Abstract
Central nervous system (CNS) malignancies are associated with poor prognosis, as well as exceptional morbidity and mortality, likely as a result of low rates of early diagnosis and limited knowledge of the tumor growth and resistance mechanisms, dissemination, and evolution in the CNS. Monitoring patients with CNS malignancies for treatment response and tumor recurrence can be challenging because of the difficulty and risks of brain biopsies and the low specificity and sensitivity of the less invasive methodologies that are currently available. Therefore, there is an urgent need to detect and validate reliable and minimally invasive biomarkers for CNS tumors that can be used separately or in combination with current clinical practices. The circulating tumor DNA (ctDNA) of cerebrospinal fluid (CSF) samples can outline the genetic landscape of entire CNS tumors effectively and is a promising, suitable biomarker, though its role in managing CNS malignancies has not been studied extensively. This review summarizes recent studies that explore the diagnostic, prognostic, and predictive roles of CSF-ctDNA as a liquid biopsy with primary and metastatic CNS malignancies.
Keywords: central nervous system (CNS) tumors, circulating tumor DNA, ctDNA, cerebrospinal fluid, CSF, liquid biopsy
Introduction
Central Nervous System (CNS) malignancies, including primary tumors of the brain or spinal cord and intracranial metastases tumors, are common worldwide and are associated with significant morbidity and mortality. The current standard methods used to diagnose and monitor CNS tumors are neuroimaging techniques, such as CT or MRI, but both methods lack sensitivity and specificity. Neuroimaging approaches provide no genetic information and little data pertaining to treatment response or disease progression.1,2 So, it is inadvisable to wait for changes in MRI/CT necessary to tailor treatment regimens while patients miss potential therapeutic opportunities.
The genomic landscape and molecular profile of a tumor are highly heterogeneous and evolve dynamically over time.3 Identifying actionable mutations and providing tailored therapies has become increasingly important. Unlike extracranial tumors, the biopsy of intracranial lesions is invasive and risky, and sampling is biased because of tumor heterogeneity. Moreover, some CNS tumors are located in the vital regions, such as the brain stem, thalamus, and spinal cord, making biopsy or surgery to obtain tumor tissues extremely difficult. As a result, there is an urgent need to seek reliable tumor biomarkers that provide real-time quantitative information regarding tumor burden and qualitative information on genetic profiles that could be used for diagnosis, prognosis, and prediction of CNS tumors.
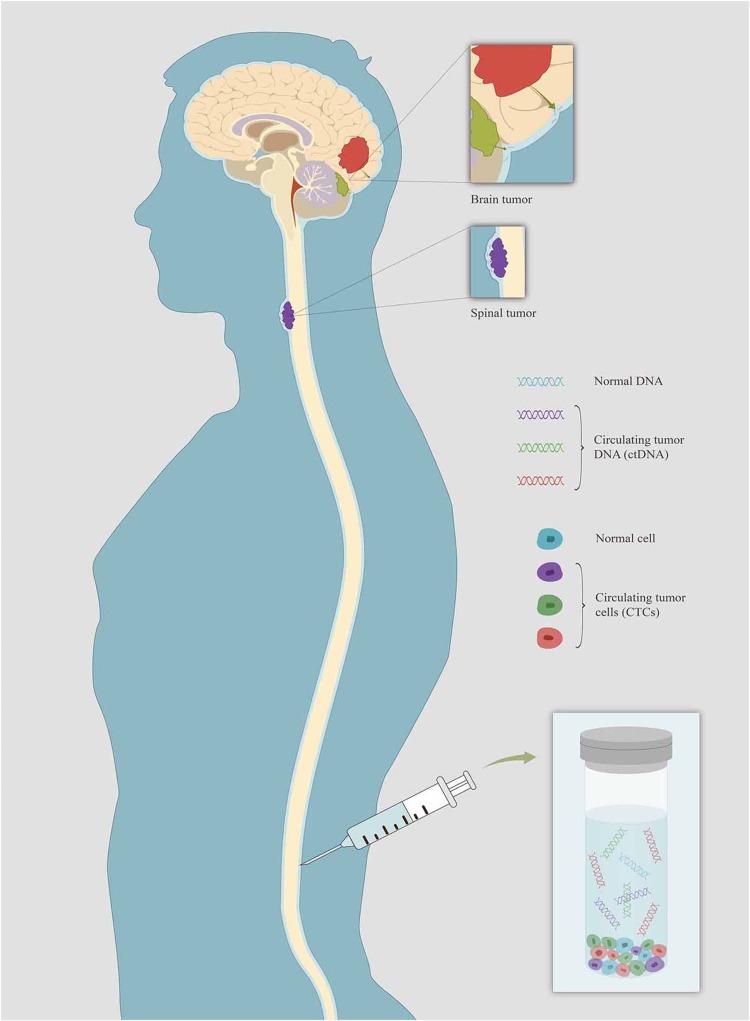
For primary or metastatic CNS tumors, the cerebrospinal fluid (CSF) is in intimate contact with tumor lesions and represents a reliable source of CNS tumor-derived circulating DNA (Figure 1). Hence, the CSF may serve as an alternative “liquid biopsy” for evaluating the ctDNA of CNS malignancies for evaluating ctDNA to characterize tumor-specific mutations, monitor tumor dynamics and genomic evolution, and assess acquired resistance mechanisms.3–6 Several past studies have reported the presence of cell-free circulating tumor DNA (ctDNA) in the CSF of patients with CNS primary tumors or metastatic lesions.7–10 This review is an overview of current studies and prospects on the application of CSF-ctDNA in the management of primary and secondary CNS malignancies, including their diagnostic, prognostic, and predictive roles (Table 1). The review also contains the biology of ctDNA and technological features.
Figure 1.
Schematic showing the source of CSF-ctDNA and CTCs from primary and metastatic CNS tumors. CSF serve as a “liquid biopsy” of CNS malignancies for evaluating ctDNA to characterize tumor-specific mutations.
Table 1.
