Abstract
Background
Genetic background affects serum urate concentration and gout risk, especially regarding these variants in the urate-transporter gene ABCG2. However, the role of epistasis between PKD2 and ABCG2 on the pathogenesis of gout is poorly understood. Here we assess this epistatic interaction in the progression from elevated serum urate to gout.
Results
We identified two epistatic interaction pairs (rs2728121: rs1481012 and rs2728121: rs2231137) were associated with urate levels in 4914 Chinese individuals (Pint = 0.018 and 0.004, respectively). Using subgroup analysis for gender and BMI, we found the degree of associations was varied by gender and BMI. The SNP pair rs2728121:rs1481012 influenced urate levels in females and overweight subjects (Pint = 0.006 and 0.022, respectively), but rs2728121:rs2231137 did in males, overweight and normal-weight subjects (Pint = 0.017, 0.047 and 0.013, respectively). Consistent results were also observed in associations between these epistatic interactions with hyperuricemia. Next, the SNP pair rs2728121:rs2231137 was identified to influence the development of gout from both hyperuricemia and healthy (Pint = 0.035 and 0.001, respectively), especially in males (Pint = 0.030 and 0.001, respectively). Furthermore, we demonstrated that interacting regions were enriched by regulatory elements. Finally, we observed a strong gene co-expression pattern between PKD2 and ABCG2 (r = 0.743, P = 5.83E-06).
Conclusion
Our findings indicate epistasis between PKD2 and ABCG2 influence serum urate concentrations, hyperuricemia and gout risk, thus providing insight into the pathogenesis of gout.
Keywords: Epistasis, Serum urate, Body mass index (BMI), Gene transcript, Gender, Enhancer
Background
Urate is the end product of purineger breakdown in humans. Elevated urate levels in the blood (hyperuricemia) can cause the deposition of monosodium urate (MSU) crystals in the joints and tissues that play a predominant role in the development of gout [1–5]. Gout is a form of acute inflammatory arthritigers [6, 7] that affects range from 0.1% to approximately 10% of individuals worldwide [8]. Although hyperuricemia can cause gout [9], not all hyperuricemia patients develop gout [10]. Generally, only a quarter of hyperuricemia patients develop gout, suggesting that hyperuricemia is necessary but not sufficient for gout development [10]. Several loci in different genes have been identified to affect serum urate concentration and gout risk [1, 10]. However, these loci only explained ~ 7% of the variance in urate levels [1, 4]. Therefore, it is necessary to identify other genetic factors contribute to the progression from elevated serum urate to hyperuricemia to gout.
PKD2 is located nearby ABCG2, which encodes a urate transporter that plays a certain role in serum urate concentrations and gout risk [4, 11]. Even though serval GWAS studies found PKD2 variants associated with serum urate and gout, none variants in the PKD2 gene were independently associated with serum urate and gout conditional on the ABCG2 variants [1–3, 12]. Based on that, PKD2 is commonly not considered as a candidate gene for serum urate and gout. This also happened for WDR1, which is adjacent to a urate transporter gene SLC2A9. Although one studies reported no relationship between SCL2A9 variants and hyperuricemia/gout [13], a number of GWAS and functional studies revealed a significant association between SCL2A9 variants and hyperuricemia/gout [1–4, 12]. A previous study showed that epistatic interactions between WDR1 and SLC2A9 regulated serum urate concentrations, suggesting the biological value of epistatic interactions in the pathogenesis of gout [14].. Therefore, we attempted to investigate a potential epistatic interaction between PKD2 and ABCG2.
Here we aimed to explore epistasis between PKD2 and ABCG2 using a four-step approach. First, the epistatic effect of loci on serum urate was investigated. Next, these identified urate-related epistatic interactions were tested in the development of hyperuricemia and gout, separately. Common heterogeneity factors, including gender, body mass index (BMI) and smoking status, were considered as covariates in the analysis. Besides, enrichment analysis was performed to provide insight into the biological function of these urate-related epistatic interactions. Finally, the interplay between PDK2 and ABCG2 transcripts was investigated. All statistical analyses were performed with adjustment for marginal effects. Through this strategy, our results suggested that epistatic interactions between PDK2 and ABCG2 contributed to serum urate and gout and that PKD2 is supposed to influence the progression from elevated serum urate to hyperuricemia to gout by epistatically interacting with ABCG2.
