Abstract
Targeted inhibitors to oncogenic kinases demonstrate encouraging clinical responses early in the treatment course; however, most patients will relapse because of target-dependent mechanisms that mitigate enzyme-inhibitor binding or through target-independent mechanisms, such as alternate activation of survival and proliferation pathways, known as adaptive resistance. Here, we describe mechanisms of adaptive resistance in FMS-like receptor tyrosine kinase (FLT3)–mutant acute myeloid leukemia (AML) by examining integrative in-cell kinase and gene regulatory network responses after oncogenic signaling blockade by FLT3 inhibitors (FLT3i). We identified activation of innate immune stress response pathways after treatment of FLT3-mutant AML cells with FLT3i and showed that innate immune pathway activation via the interleukin-1 receptor–associated kinase 1 and 4 (IRAK1/4) complex contributes to adaptive resistance in FLT3-mutant AML cells. To overcome this adaptive resistance mechanism, we developed a small molecule that simultaneously inhibits FLT3 and IRAK1/4 kinases. The multikinase FLT3-IRAK1/4 inhibitor eliminated adaptively resistant FLT3-mutant AML cells in vitro and in vivo and displayed superior efficacy as compared to current targeted FLT3 therapies. These findings uncover a polypharmacologic strategy for overcoming adaptive resistance to therapy in AML by targeting immune stress response pathways.
INTRODUCTION
The identification of oncogenic kinases and small molecules designed to target active, functionally relevant kinases has revolutionized cancer treatment. Frustratingly, although many of these targeted inhibitors initially demonstrate encouraging clinical responses, most patients relapse as a result of primary or acquired resistance. Therapy resistance occurs through target-dependent mechanisms resulting from point mutations in the kinase domain that mitigate enzyme inhibitor binding or through target-independent mechanisms, such as alternate activation of survival and proliferation pathways (1, 2). One example involves the FMS-like receptor tyrosine kinase (FLT3). Activating mutations of FLT3 result in its autophosphorylation and initiation of intracellular signaling pathways, which induce abnormal survival and proliferation of leukemic cells (3–6). One of the most common mutations in acute myeloid leukemia (AML) involves the internal tandem duplication (ITD) of FLT3, which occurs in ~25% of all cases of newly diagnosed AML and confers a particularly poor prognosis (4, 7–10). FLT3 inhibitors (FLT3i) evaluated in clinical studies as monotherapy and combination therapies have shown good initial response rates; however, patients eventually relapse with FLT3i-resistant disease (11–20). The absence of durable remission in patients treated with potent and selective FLT3i highlights the need to identify resistance mechanisms and to develop additional treatment strategies. Several mechanisms contribute to resistance to selective FLT3i, including mutations in the tyrosine kinase domain of FLT3 (20 to 50%) or activation of parallel signaling mechanisms that bypass FLT3 signaling, referred to as adaptive resistance (30 to 50%) (21–23). Furthermore, it is possible for both mechanisms to simultaneously occur in different leukemic populations within a single patient (23). Adaptive resistance of FLT3-ITD AML cells to FLT3i had been attributed to alternate activation of survival and proliferation pathways (1, 24–30). However, combined inhibition of Ras/mitogen-activated protein kinase (MAPK) or phosphatidylinositol 3-kinase (PI3K) signaling alongside FLT3 signaling blockade has not been sufficiently effective at eliminating resistant FLT3-ITD AML cells, implicating additional and/or broader mechanisms of adaptive resistance (31–42). Moreover, multidrug combination regimens present challenges, including synchronized drug exposure and/or cumulative toxicity, which often prevents dosing to therapeutically optimal exposures (43). Therefore, identification of adaptive resistance mechanisms and development of therapies that concomitantly target the primary oncogenic signaling pathway and the relevant adaptive resistance mechanism will likely yield the best clinical outcomes.
RESULTS
FLT3i induce adaptive resistance in FLT3-ITD AML
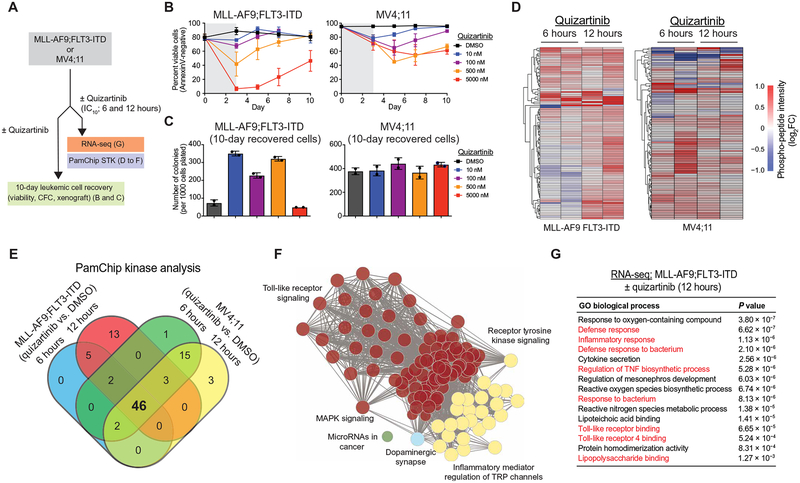
To investigate adaptive resistance to FLT3i in FLT3-ITD AML, we cultured an engineered primary CD34+ human cell line expressing MLL-AF9 and FLT3-ITD (MLL-AF9;FLT3-ITD) and an FLT3-ITD AML cell line (MV4;11) in the presence of cytokines overexpressed in the bone marrow (BM) of patients with AML, including interleukin-3 (IL-3), IL-6, stem cell factor (SCF), thrombopoietin (TPO), and FLT3 ligand (FL) (44–53). This experimental design explored primary adaptive resistance mechanisms occurring immediately after FLT3i treatment. This approach avoids the possibility of subclones acquiring on-target mutations in FLT3, as observed after chronic exposure to FLT3i (54–56). The FLT3-ITD AML cell lines were treated with increasing concentrations of AC220 (quizartinib), a selective inhibitor of FLT3 currently in phase 3 clinical evaluation (), for 72 hours and then examined for leukemic cell recovery (Fig. 1A). Quizartinib treatment at the indicated doses decreased the viability of FLT3-ITD AML cell lines relative to control-treated [dimethyl sulfoxide (DMSO)] cells as measured by AnnexinV staining (Fig. 1B). Although the FLT3-ITD AML cell lines were initially sensitive to quizartinib, FLT3-ITD AML cell lines rapidly proliferated after 3 days of quizartinib treatment (Fig. 1B). To determine whether the leukemic potential of the resistant FLT3-ITD AML cell lines is affected by quizartinib treatment, we examined leukemic progenitor function in vitro and leukemia in vivo. Adaptively resistant FLT3-ITD AML cell lines recovered 10 days after quizartinib exposure, as demonstrated by formation of leukemic cell colonies in methylcellulose (Fig. 1C). At the highest dose of quizartinib, the leukemic progenitor function was decreased, which is likely the result of more robust on-target inhibition of FLT3 and potential off-target effects of quizartinib. Furthermore, resistant MLL-AF9;FLT3-ITD cells that recovered after 30 days of repeated quizartinib exposure rapidly developed leukemia in xenografted NOD.Rag1−/−;γcnull (NRG) mice expressing human IL-3, granulocyte-macrophage colony-stimulating factor (GM-CSF), and steel factor (NRGS) at a comparable rate to parental MLL-AF9;FLT3-ITD cells (fig. S1A). Repeated exposure of the FLT3-ITD AML cell lines to quizartinib for 30 days revealed a diminished sensitivity to FLT3 inhibition at concentrations sufficient to induce cell death of parental cells (fig. S1B). FLT3 and NRAS resequencing confirmed the absence of second-site mutations (F691 and D835) in FLT3 or activating mutations (G12 and G13) in NRAS in the FLT3-ITD AML cell lines resistant to quizartinib treatment, indicating that these cell populations were relying on adaptive signaling resistance mechanisms, rather than acquired mutations (fig. S1C). Parallel studies in FLT3-ITD AML cell lines cultured under standard conditions exhibited a similar outgrowth and leukemic potential, suggesting that the presence of cytokines was not the sole mediator of the adaptive resistance (fig. S1, D and E). FLT3-ITD AML cell lines cultured under standard conditions or in the presence of cytokines remained sensitive to blockade of FLT3 signaling after treatment with quizartinib (fig. S1F), suggesting that the cellular basis of adaptive resistance to FLT3i is mediated by an alternate (non–FLT3-mediated) cell-intrinsic mechanism. Exposure of FLT3-ITD AML cells to the next-generation FLT3i gilteritinib also resulted in cells with competent outgrowth potential, indicating that adaptive resistance is not specific to quizartinib (fig. S1G). These findings are consistent with eventual failure of FLT3i in the clinic without evidence of acquired FLT3 mutations.
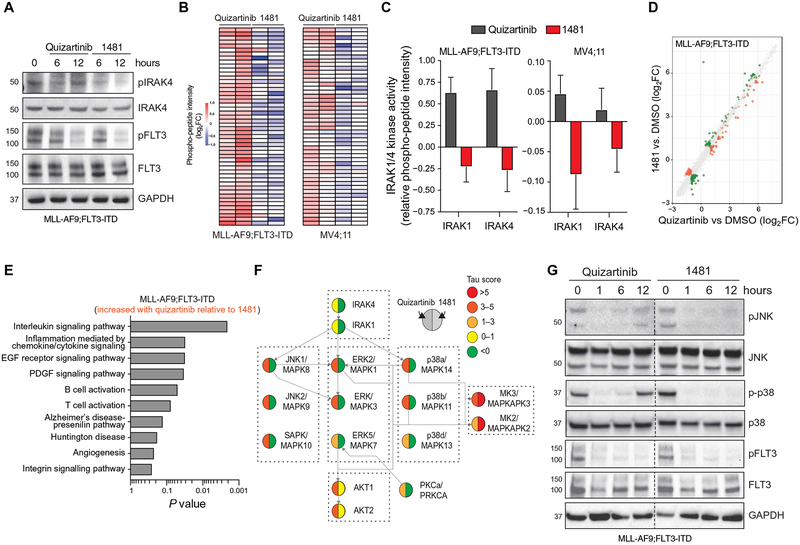
Fig. 1. FLT3-ITD AML develops adaptive resistance and activates innate immune pathways after FLT3i treatment.
(A) Overview of experimental design to evaluate adaptive resistance. CFC, colony-forming cell assay. (B) MLL-AF9;FLT3-ITD or MV4;11 cells were cultured with quizartinib for 3 days and then replated in fresh medium, and cell viability was measured by AnnexinV staining. Values are expressed as means ± SEM from three biological replicates. (C) After 10 days in liquid culture [from (B)], the remaining viable cells were plated in methylcellulose, and colony formation was determined after 7 days. Values are expressed as means ± SD from two biological replicates. (D) Serine-threonine kinase (STK) PamChip analysis was performed on protein lysates isolated from MLL-AF9;FLT3-ITD and MV4;11 cells treated with quizartinib (0.3 nM) for 6 and 12 hours. Hierarchical clustering analysis was performed on differentially phosphorylated peptides in the indicated groups relative to DMSO (two biological replicates). (E) In-cell active kinases inferred from the phosphorylated peptides (STK PamChip) are shown for each of the indicated conditions. (F) Pathway enrichment of differential in-cell kinase activity in MLL-AF9;FLT3-ITD and MV4;11 cells treated with quizartinib for 6 and 12 hours was determined using Panther. (G) Pathway enrichment of differentially expressed genes (>2-fold, P < 0.05) in MLL-AF9;FLT3-ITD cells treated with quizartinib for 12 hours was determined using ToppGene (n = 3 per group).
FLT3i induce compensatory innate immune stress responses in FLT3-ITD AML
Resistance of FLT3-ITD AML cells to FLT3i has been attributed to point mutations at or near the adenosine 5′-triphosphate (ATP)–binding domain of FLT3 and to alternate activation of survival and proliferation pathways (1, 2, 22–27, 29, 30). However, global approaches to delineate the alternate pathways contributing to adaptive resistance in FLT3-ITD AML are lacking. To define mechanisms of adaptive resistance, we examined in-cell kinase activity and gene regulatory networks in adaptively resistant FLT3-ITD AML cells (Fig. 1A). To identify active signaling cascades in adaptively resistant cells, we subjected protein lysates from MLL-AF9;FLT3-ITD and MV4;11 cells treated with quizartinib [IC10 (10% inhibitory concentration), 0.3 nM] for 6 and 12 hours in biological duplicates to peptide phosphorylation profiling using commercially available serine-threonine kinase (STK) PamChip arrays. This concentration of quizartinib was selected because it blocks FLT3-ITD signaling and results in adaptively resistant FLT3-ITD AML cells without evidence of cell death, permitting analysis of adaptive responses to FLT3 signaling blockade in the absence of detectable cytotoxic effects. The PamChip arrays generate a dataset of relative phosphorylation propensity of synthetic peptides containing known substrate recognition sites of STKs in the presence and absence of inhibitor. Unsupervised hierarchical clustering analysis using these data identified two major signaling profiles based on the identification of peptides with a relative decrease or increase in phosphorylation after quizartinib treatment for 6 and 12 hours in MLL-AF9;FLT3-ITD and MV4;11 cells (Fig. 1D and fig. S2A). The in-cell active kinases were inferred on the basis of the combination of distinct phosphorylated peptides using the database of STK-substrate pairs from PhosphoNET (www.phosphonet.ca) (table S1 in data file S1). Using a cutoff (τ > 3 × 10−3), 46 kinases were activated in both FLT3-mutant AML cell lines after 6 and 12 hours of quizartinib exposure (Fig. 1E and table S2 in data file S1). To identify the compensatory signaling networks associated with adaptive resistance to FLT3i, we performed functional annotation of the inferred active kinases common to both FLT3-mutant AML cell lines after 6 and 12 hours of quizartinib exposure (table S2). Using Panther, we identified known compensatory and stress signaling pathways, such as MAPK signaling and dopamine signaling, and several other signaling pathways, including Toll-like receptor (TLR) activation (“innate immune signaling”), that have not previously been implicated in FLT3i adaptive resistance mechanisms to therapy (Fig. 1F) (25, 26, 57).
