R835–R843, 2011. doi: 10.1152/ajpregu.00659.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross R, Goodpaster BH, Koch LG, Sarzynski MA, Kohrt WM, Johannsen NM, Skinner JS, Castro A, Irving BA, Noland RC, Sparks LM, Spielmann G, Day AG, Pitsch W, Hopkins WG, Bouchard C. Precision exercise medicine: understanding exercise response variability. Br J Sports Med 53: 1141–1153, 2019. doi: 10.1136/bjsports-2018-100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shathasivam P, Kollara A, Ringuette MJ, Virtanen C, Wrana JL, Brown TJ. Human ortholog of Drosophila Melted impedes SMAD2 release from TGF-β receptor I to inhibit TGF-β signaling. Proc Natl Acad Sci USA 112: E3000–E3009, 2015. doi: 10.1073/pnas.1504671112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva JC, Gorenstein MV, Li G, Vissers JPC, Geromanos SJ. Absolute Quantification of Proteins by LCMS E. Mol Cell Proteomics 5: 144–156, 2006. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Sollanek KJ, Burniston JG, Kavazis AN, Morton AB, Wiggs MP, Ahn B, Smuder AJ, Powers SK. Global Proteome Changes in the Rat Diaphragm Induced by Endurance Exercise Training. PLoS One 12: e0171007, 2017. doi: 10.1371/journal.pone.0171007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445, 2003. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sutter SB, Raeker MO, Borisov AB, Russell MW. Orthologous relationship of obscurin and Unc-89: phylogeny of a novel family of tandem myosin light chain kinases. Dev Genes Evol 214: 352–359, 2004. doi: 10.1007/s00427-004-0413-5. [DOI] [PubMed] [Google Scholar]
- 55.Tassi E, Biesova Z, Di Fiore PP, Gutkind JS, Wong WT. Human JIK, a novel member of the STE20 kinase family that inhibits JNK and is negatively regulated by epidermal growth factor. J Biol Chem 274: 33287–33295, 1999. doi: 10.1074/jbc.274.47.33287. [DOI] [PubMed] [Google Scholar]
- 56.Warburton DE, Nicol CW, Bredin SS. Health benefits of physical activity: the evidence. CMAJ 174: 801–809, 2006. doi: 10.1503/cmaj.051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wisløff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernström M, Rezaei K, Lee SJ, Koch LG, Britton SL. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science 307: 418–420, 2005. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 58.Wu S, Moore TM, Brough GH, Whitt SR, Chinkers M, Li M, Stevens T. Cyclic nucleotide-gated channels mediate membrane depolarization following activation of store-operated calcium entry in endothelial cells. J Biol Chem 275: 18887–18896, 2000. doi: 10.1074/jbc.M002795200. [DOI] [PubMed] [Google Scholar]
- 59.Yi Z, Bowen BP, Hwang H, Jenkinson CPL, Coletta DK, Lefort N, Bajaj M, Kashyap S, Berria R, De Filippis EA, Mandarino LJ. Global relationship between the proteome and transcriptome of human skeletal muscle. J Proteome Res 7: 3230–3241, 2008. doi: 10.1021/pr800064s. [DOI] [PMC free article] [PubMed] [Google Scholar]