Abstract
Background
Understanding the genomic landscape and immune microenvironment features of preinvasive and early invasive lung adenocarcinoma may provide critical insight and facilitate development of novel strategies for early detection and intervention.
Methods
A total of 80 tumor tissue samples and 30 paired histologically normal lung tissue samples from 30 patients with adenocarcinoma in situ (AIS) (n = 8), minimally invasive adenocarcinoma (MIA) (n = 8), and invasive adenocarcinoma (IAC) (n = 14) were subjected to multiregion whole exome sequencing and immunohistochemistry staining for CD8 and programmed death ligand 1 (PD-L1).
Results
All tumors, including AIS, exhibited evidence of genomic intratumor heterogeneity. Canonical cancer gene mutations in EGFR, erb-b2 receptor tyrosine kinase 2 gene (ERBB2), NRAS, and BRAF were exclusively trunk mutations detected in all regions within each tumor, whereas genes associated with cell mobility, gap junction, and metastasis were all subclonal mutations. EGFR mutation represented the most common driver alterations across AIS, MIA, and IAC, whereas tumor protein p53 gene (TP53) was identified in MIA and IAC but not in AIS. There was no difference in PD-L1 expression among AIS, MIA, and IAC, but the CD8 positivity rate was higher in IAC. Tumors positive for both PD-L1 and CD8 had a larger proportion of subclonal mutations.
Conclusions
Mutations in EGFR, ERBB2, NRAS, and BRAF are early clonal genomic events during carcinogenesis of lung adenocarcinoma, whereas TP53 and cell mobility, gap junction, and metastasis-related genes may be late events associated with subclonal diversification and neoplastic progression. Genomic intratumor heterogeneity and immunoediting are common and early phenomena that may have occurred before the acquisition of invasion.
Keywords: Lung adenocarcinoma, Whole exome sequencing, Genomic evolution, Pathological subtypes
Introduction
Lung adenocarcinoma (LUAD) is the most common subtype of lung cancer worldwide.1 Advances in surgery, radiotherapy, and systemic treatment have significantly improved the clinical outcome of patients with LUAD.2–4 However, a considerable proportion of patients with LUAD will experience development of resistance and succumb to this disease.5,6 Early detection and intervention are associated with better survival. Low-dose computed tomography–guided screening has improved lung cancer mortality by 20%.7 Meanwhile, the widespread of implementation of computed tomography has resulted in a drastic increase in the detection of small pulmonary nodules presenting as ground glass opacities (GGOs).8 Many of these GGOs are atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), and sometimes invasive adenocarcinoma (IAC).9 Surgical resection was reported to yield a 5-year survival rate of almost 100% for AAH, AIS, and MIA, which decreases substantially for IAC.10 Different histological patterns of IACs have been evaluated by using a comprehensive grading system to depict the pathological scenario and estimate prognosis. Lepidic and acinar predominant subtypes of IACs, which are considered to be less invasive, were demonstrated to have better prognosis than other histologic subtypes do.11 As for the definition of preinvasive LUAD such as AIS, 100% lepidic pattern should be confirmed through hematoxylin-eosin staining.10 It was proposed that AAH may progress to AIS, MIA, and eventually IAC.10,12 However, this model has very little molecular support, as very few studies have focused on the genomic evolution from preinvasive to invasive LUAD.
Large-scale sequencing studies have revealed the complex genomic landscape of NSCLC.13 Furthermore, different tumor cells (TCs) within the same tumors may have different genomic features, a phenomenon termed intratumor heterogeneity (ITH). Through use of a multiregion sequencing approach, several previous studies have delineated the ITH and evolutionary process of invasive NSCLC, including IAC.14–16 However, the ITH architecture of preinvasive LUAD has not been well defined. In this study, we depicted the ITH and early evolutionary trajectory of LUADs by multiregion whole exome sequencing (WES) of preinvasive LUAD and IACs. In addition, we sought to explore the potential role of antitumor immune surveillance during early carcinogenesis of LUAD by immunohistochemistry (IHC) analysis of CD8 and programmed death ligand 1 (PD-L1).
Methods
Study Design and Patient Cohorts
Patients who underwent surgical resection of pulmonary nodules at Guangdong Provincial Peoples’ Hospital were enrolled. Tumors that met the following criteria were included in this study: (1) pathologically confirmed early-stage LUAD with no lymph node metastasis; (2) tumor no larger than 30 mm (except for one patient with a 34-mm tumor that presented as a part-solid nodule), and (3) no components other than lepidic/acinar subtypes (Supplementary Fig. 1). Histologic subtypes were determined by two specialized lung cancer pathologists (Fig. 1A) and included AIS, MIA, lepidic predominant adenocarcinoma (LPA), and acinar predominant adenocarcinoma (APA). No AAH was found in this cohort because of the restricted guidelines for surgical resection. The study was approved by the institutional review board of Guangdong Provincial Peoples’ Hospital and Guangdong Academy of Medical Sciences, and all patients signed informed consent.
Figure 1.