In parallel, we performed RNA sequencing (RNA-seq) on MLL-AF9;FLT3-ITD cells treated with quizartinib (IC10, 0.3 nM) for 6 and 12 hours. The compensatory transcriptional response involved sets of genes that increased in relative expression after quizartinib treatment for 6 hours [n = 1286; log2fold-change (FC) > 2, P < 0.05] and 12 hours (n = 1281; log2FC > 2, P < 0.05) in MLL-AF9;FLT3-ITD cells (table S3 in data file S1). The differentially overexpressed genes at 12 hours were enriched in Gene Ontology (GO) pathways related to innate immune signaling (Fig. 1G), suggesting that compensatory activation of innate immune stress pathways provides a cytoprotective role after FLT3i treatment in FLT3-mutant AML.
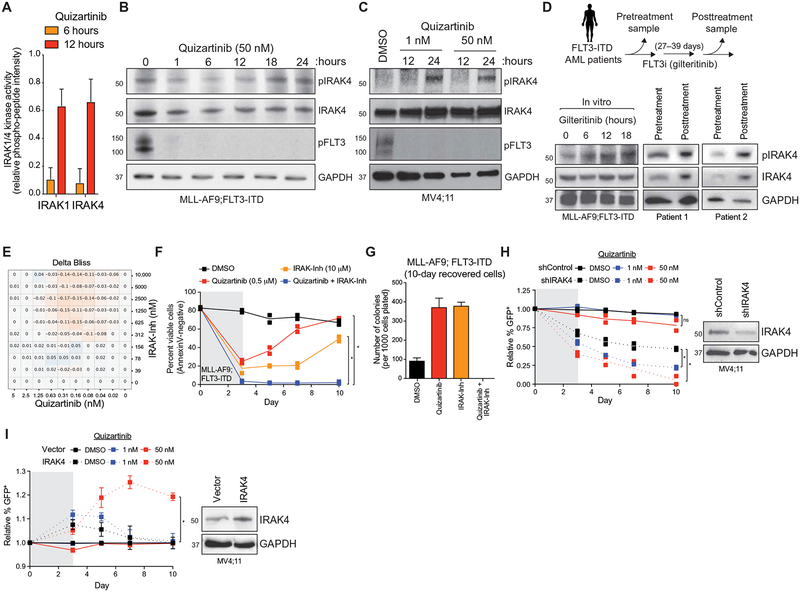
Among the critical signaling elements within the innate immune pathway are the IL-1 receptor–associated kinase 1 (IRAK1) and IRAK4, which are upstream of all signaling effectors within the innate immune pathway and are amenable to therapeutic inhibition (58–69). A more granular examination of the STK array outcomes highlights that MLL-AF9;FLT3-ITD and MV4;11 cells treated with quizartinib have increased phosphorylation of IRAK1/4-specific peptides (Fig. 2A). Orthogonal validation via immunoblotting confirmed increased phosphorylation of IRAK4 at Thr345/Ser346 after inhibition of FLT3-ITD in MLL-AF9;FLT3-ITD or MV4;11 cells by quizartinib (Fig. 2, B and C). Phosphorylation of IRAK4 was also observed after treatment with the next-generation FLT3i, gilteritinib, in MLL-AF9;FLT3-ITD cells (Fig. 2D). In these experiments, although the majority of FLT3 signaling is inhibited [as indicated by reduced pFLT3 and phosphorylated signal transducer and activator of transcription 5 (pSTAT5)] (Fig. 2B and fig. S1F), >95% of quizartinib-treated FLT3-ITD AML cells remained viable (Annexin V−), strongly suggesting that IRAK1/4 activation is an adaptive survival mechanism. The activation state of IRAK1/4 is durable, and we observed phosphorylated IRAK4 after 72 hours of quizartinib treatment in MV4;11 cells (fig. S2B). To determine whether this is an adaptive response based primarily on FLT3 blockade, the isogenic AML cell line MLL-AF9;NRASG12D, which does not depend on FLT3-ITD oncogenic signaling, was treated with quizartinib at the same dose and time points as the FLT3-ITD line. The NRAS-mutant cells did not exhibit phosphorylation of IRAK4 when treated with quizartinib, suggesting that a dependence on FLT3 signaling is needed to elicit this adaptive response (fig. S2C). These observations were extended to patients with FLT3-ITD AML enrolled in a study evaluating the efficacy of gilteritinib (study ID: 2215-CL-9100; table S4 in data file S1). As compared to peripheral blood mononuclear cells (PBMCs) obtained at diagnosis, PBMCs from two patients treated with gilteritinib for 27 and 39 days exhibited increased phosphorylated and total IRAK4 protein, comparable to gilteritinib-treated cells in vitro (Fig. 2D). As an indication of active IRAK4 signaling, we observed phosphorylated IRAK1 in MV4;11 cells after treatment with quizartinib (fig. S2D), a patient after gilteritinib treatment (fig. S2E), and BaF3 cells transduced with FLT3-ITD and treated with quizartinib (fig. S2F). These findings strongly suggest that FLT3i induce compensatory IRAK1/4 activation in FLT3-ITD AML cells in vitro and in vivo.
Fig. 2. Innate immune signaling via IRAK1/4 mediates adaptive resistance to FLT3i.
(A) IRAK1 and IRAK4 activity in MLL-AF9;FLT3-ITD cells treated with quizartinib for 6 and 12 hours is shown on the basis of the relative phosphorylation of the indicated peptides on the STK PamChip array (summary of four IRAK1- or three IRAK4-independent peptides from two biological replicates). (B) Immunoblotting of pFLT3 and pIRAK4 in MLL-AF9;FLT3-ITD cells treated with quizartinib for the indicated times. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C) Immunoblotting of pFLT3 and pIRAK4 in MV4;11 cells treated with quizartinib for the indicated times. (D) Immunoblotting of pIRAK4 and total IRAK4 in MLL-AF9;FLT3-ITD cells treated in vitro with gilteritinib (10 nM) or from the peripheral blood of patients with FLT3-ITD AML treated with gilteritinib for the indicated number of days. (E) Delta Bliss score of MLL-AF9;FLT3-ITD cells treated with the indicated concentrations of quizartinib and IRAK-Inh for 48 hours was calculated on the basis of the cellular metabolic activity using CellTiter-Glo. (F) Viability of MLL-AF9;FLT3-ITD cells treated for 3 days with DMSO (vehicle control), quizartinib (0.5 μM), IRAK-Inh (10 μM), or quizartinib and IRAK-Inh. Individual data points are shown along with the mean from two biological replicates. (G) After 10 days in liquid culture [from (F)], the remaining viable cells were plated in methylcellulose, and colony formation was determined after 7 days. Values are expressed as means ± SEM from four biological replicates. (H) MV4;11 cells expressing control shRNA (shControl-GFP) or shRNA targeting IRAK4 (shIRAK4-GFP) were treated with DMSO or quizartinib (1 or 50 nM) for 3 days. The proportion of GFP+ cells over 10 days in culture is shown relative to day 0. Individual data points are shown along with the mean from two biological replicates. *P < 0.05 (unpaired, two-tailed t test). ns, not significant. (I) MV4;11 cells expressing an empty GFP vector (vector) or a GFP vector with IRAK4 (IRAK4) were treated with DMSO or quizartinib (1 or 50 nM) for 3 days. The proportion of GFP+ cells over 10 days in culture is shown relative to day 0. Values are expressed as means ± SEM from four biological replicates. *P = 0.029 (Mann-Whitney).
To explore a potential mechanism of IRAK1/4 activation in FLT3i-treated FLT3-ITD AML cells, we examined RNA expression after quizartinib treatment of MLL-AF9;FLT3-ITD cells (Fig. 1G). Because IRAK1/4 is downstream of the TLR superfamily, we compared the expression of all TLRs before and after 6-hour quizartinib treatment. Although six of eight TLRs exhibited increased expression in MLL-AF9;FLT3-ITD cells after quizartinib treatment, only TLR9 was significantly overexpressed at 6 hours and remained elevated at 12 hours (P = 0.019, adjusted) (fig. S3, A and B). Immunoblotting of MLL-AF9;FLT3-ITD cells treated with quizartinib revealed that inhibition of FLT3-ITD increases the expression of cleaved TLR9, which has been associated with its active state (fig. S3C) (70, 71). To establish whether increased TLR9 expression and activation in quizartinib-treated FLT3-ITD AML cells result in IRAK1/4 activation, we treated MLL-AF9;FLT3-ITD simultaneously with quizartinib and a TLR9 antagonist (30 nM; ODN-INH-18). In the presence of the TLR9 antagonist, quizartinib-mediated activation of IRAK4 was suppressed as compared to MLL-AF9;FLT3-ITD cells treated only with quizartinib (fig. S3D). Increased expression of TLRs, such as TLR9, on FLT3i-treated AML cells may account for the activation of innate immune pathways in adaptively resistant FLT3-ITD AML cells.
Innate immune signaling via IRAK1/4 is required for adaptive resistance of FLT3-ITD AML to FLT3i
We next investigated whether IRAK1/4 activation is a functionally required element of adaptive resistance of FLT3-ITD AML to FLT3i. Several published inhibitors of IRAK1/4 exist, providing key tools to assess the role of IRAK1/4 in adaptive diseases (69, 72, 73). We first evaluated a pairwise matrix combination of quizartinib and a commercially available IRAK1/4 inhibitor (IRAK-Inh) in MLL-AF9;FLT3-ITD cells (74). This experiment used a 48-hour CellTiter-Glo assay format to demonstrate that the combination of quizartinib and IRAK-Inh is synergistically cytotoxic in MLL-AF9;FLT3-ITD cells (Fig. 2E). Even at low doses of quizartinib (0.3 or 0.4 nM), inhibition of IRAK1/4 decreased MLL-AF9;FLT3-ITD cell viability more than would be expected as an additive response (Fig. 2E and fig. S4A). To further confirm that inhibition of IRAK1/4 can suppress adaptive resistance to FLT3i in FLT3-ITD AML cells, we used AnnexinV staining in MLL-AF9;FLT3-ITD and MV4;11 cells treated with quizartinib (0.5 μM), IRAK-Inh (10 μM), or the combination of quizartinib and IRAK-Inh. These studies show that the combination-treated cells had significantly suppressed the outgrowth of adaptively resistant FLT3-ITD AML cells as compared to quizartinib or IRAK-Inh treatment alone (2.1% versus 71.8% or 75.3% AnnexinV-negative cells; P = 0.003) (Fig. 2F and fig. S4B). MLL-AF9;FLT3-ITD cells recovered 10 days after inhibitor exposure were also plated in methylcellulose to assess leukemic cell potential. Adaptively resistant MLL-AF9;FLT3-ITD cells treated with quizartinib or IRAK-Inh alone formed significantly more leukemic colonies as compared to parental cells (P < 0.0001; Fig. 2G). In contrast, the MLL-AF9;FLT3-ITD cells recovered after treatment with quizartinib and IRAK-Inh did not form leukemic colonies (Fig. 2G). Inhibition of IRAK1/4 with IRAK-Inh or a potent IRAK4 inhibitor (PF-06650833) alone did not affect the leukemic cell viability, leukemic progenitor activity in methylcellulose, or outgrowth of leukemic cells in liquid culture (fig. S4, B to E), indicating that targeting IRAK1/4 alone does not confer cytotoxicity in FLT3-mutant AML. To reinforce that these outcomes implicate IRAK1/4 signaling as the primary driver of adaptive resistance, we expressed short hairpin RNA (shRNA) targeting IRAK4 (shIRAK4-MV4;11) or a nontargeting control shRNA (shControl-MV4;11) in MV4;11 cells (Fig. 2H, right). After treatment with quizartinib (1 and 50 nM), the proportion of shIRAK4-MV4;11 cells was significantly reduced relative to DMSO-treated shIRAK4-MV4;11 cells (1 nM, P = 0.0035; 50 nM, P = 0.0169) or quizartinib-treated shControl-MV4;11 cells (1 nM, P = 0.0002; 50 nM, P < 0.0143) (Fig. 2H, left). Conversely, we also examined the consequences of IRAK4 overexpression on mediating adaptive resistance of MV4;11 cells to FLT3i (Fig. 2I, right). After treatment with quizartinib (1 and 50 nM), the relative proportion of IRAK4-overexpressing MV4;11 cells was significantly increased relative to DMSO-treated IRAK4-overexpressing MV4;11 cells (50 nM, P = 0.029) or quizartinib-treated control MV4;11 cells (empty vector; 50 nM, P = 0.029) (Fig. 2I, left). Together, these studies suggest that IRAK1/4 signaling is required for adaptive resistance in FLT3-mutant AML immediately after inhibition of FLT3 and that inhibition of IRAK1/4 signaling creates a synthetic lethality when combined with FLT3i.