Clinical Applications of ctDNA in Primary and Metastatic CNS Tumors
| Role | Application | Primary or Metastatic CNS Tumors | Methods | Reference |
|---|---|---|---|---|
| Diagnostic | Early detection | Medulloblastomas, ependymomas,and gliomas | WES | 10 |
| Gliomas | qPCR | 25 | ||
| Gliomas | Real-time PCR | 42 | ||
| Diffuse gliomas | Targeted exome sequencing+ddPCR | 8 | ||
| PCNSL | qPCR | 44 | ||
| BM | Digital PCR and targeted amplicon sequencing |
4 | ||
| LM | Cancer panel sequencing | 4 | ||
| LM | NGS | 61 | ||
| LM | Real-time PCR | 58 | ||
| LM | ddPCR | 7 | ||
| LM | ddPCR+NGS | 5 | ||
| LM | Real-time MS-HRM+ real-time TaqMan PCR | 62 | ||
| LM | Direct DNA sequencing | 29 | ||
| Predictive | Identification of therapeutic targets | LM | Direct DNA sequencing | 29 |
| BM | WES | 33 | ||
| BM | ARMS-PCR | 60 | ||
| CPU | CellMax SMSEQ+NGS | 68 | ||
| Identification of drug-resistant mutations | LM | NGS | 61 | |
| LM | Real-time PCR | 69 | ||
| BM | NGS | 9 | ||
| Monitoring treatment response | GBM and BMs | ddPCR | 7 | |
| Gliomas | Real-time PCR | 42 | ||
| Glioblastoma | PCR | 47 | ||
| Prognostic | Estimation of the risk for recurrence or progression | Neuroblastoma | Real-time qPCR | 10 |
| Medulloblastoma | PCR | 56 | ||
| Monitoring tumor burden | GBM and BMs | ddPCR | 7 | |
| LM | NGS | 61 | ||
| Metastatic breast cancer with BM | ddPCR+WES | 72 | ||
| Melanoma+ECD | dPCR | 32 |
Abbreviations: WES, whole exome sequencing; PCR, polymerase chain reaction; qPCR, quantitative PCR; ddPCR, droplet digital PCR; ARMS-PCR, allele refractory mutation system PCR; PCNSL, primary central nervous system lymphoma; LM, leptomeningeal metastasis; BM, brain metastasis; GBM, glioblastoma; CPU, cancer of unknown primary; NGS, next-generation exome; MS-HRM, methylation-sensitive high-resolution melting; ECD, Erdheim-Chester Disease.
Characteristics of Cell-Free Circulating DNA and Circulating Tumor DNA
Before looking at cell-free circulating tumor DNA (ctDNA), we need to first present the concept of cell-free circulating DNA (cfDNA). cfDNA consists of 70–200 base-pairs of DNA fragments released from apoptotic and necrotic cells into blood circulation or other bodily fluids, including CSF, sputum, stool, pleural fluid, urine, and saliva.11 The presence of cfDNA in the blood of healthy individuals was first reported in 1948 by Mandel and Metais.12 Leon et al then revealed for the first time, in 1977, that the concentration of cfDNA increased in the serum and plasma of pancreatic cancer patients and decreased in some patients after treatments; it was, however, not until 1994 that KRAS mutations were identified in the plasma of patients with pancreatic cancer.13,14 The subgroup of tumor-derived cfDNA, called cell-free circulating tumor DNA (ctDNA), is shed from apoptotic or necrotic cancer cells in tumor microenvironments, or from circulating tumor cells.15–18 According to reports, ctDNA accounts for only 0.01% of the total cfDNA in the circulation or up to 90%.19–21 The half-life of ctDNA is less than 1.5 hrs, providing real-time and dynamic information on tumor evolution.21 ctDNA carries information on two different changes: quantitative changes that monitor tumor burden and qualitative changes that detect tumor-specific genetic or epigenetic alterations, such as point mutations, copy number variations (CNVs), chromosomal rearrangements, and DNA methylation patterns.22 The distinction between ctDNA and normal cfDNA is based on the fact that tumor DNA harbors specific mutations that do not exist in normal DNA. These tumor-specific mutations are predominantly single base-pair substitutions.23
For patients with CNS malignancies, the detection of ctDNA in plasma is relatively insensitive.7,24–26 Less than 10% of glioma patients harbor detectable ctDNA in their plasma.26 One explanation for the low detectability of ctDNA in plasma is that the blood-brain barrier (BBB) prevents the release of ctDNA into the blood circulation and precludes the genomic characterization of CNS tumors through plasma ctDNA. Moreover, the comparatively high background of normal DNA in the plasma always dilutes tumor DNA derived from the CNS. By contrast, the CSF may serve as an optimal sampling source for CNS tumors, especially the supernatant of the CSF, likely as a result of the scarcity of cells in the CSF, which may reduce the genomic background noise caused by normal DNA.6,9,10,27 In a study performed by Pentsova et al9 63% (20 of 32) of patients with CNS metastases of solid tumors and 50% (6 of 12) of patients with primary brain tumors had high-confidence somatic mutations in the CSF assessed using next-generation sequencing (NGS). Pan et al28 also reported that mutation detection using CSF-ctDNA was more sensitive than sequencing plasma ctDNA (100% vs 38%, respectively) in brainstem glioma. More recently, tumor-specific mutations have been detected and quantified in the CSF of patients with different types of primary and metastatic CNS tumors, and these mutations have been used for clinical diagnosis and monitoring tumor burden or treatment response.4,7,9,10,24,29
Anatomical Constraints of CNS Tumors
Several studies have, in the past, demonstrated that a CSF-based liquid biopsy may not be feasible in genetically diverse, anatomically sequestered, and low-grade CNS tumors.4,7,10,30 The most important factor that is associated with CSF-ctDNA levels is anatomical sequestration. Jimenez et al described three CNS barriers – the BBB, the blood-CSF barrier of the choroid plexus and arachnoid membrane, and the CSF-parenchyma barrier of the ependymal – as potentially usable for tumor studies.31 In cases where parenchyma or arachnoid membranes surround a tumor, the diffusive ability of tumor-derived DNA to diffuse into the CSF could be limited significantly due to the presence of the blood-CSF barrier and the CSF-parenchyma barrier.4,10,30 Connolly et al30 found that ctDNA was not detected in the CSF of three patients with WHO grade II intramedullary spinal ependymoma, possibly as a result of the tumor being encased in the surrounding spinal cord parenchyma and pia mater. Wang et al10 established that all medulloblastomas, ependymomas, and high-grade gliomas directly adjoining the CSF space or the cortical surface had detectable CSF-ctDNA (100% of 21 cases; 95% CI=88–100%), whereas no ctDNA was detected in patients whose tumors were encapsulated entirely by the brain or spinal cord parenchyma. Interestingly, all four low-grade gliomas directly adjacent to the CSF reservoir did not have reliably detectable ctDNA. Wang et al revealed that analyzing ctDNA is not a dependable approach to examining and characterizing CNS tumors that are either completely encapsulated by the parenchyma or are low grade. The level of CSF-ctDNA in patients with leptomeningeal metastases (LM) is remarkably higher than that in other CNS tumors.32
Heterogeneity of CNS Metastatic Tumors
CNS metastatic tumors could harbor private and specific genetic mutations that differ from corresponding primary tumors or other extracranial metastatic lesions, termed intermetastatic heterogeneity.5,7,33 Intermetastatic heterogeneity emphasizes the importance of sequencing intracranial lesions to discover actionable mutations and provide opportunities for CNS tumors to receive targeted therapies. Intratumoral heterogeneity, in which different portions of the same tumor exhibit different genetic profiles, must be considered in addition to intermetastatic heterogeneity.34,35 A single biopsy or a surgical specimen of CNS tumors only provides a single snapshot of tumor characteristics that may not be representative of the genetic alterations of the entire tumor. In the end, the information available points to the fact that the intratumoral and the intermetastatic heterogeneity of CNS tumors need further studies.