Clinicopathologic characteristics and genomic profiling among different subtypes of lung adenocarcinoma (LUAD). (A) Pathological and corresponding radiological images of three representative pathological subtypes. Red arrow indicates the annotation of the lesion. (B) Comprehensive visualized plot based on surgical specimens of patients with adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), and invasive carcinoma (IAC) lesions. Each column represents a single patient whose specimen was examined by multiregion sequencing. Top bar graph describes the mutation number in each patient. Key characteristics, including age, sex, smoking status, and radiological pattern, are presented as a heatmap below the bar graph. Mutation recurrent rate is illustrated by bar graph to the left. Selected genes that have been reported to be correlated with lung adenocarcinoma are shown in the middle heatmap. (C) Driver alterations of EGFR, erb-b2 receptor tyrosine kinase 2 gene (ERBB2), BRAF, and NRAS are specifically presented through a heatmap. (D) Dual bar graph of stage I lung adenocarcinomas, most of which are bronchioloalveolar carcinomas from four external cohorts, reveals both somatic nucleotides variants and copy number variants of several genes with the largest proportion and driver genes. ALK, ALK receptor tyrosine kinase; ALK, ALK receptor tyrosine kinase gene; APA, acinar predominant adenocarcinoma; ARMS, amplification refractory mutation system; CDKN2A, cyclin dependent kinase inhibitor 2A gene; CDKN2B, cyclin dependent kinase inhibitor 2B gene; CKS1B, CDC29 protein kinase regulatory subunit 1B; CNA, copy number alteration; CRIPAK, cysteine rich PAK1 inhibitor gene; CRNN, cornulin gene; DMD, dystrophin gene; DROSHA, drosha ribonuclease 3 gene; ERBB2, erb-b2 receptor tyrosine kinase 2 gene; ERBB4, erb-b2 receptor tyrosine kinase 24 gene; FLG, filaggrin gene; GOLPH3, golgi phosphoprotein 3 gene; ICE1, interactor of elongation complex ELL subunit 1 gene; IHC, immunohistochemistry; IL7R, interleukin 7 receptor gene; KEAP1, kelch like ECH associated protein 1 gene; LFR, intelectin 1 gene (approved symbol is now ITLN1); LPA, lepidic predominant adenocarcinoma; LRP1B, LDL receptor related protein 1B gene; MET, MET proto-oncogene, receptor tyrosine kinase; MET, MNNG HOS Transforming gene; mGGO, mixed ground glass opacity; MIA, MIA SH3 domain containing gene; MTAP, methylthioadenosine phosphorylase gene; MUC16, mucin 16, cell surface associated gene; NBPF6, NBPF member 6 gene; NOTCH2, notch 2 gene; pGGO, pure ground glass opacity; PRB1, proline rich protein BstN1 subfamily 1 (gene/pseudogene) gene; PRDM9, R/SET domain 9 gene; PTPRD, protein tyrosine phosphatase, receptor type D gene; RB1, retinoblastoma gene; RET, ret proto-oncogene gene; RICTOR, RPTOR independent companion of MTOR complex 2 gene; RYR2, ryanodine receptor 2 gene; SDHA, succinate dehydrogenase complex flavoprotein subunit A gene; SKP2, S-phase kinase associated protein 2 gene; SPTA1, spectrin alpha, erythrocytic 1 gene; STK11, serine/threonine kinase 11 gene; SYNE2, spectrin repeat containing nuclear envelope protein 1 gene; TERT, telomerase reverse transcriptase gene; TNR, tenascin R gene; TP53, tumor protein p53 gene; TTN, titin gene; TXNDC2, thioredoxin domain containing 2 gene; USH2A, usherin gene; ZFHX4, zinc finger homeobox 4 gene; ZNF536, zinc finger protein 536 gene.
Specimen Processing and Multiregion WES
To ensure the quality of specimens meeting the requirement for next-generation sequencing at Nanjing Geneseeq Technology Inc. (≥20% viable TCs), each specimen was assessed by two lung cancer pathologists (L. Z. and X. F. P.) to confirm the histologic subtype and estimate tumor purity before DNA extraction. The median tumor purity was 70% (range 50%–80%) for the current cohort of patients with LUAD. Two spatially separated regions per tumor at least 0.3 cm apart from 20 tumors plus a single tumor region from 10 tumors were subjected to DNA extraction and WES using the Illumina HiSeq4000 system (Illumina) for a median depth of 150×. DNA from histologically normal lung tissue samples (at least 2 cm from tumor, with no cancer cells detected) from the same patients served as a germline DNA control. Details of the DNA extraction, WES, and data processing are described in the Sequencing Procedure section in the Supplementary Material.
Sequencing Data Processing and Analyses
Sequencing data were aligned with the reference human genome (build hg19) with the Burrows-Wheeler Aligner (bwa-mem).17 Subsequent BAM files were further processed for deduplication, base quality recalibration, and indel realignment using the Picard suite (http://picard.sourceforge.net/) and the Genome Analysis Toolkit.18 MuTect19 with default parameters was applied to paired normal tumor BAM files for identification of somatic single-nucleotide variants (SNVs). Somatic SNVs were filtered out if there were fewer than eight alternate reads in tumor samples, the variant allele frequency in tumor the was less than 10%, there were fewer than 20 total reads in the tumor or normal samples, or the variant was present in the 1000 Genomes Project at a frequency higher than 1%. All somatic SNVs were then annotated with use of the Oncotator tool.20 Gene-level copy number variations (CNVs) were calculated by using CNVKit.21 With use of the CNVKit algorithm, relative copy ratios for each exome were calculated by correcting imbalanced library size, GC bias, sequence repeats, and target density.
To ensure the accuracy of mutation calls from WES, we validated a subset of mutations by using a customized target panel of 139 genes to a sequencing depth of 1000×. Because there was not sufficient DNA remaining for AIS and MIA, we applied the validation in only 14 IAC specimens.
GISTIC was applied22 to identify chromosomal losses or gains across samples. Copy number gain or loss was defined as chromosomal segments with a log2 ratio of at least 0.25 (gain) or less than −0.25 (loss), respectively. Tumor mutation burden (TMB) was defined as the number of missense mutations per megabase of coding regions of genome sequenced. Please see the section Supplementary for Tumor Mutation Burden in the Supplementary Materials for details. Phylogenetic trees from each tumor were constructed by using all SNVs with the Wagner parsimony method of the software package PHYLIP (PHYLogeny Inference Package). In addition, we applied ABSOLUTE software (version 1.0.6, Absolute Software Corporation)23 to estimate tumor purity in each specimen.