Small-molecule inhibitors were generated to simultaneously target IRAK1/4 and FLT3
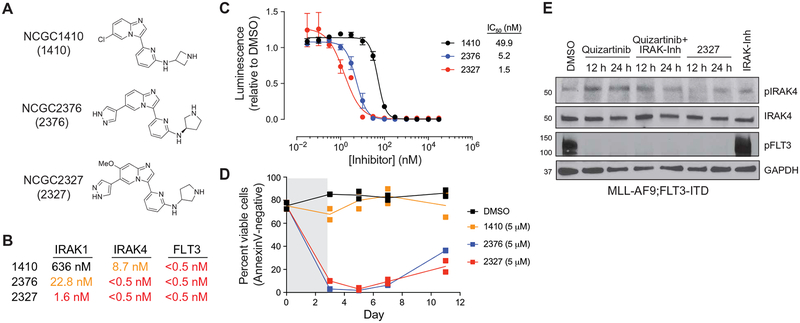
The immediate nature of IRAK1/4 activation in adaptively resistant FLT3-ITD AML cells requires concomitant inhibition of these targets to avoid compensatory signaling and cell survival. Achieving optimal multidrug combination regimens that yield extended overlapping exposure while avoiding unwanted toxicities is challenging. Therefore, we desired a small-molecule inhibitor that simultaneously targeted the FLT3 and IRAK1/4 kinases to eradicate adaptively resistant FLT3-ITD AML. Inhibition of FLT3 is a common “off-target” pharmacology for many advanced kinase inhibitors (cabozantinib, sorafenib, and ponatinib). We therefore considered previously reported IRAK1/4 inhibitors as potential starting points for the optimization of a dual FLT3/IRAK inhibitor. Among the more promising candidates were a series of 3-(pyridin-2-yl)imidazo[1,2-a]pyridines that were previously reported as selective IRAK4 inhibitors and appeared to be attractive chemical starting points for optimization as dual FLT3/IRAK inhibitors (72). Structure activity relationship exploration around this core scaffold yielded a series of small molecules that potently targeted IRAK1, IRAK4, and FLT3. In a functional biochemical assay, three of these agents (NCGC2376, NCGC2327, and NCGC1410) inhibited FLT3 at subnanomolar concentrations (IC50 < 0.5 nM) and IRAK1 and IRAK4 at low nanomolar concentrations (Fig. 3, A and B). Whereas the three compounds were equipotent FLT3i (IC50 < 0.5 nM), their relative efficacies at inhibiting FLT3-ITD AML cell viability correlated with their IRAK1/4 inhibitory potencies (Fig. 3, B and C). Thus, the most potent IRAK1/4 inhibitor NCGC2327 (IRAK1 IC50 = 1.6 nM; IRAK4 IC50 < 0.5 nM) was the most efficacious at inhibiting MLL-AF9;FLT3-ITD cell growth, and the weakest IRAK1/4 inhibitor NCGC1410 (IRAK1 IC50 = 636 nM; IRAK4 IC50 = 8.7 nM) was correspondingly less efficacious (Fig. 3C). Moreover, NCGC2327 and NCGC2376 were more effective at suppressing MLL-AF9;FLT3-ITD leukemic cell recovery relative to NCGC1410, as measured by AnnexinV staining (Fig. 3D), suggesting that potency against IRAK1/4 is a driving element for suppressing adaptively resistant FLT3-ITD AML cells. The finding that NCGC1410 retained potency against IRAK4 yet was less effective at suppressing MLL-AF9;FLT3-ITD AML cells than NCGC2376, which inhibits IRAK1 and IRAK4, argues that targeting both kinases is necessary to achieve optimal suppression of FLT3-mutant AML. NCGC2327 also effectively suppressed FLT3-ITD signaling and compensatory activation of IRAK4 in MLL-AF9;FLT3-ITD cells and was more effective at preventing compensatory activation of IRAK4 as compared to simultaneously inhibiting FLT3 and IRAK1/4 with a combination of quizartinib and IRAK-Inh (Fig. 3E). These findings indicate that the efficacy of suppressing FLT3-ITD AML with an FLT3i correlates with concomitant targeting of IRAK1 and IRAK4.
Fig. 3. Structure activity relationship of small-molecule inhibitors reveals the importance of targeting IRAK1/4 and FLT3 in FLT3+ AML.
(A) Chemical structures of NCGC1410, NCGC2376, and NCGC2327. (B) The half maximal inhibitory concentration (IC50) for NCGC1410, NCGC2376, and NCGC2327 on IRAK1, IRAK4, and FLT3 activity (Reaction Biology). (C) Metabolic activity of MLL-AF9;FLT3-ITD cells treated with NCGC1410, NCGC2376, or NCGC2327 for 72 hours as measured by CellTiter-Glo. Values are expressed as means ± SEM from three biological replicates. (D) Viability of MLL-AF9;FLT3-ITD cells treated for 3 days with DMSO (vehicle control), NCGC1410, NCGC2376, or NCGC2327. Individual data points are shown along with the mean from two biological replicates. (E) Immunoblotting of MLL-AF9;FLT3-ITD cells treated with quizartinib (50 nM), quizartinib and IRAK-Inh (10 μM), NCGC2327 (50 nM), or IRAK-Inh.
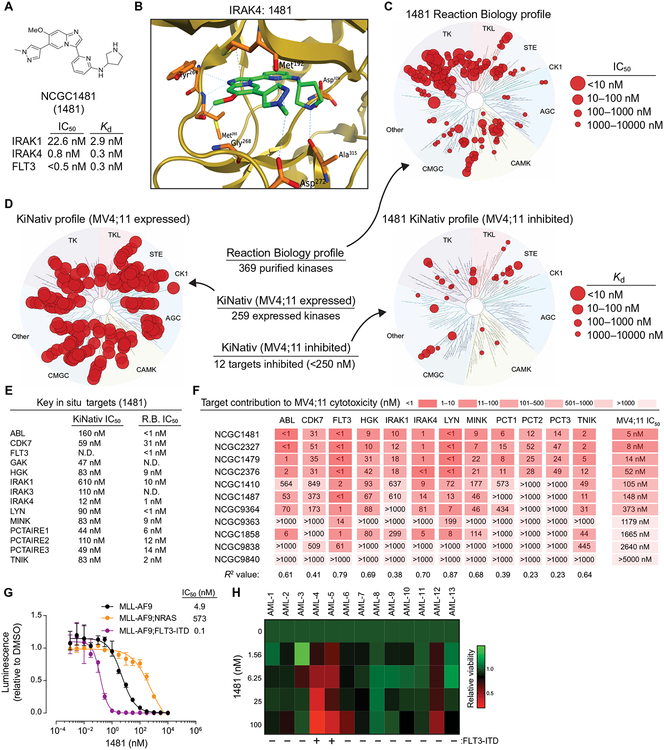
Continued optimization of these agents led to the identification of NCGC1481, which retained strong biochemical potency versus FLT3, IRAK1, and IRAK4 while also displaying acceptable pharmacokinetic properties in mice (Fig. 4A and fig. S5). NCGC1481 exhibited potent binding affinity for IRAK1 (Kd (dissociation constant) = 2.9 nM], IRAK4 (Kd = 0.3 nM), and FLT3 (Kd = 0.3 nM) and potent functional inhibition of IRAK1 (IC50 = 22.6 nM), IRAK4 (IC50 = 0.8 nM), and FLT3 (IC50 = <0.5 nM) (Fig. 4A). To gain insight into the structure-activity relationship of this agent, we obtained a high-resolution (2.1 Å) crystal structure of the NCGC1481-IRAK4 complex [Protein Data Bank (PDB): 6MOM; Fig. 4B], which demonstrates that NCGC1481 binds as a type I inhibitor (ATP-competitive binding to the active state) with excellent shape complementarity at the ATP-binding pocket. Key hydrogen bonds are established between the NCGC1481 imidazole nitrogen and a hinge domain (Met265) amide and the NCGC1481 pyrrolidine and Asp329 and Ala315 (Fig. 4B). Consistent with our data that NCGC1481 also inhibits FLT3, we have obtained a crystal structure of the NCGC1481-FLT3 complex (PDB: 6IL3). Sequence alignment and two-dimensional (2D) interaction diagrams derived from these structures highlight conserved binding elements between IRAK4 and FLT3 and show that the FLT3 ATP-binding domain is relatively more permissive, which is consistent with the fact that FLT3 is routinely found as an off-target pharmacology for multiple kinase inhibitors (fig. S6). When profiled against 369 kinases using Reaction Biology biochemical inhibition assays, NCGC1481 demonstrated modest selectivity (10-fold or greater selectivity versus more than 80% of tested kinases relative to IRAK1, IRAK4, and FLT3) with strong activity noted versus Src-family kinases and selected classes of receptor tyrosine kinases (Fig. 4C and table S5 in data file S1). Given the known caveats of relying on biochemical assays to decipher kinome selectivity, we felt that it was critical to establish the in situ kinome selectivity for this class of agents in relevant cells (75). We therefore submitted NCGC1481 to the KiNativ in situ kinase profiling platform. Examination of NCGC1481 in MV4;11 revealed a higher kinase selectivity in situ relative to the selectivity observed in biochemical assays using purified active proteins (Fig. 4, D and E, and table S6 in data file S1). Of the 259 expressed and active kinases in MV4;11 cells, only 12 were inhibited with an IC50 value of less than 250 nM (Fig. 4E). We next sought to delineate which of these kinase targets contribute to the cytotoxicity of NCGC1481. For this, we generated a series of analogs of NCGC1481 with varying potency against the 12 top kinase targets based on biochemical inhibition assays and then evaluated each analog’s cytotoxicity in MV4;11 cells (Fig. 4F). On the basis of this analysis, the more highly correlated contributing targets to the cytotoxicity of MV4;11 cells are FLT3 (R2 = 0.79), IRAK4 (R2 = 0.70), and LYN (R2 = 0.87). IRAK1 also shows correlation between potency and cytotoxicity, but the potency decline between NCGC2376 and NCGC1410 is ~35-fold, whereas the cytotoxicity decline between these two analogs is only 2-fold. Although LYN was inhibited by NCGC1481 in the biochemical assays, NCGC1481 exhibited only moderate effects on the phosphorylation status of LYN at the doses at which FLT3 and IRAK1/4 were inhibited in MV4;11 cells (fig. S7A), suggesting that inhibition of FLT3 and IRAK4 primarily contributes to the cytotoxicity of NCGC1481. Notwithstanding, most small-molecule kinase inhibitors target more than one kinase, and it is often the collective inhibition of multiple signaling nodes that contributes to the broad biological effects of a kinase inhibitor. Additional assays further demonstrated that NCGC1481 inhibited phosphorylation of FLT3 and IRAK4 and IRAK1/4-mediated nuclear factor κB (NF-κB) transcriptional activation in a time- and dose-dependent manner (fig. S7, B and C). Moreover, NCGC1481 exhibits attractive physical properties, including good aqueous solubility, cell permeability, metabolic stability, and low activity in selected in vitro–based toxicity-associated target assay (fig. S5, A and B).
Fig. 4. NCGC1481 is a potent small-molecule inhibitor of FLT3 and IRAK1/4.
(A) Chemical structure of NCGC1481 (1481). The IC50 and equilibrium dissociation constant (Kd) for NCGC1481 with IRAK1, IRAK4, and FLT3 is shown below the structure. (B) NCGC1481-binding pocket of IRAK4 from NCGC1481-IRAK4 crystal structure. IRAK4 is shown as a ribbon structure along with contact residues. NCGC1481 is shown in green and blue. (C) Reaction Biology kinome map showing selectivity of NCGC1481 across 369 purified and active kinases. (D) KiNativ in situ kinome profile of NCGC1481 in MV4;11 cells showing the expressed and active kinases (left dendrogram) and these inhibited by NCGC1481 (right dendrogram). (E) Top kinases (listed in alphabetical order) inhibited by NCGC1481 as determined by the KiNativ in situ kinome profile and the corresponding IC50 value for NCGC1481 as determined versus purified active kinases at Reaction Biology (R.B.). N.D., not determined. (F) Kinase inhibitory activity (Reaction Biology, IC50 values) for top kinase targets for selected NCGC1481 analogs with variable cytotoxicity (far right column) versus MV4;11 cells. (G) Metabolic activity of MLL-AF9, MLL-AF9;FLT3-ITD, and MLL-AF9;NRAS cells treated with the indicated concentration of NCGC1481 for 72 hours as measured by CellTiter-Glo. Values are expressed as means ± SEM from three biological replicates. (H) Viability of primary AML cells from 13 patients was determined in the presence of NCGC1481 for 48 hours by trypan blue exclusion from two biological replicates. The heat map indicates the mean value. The FLT3-ITD status of the patients is indicated below the heat map and in table S7.