The Role of CSF-ctDNA in Primary CNS Malignancies
ctDNA as a Diagnostic Biomarker
Primary CNS malignancies can be divided roughly into malignant gliomas, primary CNS lymphomas (PCNSL), medulloblastomas, and primary neuroectodermal tumors that are common in children.36 There are two minimally invasive methods for diagnosing CNS tumors – neuroimaging and CSF cytology – both of which have low sensitivity or specificity and are inadequate for the early detection of tumors.37 The evaluation of tumor-specific genetic alterations in CSF-ctDNA offers a useful diagnostic method for patients with primary CNS cancers to avoid high-risk diagnostic biopsies or surgical operations. In one study assessing 35 patients with primary CNS tumors (medulloblastomas, ependymomas, and gliomas), CSF-ctDNA was detected in 74% of the patients;10 two of four patients with brainstem lesions had detectable ctDNA in the CSF, as determined by whole-exome sequencing, demonstrating that ctDNA plays a significant role in the diagnosis of CNS tumors.
Moreover, some studies have indicated that the integrity of circulating DNA, measured as the ratio of longer to shorter DNA fragments, is higher in cancer patients than in healthy individuals.38–41 Shi et al25 revealed that the reliability of CSF-ctDNA or the ratio of long Alu repeats (Alu247) to short Alu repeats (Alu115) could serve as new markers for diagnosing and monitoring gliomas at apparently high specificity and sensitivity levels. Liu et al42 found that promoter hypermethylation in multiple genes of gliomas tissues is always accompanied by hypermethylation in the corresponding CSF-ctDNA, with 100% specificity, suggesting that a CSF-based multiple gene promoter hypermethylation analysis could serve as a potential biomarker for early diagnosis in cases where CNS tumor tissue samples are unavailable.
Diffuse gliomas are the most common primary malignant tumors of the CNS, with different subtypes and diverse prognosis. The genomic analysis of IDH1, IDH2, TP53, ATRX, TERT, H3F3A, and HIST1H3B gene mutations in the CSF-ctDNA of diffuse gliomas facilitates the molecular diagnosis and subclassification of diffuse gliomas in a minimally invasive manner, boosting the clinical management of diffuse gliomas and minimizing complex and high-risk surgical interventions.8 Diffuse midline gliomas typically arise in young children and are not surgically resectable due to their anatomic location (such as the thalamus or brainstem), which limits the diagnosis and molecular study of their tumor tissues.
Almost every patient with PCNSL undergoes invasive surgical procedures to obtain an accurate diagnosis. Nevertheless, the presence of a deep brain structure makes it difficult to obtain tumor tissues during operation, rendering the histopathological diagnosis of PNCSL difficult. Reportedly, PCNSL occurs in 2–13% of HIV-infected patients,43 and the presence of the Epstein-Barr virus (EBV) DNA in the CSF of HIV-infected patients could be used as a more reliable detection method to help diagnose HIV-associated PCNSL.44 Quantifying EBV DNA in the CSF using quantitative PCR improves its diagnostic specificity for PCNSL; however, the positive predictive value remains only 10%. Future studies need to examine the use of CSF-ctDNA in the early diagnosis of asymptomatic cancers or in the absence of tumor lesions in neuroimaging.
ctDNA as a Predictive Biomarker
Identifying Therapeutic Targets and Drug-Resistant Mutations
The concept of precision medicine has been studied and employed in the field of oncology medicine over the last decades, with the introduction of patient-tailored therapies facilitating the development of personalized patterns of tumors.45,46 A single biopsy or a surgical specimen of CNS tumors provides a single genetic signature that may miss a targetable mutation site. The discovery of novel mutations in the ctDNA derived from the CSF, therefore, provides new therapeutic targets that are not identifiable in tumor tissues. Information from ctDNA reflects the entire molecular makeup of CNS tumors, including information on both targetable mutations and drug resistance mechanisms under selective therapeutic stress.3 Analyzing ctDNA can reveal acquired drug-resistant mutations (that are only visible by neuroimaging or conventional tumor biomarker analyses in patients with CNS tumors) prior to tumor progression, and this information could lead to a switch to a more effective treatment regimen, as soon as possible, before the tumor is overburdened and incurable. At present, there are few studies on the identification of therapeutic targets and drug-resistant mutations in CNS tumors using CSF-ctDNA.
Monitoring Treatment Response
Treatment response is commonly assessed by clinical manifestations, radiographic imaging, and tumor biomarkers. Unfortunately, these methods are not very accurate in monitoring treatment response and do not provide information on the genetic alterations of tumors. One study analyzed CSF-ctDNA at various time points in six patients with primary or metastatic brain tumors and found that the mutant allelic frequencies (MAFs) of DNA decreased after surgical resection and response to systemic therapy but increased with tumor progression. These results indicate that CSF-ctDNA levels fluctuate longitudinally over time and follow the changes in brain tumor burden, providing biomarkers to monitor tumor progression and response to treatment.7
MGMT (O6-methylguanine-DNA methyltransferase) is a DNA repair protein that counteracts the cytotoxic effect of alkylating agents, such as temozolomide. Hypermethylating the MGMT gene promoter in glioblastoma could silence the expression of the MGMT protein, thus, increasing the sensitivity of tumor cells to temozolomide, and this might serve as a useful predictor of prolonged survival.47 In glioma, meanwhile, hypermethylating the MGMT gene promoter in the CSF is an independent prognostic factor of prolonged progression-free survival, likely due to patients benefiting more from alkylating agents.42 From a clinical standpoint, promoter hypermethylation in CSF samples might be a useful predictor of the chemosensitivity of tumors to alkylating agents, which could be used to predict treatment response in advance.
ctDNA as a Prognostic Biomarker
Surveilling a Minimal Residual Disease
Several studies have shown that ctDNA levels in the blood can be used to monitor minimal residual disease (MRD) after curative-intent surgery or other treatments and may determine which patients will experience recurrence.48–51 The amount of ctDNA is reportedly proportional to the residual tumor burden after curative-intent surgery for gastric, lung, and colorectal cancers.48,50,51 Per Chaudhuri et al,49 blood ctDNA analysis after the first treatment of lung cancer could help detect MRD earlier before macroscopic recurrence and could facilitate individualized adjuvant treatment in the case of the lowest disease burden.
For primary CNS tumors, except for those located in critical areas (eg, brainstem), the preferred treatment option is effective surgical resection. It is possible, currently, to predict which patients will be cured after surgical resection and which patients will have a residual disease leading to its recurrence, which depends to a large extent on postoperative pathological and neuroimaging criteria. However, these methods are not effective in identifying MRD. ctDNA could be used to select postoperative patients who can truly benefit from adjuvant therapies, avoiding unnecessary treatment for patients who have been cured and do not need such potentially toxic treatments. Still, few studies have ventured to investigate the role of ctDNA in CNS tumors in monitoring MRD after surgical resection.
Determining Recurrence or Progression
Post-treatment monitoring of patients with primary CNS tumors is challenging because alterations due to secondary effects of chemoradiation or pseudoprogression cannot be distinguished reliably from tumor recurrence using neuroimaging. Furthermore, the treatment of most primary CNS tumors includes total surgical resection with or without adjuvant radiotherapy and chemotherapy, but aggressive resection must be balanced with the risk of adjacent normal tissue injury, and recurrence occurs commonly in patients with subtotal resection.30 Surveillance neuroimaging and tumor biomarkers are commonly used to monitor recurrence or progression after subtotal resection of primary CNS tumors, but these are traditional techniques with low sensitivity and specificity that do not provide a timely detection recurrence or progression. The presence of ctDNA in the CSF may serve as a biomarker for the early detection of tumor recurrence or progression, even before neuroimaging and tumor biomarkers recognize relapsed lesions.