Histochemistry for PD-L1 and CD8 Analysis
Levels of expression of PD-L1 (determined by using SP142 antibody [Spring Bioscience, Pleasanton, CA]) and CD8 (determined by using C8/144B antibody [Gene Tech Co. Ltd., Shanghai, People’s Republic of China]) were independently scored by four pathologists (Z. L., F. P. X., Z. X., and J. S.). PD-L1 expression was quantified as the proportion of PD-L1–positive TCs or immune cells. Positive expression of PD-L1 in a given specimen was defined as at least 1% for TCs or at least 5% for immune cells. CD8 expression on lymphocytes was defined as the proportion of CD8-positive cells among nucleated cells in the stromal compartment of each specimen. The results were classified as positive (≥5%) or negative (<5%).
Detection of Pathogenic Alterations in EGFR, ALK, and MET
EGFR mutations were identified by using an amplification refractory mutation system-based kit that could detect 29 hotspot mutations (DxS EGFR Mutation Test Kit [Amoy Diagnostics, Xiamen, People’s Republic of China]). ALK receptor tyrosine kinase gene (ALK) fusion status was evaluated by IHC with a monoclonal antibody (D5F3 [Ventana/Roche Diagnostics, Mannheim, Germany]) according to the manufacturer’s instructions. The level of MNNG HOS Transforming gene (MET) amplification was assessed by c-Met/CEN7q dual-color fluorescence in situ hybridization (Vysis, Abbott Laboratories, Abbott Park, IL). Amplification was defined as (1) a MET proto-oncogene, receptor tyrosine kinase–to–centromere 7 ratio higher than 2.0, (2) an average MET gene copy number greater than 6.0 per cell, or (3) more than 10% of the TCs containing more than 15 MET proto-oncogene, receptor tyrosine kinase signals.24
External Data Sets from a Western Patient Populations
We queried the Cancer Genome Atlas,25 Broad Institute cohort,13 and Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) cohort26 for stage I LUAD with pathologically confirmed bronchioloalveolar carcinoma or mixing subtypes excluding micropapillary or solid predominant subtypes for SNV and CNV profiles. In addition, we analyzed the MSK-IMPACT cohort on the basis of response to immunotherapy.27,28
Statistical Analysis
Statistical analysis was conducted by using GraphPad Prism software (version 7.00, GraphPad Software, La Jolla, CA) and SPSS software (version 24.0, IBM Corp, Armonk, NY). Column plots, scatter dot, and box plots were generated to indicate median values and 95% confidence intervals (CIs). The chi-square, Fisher exact, and Kruskal-Wallis tests were set to calculate the significance of the differences between different subsets. All reported p values were two tailed, and significant differences were defined as those with a p value less than 0.05.
Results
Clinicopathologic Characteristics and Mutational Landscape
From January 2017 to April 2018, 208 surgically resected lung tumors were consecutively collected and eight AIS, eight MIAs, and 14 IAC (eight LPA and six APA) were included in the current study (see Supplementary Fig. 1). The clinicopathologic characteristics are summarized in Figure 1 and Supplementary Table 1. One patient with MIA was removed from subsequent analyses because he was found to have multifocal LUAD. WES yield of a total of 1012 SNVs, and 106 indels were identified (Supplementary Appendix Data) from 48 tumor samples of these 29 tumors (two spatially separated regions from eight AISs, seven MIAs, two LPAs, and two APAs plus single tumor samples from six LPAs and four APAs. Additionally, we applied deep targeted sequencing using a panel of 139 genes to DNA from 14 IAC specimens (that had sufficient DNA), and of the 117 mutations detected by targeted sequencing, 114 (97.4%) were identified by WES. According to ABSOLUTE analysis, the mean tumor purity for all samples was 59.6% (ranging from 20%–100%) (Supplementary Fig. 2). A significantly higher TMB was observed in IAC versus in MIA versus in AIS (AIS versus MIA, p = 0.04; MIA versus IAC, p = 0.015; AIS versus IAC, p < 0.001), but TMB was not associated with presence of solid components (p = 0.66) (Fig. 2C). As shown in Figure 1B and Supplementary Figure 3, EGFR was the most commonly mutated cancer gene, occurring in five of 16 AISs/MIAs and seven of 14 IACs (p = 0.46 by the Fisher exact test). On the other hand, TP53 was detected in five of 14 IACs but in only one of 16 AISs/MIAs (p = 0.04 by the chi-square test), suggesting that EGFR mutations may be early molecular events during carcinogenesis of LUAD whereas TP53 may play a more important role in acquisition of invasiveness. Otherwise, there was no significant difference in incidence of cancer gene mutations between different pathologic subtypes (Fig. 1C).
Figure 2.
Immunohistochemistry of programmed death ligand 1 (PD-L1) (tumor cells [TCs]) by SP142 and CD8-positive T cells among adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), and invasive adenocarcinoma (IAC). (A) Representative immunohistochemistry images for expression of PD-L1/CD8 in different pathological subtypes. (B) Proportion of patients with PD-L1–positive, CD8 positive, and dual-positive index. Only CD8 positivity is observed to be significantly increased in the IAC group. (C) Evaluation of tumor mutation burden (TMB) in different pathological subtypes and radiological patterns. DP, dual-positive; mut, mutation; Mb, megabase; GGO, ground glass opacity; mGGO, mixed ground glass opacity.