To demonstrate that NCGC1481 is selective for AML cells dependent on FLT3 signaling, we measured proliferation of isogenic AML cells lines derived from primary CD34+ human cord blood cells transduced with MLL-AF9 and then transduced with NRASG12D or FLT3-ITD (45). MLL-AF9;FLT3-ITD cells were highly sensitive to NCGC1481 (IC = 0.1 nM), whereas MLL-AF9;NRASG12D cells were less responsive to NCGC1481 (IC50 = 573 nM) (Fig. 4G). The parental MLL-AF9 cells exhibited an intermediate sensitivity to NCGC1481 (IC5 = 4.9 nM) because these cells are dependent on wild-type FLT3 signaling (Fig. 4G) (45). As expected, NCGC1481 suppressed the short-term proliferation of MLL-AF9;FLT3-ITD, MV4;11, and MOLM13 cells (fig. S7D). In a panel of 13 primary AML patient samples, NCGC1481 was primarily effective against FLT3-mutant AML while exhibiting minimal activity against FLT3–wild type AML (Fig. 4H and table S7 in data file S1). Collectively, these data demonstrate (i) the importance of concomitant in vitro and in situ kinase target profiling; (2) that NCGC1481 is highly effective at targeting FLT3, IRAK1, and IRAK4 in FLT3-ITD mutant AML cells; and (3) that inhibition of these targets correlates with the cytotoxicity of NCGC1481.
NCGC1481 inhibits IRAK1/4 and compensatory innate immune signaling in FLT3-ITD AML cells
The system-level differences associated with simultaneous IRAK1/4 and FLT3 inhibition versus selective inhibition of FLT3 were next examined in FLT3-ITD AML cells. We first confirmed, via immunoblotting for pIRAK4 and pFLT3, that NCGC1481 simultaneously inhibited IRAK4 and FLT3, whereas quizartinib induced activation of IRAK4 upon inhibition of FLT3 in the appropriate cell models (MLL-AF9;FLT3-ITD and MV4;11) (Fig. 5A and fig. S7B). We thereafter collected protein lysates from MLL-AF9;FLT3-ITD and MV4;11 cells treated with NCGC1481 (0.1 nM, IC10) for 6 and 12 hours and subjected these samples to PamChip peptide phosphorylation profiling. Our ability to contrast these outcomes with the data generated in the same cells after quizartinib treatment (Fig. 1) offered insight into the divergent cellular response to these agents (table S8 in data file S1). The set of peptides with increased phosphorylation intensity after 12 hours of quizartinib treatment showed decreased phosphorylation in NCGC1481-treated MLL-AF9;FLT3-ITD and MV4;11 cells (Fig. 5B). Inspection of the IRAK1/4-specific peptides revealed that IRAK1 and IRAK4 activation was also inhibited after 6 and 12 hours of NCGC1481 treatment as compared to quizartinib (Fig. 5C). We next compared the effects of NCGC1481 and quizartinib on global gene expression in FLT3-ITD AML cells at 6 and 12 hours (Fig. 5D and tables S3 and S9 in data file S1). Gene expression profiling by RNA-seq in MLL-AF9;FLT3-ITD cells treated with NCGC1481 (0.1 nM), quizartinib (0.3 nM), or DMSO showed that genes up-regulated by quizartinib, but not NCGC1481, at 12 hours were enriched for interleukin (P = 0.0021) and inflammation signaling (P = 0.030) by ToppGene analysis (Fig. 5E). Because quizartinib induced genes and kinases associated with immune signaling pathway activation, we determined whether MAPKs, which are implicated as compensatory pathways in FLT3i-treated AML cells, are regulated by IRAK1/4. On the basis of the in-cell kinase analysis, quizartinib-treated MLL-AF9;FLT3-ITD cells exhibited compensatory activation of Ras/MAPK and PI3K/AKT pathways (Fig. 5F). In contrast, NCGC1481-treated MLL-AF9;FLT3-ITD cells did not reactivate these kinases (Fig. 5F). To confirm these downstream signaling consequences, we used immunoblotting to show that quizartinib treatment resulted in increased phosphorylation of c-Jun N-terminal kinase (JNK) and p38 at 12 hours in MLL-AF9;FLT3-ITD cells. This is approximately the same time point at which phosphorylation of IRAK4 is detected (Figs. 2B and 5A). In contrast, NCGC1481-treated FLT3-ITD AML cells did not exhibit phosphorylated JNK and p38 (Fig. 5G), suggesting that IRAK1/4 activation mediates immune signaling in part via MAPKs after FLT3i treatment of FLT3-ITD AML cells. Collectively, these findings confirm that NCGC1481 inhibits compensatory IRAK1/4 activation and downstream immune pathways in FLT3-ITD AML.
Fig. 5. NCGC1481 inhibits compensatory IRAK1/4 signaling in FLT3-ITD AML cells.
(A) Immunoblotting of pFLT3 and pIRAK4 in MLL-AF9;FLT3-ITD and MV4;11 cells treated with IC10 of quizartinib or NCGC1481 for 6 and 12 hours. (B) STK PamChip analysis was performed on protein lysates isolated from MLL-AF9;FLT3-ITD and MV4;11 cells treated with IC10 of quizartinib or NCGC1481 for 12 hours. (C) IRAK1 and IRAK4 activity in MLL-AF9;FLT3-ITD and MV4;11 cells treated with IC10 of quizartinib or NCGC1481 for 12 hours (summary of four IRAK1- or three IRAK4-independent peptides from two biological replicates). (D) Differential gene expression of MLL-AF9;FLT3-ITD treated with IC10 of quizartinib or NCGC1481 for 12 hours. Individual data points represent means from biological triplicate samples. (E) Pathway enrichment of differential gene expression (RNA-seq) in MLL-AF9;FLT3-ITD cells treated with quizartinib for 12 hours was determined using ToppGene. EGF, epidermal growth factor; PDGF, platelet-derived growth factor. (F) Network map of active kinases in MLL-AF9;FLT3-ITD cells treated with quizartinib for 12 hours. Tau (τ) scores indicate activity inferred from the phosphorylated peptides (STK PamChip). The left half of the circle represents data from quizartinib-treated cells. The right half of the circle represents data from NCGC1481-treated cells. (G) Immunoblotting of MLL-AF9;FLT3-ITD cells treated with 1 nM quizartinib or NCGC1481 for the indicated times.
NCGC1481 prevents adaptive resistance of FLT3-ITD AML in vitro
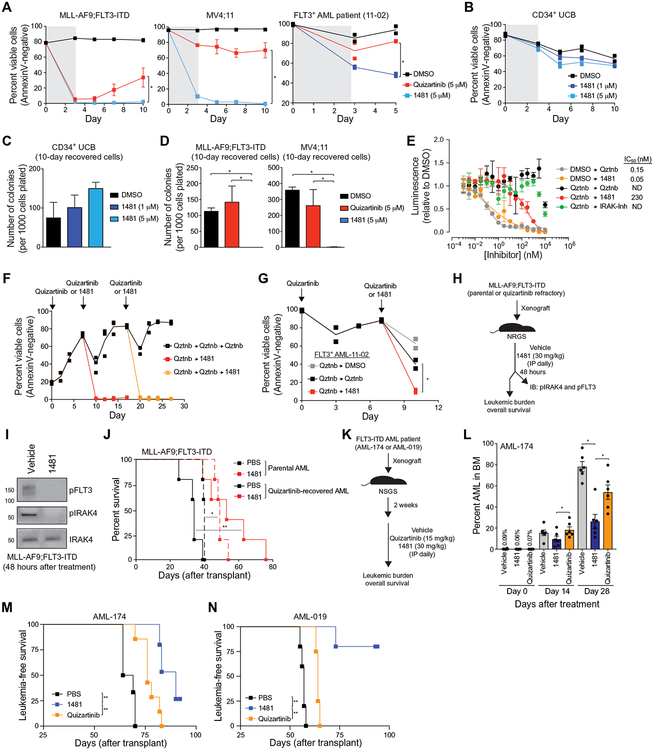
We next investigated whether NCGC1481 can suppress the adaptive resistance of FLT3-ITD AML in vitro. NCGC1481 treatment of MLL-AF9;FLT3-ITD, MV4;11, or FLT3-ITD AML patient-derived cells abolished the outgrowth of adaptively resistant FLT3-ITD AML cells as compared to quizartinib (Fig. 6A). NCGC1481 treatment did not inhibit the viability and progenitor function of normal CD34+ BM cells after 72 hours of treatment and 7 days of recovery (Fig. 6, B and C). MLL-AF9;FLT3-ITD and MV4;11 cells recovered 10 days after inhibitor exposure were plated in methylcellulose to assess leukemic cell potential. The recovered MLL-AF9;FLT3-ITD and MV4;11 cells treated with NCGC1481 did not form any leukemic colonies, whereas quizartinib-treated recovered cells maintained their leukemic potential (Fig. 6D). In a direct comparison to gilteritinib, NCGC1481 was more effective at suppressing adaptively resistant MV4;11 cells (fig. S8, A and B). We also noted that the leukemic cell potential of parental FLT3-ITD AML cells was diminished by NCGC1481 relative to quizartinib or DMSO (fig. S8C), which coincided with drug-stimulated induction of apoptosis (fig. S8D). NCGC1481 did not affect colony formation of healthy cord blood CD34+ cells or adult CD34+ BM cells at equivalent or even higher concentrations (fig. S8E).
Fig. 6. NCGC1481 prevents adaptive resistance in FLT3-ITD AML cells in vitro and in vivo.
(A) MLL-AF9;FLT3-ITD, MV4;11, or FLT3-ITD AML patient-derived cells were cultured with quizartinib or NCGC1481 for 3 days and replated in fresh medium, and cell viability was measured by AnnexinV staining. Values are expressed as means ± SEM from three to four biological replicates for cell lines and two replicates for the patient sample. *P < 0.05 (unpaired, two-tailed t test). (B) Healthy human CD34+ umbilical cord blood (UCB) cells were treated with 1481 (1 or 5 μM) for 3 days and replated in fresh medium. Cell viability was measured by AnnexinV staining. Individual data points are shown along with the mean from two biological replicates. (C) After 10 days in liquid culture [from (B)], the remaining viable CD34+ cells were plated in methylcellulose, and colony formation was determined after 14 days. Values are expressed as means ± SEM from four biological replicates. (D) After 10 days in liquid culture [from (A)], the remaining viable cells were plated in methylcellulose, and colony formation was determined after 7 days. Values are expressed as means ± SEM from four biological replicates. *P < 0.05 (unpaired, two-tailed t test). (E) MV4;11 cells were cultured with quizartinib (5 μM) for 3 days and replated in fresh medium, and cell proliferation was determined after treatment with the indicated concentration of NCGC1481 or quizartinib (Qztnb) for 72 hours. Values are expressed as means ± SEM from three biological replicates. (F) MLL-AF9;FLT3-ITD cells were cultured with quizartinib (5 μM) for 3 days, plated in fresh medium (days 0 and 7), and then replated in medium containing quizartinib (5 μM) or NCGC1481 (5 μM) at days 7 and 17. Cell viability was measured by AnnexinV staining. Individual data points are shown along with the mean from two to three biological replicates. (G) FLT3-ITD AML patient-derived cells were cultured with quizartinib (5 μM) for 3 days, plated in fresh medium, and then replated in medium containing DMSO, quizartinib (5 μM), or NCGC1481 (5 μM) at day 7. Cell viability was measured by AnnexinV staining. Individual data points are shown along with the mean from two biological replicates. (H) Overview of experimental design of xenograft studies. Parental or quizartinib refractory [from (E)] MLL-AF9;FLT3-ITD cells were intravenously injected into NRGS mice. On day 10 after transplant, the mice were intraperitoneally (IP) treated with NCGC1481 (30 mg/kg) or vehicle control daily (n = 5 mice per condition). (I) After 48 hours of treatment with NCGC1481, MLL-AF9;FLT3-ITD (GFP+) cells were isolated from the BM for immunoblot (IB) analysis. (J) Disease-free survival of NRGS mice xenografted with parental or quizartinib refractory MLL-AF9;FLT3-ITD cells and treated with NCGC1481 or vehicle (n = 5 mice per condition). *P < 0.05 and **P < 0.01 (Mantel-Cox test). (K) Overview of experimental design of xenograft studies using FLT3-ITD AML cells obtained from a patient (FLT3+ AML-174). FLT3-ITD AML cells were intravenously injected into NSGS mice. Two weeks after transplant, mice were treated with vehicle control, quizartinib (15 mg/kg), or NCGC1481 (30 mg/kg) intraperitoneally daily. (L) BM aspirates were analyzed for leukemic burden on days 0, 14, and 28 after treatment (n = 6 mice per condition). Values are expressed as means ± SEM from six individual mice. *P < 0.05 (unpaired, two-tailed t test). (M) Leukemia-free survival of NRGS mice xenografted with AML-174 patient cells and treated with quizartinib, NCGC1481, or vehicle (n = 5 to 7 mice per group). **P < 0.005 (Mantel-Cox test). (N) Leukemia-free survival of NRGS mice xenografted with AML-019 patient cells and treated with quizartinib, NCGC1481, or vehicle (n = 4 to 5 mice per group). **P < 0.005 (Mantel-Cox test).
