Summary
We studied the influence of AIDS restriction genes (ARGs) CCR5-Δ32, CCR2–64I, SDF1–3′A, IL10–5′A, CX3CR1-V249I, CX3CR1-T280M, and MDR1-C3435T and haplotypes of the CCR5 P1 promoter and RANTES variants −403A, In1.1C, 3′222C, and −28 G among HIV-1 infected patients on highly active antiretroviral therapy (HAART) in the Multicenter AIDS Cohort Study (MACS) and the Multicenter Hemophilia Cohort Study (MHCS). Our results indicate that several ARGs also influence therapy efficacy (ie, the success in viral suppression) and subsequent progression to AIDS while on HAART. CCR5-Δ32 decreased time to viral suppression (<200 HIV RNA copies/mL, relative hazard [RH] = 1.40; P = 0.008) and was protective against AIDS (RH = 0.11; P = <0.0001), whereas the CCR5 P1 haplotype was associated with delayed viral suppression (RNA <50 copies/mL, odds ratio [OR] = 0.65; P = 0.03) and accelerated time to AIDS (RH = 2.68; P = 0.02). SDF1–3′A viral suppression (OR = 0.61; P = 0.02) and accelerated AIDS (RH = 3.18; P = 0.009). Accelerated AIDS progression was also observed with the RANTES haplotype carrying RANTES-IN1.1C and RANTES-3′ 222C (P = 0.005 to 0.007). In contrast, the RANTES haplotype H1, which lacks suspected deleterious single-nucleotide polymorphisms, was protective against AIDS. CX3CR1-V249I seemed to accelerate viral suppression (RNA <50 copies/mL, OR = 1.27; P = 0.01). ARG influence after HAART suggests residual HIV-1 replication, and spread continues even in patients successfully suppressing detectable viral RNA.
Keywords: AIDS, AIDS restriction gene, highly active antiretroviral therapy, single-nucleotide polymorphism
Highly active antiretroviral therapy (HAART) has been shown to decrease AIDS incidence, sequelae, and mortality markedly among HIV-infected individuals.1 How-ever, replication-competent virus can be isolated from patients receiving HAART whose plasma viral loads have been clinically undetectable for years,2 resulting in viral rebound,3–5 emergence of drug-resistant strains,6,7 and progression to AIDS in many patients.8 Although multiple clinical factors influence HAART efficacy, including adherence, regimen, and disease stage at initiation, host genetics likely play an important role in treatment success.
AIDS restriction genes (ARGs) are genes with polymorphisms that influence HIV-1 infection and AIDS progression in untreated patients.9–16 It would be important to establish whether these polymorphisms continue to have an effect on progression once a person starts HAART. Specific haplotypes of the human leukocyte antigen (HLA) complex, which is associated with AIDS progression in untreated patients,17 have been linked to potentially fatal hypersensitivity to abacavir, a nucleoside reverse transcriptase inhibitor (NRTI) used in some HAART regimens.18,19 Previous studies of viral suppression and CD4 T-cell response to HAART have suggested that the positive and negative effects of CCR5-Δ32 and the CCR5 P1 promoter haplotype, respectively, observed before therapy, continue during HAART.4,20–22 In the case of CCR2–64I, however, no strong influence has been reported in a number of post-HAART studies,4,21,23–25 with the exception of a single study.26 Furthermore, there are inconsistencies between post-HAART studies, as for CX3CR124,27 and SDF1 (3′ A).23,24,28 The role of SDF1–3′ A in AIDS progression is complex, perhaps influencing the transition from the R5-trophic to more virulent X4-trophic viral pathways in untreated late-stage patients.29 This is potentially relevant to the HAART era because the R5-X4 transition of HIV-1 usually coincides with a rapid depletion of CD4 T lymphocytes, a common indicator for HAART initiation.
Other genes with polymorphisms that influence AIDS progression have not yet been studied in patients receiving HAART. Interleukin (IL)-10 upregulates major histocompatibility complex (MHC) classes I and II and production of immunomodulatory cytokines. IL10–5′ A is associated with more rapid AIDS progression,30,31 but its role in HAART is unknown. RANTES variants, including −403A, −28G, 3′ 222C, and In1.1C, influence R5 HIV-1 viral infection32,33 and AIDS progression before HAART through the CCR5-mediated pathway.10,34,35 Serum levels of RANTES significantly increase during HAART treatment36; however, the influence of host genetic variability in RANTES on HAART efficacy has not been studied.