Results
Epistatic interactions between PDK2 and ABCG2 affected serum urate concentrations
Four SNP pairs were tested in our study (Table 1). Of them, two SNP pairs (rs2728121:rs1481012 and rs2728121:rs2231137) were found to influence serum urate concentrations with contradictory effects (Estimate = − 14.487, Pint = 0.018 and Estimate = 9.781, Pint = 0.004, respectively) (Table 1). The two SNP pairs rs2728121:rs1481012 and rs2728121:rs2231137 could explain 0.099 and 0.164% of urate variance without conditioning on the marginal SNPs, separately (Table 1).
Table 1.
Associations between serum urate and epistatic interactions in PKD2 and ABCG2
| Chr. | SNP1 | Pos1 | A1 | SNP2 | Pos2 | A2 | Dist | LD (r2) | Estimate | Pint | Variance explained (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | rs2725215 | 88,961,571 | T | rs1481012 | 89,039,082 | G | 77,511 | 0.605 | Male | −2.309 | 0.693 | – |
| Female | −12.817 | 0.236 | – | |||||||||
| Total | −7.020 | 0.181 | – | |||||||||
| 4 | rs2725215 | 88,961,571 | T | rs2231137 | 89,061,114 | T | 99,543 | 0.116 | Male | 4.830 | 0.618 | – |
| Female | 20.178 | 0.220 | – | |||||||||
| Total | 9.306 | 0.270 | – | |||||||||
| 4 | rs2728121 | 88,997,102 | C | rs1481012 | 89,039,082 | G | 41,980 | 0.370 | Male | −6.051 | 0.379 | – |
| Female | −33.315 | 0.006 | 0.493 | |||||||||
| Total | −14.487 | 0.018 | 0.099 | |||||||||
| 4 | rs2728121 | 88,997,102 | C | rs2231137 | 89,061,114 | T | 64,012 | 0.045 | Male | 8.980 | 0.017 | 0.144 |
| Female | 9.250 | 0.171 | – | |||||||||
| Total | 9.781 | 0.004 | 0.164 |
Chr, chromosome of an SNP pair. SNP1 (SNP2), Pos1 (Pos2), name and position of the first (second) SNP. A1, effect allele of SNP1; A2, effect allele of SNP2. dist, distance in bp between two SNPs. LD (r2), linkage disequilibrium between two SNPs. Pint, P-value of the interaction between SNP pair in serum urate was calculated by linear regression adjusted age, gender, and target SNPs. Variance explained of the interaction between the SNP pair in serum urate was calculated by linear regression
Next, our results showed that these associations were significantly modified by gender (Table 1). SNP pair rs2728121:rs2231137 only affected urate (Estimate = 8.980, Pint = 0.017) in the male subgroup. In contrast, SNP pair rs2728121:rs1481012 only contributed to the concentrations of serum urate in females (Estimate = − 33.315, Pint = 0.006). Lager urate variance (0.493%) could be explained by epistatic interactions rs2728121:rs1481012 in female subgroup than in all subjects, highlighting the role of gender on associations between these epistasis and serum urate concentrations.
Epistatic interactions of PDK2 and ABCG2 affected the development of hyperuricemia and gout
Given that elevated serum urate is a critical risk factor for the development of hyperuricemia and gout, we further assessed the contributions of the two identified urate-related epistatic interactions on hyperuricemia and gout risk. Regarding hyperuricemia, SNP pair rs2728121:rs2231137 significantly increased hyperuricemia risk (Estimate = 0.193, Pint = 0.009). In essence, SNP pair rs2728121:rs2231137 only affected the risk of hyperuricemia in males (Estimate = 0.206, Pint = 0.018), but did not in females (Pint = 0.427). In contrast, SNP pair rs2728121:rs1481012 decreased the risk of hyperuricemia in females (Estimate = − 0.648, Pint = 0.017) (Table 2), but did not in males (Pint = 0.483). All the above findings are pretty consistent with the results in serum urate.
Table 2.