Given the efficacy of NCGC1481 in suppressing adaptive resistance in naïve FLT3-ITD AML cells, we asked whether NCGC1481 remained effective at inhibiting adaptively resistant FLT3-ITD AML cells after treatment with quizartinib. Prior exposure of MV4;11 cells to quizartinib for 3 days followed by 7 days of recovery in fresh medium resulted in diminished sensitivity to re-exposure with quizartinib even at increased concentrations (IC50 = not achieved) (Fig. 6E). In contrast, NCGC1481 remained effective at eliminating adaptively resistant FLT3-ITD AML cells after primary exposure to quizartinib (IC50 = 230 nM) (Fig. 6E). This effect was mediated by inhibition of FLT3 and IRAK1/4, and the IRAK-Inh alone was insufficient for suppressing the viability of adaptively resistant FLT3-ITD AML (Fig. 6E). As an orthogonal approach, after 3 days of exposure to quizartinib (5 μM) followed by 7 days of recovery, MLL-AF9;FLT3-ITD cells were either exposed to quizartinib (5 μM) or NCGC1481 (5 μM). After 10 days, the recovered MLL-AF9;FLT3-ITD cells were treated again with quizartinib or NCGC1481. Although repeated exposure of MLL-AF9;FLT3-ITD cells to quizartinib resulted in diminished sensitivity to quizartinib at concentrations sufficient to induce cell death of parental cells, NCGC1481 remained effective at eliminating adaptively resistant FLT3-ITD AML cells in culture after the primary or secondary exposure to quizartinib (Fig. 6F). To assess whether a similar effect of NCGC1481 was observed in FLT3-ITD AML patient-derived cells, we treated primary FLT3-ITD AML cells with quizartinib (5 μM), which resulted in an acute drop in viable cells that began to recover after day 3. After 7 days, the recovered FLT3-ITD AML cells were treated again with quizartinib (5 μM) or NCGC1481 (5 μM). In this setting, primary FLT3-ITD AML cells were moderately sensitive (relative to vehicle-treated cells) to quizartinib retreatment, whereas treatment with NCGC1481 significantly reduced the number of viable FLT3-ITD AML cells (P = 0.027; Fig. 6G).
NCGC1481 effectively targets resistant FLT3-ITD AML xenografts
Last, we assessed the in vivo antileukemic activity of NCGC1481. NCGC1481 exhibits suitable pharmacokinetic properties in mice for once-daily intraperitoneal dosing and does not result in hematologic toxicity (fig. S5, C to E). The antileukemic activity of NCGC1481 was initially assessed on parental and quizartinib-refractory MLL-AF9;FLT3-ITD cells intravenously injected into NRGS mice, which develop aggressive disseminated AML (Fig. 6H) (76, 77). After 48 hours, phosphorylated IRAK4 and FLT3 were lower in MLL-AF9;FLT3-ITD cells isolated from mice treated with NCGC1481 (30 mg/kg per day) as compared to mice receiving vehicle (Fig. 6I). At the same dose, NCGC1481 administration significantly extended the overall median survival of mice xenografted with parental MLL-AF9;FLT3-ITD cells from 40 to 49 days (P = 0.0026; Fig. 6J). NCGC1481 also significantly extended the overall median survival of mice xenografted with quizartinib-recovered MLL-AF9;FLT3-ITD cells from 34 to 53 days (P = 0.0068; Fig. 6J). Mice were euthanized when they exhibited physical symptoms of leukemia such as reduced motility, rough coat, and hunched posture. At the time of euthanasia, leukemic burden [percent green fluorescent protein (GFP)] in the BM (88.4 ± 5.7% versus 70.1 ± 12.8%) and spleen (79.3 ± 8.2% versus 51.4 ± 19%) was reduced after treatment with NCGC1481 as compared to vehicle-treated mice (fig. S9A). In a second approach, we compared the antileukemic effects of NCGC1481 and quizartinib using FLT3-mutant AML cells from patients with refractory leukemia (AML-174 and AML-019). NSGS mice were intravenously xenografted with AML-174 or AML-019 cells and then dosed with NCGC1481 (30 mg/kg per day), quizartinib (15 mg/kg per day), or vehicle (Fig. 6K). These doses were chosen to correct for the disparity in exposure concentration and half-life (T1/2) (NCGC1481: AUClast = 6750 hours × ng/ml and T1/2 = 4.2 hours; quizartinib: AUClast = 155 hours × μg/ml and T1/2 = 5.7 hours; fig. S5C). After confirming engraftment of AML-174 cells (day 0) and then after 14 days of treatment, NCGC1481 afforded a 38 and 48% reduction in leukemic burden in the BM as compared to mice receiving vehicle or quizartinib, respectively (Fig. 6L). After 28 days of treatment, mice that were treated with NCGC1481 exhibited a 66 and 50% reduction in leukemic burden in the BM as compared to mice receiving vehicle or quizartinib, respectively (Fig. 6L). Moreover, the frequency of CD34+ AML cells in the BM (P = 0.0011) and spleen (P = 0.00016) was significantly reduced after NCGC1481 administration as compared to control mice (fig. S9B), indicating that NCGC1481 has an effect on disease-propagating AML cells. In this model, NCGC1481 significantly extended the overall median survival to 90 days compared to 66.5 days for mice receiving vehicle (P = 0.0016) or 76 days for mice receiving quizartinib (P = 0.0097) (Fig. 6M). For mice xenografted with AML-019 cells, NCGC1481 administration also significantly extended the overall median survival of mice (P = 0.0021; Fig. 6N). As compared to vehicle (median survival = 57 days) or quizartinib (median survival = 64 days), most of the mice treated with NCGC1481 did not succumb to leukemia beyond 94 days of treatment, at which time the experiment was terminated (Fig. 6N). Last, we wanted to determine whether NCGC1481 can reduce the leukemic burden of mice treated with quizartinib. NSGS mice were intravenously xenografted with AML-174 cells and then dosed with quizartinib (15 mg/kg per day) for 43 days and then either continued on quizartinib or switched to NCGC1481 for an additional 14 days (fig. S10A). After 43 days of quizartinib treatment, the leukemic burden in the mice was approximately 18% (fig. S10B). For the mice that continued on quizartinib treatment, the leukemic burden in the BM increased from 18 to 54% (fig. S10B), whereas for the mice that were switched to NCGC1481, the leukemic burden increased only from 17 to 43% (fig. S10B). The mice that were switched to NCGC1481 did not exhibit a significant change in the CD34+ leukemic stem cell population as compared to mice that continued on quizartinib treatment (P = 0.12; fig. S10C).
DISCUSSION
Targeted inhibitors to oncogenic kinases initially demonstrate encouraging clinical responses; however, most patients relapse because of target-dependent and target-independent mechanisms. Monotherapy and combination therapies have shown good initial response rates to FLT3i in clinical studies for FLT3-mutant leukemia; however, patients eventually relapse with FLT3i-resistant clones (11–20). The absence of durable remission in patients with FLT3-mutant leukemia treated with potent and selective FLT3i establishes the need to identify resistance mechanisms and develop additional treatment strategies. Here, we identified mechanisms of adaptive resistance to targeted inhibitors in AML associated with activating FLT3 mutations. Our results suggest that FLT3i adaptive resistance occurs through compensatory activation of innate immune stress pathways in FLT3-mutant AML and that inhibition of IRAK1/4 with FLT3i targets the emergence of adaptively resistant mutant clones. Specifically, activation of IRAK1/4 in FLT3i-treated AML restored Ras/MAPK signaling along with NF-κB, which represents a major mechanism of resistance after tyrosine kinase inhibition (1, 24–26, 78).
Cellular stress responses are survival mechanisms activated by cells. Although stress pathways have been extensively characterized, recent studies have shown that proteins in cellular stress responses interact with and regulate signaling intermediates involved in the activation of immune-related pathways (79). Although we report that FLT3i treatment resulted in TLR9 overexpression and IRAK1/4 activation, the precise mechanism of innate immune signaling and specifically IRAK1/4 activation after targeted therapy is not resolved and may involve various cellular stress response pathways. Cellular stresses associated with FLT3i treatment, such as oxidative stress, heat shock, unfolded protein, and DNA damage responses have been independently shown to activate innate immune signaling, albeit by distinct mechanisms (67, 80–87). In addition to overexpression of certain TLRs, gene expression analysis of FLT3i-treated cells also revealed overexpression of TLR ligands and inflammatory cytokines. In such conditions, fractional cell death and/or cellular stress after FLT3i treatment can result in the release of inflammatory mediators that subsequently induce innate immune signaling and IRAK1/4 activation, such as via TLR9. Therefore, one potential mechanism of compensatory activation of the innate immune stress pathway in FLT3i-resistant AML subclones is through paracrine and autocrine activation of IRAK1/4. We also observed a modest, yet consistent, increase in IRAK4 expression after prolonged FLT3i treatment of FLT3-mutant AML cells in vitro and in vivo. Consistent with the idea that increased IRAK4 expression correlates with adaptive resistance, retroviral overexpression of IRAK4 decreased the sensitivity of FLT3-ITD AML cells to FLT3i. Thus, another potential mechanism of compensatory activation of the innate immune stress pathway in FLT3i-resistant AML subclones may occur as a result of IRAK1/4 overexpression.
There are ongoing efforts to suppress activation of parallel signaling pathways, such as MAPK kinase and extracellular signal–regulated kinase, after prolonged exposure to targeted therapies (31–33, 36, 37, 42, 88). Ras/MAPK signaling is responsible for adaptively resistant FLT3-mutant AML; however, targeting only a single arm of the signaling cascade has yielded limited clinical benefit (). Because IRAK1/4 complex is upstream of Ras/MAPK and NF-κB (58), we posit that targeting IRAK1/4 will yield more durable inhibition of bypass signaling cascades, prevent adaptive resistance, and result in improved therapeutic efficacy in FLT3-mutant AML. Although IRAK1 or IRAK4 inhibition has been explored in myelodysplastic syndrome, AML, T cell acute lymphoblastic leukemia, and lymphoma, albeit with limited efficacy, we present evidence that innate immune pathway activation via IRAK1 and IRAK4 is essential for adaptive resistance to therapy (64, 66, 69, 73, 89, 90). Our data suggest that concomitant targeting of IRAK1 or IRAK4, alongside FLT3, is the most effective means to overcome the adaptive resistance incurred when targeting FLT3. Gilteritinib, which exhibits modest inhibition of IRAK1/4 at higher concentrations, is less effective at suppressing adaptive resistance to therapy in FLT3-mutant AML as compared to NCGC1481, which potently targets IRAK1 and IRAK4. To overcome the current limitations of FLT3i, we report a polypharmacologic strategy and a multitarget small molecule with potent activity against the IRAK1/4 complex and FLT3, which suppresses adaptive resistance to therapy in FLT3-mutant AML by targeting inflammatory stress response pathways.
Given the restrictions of using patient-derived samples for functional studies, it was necessary to confirm the adaptive resistance mechanism via innate immune pathway activation and validate NCGC1481 in independent FLT3-mutant AML cell lines. However, cell lines may not accurately represent the complexity of adaptive resistance mechanisms to FLT3i in patients, arguing for cautious interpretation of these data. Furthermore, the current experimental design is limited to adaptive resistance mechanisms occurring immediately after FLT3i treatment, and it should be noted that adaptive resistance mechanisms after prolonged FLT3i treatment may differ.
Here, we identified activation of innate immune stress response pathways after treatment of FLT3-mutant AML cells with FLT3i. Although further studies are needed, our study demonstrates that therapies that simultaneously inhibit FLT3 signaling and compensatory IRAK1/4 activation have the potential to improve the therapeutic efficacy in patients with FLT3-mutant AML. We demonstrate that inflammatory stress response pathways contribute to adaptive resistance in FLT3-mutant AML and propose that this mechanism may extend to other malignant cells undergoing a stress-induced response to therapy.
MATERIALS AND METHODS
Study design
The first objective of this study was to find target-independent mechanisms of resistance, such as alternate activation of survival and proliferation pathways (adaptive resistance), to FLT3 inhibition in FLT3-mutant AML by performing an integrative in-cell kinase (PamChip kinase array) and gene regulatory network (RNA-seq) analysis. The second objective was to identify an inhibitor with the potential of suppressing FLT3-ITD and the pathway contributing to adaptive resistance in FLT3-mutant AML. To overcome adaptive resistance to FLT3 inhibition, we synthesized a series of small molecules to inhibit the compensatory pathway activation contributing to adaptive resistance (via IRAK1/4) and FLT3 in FLT3-mutant AML. The chemical starting points for optimization of IRAK1/4 and FLT3 small-molecule inhibitors were based on the 3-(pyridin-2-yl)imidazo[1,2-a] pyridines that were previously reported as selective IRAK4 inhibitors. The potency and selectivity of the inhibitors were determined by biochemical binding and inhibitory assays and in situ kinase profiling. The optimized small-molecule inhibitor (NCGC1481) was confirmed by cocrystallography to bind IRAK4 in an inactive confirmation. In-cell kinase (PamChip kinase array) assays, immunoblotting, and gene expression profiling confirmed that NCGC1481 simultaneously suppresses FLT3 and IRAK1/4 in FLT3-mutant AML. The therapeutic benefit of targeting IRAK1/4 and FLT3 in FLT3-mutant AML with NCGC1481 as compared to a selective FLT3i was confirmed in human cell lines and patient-derived samples in vitro and in vivo. Existing deidentified cryopreserved samples were used for the study without age or gender preferences. Investigators and data analyzers were blinded for the evaluation of NCGC1481 in primary patient-derived AML samples in vitro. Mouse experiments were planned in an effort to provide 60 to 80% power for a target effect size of 1.2 to 1.5 (effect size = |mean difference|/SD). All mice were randomly allocated into experimental groups. For all other experiments, at least two independent biological replicates were performed and used in the sample calculation. No data were excluded from the studies.