We next queried the mutational profiles of 70 stage I LUADs with a lepidic predominant growth pattern from the Cancer Genome Atlas,25 Broad Institute cohort,13 and MSK-IMPACT cohort.26 As expected, there was significantly less EGFR mutation (17.1% versus 43.3% [p = 0.0013 by the chi-square test]) and significantly more TP53 mutation (40.0% versus 10.0% [p = 0.0003 by the chi-square test]) in the Western patient population. In addition, KRAS and ALK alterations also appeared to be more common than in our cohort (Fig. 1D). Taken together, these results may reflect the potential different ethnic background and exposure history between Chinese and Western patient populations.29
Intertumoral Heterogeneity and Mutation Spectrum
Extensive ITH and intertumoral heterogeneity was observed in this cohort of early-stage LUAD. In total, there were 303, 207, and 484 SNVs identified in the AIS, MIA, and IAC specimens, respectively. Analysis of 19 LUADs with multiregion WES revealed a high level of genomic ITH in both preinvasive and early invasive LUAD, with an average private mutation detected in one of the two tumor regions of 68.25% for AIS, 70.3% for MIA, and 75.36% for IAC (Fig. 3A and B). Overall, the mutation spectrum was similar among the different histologic subtypes, except for a lower number of C>T transitions in patients with IAC than in patients with AIS or MIA (p = 0.0324) (Fig. 3C), likely because of more smokers in the group of patients with IAC (no smokers in the group with AIS, 14.3% in the group with MIA, and 21.4% in the group of patients with IAC).
Figure 3.
Intertumoral and intratumoral heterogeneity among different pathological subtypes. (A) Venn plots indicated intratumoral heterogeneity of somatic mutations within different pathological subtypes. (B) Venn plot of intertumoral heterogeneity of somatic mutations and tables showing mutated genes exclusively shared in the adenocarcinoma in the situ (AIS) and minimally invasive adenocarcinoma (MIA) group and the MIA or invasive adenocarcinoma (IAC) group, respectively. (C) Mutational spectra of different pathological subtypes. Box plot shows increasing discrimination of shared and unique mutations through linear pathological evolvement. (D) Visualized GISTIC analysis of copy number variants (CNVs) among three pathological subtypes and lung cancer–related genes are presented. Genes in red represent gain of copy number variants whereas genes in blue represent loss of copy number variants. AC092850.1, LOC100128002; ACAN, aggrecan gene; ALK, ALK receptor tyrosine kinase gene; ANKRD188; ankyrin repeat domain 1; ARVCF, ARVCF delta catenin family member gene; BAG1, BCL2 associated athanogene 1 gene; CENPB, centromere protein B gene; CHCD10, coiled-coil-helix-coiled-coil-helix domain containing 10 gene; CNTN1, contactin 1 gene; CSMD3, CUB and Sushi multiple domains 3 gene; CYP11A1, cytochrome P450 family 11 subfamily A member 1 gene; DNAH9, dynein axonemal heavy chain 9 gene; EMR2, adhesion G protein-coupled receptor E2 gene (this is the previous symbol; the current approved symbol is ADGRE2); ERBB2, erb-b2 receptor tyrosine kinase 2 gene; ERVW-1, endogenous retrovirus group W member 1, envelope gene; FAM230A, family with sequence similarity 230 member A gene; FGFR3, fibroblast growth factor receptor 3 gene; FGFR4, fibroblast growth factor receptor 3 gene; FGFRL1, fibroblast growth factor receptor like 1 gene; FMN2, formin 2 gene; FOLR3, folate receptor 3 gene; GOLGA6L2, golgin A6 family like 2 gene; GOLGA6L18, golgin A6 family like 10 gene (this is the previous symbol; the current approved symbol is GOLGA6L10); GOLGA8J, golgin A8 family member J gene; HDLBP, high density lipoprotein binding protein gene; IGHG1, immunoglobulin heavy chain constant gamma 1 (G1m marker) gene; IGSF3, immunoglobulin superfamily member 3 gene; KDM4E, lysine demethylase 4E gene; KIF21A, kinesin family member 21A gene; KIF25, kinesin family member 25 gene; MDC1, mediator of DNA damage checkpoint 1 gene; MDM4, MDM4 regulator of p53 gene; MED1, mediator complex subunit 1 gene; MTOR, mechanistic target of rapamycin kinase gene; MYCL1, v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog gene; MYH1, myosin heavy chain 1 gene; MYH6, myosin heavy chain 6 gene; NOTCH1, notch 1 gene; OBP2B, odorant binding protein 2B gene; OR10G9, olfactory receptor family 10 subfamily G member 9 gene; OR2T33, olfactory receptor family 2 subfamily T member 33 gene; PARP4, poly(ADP-ribose) polymerase family member 4 gene; PCDH19, protocadherin 1 gene; PI4KA, phosphatidylinositol 4 kinase alpha gene; PRB2, proline rich protein BstN1 subfamily 1 gene; PRG4, proteoglycan 4 gene; PRR21, proline rich 21 gene; RIPK4, receptor interacting serine/threonine kinase 4 gene; SAMD9L, sterile alpha motif domain containing like 9 gene; SERPINI1, serpin family I member 1 gene; SOX1, SRY-box transcription factor 1 gene; SPRY4, sprouty RTK signaling antagonist 4 gene; STK11, serine/threonine kinase 11 gene; TERT, telomerase reverse transcriptase gene; TMEM259, transmembrane protein 259 gene; TP53, tumor protein p53 gene; TTF1, transcription termination factor 1 gene; TTN, titin gene; UGT2B11, UDP glucuronosyltransferase family 2 member B11 gene; VCX3B, variable change X-linked 3B gene; XIRP1, xin actin binding repeat containing 1 gene; ZFHX3, zinc finger homeobox 3 gene; ZP3, zona pellucida glycoprotein 3 gene.