Associations between urate-related epistatic interactions and hyperuricemia/gout
| SNP1 | SNP2 | Hyperuricemia Vs. Control | Gout Vs. Hyperuricemia | Gout Vs. Control | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | SE | Pint | Estimate | SE | Pint | Estimate | SE | Pint | |||
| rs2728121 | rs1481012 | Male | −0.108 | 0.153 | 0.483 | 0.188 | 0.245 | 0.444 | 0.077 | 0.258 | 0.765 |
| Female | −0.648 | 0.272 | 0.017 | −0.528 | 0.947 | 0.577 | −0.897 | 0.835 | 0.282 | ||
| Total | −0.254 | 0.130 | 0.051 | 0.165 | 0.234 | 0.482 | −0.004 | 0.239 | 0.987 | ||
| rs2728121 | rs2231137 | Male | 0.206 | 0.087 | 0.018 | 0.338 | 0.156 | 0.030 | 0.524 | 0.161 | 0.001 |
| Female | 0.118 | 0.148 | 0.427 | 0.100 | 0.536 | 0.852 | 0.229 | 0.523 | 0.661 | ||
| Total | 0.193 | 0.074 | 0.009 | 0.313 | 0.149 | 0.035 | 0.480 | 0.149 | 0.001 | ||
SNP1 (SNP2), name of the first (second) SNP. SE, standard error. Pint, P-value of the interaction between the SNP pair in hyperuricemia and gout were calculated by logistic regression adjusted age, gender, and target SNPs
For the development from hyperuricemia to gout, SNP pair rs2728121:rs2231137 played an important role (Estimate = 0.313, Pint = 0.035), especially in males (Estimate = 0.338, Pint = 0.030) (Table 2). However, SNP pair rs2728121:rs1481012 did not contribute to this progression in neither males nor females (Pint = 0.444 and Pint = 0.577, respectively). Because SNP pair rs2728121:rs2231137 influence all progressions from elevated serum urate to gout, it had a definitive effect in the pathogenesis of gout (Estimate = 0.480, Pint = 0.001), especially regarding in males (Estimate = 0.524, Pint = 0.001).
Associations between epistatic interactions and serum urate in BMI and smoking subgroups
Our previous studies have suggested that body mass index (BMI) and cigarette smoking could modify urate levels [4, 9, 15]. But their impact on the degree of associations between epistasis and serum urate was not determined. Therefore, we further performed subgroup analyses for BMI and smoking status in our study.
For the subgroups of BMI, SNP pair rs2728121:rs2231137 was identified to be associated with urate in normal (Estimate = 11.456, Pint = 0.013) and overweight individuals (Estimate = 9.844, Pint = 0.047) (Table 3). SNP pair rs2728121:rs1481012 only affected the concentrations of serum urate in overweight subgroup (Estimate = − 21.702, Pint = 0.022). However, no significant associations were observed in any subgroups of smoking status (all Pint > 0.05) (Table 3).
Table 3.
Associations between epistatic interactions and serum urate in subgroups of BMI and smoking status
| SNP1 | SNP2 | Subgroup-1 | Subgroup-2 | Subgroup-3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | SE | Pint | Estimate | SE | Pint | Estimate | SE | Pint | |||
| rs2728121 | rs1481012 | BMI | 19.794 | 41.811 | 0.637 | −10.483 | 8.306 | 0.207 | −21.702 | 9.477 | 0.022 |
| Smoke | −25.467 | 15.210 | 0.095 | −7.751 | 10.478 | 0.460 | −9.196 | 8.343 | 0.271 | ||
| rs2728121 | rs2231137 | BMI | 24.512 | 22.948 | 0.288 | 11.456 | 4.620 | 0.013 | 9.844 | 4.962 | 0.047 |
| Smoke | 9.972 | 8.385 | 0.235 | 7.106 | 5.223 | 0.174 | 7.592 | 4.608 | 0.100 | ||
SNP1 (SNP2), name of the first (second) SNP. SE, standard error. Pint, P-value of the interaction between the SNP pair in serum urate was calculated by linear regression adjusted age, gender and target SNPs. Subgroup of BMI: 1, Underweight (BMI < 18.5); 2, Normal (18.5 ≦ BMI < 25); 3, Overweight (BMI ≧ 25). Subgroup of smoke: 1, non-smokers; 2, former smokers; 3, current smokers
Colocation of interacting regions with regulatory elements
By annotating urate-related interacting regions with chromatin state, we found several strong and weak enhancers were located at these regions, especially at gene PKD2 (Additional file 1: Fig. S1 and Additional file 2: Figure S2). Consistent with the above result, transcription factors and DNase binding sites, were also observed at the PKD2 gene region (Additional file 1: Figure S1 and Additional file 2: Figure S2). Our findings supposed the potential regulatory effects across these interacting regions, especially in PKD2. Besides, ABCG2 has been identified as a urate transporter in previous studies that played a key role in serum urate and gout [1, 11, 16]. Above all, we hypothesized that PKD2 can influence serum urate by epistatically interacting with ABCG2.
