Abstract
Background
Campylobacter and Salmonella, particularly non-typhoidal Salmonella, are important bacterial enteric pathogens of humans which are often carried asymptomatically in animal reservoirs. Bacterial foodborne infections, including those derived from meat, are associated with illness and death globally but the burden is disproportionately high in Africa. Commercial meat production is increasing and intensifying in many African countries, creating opportunities and threats for food safety.
Methods
Following Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines, we searched six databases for English language studies published through June 2016, that reported Campylobacter or Salmonella carriage or infection prevalence in food animals and contamination prevalence in food animal products from African countries. A random effects meta-analysis and multivariable logistic regression were used to estimate the species-specific prevalence of Salmonella and Campylobacter and assess relationships between sample type and region and the detection or isolation of either pathogen.
Results
Seventy-three studies reporting Campylobacter and 187 studies reporting Salmonella across 27 African countries were represented. Adjusted prevalence calculations estimate Campylobacter detection in 37.7% (95% CI 31.6–44.3) of 11,828 poultry samples; 24.6% (95% CI 18.0–32.7) of 1975 pig samples; 17.8% (95% CI 12.6–24.5) of 2907 goat samples; 12.6% (95% CI 8.4–18.5) of 2382 sheep samples; and 12.3% (95% CI 9.5–15.8) of 6545 cattle samples. Salmonella were detected in 13.9% (95% CI 11.7–16.4) of 25,430 poultry samples; 13.1% (95% CI 9.3–18.3) of 5467 pig samples; 9.3% (95% CI 7.2–12.1) of 2988 camel samples; 5.3% (95% CI 4.0–6.8) of 72,292 cattle samples; 4.8% (95% CI 3.6–6.3) of 11,335 sheep samples; and 3.4% (95% CI 2.2–5.2) of 4904 goat samples. ‘External’ samples (e.g. hide, feathers) were significantly more likely to be contaminated by both pathogens than ‘gut’ (e.g. faeces, cloaca) while meat and organs were significantly less likely to be contaminated than gut samples.
Conclusions
This study demonstrated widespread prevalence of Campylobacter species and Salmonella serovars in African food animals and meat, particularly in samples of poultry and pig origin. Source attribution studies could help ascertain which food animals are contributing to human campylobacteriosis and salmonellosis and direct potential food safety interventions.
Keywords: Africa, Campylobacter, Food animals, Food safety, Prevalence, Salmonella
Highlights
-
•
Campylobacter prevalence data was compiled from 14 African countries.
-
•
Salmonella prevalence data was compiled from 27 African countries.
-
•
Campylobacter and Salmonella were most prevalent in poultry and pig samples.
-
•
C. jejuni was the most predominant Campylobacter species except in pigs where C. coli predominates.
-
•
S. enterica serovar Typhimurium was the most commonly identified Salmonella serovar.
1. Introduction
Campylobacter and non-typhoidal Salmonella (NTS) are bacterial enteric pathogens associated with food animal reservoirs. They are transmitted to humans predominantly by contaminated food and water. Foodborne zoonoses, including those caused by Campylobacter and NTS, are recognised by the World Health Organization (WHO) as important causes of human illness and death worldwide (Havelaar et al., 2015). It is estimated that Campylobacter are responsible for >95 million foodborne illnesses and >21,000 deaths and NTS for >78 million foodborne illnesses and >59,000 deaths globally (Havelaar et al., 2015). The burden of bacterial foodborne disease, including disease caused by Campylobacter and NTS, is disproportionately higher in African regions compared with other parts of the world, with the number of Disability Adjusted Life Years (DALYs) per 100,000 exceeding that of other global regions (Havelaar et al., 2015).
While Campylobacter and NTS infections are usually self-limiting in healthy humans, complicated disease may develop in some. Bacteraemia and immunological syndromes such as reactive arthritis have been linked to both pathogens (Carter and Hudson, 2009; Sandhu and Paul, 2014) while Guillain-Barré syndrome is associated with Campylobacter infection (Esan et al., 2017, World Health Organization (WHO), 2012). For both Campylobacter and NTS, bacteraemia is more common among children, the elderly, and immunocompromised persons, especially those with HIV/AIDS (Food and Drug Administration (FDA), 2012). Almost 70% of the global HIV burden is in sub-Saharan Africa (Joint United Nations (UN) Programme on HIV/AIDS, 2017). An increased risk of NTS bacteraemia has also been linked with recent or current malaria, and malnutrition (Feasey et al., 2012).
Campylobacter are usually a non-pathogenic component of the gastrointestinal tract microbiota of livestock such as cattle, pigs, and sheep, with poultry considered to be the major reservoir, particularly of C. jejuni (Sahin et al., 2002; Skarp et al., 2016). Similarly, NTS have been isolated from the gastrointestinal tract of birds and mammals, including poultry and livestock (Barrow et al., 1988; Ellis, 1969; Gay et al., 1994). The transmission of potentially pathogenic Campylobacter or NTS from animal hosts to humans is predominantly via faecally contaminated food and water. However, humans may be infected by contact with live animals and environments contaminated with animal faeces and subsequent incidental ingestion of pathogens (Cummings et al., 2012, World Health Organisation (WHO), 2017). During processing for meat, animal gut microbiota and associated pathogens from the caecum, cloaca, intestines, or faeces may be transferred directly to the carcass or meat and organ surfaces. Transfer may also be indirect, such as from human hands and equipment. Both pathways may result in transfer of gastro-intestinal flora to meat and organ surfaces or to external animal surfaces such as feathers, fleece, hide, and skin. Such transfer is associated with increased risk for human infection from consumption of contaminated meat and cross-contamination of other foods. Uncooked produce may also be a direct source of human infection with zoonotic pathogens through contamination by animal faeces or untreated irrigation water (Food and Drug Administration (FDA), 2012).
Meat production is central to livelihoods in many African countries, with meat from livestock and poultry being a key protein source in subsistence communities (OECD/FAO, 2016). In many low-resource settings, industrialisation, urbanisation, and the shift from planned to market economies are leading to rapid changes in the way that food is produced, distributed, sold, and consumed (Carron et al., 2018; Grace, 2017). Such market-driven changes within agricultural production towards wider distribution networks, centralised processing, larger-scale and more intensive systems, have been linked to the emergence of zoonotic diseases (Jones et al., 2013) and the potential impact on food safety within low- and middle-income countries is increasingly recognised (World Health Organisation (WHO) Regional Office for Africa, 2017). Data on key bacterial pathogens in the meat production pathway in Africa are limited and are not currently available in aggregate form. To inform food safety policy and to identify data gaps, we undertook a systematic review on Campylobacter and Salmonella prevalence in food animals and meat in Africa.
2. Methods
2.1. Study design and systematic review protocol
References were sought and identified following the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines (Moher et al., 2009) (Supplementary File 1 checklist). Studies were searched in African Index Medicus (AIM), CABI Global Health, Proquest Health and Medicine, PubMed, Scopus, and Web of Science. Search terms are listed in Table 1 and no date restrictions were applied. The last search took place on 21 July 2016.
Table 1.
Full search strategies for database searches, date searched, database name, and number of articles retrieved systematic review of prevalence of Campylobacter and Salmonella in African food animals and meat, 1953–2016.
| Date Search performed | Database | Number of articles retrieved | Search string/terms and limits |
|---|---|---|---|
| 11 Jul 16 | Africa Index Medicus | 46 | Campylobacter OR Salmonella |
| 21 Jul 16 | CABI Global Health | 2251 | All = (Search #1) AND All = (Search #2) AND All = (Search #3) |
| 11 Jul 16 | ProQuest | 774 | Anywhere = (Search #1) AND All = (Search #2) AND All = (Search #3) |
| 11 Jul 16 | PubMed | 1423 | All = (Search #1) AND All = (Search #2) AND All = (Search #3) |
| 11 Jul 16 | Scopus | 2059 | Abstract, Title, Keyword = (Search #1) AND Abstract, Title, Keyword = (Search #2) AND Abstract, Title, Keyword = (Search #3) in the categories of Life Sciences and Health Sciences |
| 11 Jul 16 | Web of Science | 1051 | TOPIC = (Search #1) AND TOPIC = (Search #2) AND TOPIC = (Search #3) |
| Where: | |||
| Search #1 | (Campylobacter*) OR (Salmonell*) | ||
| Search #2 | (cattle) OR (cow) OR (bull) OR (beef) OR (heifer) OR (steer) OR (bovine) OR (calf) OR (calves) OR (sheep) OR (mutton) OR (hogget) OR (lamb) OR (ovine) OR (goat) OR (caprine) OR (chicken) OR (avian) OR (poultry) OR (hen) OR (chick) OR (broiler) OR (layer) OR (pork) OR (porcine) OR (pig) OR (camel) OR (offal) OR (food) OR (meat) | ||
| Search #3 | (Africa*) OR (algeria) OR (angola) OR (benin) OR (botswana) OR (burkina faso) OR (burundi) OR (cameroon) OR (cape verde) OR (central african republic) OR (chad) OR (comoros) OR (congo) OR (cote d'ivoire) OR (ivory coast) OR (democratic republic of the congo) OR (zaire) OR (djibouti) OR (egypt) OR (equatorial guinea) OR (eritrea) OR (ethiopia) OR (gabon) OR (gambia) OR (ghana) OR (guinea) OR (guinea-bissau) OR (kenya) OR (lesotho) OR (liberia) OR (libya) OR (madagascar) OR (malawi) OR (mali) OR (mauritania) OR (mauritius) OR (mayotte) OR (morocco) OR (mozambique) OR (namibia) OR (niger) OR (nigeria) OR (reunion) OR (rwanda) OR (saint helena) OR (sao tome and principe) OR (senegal) OR (seychelles) OR (sierra leone) OR (somalia) OR (south africa) OR (south sudan) OR (sudan) OR (swaziland) OR (tanzania) OR (togo) OR (tunisia) OR (uganda) OR (western sahara) OR (zambia) OR (zimbabwe) |
2.2. Search strategy
Article titles and abstracts were reviewed for suitability for inclusion by KMT. They were selected for full text review if the studies investigated Campylobacter or Salmonella, reported on samples collected from food animals, their organs or meat, and data collection took place in African regions or countries as defined by the United Nations (UN) statistics division (United Nations (UN) Statistics Division, 2016). Full text articles were reviewed independently by two authors (KMT, WdG) to determine if each article met pre-determined inclusion and exclusion criteria (Supplementary File 2). Articles were included for full text review if the full text article could be retrieved, if it reported primary data, if the article reported isolation by culture or detection by PCR of Campylobacter or Salmonella, in individual food animals or meat products, regardless of laboratory methods used, if the prevalence of contamination with either pathogen could be calculated from information available in the paper, and if the materials tested represented ‘gut’, ‘external’, or meat and organ samples from individual animals. Gut samples were defined as caecal, intestinal, cloacal contents and faeces, which have the potential to contaminate ‘external’ and meat and organ samples from individual animals. External samples were defined as those from the exterior of individual animals, including hide, skin, or feathers. Meat and organ samples included bile, lymph nodes, raw organs, or meat. Serological studies were excluded. Studies were excluded if the numerator (i.e. number positive) and denominator (i.e. number tested) information were not reported at the species and sample type level. Studies were excluded if samples were frozen at the time of sample collection to avoid underestimates of prevalence data, particularly of Campylobacter, as a result of loss of viable cells due to freezing. Studies were excluded if they were solely from sick animals, from free-living wildlife, or from farmed game animals. Studies were also excluded if they were in a language other than English. When required, a third author (JAC) served as tiebreaker, independently reviewing articles to resolve disagreement between the two primary reviewers.
2.3. Data extraction
From each included article, we extracted information on source food animal species, sample type, the total number of samples tested. The number of Campylobacter or Salmonella isolated or detected was extracted to determine pathogen prevalence. Sample location data, including UN statistics division African geographic region (United Nations (UN) Statistics Division, 2016), country, administrative level, city, town or village, and Global Positioning System (GPS) coordinates were extracted from articles when reported. Where available, data were recorded on the prevalence of individual Campylobacter species, and Salmonella enterica serovars or serogroups, as per standards of the WHO Collaborating Centre for Reference and Research on Salmonella (Grimont and Weill, 2007). During data extraction, elements of sample selection, sample handling, and laboratory methods were noted, including length of study, conditions and time of transport to laboratory, amount of sample tested, media used, and temperature and gas conditions of incubation. A formal bias assessment was established (Supplementary Table 1), assigning low (L), moderate (M), high (H), unknown (U), implied (I), yes (Y), no (N), or not applicable (NA) to each potential introduction of bias. The bias elements considered in the formal assessment relating to sample selection and handling were study length, temperature and time of transport to the laboratory. The bias elements relating to laboratory testing were amount of sample tested, type of isolation or detection methods, incubation conditions, and the quality of the serotyping methods used. An overall assessment of low, moderate or high risk of bias was assigned to each included article.
2.4. Data analysis
Prevalence estimates were calculated from pooled data for each pathogen by livestock species, geographic region and sample type, and for each geographic region and sample type by livestock species. An inverse variance approach with study ID as a random effect was used to derive weighted prevalence estimates (Marín-Martínez and Sánchez-Meca, 2010). In the presence of small numbers of total samples for some studies, the logit transformation was used (Barendregt et al., 2013). Between study heterogeneity was quantified using the I2 statistic, which provides an estimate of the percentage of total variation between studies that is due to prevalence differences rather than to chance variation (Higgins et al., 2003). Eighty percent prediction intervals (80% PI) were derived to provide an estimate of the interval between which the prevalence of a future study of Campylobacter or Salmonella prevalence could be expected to fall with 80% probability (IntHout et al., 2016). Weighted average prevalence estimates, their 95% confidence intervals (95% CI), 80% PI and the I2 statistic were derived using the meta package (Schwarzer, 2007) in the R statistical environment, version 3.4.2. (http://cran.r-project.org/).
Mixed effects logistic regression was used to explore predictors of Campylobacter and Salmonella infection or colonisation and contamination. Publication ID was included as a random effect and geographic region, sample type (i.e. ‘gut’, ‘external’, ‘meat and organ’) and species type were included as fixed effects. Data were then disaggregated by host species and separate mixed effects logistic regression models constructed for each pathogen for camels, cattle, goats, pigs, poultry, and sheep. Host species-specific models included publication ID as a random effect and sample type and geographic region as fixed effects. Where data were available, Northern Africa was used as the referent region and ‘gut’ as the baseline sample type. Deviations are stated when chosen referent baselines did not have data. Poultry was used as the baseline for species type and compared to pigs and a combined category containing all other species (buffalo, camels, cattle, goats, and sheep). The overall contribution of each fixed effect to model fit was assessed using a likelihood ratio test. In addition to the I2 statistic described above, between study heterogeneity in prevalence was quantified for the overall logistic regression model and species-specific logistic regression models using the median odds ratio (MOR). This statistic represents the median value of the odds ratio when comparing group (study) level residuals from randomly selected pairs of samples from different studies (Larsen and Merlo, 2005). It can be considered to provide an indication of the magnitude of the difference in odds of animal infection or carriage or sample contamination when comparing two studies: where there is little between study variation, the MOR would be close to one. Mixed effects logistic regression models were constructed using the lme4 package (Bates et al., 2015) in R.
3. Results
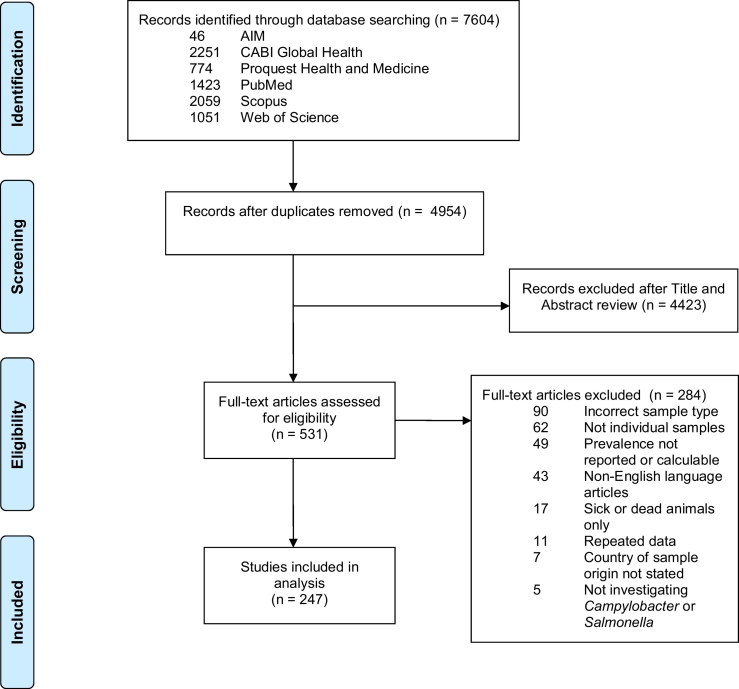
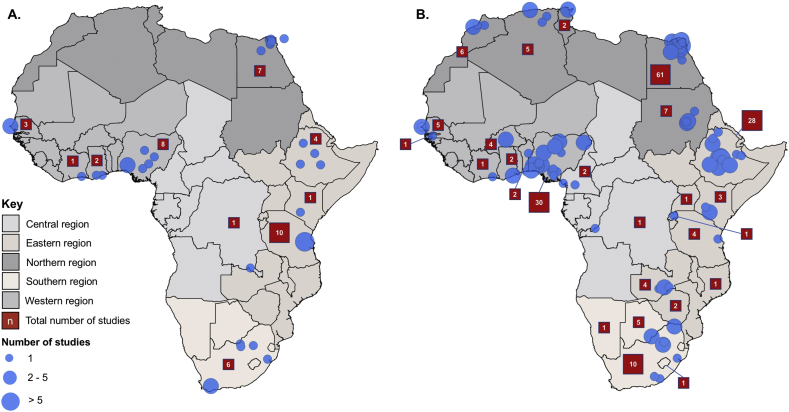
After removing duplicate articles from the searches of six selected databases, 4954 articles were available for title and abstract screening. Of these 531 (10.7%) were identified as potentially relevant and 247 (5.0%) were eligible for inclusion after full text review (Fig. 1). Sixty articles from 14 countries reported prevalence data on Campylobacter, 174 articles from 27 countries reported prevalence data on Salmonella, and 13 articles from eight countries reported prevalence data on both pathogens. No prevalence data were excluded as a result of the quality assessment. Table 2 shows the decades in which the sampling began in each study. The geographic location of included studies is represented in Fig. 2. The number of studies from each country and the animal species investigated are listed in Table 3.
Fig. 1.
PRISMA flowchart showing identification, screening, and selection of eligible articles for inclusion in systematic review, 1953–2016.
Table 2.
Numbers of articles on Campylobacter or Salmonella and Campylobacter and Salmonella prevalence included in the systematic review by year of when sampling and testing began.
| Years | Campylobacter | Salmonella | Campylobacter and Salmonella |
|---|---|---|---|
| 1951–1959 | 0 | 4 | 0 |
| 1960–1969 | 0 | 6 | 0 |
| 1970–1979 | 0 | 3 | 0 |
| 1980–1989 | 0 | 2 | 1 |
| 1990–1999 | 1 | 8 | 0 |
| 2000–2009 | 24 | 41 | 3 |
| 2010–2016 | 10 | 34 | 0 |
| Unspecified | 25 | 76 | 9 |
| TOTAL | 60 | 174 | 13 |
Fig. 2.
Map showing location of included studies describing (A) Campylobacter and (B) Salmonella contamination prevalence where geographic information was available (blue circles), 1953–2016. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Studies included in systematic review of the prevalence of Campylobacter and Salmonella in African food animals and animal food products by region, country, and animal species, 1953–2016.