Next, we delineated the CNV profiles of this cohort of patients with LUAD and we observed a progressive increase in CNVs from AIS to MIA and IAC (Fig. 3D). CNVs in AIS involved genes related to cell adhesion and growth,30,31 including significantly higher copy number gain of EGFR, whereas in MIA and IAC, CNVs more commonly affected genes associated with metastasis and invasion32–36 (see Fig. 3D). Pathway analysis based on CNV and SNV demonstrated distinguishable pathway enrichment in AIS or MIA versus in IAC, and some of the common pathways identified in IAC were related to cell mobility (e.g., sarcomere, contractile fiber, actin filament) and oxytocin signaling (Supplementary Fig. 4).
Evolutionary Trajectory and Tumor Clonal Architecture
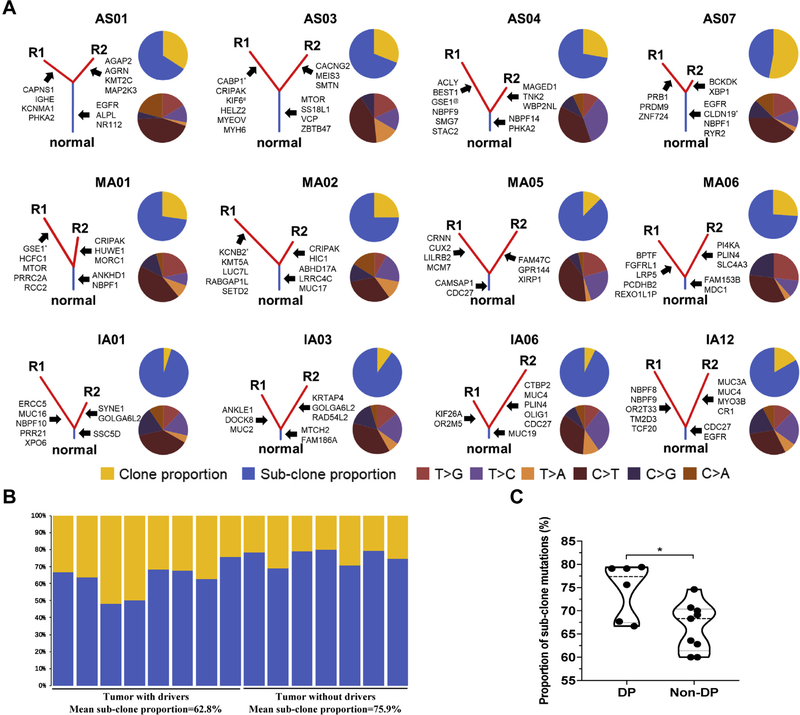
We next leveraged the multiregion sequencing data and constructed phylogenetic trees to depict the evolutionary trajectory of preinvasive and early invasive LUAD of different histologic subtypes (Fig. 4A). Canonical cancer gene mutations in EGFR, erb-b2 receptor tyrosine kinase 2 (ERBB2), NRAS, and BRAF were found exclusively as trunk mutations, whereas genes associated with cell mobility, gap junction, and metastasis, such as FSHD region gene 1 gene (FRG1), dedicator of cytokinesis 7 (DOCK7), SH3 domain binding protein 1 (SH3BP1), and gap junction protein alpha 1 (GJA1), were all subclonal mutations. ITH and branched evolution were obvious in all pathologic subtypes, with 18% to 95% of mutations being subclonal. Of particular interest, tumors without cancer gene mutations in EGFR, BRAF, NRAS, and ERBB2 showed a higher proportion of subclonal mutations (65.2% versus 79.9% [p = 0.002]) (Fig. 4B).
Figure 4.
Tumor evolutionary architecture and subgroup analysis among the different pathological subtypes. (A) Established phylogenetic tree of each pathological subtype. Blue bars represent the truncal mutations and red bars represent branched mutations, with the length of bars representing the proportion of mutated genes in each part. Some of the cancer-related genes are pointed out right next to corresponding parts. The upper pie charts show the proportion between clonal (yellow) and subclonal mutations (blue). The lower pie charts indicate the proportion of mutational spectrums, with R1 indicating region 1 and R2 indicating region 2. (B) Bar plots show the proportion of clone or subclone in tumors with or without driver genes. (C) Subgroup analysis regarding expression of CD8 and programmed death ligand 1 (PD-L1). ABHD17A, abhydrolase domain containing 17A gene; ACL, ATP-citrate lyase; AGAP2, ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 gene; AGRN, agrin gene; ALPL, alkaline phosphatase, biomineralization associated gene; ANKHD1, ankyrin repeat and KH domain containing 1 gene; ANLKE1, ankyrin repeat and LEM domain containing 1 gene; BCKDK, branched chain keto acid dehydrogenase kinase gene; BEST1, bestrophin 1 gene; BPTF, bromodomain PHD finger transcription factor gene; CABP1, alcium binding protein 1 gene; CACNG2, calcium voltage-gated channel auxiliary subunit gamma 2 gene; CAMSAP1, calmodulin regulated spectrin associated protein 1 gene; CAPNS1, calpain small subunit 1 gene; CDC27, cell division cycle 27 gene; CLDN19, claudin 19 gene; CR1, complement C3b/C4b receptor 1 (Knops blood group) gene; CRIPAK, cysteine rich PAK1 inhibitor gene; CRNN, cornulin gene; CTBP2, C-terminal binding protein 2 gene; CUX2, cut-like homeobox 2 gene; DOCK8, dedicator of cytokinesis 8 gene; DP, dual-positive; ERCC5, ERCC excision repair 5, endonuclease gene; FAM47C, family with sequence similarity 47 member C gene; FAM153B, family with sequence similarity 153 member B gene; FAM186A, family with sequence similarity 186 member A gene; FGFRL1, fibroblast growth factor receptor like 1 gene; GOLGA6L2, golgin A6 family like 2 gene; GPR144, adhesion G protein-coupled receptor D2 gene (this is the previous symbol; current approved symbol is ADGRD2); GSE1, Gse1 coiled-coil protein gene; H1C1, HIC ZBTB transcriptional repressor 1 gene; HCFC1, host cell factor C1 gene; HELZ2, helicase with zinc finger 2 gene; HUWE1, HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 gene; IGHE, immunoglobulin heavy constant epsilon gene; KCNB2, potassium voltage-gated