To further explore potential regulatory effects at the PKD2 region, two sets of SNPs were tested using enhancer enrichment analysis (Table 4). Regarding urate-related SNPs identified in a recent genome-wide association study [1], we found a significant enrichment in enhancer regions in aHuvec cell line (3.6-fold enrichment, P = 0.012), especially for strong enhancers (5.7-fold enrichment, P = 0.005) (Table 4). For all loci in PKD2, distinct enrichment for these loci in enhancers were observed across various cell lines (H1, HepG2, Huvec, HSMM, NHLF, HMEC, GM12878, and NHEK cell lines) (Table 4). Our findings provided a piece of supporting evidence that regulatory factors in PKD2 control the concentrations of serum urate by epistatic interactions with ABCG2.
Table 4.
Enhancer enrichment analysis of loci in PKD2
| Set | Cell type | All enhancers | Strongest enhancers | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID | Description | Obs | Exp | Fold | P | Obs | Exp | Fold | P | |
| 1 | Huvec | umbilical vein endothelial cells | 5 | 1.4 | 3.6 | 0.012 | 4 | 0.7 | 5.7 | 0.005 |
| 2 | H1 | H1 cell line | 4 | 10.5 | 0.4 | 0.994 | 4 | 0.9 | 4.5 | 0.013 |
| HepG2 | hepatocelluar carcinoma | 24 | 9.3 | 2.6 | 2.50E-05 | 24 | 3.1 | 7.8 | < 1.0E-06 | |
| Huvec | umbilical vein endothelial cells | 41 | 11.4 | 3.6 | < 1.0E-06 | 29 | 5.7 | 5.1 | < 1.0E-06 | |
| HSMM | skeletal muscle myoblasts | 30 | 12.9 | 2.3 | 1.80E-05 | 4 | 5.7 | 0.7 | 0.818 | |
| NHLF | lung fibrolasts | 43 | 11.6 | 3.7 | < 1.0E-06 | 15 | 4.4 | 3.4 | 4.20E-05 | |
| HMEC | mammary epithelial cells | 36 | 15 | 2.4 | 1.00E-06 | 10 | 5.7 | 1.7 | 0.065 | |
| GM12878 | B-lymohocyte, lymphoblastoid | 20 | 11.8 | 1.7 | 0.016 | 13 | 4.3 | 3 | 4.36E-04 | |
| NHEK | epidermal keratinocytes | 30 | 13 | 2.3 | 2.00E-05 | 17 | 5.7 | 3 | 7.40E-05 | |
Set one was urate-related SNPs identified in a recent genome-wide association study. Set two was all SNPs in the gene region of PKD2
The interplay between PDK2 and ABCG2 transcripts
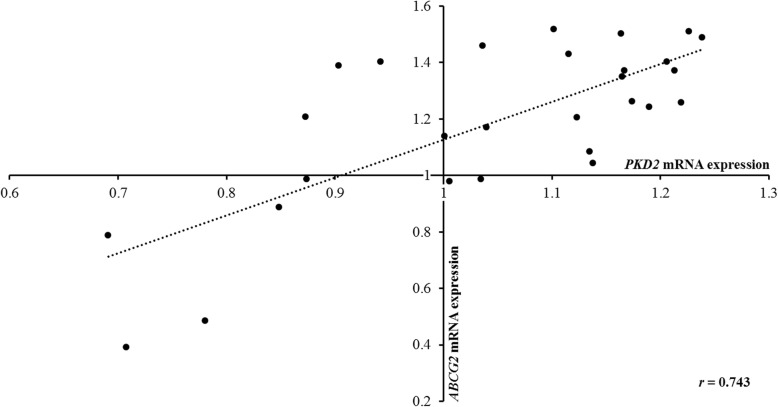
To further confirm these urate-related epistatic interactions between PKD2 and ABCG2, gene transcripts of PKD2 and ABCG2 were investigated. The result indicated a strong positive correlation between PKD2 with ABCG2 transcripts (r = 0.743, P = 5.83E-06) (Fig. 1), thus supporting our hypothesis that PKD2 can affect serum urate by epistatically interacting with ABCG2.