3.1. Campylobacter
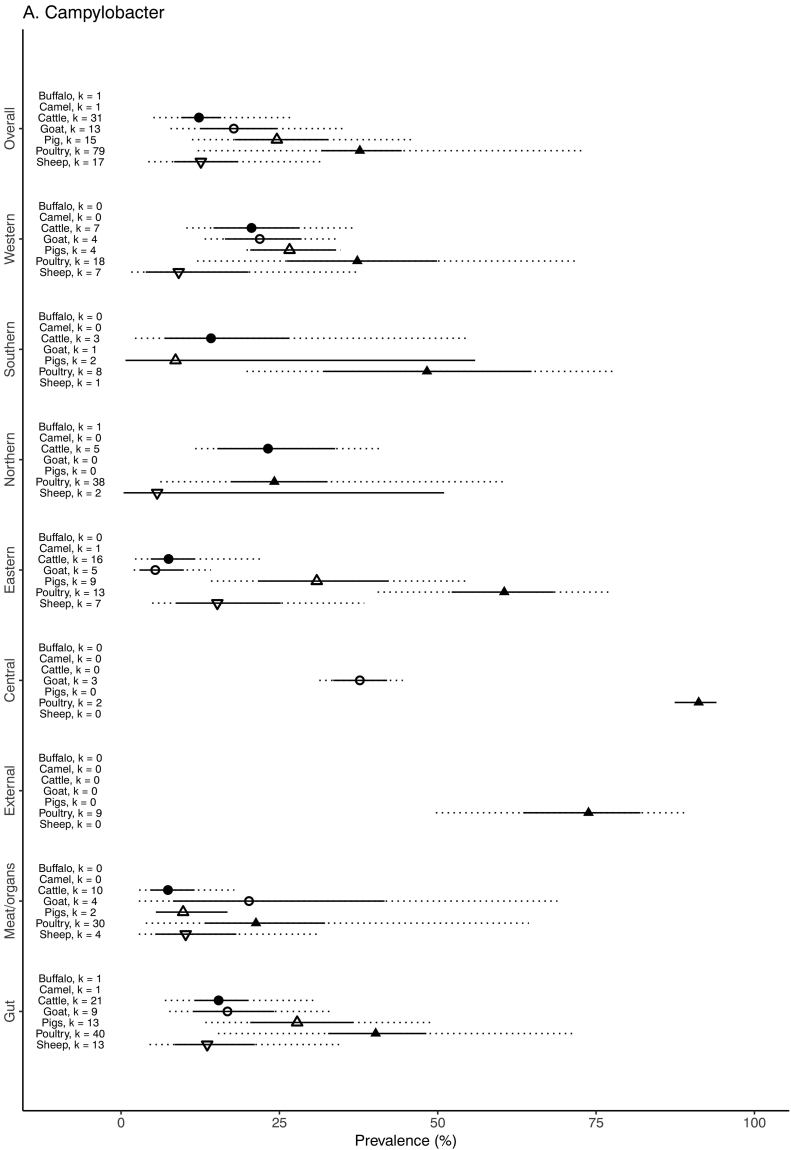
The unadjusted prevalence of Campylobacter by animal species among various sample types for each African region by source animal species is shown in Table 4. The weighted prevalence of Campylobacter by animal species, sample type, and geographic region is summarized in Fig. 3. The prevalence was highest in poultry samples (37.7%, 95% CI 31.6–44.3, 80% PI 12.1–72.7) followed by pig samples (24.6%, 95% CI 18.0–32.7, 80% PI 11.2–45.9), goat samples (17.8%, 95% CI 12.6–24.5, 80% PI 7.8–35.6), sheep samples (12.6%, 95% CI 8.4–18.5, 80% PI 4.3–31.7), and cattle samples (12.3%, 95% CI 9.5–15.8, 80% PI 5.1–26.7). One study reported attempted isolation or detection of Campylobacter from camels and buffalo, with zero positive cases for both species. Fig. 4 shows the breakdown of Campylobacter species between food animals. For all animal samples, Campylobacter were significantly less likely to be isolated or detected from meat or organ samples than from gut samples (OR = 0.67, 95% CI 0.56–0.80), and significantly more likely to be isolated or detected from external samples than gut samples (OR = 1.77, 95% CI 1.19–2.65) (Table 5). With adjustment for sample and species type, the odds of contamination of food animal samples with Campylobacter from Central Africa was significantly higher than the referent region of Northern Africa (OR = 9.06, 95% CI 1.92–42.70). The odds of contamination were significantly lower in samples from pigs (OR = 0.60, 95% CI 0.49–0.73) and all other species (OR = 0.29, 95% CI 0.25–0.33) when compared to samples from poultry. There was evidence that the inclusion of region, sample and species type as fixed effects improved model fit (Χ2 = 10.6, p = 0.03; Χ2 = 32.8, p ≤0.001; Χ2 = 332.7, p ≤0.001, respectively).
Table 4.
Combined unadjusted Campylobacter prevalence and species for food animal type and various sample types, 1979–2015.
| Host animal type | Sample stage | Sample type | No. of samples tested | No. of samples positive (%) |
Campylobacter species when isolates were typed from positive samples |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C. jejuni (%) | C. coli (%) | Other C. spp.a (%) | |||||||||
| Buffalo | Gut | Intestinal contents | 55 | 0 | (0.0) | – | – | – | – | – | – |
| TOTAL | 55 | 0 | (0.0) | – | – | – | – | – | – | ||
| Camel | Gut | Faeces/Rectal swab | 3 | 0 | (0.0) | – | – | – | – | – | – |
| TOTAL | 3 | 0 | (0.0) | – | – | – | – | – | – | ||
| Cattle | Gut | Faeces/Rectal swab | 4957 | 733 | (14.8) | 512 | (68.7) | 187 | (25.1) | 46 | (6.2) |
| Gut | Intestinal contents | 80 | 4 | (5.0) | NT | – | NT | – | NT | – | |
| Meat/organ | Carcass | 827 | 55 | (6.7) | 37 | (67.3) | 16 | (29.1) | 2 | (3.6) | |
| Meat/organ | Gallbladder | 100 | 12 | (12.0) | 0 | (0.0) | 12 | (100) | 0 | (0.0) | |
| Meat/organ | Liver | 30 | 8 | (26.7) | 5 | (62.5) | 3 | (37.5) | 0 | (0.0) | |
| Meat/organ | Meat | 521 | 25 | (4.8) | 21 | (84.0) | 4 | (16.0) | 0 | (0.0) | |
| Meat/organ | Tripe | 30 | 1 | (3.3) | NT | – | NT | – | NT | – | |
| TOTAL | 6545 | 838 | (12.8) | 575 | (68.0) | 222 | (26.3) | 48 | (5.7) | ||
| Goat | Gut | Faeces/Rectal swab | 2372 | 472 | (19.9) | 283 | (59.0) | 149 | (31.0) | 48 | (10.0) |
| Meat/organ | Carcass | 180 | 17 | (9.4) | 12 | (70.6) | 5 | (29.4) | 0 | (0.0) | |
| Meat/organ | Meat | 269 | 80 | (29.7) | 17 | (20.7) | 65 | (79.3) | 0 | (0.0) | |
| Meat/organ | Stomachs | 86 | 32 | (91.3) | 7 | (21.9) | 25 | (78.1) | 0 | (0.0) | |
| TOTAL | 2907 | 601 | (20.7) | 319 | (53.1) | 244 | (40.6) | 48 | (8.0) | ||
| Pigs | Gut | Caeca/Intestine | 454 | 59 | (13.0) | 36 | (61.0) | 23 | (39.0) | 0 | (0.0) |
| Gut | Faeces/Rectal swab | 1408 | 467 | (33.2) | 83 | (17.8) | 354 | (76.1) | 28 | (6.0) | |
| Meat/organ | Carcass | 66 | 7 | (10.6) | 6 | (85.7) | 1 | (14.3) | 0 | (0.0) | |
| Meat/organ | Meat | 47 | 4 | (8.5) | 1 | (25.0) | 2 | (50.0) | 1 | (25.0) | |
| TOTAL | 1975 | 537 | (27.2) | 126 | (23.6) | 380 | (71.0) | 29 | (5.4) | ||
| Poultry | Gut | Caeca | 1902 | 759 | (39.9) | 442 | (71.9) | 161 | (26.2) | 12 | (2.0) |
| Gut | Faeces/Cloaca swab | 5548 | 2457 | (44.3) | 1893 | (82.6) | 276 | (12.0) | 122 | (5.3) | |
| Meat/organ | Carcass | 653 | 335 | (51.3) | 130 | (55.3) | 97 | (41.3) | 8 | (3.4) | |
| Meat/organ | Gallbladder or bile | 65 | 3 | (4.6) | 3 | (100) | 0 | (0.0) | 0 | (0.0) | |
| Meat/organ | Giblet | 37 | 15 | (40.5) | 5 | (100) | 0 | (0.0) | 0 | (0.0) | |
| Meat/organ | Gizzard | 250 | 43 | (17.2) | 43 | (100) | 0 | (0.0) | 0 | (0.0) | |
| Meat/organ | Heart | 250 | 11 | (4.4) | 11 | (100) | 0 | (0.0) | 0 | (0.0) | |
| Meat/organ | Kidney | 25 | 2 | (8.0) | 2 | (100) | 0 | (0.0) | 0 | (0.0) | |
| Meat/organ | Liver | 300 | 75 | (25.0) | 68 | (100) | 0 | (0.0) | 0 | (0.0) | |
| Meat/organ | Meat | 1193 | 257 | (21.5) | 132 | (71.4) | 10 | (5.4) | 43 | (23.2) | |
| Meat/organ | Spleen | 200 | 17 | (8.5) | 17 | (100) | 0 | (0.0) | 0 | (0.0) | |
| External | Skin | 1405 | 948 | (67.5) | 346 | (47.7) | 341 | (47.0) | 38 | (5.2) | |
| TOTAL | 11,828 | 4922 | (41.6) | 3092 | (73.6) | 885 | (21.1) | 223 | (5.3) | ||
| Sheep | Gut | Faeces/Rectal swab | 1430 | 248 | (17.3) | 148 | (59.7) | 87 | (35.1) | 13 | (5.2) |
| Gut | Intestinal contents | 300 | 17 | (5.7) | 11 | (64.7) | 4 | (23.5) | 2 | (11.8) | |
| Meat/organ | Carcass | 288 | 38 | (13.2) | 31 | (81.6) | 7 | (18.4) | 0 | (0.0) | |
| Meat/organ | Gallbladder | 250 | 10 | (4.0) | 8 | (80.0) | 1 | (10.0) | 1 | (10.0) | |
| Meat/organ | Meat | 114 | 12 | (10.5) | 10 | (83.3) | 2 | (16.7) | 0 | (0.0) | |
| TOTAL | 2382 | 325 | (13.6) | 208 | (64.0) | 101 | (31.1) | 16 | (4.9) | ||
Where “Other” Campylobacter species include: C. faecalis, C. fetus, C. hyointesinalis, C. lari, C. sputorum, C. upsaliensis.
Fig. 3.
Forest plot with adjusted prevalence estimates for Campylobacter in food animals and meat for each animal species, sample type and African region, 1953–2016. 95% confidence intervals shown in solid line. 80% prediction intervals shown with dotted line. Adjusted prevalence not estimated when number of studies (k) < 2, 1979–2015.
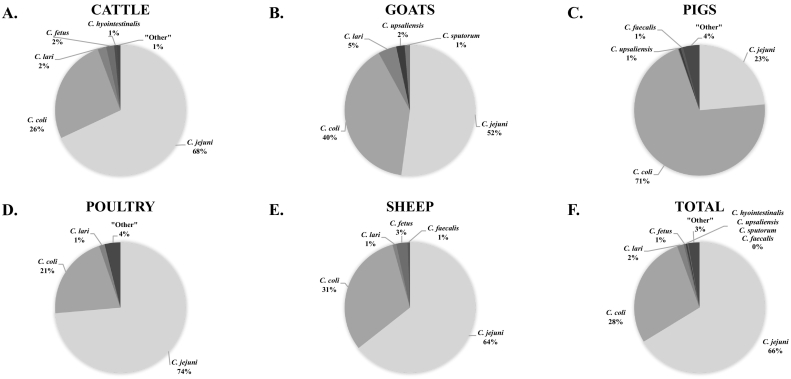
Fig. 4.
Pie graphs showing the breakdown of Campylobacter species reported when typed from (A) Cattle, (B) Goats, (C) Pigs, (D), Poultry, (E) Sheep and (F) all African food animal species combined, 1953–2016.
Table 5.
Odds Ratios (OR) with 95% Confidence intervals (CI) for fixed effects from multi-level multivariable logistic regression models for Campylobacter and Salmonella prevalence for sample types, 1953–2016.
| Model | Fixed effects | Campylobacter |
Salmonella |
||
|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | ||
| All | Region | ||||
| Northern | Baseline | Baseline | |||
| Central | 9.06 | (1.92–42.70)⁎ | 6.64 | (1.37–32.24)⁎ | |
| Eastern | 1.23 | (0.61–2.46) | 1.36 | (0.91–2.05) | |
| Southern | 2.08 | (0.81–5.39) | 1.35 | (0.69–2.68) | |
| Western | 1.59 | (0.79–3.21) | 1.79 | (1.10–2.92)⁎ | |
| Sample type | |||||
| Gut | Baseline | Baseline | |||
| External | 1.77 | (1.19–2.65)⁎ | 1.77 | (1.44–2.18)⁎ | |
| Internal | 0.67 | (0.56–0.80)⁎ | 0.86 | (0.79–0.95)⁎ | |
| Species type | |||||
| Poultry | Baseline | Baseline | |||
| Pigs | 0.60 | (0.49–0.73)⁎ | 0.82 | (0.63–1.07) | |
| All other | 0.29 | (0.25–0.33)⁎ | 0.47 | (0.42–0.54)⁎ | |
| Cattle | Region | ||||
| Northern | Baseline | Baseline | |||
| Central | – | 6.53 | (0.66–64.87) | ||
| Eastern | 0.60 | (0.19–1.86) | 1.07 | (0.53–2.17) | |
| Southern | 0.96 | (0.19–4.76) | 0.57 | (0.18–1.83) | |
| Western | 1.45 | (0.42–5.02) | 1.88 | (0.77–4.61) | |
| Sample type | |||||
| Gut | Baseline | Baseline | |||
| External | – | 8.11 | (5.5–11.96)⁎ | ||
| Internal | 0.40 | (0.26–0.61)⁎ | 0.97 | (0.81–1.17) | |
| Goats | Region | ||||
| Northern | – | – | Baseline | ||
| Central | 10.3 | (7.0–15.1)⁎ | 17.3 | (0.65–461.6) | |
| Eastern | Baseline | 4.8 | (0.74–31.8) | ||
| Southern | 8.7 | (5.3–14.1)⁎ | 0.45 | (0.02–12.8) | |
| Western | 5.5 | (3.7–8.1)⁎ | 3.4 | (0.5–22.5) | |
| Sample type | |||||
| Gut | Baseline | Baseline | |||
| External | – | – | 0.91 | (0.23–3.6) | |
| Internal | 1.5 | (1.1–2.1)⁎ | 1.3 | (0.68–2.6) | |
| Sheep | Region | ||||
| Northern | Baseline | Baseline | |||
| Central | – | – | 6.5 | (1.4–30.3)⁎ | |
| Eastern | 2.4 | (0.42–13.8) | 1.1 | (0.55–2.2) | |
| Southern | 7.1 | (0.63–79.5) | 0.25 | (0.07–0.98)⁎ | |
| Western | 2.2 | (0.35–13.6) | 3.1 | (1.3–7.1)⁎ | |
| Sample type | |||||
| Gut | Baseline | Baseline | |||
| External | – | – | 1.4 | (0.93–2.0) | |
| Internal | 1.27 | (0.78–2.1) | 0.85 | (0.4–2.1) | |
| Pigs | Region | ||||
| Northern | – | Baseline | |||
| Central | – | 10.92 | (1.36–87.54)⁎ | ||
| Eastern | Baseline | 2.67 | (0.53–13.44) | ||
| Southern | 0.17 | (0.06–0.52)⁎ | 2.56 | (0.39–16.70) | |
| Western | 0.42 | (0.16–1.15) | 3.46 | (0.67–18.03) | |
| Sample type | |||||
| Gut | Baseline | Baseline | |||
| External | – | – | |||
| Internal | 0.19 | (0.09–0.40)⁎ | 1.07 | (0.83–1.39) | |
| Poultry | Region | ||||
| Northern | Baseline | Baseline | |||
| Central | 13.3 | (4.35–40.74)⁎ | 3.53 | (0.42–29.93) | |
| Eastern | 1.87 | (0.73–4.83) | 1.12 | (0.45–2.81) | |
| Southern | 2.89 | (0.78–10.68) | 2.11 | (0.63–7.06) | |
| Western | 1.45 | (0.56–3.71) | 1.81 | (0.86–3.80) | |
| Sample type | |||||
| Gut | Baseline | Baseline | |||
| External | 1.57 | (1.03–2.39)⁎ | 0.73 | (0.55–0.98)⁎ | |
| Internal | 0.55 | (0.41–0.73)⁎ | 0.45 | (0.39–0.53)⁎ | |
p < 0.05.
Between study heterogeneity on the basis of the I2 statistic was high (>90%) for all livestock species, except cattle for which a relatively small proportion of the between study variation (15.8%) was estimated to be due to prevalence differences rather than chance variation. This is supported by values for MOR, which suggest that when comparing pairs of samples from two randomly selected studies, the odds of Campylobacter sample contamination in the study reporting higher prevalence would, in median, be 3.2 times the odds of sample contamination in the lower prevalence study, with control for region, sample type, and species type. For the species-specific models, between study heterogeneity on the basis of MOR was notably high for poultry, in which the estimated median difference in odds of sample contamination between a low and high prevalence study was 4.1 (Table 6).
Table 6.
Inter-study heterogeneity based on the I2 statistic and median odds ratio (MOR) for Campylobacter and Salmonella for all included studies (overall) and for included studies for each African food animal species for included studies, 1953–2016.
| Campylobacter |
Salmonella |
|||
|---|---|---|---|---|
| I2 | MOR | I2 | MOR | |
| Overall | 96.7% | 3.2 | 98.3% | 3.7 |
| Camels | – | – | 72.9% | 3.0 |
| Cattle | 15.8% | 2.2 | 97.1% | 4.5 |
| Goats | 92.1% | n.d. | 80.6% | 3.6 |
| Pigs | 90.4% | 1.8 | 93.5% | 2.7 |
| Poultry | 96.9% | 4.1 | 95.3% | 4.0 |
| Sheep | 90.8% | 2.4 | 77.6% | 1.7 |
3.1.1. Poultry
The adjusted prevalence on the basis of 7450 poultry gut samples was 40.2% (95% CI 32.7–48.2), 73.8% (95% CI 63.5–82.0) in 1405 poultry external samples, and 21.3% (95% CI 13.3–32.2), among 2973 meat or organ samples (Fig. 3). On the basis of the multivariable model, Campylobacter were significantly less likely to be isolated or detected in meat or organ samples than gut samples (OR = 0.55, 95% CI 0.41–0.73), but significantly more likely to be isolated or detected in external samples than gut samples (OR = 1.57, 95% CI 1.03–2.39) (Table 5). The adjusted Campylobacter prevalence was highest in Central Africa (91.2, 95% CI 87.4–94.0) and lowest in Northern Africa (24.2%, 95% CI 17.3–32.6) (Fig. 3). On the basis of the multivariable model, Campylobacter were significantly more likely to be isolated or detected in poultry samples from Central Africa than the referent region of Northern Africa (OR = 13.3, 95% CI 4.35–40.74) (Table 5). However, there was no evidence that the inclusion of region as a fixed effect improved model fit (Χ2 = 7.4, p = 0.11). There was strong evidence for an improvement in model fit with the inclusion of sample type (Χ2 = 28.1, p ≤0.001).
Among the 4922 poultry samples from which Campylobacter was isolated or detected, 4200 (85.7%) isolates were speciated. Of 4200 speciated isolates, 3092 (73.6%) were C. jejuni, 885 (21.1%) were C. coli, 152 (3.6%) were reported as ‘other’, 56 (1.3%) were C. lari, 10 (0.2%) were C. upsaliensis, and five (0.1%) were C. fetus (Table 4).
3.1.2. Pigs
The adjusted prevalence on the basis of 1862 pig gut samples was 27.8% (95% CI 20.4–36.7). No pig external samples were tested for Campylobacter. Among 113 pig meat or organ samples, the adjusted prevalence was 9.8% (95% CI 5.5–16.8) (Fig. 3). Campylobacter were significantly less likely to be isolated or detected from pig meat or organ samples than from pig gut samples (OR = 0.19, 95% CI 0.09–0.40) (Table 5). The adjusted Campylobacter prevalence was 30.9% (95% CI 21.6–42.1) for samples from Eastern Africa, 26.6% (95% CI 20.4–33.8) for samples from Western Africa, and 8.6% (95% CI 0.7–55.9) for samples from Southern Africa (Fig. 3). No studies from Central or Northern Africa reported on Campylobacter prevalence in pigs. When adjusted for sample type, the prevalence of Campylobacter in samples from Southern Africa was significantly lower (OR = 0.17, 95% CI 0.06–1.15) than the referent region of Eastern Africa (Table 5). There was evidence that the inclusion of region and sample type improved model fit (Χ2 = 7.7, p = 0.02; Χ2 = 24.0, p ≤0.001, respectively).
Among the 537 pig samples from which Campylobacter was isolated or detected, 535 isolates (99.6%) were speciated. Of 535 speciated isolates, 126 (23.6%) were C. jejuni, 380 (71.1%) were C. coli, 22 (4.1%) were reported as ‘other’, three (0.6%) were C. hyointestinalis, three (0.6%) were C. faecalis, and one (0.2%) was C. lari (Table 4).