channel subfamily B member 2 gene; KCNMA1, potassium calcium-activated channel subfamily M alpha 1 gene; KIF26A, kinesin family member 2 gene; KIF6, kinesin family member 6 gene; KMT2C, lysine methyltransferase 2C gene; KMT5A, lysine methyltransferase 5A gene; KRTAP4, keratin associated protein 4; LILRB2, leukocyte immunoglobulin like receptor B2 gene; LRP5, LDL receptor related protein 5 gene; LRRC4C, leucine rich repeat containing 4C gene; LUC7L, LUC7 like gene; MAGED1, MAGE family member D1 gene; MAP2K3, mitogen-activated protein kinase kinase 3 gene; MCM7, minichromosome maintenance complex component 7 gene; MDC1, mediator of DNA damage checkpoint 1 gene; MEIS3, MEIS3 gene; MORC1, MORC family CW-type zinc finger 1 gene; MTCH2, mitochondrial carrier 1 gene; MTOR, mechanistic target of rapamycin kinase gene; MUC2, mucin 2, oligomeric mucus/gel-forming gene; MUC3A, mucin 3A, cell surface associated gene; MUC4, mucin 4, cell surface associated gene; MUC16, mucin 16, cell surface associated gene; MUC17, mucin 17, cell surface associated gene; MUC19, mucin 19, oligomeric gene; MYO3B, myosin IIIB gene; NR112, nuclear receptor 1; NBPF1, NBPF member 1 gene; NBPF9, NBPF member 9 gene; NBPF10, NBPF member 10 gene; NBPF14, NBPF member 14 gene; OLIG1, oligodendrocyte transcription factor 1 gene; OR2M5, olfactory receptor family 2 subfamily M member 5 gene; OR2T33, olfactory receptor family 2 subfamily T member 33 gene; PCDHB2, protocadherin beta 2 gene; PHKA2, phosphorylase kinase regulatory subunit alpha 2 gene; PI4KA, phosphatidylinositol 4 kinase alpha gene; PLIN4, perilipin 4 gene; PRB1, proline rich protein BstN1 subfamily 1 (gene/pseudogene) gene; PRDM9, PR/SET domain 9 gene; PRR21, proline rich 21 gene; PRRC2A, proline rich coiled-coil 2A gene; RABGAP1L, RAB GTPase activating protein like gene; RAD54L2, RAD54 like 2 gene; RCC2, regulator of chromosome condensation 2 gene; REXO1l1P, REXO1 like, pseudogene gene; RYR2, ryanodine receptor 2 gene; SETD2, SET domain containing 2 gene; SLC4A3, solute carrier family 4 member 3 gene; SMG7, SMG7 nonsense mediated mRNA decay factor gene; SMTN, smoothelin gene; SS18L1, SS18L1 subunit of BAF chromatin remodeling complex gene; SSC5D, scavenger receptor cysteine rich family member with 5 domains gene; STAC2, SH3 and cysteine rich domain 2 gene; SYNE1, spectrin repeat containing nuclear envelope protein 1 gene; TCF20, transcription factor 20 gene; TM2D3, TM2 domain containing 3 gene; TNK2, tyrosine kinase non receptor 2 gene; VCP, vocal cord paralysis gene; WBP2NL, WBP2 N-terminal like gene; XBP1, X-box binding protein 1 gene; XIRP1, xin actin binding repeat containing 1 gene; XPO6, exportin 6 gene; ZBTB47, zinc finger and BTB domain containing 47 gene; ZNF724, zinc finger protein 724 gene.KRTAP4 refers to Keratin Associated Protein 4nuclear receptor 1NR nuclear receptor 1
Immune-Relevant Index Analysis
We next performed IHC for the expression of CD8 and PD-L1 to assess the role of T-cell immune surveillance in eight AIS, seven MIA, and 14 IAC specimens. As shown in Figure 2A, 10 of 15 AIS or MIA specimens (66.7%) had positive PD-L1 expression, as did nine of 14 IAC specimens (64.3%). On the other hand, the results of testing for CD8 was positive in eight of 16 AIS or MIA specimens (50.0%) versus in 10 of 14 IAC specimens (71.4%) (p = 0.28 by Fisher’s exact test), indicating a higher level of T-cell infiltration in IAC in this cohort of patients with LUAD. Expression of PD-L1 in AIS or MIA was similar to that in IAC (p = 0.44), whereas expression of CD8 was higher in IAC (p = 0.004 by the Man-Whitney test) (Fig. 2B). We next grouped these preinvasive and early invasive LUADs into four immune subgroups based on PD-L1 and CD8, as previously proposed37: adaptive resistance (CD8-positive/PD-L1–positive), immune tolerance (CD8-positive/PD-L1–negative), immune ignorance (CD8-negative/PD-L1–negative), and intrinsic induction (CD8-negative/PD-L1–positive), but we did not observe tumors of any certain histologic subtype to be enriched in any immune subgroups (Supplementary Table 2) on account of limited sample size. However, tumors from the adaptive resistance immune subgroup (CD8-positive/PD-L1–positive) had a significantly higher proportion subclonal mutations (p = 0.0454) (Fig. 4C).