Cell lines, patient samples, and culture conditions
MLL-AF9 FLT3-ITD and MLL-AF9 NRASG12D cell lines, provided by J.C.M., were cultured in Iscove’s Dulbecco’s modified Eagle’s medium (Corning Cell Grow, catalog no. 10–016-CV) with 20% fetal bovine serum (FBS) (Atlanta Biologicals, catalog no. S11550) and 1% penicillin-streptomycin (P/S) (HyClone, catalog no. SV30010) (45). MV4;11 cell line was provided by L. Grimes [Cincinnati Children’s Hospital Medical Center (CCHMC), Cincinnati, OH] and purchased from American Type Culture Collection (ATCC) (catalog no. CRL-9591). The cells were cultured in RPMI 1640 medium with 10% FBS and 1% P/S. MOLM13 cell line, purchased from AddexBio (catalog no. C0003003), was cultured in RPMI 1640 medium (HyClone, catalog no. SH30027.01) with 20% FBS and 1% P/S. THP1-Blue NF-κB reporter cells were obtained from InvivoGen (catalog no. thp-nfkb) and grown according to the manufacturer’s instructions. BaF3 cells were provided by M. Azam (CCHMC, Cincinnati, OH) and purchased from ATCC (catalog no. HB-283). They were cultured in RMPI 1640 medium with 10% FBS, 1% P/S, and recombinant murine IL-3 (PeproTech, catalog no. 213-12-50UG) (all at 10 ng/ml). Human CD34+ umbilical cord blood, human CD34+ BM, and human normal whole BM were obtained from the Translational Research Development Support Laboratory of Cincinnati Children’s Hospital under an approved Institutional Review Board protocol. These cells were maintained in StemSpan serum-free expansion medium (STEMCELL Technologies, catalog no. 09650) supplemented with recombinant human SCF (10 ng/ml; PeproTech, catalog no. 300-07-50UG), recombinant human TPO (PeproTech, catalog no. 300-18-50UG), recombinant human FLT3 ligand (FL) (PeproTech, catalog no. 300-19-50UG), recombinant human IL-3 (PeproTech, catalog no. 200-03-50UG), and recombinant human IL-6 (PeproTech, catalog no. 200-06-50UG). AML primary patient samples were obtained with written informed consent and approved by the Institutional Review Board of CCHMC. These samples had been obtained within the framework of routine diagnostic BM aspirations after written informed consent in accordance with the Declaration of Helsinki. AML-019 was purchased from the Public Repository of Xenografts (catalog no. DFAM-16835-V1). Analysis of short tandem repeat loci was performed on all cell lines when received and after experimentation was complete. All cell lines were routinely tested and were confirmed to be negative for mycoplasma.
Lenti- and retroviral infections
The pLKO.1 (Open BioSystems) constructs were obtained from the Viral Vector Core at CCHMC and used to express shCTL and shIRAK4 (TRCN0000002065). Puromycin resistance gene was replaced by GFP. The pGreenFire1-NF-κB (EF1α-puro) lentivector was purchased from System Biosciences (catalog no. TR012VA-P). Flag-IRAK4 in pMSCV-pGK-GFP was designed as previously described (90). Cells were transduced as previously described (91).
RNA sequencing
RNA was isolated using Quick-RNA MiniPrep (Zymo Research, catalog no. R1055) from MLL-AF9;FLT-ITD cells treated with DMSO, quizartinib (0.3 nM), or NCGC1481 (0.1 nM) for 6 and 12 hours in biological triplicates. RNA libraries were prepared according to the Illumina TruSeq Stranded mRNA (polyA capture) library protocol by the DNA Sequencing and Genotyping Core at CCHMC. The data discussed in this publication have been deposited in National Center for Biotechnology Information’s (NCBI’s) Gene Expression Omnibus (GEO) and are accessible through GEO series accession no. GSE121272.
Quantitative high-throughput drug screening
MLL-AF9;FLT3-ITD cells were grown in Iscove’s modified Dulbecco’s medium (IMDM) (Thermo Fisher Scientific, catalog no. 12440-061) supplemented with 20% FBS (STEMCELLTechnologies, catalog no. 06100), 1% P/S (Thermo Fisher Scientific, catalog no. 15140122) with recombinant human SCF (PeproTech, catalog no. 300-07-50UG), recombinant human TPO (PeproTech, catalog no. 300-18-50UG), recombinant human FLT3 ligand (FL) (PeproTech, catalog no. 300-19-50UG), recombinant human IL-3 (PeproTech, catalog no. 200-03-50UG), and recombinant human IL-6 (PeproTech, catalog no. 200-06-50UG) (all at 10 ng/ml). MLL-AF9;FLT3-ITD cells were maintained in IMDM supplemented with 20% FBS and no growth factors. Cells were plated at a density of 500 cells per well in 5 μl of complete growth medium in 1536-well white tissue culture assay plates (Greiner). Compounds (23 nl) were then added to each assay plate using a PinTool Dispenser (Kalypsys). Plates were then covered with a stainless steel gasketed lid and placed into an incubator in 37°C and 5% CO2 for 48 hours. After this incubation, 3 μl of CellTiter-Glo reagent was added to each well and then incubated for 15 min at room temperature. Luminescence readings were taken using a ViewLux (PerkinElmer) with clear filter and a 2-s exposure time. Curve fitting was done using a four-parameter Hill slope equation (GraphPad Prism 7).
STK array and analysis
MLL-AF9 FLT3-ITD or MV4–11 cells were treated for 6 or 12 hours with quizartinib (0.3 nM), NCGC-1481 (0.1 nM), or DMSO. Whole-cell lysates were prepared according to PamGene instructions (protocol 1160). PamChip STK array was performed by PamGene. From 144 nonredundant peptides, individual peptide phosphorylation intensities were normalized to DMSO control and log-transformed. Peptides determined to have significant increased or decreased phosphorylation (P < 0.05) were used to infer active STKs. The database of potential upstream STKs was downloaded from PhosphoNET (www.phosphonet.ca). A given STK-substrate pair was considered highly probable if its Kinexus predictor score (v2) was greater than 300. The PamChip peptide data were integrated with the PhosphoNET kinase-substrate network to calculate kinase specificity scores (using 2000 permutations across target peptides) and the kinase scores (using 2000 permutations across sample labels). The kinases were then prioritized on the basis of the sum of the two scores.
Pathway and network analyses
Of the inferred kinases showing increased activity in the PamGene assay in AC200 relative to DMSO, 46 were common to both the MLL-AF9;FLT3-ITD and MV4–11 cells lines at both 6 and 12 hours. These kinases were analyzed using ToppFun in the ToppGene Suite (https://toppgene.cchmc.org/) to determine enriched signaling pathways. ClueGo (v2.5.1) was used to create the network map (ontology = GO biological processes, network specificity = medium; P < 0.05).
Xenografts
NRGS (NOD.Rag1−/−;γcnull; hIL-3, hGM-CSF, hSF) mice were provided by J.C.M. (92). NSGS mice were purchased from the CCHMC Comprehensive Mouse and Cancer Core. For xenotransplantation, MLL-AF9;FLT3-ITD CD34+ cells (2 × 105 per mouse) or AML patient cells (2 × 106 per mouse) were intravenously injected into the tail veins of NRGS or NSG animals, as indicated in the figure legends. Mice were monitored by BM aspirate and observed for physical attributes of disease, such as limb paralysis, fatigue, and rough fur. NCGC-1481 and quizartinib were prepared in DMSO and further dissolved in sterile phosphate-buffered saline. Animals were intraperitoneally injected with NCGC-1481 (30 mg/kg) or quizartinib (15 mg/kg) 5× weekly. All mice were bred, housed, and handled in the Association for Assessment and Accreditation of Laboratory Animal Care–accredited animal facility of CCHMC. The study is compliant with all relevant ethical regulations regarding animal research.
Statistical analysis
The numbers of animals, cells, and experimental replicates can be found in the figure legends. Differences among multiple groups were assessed by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison post test for all possible comparisons. Comparison of two groups was performed using a Student’s t test (unpaired, two-tailed) or Mann-Whitney test when sample size allowed. Significance was set at P < 0.05. Unless otherwise specified, results are depicted as the means ± SEM. D’Agostino and Pearson and Shapiro-Wilk tests were performed to assess data distributions. For Kaplan-Meier analysis, Mantel-Cox test was used. Data were analyzed and plotted using GraphPad Prism 7 software. Original data are provided in data file S2.
Supplementary Material
Acknowledgments:
We thank J. Bailey and V. Summey for assistance with transplantations (Comprehensive Mouse and Cancer Core at CCHMC). We thank the Viral Vector Core and DNA sequencing and Genotyping Core at CCHMC for their assistance. We thank G. Rhyasen for thoughtful discussions and insight.
Funding: This work was supported by Cincinnati Children’s Hospital Research Foundation, Cancer Free Kids, Leukemia Lymphoma Society, and National Institutes of Health (R35HL135787, RO1DK102759, and RO1DK113639) grants to D.T.S. and by the Intramural Research Programs of the National Center for Advancing Translational Sciences and the National Cancer Institute to C.J.T. D.T.S. is a Leukemia Lymphoma Society Scholar. K.M. is supported by a National Institutes of Health Research Training and Career Development Grant (F31CA217140).
Footnotes
SUPPLEMENTARY MATERIALS
stm.sciencemag.org/cgi/content/full/11/508/eaaw8828/DC1
Fig. S1. FLT3+ AML develop adaptive resistance to FLT3i.
Fig. S2. Adaptively resistant FLT3+ AML exhibit increased IRAK1/4 activation.
Fig. S3. Quizartinib induces TLR9-mediated activation of IRAK4.
Fig. S4. Inhibition of IRAK1/4 sensitizes FLT3+ AML to quizartinib.
Fig. S5. NCGC1481 exhibits promising physicochemical and in vivo pharmacokinetic properties.
Fig. S6. 2D interaction diagrams for NCGC1481 bound to FLT3 and IRAK4.
Fig. S7. NCGC1481 inhibits compensatory IRAK1/4 activation and adaptive resistance of FLT3-ITD AML.
Fig. S8. NCGC1481 prevents adaptive resistance of FLT3-ITD AML cells in vitro and has minimal effects on normal hematopoietic cells.
Fig. S9. NCGC1481 reduces the leukemic burden of FLT3-ITD AML.
Fig. S10. NCGC1481 reduces the leukemic burden of FLT3-ITD AML after quizartinib treatment. Data file S1 contains the following tables:
Table S1. Peptide phosphorylation in the PamChip serine-threonine in-cell kinase array.
Table S2. Top active kinases inferred from the PamChip in-cell kinase assay.
Table S3. Gene expression analysis of FLT3-ITD AML treated with FLT3i.
Table S4. AML patients evaluated with gilteritinib.
Table S5. Reaction Biology profile of NCGC1481.
Table S6. KiNativ profile of NCGC1481 in MV4;11 lysate.
Table S7. Characteristics of patients with AML.
Table S8. Peptide phosphorylation in the PamChip serine-threonine in-cell kinase array with NCGC1481 treatment.
Table S9. Gene expression analysis of FLT3-ITD AML treated with NCGC1481.
Data file S2. Original data.
Competing interests: C.J.T. M.W., J.-K.J., K.M., and D.T.S. are inventors on the following patent PCT/US2017/047088: Compounds, compositions, methods for treating disease, and methods for preparing compounds. K.M., M.M.W., J.-K.J., C.J.T., and D.T.S. are inventors on a patent application (WO/2018/038988) jointly held by the CCHMC and the U.S. National Institutes of Health covering chemical composition, methods of preparation and methods of treatment for small molecules reported in this manuscript including NCGC1481. D.T.S. has received support from Celgene and honoraria from Curis Inc. R.L.L. is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics, and IsoPlexis, which each include an equity interest. R.L.L. also received research support from and consulted for Celgene and Roche, received research support from Prelude Therapeutics, consulted for Incyte, Novartis, MorphoSys, and Janssen, and received honoraria from Lilly and Amgen for invited lectures and from Gilead for grant reviews. All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. The RNA-seq data discussed in this publication have been deposited in NCBI’s GEO and are accessible through GEO series accession number GSE121272. Compounds can be made available in limited supply through material transfer agreement with CCHMC. All other reagents described are available upon request or can be purchased from the vendors.