3.1.3. Goats
The adjusted Campylobacter prevalence on the basis of 2372 goat gut samples was 16.8% (95% CI 11.4–24.6). No goat external samples were tested. Among 535 goat meat or organ samples, the adjusted prevalence was 20.2% (95% CI 8.2–41.6) (Fig. 3) After controlling for region, goat meat or organ samples were significantly more likely to be contaminated than gut samples (OR = 1.5, 95% CI 1.1–2.1) (Table 5). The adjusted prevalence of Campylobacter contamination was highest in the Central region (37.7%, 95% CI 33.5–42.0) and lowest in the Eastern region (5.4%, 95% CI 2.9–9.9) (Fig. 3). No samples were collected from the Northern region. There were significant differences in the odds of Campylobacter contamination when comparing samples from Central, Southern and Western regions with the Eastern baseline (Table 5). There was evidence that the inclusion of region and sample type as fixed effects improved model fit (Χ2 = 23.3, p ≤0.001; Χ2 = 6.3, p = 0.01, respectively).
For Campylobacter from goats, 616 isolates were speciated from 601 samples. Of these 616 isolates, 321 (52.1%) were C. jejuni, 246 (39.9%) were C. coli, 28 (4.5%) were C. lari, 13 (2.1%) were C. upsaliensis, and eight (1.3%) were C. sputorum (Table 4).
3.1.4. Sheep
The adjusted prevalence of Campylobacter contamination in 1730 sheep gut samples was 13.6%, (95% CI 8.5–21.1) and 10.2% (95% CI 5.4–18.2) on the basis of 652 meat or organ samples (Fig. 3). No external samples were reported. There was no evidence of a difference between the odds of contamination between sample types in the multivariable regression (Table 5). The adjusted prevalence of Campylobacter contamination was highest in the Southern region (30.0%, 95% CI 25.1–35.4) and lowest in the Northern region (5.7%, 95% CI 0.4–51.0) (Fig. 3). There was no evidence that including region or sample type improved model fit (Χ2 = 2.5, p = 0.48; Χ2 = 0.9, p = 0.35, respectively).
For Campylobacter from sheep, 323 isolates were speciated. Of these 323 isolates, 208 (64.4%) were C. jejuni, 101 (31.3%) were C. coli, nine (2.8%) were C. fetus, three (0.9%) were C. lari, and two (0.6%) were C. faecalis (Table 4).
3.1.5. Cattle
The adjusted prevalence derived from 5037 cattle gut samples was 15.4% (95% CI 11.7–20.0, 80% PI 6.9–30.8) compared to 7.4% (95% CI 4.6–11.6, 80% PI 2.9–17.9) from 1508 meat or organ samples (Fig. 3). No cattle external samples were tested for Campylobacter. On the basis of the cattle-specific multivariable model, Campylobacter were significantly less likely to be isolated or detected from meat or organ samples than from gut samples (OR = 0.40, 95% CI 0.26–0.61) (Table 5). The adjusted prevalence of contamination was highest in Northern Africa (23.2%, 95% CI 15.2–33.8) and lowest in Eastern Africa (7.5%, 95% CI 4.7–11.7) (Fig. 3), but there was no evidence of a difference in the odds of Campylobacter contamination between samples from Northern Africa and any other region from the multivariable model (Table 5). No studies from Central Africa reported on Campylobacter prevalence in cattle samples. There was evidence that the inclusion of sample type as a fixed effect improved model fit (Χ2 = 18.6, p ≤0.001), but no evidence for geographic region (Χ2 = 4.0, p = 0.26).
For Campylobacter from cattle, 845 isolates were speciated. Of these 845 isolates, 575 (68.0%) were C. jejuni, 222 (26.3%) were C. coli, 18 (2.1%) were C. lari, 17 (2.0%) were C. fetus, eight (0.9%) were reported as ‘other’, and five (0.6%) were C. hyointestinalis (Table 4).
3.2. Salmonella
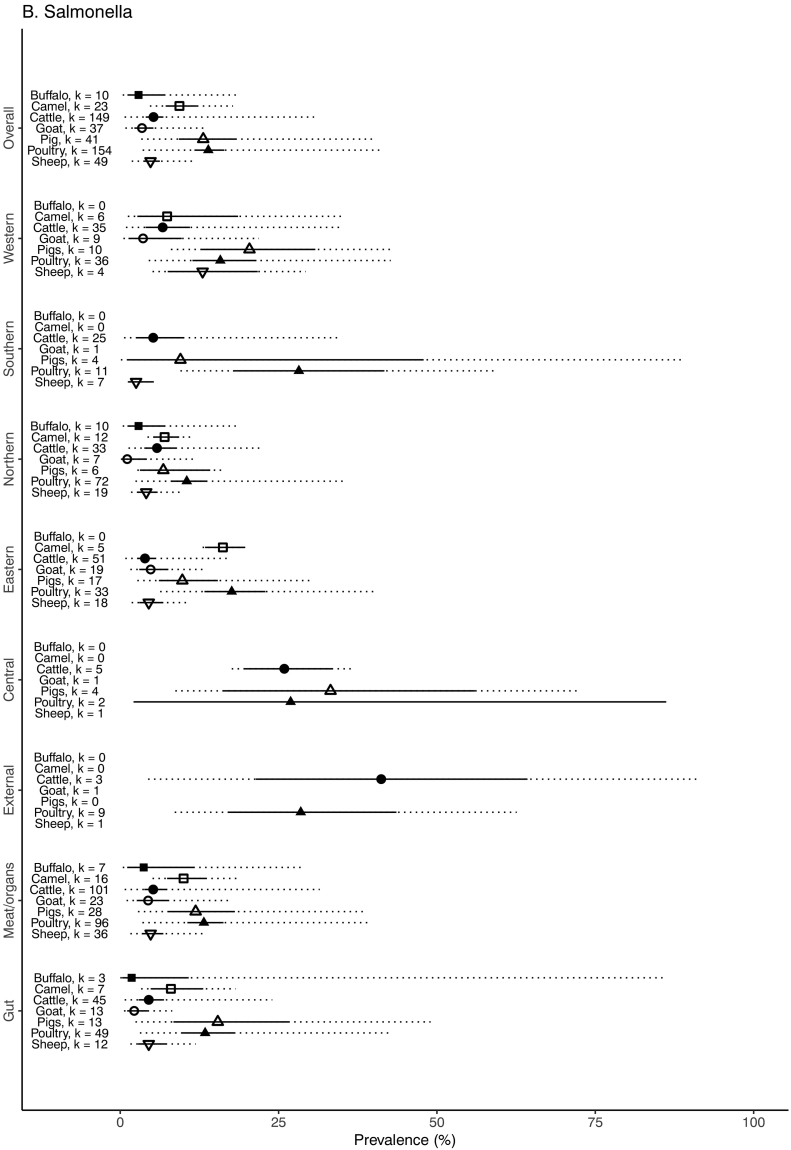
The unadjusted prevalence of Salmonella and serovar composition in the various sample types for each African region for each host animal species is shown in Table 7. The weighted prevalence of Salmonella by animal species, sample type, and geographic region is summarized in Fig. 5. The prevalence of Salmonella was highest in poultry samples (13.9% (95% CI 11.7–16.4, 80% PI 3.5–41.5)), followed by pig samples (13.1% (95% CI 9.3–18.3, 80% PI 3.3–39.9)), camel samples (9.3% (95% CI 7.2–12.1, 80% PI 4.7–17.8)), cattle samples (5.3% (95% CI 4.0–6.8, 80% PI 4.0–6.8)), sheep samples (4.8% (95% CI 3.6–6.3, 80% PI 1.8–12.0)), goat samples (3.4% (95% CI 2.2–5.2, 80% PI 2.2–5.2)) and buffalo samples (2.9% (95% 1.1–7.1, 80% PI 1.1–7.1)). Salmonella were significantly more likely to be detected in samples from Central and Western Africa compared to the referent region of North Africa (OR = 6.64, 95% CI 1.37–32.24 and OR = 1.79, 95% CI 1.01–2.92, respectively). Samples from pigs and all other species were less likely to be found to be contaminated than samples from poultry, but this was only significant in the case of the all other species category (OR = 0.47, 95% CI 0.42–0.54) (Table 5). There was weak evidence that the inclusion of region improved model fit (Χ2 = 8.9, p = 0.06) with stronger evidence for an improvement with the inclusion of sample type (Χ2 = 48.9, p ≤0.001) and species type (Χ2 = 134.2, p ≤0.001). Supplementary Table 2 details all reported Salmonella serovars quantified in each animal species by region.
Table 7.
Combined unadjusted Salmonella prevalence and three most numerous serovars for food animal type and various sample types, 1953–2016.
| Animal species | Sample stage | Sample type | No. of samples tested | No. of samples positive (%) | Number of isolates typed to serovara (%) | Ranking of most commonly typed Salmonella enterica serovars |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
2 |
3 |
|||||||||||
| Serovar name | No. (%) |
Serovar name | No. (%) |
Serovar name | No. (%) |
||||||||
| Buffalo | Gut | Faeces/Rectal swab | 2237 | 53 | (2.4) | ||||||||
| Meat/organ | Bile | 900 | 12 | (1.3) | |||||||||
| Meat/organ | Lymph node | 999 | 8 | (0.8) | |||||||||
| Meat/organ | Meat | 88 | 10 | (11.4) | |||||||||
| TOTAL | 4224 | 83 | (2.0) | 83 | (100) | Typhimurium | 35 (42.2) | Dublin | 13 (15.7) | Anatum | 8 (9.6) | ||
| Camel | Gut | Faeces/Rectal swab | 811 | 33 | (4.1) | ||||||||
| Gut | Intestine | 450 | 32 | (7.1) | |||||||||
| Meat/organ | Liver | 269 | 20 | (7.4) | |||||||||
| Meat/organ | Lymph node | 886 | 95 | (10.7) | |||||||||
| Meat/organ | Meat | 403 | 54 | (13.4) | |||||||||
| Meat/organ | Spleen | 169 | 24 | (14.2) | |||||||||
| TOTAL | 2988 | 258 | (8.6) | 251 | (97.3) | Saintpaul | 49 (19.5) |
Typhimurium | 27 (10.8) |
Branderup | 26 (10.4) |
||
| Cattle | Gut | Faeces/Rectal swab | 8683 | 517 | (6.0) | ||||||||
| Gut | Intestine | 3090 | 143 | (4.6) | |||||||||
| Meat/organ | Carcass | 6019 | 162 | (2.7) | |||||||||
| Meat/organ | Gallbladder/bile | 6558 | 135 | (2.1) | |||||||||
| Meat/organ | Heart | 37 | 1 | (2.7) | |||||||||
| Meat/organ | Kidney | 126 | 6 | (4.8) | |||||||||
| Meat/organ | Liver | 1248 | 40 | (3.2) | |||||||||
| Meat/organ | Lymph node | 3835 | 139 | (3.6) | |||||||||
| Meat/organ | Meat | 23,998 | 1050 | (4.4) | |||||||||
| Meat/organ | Meat rinse | 4396 | 19 | (0.4) | |||||||||
| Meat/organ | Spleen | 42 | 1 | (2.4) | |||||||||
| Meat/organ | Tongue | 13,680 | 199 | (1.5) | |||||||||
| Meat/organ | Tripe | 403 | 23 | (5.7) | |||||||||
| External | Hide | 177 | 74 | (41.8) | |||||||||
| TOTAL | 72,292 | 2509 | (3.5) | 1637 | (65.2) | Typhimurium | 289 (17.7) |
Anatum | 124 (7.6) |
Enteritidis | 120 (7.3) |
||
| Goat | Gut | Faeces/Rectal swab | 1411 | 30 | (2.1) | ||||||||
| Gut | Intestine | 685 | 5 | (0.7) | |||||||||
| Meat/organ | Carcass | 309 | 49 | (15.9) | |||||||||
| Meat/organ | Gallbladder/bile | 625 | 0 | (0.0) | |||||||||
| Meat/organ | Liver | 160 | 3 | (1.9) | |||||||||
| Meat/organ | Lymph node | 911 | 33 | (3.6) | |||||||||
| Meat/organ | Meat | 583 | 24 | (4.1) | |||||||||
| Meat/organ | Spleen | 160 | 2 | (1.3) | |||||||||
| External | Skin | 60 | 3 | (5.0) | |||||||||
| TOTAL | 4904 | 149 | (3.0) | 47 | (32.4) | Typhimurium | 18 (12.4) |
Typhi | 5 (3.4) |
Amersfoort Poona Derby “Other”/UT |
2 (1.4) |
||
| Pig | Gut | Caeca/Intestine | 510 | 92 | (18.0) | ||||||||
| Gut | Faeces/Rectal swab | 2074 | 372 | (17.9) | |||||||||
| Meat/organ | Carcass | 371 | 22 | (5.9) | |||||||||
| Meat/organ | Gallbladder/bile | 660 | 4 | (0.6) | |||||||||
| Meat/organ | Heart | 1 | 0 | (0.0) | |||||||||
| Meat/organ | Kidney | 4 | 0 | (0.0) | |||||||||
| Meat/organ | Liver | 121 | 18 | (14.9) | |||||||||
| Meat/organ | Lymph node | 1184 | 173 | (14.6) | |||||||||
| Meat/organ | Meat | 425 | 51 | (12.0) | |||||||||
| Meat/organ | Tongue | 117 | 30 | (25.6) | |||||||||
| TOTAL | 5467 | 762 | (13.9) | 478 | (62.7) | Hadar | 85 (17.9) |
Group O:2 | 48 (10.0) |
Saintpaul | 46 (9.6) |
||
| Poultry | Gut | Caeca/Intestine | 1997 | 360 | (18.0) | ||||||||
| Gut | Faeces/Cloaca swab | 10,060 | 1321 | (13.1) | |||||||||
| Meat/organ | Carcass | 1752 | 335 | (19.1) | |||||||||
| Meat/organ | Gallbladder | 279 | 19 | (6.8) | |||||||||
| Meat/organ | Giblets | 42 | 18 | (42.9) | |||||||||
| Meat/organ | Gizzard | 876 | 267 | (30.5) | |||||||||
| Meat/organ | Heart | 452 | 38 | (8.4) | |||||||||
| Meat/organ | Kidney | 90 | 0 | (0.0) | |||||||||
| Meat/organ | Liver | 1852 | 180 | (9.7) | |||||||||
| Meat/organ | Meat | 6578 | 712 | (27.3) | |||||||||
| Meat/organ | Spleen | 425 | 20 | (4.7) | |||||||||
| External | Feathers | 120 | 61 | (50.8) | |||||||||
| External | Skin | 907 | 284 | (31.3) | |||||||||
| TOTAL | 25,430 | 3615 | (14.2) | 2098 | (58.0) | Enteritidis | 464 (22.1) |
Typhimurium | 311 (14.8) |
Typhi | 174 (8.3) |
||
| Sheep | Gut | Faeces/Rectal swab | 2370 | 97 | (4.1) | ||||||||
| Gut | Intestine | 1797 | 28 | (1.6) | |||||||||
| Meat/organ | Carcass | 381 | 25 | (6.6) | |||||||||
| Meat/organ | Gallbladder/bile | 2305 | 31 | (1.3) | |||||||||
| Meat/organ | Heart | 207 | 0 | (0.0) | |||||||||
| Meat/organ | Kidney | 9 | 0 | (0.0) | |||||||||
| Meat/organ | Liver | 190 | 4 | (2.1) | |||||||||
| Meat/organ | Lymph node | 2817 | 71 | (2.5) | |||||||||
| Meat/organ | Meat | 936 | 68 | (7.3) | |||||||||
| Meat/organ | Spleen | 181 | 1 | (0.6) | |||||||||
| External | Skin | 142 | 7 | (4.9) | |||||||||
| TOTAL | 11,335 | 332 | (2.9) | 165 | (49.7) | Typhimurium | 68 (20.5) | Enteritidis | 14 (4.2) | Eastbourne | 11 (3.3) | ||
UT = Untypeable.
Serovars were excluded from number of isolates typed to serovar if numbers of isolates identified were not stated.
Fig. 5.
Forest plot with adjusted prevalence estimates for Salmonella in food animals and meat for each animal species, sample type and African region, 1953–2016. 95% confidence intervals shown in solid line. 80% prediction intervals shown with dotted line. Adjusted prevalence not estimated when number of studies (k) < 2, 1953–2016.
Between study heterogeneity was high for all studies, and above 90% for cattle, pigs, and poultry. Values of MOR suggest that a sample collected in a higher prevalence study would have, in median, around 3.7 times the odds of Salmonella sample contamination than a sample collected from a lower prevalence study, with control for region, sample type, and species type. For the species-specific models, between study heterogeneity on the basis of MOR was notably high for cattle and poultry, in which the estimated median difference in odds of sample contamination between a low and high prevalence study was 4.5 and 4.0, respectively (Table 6).
3.2.1. Poultry
The adjusted prevalence on the basis of 12,057 poultry gut samples was 13.4% (95% CI 9.8–18.0), 28.5% (95% CI 17.1–43.6) on the basis of 1027 external samples, and 13.2% (95% CI 10.6–16.3) on the basis of 12,346 poultry meat or organ samples. Salmonella were significantly less likely to be isolated or detected from both external (OR = 0.73, 95% CI 0.55–0.98) and meat or organ samples (OR = 0.45, 95% CI 0.39–0.53) when compared with the poultry gut sample baseline (Table 5). The adjusted prevalence in poultry samples was highest in the Southern region (28.2%, 95% CI 17.8–41.7) and lowest in the Northern region (10.5%, 95% CI 8.0–13.6) (Fig. 5). There was no evidence for improvement in model fit with the inclusion of region (Χ2 = 4.1, p = 0.4), but strong evidence for improvement with the inclusion of sample type (Χ2 = 97.1, p ≤0.001).
In total, Salmonella was isolated or detected in 3615 poultry samples and 2236 (61.9%) were serotyped. Among the 2236 isolates serotyped from poultry samples, 464 (20.8%) were Salmonella enterica serovar Enteritidis, 311 (13.9%) were Salmonella enterica serovar Typhimurium, and 174 (7.8%) were Salmonella enterica serovar Typhi (Table 7).
3.2.2. Pigs
The adjusted Salmonella prevalence on the basis of 2584 pig gut samples was 15.4% (95% CI 8.4–26.6). No pig external samples were tested for Salmonella. Among 2883 pig meat or organ samples, the adjusted prevalence was 11.9% (95% CI 7.6–18.1) (Fig. 5). There was no evidence for differences in the odds of Salmonella contamination when comparing pig gut and meat or organ samples. The adjusted Salmonella prevalence was highest in Central Africa (33.2% (95% CI 16.3–56.0)) and lowest in Northern Africa (6.8% (95% CI 3.1–14.2)). The odds of Salmonella contamination were significantly higher in the Central region than the Northern region baseline (OR = 10.9 (95% CI 1.36–87.5)) (Table 5). There was no evidence for improvement in model fit with the inclusion of either region (Χ2 = 4.8, p = 0.3) or sample type (Χ2 = 0.29, p = 0.59).
In total, Salmonella was isolated or detected in 762 pig samples and 428 (56.2%) of these were serotyped. Among the 494 isolates serotyped from pig samples, 85 (19.9%) were Salmonella enterica serovar Hadar, 46 (10.7%) were Salmonella enterica Saintpaul and 40 (9.3%) Salmonella enterica serovar Eastbourne (Table 7).
3.2.3. Cattle
The adjusted Salmonella prevalence on the basis of 11,773 cattle gut samples was 4.5% (95% CI 2.9–6.9), 5.2% on the basis of 60,342 meat or organ samples, and 41.2% on the basis of 177 external samples (Fig. 5). Salmonella were significantly more likely to be isolated or detected from cattle external samples than cattle gut samples (OR = 8.11, 95% CI 5.5–11.96) (Table 5). The prevalence in cattle samples was highest in the Central region (25.9%, 95% CI 19.6–33.4) and lowest in the Eastern region (3.9%, 95% CI 2.7–5.5) (Fig. 5). There was no evidence for improvement in model fit with the inclusion of region (Χ2 = 6.0, p = 0.2), but strong evidence for improvement with the inclusion of sample type (Χ2 = 112.1, p ≤0.001).
In total, Salmonella was isolated or detected in 2509 cattle samples and 1750 (69.7%) of those were serotyped. Among the 1750 isolates serotyped from cattle, 289 (16.5%) were Salmonella enterica serovar Typhimurium, 124 (7.1%) were Salmonella enterica serovar Anatum, and 120 (6.9%) were Salmonella enterica serovar Enteritidis (Table 7).