Glioblastoma (GBM) is the most widespread and devastating primary malignant brain tumor in adults.52 One of the characteristics of GBM is that the tumors are likely to recur.7 However, Woodworth et al53 found, after pathological examination of repeated resected specimen, that approximately 30% of patients with GBM who underwent a repeat resection due to suspected recurrence exhibited necrosis, scarring, or treatment-related changes rather than a recurrence of the disease. Thus, CSF-ctDNA can provide a minimally invasive method to identify true recurrence from treatment-related changes by assessing the genomic alterations of the suspected relapsed GBM.
A previous study suggested that MYCN amplification is strongly associated with tumor progression and is a significant risk factor for CNS recurrence.54 Kimoto et al55 presented the case of a 1-year-old female with stage-4 neuroblastoma after MYCN amplification in CSF-ctDNA. Complete remission was achieved after a series of effective therapies. However, the MYCN copy number in CSF-ctDNA was revealed to be high nine months later, pointing to tumor relapse in the CNS. Since the relapse of neuroblastoma is difficult to detect, changes in MYCN copy number in the CSF-ctDNA could be used as a prognostic biomarker in neuroblastoma. Per Wong et al’s56 assessment of a case involving a medulloblastoma patient, mitochondrial DNA (mtDNA) mutation was detected in the CSF one month after the completion of radiation therapy, and although there was no sign of disease progression in the patient’s MRI at the time, they noted a recurrence five months later. This case shows that by analyzing ctDNA in the CSF, relapses could be detected several months earlier than when using conventional surveillance, like MRI. Hence, CSF-ctDNA can be used for screening and catching CNS tumors before symptoms appear or neuroimaging is employed when chances for a cure are best. Moreover, early detection of tumor recurrence or progression can help avoid the unnecessary toxic effects of therapies doomed to fail and would switch to alternative regimens.
Monitoring Tumor Burden
Radiographic imaging and tumor biomarkers are commonly used in the clinical monitoring of tumor burden and tumor management. However, CNS malignancies lack reliable tumor biomarkers, which often lack specificity. CSF-ctDNA could be used as a minimally invasive technique to monitor tumor burden. The half-life of ctDNA is less than 1.5 hrs, and tumor changes can be assessed within hours instead of weeks to months.21 Changes in ctDNA may take several weeks to months, predating changes in neuroimaging or protein biomarkers.19,21 Mattos-Arruda et al7 established that the amount of ctDNA decreased with surgical resection and increased with tumor progression. So, CSF-ctDNA could be used to monitor tumor burden longitudinally.
The Role of CSF-ctDNA in Metastatic CNS Malignancies
ctDNA as a Diagnostic Biomarker
CNS metastases are devastating neurological complications of tumors; they are associated with significant morbidity and mortality. The most common site of CNS metastases is the brain parenchyma, followed by the leptomeningeal and cranial nerves.36 The diagnosis of CNS metastases is usually based on clinical presentation, primary malignant tumor, neuroimaging (CT or MRI), and brain tumor biopsies or CSF cytology. However, these methods are limited in their ability due to insufficient sensitivity or specificity.
LM is an unfavorable complication of tumors that needs to be diagnosed in the early stages of a disease. The diagnostic criteria for LM are a positive result on brain MRI and/or CSF cytology, but MRI and CSF cytology have limited sensitivity and specificity.5,57 Several studies have suggested in the past that the analysis of tumor-specific DNA in the CSF provides a useful biomarker for facilitating and supplementing the diagnosis of LM, especially in clinically suspected LM cases with negative CSF cytology or MRI; the analysis of CSF genomic mutations is apparently a more sensitive technique for diagnosing LM than CSF cytology or MRI.4,7,29,58–62 Pan et al4 identified seven somatic mutations from the CSF of an LM patient that were consistent with genetic alterations of the primary tumor. On their part, Li et al61 detected EGFR mutations in 100% of CSF samples from 26 EGFR-mutated non-small cell lung cancer (NSCLC) patients diagnosed with LM. Another study revealed that EGFR mutations in the CSF were detected in 5 of 16 LM patients (31%) with negative CSF cytology.29 In another case with suspected LM from EGFR-mutated lung adenocarcinoma, EGFR mutations were spotted in the ctDNA of the CSF, although CSF cytology was negative.63 Ballester et al5 and Shingyoji et al29 also revealed that a CSF-ctDNA analysis, in combination with MRI and CSF-cytology, could improve diagnosis, detect genetic mutations, and monitor the tumor burden of melanoma with LM.
ctDNA as a Predictive Biomarker
Identifying Therapeutic Targets
The choice of treatment regimens for patients with brain metastases (BMs) are based primarily on the information regarding the genetic profile of the primary tumor. Several past investigations have pointed out that intracranial metastases can gain new oncogenic mutations that are different from those in the corresponding primary tumors and other extracranial metastases.29,33,60,64–67 In the past, EGFR mutations have been identified in the CSF of four LM patients but not in the primary or metastatic lesions.29 The analysis of EGFR mutations in primary or metastatic lesions may also be inadequate to guide the use of Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors (EGFR-TKIs) in LM. Similar results were expressed in a study on lung adenocarcinoma patients with BMs,78 in which EGFR mutations were identified in the CSF of two patients but were not present in the matched primary tumors, further demonstrating that CNS metastases can harbor specific mutations that differ from those in primary tumors. The two patients were, as a result, subjected to EGFR-TKI treatments with subsequent tumor remission. So, it is important to identify specific genomic mutations in the CSF and provide novel targeted therapeutic agents against these mutations in the treatment of BMs. Huang et al68 identified somatic mutations in the CSF of a patient with metastatic brain adenocarcinoma of an unknown primary tumor using the SMSEQ NGS assay, and the patient was given tailored treatments that led to clinical remission. To put it briefly, a CSF-ctDNA analysis helps with the characterization of tumor mutational profiles that may refine the treatment protocols further and tailor the tumor management plan.