Fig. 1.
The gene co-expression pattern between PDK2 and ABCG2
Discussion
Our study explored the effect of epistatic interactions between PKD2 and ABCG2 on the progression from elevated serum urate to gout. Here we identified two SNP pairs (rs2728121:rs1481012 and rs2728121:rs2231137) significantly associated with serum urate, hyperuricemia or gout. The role of these two identified urate-related epistatic interactions was investigated in detail, including subgroup analyses for heterogeneity factors (such as gender, BMI and smoking status), enrichment analysis and gene co-expression analysis.
ABCG2 gene encodes a urate transporter that influences serum urate concentrations and gout risk. It has been proved to be one of the strongest risk factors for the development of gout [1, 11]. However, there are conflicting results regarding the association between PKD2 and urate/gout. Recent studies suggested that PKD2 is associated with urate and gout [17], but contrary results were observed in other studies [3]. Furthermore, no functional experiments are suggesting the contribution of PKD2 on serum urate and gout to date. Therefore, we performed a systemic analysis to explore the role of PKD2 on the development of hyperuricemia and gout.
Our observations showed that two SNP pairs (rs2728121:rs1481012 and rs2728121:rs2231137) were associated with serum urate concentrations or hyperuricemia risk. SNP pair rs2728121: rs2231137 was also identified to be associated with the development of gout from both hyperuricemia and healthy (Tables 1 and 2). These results revealed a potential mechanism for the biological role of PKD2 in serum urate and gout. SNP pair rs2728121:rs1481012 can explain 0.099% of urate variance, and even more in females (0.493%). Whereas SNP pair rs2728121: rs2231137 explains 0.164% of the variance (Table 1). These results suggested that epistasis may be an additional way to solve the ‘missing heritability’ problem. Additionally, consistent with our previous results [4, 9, 15], heterogeneity factors, such as gender and BMI, modified the degree of associations between epistatic interactions and serum urate/gout (Table 3).
The potential mechanism of epistatic interactions between PKD2 and ABCG2 genes were analyzed by enrichment analysis and gene co-expression analysis. We supposed that PKD2 can indirectly influence the development of gout, by interacting with ABCG2 since no direct role of PKD2 have been identified until now. Enrichment analysis showed that regulatory factors were enriched in PKD2, suggesting the potential regulatory function of PKD2. To further confirm this result, gene transcripts of these two genes were examined. The result indicated a positive correlation between PDK2 and ABCG2 gene expression, further supporting our hypothesis that PKD2 can influence serum urate by epistatically interacting with ABCG2.
Conclusions
Our results uncover that epistatic interactions between PKD2 and ABCG2 influence all progressions from elevated serum urate to gout. Furthermore, the degree of association has been found to vary with gender and BMI.
Methods and materials
Study subjects
This study was approved by the Ethical Committees of the School of Life Sciences of Fudan University (approval number of 140) and conducted following the principles of the Declaration of Helsinki. All subjects in this study provided written informed consent. All 582 gout patients enrolled in this study were diagnosed with gout (OMIM: #138900) following the American College of Rheumatology diagnostic criteria [18]. These gout patients did not use any urate-lowering drugs for two weeks before sample collection. Their clinical information was collected at Changhai Hospital, Taixing People’s Hospital, and Taizhou People’s Hospital.