REFERENCES AND NOTES
- 1.Chu SH, Small D, Mechanisms of resistance to FLT3 inhibitors. Drug Resist. Updat 12, 8–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larrosa-Garcia M, Baer MR, FLT3 inhibitors in acute myeloid leukemia: Current status and future directions. Mol. Cancer Ther 16, 991–1001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, Lippke J, Saxena K, The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol. Cell 13, 169–178 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Gilliland DG, Griffin JD, The roles of FLT3 in hematopoiesis and leukemia. Blood 100, 1532–1542 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Grafone T, Palmisano M, Nicci C, Storti S, An overview on the role of FLT3-tyrosine kinase receptor in acute myeloid leukemia: Biology and treatment. Oncol. Rev 6, e8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drexler HG, Quentmeier H, FLT3: Receptor and ligand. Growth Factors 22, 71–73 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Kottaridis PD, Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, Walker H, Wheatley K, Bowen DT, Burnett AK, Goldstone AH, Linch DC, The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: Analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood 98, 1752–1759 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Thiede C, Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, Wermke M, Bornhäuser M, Ritter M, Neubauer A, Ehninger G, Illmer T, Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 99, 4326–4335 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Whitman SP, Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, Carroll AJ, Mrózek K, Vardiman JW, George SL, Kolitz JE, Larson RA, Bloomfield CD, Caligiuri MA, Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A cancer and leukemia group B study. Cancer Res. 61, 7233–7239 (2001). [PubMed] [Google Scholar]
- 10.Boissel N, Cayuela JM, Preudhomme C, Thomas X, Grardel N, Fund X, Tigaud I, Raffoux E, Rousselot P, Sigaux F, Degos L, Castaigne S, Fenaux P, Dombret H, Prognostic significance of FLT3 internal tandem repeat in patients with de novo acute myeloid leukemia treated with reinforced courses of chemotherapy. Leukemia 16, 1699–1704 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Cortes J, Perl AE, Döhner H, Kantarjian H, Martinelli G, Kovacsovics T, Rousselot P, Steffen B, Dombret H, Estey E, Strickland S, Altman JK, Baldus CD, Burnett A, Krämer A, Russell N, Shah NP, Smith CC, Wang ES, Ifrah N, Gammon G, Trone D, Lazzaretto D, Levis M, Quizartinib, an FLT3 inhibitor, as monotherapy in patients with relapsed or refractory acute myeloid leukaemia: An open-label, multicentre, single-arm, phase 2 trial. Lancet Oncol. 19, 889–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes JE, Kantarjian H, Foran JM, Ghirdaladze D, Zodelava M, Borthakur G, Gammon G, Trone D, Armstrong RC, James J, Levis M, Phase I study of quizartinib administered daily to patients with relapsed or refractory acute myeloid leukemia irrespective of FMS-like tyrosine kinase 3-internal tandem duplication status. J. Clin. Oncol 31, 3681–3687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes JE, Tallman MS, Schiller GJ, Trone D, Gammon G, Goldberg SL, Perl AE, Marie J-P, Martinelli G, Kantarjian HM, Levis MJ, Phase 2b study of 2 dosing regimens of quizartinib monotherapy in FLT3-ITD-mutated, relapsed or refractory AML. Blood 132, 598–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiedler W, Kayser S, Kebenko M, Janning M, Krauter J, Schittenhelm M, Götze K, Weber D, Göhring G, Teleanu V, Thol F, Heuser M, Döhner K, Ganser A, Döhner H, Schlenk RF, A phase I/II study of sunitinib and intensive chemotherapy in patients over 60 years of age with acute myeloid leukaemia and activating FLT3 mutations. Br. J. Haematol 169, 694–700 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Fiedler W, Serve H, Döhner H, Schwittay M, Ottmann OG, O’Farrell A-M, Bello CL, Allred R, Manning WC, Cherrington JM, Louie SG, Hong W, Brega NM, Massimini G, Scigalla P, Berdel WE, Hossfeld DK, A phase 1 study of SU11248 in the treatment of patients with refractory or resistant acute myeloid leukemia (AML) or not amenable to conventional therapy for the disease. Blood 105, 986–993 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Fischer T, Stone RM, DeAngelo DJ, Galinsky I, Estey E, Lanza C, Fox E, Ehninger G, Feldman EJ, Schiller GJ, Klimek VM, Nimer SD, Gilliland DG, Dutreix C, Huntsman-Labed A, Virkus J, Giles FJ, Phase IIB trial of oral Midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J. Clin. Oncol 28, 4339–4345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Man CH, Fung TK, Ho C, Han HHC, Chow HCH, Ma ACH, Choi WWL, Lok S, Cheung AMS, Eaves C, Kwong YL, Leung AYH, Sorafenib treatment of FLT3-ITD+ acute myeloid leukemia: Favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood 119, 5133–5143 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Perl AE, Altman JK, Cortes J, Smith C, Litzow M, Baer MR, Claxton D, Erba HP, Gill S, Goldberg S, Jurcic JG, Larson RA, Liu C, Ritchie E, Schiller G, Spira AI, Strickland SA, Tibes R, Ustun C, Wang ES, Stuart R, Röllig C, Neubauer A, Martinelli G, Bahceci E, Levis M, Selective inhibition of FLT3 by gilteritinib in relapsed or refractory acute myeloid leukaemia: A multicentre, first-in-human, open-label, phase 1–2 study. Lancet Oncol. 18, 1061–1075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stone RM, DeAngelo DJ, Klimek V, Galinsky I, Estey E, Nimer SD, Grandin W, Lebwohl D, Wang Y, Cohen P, Fox EA, Neuberg D, Clark J, Gilliland DG, Griffin JD, Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood 105, 54–60 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Döhner K, Marcucci G, Lo-Coco F, Klisovic RB, Wei A, Sierra J, Sanz MA, Brandwein JM, de Witte T, Niederwieser D, Appelbaum FR, Medeiros BC, Tallman MS, Krauter J, Schlenk RF, Ganser A, Serve H, Ehninger G, Amadori S, Larson RA, Döhner H, Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N. Engl. J. Med 377, 454–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith CC, Wang Q, Chin C-S, Salerno S, Damon LE, Levis MJ, Perl AE, Travers KJ, Wang S, Hunt JP, Zarrinkar PP, Schadt EE, Kasarskis A, Kuriyan J, Shah NP, Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 485, 260–263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarado Y, Kantarjian HM, Luthra R, Ravandi F, Borthakur G, Garcia-Manero G, Konopleva M, Estrov Z, Andreeff M, Cortes JE, Treatment with FLT3 inhibitor in patients with FLT3-mutated acute myeloid leukemia is associated with development of secondary FLT3–tyrosine kinase domain mutations. Cancer 120, 2142–2149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CC, Paguirigan A, Jeschke GR, Lin KC, Massi E, Tarver T, Chin C-S, Asthana S, Olshen A, Travers KJ, Wang S, Levis MJ, Perl AE, Radich JP, Shah NP, Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood 130, 48–58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piloto O, Wright M, Brown P, Kim K-T, Levis M, Small D, Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood 109, 1643–1652 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siendones E, Barbarroja N, Torres LA, Buendía P, Velasco F, Dorado G, Torres A, López-Pedrera C, Inhibition of Flt3-activating mutations does not prevent constitutive activation of ERK/Akt/STAT pathways in some AML cells: A possible cause for the limited effectiveness of monotherapy with small-molecule inhibitors. Hematol. Oncol 25, 30–37 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Bruner JK, Ma HS, Li L, Qin ACR, Rudek MA, Jones RJ, Levis MJ, Pratz KW, Pratilas CA, Small D, Adaptation to TKI treatment reactivates ERK signaling in tyrosine kinase–driven leukemias and other malignancies. Cancer Res. 77, 5554–5563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandarlapaty S, Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov. 2, 311–319 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Oosterwijk JG, Buelow DR, Drenberg CD, Vasilyeva A, Li L, Shi L, Wang Y-D, Finkelstein D, Shurtleff SA, Janke LJ, Pounds S, Rubnitz JE, Inaba H, Pabla N, Baker SD, Hypoxia-induced upregulation of BMX kinase mediates therapeutic resistance in acute myeloid leukemia. J. Clin. Invest 128, 369–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindblad O, Cordero E, Puissant A, Macaulay L, Ramos A, Kabir NN, Sun J, Vallon-Christersson J, Haraldsson K, Hemann MT, Borg Å, Levander F, Stegmaier K, Pietras K, Rönnstrand L, Kazi JU, Aberrant activation of the PI3K/mTOR pathway promotes resistance to sorafenib in AML. Oncogene 35, 5119–5131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green AS, Maciel TT, Hospital M-A, Yin C, Mazed F, Townsend EC, Pilorge S, Lambert M, Paubelle E, Jacquel A, Zylbersztejn F, Decroocq J, Poulain L, Sujobert P, Jacque N, Adam K, So JCC, Kosmider O, Auberger P, Hermine O, Weinstock DM, Lacombe C, Mayeux P, Vanasse GJ, Leung AY, Moura IC, Bouscary D, Tamburini J, Pim kinases modulate resistance to FLT3 tyrosine kinase inhibitors in FLT3-ITD acute myeloid leukemia. Sci. Adv 1, e1500221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishioka C, Ikezoe T, Yang J, Takeshita A, Taniguchi A, Komatsu N, Togitani K, Koeffler HP, Yokoyama A, Blockade of MEK/ERK signaling enhances sunitinib-induced growth inhibition and apoptosis of leukemia cells possessing activating mutations of the FLT3 gene. Leuk. Res 32, 865–872 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Borthakur G, Gao C, Chen Y, Mu H, Ruvolo VR, Nomoto K, Zhao N, Konopleva M, Andreeff M, The dual MEK/FLT3 inhibitor E6201 exerts cytotoxic activity against acute myeloid leukemia cells harboring resistance-conferring FLT3 mutations. Cancer Res. 76, 1528–1537 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griessinger E, Frelin C, Cuburu N, Imbert V, Dageville C, Hummelsberger M, Sirvent N, Dreano M, Peyron J-F, Preclinical targeting of NF-κB and FLT3 pathways in AML cells. Leukemia 22, 1466–1469 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Jordheim LP, Plesa A, Dreano M, Cros-Perrial E, Keime C, Herveau S, Demangel D, Vendrell JA, Dumontet C, Sensitivity and gene expression profile of fresh human acute myeloid leukemia cells exposed ex vivo to AS602868. Cancer Chemother. Pharmacol 68, 97–105 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Wang F, Liu Z, Zeng J, Zhu H, Li J, Cheng X, Jiang T, Zhang L, Zhang C, Chen T, Liu T, Jia Y, Metformin synergistically sensitizes FLT3-ITD-positive acute myeloid leukemia to sorafenib by promoting mTOR-mediated apoptosis and autophagy. Leuk. Res 39, 1421–1427 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Minson KA, Smith CC, DeRyckere D, Libbrecht C, Lee-Sherick AB, Huey MG, Lasater EA, Kirkpatrick GD, Stashko MA, Zhang W, Jordan CT, Kireev D, Wang X, Frye SV, Earp HS, Shah NP, Graham DK, The MERTK/FLT3 inhibitor MRX-2843 overcomes resistance-conferring FLT3 mutations in acute myeloid leukemia. JCI Insight 1, e85630 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keegan K, Li C, Li Z, Ma J, Ragains M, Coberly S, Hollenback D, Eksterowicz J, Liang L, Weidner M, Huard J, Wang X, Alba G, Orf J, Lo M-C, Zhao S, Ngo R, Chen A, Liu L, Carlson T, Quéva C, McGee LR, Medina J, Kamb A, Wickramasinghe D, Dai K, Preclinical evaluation of AMG 925, a FLT3/CDK4 dual kinase inhibitor for treating acute myeloid leukemia. Mol. Cancer Ther 13, 880–889 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Ma S, Yang L-L, Niu T, Cheng C, Zhong L, Zheng M-W, Xiong Y, Li L-L, Xiang R, Chen L-J, Zhou Q, Wei Y-Q, Yang S-Y, SKLB-677, an FLT3 and Wnt/β-catenin signaling inhibitor, displays potent activity in models of FLT3-driven AML. Sci. Rep 5, 15646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson EA, Walker SR, Xiang M, Weisberg E, Bar-Natan M, Barrett R, Liu S, Kharbanda S, Christie AL, Nicolais M, Griffin JD, Stone RM, Kung AL, Frank DA, The STAT5 inhibitor pimozide displays efficacy in models of acute myelogenous leukemia driven by FLT3 mutations. Genes Cancer 3, 503–511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Natarajan K, Xie Y, Burcu M, Linn DE, Qiu Y, Baer MR, Pim-1 kinase phosphorylates and stabilizes 130 kDa FLT3 and promotes aberrant STAT5 signaling in acute myeloid leukemia with FLT3 internal tandem duplication. PLOS ONE 8, e74653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohi MG, Boulton C, Gu T-L, Sternberg DW, Neuberg D, Griffin JD, Gilliland DG, Neel BG, Combination of rapamycin and protein tyrosine kinase (PTK) inhibitors for the treatment of leukemias caused by oncogenic PTKs. Proc. Natl. Acad. Sci. U.S.A 101, 3130–3135 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weisberg E, Liu Q, Zhang X, Nelson E, Sattler M, Liu F, Nicolais M, Zhang J, Mitsiades C, Smith RW, Stone R, Galinsky I, Nonami A, Griffin JD, Gray N, Selective Akt inhibitors synergize with tyrosine kinase inhibitors and effectively override stroma-associated cytoprotection of mutant FLT3-positive AML cells. PLOS ONE 8, e56473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez JS, Banerji U, Combine and conquer: Challenges for targeted therapy combinations in early phase trials. Nat. Rev. Clin. Oncol 14, 57–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueda T, Tsuji K, Yoshino H, Ebihara Y, Yagasaki H, Hisakawa H, Mitsui T, Manabe A, Tanaka R, Kobayashi K, Ito M, Yasukawa K, Nakahata T, Expansion of human NOD/SCID-repopulating cells by stem cell factor, Flk2/Flt3 ligand, thrombopoietin, IL-6, and soluble IL-6 receptor. J. Clin. Invest 105, 1013–1021 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wunderlich M, Mulloy JC, Model systems for examining effects of leukemia-associated oncogenes in primary human CD34+ cells via retroviral transduction. Methods Mol. Biol 538, 263–285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA, Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science 322, 1861–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Corazza F, Hermans C, D’Hondt S, Ferster A, Kentos A, Benoît Y, Sariban E, Circulating thrombopoietin as an in vivo growth factor for blast cells in acute myeloid leukemia. Blood 107, 2525–2530 (2006). [DOI] [PubMed] [Google Scholar]