Detecting Drug-Resistant Mutations in Real-Time
The analyses of CSF-ctDNA to detect secondary, acquired drug resistance have already been carried out in samples of patients with metastatic CNS tumors before.9,60,61,69,70 The penetration ability of targeted agents is restricted significantly by the presence of the BBB. Lower concentration and stimulation of TKIs in the CSF leads to differences in selective pressure, resulting in different mechanisms of resistance in the CNS and peripheral system.61 The EGFR T790M mutation is the most common cause of acquired resistance in the CNS,71 but much lower frequencies of T790M in CSF or CNS lesions have been reported.60,69,70 The occurrence of this phenomenon gives rise to the existence of other specific resistance mechanisms in the CSF. Li et al61 reported that multiple copy number variations (CNVs) of MET, KRAS, and ERBB2, and the loss of the heterozygosity (LOH) of TP53 in CSF-ctDNA might be the signal of a potential metastasis and acquired resistance mechanisms of LM in EGFR-mutant NSCLC. Nanjo and colleagues61 showed that a MET copy number gain in the CSF is associated with LM’s resistance to gefitinib, but the combination of EGFR-TKIs and the MET inhibitor, crizotinib, relieved LM considerably. In a cohort of 12 patients whose CNS disease progressed when receiving TKIs targeted therapies (EGFR, ALK, HER2, or BRAF), four patients were revealed to have drug-resistant mutations in CSF-ctDNA that were absent in other metastatic tumor tissues.9 These findings suggest that regularly monitoring CSF-ctDNA to identify drug-resistant mutations early gives room for the provision of tailored treatments that target these resistant mutations when progression has not been determined by imaging yet.
ctDNA as a Prognostic Biomarker
Monitoring Tumor Burden
There is an obvious need for sensitive and specific markers to monitor the tumor dynamics of metastatic CNS lesions. Li et al’s61 suggested that the CSF-ctDNA is a promising biomarker that could help reveal dynamic changes in LM tumor burden throughout treatment. Another report established that CSF-ctDNA levels in a HER2-positive metastatic breast cancer patient with BMs matched tumor burden variations and might be more informative and sensitive than traditional imaging.72 Momtaz et al32 used digital PCR to quantify tumor-derived cfDNA in the CSF of patients with BRAF V600E or K-mutated melanoma (N=8) or BRAF V600E mutated Erdheim-Chester Disease (ECD) (N=3) with suspected CNS involvement and found that the level of CSF-ctDNA reflected tumor burden.
Technologies for ctDNA Detection
Considerable technology progress in sequencing and analyzing ctDNA has been made in recent years. Several studies have demonstrated that high throughput sequencing approaches can detect CNS tumor-derived cfDNA in the CSF.4,6 At present, there are two main categories of techniques for detecting ctDNA: targeted methods for evaluating the presence of known point mutations, and next-generation sequencing (NGS) or whole-genome sequencing (WGS) for sequencing the whole genome or pre-identified genomic regions of interest.27,73–81 The most commonly used ctDNA analysis methods are based on digital polymerase chain reaction (PCR) and NGS, which allow for the assessment of point mutations, copy number variations (CNVs), and chromosomal rearrangements.20,23
Targeted Approaches for the Analysis CSF-ctDNA
A targeted approach for analyzing ctDNA is an approach that uses known mutation information.82–85 Such methods extract ctDNAs that correspond to probes or primers containing known mutation sequences. Several PCR-based technologies are used commonly for ctDNA analysis, including ARMS (amplification refractory mutation system), BEAMing (beads, emulsion, amplification, and magnetics), and droplet digital PCR (ddPCR).76,79,80,86–89
Examples of the use of targeted approaches in tumor mutations exist. Pan et al4 applied digital PCR, targeted amplicon sequencing, and cancer panel sequencing to characterize tumor mutations in the CSF samples collected from seven patients with primary and metastatic brain tumors. The first two methods, as presented above, were used to evaluate the presence of known driver mutations to monitor brain metastasis, while the third helped to characterize the somatic mutation profile from the CSF total DNA globally; 7 somatic mutations were identified from the CSF of a patient with suspected LM. Another study, by Wang et al10 used known tumor-specific mutations as primers and a targeted sequencing method; 74% of 35 CSF samples had detectable levels of ctDNA. Ballester et al5 recently reported the use of ddPCR in combination with NGS to detect melanoma-associated mutations in the CSF-ctDNA of melanoma patients with LM, and approximately 30% of the CSF-cytology negative samples were positive for the presence of mutant DNA in the CSF after ddPCR.
Untargeted Approaches for the Analysis CSF-ctDNA
Untargeted approaches, such as WGS (whole-genome sequencing) or WES (whole-exome sequencing) aim to screen whole genomes or whole exomes and discover new genomic aberrations, enabling the discovery of novel mutations that do not require prior knowledge of specific mutations.27,79,81,90 Novel NGS-based ctDNA detection methods, including Safe-Seq (safe sequencing system), TAm-Seq (tagged amplicon deep sequencing), CAPP-Seq (cancer-personalized profiling by deep sequencing), and Ampli-Seq, have improved the detection sensitivity in identifying mutations,74,76,91–93 and there are examples of this.
In 2016, Pentsova et al9 sequenced 341 cancer-associated genes in CSF samples obtained from 53 patients with suspected or known CNS malignancies. Using NGS, they detected high-confidence somatic alterations in 63% (20 of 32) of the patients with CNS metastases of solid tumors, 50% (six of 12) of the patients with primary brain tumors, and 0% (zero of nine) of patients without CNS cancers. However, NGS-based methods are relatively more expensive than digital PCR and cannot be applied readily to monitor tumors, and they also have longer turnaround times.
Future Perspectives
Immunotherapy has finally entered the treatment phase of CNS tumors.94–100 However, there are no reliable biomarkers to screen appropriate populations and predict immunotherapy responses in CNS tumors. Tumor mutational burden (TMB), as detected by tissue NGS, is a potential predictor of immunotherapy response.101 Still, tissue biopsy of CNS tumors is invasive, making it non-ideal. A recent study published in Clinical Cancer Research confirmed that there is a correlation between high alteration number detection in ctDNA and improved response following immunotherapy;102 supporting the use of ctDNA in place of tissue biopsy. Weiss et al103 also lent their support to the use of ctDNA, revealing that the copy number variation in cell-free DNA correlated with response to immunotherapy. According to forecasts from previous studies, blood-derived ctDNA is expected to be a non-invasive predictive marker for immunotherapy soon. These forecasts seem imminent, as there is an urgent need to evaluate the value of CSF-ctDNA in CNS tumors to determine if it can be a good biomarker for assessing immune checkpoint inhibitor responses.
Up to 10% of patients treated with PD-1 antibodies will develop pseudoprogression, which occurs after the initial development of new lesions or an increase in the size of the target lesions.104 The confirmation of pseudoprogression remains a continuing challenge despite the development of immune-related response criteria (irRC).105 Cohen et al98 revealed that a patient with progressive brain metastases who presented mental issues and was treated with a single cycle of pembrolizumab showed changes 11 days later. An MRI of his brain showed the progression of CNS lesions, but histopathology results of the resected lesions revealed that all the changes might have been inflammatory effects rather than true tumor progression. Given the increasing use of immune checkpoint inhibitors in patients with brain metastases from melanoma and other diseases, early recognition of this unique response pattern can prevent the premature termination of a potentially effective treatment in the management of these patients. Following this line of understanding, Lee et al104 assessed the feasibility of using plasma ctDNA to differentiate pseudoprogression from the true progression of disease in patients with metastatic melanoma treated with anti-programmed cell death-1 antibodies. The sensitivity of ctDNA in predicting pseudoprogression from true progression was 90% (95% CI, 68–99%), and the specificity was 100% (95% CI, 60–100%). Nevertheless, given its rarity, the application of CSF-ctDNA to identify the pseudoprogression of CNS tumors in the treatment of immunotherapy requires further studies.