Genomic Clustering of Early-Stage LUAD
Currently, the diagnosis of AIS, MIA, and IAC is based on morphologic assessment, which may not always be able to accurately capture the biology of tumors. We therefore performed unsupervised genomic clustering regardless of histologic subtype. As shown in Supplementary Figure 5, different regions from the same tumors almost always clustered together (26 of 29 for SNVs and 25 of 29 for CNVs), suggesting more intertumor heterogeneity than with ITH. According to either SNV- or CNV-based clustering, clustering tended to occur in IAC whereas intertwining occurred in AIS and MIA, highlighting the limitations of morphology-based histologic classifications. We next attempted to explore whether the genomic features (SNV burden, CNV burden, and SNV or CNV of particular cancer genes) were associated with recurrence-free survival, which is a more definitive surrogate for cancer biology than histologic subtype is, but we did not find any significant correlation, likely because of the small sample size, short follow-up time (median follow-up time 16.6 months), and the fact that most of these patients have been doing well with no recurrence (only one patient in the IAC group had disease recurrence).
Discussion
Previous studies had reported extensive ITH in NSCLC that may lead to a distinct response to treatment and poor prognosis.15,38 In this study, we performed multiregion sequencing through WES in a cohort of preinvasive and early invasive LUAD and demonstrated profound genomic ITH even at the stage of AIS and MIA. Consistent with previous reports,15,38 canonical cancer gene mutations in EGFR, ERBB2, NRAS, and BRAF were always truncal as early as in the AIS stage, suggesting that these mutations are very early genomic events before acquisition of invasiveness. In addition, we observed that some genes associated with cell mobility, gap junctions, and metastasis (for example, FRG1, DOCK7, SH3BP1, and GJA1) were all subclonal, implying their roles in subclonal diversification. TP53 genes were found only in MIA and IAC and not in AIS, suggesting that they are relatively later molecular events driving tumor progression. A distinct mutational profile was captured in different branches, indicating parallel evolution as the predominant evolutionary pattern in early-stage LUAD. The observed TMB and proportion of truncal mutations were much lower than in previous reports.15,38 One potential explanation is that most of the patients in the current study were female never-smokers, which is typical for the population of Chinese patients with pulmonary nodules. In addition, different genetic background may also be a contributing factor, as evidenced by the different cancer gene mutations in our cohort compared with in the Western patient population (see Fig. 1D). On the basis of the fact that most of the patients enrolled in this study were female and nonsmokers, it may potentially confound the consequence for prevalence of transitions over transversions.
Antitumor immune surveillance plays a critical role during carcinogenesis and postsurgical recurrence. We observed PD-L1 positivity in AIS, MIA, and IAC, suggesting that immunoediting has started before the invasion. However, expression of CD8 appeared to be higher in IAC, which is consistent with the results of a recent study39 implying invasion; thus, exposure to neoantigen may enhance antitumor T-cell response. The immune subgroups based on CD8 and PD-L137 may provide critical additional information on the tumor immune landscape. Likely because of small sample size, we did not observe any association between histologic subtype and immune subgroups (see Supplementary Table 2). However, tumors from the adaptive resistance immune subgroup (CD8-positive/PD-L1–positive) may have more complex genomic ITH, which was reported to be associated with postsurgical recurrence of early-stage NSCLC.15,16,40,41
The current diagnoses of AAH, AIS, MIA, and IAC are based on morphologic assessment, which may not fully capture the underlying biology of these lesions. Indeed, unsupervised clustering based on genomic features regardless of histologic subtype demonstrated that in our cohort, most AISs and MIAs were intertwined, although IAC tended to be separate from preinvasive and minimally invasive tumors (see Supplementary Fig. 4). Therefore, more definitive end points, such as recurrence-free survival and overall survival, should be integrated with molecular features to define the molecular subtypes of these lesions in future studies.
Another major caveat of the current study and most, if not all, of the previous studies is that all the analyses were based on resected tumors, which offered a single molecular snapshot of the evolutionary process of a given lesion. There is an assumption of a linear model of pathological progression from AIS to MIA and eventually to IAC. However, whether all AIS progress to MIA or IAC and whether each IAC follows the evolutionary trajectory from AIS to MIA and IAC is unknown. These questions can be addressed only by longitudinal biopsies over the course of disease progression, which is impractical in clinical practice. Clinical trials requiring longitudinal biopsies and rapidly developing technologies such as liquid biopsy and radiomics may potentially offer new insight into the evolutionary processes.
In summary, we have demonstrated that genomic heterogeneity may have emerged at the very early phase of lung carcinogenesis, after acquisition of many canonical cancer gene mutations. On the other hand, some genes associated with cell mobility, adhesion, or gap junctions (such as FRG1, DOCK7, SH3BP1, and GJA1) may be potential drivers for subclonal diversification and acquisition of invasiveness. The proportion of subclonal mutation was higher in tumors from the adaptive resistance immune subgroup (CD8-positive/PD-L1–positive), implying complicated interaction between TCs with particular molecular features and host antitumor immune surveillance.