3.2.4. Sheep
The adjusted Salmonella prevalence on the basis of 4167 sheep gut samples was 4.5% (95% CI 2.8–7.2), and 4.8% (95% CI 3.4–6.8) on the basis of 7026 meat or organ samples (Fig. 5). A single study reported a prevalence of 4.9% for sheep external samples (n = 142). There was no evidence for a difference in the odds of contamination by sample type (Table 5). Like goats, the adjusted prevalence was highest in sheep samples from the Central region (25.0%, based on a single study) and lowest in the Southern region (2.5%, 95% CI 1.2–5.3) (Fig. 5). The odds of sample contamination were significantly elevated in Central (OR = 6.5, 95% CI 1.4–30.3) and Western regions (OR = 3.1, 95% CI 1.3–7.1), and significantly reduced in the Southern region (OR = 0.25, 95% CI 0.07–0.98) (Table 5). There was no evidence for improvement in model fit with the inclusion of sample type (Χ2 = 4.9, p = 0.29), but strong evidence for region (Χ2 = 14.4, p = 0.006).
In total, Salmonella was isolated or detected in 332 sheep samples and 165 (49.7%) of these were serotyped. Among the 165 isolates serotyped from sheep samples, 69 (41.8%) were Salmonella enterica serovar Typhimurium, 14 (8.5%) were Salmonella enterica Enteritidis and 11 (6.7%) Salmonella enterica serovar Eastbourne (Table 7).
3.2.5. Goats
The adjusted Salmonella prevalence on the basis of 2096 goat gut samples was 2.2% (95% CI 1.1–4.3). Among 2748 goat meat or organ samples, the adjusted prevalence was 4.4% (95% CI 2.6–7.5) (Fig. 5). A single study reported on Salmonella detection in goat external samples. The adjusted prevalence was highest in the Central region (16.7%) and lowest in the Southern region (0.4%) (both based on a single study). There was no evidence of a difference in the odds of contamination by sample type or geographic region on the basis of a multivariable model (Table 3, Table 4). There was also no evidence for an improvement in model fit following inclusion of either region (Χ2 = 4.9, p = 0.29) or sample type (Χ2 = 0.9, p = 0.6) as fixed effects.
In total, Salmonella was isolated or detected in 145 goat samples and 47 (32.4%) of these were serotyped or serogrouped. Among the 47 isolates serotyped from goat samples, 19 (40.4%) were Salmonella enterica serovar Typhimurium, five (10.6%) were Salmonella enterica Typhi and two (4.3%) each of Salmonella enterica serovar Amersfoort, Salmonella enterica serovar Derby, Salmonella enterica serovar Poona, and untypeable Salmonella species (Table 7).
3.3. Typhoidal Salmonella
Salmonella enterica serovar Typhi isolates were reported from 201 samples in nine studies from Eastern, Northern, and Western African regions. Among the 201 Salmonella enterica serovar Typhi isolated or detected, 165 (82.1%) were from poultry gut samples from one study and 36 (17.9%) were from cattle, goat, pig, poultry, and sheep meat or organ samples.
Salmonella enterica serovar Paratyphi A were reported from 40 samples in five studies in Northern and Western African regions. Among the 40 Salmonella enterica serovar Paratyphi A isolated or detected, 33 (82.5%) were isolated from cattle, and poultry gut samples and 7 (17.5%) were isolated from cattle, poultry, and sheep meat or organ samples.
3.4. Risk of bias
Forty four of 246 studies (17.9%) had an overall low risk of bias, 178 (72.4%) had an overall moderate risk of bias, and 24 (9.8%) had an overall high risk of bias (S1 file - B). In regards to sample selection and transport, the number of days, months, or years over which a particular study took place was specified in 127 (51.6%) studies. The time from sampling to laboratory was specified as less than 4 h in 54 (22.0%) studies and unspecified in 181 (73.6%) studies. Temperature during transport was at refrigeration temperatures in 25 (10.2%) studies, chilled in 82 (33.3%) studies, and unspecified in 137 (55.7%) of studies. With regards to laboratory testing, the amount of any individual sample tested was specified in 182 (74.0%) of 246 studies. Campylobacter or Salmonella specific liquid and solid media were used in 150 (61.0%) of studies. Campylobacter were incubated microaerophilically or in a candle jar in 61 (83.6%) of the 73 studies focusing on that organism. Campylobacter and Salmonella were specified as incubated between 35 °C and 42 °C in 48 (65.8%) of 73 and 120 (64.5%) of 186 studies, respectively. The speciation methods of Campylobacter isolates were deemed to have low or moderate risk of bias in 50 (68.5%) of 73 studies. The typing methods for Salmonella isolates was deemed to have low or moderate risk of bias in 91 (48.9%) of 186 studies.
Forty-three (8.3%) of 518 articles were excluded at the full text screening stage for being in a language other than English.
4. Discussion
Our systematic review has demonstrated widespread prevalence of Campylobacter species and Salmonella serovars in food animal species or meat across Africa. Both Campylobacter and Salmonella were most prevalent among samples from poultry and pigs, and were less common among samples from ruminant livestock and other animals. C. jejuni was the most predominant Campylobacter species and Salmonella enterica serovar Typhimurium was the most commonly identified Salmonella serovar in African food animals and meat products.
Campylobacter and Salmonella were present in animal gut, external, and meat or organ animal samples, but patterns of carriage or contamination varied among host species. Pathogen contamination during slaughter and meat processing may be more important in some animal species than others. The finding that Campylobacter were more likely to be isolated or detected from poultry external samples than poultry gut samples may result from contamination of feathers and skin by gut contents of many different animals, both while alive and during slaughter. On-farm environmental sources, both inside and outside housing, waterways and wildlife, including wild birds, have been identified as sources of Campylobacter (Agunos et al., 2014). In contrast, Salmonella were significantly more likely to be isolated or detected in poultry gut samples than poultry external and meat or organ samples. This may be, in part, due to relative concentration of carriage of the two pathogens in poultry, which a report has shown to be >5 log cfu/g greater for Campylobacter than Salmonella in poultry faecal samples (FSANZ and the South Australian Research and Development Institute, 2010). Environmental contamination of cattle hides from multiple sources may explain why Salmonella were significantly more likely to be isolated or detected from cattle external samples than cattle gut samples. No cattle external samples were tested for Campylobacter to compare these findings. Campylobacter were significantly more likely to be isolated or detected from cattle gut samples than cattle meat or organ samples, while Salmonella were significantly more likely to be isolated or detected from goat meat or organ samples than goat gut samples. Campylobacter are micro-aerophilic organisms and sensitive to extra-intestinal environments (Cools et al., 2005). From extra-intestinal samples types, Campylobacter cells may be present but damaged and therefore not culturable in the laboratory. The difference for Salmonella isolation and detection between goats and cattle may be a result of differences in the slaughter and production, differences in sample consistency (particularly faecal samples) or handling of meat products. Goats are more easily hung for dressing than cattle, likely decreasing floor-to-carcass contact and therefore faecal contamination. Larger slaughter facilities in Africa have been shown to have greater incidence of Salmonella, at all stages of the slaughter process, than smaller facilities (Hassanien et al., 2006).
We have shown that Campylobacter were significantly more likely to be isolated or detected in Central African food animal samples than those from other African regions following adjustment for types of samples tested. The prevalence of Campylobacter detection was particularly high among Central African poultry and was significantly higher than that of other regions (Fig. 3). Salmonella were significantly more likely to be isolated or detected from Central and Western African food animal samples than the referent region of Northern Africa. Unlike for Campylobacter, no single food animal species contributed to the higher Salmonella prevalence in Central and Western Africa. Rather, it seems that the Salmonella prevalence across all food animal species drove the regional differences. The finding that Southern African studies isolated or detected significantly more Campylobacter in goat and sheep samples but significantly less Campylobacter in pig samples, than the referent region of Northern Africa, reinforces that the contamination or carriage may vary between animal species within the same region.
Campylobacter jejuni was the predominant Campylobacter species isolated from food animals in Africa. However, goat and sheep, and poultry samples from Central Africa and pig faecal samples and pork regardless of region of origin, had a higher prevalence of C. coli. The observation that C. coli predominates in pig and pork samples agrees with studies from North and South America (Cummings et al., 2018; Modolo et al., 1999; Varela et al., 2007), and Europe (Boes et al., 2005; Kempf et al., 2017) with varying predominance in Asia (Haruna et al., 2013; Carrique-Mas et al., 2014). Thermophilic Campylobacter species, C. jejuni, C. coli, C. lari, and C. upsaliensis accounted for the majority of campylobacters isolated. Thermophilic campylobacters cause the majority of human Campylobacter infection but non-thermophilic species also cause human illness (Lastovica and le Roux, 2000). C. sputorum, C. hyointestinalis, C. faecalis, and C. fetus are non-thermophilic Campylobacter species, and were identified in low numbers from African food animals. For non-thermophilic species underestimation is particularly likely where studies use an elevated temperature for isolation.
Campylobacter were not recovered from African camel samples. Campylobacter have been recovered from camel faecal and meat samples elsewhere (Mohammed et al., 2015; Rahimi et al., 2010). As so few camel samples were tested for Campylobacter (n = 3), there is a need for more sampling of camels in Africa.
Salmonella enterica serovar Typhimurium was the most commonly identified serovar in African food animals in our review. This contrasts with findings in a report from the WHO Global Salm-Surv database (Galanis et al., 2006); a now discontinued web-based country databank of Salmonella serovars from human and non-human sources. In the WHO report, Salmonella enterica serovar Typhimurium was not in the top five Salmonella serovars submitted from non-human sources from African countries between 2000 and 2002. Instead, Salmonella enterica serovar Anatum (16%), Salmonella enterica serovar Enteritidis (16%), Salmonella enterica serovar Corvalis (8%), Salmonella enterica serovar Amsterdam (8%), and Salmonella enterica serovar Braenderup (8%) were the most common from non-human sources across the continent (Galanis et al., 2006). This may result in bias of reporting of non-human Salmonella strains isolates to the databank, particularly if submissions came from sick animals by way of veterinary laboratories. It should be noted that the Global Salm-Surv database had only three African countries contributing non-human data, compared to 27 countries represented in our analyses. The Global Salm-Surv database was also more restricted in time, reporting contributed data between 2000 and 2002, compared to between 1953 and 2016 for our analyses.
Common Salmonella enterica serovars from pigs varied from those isolated from other food animals. Salmonella enterica serovar Hadar, Salmonella enterica serovar Saintpaul, and Salmonella enterica serovar Eastbourne were the three most frequently identified serovars. The low proportion of porcine Salmonella enterica serovar Typhimurium isolates contrasts with reports from Europe (Animal and Plant Health Agency (APHA), 2014, Hald et al., 2003, Pires et al., 2011), North America (Letellier et al., 1999), South America (Kich et al., 2011) and Asia (Amal et al., 2014; Thai et al., 2012) where Salmonella enterica serovar Typhimurium is a common serovar in pig and pig products.
Typhoidal and Paratyphoidal Salmonella enterica serovars Typhi, Paratyphi A, Paratyphi B, with the exception of the biovar Java, and Paratyphi C are not known to have animal or environmental reservoirs (Centers for Disease Control and Prevention (CDC), 2017). One Salmonella isolate reported as Salmonella enterica serovar Paratyphi B was from a study published in 1953, prior to Salmonella enterica serovar Java first being redefined as a biovar of Salmonella enterica serovar Paratyphi B in 1988 by Le Minor (Le Minor, 1988). The majority of the Salmonella enterica serovar Typhi reported as isolated or detected was from a single Western African poultry study whose typing methods were questionable and considered high risk for bias. However, three other studies from Western Africa reported Salmonella enterica serovar Typhi in poultry. Of 11 studies reporting the isolation of Salmonella enterica serovar Typhi from meat pathway samples, one (9.1%) reported farm production type, slaughter facility size, or characteristics of meat retail facilities. The isolation of Salmonella enterica serovar Typhi and Salmonella enterica serovar Paratyphi from gut, external and meat or organ sample types likely suggests contamination from human faeces indicating serious breaches in sample collection and processing, or extremely poor farm or slaughter facility hygiene practices.
We have shown that African food animals and meat may be an important source of campylobacteriosis and salmonellosis as has been shown worldwide (Barrow et al., 1988; Ellis, 1969; Sahin et al., 2002; Skarp et al., 2016). Source attribution studies have implicated chicken as a cause of >70% of human campylobacteriosis cases in studies from Germany (Rosner et al., 2017) and Switzerland (Kittl et al., 2013). Other studies report chicken consumption to be the predominant source of campylobacteriosis in other parts of Europe, Canada and New Zealand (Bessell et al., 2012; Boysen et al., 2011; Levesque et al., 2013; Mughini Gras et al., 2012; Ravel et al., 2017; Smid et al., 2013; Mullner et al., 2009). Ruminants appear to be the source of ~15–30% human campylobacteriosis in some studies, but as low as 1% in Germany (Rosner et al., 2017). Poultry was the most common source of foodborne salmonellosis in both the US and Japan where ~70% of human cases were attributable to chicken, turkey and egg products (Guo et al., 2011; Pires et al., 2014; Toyofuku and Hald, 2011). Poultry was similarly found to be the most important source of human salmonellosis in Europe. According to a European Food Safety Authority (EFSA) report, poultry was the greatest food animal contributor to human illness, with 51.2% of cases attributed to laying hens, broilers, and turkeys. We have shown that poultry in Africa have a higher prevalence of both Campylobacter and Salmonella than other food animals and meat suggesting that poultry may be an important source of human illness in Africa too. The second most common attributable source of illness in both the EU and Japan were pigs with 26.9% and 5.3% of cases respectively (Pires et al., 2011; Pires et al., 2014). Beef was the second most common attributable source in the US with 29% of cases (Guo et al., 2011).
While source attribution studies have been performed in some parts of the world, few are available for Campylobacter and Salmonella in Africa (Mather et al., 2015). It is important to ascertain whether or not increasing scales and intensification of meat production are contributing to human disease in Africa. The presence of Campylobacter and Salmonella in food animals and meat products may or may not indicate human disease risk. As we have shown, Campylobacter species and Salmonella serovars identified, vary between animal species. Without a more in depth look at the link between animal and human isolates, such as whole genome sequencing and patterns of human exposure, including raw meat handling and cooking practices, we have less information about whether or not Campylobacter and Salmonella from African food animals are contributing to human disease.
Foodborne diseases are ‘widespread and represent significant threats to health and economies of countries’ according to the WHO Regional Office for Africa (World Health Organisation (WHO) Regional Office for Africa, 2017). It has been shown that implementation of greater hygiene measures in slaughter systems has been successful in reducing the numbers of cases of salmonellosis in Europe (European Food Safety Authority (EFSA): European Centre for Disease Prevention Control, 2012) and campylobacteriosis in New Zealand (Cressey and Lake, 2011). There are many challenges to improving food safety in Africa. Some are logistical, such as reliable access to electricity and safe water, others are social, such as population changes, and some are environmental, such as extreme weather conditions (Food and Agriculture Organization of the United Nations (FAO) and World Health Organization (WHO), 2005). Where prevalence data show that end product samples are highly contaminated with Campylobacter or Salmonella, as was the case for poultry meat and organs, local and national policy makers and enforcers may be able to more effectively develop control measures to reduce potential pathogens in the food chain.
4.1. Limitations of the study
Our formal bias assessment determined the overall risk of bias from sample selection, sample transport and laboratory methods used. One hundred and eighty-two (73.7%) of 247 studies did not specify sample transport time, and 138 (55.9%) did not specify temperature of sample transport, both of which can affect the survival of pathogen before testing.
Most of the included studies either began testing samples in the years between 2000 and 2016 or did not specify when the study began. The analyses are therefore weighted more heavily on more recent studies as opposed to results uniformly spread over the years since 1951 when earliest sampling was stated to have taken place.
Laboratory methods varied between studies limiting direct comparability between analyses. Sixty-four (26.0%) of 247 studies did not specify the amount of sample tested. The weight of sample tested has been shown to influence the estimates of prevalence of Salmonella (Funk et al., 2000). Forty (16.2%) of 247 studies did not specify culture methods used. Among the 73 Campylobacter studies that used culture techniques for isolation, nine different broths were used as selective enrichment and 14 different agars were used. Among the 177 Salmonella studies that used culture techniques, eight different primary enrichment broths, six different selective enrichment broths, and 24 different agars were used for isolation. Speciating emerging Campylobacter requires tests beyond basic phenotypic and biochemical assays. The prevalence of these Campylobacter species is therefore likely underestimated. Prevalence estimates were not corrected for test sensitivity and specificity, implying that the true prevalence in each study is likely to differ from the apparent prevalence, and that the magnitude of this difference may be affected by sampling and testing methodology in an unquantified manner. No study was excluded due to chosen sampling or testing methodologies. Potential biases involving study size and sample type were adjusted for in the data analysis.
4.2. Conclusion
For many subsistence households and communities in many African countries, meat is a key protein source and livestock and poultry production is central to people's livelihoods. As market-driven changes occur within agricultural production towards wider distribution networks, centralised processing, and more large-scale and intensive systems, have been linked to the emergence of zoonotic diseases. Both Campylobacter and Salmonella were most prevalent among samples from poultry and pigs, and were less common among samples from ruminant livestock species. C. jejuni was the predominant Campylobacter species and Salmonella enterica serovar Typhimurium was the most commonly identified Salmonella serovar in African food animals and animal products. The presence of Salmonella enterica serovar Typhi and Salmonella enterica serovar Paratyphi in samples of food animal origin is particularly troubling and strongly indicates the need for increased hygiene measures to ensure food animals are not exposed to human faeces and human faeces do not contaminate the community meat supply.
The high prevalence of these organisms in livestock and poultry, their important role as human pathogens, and lack of evidence on which animal hosts contribute most to human illness in Africa, indicate source attribution studies would be a useful tool to more definitively identify priorities for food safety interventions.
The following are the supplementary data related to this article.
PRISMA checklist.
Inclusion and exclusion criteria.
Bias assessment of all included studies.
Names and number of isolates for all Salmonella enterica serovars named in included articles according to African region.
Author contributions
All authors have contributed the concept or design of the work. KMT organised and populated the database. KMT wrote the first draft and subsequent revisions of the manuscript. WdG performed the meta-analysis. JAC, RNZ, NPF, JJB, SC, MAD, GB, ESS, JB, and WdG made substantial contributions to the manuscript. All authors contributed to manuscript revisions, read and approved the submitted version.
Funding
The authors were funded by the UK Biotechnology and Biological Sciences Research Council, Department for International Development, UK, UK Economic and Social Research Council, UK Medical Research Council, UK Natural Environment Research Council and UK Defence Science and Technology Laboratory, under the Zoonoses and Emerging Livestock Systems (ZELS) programme (grant numbers BB/L017679/1 and BB/L018926/1). The funders were not involved in study design, analysis or interpretation of data, writing or decision to submit for publication.
Declaration of competing interest
The authors declare that the submitted work was conducted in the absence of any commercial or financial relationships that could be construed as a conflict of interest.
Acknowledgements
The authors would like to thank the University of Otago Distance Library Service for sourcing full text articles not available online.