Furthermore, 4332 individuals without a history of gout were enrolled from the Taizhou Longitudinal Study [19]. Among them, 1387 subjects were considered hyperuricemia patients due to their high serum urate levels (> 417 umol/L) [20]. The other individuals were treated as healthy controls. All 4332 subjects were divided into subgroups according to their body mass index (BMI) and smoking status recorded in questionnaires. Three BMI subgroups (underweight: BMI < 18.5; normal-weight: 18.50 ≤ BMI < 25; overweight: BMI ≥ 25) were defined following the categories of the World Health Organization (WHO) [4, 15]. Besides, smoking status categories included non-smoker, former smoker, and current smoker. The detailed characteristics of these participants were illustrated in Additional file 3: Table S1.
Target loci selected in ABCG2
Here we leveraged ABCG2 SNPs rs2231137 and rs1481012 in our analysis. The SNP rs2231137 is a missense variant that strongly linked with serum urate and gout [1, 4]. Another SNP rs1481012 is strongly LD with rs2231142 (r2 = 1 in Eastern Asian), which is a significant gout-related loci [11, 21]. Due to the complexity of the sequence region for rs2231142, it was difficult to detect its genotype by using SNaPshot (TianHao, China). Therefore, we defined rs1481012 as a replacement of rs2231142 in our further analysis. Besides, the two SNPs are independent of each other (r2 = 0.186 in Eastern Asian) and other ABCG2 SNPs highly LD (r2 > 0.6) with the two loci were also excluded. Finally, the two SNPs are common variants in the Chinese population with a minor allele frequency (MAF) > 0.2. Altogether, we selected the two SNPs in our study.
SNP genotype and real-time qPCR
Peripheral blood was collected from all participants in this study. DNA was extracted from blood samples according to standard procedures. To measure gene expression, we randomly collected RNA samples from 58 male subjects. These subjects were a subset of 2945 healthy individuals enrolled in genotype analysis. Detailed information for DNA and RNA processing have been described in our previous study [4]. Genotyping of target SNPs in PKD2 (rs2725215 and rs2728121) and ABCG2 (rs2231137 and rs1481012) was performed by SNaPshot (TianHao, China). Gene transcript was tested by real-time qPCR.
Enrichment analysis
Two sets of loci in PKD2 were used in our enrichment analysis. One was comprised of urate-related SNPs identified in a recent genome-wide association study [1]. Another contained all SNPs in the gene region of PKD2. We performed cell-type-specific enhancer enrichment analysis in both two SNP sets using HaploReg (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) [22]. For each set of SNPs, the overlap of SNPs with ENCODE [23] cell-type-specific enhancers was compared to all 1000 Genomes variants with a minor allele frequency above 5%. The fold enrichment and P-value were calculated using a binomial test. SNPnexus [24] and UCSC genome browser [25] was used to annotate these loci. Enlight (http://enlight.wglab.org) was applied to draw gene-regional plots and show epigenetic modifications in PKD2 [26]. Additionally, linkage disequilibrium (LD) relationships between target SNPs were analyzed using SNAP (http://archive.broadinstitute.org/mpg/snap/index.php) [27].
Statistical analysis
P-values for associations between target SNP pairs with serum urate concentrations were calculated by linear regression with adjustment for age, gender, and marginal effects. The variance of serum urate explained by each SNP pair was calculated by linear regression. All P-values for associations between SNP pairs with hyperuricemia and gout were measured by logistic regression adjusted for age, gender and marginal effects. Subgroup analyses for gender, BMI and smoking status were also performed in our study. Moreover, the relationship between PKD2 and ABCG2 transcripts was investigated. P values of < 0.05 were considered statistically significant. All statistical analyses in this study were processed using R (Version 3.0.2: www.r-project.org/).
Supplementary information
Additional file 1: Figure S1. Chromatin state analysis of PKD2 and ABCG2 genes by Enlight.
Additional file 2: Figure S2. Chromatin state analysis of PKD2 and ABCG2 genes by the UCSC genome browser.
Additional file 3: Table S1. Characteristics of all participants in our study. HUA, hyperuricemia. The data are shown as the mean (SD).
Acknowledgments
Computational support was provided by the High-End Computing Center located at Fudan University.
Consent to participate
All subjects in this study provided written informed consent.