Supplementary Material
Acknowledgments
This study was supported by the National Key R&D Program of China (grant 2016YFC1303800 [to Dr. Zhou]), Special Fund of Public Interest by the National Health and Family Control Committee (grant 201402031 [to Dr. Wu]), National Natural Science Foundation (grant 81872510 to [Dr. Zhong]), National Cancer Institute of the National Institute of Health Research (project grant R01CA234629-01 to [Dr. Jianjun Zhang]), and an American Association for Cancer Research-Johnson & Johnson Lung Cancer Innovation Science Grant (grant 18-90-52-ZHAN [to Dr. Jianjun Zhang]). Dr. Chao Zhang, Dr. Zhong, and Dr. Wu contributed to study design. Mrs. Xie, Mrs. Su, Mr. Xu, and Dr. Li contributed to pathological and immunohistochemistry evaluation. Dr. Chao Zhang, Dr. Xu-Chao Zhang, Dr. Wang, and Dr. Shao contributed to data bioinformatic analysis. Dr. Chao Zhang, Dr. Dong, Dr. Nie, Dr. Zhou, Dr. Yang, and Dr. Yang contributed to data collection. Dr. Chao Zhang and Dr. Zhong contributed to drafting of the article. Dr. Zhang participated in data analysis and interpretation and led the article’s revision. All authors read and approved the article. We thank all the contributing authors for their great effort on this article.
Disclosure: Dr. Wu received research funding from Roche and speech honoraria from AstraZeneca, Roche, Eli Lilly, Pfizer, and Sanofi, and he was a research consultant for AstraZeneca. Dr. Zhang received personal fees from BMS, AstraZeneca, Geneplus, OrigMed, and Innovent and a grant from Merckoutside the submitted work. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Data
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology at www.jto.org and at https://doi.org/10.1016/j.jtho.2019.07.031.
References
- 1.Bray F, Ferlya J, Soerjomataram I, Siegel RL, Torre LA, Jemal L. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Wright G, Manser RL, Byrnes G, Hart D, Campbell DA. Surgery for non-small cell lung cancer: systematic review and meta-analysis of randomised controlled trials. Thorax. 2006;61:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. [DOI] [PubMed] [Google Scholar]
- 4.Gettinger S, Horn L, Jackman D, et al. Five-year follow-up of nivolumab in previously treated advanced non-small-cell lung cancer: results from the CA209–003 study. J Clin Oncol. 2018;36:1675–1684. [DOI] [PubMed] [Google Scholar]
- 5.Hunter KW, Amin R, Deasy S, Ha NH, Wakefield L. Genetic insights into the morass of metastatic heterogeneity. Nat Rev Cancer. 2018;18:211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang DC, Wang W, Zhu B, Wang X, et al. Lung cancer heterogeneity and new strategies for drug therapy. Annu Rev Pharmacol Toxicol. 2018;58:531–546. [DOI] [PubMed] [Google Scholar]
- 7.National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bongiolatti S, Corzani R, Borgianni S, et al. Long-term results after surgical treatment of the dominant lung adenocarcinoma associated with ground-glass opacities. J Thorac Dis. 2018;10:4838–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Travis WD, Brambilla E, Noguchi M, et al. Diagnosis of lung adenocarcinoma in resected specimens: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch Pathol Lab Med. 2013;137:685–705. [DOI] [PubMed] [Google Scholar]
- 10.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung JJ, Yeh YC, Jeng WJ, et al. Predictive value of the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification of lung adenocarcinoma in tumor recurrence and patient survival. J Clin Oncol. 2014;32:2357–2364. [DOI] [PubMed] [Google Scholar]
- 12.Inamura K Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swanton C Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nahar R, Zhai W, Zhang T, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun. 2018;9:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36:E2423–E2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12, e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan EJ, Kim HR, Arcila ME, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang XC, Wang J, Shao GG, et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun. 2019;10:1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikawa K, Mizusaki A, Takanashi M, et al. PRG4 expression in myxoid liposarcoma maintains tumor cell growth through suppression of an antitumor cytokine IL-24. Biochem Biophys Res Commun. 2017;485:209–214. [DOI] [PubMed] [Google Scholar]
- 31.Pan W, Sun K, Tang K, et al. Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion. J Biol Chem. 2018;293:1944–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R, Song C. Loss of CSMD1 or 2 may contribute to the poor prognosis of colorectal cancer patients. Tumour Biol. 2014;35:4419–4423. [DOI] [PubMed] [Google Scholar]
- 33.Matsuda Y, Mira K, Yamane J, et al. SERPINI1 regulates epithelial-mesenchymal transition in an orthotopic implantation model of colorectal cancer. Cancer Sci. 2016;107:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan J, Ojo D, Kapoor A, et al. Neural cell adhesion protein CNTN1 promotes the metastatic progression of prostate cancer. Cancer Res. 2016;76:1603–1614. [DOI] [PubMed] [Google Scholar]
- 35.Chen N, He S, Geng J, et al. Overexpression of contactin 1 promotes growth, migration and invasion in Hs578T breast cancer cells. BMC Cell Biol. 2018;19:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roper N, Gao S, Maity S, et al. APOBEC mutagenesis and copy-number alterations are drivers of proteogenomic tumor evolution and heterogeneity in metastatic thoracic tumors. Cell Rep. 2019;26:2651–2666.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 2015;75:2139–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. [DOI] [PubMed] [Google Scholar]
- 39.Krysan K, Tran LM, Grimes BS, et al. The immune contexture associates with the genomic landscape in lung adenomatous premalignancy [e-pub ahead of print]. Cancer Res. 10.1158/0008-5472.CAN-19-0153 Accessed May 29, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee AJ, Endesfelder D, Rowan AJ, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.