References
- a Mpalang R.K., Boreux R., Melin P., Akir Ni Bitiang K., Daube G., De Mol P. Prevalence of Campylobacter among goats and retail goat meat in Congo. J. Infect. Dev. Ctries. 2014;8:168–175. doi: 10.3855/jidc.3199. [DOI] [PubMed] [Google Scholar]
- Abbassi-Ghozzi I., Jaouani A., Hammami S., Martinez-Urtaza J., Boudabous A., Gtari M. Molecular analysis and antimicrobial resistance of Salmonella isolates recovered from raw meat marketed in the area of “Grand Tunis”, Tunisia. Pathol. Biol. (Paris) 2012;60:e49–e54. doi: 10.1016/j.patbio.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Abd el-Aziz A.S., Elmossalami M.K., el-Neklawy E. Bacteriological characteristics of dressed young pigeon (squabs) Columba livia domesticus. Nahrung. 2002;46:51–53. doi: 10.1002/1521-3803(20020101)46:1<51::AID-FOOD51>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Abd El-Ghany W.A., El-Shafii S.S.A., Hatem M.E. A survey on Salmonella species isolated from chicken flocks in Egypt. Asian J. Anim. Vet. Adv. 2012;7:489–501. [Google Scholar]
- Abd-Elghany S.M., Sallam K.I., Abd-Elkhalek A., Tamura T. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol. Infect. 2015;143:997–1003. doi: 10.1017/S0950268814001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdellah C., Fouzia R.F., Abdelkader C., Rachida S.B., Mouloud Z. Occurrence of Salmonella in chicken carcasses and giblets in Meknès-Morocco. Pak. J. Nutr. 2008;7:231–233. [Google Scholar]
- Abdel-Maksoud M., Abdel-Khalek R., El-Gendy A., Gamal R.F., Abdelhady H.M., House B.L. Genetic characterisation of multidrug-resistant Salmonella enterica serotypes isolated from poultry in Cairo, Egypt. Afr. J. Lab. Med. 2015;4 doi: 10.4102/ajlm.v4i1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abebe M., Tafese B. Zoonotic bacterial pathogens isolated from food of bovine in selected Woredas of Tigray, Ethiopia. World Appl. Sci. J. 2014;31:1864–1868. [Google Scholar]
- Abou El Hassan D.G. Neonatal diarrhoea in lambs and goat kids. Vet. Med. J. Giza. 1996;44:371–380. [Google Scholar]
- Abrahams C.A., Agbodaze D., Nakano T., Afari E.A., Longmatey H.E. Prevalence and antibiogram of Campylobacter jejuni in domestic animals in rural Ghana. Arch. Environ. Health. 1990;45:59–62. doi: 10.1080/00039896.1990.9935926. [DOI] [PubMed] [Google Scholar]
- Acha S.J., Kuhn I., Jonsson P., Mbazima G., Katouli M., Mollby R. Studies on calf diarrhoea in Mozambique: prevalence of bacterial pathogens. Acta Vet. Scand. 2004;45:27–36. doi: 10.1186/1751-0147-45-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis Z., Kebede N., Worku Z., Gezahegn H., Yirsaw A., Kassa T. Prevalence and antimicrobial resistance of Salmonella isolated from lactating cows and in contact humans in dairy farms of Addis Ababa: a cross sectional study. BMC Infect. Dis. 2011;11 doi: 10.1186/1471-2334-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adekeye J.O., Abdu P.A., Bawa E.K. Campylobacter fetus subsp. jejuni in poultry reared under different management systems in Nigeria. Avian Dis. 1989;33:801–803. [PubMed] [Google Scholar]
- Adesiji Y.O., Alli O.T., Adekanle M.A., Jolayemi J.B. Prevalence of Arcobacter, Escherichia coli, Staphylococcus aureus and Salmonella species in retail raw chicken, pork, beef and goat meat in Osogbo, Nigeria. Sierra Leone J. Biomed. Res. 2011;3 [Google Scholar]
- Adesiyun A.A., Oni O.O. Prevalence and antibiograms of Salmonellae in slaughter cattle, slaughter areas and effluents in Zaria abattoir, Nigeria. J. Food Prot. 1989;52:232–235. doi: 10.4315/0362-028X-52.4.232. [DOI] [PubMed] [Google Scholar]
- Adeyanju G.T., Ishola O. Salmonella and Escherichia coli contamination of poultry meat from a processing plant and retail markets in Ibadan, Oyo State, Nigeria. SpringerPlus. 2014;3 doi: 10.1186/2193-1801-3-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adu-Gyamfi A., Torgby-Tetteh W., Appiah V. Microbiological quality of chicken sold in Accra and determination of D10-value of E. coli. Food Nutr. Sci. 2012;3:693–698. [Google Scholar]
- Agunos A., Waddell L., Léger D., Taboada E. A systematic review characterizing on-farm sources of Campylobacter spp. for broiler chickens. PLoS One. 2014;9:e104905. doi: 10.1371/journal.pone.0104905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A.M., Shimamoto T. Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157:H7 and Shigella spp. from meat and dairy products in Egypt. Int. J. Food Microbiol. 2014;168:57–62. doi: 10.1016/j.ijfoodmicro.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Ahmed H.A., Ibrahim A.F., Hussein M.A., El-Bayomi R.M. ERIC-PCR fingerprinting of some S. Typhimurium isolates from chicken and humans with reference to the microbiological quality of retail chicken meat in Dakahlia, Egypt. Glob. Vet. 2014;13:95–104. [Google Scholar]
- Ahmed H.A., El-Hofy F.I., Shafik S.M., Abdelrahman M.A., Elsaid G.A. Characterization of virulence-associated genes, antimicrobial resistance genes, and class 1 integrons in Salmonella enterica serovar Typhimurium isolates from chicken meat and humans in Egypt. Foodborne Pathog. Dis. 2016;13:281–288. doi: 10.1089/fpd.2015.2097. [DOI] [PubMed] [Google Scholar]
- Ajayi A.O., Egbebi A.O. Antibiotic sucseptibility of Salmonella Typhi and Klebsiella pneumoniae from poultry and local birds in ado-Ekiti, Ekiti-State, Nigeria. Ann. Biol. Res. 2011;2:431–437. [Google Scholar]
- Akam A., Khelef D., Kaidi R., Lafri M., Rahal K., Chirila F., Cozma V. Frequency of Cryptosporidium parvum, Escherichia coli K99 and Salmonella spp. isolated from calves in six breeding farms from Mitidja, Algeria. Bulletin UASVM Animal Science and Biotechnologies. 2004;61:287–288. [Google Scholar]
- Akoachere J.-F.T.K., Tanih N.F., Ndip L.M., Ndip R.N. Phenotypic characterization of Salmonella Typhimurium isolates from food-animals and abattoir drains in Buea, Cameroon. J. Health Popul. Nutr. 2009;27:612–618. doi: 10.3329/jhpn.v27i5.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akwuobu C.A., Oboegbulem S.I., Ofukwu R.A. Characterization and antibiogram of local isolates of Campylobacter species from chicken in Nsukka area, Southeast Nigeria. Am.-Eurasian J. Sustain. Agric. 2010;4:117–121. [Google Scholar]
- Alao F.O., Kester C.T., Gbagba B.K., Fakilede F.K. Comparison of prevalence and antimicrobial sensitivity of Salmonella Typhimurium in apparently healthy cattle and goat in Sango-Ota, Nigeria. Internet J. Microbiol. 2012;10 [Google Scholar]
- Alemayehu D., Molla B., Muckle A. Prevalence and antimicrobial resistance pattern of Salmonella isolates from apparently healthy slaughtered cattle in Ethiopia. Trop. Anim. Health Prod. 2003;35:309–319. doi: 10.1023/a:1025189204496. [DOI] [PubMed] [Google Scholar]
- Alemu S., Zewde B.M. Prevalence and antimicrobial resistance profiles of Salmonella enterica serovars isolated from slaughtered cattle in Bahir Dar, Ethiopia. Trop. Anim. Health Prod. 2012;44:595–600. doi: 10.1007/s11250-011-9941-y. [DOI] [PubMed] [Google Scholar]
- Al-Hazmi M., Al-Arfaj A., Mostafa A., Ihab M. Molecular detection of Salmonella enterica serovar Enteritidis in chicken-related samples collected from Egypt. Life Sci. J. 2013;10:2645–2649. [Google Scholar]
- Ali F.H.M., Ouf J.M. Evaluation of the hygienic status of broiler parts with and without skin in retail markets in Egypt. Vet. Med. J. Giza. 2005;53:611–623. [Google Scholar]
- Aliaa S.O., Ibrahim A.M., Tolba K., Atalla O.A. Bacterial evaluation of fresh and frozen beef and buffalo meat. Glob. J. Agric. Food Safety Sci. 2014;1:371–381. [Google Scholar]
- Amal A.A.S., Nassif M.R.M., Dalia Y.Y.M. Follow up of some foodborne bacterial pathogens on hides and meat of bovine carcasses. Glob. J. Agric. Food Safety Sci. 2014;1:199–213. [Google Scholar]
- Amara A., Badou M., Faid M., Bouzoubaa K. Microbial contamination of poultry slaughtered in traditional shops in Morocco. Microbiologie, Aliments, Nutrition. 1994;12:323–327. [Google Scholar]
- Amin H.S., El-Rahman A.E.A.A. Molecular characterization of Salmonella enterica isolated from chicken meat and its products by multiplex PCR. Alex. J. Vet. Sci. 2015;46:155–160. [Google Scholar]
- Ammar A.M.A., Ahmed Y.A.E., Asawy A.M.I., Ibrahim A.A. Bacteriological studies on Salmonella Enteritidis isolated from different sources in Dakhlia Governorate. Assiut Vet. Med. J. 2009;56:125–135. [Google Scholar]
- Ammar A., Alloui N., Bennoune O., Kassah-Laouar A. Survey of Salmonella serovars in broilers and laying breeding reproducers in East of Algeria. J. Infect. Dev. Ctries. 2010;4:103–106. doi: 10.3855/jidc.562. [DOI] [PubMed] [Google Scholar]
- Animal and Plant Health Agency (APHA) Chapter 4: Reports of Salmonella in Pigs. 2014. Salmonella in livestock production in Great Britain, 2014. [Google Scholar]
- Aragaw K., Molla B., Muckle A., Cole L., Wilkie E., Poppe C., Kleer J., Hildebrandt G. The characterization of Salmonella serovars isolated from apparently healthy slaughtered pigs at Addis Ababa abattoir, Ethiopia. Prev. Vet. Med. 2007;82:252–261. doi: 10.1016/j.prevetmed.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Aragaw K., Terefe L., Abera M. Prevalence of Salmonella infection in intensive poultry farms in Hawassa and isolation of Salmonella species from sick and dead chickens. Ethiopian Veterinary Journal. 2010;14:115–124. [Google Scholar]
- Ashenafi M. Microbial flora and incidence of some food-borne pathogens on fresh raw beef from butcher's shops in Awassa, Ethiopia. Bull. Anim. Health Prod. Afr. 1994;42:273–277. [Google Scholar]
- Awadallah M.A.I., Ahmed H.A., El-Gedawy A.A., Saad A.M. Molecular identification of C. jejuni and C. coli in chicken and humans, at Zagazig, Egypt, with reference to the survival of C. jejuni in chicken meat at refrigeration and freezing temperatures. Food Res. Int. 2014;21:1801–1812. [Google Scholar]
- Bada-Alambedji R., Fofana A., Seydi M., Akakpo A.J. Antimicrobial resistance of Salmonella isolated from poultry carcasses in Dakar (Senegal) Braz. J. Microbiol. 2006;37:510–515. [Google Scholar]
- Barendregt J.J., Doi S.A., Lee Y.Y. Meta-analysis of prevalence. Epidemiol Community Health. 2013;67:974–978. doi: 10.1136/jech-2013-203104. [DOI] [PubMed] [Google Scholar]
- Barrow P.A., Simpson J.M., Lovell M.A. Intestinal colonisation in the chicken by food-poisoning Salmonella serotypes; microbial characteristics associated with faecal excretion. Avian Pathol. 1988;17:571–588. doi: 10.1080/03079458808436478. [DOI] [PubMed] [Google Scholar]
- Bata S.I., Karshima N.S., Yohanna J., Dashe M., Pam V.A., Ogbu K.I. Isolation and antibiotic sensitivity patterns of Salmonella species from raw beef and quail eggs from farms and retail outlets in Jos, Plateau State, Nigeria. J. Vet. Med. Anim. Health. 2016;8:29–34. [Google Scholar]
- Bates D., Mächler M., Bolker B., Walker S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015;67 [Google Scholar]
- Bawa I.H., Bsadjo T.G., Bagré T.S., Bouda S.C., Konaté A., Bako E., Kagambèga A., Zongo C., Somda M., Savadogo A., Traoré A.S., Barro N. Antimicrobial susceptibility of Salmonella enterica strains isolated from raw beef, mutton and intestines sold in Ouagadougou, Burkina Faso. J. Appl. Biosci. 2015;95:8966–8972. [Google Scholar]
- Bekele B., Ashenafi M. Distribution of drug resistance among enterococci and Salmonella from poultry and cattle in Ethiopia. Trop. Anim. Health Prod. 2010;42:857–864. doi: 10.1007/s11250-009-9499-0. [DOI] [PubMed] [Google Scholar]
- Beshatu F., Fanta D., Aklilu F., Getachew T., Nebyu M. Prevalence and antimicrobial susceptibility of Salmonella isolates from apparently healthy slaughtered goats at Dire Dawa municipal abattoir, eastern Ethiopia. J. Microbiol. Antimicrob. 2015;7:1–5. [Google Scholar]
- Bessell P.R., Rotariu O., Innocent G.T., Smith-Palmer A., Strachan N.J., Forbes K.J., Cowden J.M., Reid S.W., Matthews L. Using sequence data to identify alternative routes and risk of infection: a case-study of Campylobacter in Scotland. BMC Infect. Dis. 2012;12:80. doi: 10.1186/1471-2334-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester L.A., Essack S.Y. Observational study of the prevalence and antibiotic resistance of Campylobacter spp. from different poultry production systems in KwaZulu-Natal, South Africa. J. Food Prot. 2012;75:154–159. doi: 10.4315/0362-028X.JFP-11-237. [DOI] [PubMed] [Google Scholar]
- Boes J., Nersting L., Nielsen E.M., Kranker S., Enoe C., Wachmann H.C., Baggesen D.L. Prevalence and diversity of Campylobacter jejuni in pig herds on farms with and without cattle or poultry. J. Food Prot. 2005;68:722–727. doi: 10.4315/0362-028x-68.4.722. [DOI] [PubMed] [Google Scholar]
- Boko C.K., Kpodekon T.M., Duprez J.N., Imberechts H., Taminiau B., Bertrand S., Mainil J.G. Identification and typing of Salmonella enterica serotypes isolated from guinea fowl (Numida meleagris) farms in Benin during four laying seasons (2007 to 2010) Avian Pathol. 2013;42:1–8. doi: 10.1080/03079457.2012.751484. [DOI] [PubMed] [Google Scholar]
- Bouchrif B., Paglietti B., Murgia M., Piana A., Cohen N., Ennaji M.M., Rubino S., Timinouni M. Prevalence and antibiotic-resistance of Salmonella isolated from food in Morocco. J. Infect. Dev. Ctries. 2009;(1):3. doi: 10.3855/jidc.103. [DOI] [PubMed] [Google Scholar]
- Boysen L., Vigre H., Rosenquist H. Seasonal influence on the prevalence of thermotolerant Campylobacter in retail broiler meat in Denmark. Food Microbiol. 2011;28:1028–1032. doi: 10.1016/j.fm.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Cardinale E., Dromigny J.A., Tall F., Ndiaye M., Konte M., Perrier-Gros-Claude J.D. Fluoroquinolone susceptibility of Campylobacter strains, Senegal. Emerg. Infect. Dis. 2003;9:1479–1481. doi: 10.3201/eid0911.020693. [DOI] [PubMed] [Google Scholar]
- Cardinale E., Gros-Claude J.D.P., Tall F., Cissé M., Guèye E.F., Salvat G. Prevalence of Salmonella and Campylobacter in retail chicken carcasses in Senegal. Rev. Elev. Med. Vet. Pays Trop. 2003;56:13–16. [Google Scholar]
- Cardinale E., Rose V., Perrier Gros-Claude J.D., Tall F., Rivoal K., Mead G., Salvat G. Genetic characterization and antibiotic resistance of Campylobacter spp. isolated from poultry and humans in Senegal. J. Appl. Microbiol. 2006;100:209–217. doi: 10.1111/j.1365-2672.2005.02763.x. [DOI] [PubMed] [Google Scholar]
- Carrique-Mas J.J., Bryant J.E., Cuong N.V., Hoang N.V., Campbell J., Hoang N.V., Dung T.T., Duy D.T., Hoa N.T., Thompson C., Hien V.V., Phat V.V., Farrar J., Baker S. An epidemiological investigation of Campylobacter in pig and poultry farms in the Mekong delta of Vietnam. Epidemiol. Infect. 2014;142:1425–1436. doi: 10.1017/S0950268813002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carron M., Chang Y.-M., Momanyi K., Akoko J., Kiiru J., Bettridge J., Chaloner G., Rushton J., O’Brien S., Williams N., Fèvre E.M., Häsler B. Campylobacter, a zoonotic pathogen of global importance: prevalence and risk factors in the fast-evolving chicken meat system of Nairobi, Kenya. PLoS Negl. Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter J.D., Hudson A.P. Reactive arthritis: clinical aspects and medical management. Rheum. Dis. Clin. N. Am. 2009;35:21–44. doi: 10.1016/j.rdc.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Chapter 3: Typhoid & Paratyphoid Fever. Travelers’ Health; 2017. Infectious diesases related to travel. [Google Scholar]
- Chambers P.G. Salmonellae in Rhodesia: sources and serotypes of some isolates from abattoirs, domestic animals, birds and man. J. S. Afr. Vet. Assoc. 1977;48:241–244. [PubMed] [Google Scholar]
- Chanyalew Y., Asrat D., Amavisit P., Loongyai W. Prevalence and antimicrobial susceptibility of thermophilic Campylobacter isolated from sheep at Debre Birhan, North-Shoa, Ethiopia. Kasetsart J. Nat. Sci. 2013;47:551–560. [Google Scholar]
- Cohen N., Ennaji H., Hassar M., Karib H. The bacterial quality of red meat and offal in Casablanca (Morocco) Mol. Nutr. Food Res. 2006;50:557–562. doi: 10.1002/mnfr.200500180. [DOI] [PubMed] [Google Scholar]
- Cohen N., Ennaji H., Bouchrif B., Hassar M., Karib H. Comparative study of microbiological quality of raw poultry meat at various seasons and for different slaughtering processes in Casablanca (Morocco) J. Appl. Poult. Res. 2007;16:502–508. [Google Scholar]
- Collard P., Sen R. Isolation of Salmonellae from cattle in Ibadan. West Afr. Med. J. 1956;5:118–120. [PubMed] [Google Scholar]
- Cools I., Uyttendaele M., Cerpentier J., D’Haese E., Nelis H.J., Debevere J. Persistence of Campylobacter jejuni on surfaces in a processing environment and on cutting boards. Lett. Appl. Microbiol. 2005;40:418–423. doi: 10.1111/j.1472-765X.2005.01694.x. [DOI] [PubMed] [Google Scholar]
- Cooper R., Shires K., Lastovica A.J., Elisha B.G. Molecular epidemiology of fluoroquinolone-resistant and susceptible Campylobacter jejuni isolates. South Afr. J. Epidemiol. Infect. 2001;16:57–61. [Google Scholar]
- Cressey P., Lake R.J. MPI Technical Paper no: 2012/11. 2011. Estimated incidence of foodborne illness in New Zealand: Application of overseas models and multipliers. [Google Scholar]
- Cummings K.J., Warnick L.D., Davis M.A., Eckmann K., Grohn Y.T., Hoelzer K., MacDonald K., Root T.P., Siler J.D., McGuire S.M., Wiedmann M., Wright E.M., Zansky S.M., Besser T.E. Farm animal contact as risk factor for transmission of bovine-associated Salmonella subtypes. Emerg. Infect. Dis. 2012;18:1929–1936. doi: 10.3201/eid1812.110831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings K.J., Rodriguez-Rivera L.D., McNeely I., Suchodolski J.S., Mesenbrink B.T., Leland B.R., Bodenchuk M.J. Fecal shedding of Campylobacter jejuni and Campylobacter coli among feral pigs in Texas. Zoonoses Public Health. 2018;65:215–217. doi: 10.1111/zph.12390. [DOI] [PubMed] [Google Scholar]
- Dabassa A. Evaluation of home slaughtered meat quality used for human consumption at household and food seller house in Jimma. J. Med. Sci. 2013;13:779–784. [Google Scholar]
- Dabassa A., Bacha K. The prevalence and antibiogram of Salmonella and Shigella isolated from abattoir, Jimmarjruth West Ethiopia. IJPBR. 2012;3:143–148. [Google Scholar]
- Dadi L., Asrat D. Prevalence and antimicrobial susceptibility profiles of thermotolerant Campylobacter strains in retail raw meat products in Ethiopia. Ethiop. J. Health Dev. 2008;22(2):195–200. [Google Scholar]