- 48.De Waele M, Renmans W, Gucht KV, Jochmans K, Schots R, Otten J, Trullemans F, Lacor P, Van Riet I, Growth factor receptor profile of CD34+ cells in AML and B-lineage ALL and in their normal bone marrow counterparts. Eur. J. Haematol 66, 178–187 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Dong-Feng Z, Ting L, Yong Z, Cheng C, Xi Z, Pei-Yan K, The TPO/c-MPL pathway in the bone marrow may protect leukemia cells from chemotherapy in AML Patients. Pathol. Oncol. Res 20, 309–317 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Drexler HG, Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells. Leukemia 10, 588–599 (1996). [PubMed] [Google Scholar]
- 51.Lisovsky M, Estrov Z, Zhang X, Consoli U, Sanchez-Williams G, Snell V, Munker R, Goodacre A, Savchenko V, Andreeff M, Flt3 ligand stimulates proliferation and inhibits apoptosis of acute myeloid leukemia cells: Regulation of Bcl-2 and Bax. Blood 88, 3987–3997 (1996). [PubMed] [Google Scholar]
- 52.Sanchez-Correa B, Bergua JM, Campos C, Gayoso I, Arcos MJ, Bañas H, Morgado S, Casado JG, Solana R, Tarazona R, Cytokine profiles in acute myeloid leukemia patients at diagnosis: Survival is inversely correlated with IL-6 and directly correlated with IL-10 levels. Cytokine 61, 885–891 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Tao M, Li B, Nayini J, Andrews CB, Huang R-W, Devemy E, Song S, Venugopal P, Preisler HD, SCF, IL-1β, IL-1ra and GM-CSF in the bone marrow and serum of normal individuals and of AML and CML patients. Cytokine 12, 699–707 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Man CH, Lam SSY, Sun MKH, Chow HCH, Gill H, Kwong YL, Leung AYH, A novel tescalcin-sodium/hydrogen exchange axis underlying sorafenib resistance in FLT3-ITD+ AML. Blood 123, 2530–2539 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Smith CC, Lasater EA, Zhu X, Lin KC, Stewart WK, Damon LE, Salerno S, Shah NP, Activity of ponatinib against clinically-relevant AC220-resistant kinase domain mutants of FLT3-ITD. Blood 121, 3165–3171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W, Gao C, Konopleva M, Chen Y, Jacamo RO, Borthakur G, Cortes JE, Ravandi F, Ramachandran A, Andreeff M, Reversal of acquired drug resistance in FLT3-mutated acute myeloid leukemia cells via distinct drug combination strategies. Clin. Cancer Res 20, 2363–2374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boyd AL, Aslostovar L, Reid J, Ye W, Tanasijevic B, Porras DP, Shapovalova Z, Almakadi M, Foley R, Leber B, Xenocostas A, Bhatia M, Identification of chemotherapy-induced leukemic-regenerating cells reveals a transient vulnerability of human AML recurrence. Cancer Cell 34, 483–498.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Akira S, Takeda K, Toll-like receptor signalling. Nat. Rev. Immunol 4, 499–511 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Beverly LJ, Starczynowski DT, IRAK1: Oncotarget in MDS and AML. Oncotarget 5, 1699–1700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown J, Wang H, Hajishengallis GN, Martin M, TLR-signaling networks: An integration of adaptor molecules, kinases, and cross-talk. J. Dent. Res 90, 417–427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaudhary D, Robinson S, Romero DL, Recent advances in the discovery of small molecule inhibitors of interleukin-1 receptor-associated kinase 4 (IRAK4) as a therapeutic target for inflammation and oncology disorders. J. Med. Chem 58, 96–110 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Dussiau C, Trinquand A, Lhermitte L, Latiri M, Simonin M, Cieslak A, Bedjaoui N, Villarèse P, Verhoeyen E, Dombret H, Ifrah N, Macintyre E, Asnafi V, Targeting IRAK1 in T-cell acute lymphoblastic leukemia. Oncotarget 6, 18956–18965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flannery S, Bowie AG, The interleukin-1 receptor-associated kinases: Critical regulators of innate immune signalling. Biochem. Pharmacol 80, 1981–1991 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Hosseini MM, Kurtz SE, Abdelhamed S, Mahmood S, Davare MA, Kaempf A, Elferich J, McDermott JE, Liu T, Payne SH, Shinde U, Rodland KD, Mori M, Druker BJ, Singer JW, Agarwal A, Inhibition of interleukin-1 receptor-associated kinase-1 is a therapeutic strategy for acute myeloid leukemia subtypes. Leukemia 32, 2374–2387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janssens S, Beyaert R, Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol. Cell 11, 293–302 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Li Z, Younger K, Gartenhaus R, Joseph AM, Hu F, Baer MR, Brown P, Davila E, Inhibition of IRAK1/4 sensitizes T cell acute lymphoblastic leukemia to chemotherapies. J. Clin. Invest 125, 1081–1097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moresco EMY, LaVine D, Beutler B, Toll-like receptors. Curr. Biol 21, R488–R493 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Patra MC, Choi S, Recent progress in the molecular recognition and therapeutic importance of interleukin-1 receptor-associated kinase 4. Molecules 21, E1529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rhyasen GW, Bolanos L, Fang J, Jerez A, Wunderlich M, Rigolino C, Mathews L, Ferrer M, Southall N, Guha R, Keller J, Thomas C, Beverly LJ, Cortelezzi A, Oliva EN, Cuzzola M, Maciejewski JP, Mulloy JC, Starczynowski DT, Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell 24, 90–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi G-P, Chapman HA, Barton GM, The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guerrier T, Pochard P, Lahiri A, Youinou P, Pers J-O, Jamin C, TLR9 expressed on plasma membrane acts as a negative regulator of human B cell response. J. Autoimmun 51, 23–29 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Powers JP, Li S, Jaen JC, Liu J, Walker NPC, Wang Z, Wesche H, Discovery and initial SAR of inhibitors of interleukin-1 receptor-associated kinase-4. Bioorg. Med. Chem. Lett 16, 2842–2845 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Singer JW, Fleischman A, Al-Fayoumi S, Mascarenhas JO, Yu Q, Agarwal A, Inhibition of interleukin-1 receptor-associated kinase 1 (IRAK1) as a therapeutic strategy. Oncotarget 9, 33416–33439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buckley GM, Fosbeary R, Fraser JL, Gowers L, Higueruelo AP, James LA, Jenkins K, Mack SR, Morgan T, Parry DM, Pitt WR, Rausch O, Richard MD, Sabin V, IRAK-4 inhibitors. Part III: A series of imidazo[1,2-a]pyridines. Bioorg. Med. Chem. Lett 18, 3656–3660 (2008). [DOI] [PubMed] [Google Scholar]
- 75.Graves LM, Litchfield DW, “Going KiNativ”: Probing the Native Kinome. Chem. Biol 18, 683–684 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Wunderlich M, Chou F-S, Link KA, Mizukawa B, Perry RL, Carroll M, Mulloy JC, AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia 24, 1785–1788 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wunderlich M, Mizukawa B, Chou F-S, Sexton C, Shrestha M, Saunthararajah Y, Mulloy JC, AML cells are differentially sensitive to chemotherapy treatment in a human xenograft model. Blood 121, e90–e97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang J, Li L, Friedman AD, Small D, Paz-Priel I, Canonical NF-kB signalling is a potential target in FLT3/ITD AML. Blood 120, 2447 (2012). [Google Scholar]
- 79.Muralidharan S, Mandrekar P, Cellular stress response and innate immune signaling: Integrating pathways in host defense and inflammation. J. Leukoc. Biol 94, 1167–1184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gregory MA, D’Alessandro A, Alvarez-Calderon F, Kim J, Nemkov T, Adane B, Rozhok AI, Kumar A, Kumar V, Pollyea DA, Wempe MF, Jordan CT, Serkova NJ, Tan AC, Hansen KC, DeGregori J, ATM/G6PD-driven redox metabolism promotes FLT3 inhibitor resistance in acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A 113, E6669–E6678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang A, Ju H-Q, Liu K, Zhan G, Liu D, Wen S, Garcia-Manero G, Huang P, Hu Y, Metabolic alterations and drug sensitivity of tyrosine kinase inhibitor resistant leukemia cells with a FLT3/ITD mutation. Cancer Lett. 377, 149–157 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Ishii KJ, Akira S, Innate immune recognition of, and regulation by, DNA. Trends Immunol. 27, 525–532 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Janssens S, Pulendran B, Lambrecht BN, Emerging functions of the unfolded protein response in immunity. Nat. Immunol 15, 910–919 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levis M, Allebach J, Tse K-F, Zheng R, Baldwin BR, Douglas Smith B, Jones-Bolin S, Ruggeri B, Dionne C, Small D, A FLT3-targeted tyrosine kinase inhibitor is cytotoxic to leukemia cells in vitro and in vivo. Blood 99, 3885–3891 (2002). [DOI] [PubMed] [Google Scholar]
- 85.Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, Resnick MA, The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLOS Genet. 7, e1001360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schaefer L, Complexity of danger: The diverse nature of damage-associated molecular patterns. J. Biol. Chem 289, 35237–35245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zarrinkar PP, Gunawardane RN, Cramer MD, Gardner MF, Brigham D, Belli B, Karaman MW, Pratz KW, Pallares G, Chao Q, Sprankle KG, Patel HK, Levis M, Armstrong RC, James J, Bhagwat SS, AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML). Blood 114, 2984–2992 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang A, Wu H, Chen C, Hu C, Qi Z, Wang W, Yu K, Liu X, Zou F, Zhao Z, Wu J, Liu J, Liu F, Wang L, Stone RM, Galinksy IA, Griffin JD, Zhang S, Weisberg EL, Liu J, Liu Q, Dual inhibition of AKT/FLT3-ITD by A674563 overcomes FLT3 ligand-induced drug resistance in FLT3-ITD positive AML. Oncotarget 7, 29131–29142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelly PN, Romero DL, Yang Y, Shaffer III AL, Chaudhary D, Robinson S, Miao W, Rui L, Westlin WF, Kapeller R, Staudt LM, Selective interleukin-1 receptor–associated kinase 4 inhibitors for the treatment of autoimmune disorders and lymphoid malignancy. J. Exp. Med 212, 2189–2201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith MA, Choudhary GS, Pellagatti A, Choi K, Bolanos LC, Bhagat TD, Gordon-Mitchell S, Von Ahrens D, Pradhan K, Steeples V, Kim S, Steidl U, Walter M, Fraser IDC, Kulkarni A, Salomonis N, Komurov K, Boultwood J, Verma A, Starczynowski DT, U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol 21, 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang J, Rhyasen G, Bolanos L, Rasch C, Varney M, Wunderlich M, Goyama S, Jansen G, Cloos J, Rigolino C, Cortelezzi A, Mulloy JC, Oliva EN, Cuzzola M, Starczynowski DT, Cytotoxic effects of bortezomib in myelodysplastic syndrome/acute myeloid leukemia depend on autophagy-mediated lysosomal degradation of TRAF6 and repression of PSMA1. Blood 120, 858–867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goyama S, Wunderlich M, Mulloy JC, Xenograft models for normal and malignant stem cells. Blood 125, 2630–2640 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Stones CJ, Kim JE, Joseph WR, Leung E, Marshall ES, Finlay GJ, Shelling AN, Baguley BC, Comparison of responses of human melanoma cell lines to MEK and BRAF inhibitors. Front. Genet 4, 66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mott BT, Eastman RT, Guha R, Sherlach KS, Siriwardana A, Shinn P, McKnight C, Michael S, Lacerda-Queiroz N, Patel PR, Khine P, Sun H, Kasbekar M, Aghdam N, Fontaine SD, Liu D, Mierzwa T, Mathews-Griner LA, Ferrer M, Renslo AR, Inglese J, Yuan J, Roepe PD, X.-z. Su, C. J. Thomas, High-throughput matrix screening identifies synergistic and antagonistic antimalarial drug combinations. Sci. Rep 5, 13891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Molecular Operating Environment (MOE) (Chemical Computing Group ULC, Montreal, QC, Canada, 2017). [Google Scholar]
- 96.Clark AM, Labute P, 2D depiction of protein–ligand complexes. J. Chem. Inf. Model 47, 1933–1944 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Needleman SB, Wunsch CD, A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol 48, 443–453 (1970). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.