Abbreviations
- ABCG2
ATP-binding cassette subfamily G member 2
- BMI
Body mass index
- LD
Linkage disequilibrium
- MSU
Monosodium urate
- OMIM
Online Mendelian Inheritance in Man
- OR
Odds ratio
- Pint
P values for epistatic interactions
- PKD2
Polycystin-2
- RNA
Ribonucleic acid
- SLC2A9
Solute Carrier Family 2 Member 9
- SNP
Single nucleotide polymorphism
- WDR1
WD Repeat Domain 1
- WHO
World Health Organization
Authors’ contributions
Conceptualization, J.W., and Z.D.; Formal analysis, Z.D.; Methodology, Z.D.; Investigation: Z.D.; Data Curation, J.Z., D.Z., C.Y., Y.M., H.H., H.J., S.J., and Y.L.; Writing - Original Draft Preparation, Z.D.; Writing - Review & Editing, J.W., H.Z., and L.J.
Funding
This work was supported by grants from the “Five New” Translational Project on Specialist Diseases of Shanghai Shenkang Hospital Development Center (16CR3012A), the Program for 2012 Outstanding Medical Academic Leader for Hejian Zou, Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), and International S&T Cooperation Program of China (2013DFA30870).
Availability of data and materials
The datasets used and/or analysis in the current study can be obtained from the corresponding author according to reasonable requirements.
Ethics approval
All procedures followed were by the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients included in the study.
Consent for publication
The manuscript is approved by all authors for publication.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hejian Zou, Email: hjzou@fudan.edu.cn.
Jiucun Wang, Email: jcwang@fudan.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s41065-020-0116-6.
References
- 1.Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet [Internet]. 2012;45:145–154. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3663712&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed]
- 2.Yang Q, Köttgen A, Dehghan A, Smith AV, Glazer NL, Chen MH, et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dehghan A, Köttgen A, Yang Q, Hwang S-J, Kao WL, Rivadeneira F, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Z, Zhou J, Jiang S, Li Y, Zhao D, Yang C, et al. Effects of multiple genetic loci on the pathogenesis from serum urate to gout. Sci Rep [Internet]. Nature Publishing Group; 2017 [cited 2017 Apr 23];7:43614. Available from: http://www.nature.com/articles/srep43614 [DOI] [PMC free article] [PubMed]
- 5.Perez-Ruiz F, Dalbeth N, Bardin T. A Review of Uric Acid, Crystal Deposition Disease, and Gout. Adv. Ther. 2014:31–41. [DOI] [PMC free article] [PubMed]
- 6.Dong Z, Li Y, Zhou J, Jiang S, Wang Y, Chen Y, et al. Copy number variants of ABCF1, IL17REL, and FCGR3A are associated with the risk of gout. Protein Cell [Internet]. Higher Education Press; 2017 [cited 2017 Apr 23];1–4. Available from: http://link.springer.com/10.1007/s13238-017-0401-y [DOI] [PMC free article] [PubMed]
- 7.Dong Z. Wang J. Gout and Hyperuricemia Copy number variation and gout : the next frontier. 2015;2:42–49. [Google Scholar]
- 8.Kuo C-F, Grainge MJ, Zhang W, Doherty M. Global epidemiology of gout: prevalence, incidence and risk factors. Nat Rev Rheumatol [Internet] 2015;11:649–662. doi: 10.1038/nrrheum.2015.91. [DOI] [PubMed] [Google Scholar]
- 9.Dong Z, Zhao D, Yang C, Zhou J, Qian Q, Ma Y, et al. Common variants in LRP2 and COMT genes affect the susceptibility of gout in a chinese population. PLoS One. 2015;10. [DOI] [PMC free article] [PubMed]
- 10.Merriman TR. An update on the genetic architecture of hyperuricemia and gout. Arthritis Res Ther [Internet]. 2015;17:98. Available from: http://arthritis-research.com/content/17/1/98 [DOI] [PMC free article] [PubMed]
- 11.Dong Z, Guo S, Yang Y, Wu J, Guan M, Zou H, et al. Association between ABCG2 Q141K polymorphism and gout risk affected by ethnicity and gender: a systematic review and meta-analysis. Int J Rheum Dis. 2015;18:382–391. doi: 10.1111/1756-185X.12519. [DOI] [PubMed] [Google Scholar]
- 12.Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, Perola M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5. [DOI] [PMC free article] [PubMed]