- Daniyan S.Y. Microbiological quality of pork meat from local mammy market in Niger State, Nigeria. AU J. Tech. 2011;14:229–231. [Google Scholar]
- Dione M.M., Ieven M., Garin B., Marcotty T., Geerts S. Prevalence and antimicrobial resistance of Salmonella isolated from broiler farms, chicken carcasses, and street-vended restaurants in Casamance, Senegal. J. Food Prot. 2009;72:2423–2427. doi: 10.4315/0362-028x-72.11.2423. [DOI] [PubMed] [Google Scholar]
- Dione M.M., Ikumapayi U.N., Saha D., Mohammed N.I., Geerts S., Ieven M., Adegbola R.A., Antonio M. Clonal differences between non-Typhoidal Salmonella (NTS) recovered from children and animals living in close contact in The Gambia. PLoS Negl. Trop. Dis. 2011;5 doi: 10.1371/journal.pntd.0001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguale T., Engidawork E., Gebreyes W.A., Asrat D., Alemayehu H., Medhin G., Johnson R.P., Gunn J.S. Fecal prevalence, serotype distribution and antimicrobial resistance of Salmonellae in dairy cattle in central Ethiopia. BMC Microbiol. 2016;16 doi: 10.1186/s12866-016-0638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisa M.I., Mohamed A.A. Role of enteric pathogens in enteritis in lambs, goat kids and children and their zoonotic importance. Vet. Med. J. Giza. 2004;52:41–59. [Google Scholar]
- Eissa W.M.M., Moussa M.M., Ahmed M.I. Bacteriological status of chicken carcasses from poultry abattoirs as compared with private shops. Glob. J. Agric. Food Safety Sci. 2014;1:307–316. [Google Scholar]
- Ejeta G., Molla B., Alemayehu D., Muckle A. Salmonella serotypes isolated from minced meat beef, mutton and pork in Addis Ababa, Ethiopia. Rev. Med. Vet. 2004;155:547–551. [Google Scholar]
- Elegbe I.A. Campylobacter infections: domestic cows as a possible source of infection in Nigeria. Med. Lab. Sci. 1983;40:145–147. [PubMed] [Google Scholar]
- Elegbe I.A., Juba A., Adebayo J.O. Species and serotypes of Campylobacter from domestic animals in Nigeria. J. Diarrhoeal Dis. Res. 1987;5:97–101. [PubMed] [Google Scholar]
- El-Gamal A.M., El-Bahi E.F. Molecular characterization of rectal carriage of E. coli O157: H7 and Salmonella spp. in feedlot animals and its effects on carcasses contamination. Alex. J. Vet. Sci. 2016;48:42–49. [Google Scholar]
- El-Gamal Galal B., Refaie R.S., El-Ailla A.A.A. Occurrence of Campylobacter in poultry carcasses. Assiut Vet. Med. J. 1992;26:110–113. [Google Scholar]
- El-Jakee J., Ata N.S., Hakim A.S., Syame S.M., Omara S.T. Prevalence of virulence genes and antimicrobial resistance patterns of Campylobacter species isolated from chicken in Egypt. Asian J. Poultry Sci. 2015;9:250–261. [Google Scholar]
- Ellis E.M. Salmonella reservoirs in animals and feeds. J. Am. Oil Chem. Soc. 1969;46:227–229. doi: 10.1007/BF02544802. [DOI] [PubMed] [Google Scholar]
- Elmossalami E., Yassein N.A. Salmonellae in slaughtered camels. Arch. Leb. 1994;45:118–120. [Google Scholar]
- El-Naker Y.F.I., El-Sawalhy A.A., Youssef M.A.A., Zeidan S.M. Some studies on neonatal calf diarrhea in Egypt. Part 1: causative agents and some epidemiological aspects. Bull. Anim. Health Prod. Afr. 2008;56:161–190. [Google Scholar]
- El-Shibiny A.A., Shiha A.N., Ghoneim S.I. Occurrence and enumeration of campylobacters before and after processing of conventional chickens in Egypt. World J. Dairy Food Sci. 2012;7:222–228. [Google Scholar]
- El-Tras W.F., Tayel A.A., Samir A. Potential zoonotic pathways of Salmonella Enteritidis in laying farms. Vector Borne Zoonotic Dis. 2010;10:739–742. doi: 10.1089/vbz.2009.0080. [DOI] [PubMed] [Google Scholar]
- El-Tras W.F., Holt H.R., Tayel A.A., El-Kady N.N. Campylobacter infections in children exposed to infected backyard poultry in Egypt. Epidemiol. Infect. 2015;143:308–315. doi: 10.1017/S095026881400096X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esan O.B., Pearce M., van Hecke O., Roberts N., Collins D.R.J., Violato M., McCarthy N., Perera R., Fanshawe T.R. Factors sssociated with dequelae of Campylobacter and non-typhoidal Salmonella infections: a systematic review. EBioMedicine. 2017;15:100–111. doi: 10.1016/j.ebiom.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority (EFSA), European Centre for Disease Prevention Control The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J. 2012:10. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewnetu D., Mihret A. Prevalence and antimicrobial resistance of Campylobacter isolates from humans and chickens in Bahir Dar, Ethiopia. Foodborne Pathog. Dis. 2010;7:667–670. doi: 10.1089/fpd.2009.0433. [DOI] [PubMed] [Google Scholar]
- Falade S., Ehizokhale M.U. Salmonella and Escherichia coli strains isolated from poultry in Ibadan, Nigeria. Bull. Anim. Health Prod. Afr. 1981;29:99–101. [PubMed] [Google Scholar]
- Farrag H., El-Afify A. Salmonella in apparently normal camels. J. Egypt Med. Assoc. 1956;39:698–699. [Google Scholar]
- Farrag H., El-Affifi A., Zaki O. Salmonella in apparently normal pigs. J. Egypt Med. Assoc. 1954;37:1110–1113. [PubMed] [Google Scholar]
- Farrag H.F., El-Garhy M.T., Fahmy A.N. Bacteriological and histological studies of the gall bladder of pigs slaughtered in Cairo and used for human consumption with special reference to Salmonella food poisoning. J. Egypt Med. Assoc. 1962;45:310–316. [PubMed] [Google Scholar]
- Fashae K., Ogunsola F., Aarestrup F.M., Hendriksen R.S. Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria. J. Infect. Dev. Ctries. 2010;4:484–494. doi: 10.3855/jidc.909. [DOI] [PubMed] [Google Scholar]
- Feasey N.A., Dougan G., Kingsley R.A., Heyderman R.S., Gordon M.A. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd T.M., Baranski J.R., El-Gannani M. Recovery of human enteric pathogens on meat from butcher shops in Cairo, Egypt. J. Infect. Dis. 1953;92:224–227. doi: 10.1093/infdis/92.3.224. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAO), World Health Organization (WHO) 2005. FAO/WHO Regional Conference of Food Safety for Africa. Final Report. [Google Scholar]
- Food and Drug Administration (FDA) Second edition. 2012. Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins. [Google Scholar]
- FSANZ and the South Australian Research and Development Institute . 2010. Baseline Survey on the Prevalence and Concentration of Salmonella and Campylobacter in Chicken Meat On-farm and at Primary Processing. [Google Scholar]
- Funk J.A., Davies P.R., Nichols M.A. The effect of fecal sample weight on detection of Salmonella enterica in swine feces. J. Vet. Diagn. Investig. 2000;12:412–418. doi: 10.1177/104063870001200504. [DOI] [PubMed] [Google Scholar]
- Gaedirelwe O.G., Sebunya T.K. The prevalence and antibiotic susceptibility of Salmonella spp. in poultry and ostrich samples from slaughter houses in Gaborone, Botswana. J. Anim. Vet. Adv. 2008;7:1151–1154. [Google Scholar]
- Galanis E., Lo Fo Wong D.M., Patrick M.E., Binsztein N., Cieslik A., Chalermchikit T., Aidara-Kane A., Ellis A., Angulo F.J., Wegener H.C. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg. Infect. Dis. 2006;12:381–388. doi: 10.3201/eid1203.050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garedew L., Hagos Z., Addis Z., Tesfaye R., Zegeye B. Prevalence and antimicrobial susceptibility patterns of Salmonella isolates in association with hygienic status from butcher shops in Gondar town, Ethiopia. Antimicrob. Resist. Infect. Control. 2015;4 doi: 10.1186/s13756-015-0062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin B., Gouali M., Wouafo M., Perchec A.M., Thu P.M., Ravaonindrina N., Urbès F., Gay M., Diawara A., Leclercq A., Rocourt J., Pouillot R. Prevalence, quantification and antimicrobial resistance of Campylobacter spp. on chicken neck-skins at points of slaughter in 5 major cities located on 4 continents. Int. J. Food Microbiol. 2012;157:102–107. doi: 10.1016/j.ijfoodmicro.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Gashe B.A., Mpuchane S. Prevalence of salmonellae on beef products at butcheries and their antibiotic resistance profiles. J. Food Sci. 2000;65:880–883. [Google Scholar]
- Gay J.M., Rice D.H., Steiger J.H. Vol. 57. 1994. Prevalence of Fecal Salmonella Shedding by Cull Dairy Cattle Marketed in Washington State; pp. 195–197. [DOI] [PubMed] [Google Scholar]
- Gblossi Bernadette G., Eric Essoh A., Elise Solange K.N., Natalie G., Souleymane B., Lamine Sébastien N., Mireille D. Prevalence and antimicrobial resistance of thermophilic Campylobacter isolated from chicken in Côte d’Ivoire. Int. J. Microbiol. 2012 doi: 10.1155/2012/150612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebeyehu A., Yousuf M., Sebsibe A. Evaluation of microbial load of beef of arsi cattle in Adama town, Oromia, Ethiopia. J. Food Process. Technol. 2013;4:234. [Google Scholar]
- Gharieb R.M., Tartor Y.H., Khedr M.H. Non-Typhoidal Salmonella in poultry meat and diarrhoeic patients: prevalence, antibiogram, virulotyping, molecular detection and sequencing of class I integrons in multidrug resistant strains. Gut Pathog. 2015;7:34. doi: 10.1186/s13099-015-0081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim N.H., Mohamed W.W., Hussein M.K. Public health importance of entero-toxigenic and multi-drug resistant Salmonella serotypes from poultry meat in Egypt. Glob. Vet. 2015;14:813–818. [Google Scholar]
- Gitter M., Brand T.F. Salmonella survey in Kenya abattoirs. Trop. Anim. Health Prod. 1970;2:18–22. [Google Scholar]
- Gopo J.M., Banda G.N. Occurrence of Salmonella on meat and products in an ostrich abattoir as determined with a DNA probe. S. Afr. J. Anim. Sci. 1997;27:1–6. [Google Scholar]
- Goualie G., Karou A.G.T., Bakayoko S., Dosso M., Diopoh J.K. Prevalence of Campylobacter in raw chicken products in Abidjan, cote d’Ivoire. Zoonoses Public Health. 2007;54:74. [Google Scholar]
- Grace D. ILRI; Nairobi, Kenya: 2017. Food Safety in Developing Countries: Research Gaps and Opportunities. White Paper. [Google Scholar]
- Grimont P.A.D., Weill F.X. WHO Collaborating Centre for Reference and Research on Salmonella. 2007. Antigenic formulae of the Salmonella serovars. [Google Scholar]
- Guergueb N., Alloui N., Ayachi A., Bennoune O. Effect of slaughterhouse hygienic practices on the bacterial contamination of chicken meat. Sci. J. Vet. Adv. 2014;3:71–76. [Google Scholar]
- Guo C., Hoekstra R.M., Schroeder C.M., Pires S.M., Ong K.L., Hartnett E., Naugle A., Harman J., Bennett P., Cieslak P., Scallan E., Rose B., Holt K.G., Kissler B., Mbandi E., Roodsari R., Angulo F.J., Cole D. Application of Bayesian techniques to model the burden of human salmonellosis attributable to U.S. food commodities at the point of processing: adaptation of a Danish model. Foodborne Pathog. Dis. 2011;8:509–516. doi: 10.1089/fpd.2010.0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hag Elsafi H.E., Nor Elmadiena M.M., El Hussein A.A., M Siddig M.A., A Muckle C., Cole L., Wilkie E., Mistry K. Salmonella Umbadah: A new Salmonella serovar isolated from cattle in Sudan. Trop. Anim. Health Prod. 2009;41:1605–1606. doi: 10.1007/s11250-009-9353-4. [DOI] [PubMed] [Google Scholar]
- Hald T., Wingstrand A., Swanenburg M., von Altrock A., Thorberg B.M. The occurrence and epidemiology of Salmonella in European pig slaughterhouses. Epidemiol. Infect. 2003;131:1187–1203. doi: 10.1017/s0950268803001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., El-Sawah H., Sherif I., Yousef M., Hikik M. Salmonella of the mesentric lymph nodes of slaughtered cattle, buffaloes and camels. J. Arab Vet. Met. Assoc. 1963;23:272–277. [Google Scholar]
- Hang’ombe B.M., Sharma N.R., Skjerve E., Tuchili L.M. Isolation of bacteria during processing of chicken carcasses for the market in Lusaka, Zambia. Vet. Arh. 1999;69:191–197. [Google Scholar]
- Haruna M., Sasaki Y., Murakami M., Mori T., Asai T., Ito K., Yamada Y. Prevalence and antimicrobial resistance of Campylobacter isolates from beef cattle and pigs in Japan. J. Vet. Med. Sci. 2013;75:625–628. doi: 10.1292/jvms.12-0432. [DOI] [PubMed] [Google Scholar]
- Hassanain N.A. Antimicrobial resistant Campylobacter jejuni isolated from humans and animals in Egypt. Glob. Vet. 2011;6:195–200. [Google Scholar]
- Hassanien A.S., Adiarose H., Talaat M.M., Nouman T.M. 2006. Contamination of Beef Carcasses During Slaughter in Two Egyptian Slaughterhouses. [Google Scholar]
- Hassb-Elnaby G.R., Abdelaziz A.H.B., Sobhy E.E. Serological and electrophoretic characterization of Salmonella species isolated from beef meat. Alex. J. Vet. Sci. 2011;32:1–11. [Google Scholar]
- Havelaar A.H., Kirk M.D., Torgerson P.R., Gibb H.J., Hald T., Lake R.J., Praet N., Bellinger D.C., de Silva N.R., Gargouri N., Speybroeck N., Cawthorne A., Mathers C., Stein C., Angulo F.J., Devleesschauwer B., World Health Organization Foodborne Disease Burden Epidemiology Reference, G World health organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 2015;12:e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.P.T., Thompson S.G., Deeks J.J., Altman D.G. Vol. 327. 2003. Measuring Inconsistency in Meta-analyses; pp. 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes F.A., Adu-Gyamfi A., Appiah V. Microbiological and parasitological quality of local beef retailed in Accra and radiation sensitivity of Salmonella sp. Int. J. Curr. Microbiol. Appl. Sci. 2015;4:86–96. [Google Scholar]
- Hummel P.H. Isolation of salmonellae from cattle at Dar es Salaam. Bull. Epizoot. Dis. Afr. 1974;22:109–113. [PubMed] [Google Scholar]
- Ibrahim A.E. Isolation of Salmonella from the bile of meat animals. Bull. Epizoot. Dis. Afr. 1974;22:234–237. [PubMed] [Google Scholar]
- Ibrahim M.A. Prevalence of Salmonella species in minced beef and meat handlers and their drug resistance. Alex. J. Vet. Sci. 2016;49:122–128. [Google Scholar]
- Ikwap K., Erume J., Owiny D.O., Nasinyama G.W., Melin L., Bengtsson B., Lundeheim N., Fellström C., Jacobson M. Salmonella species in piglets and weaners from Uganda: prevalence, antimicrobial resistance and herd-level risk factors. Prev. Vet. Med. 2014;115:39–47. doi: 10.1016/j.prevetmed.2014.03.009. [DOI] [PubMed] [Google Scholar]
- IntHout J., Ioannidis J.P.A., Rovers M.M., Goeman J.J. Vol. 6. 2016. Plea for Routinely Presenting Prediction Intervals in Meta-analysis; p. e010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iroha I.R., Ugbo E.C., Ilang D.C., Oji A.E., Ayogu T.E. Bacteria contamination of raw meat sold in Abakaliki, Ebonyi State Nigeria. J. Public Health Epidemiol. 2011;3:49–53. [Google Scholar]
- Isogai E., Silungwe M., Sinkala P., Chisenga C., Mubita C., Syakalima M., Hang’ombeb B.M., Makungu C., Yabe J., Simuunzab M., Nambota A., Isogai H., Fukushi H., Yasuda J. Rapid detection of Salmonella on commercial carcasses by using isothermal and chimeric primer-initiated amplification of nucleic acids (ICAN)-enzyme-linked immunosorbent assay (ELISA) in Zambia. Int. J. Appl. Res. Vet. 2005;3:367–371. [Google Scholar]
- Iwu C.J., Iweriebor B.C., Obi L.C., Basson A.K., Okoh A.I. Multidrug-resistant Salmonella isolates from swine in the Eastern Cape Province, South Africa. J. Food Prot. 2016;79:1234–1239. doi: 10.4315/0362-028X.JFP-15-224. [DOI] [PubMed] [Google Scholar]
- Jacob P., Mdegela R.H., Nonga H.E. Comparison of Cape Town and Skirrow’s Campylobacter isolation protocols in humans and broilers in Morogoro, Tanzania. Trop. Anim. Health Prod. 2011;43:1007–1013. doi: 10.1007/s11250-011-9799-z. [DOI] [PubMed] [Google Scholar]
- Jajere S.M., Adamu N.B., Atsanda N.N., Onyilokwu S.A., Gashua M.M., Hambali I.U., Mustapha F.B. Prevalence and antimicrobial resistance profiles of Salmonella isolates in apparently healthy slaughtered food animals at Maiduguri central abattoir, Nigeria. Asian Pac. J. Trop. Dis. 2015;5:996–1000. [Google Scholar]
- Jiwa S.F.H., Kazwala R.R., Namahungu E. Prevalence of Campylobacter spp. in clinically normal goats kept under various management systems in urban Tanzania. Small Rumin. Res. 1994;15:97–100. [Google Scholar]
- Joint United Nations (UN) Programme on HIV/AIDS . 2017. UNAIDS Data 2017. [Google Scholar]
- Jones B.A., Grace D., Kock R., Alonso S., Rushton J., Said M.Y., McKeever D., Mutua F., Young J., McDermott J., Pfeiffer D.U. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8399–8404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker A., Picard J.A. Antimicrobial susceptibility in thermophilic Campylobacter species isolated from pigs and chickens in South Africa. J. S. Afr. Vet. Assoc. 2010;81:228–236. doi: 10.4102/jsava.v81i4.153. [DOI] [PubMed] [Google Scholar]
- Kagambèga A., Haukka K., Siitonen A., Traore A.S., Barro N. Prevalence of Salmonella enterica and the hygienic indicator Escherichia coli in raw meat at markets in Ouagadougou, Burkina Faso. J. Food Prot. 2011;74:1547–1551. doi: 10.4315/0362-028X.JFP-11-124. [DOI] [PubMed] [Google Scholar]
- Kagambèga A., Barro N., Traore A.S., Siitonen A., Haukka K. Characterization of Salmonella enterica and detection of the virulence genes specific to diarrheagenic Escherichia coli from poultry carcasses in Ouagadougou, Burkina Faso. Foodborne Pathog. Dis. 2012;9:589–593. doi: 10.1089/fpd.2011.1071. [DOI] [PubMed] [Google Scholar]
- Kagambèga A., Lienemann T., Aulu L., Traoré A.S., Barro N., Siitonen A., Haukka K. Prevalence and characterization of Salmonella enterica from the feces of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolates. BMC Microbiol. 2013;13 doi: 10.1186/1471-2180-13-253. (11 November 2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapondorah T.L., Sebunya T.K. Occurrence of Salmonella species in raw chicken livers purchased from retail shops in Gaborone, Botswana. J. Anim. Vet. Adv. 2007;6:87–89. [Google Scholar]