Limitations
While ctDNA testing is being researched extensively and developed, it remains very expensive. The detection assays are also time-consuming and require complex skills and specialized equipment. Additionally, the acquisition of results takes forever. In contrast, CSF cytology and neuroimaging are far more simple and can be performed within a few hours. Also, although the extraction of CSF through lumbar puncture is less invasive than surgery, lumbar puncture is not always feasible in the case of patients with bulky tumors that obstruct cerebrospinal fluid flow or elevate intracranial pressure. A lumbar puncture can lead to cerebral herniation, potentially hampering the use of CSF-ctDNA in patients with symptoms of hydrocephalus or elevated intracranial pressure. A recent paper published in Science unearthed somatic mutations in healthy tissues, some of them known in cancer cells, which poses a challenge to the applicability of CSF. DNA-based NGS also has limitations in detecting fusion genes and breakpoint genes at long intron areas or at complex intronic sequences. Moreover, a large panel NGS or whole-exome NGS usually requires high purity and high input DNA. One should be cautious when interpreting final bioinformatic data under extreme low DNA purity, particularly for genes with low mutant allele frequency. A CSF-based liquid biopsy may not be suitable for CNS tumors that are encased in the brain parenchyma and lack any association with the CSF. Nevertheless, even with their current limitations, circulating tumor DNAs have shown tremendous clinical and research potential in many types of CNS tumors and, if expertly explored, may become the answer to most problems faced when handling CNS tumors.
Conclusions
Finding minimally invasive techniques for the molecular analysis of primary and metastatic CNS tumors has been a daunting task. The high risk of biopsies or neurosurgical procedures and the low sensitivity and specificity of current neuroimaging modalities highlight these challenges. There is light at the end of the tunnel, though, with the analysis of ctDNA from the CSF of central nervous system tumors presenting a feasible solution and, seemingly, more accurate findings. Unlike the invasive means of acquiring tumor tissue for analysis, CSF-ctDNA could be obtained via lumbar puncture. It has the advantage of representing tumor-specific genomic information frequently and longitudinally, which could help physicians to tailor treatment according to individual molecular characteristics along the pathway of the disease. This review summarizes the application of CSF-ctDNA in the diagnosis, prognostication, and prediction of different primary and metastatic CNS tumors.
In conclusion, we believe that a CSF-ctDNA analysis could be combined with other techniques, including CSF cytology examination, neuroimaging, and clinical tumor biomarkers, in the management of CNS tumors. More studies are warranted to determine the role of CSF-ctDNA in tumor types beyond those that have been studied and to assess other nucleic acids in the CSF that may be better in the management of CNS tumors. Furthermore, extensive prospective trials are necessary to determine the extent to which the CSF-ctDNA might be informative regarding actionable genetic alterations in primary or metastatic CNS tumors.
Funding Statement
This work was supported by the Key Research and Development Program of China (grant number 2018YFC1313201).
Author Contributions
All authors contributed to data analysis, drafting, and revising the article, gave final approval for the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Baldock AL, Yagle K, Born DE, et al. Invasion and proliferation kinetics in enhancing gliomas predict idh1 mutation status. Neuro Oncol. 2014;16:779–786. doi: 10.1093/neuonc/nou027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi C, Ganji SK, DeBerardinis RJ, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in idh-mutated patients with gliomas. Nat Med. 2012;18:624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14 [DOI] [PubMed] [Google Scholar]
- 4.Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballester LY, Glitza Oliva IC, Douse DY, et al. Evaluating circulating tumor DNA from the cerebrospinal fluid of patients with melanoma and leptomeningeal disease. J Neuropathol Exp Neurol. 2018;77:628–635. doi: 10.1093/jnen/nly046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly ID, Li Y, Gephart MH, Nagpal S. The “liquid biopsy”: the role of circulating DNA and rna in central nervous system tumors. Curr Neurol Neurosci Rep. 2016;16:25. doi: 10.1007/s11910-016-0629-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Mattos-arruda L, Mayor R, Ng CK, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Ricarte F, Mayor R, Martinez-Saez E, et al. Molecular diagnosis of diffuse gliomas through sequencing of cell-free circulating tumor DNA from cerebrospinal fluid. Clin Cancer Res. 2018;24:2812–2819. doi: 10.1158/1078-0432.CCR-17-3800 [DOI] [PubMed] [Google Scholar]
- 9.Pentsova EI, Shah RH, Tang J, et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol. 2016;34:2404–2415. doi: 10.1200/JCO.2016.66.6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Springer S, Zhang M, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112:9704–9709. doi: 10.1073/pnas.1511694112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. [DOI] [PubMed] [Google Scholar]
- 12.Mandel P, Metais P. [Not available]. C R Seances Soc Biol Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 13.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 14.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3:67–71. [PubMed] [Google Scholar]
- 15.Schwarzenbach H, Alix-Panabieres C, Muller I, et al. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin Cancer Res. 2009;15:1032–1038. doi: 10.1158/1078-0432.CCR-08-1910 [DOI] [PubMed] [Google Scholar]
- 16.van der Vaart M, Pretorius PJ. Circulating DNA. Its origin and fluctuation. Ann N Y Acad Sci. 2008;1137:18–26. doi: 10.1196/nyas.2008.1137.issue-1 [DOI] [PubMed] [Google Scholar]
- 17.Schwarzenbach H, Pantel K, Kemper B, et al. Comparative evaluation of cell-free tumor DNA in blood and disseminated tumor cells in bone marrow of patients with primary breast cancer. Breast Cancer Res. 2009;11:R71. doi: 10.1186/bcr2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 19.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Bettegowda C. Applications of DNA-based liquid biopsy for central nervous system neoplasms. J Mol Diagn. 2017;19:24–34. doi: 10.1016/j.jmoldx.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 21.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Han X, Yu X, et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res. 2018;37:213. doi: 10.1186/s13046-018-0893-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz LA Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Pan W, Connolly ID, et al. Tumor DNA in cerebral spinal fluid reflects clinical course in a patient with melanoma leptomeningeal brain metastases. J Neurooncol. 2016;128:93–100. doi: 10.1007/s11060-016-2081-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi W, Lv C, Qi J, et al. Prognostic value of free DNA quantification in serum and cerebrospinal fluid in glioma patients. J Mol Neurosci. 2012;46:470–475. doi: 10.1007/s12031-011-9617-0 [DOI] [PubMed] [Google Scholar]