- 13.Hurba O, Mancikova A, Krylov V, Pavlikova M, Pavelka K, Stiburková B. Complex analysis of urate transporters SLC2A9, SLC22A12 and functional characterization of non-synonymous allelic variants of GLUT9 in the Czech population: no evidence of effect on hyperuricemia and gout. PLoS One. 2014. [DOI] [PMC free article] [PubMed]
- 14.Wei WH, Guo Y, Kindt ASD, Merriman TR, Semple CA, Wang K, et al. Abundant local interactions in the 4p16.1 region suggest functional mechanisms underlying SLC2A9 associations with human serum uric acid. Hum Mol Genet. 2014;23:5061–5068. doi: 10.1093/hmg/ddu227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, Dong Z, Zhou J, Ma Y, Pu W, Zhao D, et al. Common UCP2 variants contribute to serum urate concentrations and the risk of hyperuricemia. Sci Rep [Internet]. Nature Publishing Group; 2016 [cited 2017 may 5];6:27279. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27273589. [DOI] [PMC free article] [PubMed]
- 16.Woodward OM, Köttgen A, Coresh J, Boerwinkle E, Guggino WB, Köttgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci U S A. 2009;106:10338–10342. doi: 10.1073/pnas.0901249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Liu K, Ma L, Liu K, Shi X, Zhang Y, et al. Associations of gout with polymorphisms in SLC2A9, WDR1, CLNK, PKD2, and ABCG2 in Chinese Han and Tibetan populations. Int J Clin Exp Pathol [Internet]. 2016 [cited 2017 May 5];9:7503–7517. Available from: www.ijcep.com
- 18.Wallace SL, Robinson H, Masi AT, Decker JL, Mccarty DJ, Y?? T ???F. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum 1977;20:895–900. [DOI] [PubMed]
- 19.Wang X, Lu M, Qian J, Yang Y, Li S, Lu D, et al. Rationales, design and recruitment of the Taizhou Longitudinal Study. BMC Public Health [Internet]. 2009 [cited 2017 Apr 23];9:223. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19589173. [DOI] [PMC free article] [PubMed]
- 20.Khanna D, Fitzgerald JD, Khanna PP, Bae S, Singh MK, Neogi T, et al. 2012 American college of rheumatology guidelines for management of gout. Part 1: systematic nonpharmacologic and pharmacologic therapeutic approaches to hyperuricemia. Arthritis Care Res. 2012;64:1431–1446. doi: 10.1002/acr.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Z, Zhou J, Xu X, Jiang S, Li Y, Zhao D, et al. Genetic variants in two pathways influence serum urate levels and gout risk: a systematic pathway analysis. Sci Rep [Internet]. Nature Publishing Group; 2018 [cited 2018 Apr 18];8:3848. Available from: http://www.nature.com/articles/s41598-018-21858-0 [DOI] [PMC free article] [PubMed]
- 22.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40. [DOI] [PMC free article] [PubMed]
- 23.Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, et al. ENCODE whole-genome data in the UCSC Genome Browser: Update 2012. Nucleic Acids Res 2012; [DOI] [PMC free article] [PubMed]
- 24.Dayem Ullah AZ, Lemoine NR, Chelala C. A practical guide for the functional annotation of genetic variations using SNPnexus. Br bioinform [internet]. 2013;14:437–47. Available from. http://www.ncbi.nlm.nih.gov/pubmed/23395730. [DOI] [PubMed]
- 25.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res [Internet] 2002;12:996–1006. Available from: http://www.ncbi.nlm.nih.gov/pubmed/186604%5Cn http://www.genome.org/cgi/doi/10.1101/gr.229102 [DOI] [PMC free article] [PubMed]
- 26.Guo Y, Conti DV, Wang K. Enlight: web-based integration of GWAS results with biological annotations. Bioinformatics. 2015;31:275–276. doi: 10.1093/bioinformatics/btu639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, De Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Chromatin state analysis of PKD2 and ABCG2 genes by Enlight.
Additional file 2: Figure S2. Chromatin state analysis of PKD2 and ABCG2 genes by the UCSC genome browser.
Additional file 3: Table S1. Characteristics of all participants in our study. HUA, hyperuricemia. The data are shown as the mean (SD).
Data Availability Statement
The datasets used and/or analysis in the current study can be obtained from the corresponding author according to reasonable requirements.