- Kashoma I.P., Kassem I., Kumar A., Kessy B.M., Gebreyes W., Kazwala R.R., Rajashekara G. Antimicrobial resistance and genotypic diversity of Campylobacter isolated from pigs, dairy, and beef cattle in Tanzania. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.01240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashoma I.P., Kassem I., John J., Kessy B.M., Gebreyes W., Kazwala R.R., Rajashekara G. Prevalence and antimicrobial resistance of Campylobacter isolated from dressed beef carcasses and raw milk in Tanzania. Microb. Drug Resist. 2016;22:40–52. doi: 10.1089/mdr.2015.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa T., Gebre-Selassie S., Asrat D. Antimicrobial susceptibility patterns of thermotolerant Campylobacter strains isolated from food animals in Ethiopia. Vet. Microbiol. 2007;119:82–87. doi: 10.1016/j.vetmic.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Kempf I., Kerouanton A., Bougeard S., Nagard B., Rose V., Mourand G., Osterberg J., Denis M., Bengtsson B.O. Campylobacter coli in organic and conventional pig production in France and Sweden: prevalence and antimicrobial resistance. Front. Microbiol. 2017;8:955. doi: 10.3389/fmicb.2017.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadr A.M., Haggag Y.N., Khaleil S.A. Prevalence of Campylobacter jejuni and Campylobacter coli in calves and lambs with and without diarrhea and their public health importance. Assiut Vet. Med. J. 2006;52:179–190. [Google Scholar]
- Khalafalla F.A. Campylobacter jejuni in poultry giblets. Zentralbl. Veterinarmed. B. 1990;37:31–34. doi: 10.1111/j.1439-0450.1990.tb01023.x. [DOI] [PubMed] [Google Scholar]
- Khalafalla F.A., Abdel-Atty N.S., Abdel-Wanis S.A., Hanafy A.S. Food poisoning microorganisms in chicken broiler meat. Glob. Vet. 2015;14:211–218. [Google Scholar]
- Khalifa N.O., Afify J.S.A., Rabie N.S. Zoonotic and molecular characterizations of Campylobacter jejuni and Campylobacter coli isolated from beef cattle and children. Glob. Vet. 2013;11:585–591. [Google Scholar]
- Khallaf M., Ameur N., Terta M., Lakranbi M., Senouci S., Ennaji M.M. Prevalence and antibiotic-resistance of Salmonella isolated from chicken meat marketed in Rabat, Morocco. IJIAS. 2014;6:1123–1128. [Google Scholar]
- Khan A.Q. Salmonella infections in birds in the Sudan. Bull. Epizoot. Dis. Afr. 1970;18:207–212. [PubMed] [Google Scholar]
- Khan A.Q. Salmonella infections in cattle in the Sudan. Bull Epizoot Dis Afr. 1970;18:13–18. [PubMed] [Google Scholar]
- Khan A.Q. Salmonella infections in healthy sheep and goats in the Sudan. Bull. Epizoot. Dis. Afr. 1970;18:117–122. [PubMed] [Google Scholar]
- Kich J.D., Coldebella A., Mores N., Nogueira M.G., Cardoso M., Fratamico P.M., Call J.E., Fedorka-Cray P., Luchansky J.B. Prevalence, distribution, and molecular characterization of Salmonella recovered from swine finishing herds and a slaughter facility in Santa Catarina, Brazil. Int. J. Food Microbiol. 2011;151:307–313. doi: 10.1016/j.ijfoodmicro.2011.09.024. [DOI] [PubMed] [Google Scholar]
- Kikuvi G.M., Ombui J.N., Mitema E.S., Schwarz S. Antimicrobial resistance in Salmonella serotypes isolated from slaughter animals in Kenya. East Afr. Med. J. 2007;84:233–239. doi: 10.4314/eamj.v84i5.9531. [DOI] [PubMed] [Google Scholar]
- Kittl S., Heckel G., Korczak B.M., Kuhnert P. Source attribution of human Campylobacter isolates by MLST and Fla-typing and association of genotypes with quinolone resistance. PLoS One. 2013;8:e81796. doi: 10.1371/journal.pone.0081796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komba E.V.G., Mdegela R.H., Msoffe P.L.M., Matowo D.E., Maro M.J. Occurrence, species distribution and antimicrobial resistance of thermophilic Campylobacter isolates from farm and laboratory animals in Morogoro, Tanzania. Vet. World. 2014;7:559–565. [Google Scholar]
- Kpodekon M.T., Goussanou J.S.E., Attakpa E.Y., Boko C.K., Ahounou S.G., Salifou C.F., Tougan U.P., Youssao I.A.K. Evaluation of macroscopic and microbiological hazards of indigenous pork consumption in south of Benin. Int. J. Curr. Microbiol. Appl. Sci. 2013;2:98–109. [Google Scholar]
- Kuroda K., Suzuki R., Ihara K., Miyagi H., Watanabe H., Sato K., Hang’ombe B.M., Mubita C., Isogai N., Mulenga E., Moonga L., Isogai H., Fukuda T., Yoneyama H., Isogai E. Detection of virulence genes of Escherichia coli and Salmonella spp. from fecal samples of Kafue lechwe (Kobus leche kafuensis) and pastoral cattle in the interface areas of Zambia. Afr. J. Microbiol. Res. 2013;7:504–508. [Google Scholar]
- Kusiluka L.J.M., Karimuribo E.D., Mdegela R.H., Luoga E.J., Munishi P.K.T., Mlozi M.R.S., Kambarage D.M. Prevalence and impact of water-borne zoonotic pathogens in water, cattle and humans in selected villages in Dodoma rural and Bagamoyo districts, Tanzania. Phys. Chem. Earth. 2005;30:818–825. [Google Scholar]
- Kwaga J.K. Prevalence of salmonellae in camels in Nigeria. Vet. Rec. 1985;117:291. doi: 10.1136/vr.117.11.291. [DOI] [PubMed] [Google Scholar]
- Larsen K., Merlo J. Appropriate assessment of neighborhood effects on individual health: integrating random and fixed effects in multilevel logistic regression. Am. J. Epidemiol. 2005;161:81–88. doi: 10.1093/aje/kwi017. [DOI] [PubMed] [Google Scholar]
- Lastovica A.J., le Roux E. Efficient isolation of campylobacteria from stools. J. Clin. Microbiol. 2000;38:2798–2799. doi: 10.1128/jcm.38.7.2798-2799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Minor L. Typing of Salmonella species. Eur. J. Clin. Microbiol. Infect. Dis. 1988;7:214–218. doi: 10.1007/BF01963091. [DOI] [PubMed] [Google Scholar]
- Letellier A., Messier S., Paré J., Ménard J., Quessy S. Distribution of Salmonella in swine herds in Québec. Vet. Microbiol. 1999;67:299–306. doi: 10.1016/s0378-1135(99)00049-8. [DOI] [PubMed] [Google Scholar]
- Levesque S., Fournier E., Carrier N., Frost E., Arbeit R.D., Michaud S. Campylobacteriosis in urban versus rural areas: a case-case study integrated with molecular typing to validate risk factors and to attribute sources of infection. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi Z.S., Kamel H.M. Salmonellae from abattoir slaughtered buffaloes. J. Arab Vet. Met. Assoc. 1964;24:145–150. [Google Scholar]
- Lotfi Z.S., Kamel H.M. Salmonellae from apparently healthy sheep slaughtered at Cairo abattoir. J. Arab Vet. Met. Assoc. 1964;24:89–94. [Google Scholar]
- Mabote K.I., Mbewe M., Ateba C.N. Prevalence of Campylobacter contamination in fresh chicken meat and milk obtained from markets in the North-west Province, South Africa. J. Hum. Ecol. 2011;36:23–28. [Google Scholar]
- Madoroba E., Kapeta D., Gelaw A.K. Salmonella contamination, serovars and antimicrobial resistance profiles of cattle slaughtered in South Africa. Onderstepoort J. Vet. Res. 2016:83. doi: 10.4102/ojvr.v83i1.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahangaiko M., Mabi N., Bakana M., Nyongombe U. Food contamination with Salmonella and human health in Kinshasa City, Democratic Republic of Congo (DRC) J. Appl. Biosci. 2015;94:8809–8814. [Google Scholar]
- Mahmoud Y.E., Hamouda S.N. Quality evaluation of poultry meat carcass in El-Gharbia governorate markets. Assiut Vet. Med. J. 2006;52:31–43. [Google Scholar]
- Marín-Martínez F., Sánchez-Meca J. Vol. 70. 2010. Weighting by Inverse Variance or by Sample Size in Random-effects Meta-analysis; pp. 56–73. [Google Scholar]
- Mather A.E., Vaughan T.G., French N.P. Molecular approaches to understanding transmission and source attribution in nontyphoidal Salmonella and their application in Africa. Clin. Infect. Dis. 2015;61:S259–S265. doi: 10.1093/cid/civ727. [DOI] [PubMed] [Google Scholar]
- Mathole M.A., Muchadeyi F.C., Mdladla K., Malatji D.P., Dzomba E.F., Madoroba E. Presence, distribution, serotypes and antimicrobial resistance profiles of Salmonella among pigs, chickens and goats in South Africa. Food Control. 2017;72:219–224. [Google Scholar]
- Mdegela R.H., Yongolo M.G.S., Minga U.M., Olsen J.E. Molecular epidemiology of Salmonella Gallinarum in chickens in Tanzania. Avian Pathol. 2000;29:457–463. doi: 10.1080/030794500750047216. [DOI] [PubMed] [Google Scholar]
- Mdegela R.H., Nonga H.E., Ngowi H.A., Kazwala R.R. Prevalence of thermophilic Campylobacter infections in humans, chickens and crows in Morogoro, Tanzania. J. Vet. Med. B Infect. Dis Vet. Public Health. 2006;53:116–121. doi: 10.1111/j.1439-0450.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- Mdegela R.H., Nonga H.E., Chuma I. Prevalence of thermophilic campylobacters in humans, cattle and poultry and definitive identification of Campylobacter jejuni by PCR in Morogoro, Tanzania. Zoonoses Public Health. 2007;54:46–47. [Google Scholar]
- Mdegela R.H., Laurence K., Jacob P., Nonga H.E. Occurrences of thermophilic Campylobacter in pigs slaughtered at Morogoro slaughter slabs, Tanzania. Trop. Anim. Health Prod. 2011;43:83–87. doi: 10.1007/s11250-010-9657-4. [DOI] [PubMed] [Google Scholar]
- Meara P.J., Melmed L.N., Cook R.C. Microbiological investigation of meat wholesale premises and beef carcases in Johannesburg. J. S. Afr. Vet. Assoc. 1977;48:255–260. [PubMed] [Google Scholar]
- Messad S., Hamdi T.M., Bouhamed R., Ramdani-Bouguessa N., Tazir M. Frequency of contamination and antimicrobial resistance of thermotolerant Campylobacter isolated from some broiler farms and slaughterhouses in the region of Algiers. Food Control. 2014;40:324–328. [Google Scholar]
- Mezali L., Hamdi T.M. Prevalence and antimicrobial resistance of Salmonella isolated from meat and meat products in Algiers (Algeria) Foodborne Pathog. Dis. 2012;9:522–529. doi: 10.1089/fpd.2011.1032. [DOI] [PubMed] [Google Scholar]
- Miller A.S. Salmonellosis in Botswana. I. Incidence in cattle. J. Hyg. 1971;69:491–496. doi: 10.1017/s0022172400021744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira E.K.I., Eskandar A.A. Bacteriological assessment of freshly slaughtered chicken and a trial for improvement. Assiut Vet. Med. J. 2007;53:88–101. [Google Scholar]
- Missohou A., Mbodj M., Zanga D., Niang S., Sylla K.S.B., Seydi M., Cissé O., Seck W.S. 2009. Analysis of Microbiological and Chemical Quality of Poultry Meat in the Vicinity of the Mbeubeuss Landfill in Malika (Senegal), Beekbergen. [DOI] [PubMed] [Google Scholar]
- Moawad R.K., Mohamed G.F., Ashour M.M.S., El-Hamzy E.M.A. Chemical composition, quality characteristics and nutritive value of goat kids meat from Egyptian Baladi breed. J. Appl. Sci. Res. 2013;9:5048–5060. [Google Scholar]
- Modolo J.R., Margato G.F., Gottschalk A.F., Lopes C.A. Incidence of Campylobacter in pigs with and without diarrhea. Rev. Microbiol. 1999;30:19–21. [Google Scholar]
- Mohamed S.R., Dapgh A.N. Bacteriological studies on Salmonella Typhimurium isolated from different sources. Vet. Med. J. Giza. 2007;55:329–340. [Google Scholar]
- Mohamed I.A.H., Hessain A.M., Al-Arfaj A.A., Moussa I.M. Molecular detection of Salmonella enterica serovars Typhimurium and Enteritidis in diarrheic calves. J. Food Agric. Environ. 2014;12:192–194. [Google Scholar]
- Mohammed M.E.H., Hart C.A., Kadden O.R. Viruses and bacteria associated with neonatal camel calf diarrhea in Eastern Sudan. Emir. J. Food Agric. 2003;15:56–62. [Google Scholar]
- Mohammed H.O., Stipetic K., Salem A., McDonough P., Chang Y.F., Sultan A. Risk of Escherichia coli O157:H7, non-O157 shiga toxin-producing Escherichia coli, and Campylobacter spp. in food animals and their products in Qatar. J. Food Prot. 2015;78:1812–1818. doi: 10.4315/0362-028X.JFP-14-596. [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6 [PMC free article] [PubMed] [Google Scholar]
- Molla B., Mesfin A. A survey of Salmonella contamination in chicken carcass and giblets in Central Ethiopia. Rev. Med. Vet. 2003;154:267–270. [Google Scholar]
- Molla B., Kleer J., Sinell H.J. Antibiotic resistance pattern of foodborne Salmonella isolates in Addis Ababa (Ethiopia) Berl. Munch. Tierarztl. Wochenschr. 1999;112:41–43. [PubMed] [Google Scholar]
- Molla B., Alemayehu D., Salah W. Sources and distribution of Salmonella serotypes isolated from food animals, slaughterhouse personnel and retail meat products in Ethiopia: 1997-2002. Ethiop. J. Health Dev. 2003;17:63–70. [Google Scholar]
- Molla B., Mohammed A., Salah W. Salmonella prevalence and distribution of serotypes in apparently healthy slaughtered camels (Camelus dromedarius) in Eastern Ethiopia. Trop. Anim. Health Prod. 2004;36:451–458. doi: 10.1023/b:trop.0000035013.01459.c9. [DOI] [PubMed] [Google Scholar]
- Molla B., Berhanu A., Muckle A., Cole L., Wilkie E., Kleer J., Hildebrandt G. Multidrug resistance and distribution of Salmonella serovars in slaughtered pigs. J. Vet. Med. B Infect. Dis Vet. Public Health. 2006;53:28–33. doi: 10.1111/j.1439-0450.2006.00900.x. [DOI] [PubMed] [Google Scholar]
- Molla W., Molla B., Alemayehu D., Muckle A., Cole L., Wilkie E. Occurrence and antimicrobial resistance of Salmonella serovars in apparently healthy slaughtered sheep and goats of central Ethiopia. Trop. Anim. Health Prod. 2006;38:455–462. doi: 10.1007/s11250-006-4325-4. [DOI] [PubMed] [Google Scholar]
- Montwedi M., Ateba C.N. Use of the cdt gene specific PCR in determining virulence properties of Campylobacter jejuni isolated from chicken meat samples obtained in some supermarkets in Mafikeng, NWP. South Africa. Life Sci. J. 2012;9:2696–2701. [Google Scholar]
- Motsoela C., Collison E.K., Gashe B.A. Prevalence of Salmonella in two Botswana abattoir environments. J. Food Prot. 2002;65:1869–1872. doi: 10.4315/0362-028x-65.12.1869. [DOI] [PubMed] [Google Scholar]
- Mousa M.M., Awad H.A., Yassien N.M., Gouda H.I. Microbial quality of some meat-products. Vet. Med. J. Giza. 1993;41:59–62. [Google Scholar]
- Moussa I.M., Aleslamboly Y.S., Al-Arfaj A.A., Hessain A.M., Gouda A.S., Kamal R.M. Molecular characterization of Salmonella virulence genes isolated from different sources relevant to human health. J. Food Agric. Environ. 2013;11:197–201. [Google Scholar]
- Moussa I.M., Hessain A.M., Gassem M.A., Al-Arfaj A.A., Mohamed I.A. Genotyping of Salmonella enterica collected from poultry farms located in Cairo, Egypt by multiplex-PCR. J. Food Agric. Environ. 2014;12:195–198. [Google Scholar]
- Mughini Gras L., Smid J.H., Wagenaar J.A., de Boer A.G., Havelaar A.H., Friesema I.H., French N.P., Busani L., van Pelt W. Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: a combined case-control and source attribution analysis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner P., Spencer S.E., Wilson D.J., Jones G., Noble A.D., Midwinter A.C., Collins-Emerson J.M., Carter P., Hathaway S., French N.P. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect. Genet. Evol. 2009;9:1311–1319. doi: 10.1016/j.meegid.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Muluneh G., Kibret M. Salmonella spp. and risk factors for the contamination of slaughtered cattle carcass from a slaughterhouse of Bahir Dar town, Ethiopia. Asian Pac. J. Trop. Dis. 2015;5:130–135. [Google Scholar]
- Nabawy E.E., Gharieb R.M.A., Tharwat A.E. Occurrence, phenotypic and genotypic characterization of multidrug resistant zoonotic bacteria isolated from poultry slaughterhouses. Jpn J. Vet. Res. 2016;64:S173–S180. [Google Scholar]
- Nassar A.M., El-Ela A.A. Prevalence of some food poisoning pathogens in squabs and wooden pigeons carcases in Assiut governorate. Assiut Vet. Med. J. 2000;43:209–218. [Google Scholar]
- Nawar E.M., Khedr A.M. Molecular studies on Salmonella species isolated from chicken. Alex. J. Vet. Sci. 2014;43:58–64. [Google Scholar]
- Ngoma M., Pandey G.S., Suzuki A., Sato G., Chimana H. Prevalence of Salmonella in apparently healthy slaughtered cattle and pigs in Zambia. Indian J. Anim. Sci. 1996;35:197–200. [Google Scholar]
- Ngulukun S.S., Oboegbulem S.I., Fagbamila I.O., Emennaa P.E., Ankeli P.I., Ardzard S.S., Okeke L.A., Ajayi O.T., Usman M., Muhammed M.J., Odugbo M.O., Okewole P.A. Isolation of thermophilic Campylobacter species from Japanese quails (Coturnix coturnix) in Vom, Nigeria. Vet. Rec. 2010;166:147–148. doi: 10.1136/vr.b4787. [DOI] [PubMed] [Google Scholar]
- Ngulukun S.S., Oboegbulem S.I., Fagbamila I.O., Bertu W., Odugbo M.O. Prevalence and molecular characterization of thermophilic Campylobacter species isolated from cattle in Plateau State. Nigeria. Niger. Vet. J. 2011;32:349–356. [Google Scholar]
- van Nierop W., Duse A.G., Marais E., Aithma N., Thothobolo N., Kassel M., Stewart R., Potgieter A., Fernandes B., Galpin J.S., Bloomfield S.F. Contamination of chicken carcasses in Gauteng, South Africa, by Salmonella, Listeria monocytogenes and Campylobacter. Int. J. Food Microbiol. 2005;99:1–6. doi: 10.1016/j.ijfoodmicro.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Nigatu S., Abebe M., Tesfaye R., Garedew L. Prevalence and drug sensitivity pattern of Campylobacter jejuni isolated from cattle and poultry in and around Gondar town, Ethiopia. Glob. Vet. 2015;14:43–47. [Google Scholar]
- Niyonzima E., Ongol M.P., Brostaux Y., Koulagenko N.K., Daube G., Kimonyo A., Sindic M. Daily intake and bacteriological quality of meat consumed in the households of Kigali, Rwanda. Food Control. 2016;69:108–114. [Google Scholar]
- Nonga H.E., Muhairwa A.P. Prevalence and antibiotic susceptibility of thermophilic Campylobacter isolates from free range domestic duck (Cairina moschata) in Morogoro municipality, Tanzania. Trop. Anim. Health Prod. 2010;42:165–172. doi: 10.1007/s11250-009-9401-0. [DOI] [PubMed] [Google Scholar]
- Nonga H.E., Sells P., Karimuribo E.D. Occurrences of thermophilic Campylobacter in cattle slaughtered at Morogoro municipal abattoir, Tanzania. Trop. Anim. Health Prod. 2010;42:73–78. doi: 10.1007/s11250-009-9387-7. [DOI] [PubMed] [Google Scholar]
- Nossair M.A., Khaled K., El-Shabasy N.A., Samaha I.A. Detection of some enteric pathogens in retailed meat. Alex. J. Vet. Sci. 2015;44:67–73. [Google Scholar]
- Nouichi S., Hamdi T.M. Superficial bacterial contamination of ovine and bovine carcasses at El-Harrach slaughterhouse (Algeria) Eur. J. Sci. Res. 2009;38:474–485. [Google Scholar]
- Nwachukwu N.C., Orji F.A., Madu C.N. Antibiotic-resistant Salmonella species in pork on display for sale in Umuahia, Abia state, Nigeria. Res. J. Agric. & Biol. Sci. 2010;6:750–753. [Google Scholar]
- Nyamakwere F., Muchenje V., Mushonga B., Makepe M., Mutero G. Assessment of Salmonella, Escherichia coli, Enterobacteriaceae and aerobic colony counts contamination levels during the beef slaughter process. J. Food Safety. 2016;36(4):548–556. [Google Scholar]
- Nyeleti C., Hildebrandt G., Kleer J., Molla B. Prevalence of Salmonella in Ethiopian cattle and minced beef. Berl. Munch. Tierarztl. Wochenschr. 2000;113:431–434. [PubMed] [Google Scholar]
- Nzouankeu A., Ngandjio A., Ejenguele G., Njine T., Wouafo M.N. Multiple contaminations of chickens with Campylobacter, Escherichia coli and Salmonella in Yaounde (Cameroon) J. Infect. Dev. Ctries. 2010;4:583–586. doi: 10.3855/jidc.1019. [DOI] [PubMed] [Google Scholar]