- 26.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmirotta R, Lovero D, Cafforio P, et al. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:1758835918794630. doi: 10.1177/1758835918794630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan C, Diplas BH, Chen X, et al. Molecular profiling of tumors of the brainstem by sequencing of csf-derived circulating tumor DNA. Acta Neuropathol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shingyoji M, Kageyama H, Sakaida T, et al. Detection of epithelial growth factor receptor mutations in cerebrospinal fluid from patients with lung adenocarcinoma suspected of neoplastic meningitis. J Thorac Oncol. 2011;6:1215–1220. doi: 10.1097/JTO.0b013e318219aaae [DOI] [PubMed] [Google Scholar]
- 30.Connolly ID, Li Y, Pan W, et al. A pilot study on the use of cerebrospinal fluid cell-free DNA in intramedullary spinal ependymoma. J Neurooncol. 2017;135:29–36. doi: 10.1007/s11060-017-2557-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jimenez AJ, Dominguez-Pinos MD, Guerra MM, Fernandez-Llebrez P, Perez-Figares JM. Structure and function of the ependymal barrier and diseases associated with ependyma disruption. Tissue Barriers. 2014;2:e28426. doi: 10.4161/tisb.28426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Momtaz P, Pentsova E, Abdel-Wahab O, et al. Quantification of tumor-derived cell free DNA(cfdna) by digital pcr (digpcr) in cerebrospinal fluid of patients with brafv600 mutated malignancies. Oncotarget. 2016;7:85430–85436. doi: 10.18632/oncotarget.13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brastianos PK, Carter SL, Santagata S, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell rna-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker NR, Hudson AL, Khong P, et al. Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci Rep. 2016;6:22477. doi: 10.1038/srep22477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verheul C, Kleijn A, Lamfers MLM. Cerebrospinal fluid biomarkers of malignancies located in the central nervous system. Handb Clin Neurol. 2017;146:139–169. [DOI] [PubMed] [Google Scholar]
- 37.Weston CL, Glantz MJ, Connor JR. Detection of cancer cells in the cerebrospinal fluid: current methods and future directions. Fluids Barriers CNS. 2011;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umetani N, Giuliano AE, Hiramatsu SH, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24:4270–4276. doi: 10.1200/JCO.2006.05.9493 [DOI] [PubMed] [Google Scholar]
- 39.Wang BG, Huang HY, Chen YC, et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966–3968. [PubMed] [Google Scholar]
- 40.Umetani N, Kim J, Hiramatsu S, et al. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative pcr for alu repeats. Clin Chem. 2006;52:1062–1069. doi: 10.1373/clinchem.2006.068577 [DOI] [PubMed] [Google Scholar]
- 41.Gao YJ, He YJ, Yang ZL, et al. Increased integrity of circulating cell-free DNA in plasma of patients with acute leukemia. Clin Chem Lab Med. 2010;48:1651–1656. doi: 10.1515/CCLM.2010.311 [DOI] [PubMed] [Google Scholar]
- 42.Liu BL, Cheng JX, Zhang W, et al. Quantitative detection of multiple gene promoter hypermethylation in tumor tissue, serum, and cerebrospinal fluid predicts prognosis of malignant gliomas. Neuro Oncol. 2010;12:540–548. doi: 10.1093/neuonc/nop064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cinque P, Scarpellini P, Vago L, Linde A, Lazzarin A. Diagnosis of central nervous system complications in hiv-infected patients: cerebrospinal fluid analysis by the polymerase chain reaction. AIDS. 1997;11:1–17. doi: 10.1097/00002030-199701000-00003 [DOI] [PubMed] [Google Scholar]
- 44.Corcoran C, Rebe K, van der Plas H, Myer L, Hardie DR. The predictive value of cerebrospinal fluid epstein-barr viral load as a marker of primary central nervous system lymphoma in hiv-infected persons. J Clin Virol. 2008;42:433–436. doi: 10.1016/j.jcv.2008.03.017 [DOI] [PubMed] [Google Scholar]
- 45.Hodson R. Precision medicine. Nature. 2016;537:S49. doi: 10.1038/537S49a [DOI] [PubMed] [Google Scholar]
- 46.McCarthy JJ, McLeod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med. 2013;5:189sr4. doi: 10.1126/scitranslmed.3005785 [DOI] [PubMed] [Google Scholar]
- 47.Hegi ME, Diserens AC, Gorlia T, et al. Mgmt gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- 48.Kim K, Shin DG, Park MK, et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfdna after surgical resection. Ann Surg Treat Res. 2014;86:136–142. doi: 10.4174/astr.2014.86.3.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.CD-17-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006 [DOI] [PubMed] [Google Scholar]
- 51.Frattini M, Gallino G, Signoroni S, et al. Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett. 2008;263:170–181. doi: 10.1016/j.canlet.2008.03.021 [DOI] [PubMed] [Google Scholar]
- 52.Touat M, Duran-Pena A, Alentorn A, Lacroix L, Massard C, Idbaih A. Emerging circulating biomarkers in glioblastoma: promises and challenges. Expert Rev Mol Diagn. 2015;15:1311–1323. doi: 10.1586/14737159.2015.1087315 [DOI] [PubMed] [Google Scholar]
- 53.Woodworth GF, Garzon-Muvdi T, Ye X, Blakeley JO, Weingart JD, Burger PC. Histopathological correlates with survival in reoperated glioblastomas. J Neurooncol. 2013;113:485–493. doi: 10.1007/s11060-013-1141-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matthay KK, Brisse H, Couanet D, et al. Central nervous system metastases in neuroblastoma: radiologic, clinical, and biologic features in 23 patients. Cancer. 2003;98:155–165. doi: 10.1002/cncr.v98:1 [DOI] [PubMed] [Google Scholar]
- 55.Kimoto T, Inoue M, Tokimasa S, et al. Detection of mycn DNA in the cerebrospinal fluid for diagnosing isolated central nervous system relapse in neuroblastoma. Pediatr Blood Cancer. 2011;56:865–867. doi: 10.1002/pbc.v56.5 [DOI] [PubMed] [Google Scholar]
- 56.Wong LJ, Lueth M, Li XN, Lau CC, Vogel H. Detection of mitochondrial DNA mutations in the tumor and cerebrospinal fluid of medulloblastoma patients. Cancer Res. 2003;63:3866–3871. [PubMed] [Google Scholar]
- 57.Chamberlain MC. Leptomeningeal metastasis. Semin Neurol. 2010;30:236–244. doi: 10.1055/s-0030-1255220 [DOI] [PubMed] [Google Scholar]
- 58.Sasaki S, Yoshioka Y, Ko R, et al. Diagnostic significance of cerebrospinal fluid egfr mutation analysis for leptomeningeal metastasis in non-small-cell lung cancer patients harboring an active egfr mutation following gefitinib therapy failure. Respir Investig. 2016;54:14–19. doi: 10.1016/j.resinv.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 59.Swinkels DW, de Kok JB, Hanselaar A, Lamers K, Boerman RH. Early detection of leptomeningeal metastasis by pcr examination of tumor-derived k-ras DNA in cerebrospinal fluid. Clin Chem. 2000;46:132–133. [PubMed] [Google Scholar]
- 60.Yang H, Cai L, Zhang Y, et al. Sensitive detection of egfr mutations in cerebrospinal fluid from lung adenocarcinoma patients with brain metastases. J Mol Diagn. 2014;16:558–563. doi: 10.1016/j.jmoldx.2014.04.008 [DOI] [PubMed] [Google Scholar]
- 61.Li YS, Jiang BY, Yang JJ, et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of egfr-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Ann Oncol. 2018;29:945–952. doi: 10.1093/annonc/mdy009 [DOI] [PubMed] [Google Scholar]