- Oboegbulem S.I., Muogbo E.N. A survey of salmonellae in trade cattle slaughtered at Nsukka abattoir. Int. J. Zoonoses. 1981;8:107–110. [PubMed] [Google Scholar]
- OECD/FAO . Agriculture in sub-Saharan Africa: prospects and challenges for the next decade. In: Publishing O., editor. OECD-FAO Agricultural Outlook 2016–2025. 2016. (Paris) [Google Scholar]
- Ofukwu R.A., Okoh A.E.J., Akwuobu C.A. Prevalence of Campylobacter jejuni in duck faeces around drinking water sources in Makurdi, north-central Nigeria. Sokoto J. Vet. Sci. 2008;7:26–30. [Google Scholar]
- Okunlade A.O., Ogunleye A.O., Jeminlehin F.O., Ajuwape A.T.P. Occurrence of Campylobacter species in beef cattle and local chickens and their antibiotic profiling in Ibadan, Oyo State, Nigeria. Afr. J. Microbiol. Res. 2015;9:1473–1479. [Google Scholar]
- Ola Ojo M. A survey of Salmonellae in goats and dogs in Nigeria. Bull. Epizoot. Dis. Afr. 1974;22:33–34. [PubMed] [Google Scholar]
- Olatoye O.I. Antibiotics use and resistance patterns of Salmonella species in poultry from Ibadan, Nigeria. Trop. Vet. 2011;29:28–35. [Google Scholar]
- Olayemi A.B., Gaultney J.B., Belino E.D. Isolation of Salmonella from cattle slaughtered at a Zaria abattoir. Niger. Med. J. 1979;9:707–710. [PubMed] [Google Scholar]
- Olubunmi P.A., Adeniran M.O.A. Isolation of Campylobacter species from man and domestic animals in the western part of Nigeria. Bull. Anim. Health Prod. Afr. 1986;34:224–228. [Google Scholar]
- Oluyege A.O., Ojo-Bola O. Prevalence of non-typhoidal Salmonella among HIV/AIDS patients and poultry chicken in Ekiti State. Br. Microbiol. Res. J. 2015;6:113–118. [Google Scholar]
- Omara S.T., El-Fadaly H.A., Barakat A.M.A. Public health hazard of zoonotic Campylobacter jejuni reference to Egyptian regional and seasonal variations. Res. J. Microbiol. 2015;10:343–354. [Google Scholar]
- Onyekaba C.O., Njoku H.O. Bacteria and helminth isolates from bile and faeces of zebu cattle slaughtered for human consumption in the Niger Delta areas of Nigeria. Ann. Trop. Med. Parasitol. 1986;80:421–424. doi: 10.1080/00034983.1986.11812042. [DOI] [PubMed] [Google Scholar]
- Orji M.U., Onuigbo H.C., Mbata T.I. Isolation of Salmonella from poultry droppings and other environmental sources in Awka, Nigeria. Int. J. Infect. Dis. 2005;9:86–89. doi: 10.1016/j.ijid.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Osano O., Arimi S.M. Retail poultry and beef as sources of Campylobacter jejuni. East Afr. Med. J. 1999;76:141–143. [PubMed] [Google Scholar]
- Osman K.M., Yousef A.M.M., Aly M.M., Radwan M.I. Salmonella spp. infection in imported 1-day-old chicks, ducklings, and turkey poults: a public health risk. Foodborne Pathog. Dis. 2010;7:383–390. doi: 10.1089/fpd.2009.0358. [DOI] [PubMed] [Google Scholar]
- Osman K.M., Marouf S.H., Erfan A.M., AlAtfeehy N. Salmonella enterica in imported and domestic day-old turkey poults in Egypt: repertoire of virulence genes and their antimicrobial resistance profiles. Rev. Sci. Tech. 2014;33:1017–1026. doi: 10.20506/rst.33.3.2338. [DOI] [PubMed] [Google Scholar]
- Osman K.M., Marouf S.H., Mehana O.A., AlAtfeehy N. Salmonella enterica serotypes isolated from squabs reveal multidrug resistance and a distinct pathogenicity gene repertoire. Rev. Sci. Tech. 2014;33:997–1006. doi: 10.20506/rst.33.3.2336. [DOI] [PubMed] [Google Scholar]
- Osman K.M., Marouf S.H., Zolnikov T.R., AlAtfeehy N. Isolation and characterization of Salmonella enterica in day-old ducklings in Egypt. Pathog. Glob. Health. 2014;108:37–48. doi: 10.1179/2047773213Y.0000000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaru M.M.M., Nsengwa G.R.M., Wagstaff L. Animal salmonellosis in southern Tanzania. Bull. Anim. Health Prod. Afr. 1990;38:199–201. [Google Scholar]
- Oueslati W., Rjeibi M.R., Mhadhbi M., Jbeli M., Zrelli S., Ettriqui A. Prevalence, virulence and antibiotic susceptibility of Salmonella spp. strains, isolated from beef in Greater Tunis (Tunisia) Meat Sci. 2016;119:154–159. doi: 10.1016/j.meatsci.2016.04.037. [DOI] [PubMed] [Google Scholar]
- Ouf J.M. Microbiological evaluation and mycotoxin residues in some frozen camel’s meat products. Vet. Med. J. Giza. 2004;52:213–230. [Google Scholar]
- Phagoo L., Neetoo H. Antibiotic resistance of Salmonella in poultry farms of Mauritius. JWPR. 2015;5:42–47. [Google Scholar]
- Pires S.M., Knegt L., Hald T. Vol. 8. 2011. Estimation of the Relative Contribution of Different Food and Animal Sources to Human Salmonella Infections in the European Union; p. 184E. [Google Scholar]
- Pires S.M., Vieira A.R., Hald T., Cole D. Source attribution of human salmonellosis: an overview of methods and estimates. Foodborne Pathog. Dis. 2014;11:667–676. doi: 10.1089/fpd.2014.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior B.A., Badenhorst L. Incidence of salmonellae in some meat products. S. Afr. Med. J. 1974;48:2532–2537. [PubMed] [Google Scholar]
- Rady E.M., Ibrahim H.A., Samaha I.A. Enteropathogenic bacteria in some poultry meat products. Alex. J. Vet. Sci. 2011;33:175–180. [Google Scholar]
- Rahimi E., Ameri M., Kazemeini H.R. Prevalence and antimicrobial resistance of Campylobacter species isolated from raw camel, beef, lamb, and goat meat in Iran. Foodborne Pathog. Dis. 2010;7:443–447. doi: 10.1089/fpd.2009.0421. [DOI] [PubMed] [Google Scholar]
- Raji M.A., Adekeye J.O., Kwaga J.K.P., Bale J.O.O. Bioserogroups of Campylobacter species isolated from sheep in Kaduna State, Nigeria. Small Rumin. Res. 2000;37:215–221. doi: 10.1016/s0921-4488(00)00125-5. [DOI] [PubMed] [Google Scholar]
- Ramadan, H., Jackson, C. and Hinton, A., Jr., 2015: Screening and rapid identification of Campylobacter spp. DNA by flaA PCR based method on chicken and human fecal samples in Egypt. Int. J. Poult. Sci., 14, 252–256.
- Randa G.A.S., Eman S.E., Tolba K. Bacteriological status of chicken skeleton and legs sold in local markets. Glob. J. Agric. Food Safety Sci. 2014;1:237–250. [Google Scholar]
- Raufu I., Hendriksen R.S., Ameh J.A., Aarestrup F.M. Occurrence and characterization of Salmonella Hiduddify from chickens and poultry meat in Nigeria. Foodborne Pathog. Dis. 2009;6:425–430. doi: 10.1089/fpd.2008.0150. [DOI] [PubMed] [Google Scholar]
- Raufu I., Bortolaia V., Svendsen C.A., Ameh J.A., Ambali A.G., Aarestrup F.M., Hendriksen R.S. The first attempt of an active integrated laboratory-based Salmonella surveillance programme in the north-eastern region of Nigeria. J. Appl. Microbiol. 2013;115:1059–1067. doi: 10.1111/jam.12304. [DOI] [PubMed] [Google Scholar]
- Ravel A., Hurst M., Petrica N., David J., Mutschall S.K., Pintar K., Taboada E.N., Pollari F. Source attribution of human campylobacteriosis at the point of exposure by combining comparative exposure assessment and subtype comparison based on comparative genomic fingerprinting. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refai M., Gad El-Said W., Osman K., Lotfi Z.S., Safwat E., Ellias S. Salmonella in slaughtered camels in Egypt. Zagazig Vet. J. 1984;9:266–276. [Google Scholar]
- Rene K.A., Adjehi D., Timothee O., Tago K., Marcelin D.K., Ignace-Herve M.E. Serotypes and antibiotic resistance of Salmonella spp. isolated from poultry carcass and raw gizzard sold in markets and catering in Abidjan, Côte d’Ivoire. Int. J. Curr. Microbiol. Appl. Sci. 2014;3:764–772. [Google Scholar]
- Richardson N.J., Koornhof H.J. Campylobacter infections in Soweto. S. Afr. Med. J. 1979;55:73–74. [PubMed] [Google Scholar]
- Richardson N.J., Burnett G.M., Koornhof H.J. A bacteriological assessment of meat, offal and other possible sources of human enteric infections in a bantu township. J. Hyg. 1968;66:365–375. doi: 10.1017/s0022172400041231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner B.M., Schielke A., Didelot X., Kops F., Breidenbach J., Willrich N., Golz G., Alter T., Stingl K., Josenhans C., Suerbaum S., Stark K. A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014. Sci. Rep. 2017;7:5139. doi: 10.1038/s41598-017-05227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackey B.A., Mensah P., Collison E., Sakyi-Dawson E. Campylobacter, Salmonella, Shigella and Escherichia coli in live and dressed poultry from metropolitan Accra. Int. J. Food Microbiol. 2001;71:21–28. doi: 10.1016/s0168-1605(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Sahin O., Morishita T.Y., Zhang Q. Campylobacter colonization in poultry: sources of infection and modes of transmission. Anim. Health Res. Rev. 2002;3:95–105. doi: 10.1079/ahrr200244. [DOI] [PubMed] [Google Scholar]
- Salihu M.D., Abdulkadir J.U., Oboegbulem S.I., Egwu G.O., Magaji A.A., Lawal M., Hassan Y. Isolation and prevalence of Campylobacter species in cattle from Sokoto State. Nigeria. Vet. Ital. 2009;45:501–505. [PubMed] [Google Scholar]
- Salihu M.D., Junaidu A.U., Oboegbulem S.I., Egwu G.O., Tambuwal F.M., Yakubu Y. Prevalence of Campylobacter species in apparently healthy goats in Sokoto state (Northwestern) Nigeria. Afr. J. Microbiol. Res. 2009;3:572–574. [Google Scholar]
- Salihu M.D., Junaidu A.U., Magaji A.A., Yakubu Y. Prevalence and antimicrobial resistance of thermophilic Campylobacter isolates from commercial broiler flocks in Sokoto, Nigeria. Res. J. Vet. Sci. 2012;5:51–58. [Google Scholar]
- Sallam K.I., Mohammed M.A., Hassan M.A., Tamura T. Prevalence, molecular identification and antimicrobial resistance profile of Salmonella serovars isolated from retail beef products in Mansoura, Egypt. Food Control. 2014;38:209–214. [Google Scholar]
- Samaha H.A., Draz A.A., Haggg Y.N., Abdou E.M. Small ruminants as a reservoir of certain bacterial and mycotic pathogens to man. Assiut Vet. Med. J. 2002;46:22–35. [Google Scholar]
- Samaha I.A., Ibrahim H.A., Aboukahf H.A. Enterobacteriaceae in retailed meat. Alex. J. Vet. Sci. 2011;34:1–9. [Google Scholar]
- Samaha I.A., Ibrahim H.A.A., Hamada M.O. Isolation of some enteropathogens from retailed poultry meat in Alexandria province. Alex. J. Vet. Sci. 2012;37:17–22. [Google Scholar]
- Sandhu B.K., Paul S.P. Irritable bowel syndrome in children: pathogenesis, diagnosis and evidence-based treatment. World J. Gastroenterol. 2014;20:6013–6023. doi: 10.3748/wjg.v20.i20.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharawe H.I., Ibrahim H.M., Safwat E. Studies on some Salmonella serovars isolated from slaughtered imported camels. Vet. Med. J. Giza. 2009;57:23–33. [Google Scholar]
- Schwarzer G. meta: an R package for meta-analysis. R News. 2007;7:40–45. [Google Scholar]
- Sen R., Collard P. Isolation of Salmonellae from fowls in Ibadan. West Afr. Med. J. 1957;6:15–17. [PubMed] [Google Scholar]
- Sen R., Collard P. Isolation of Salmonellae from healthy pigs in Ibadan. West Afr. Med. J. 1957;6:64–67. [PubMed] [Google Scholar]
- Shaltout F.A., Abdel-Aziz A.M. Salmonella enterica serovar Enteritidis in poultry meat and their epidemiology. Vet. Med. J. Giza. 2004;52:429–436. [Google Scholar]
- Shilangale R.P., Kaaya G.P., Chimwamurombe P.M. Prevalence and characterization of Salmonella isolated from beef in Namibia. European J. Nutr. Food Saf. 2015;5:267–274. [Google Scholar]
- Sibhat B., Zewde B.M., Zerihun A., Muckle A., Cole L., Boerlin P., Wilkie E., Perets A., Mistry K., Gebreyes W.A. Salmonella serovars and antimicrobial resistance profiles in beef cattle, slaughterhouse personnel and slaughterhouse environment in Ethiopia. Zoonoses Public Health. 2011;58:102–109. doi: 10.1111/j.1863-2378.2009.01305.x. [DOI] [PubMed] [Google Scholar]
- Skarp C.P., Hanninen M.L., Rautelin H.I. Campylobacteriosis: the role of poultry meat. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2016;22:103–109. doi: 10.1016/j.cmi.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Smid J.H., Mughini Gras L., de Boer A.G., French N.P., Havelaar A.H., Wagenaar J.A., van Pelt W. Practicalities of using non-local or non-recent multilocus sequence typing data for source sttribution in space and time of human campylobacteriosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0055029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.I., Bamidele M., Goodluck H.A., Fowora M.N., Omonigbehin E.A., Opere B.O., Aboaba O.O. Antimicrobial susceptibilities of Salmonellae isolated from food handlers and cattle in Lagos, Nigeria. Int. J. Health Res. 2009;2:189–193. [Google Scholar]
- Smith S., Braun S., Akintimehin F., Fesobi T., Bamidele M., Coker A., Monecke S., Ehricht R. Serogenotyping and antimicrobial susceptibility testing of Salmonella spp. isolated from retail meat samples in Lagos, Nigeria. Mol. Cell. Probes. 2016;30(4):189–194. doi: 10.1016/j.mcp.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Stevens A., Kaboré Y., Perrier-Gros-Claude J.D., Millemann Y., Brisabois A., Catteau M., Cavin J.F., Dufour B. Prevalence and antibiotic-resistance of Salmonella isolated from beef sampled from the slaughterhouse and from retailers in Dakar (Senegal) Int. J. Food Microbiol. 2006;110:178–186. doi: 10.1016/j.ijfoodmicro.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Tafida S.Y., Kabir J., Kwaga J.K.P., Bello M., Umoh V.J., Yakubu S.E., Nok A.J., Hendriksen R. Occurrence of Salmonella in retail beef and related meat products in Zaria, Nigeria. Food Control. 2013;32:119–124. [Google Scholar]
- Tanih N.F., Sekwadi E., Ndip R.N., Bessong P.O. Detection of pathogenic Escherichia coli and Staphylococcus aureus from cattle and pigs slaughtered in abattoirs in Vhembe District, South Africa. Sci. World J. 2015 doi: 10.1155/2015/195972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teklu A., Negussie H. Assessment of risk factors and prevalence of Salmonella in slaughtered small ruminants and environment in an export abattoir, Modjo, Ethiopia. Am. Eurasian J. Agric. Environ. Sci. 2011;10:992–999. [Google Scholar]
- Thai T.H., Hirai T., Lan N.T., Yamaguchi R. Antibiotic resistance profiles of Salmonella serovars isolated from retail pork and chicken meat in North Vietnam. Int. J. Food Microbiol. 2012;156:147–151. doi: 10.1016/j.ijfoodmicro.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Tibaijuka B., Molla B., Hildebrandt G., Kleer J., Salah W. Antimicrobial resistance to salmonellae isolated from retail raw chicken meat and giblets in Ethiopia. Bull. Anim. Health Prod. Afr. 2002;50:86–95. [Google Scholar]
- Toyofuku H., Hald T. Salmonella source attribution in Japan by a microbiological subtyping approach. Ecohealth. 2011:7. [Google Scholar]
- Turkson P.K., Lindqvist K.J., Kapperud G. Isolation of Campylobacter spp. and Yersinia enterocolitica from domestic animals and human patients in Kenya. APMIS. 1988;96:141–146. [PubMed] [Google Scholar]
- Uaboi-Egbenni P.O., Okolie P.N., Adesanya O.D., Omonigbehin E., Sobande A.O. Epidemiological studies of the incidence of pathogenic Campylobacter spp. amongst animals in Lagos metropolis. Afr. J. Biotechnol. 2008;7:2952–2956. [Google Scholar]
- Uaboi-Egbenni P.O., Bessong P.O., Samie A., Obi C.L. Campylobacteriosis in sheep in farm settlements in the Vhembe District of South Africa. Afr. J. Microbiol. Res. 2010;4:2109–2117. [Google Scholar]
- Uaboi-Egbenni P.O., Bessong P.O., Samie A., Obi C.L. Prevalence and antimicrobial susceptibility profiles of Campylobacter jejuni and coli isolated from diarrheic and non-diarrheic goat faeces in Venda region, South Africa. Afr. J. Biotechnol. 2011;10:14116–14124. [Google Scholar]
- Uaboi-Egbenni P.O., Bessong P.O., Samie A., Obi C.L. Prevalence, haemolysis and antibiograms of campylobacters isolated from pigs from three farm settlements in Venda region, Limpopo province, south. Afr. J. Biotechnol. 2011;10:703–711. [Google Scholar]
- Uaboi-Egbenni P.O., Bessong P.O., Samie A., Obi C.L. Potentially pathogenic Campylobacter species among farm animals in rural areas of Limpopo province, South Africa: A case study of chickens and cattles. Afr. J. Microbiol. Res. 2012;6:2835–2843. [Google Scholar]
- United Nations (UN) Statistics Division . 2016. Standard Country or Area Codes for Statistical Use (M49): Geographic Regions. [Google Scholar]
- Varela N.P., Friendship R.M., Dewey C.E. Prevalence of Campylobacter spp. isolated from grower-finisher pigs in Ontario. Can Vet J. 2007;48:515–517. [PMC free article] [PubMed] [Google Scholar]
- Wesonga S.M., Muluvi G.M., Okemo P.O., Kariuki S. Antibiotic resistant Salmonella and Escherichia coli isolated from indigenous Gallus domesticus in Nairobi, Kenya. East Afr. Med. J. 2010;87:205–210. [PubMed] [Google Scholar]
- Woldemariam E., Molla B., Alemayehu D., Muckle A. Prevalence and distribution of Salmonella in apparently healthy slaughtered sheep and goats in Debre Zeit, Ethiopia. Small Rumin. Res. 2005;58:19–24. [Google Scholar]
- Woldemariam T., Asrat D., Zewde G. Prevalence of thermophilic Campylobacter species in carcasses from sheep and goats in an abattoir in Debre Zeit area, Ethiopia. Ethiop. J. Health Dev. 2009;23:229–233. [Google Scholar]
- World Health Organisation (WHO) Zoonoses and Veterinary Public Health. 2017. Foodborne Zoonoses. (accessed 28 June 2017) [Google Scholar]
- World Health Organisation (WHO) Regional Office for Africa . 2017. Food Safety and Nutrition Food Law Guidelines. [Google Scholar]
- World Health Organization (WHO) 2012. The Global View of Campylobacteriosis: Report of an Expert Consultation. [Google Scholar]
- Yagoub I.A., Mohamed T.E. Isolation and identification of Salmonella from chickens in Khartoum province of Sudan. Br. Vet. Rec. 1987;143:537–540. doi: 10.1016/0007-1935(87)90043-1. [DOI] [PubMed] [Google Scholar]
- Yizengaw H.A. Isolation, identification and antimicrobial susceptibility testing of Salmonella from selected poultry farms in Debre Zeit. Value Health. 2015;18:A542. [Google Scholar]
- Zahran R., El-Behiry A. Prevalence, molecular identification and virulence attributes of Salmonella serovars isolated from feces of diarrheic cow and buffalo-calves. Int. J. Curr. Microbiol. Appl. Sci. 2014;3:9–27. [Google Scholar]
- Zishiri O.T., Mkhize N., Mukaratirwa S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J. Vet. Res. 2016;83:e1–e11. doi: 10.4102/ojvr.v83i1.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
Inclusion and exclusion criteria.
Bias assessment of all included studies.
Names and number of isolates for all Salmonella enterica serovars named in included articles according to African region.