- 62.Bougel S, Lhermitte B, Gallagher G, de Flaugergues JC, Janzer RC, Benhattar J. Methylation of the htert promoter: a novel cancer biomarker for leptomeningeal metastasis detection in cerebrospinal fluids. Clin Cancer Res. 2013;19:2216–2223. doi: 10.1158/1078-0432.CCR-12-1246 [DOI] [PubMed] [Google Scholar]
- 63.Peng M, Chen C, Hulbert A, Brock MV, Yu F. Non-blood circulating tumor DNA detection in cancer. Oncotarget. 2017;8:69162–69173. doi: 10.18632/oncotarget.v8i40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saunus JM, Quinn MC, Patch AM, et al. Integrated genomic and transcriptomic analysis of human brain metastases identifies alterations of potential clinical significance. J Pathol. 2015;237:363–378. doi: 10.1002/path.4583 [DOI] [PubMed] [Google Scholar]
- 66.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paik PK, Shen R, Won H, et al. Next-generation sequencing of stage iv squamous cell lung cancers reveals an association of pi3k aberrations and evidence of clonal heterogeneity in patients with brain metastases. Cancer Discov. 2015;5:610–621. doi: 10.1158/2159-8290.CD-14-1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang WT, Lu NM, Hsu WY, et al. Csf-ctdna smseq analysis to tailor the treatment of a patient with brain metastases: a case report. Case Rep Oncol. 2018;11:68–74. doi: 10.1159/000486568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nanjo S, Arai S, Wang W, et al. Met copy number gain is associated with gefitinib resistance in leptomeningeal carcinomatosis of egfr-mutant lung cancer. Mol Cancer Ther. 2017;16:506–515. doi: 10.1158/1535-7163.MCT-16-0522 [DOI] [PubMed] [Google Scholar]
- 70.Hata A, Katakami N, Yoshioka H, et al. Spatiotemporal t790m heterogeneity in individual patients with egfr-mutant non-small-cell lung cancer after acquired resistance to egfr-tki. J Thorac Oncol. 2015;10:1553–1559. doi: 10.1097/JTO.0000000000000647 [DOI] [PubMed] [Google Scholar]
- 71.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to egfr inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siravegna G, Geuna E, Mussolin B, et al. Genotyping tumour DNA in cerebrospinal fluid and plasma of a her2-positive breast cancer patient with brain metastases. ESMO Open. 2017;2:e000253. doi: 10.1136/esmoopen-2017-000253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014 [DOI] [PubMed] [Google Scholar]
- 74.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726 [DOI] [PubMed] [Google Scholar]
- 75.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–661. doi: 10.1158/2159-8290.CD-13-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genom Proteom Bioinf. 2017;15:59–72. doi: 10.1016/j.gpb.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parsons HA, Beaver JA, Park BH. Circulating plasma tumor DNA. Adv Exp Med Biol. 2016;882:259–276. [DOI] [PubMed] [Google Scholar]
- 80.Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med. 2017;15:75. doi: 10.1186/s12916-017-0840-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 82.Chen WW, Balaj L, Liau LM, et al. Beaming and droplet digital pcr analysis of mutant idh1 mrna in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. Beaming: single-molecule pcr on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–559. doi: 10.1038/nmeth898 [DOI] [PubMed] [Google Scholar]
- 84.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112–123. doi: 10.1373/clinchem.2014.222679 [DOI] [PubMed] [Google Scholar]
- 87.Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, kras, and braf mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18:1177–1185. doi: 10.1158/1078-0432.CCR-11-0564 [DOI] [PubMed] [Google Scholar]
- 88.Taly V, Pekin D, Benhaim L, et al. Multiplex picodroplet digital pcr to detect kras mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722–1731. doi: 10.1373/clinchem.2013.206359 [DOI] [PubMed] [Google Scholar]
- 89.Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem. 2017;41:755–768. doi: 10.1159/000458736 [DOI] [PubMed] [Google Scholar]
- 90.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065 [DOI] [PubMed] [Google Scholar]
- 91.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rothe F, Laes JF, Lambrechts D, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol. 2014;25:1959–1965. doi: 10.1093/annonc/mdu288 [DOI] [PubMed] [Google Scholar]
- 93.Narayan A, Carriero NJ, Gettinger SN, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012;72:3492–3498. doi: 10.1158/0008-5472.CAN-11-4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Silk AW, Bassetti MF, West BT, Tsien CI, Lao CD. Ipilimumab and radiation therapy for melanoma brain metastases. Cancer Med. 2013;2:899–906. doi: 10.1002/cam4.2013.2.issue-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Margolin K. Ipilimumab in a Phase II trial of melanoma patients with brain metastases. Oncoimmunology. 2012;1:1197–1199. doi: 10.4161/onci.20687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mathew M, Tam M, Ott PA, et al. Ipilimumab in melanoma with limited brain metastases treated with stereotactic radiosurgery. Melanoma Res. 2013;23:191–195. doi: 10.1097/CMR.0b013e32835f3d90 [DOI] [PubMed] [Google Scholar]
- 97.Margolin K, Ernstoff MS, Hamid O, et al. Ipilimumab in patients with melanoma and brain metastases: an open-label, Phase 2 trial. Lancet Oncol. 2012;13:459–465. doi: 10.1016/S1470-2045(12)70090-6 [DOI] [PubMed] [Google Scholar]
- 98.Cohen JV, Alomari AK, Vortmeyer AO, et al. Melanoma brain metastasis pseudoprogression after pembrolizumab treatment. Cancer Immunol Res. 2016;4:179–182. doi: 10.1158/2326-6066.CIR-15-0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Knisely JP, Yu JB, Flanigan J, Sznol M, Kluger HM, Chiang VL. Radiosurgery for melanoma brain metastases in the ipilimumab era and the possibility of longer survival. J Neurosurg. 2012;117:227–233. doi: 10.3171/2012.5.JNS111929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tazi K, Hathaway A, Chiuzan C, Shirai K. Survival of melanoma patients with brain metastases treated with ipilimumab and stereotactic radiosurgery. Cancer Med. 2015;4:1–6. doi: 10.1002/cam4.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Campesato LF, Barroso-Sousa R, Jimenez L, et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to pd-1 blockade in clinical practice. Oncotarget. 2015;6:34221–34227. doi: 10.18632/oncotarget.v6i33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khagi Y, Goodman AM, Daniels GA, et al. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res. 2017;23:5729–5736. doi: 10.1158/1078-0432.CCR-17-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weiss GJ, Beck J, Braun DP, et al. Tumor cell-free DNA copy number instability predicts therapeutic response to immunotherapy. Clin Cancer Res. 2017;23:5074–5081. doi: 10.1158/1078-0432.CCR-17-0231 [DOI] [PubMed] [Google Scholar]
- 104.Lee JH, Long GV, Menzies AM, et al. Association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with anti-programmed cell death 1 antibodies. JAMA Oncol. 2018;4:717–721. doi: 10.1001/jamaoncol.2017.5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wolchok JD, Hoos A, O’Day S, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15:7412–7420. doi: 10.1158/1078-0432.CCR-09-1624 [DOI] [PubMed] [Google Scholar]