In addition to genes associated with AIDS progression, variability of non-nucleoside reverse transcriptase inhibitor (NNRTI) and protease inhibitor (PI) plasma drug levels among patients receiving HAART suggests there may be genetic differences in drug metabolism that could lead to poor efficacy.37 One polymorphism in the multidrug resistance gene MDR1-C3435T alters gene activity and intestinal absorption of PI drugs.38 Plasma levels of antiretroviral drugs influence their efficacy39,40; however, cohort studies have been inconclusive on the effects of MDR1-C3435T on HAART success.41–44
Herein, we examined genotype and haplotype associations of 6 genes known to influence AIDS progression in untreated patients, including the polymorphisms CCR5-Δ32, CCR2–64I, SDF1–3′ A, CX3CR1-V249I, CX3CR1-T280M, and IL10–5′ A; the CCR5 P1 haplotype (+.P1.+); and 4 RANTES haplotypes based on variants −403A, −28G, 3′ 222C, and In1.1C, among patients receiving HAART in the Multicenter AIDS Cohort Study (MACS) and the Multicenter Hemophilia Cohort Study (MHCS). In addition, we evaluated MDR1-C3435T because of its possible role in drug metabolism. Our goal was to detect measurable genetic associations with 3 outcomes of HAART: (1) HIV-1 suppression, (2) AIDS progression, and (3) CD4 cell count trajectory.
METHODS
Clinical Data
Data for this study were collected from European-American men in the MACS and the MHCS. The MACS is an ongoing prospective study of HIV-1 infection among homosexual and bisexual men conducted at sites in Baltimore, Chicago, Pittsburgh, and Los Angeles. History and methods of data collection in the cohort have been described elsewhere.45 The MHCS is a study of persons with hemophilia who were infected with HIV through contaminated plasma clotting factor therapy.46 Our study was limited to European Americans to avoid confounding with racial background.
HAART was defined as (1) 2 or more NRTIs in combination with at least 1 PI or 1 NNRTI; (2) 1 NRTI in combination with at least 1 PI and at least 1 NNRTI (6%); (3) a regimen containing ritonavir and saquinavir in combination with 1 NRTI and no NNRTIs; and (4) an abacavir- or tenofovir-containing regimen of 3 or more NRTIs in the absence of PIs and NNRTIs. Combinations of zidovudine (AZT) and stavudine (d4T) with a PI or an NNRTI were not considered HAART.47 Patients in the MHCS initiated HAART between 1995 and 1999, and data collection concluded in 2000. In the MACS, the date of initiating HAARTwas defined as the midpoint between the last visit without HAART and the first visit at which HAART drugs had been administered. Only patients with <1 year between these visits were included in the study. The MACS estimated HAART start dates were between 1995 and 2003; however, 95% of the MACS participants were enrolled by the end of 2000, the end of the MHCS study. MACS data collection for the current study ended in 2003.
Genotyping
Genotyping was done using the TaqMan Assay (Perkin-Elmer, Wattham, MA) in an ABI Prism 7700 Sequence detector (Applied Biosystems, Foster City, CA), with the exception of RANTES-3′ 222C, which was identified by polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP).10 CCR5 P1 promoter haplotype (+.P1.+) was defined as the presence of single- nucleotide polymorphisms (SNPs) CCR5–627C in the absence of CCR264I and CCR5-Δ32.48 RANTES haplotype constructs contain SNPs from the RANTES promoter (−403G/A, −28C/G), the intron 1 element (In1.1T/C), and 3′ 222,10 as shown in Supplemental Figure 1 (Supplemental materials are available via the Article Plus feature at www.jaids.com. You may locate this article, then click on the Article Plus link on the right).
Statistical Analysis
We explored genetic association with 3 HAART endpoints in this study: (1) viral suppression, (2) AIDS progression, and (3) trajectory of CD4 cell count after treatment. Genetic associations with viral suppression and AIDS progression were assessed with dichotomous categoric analyses using the Fisher exact test and with Cox proportional hazards models using the log-likelihood ratio test. CD4 lymphocyte levels after initiation of HAART were analyzed with linear mixed-effects models, as described elsewhere in this article. All analyses were performed in SAS, version 9.1.3 (SAS Institute, Cary, NC). To control for HIV disease stage at the time of HAART initiation, all Cox proportional hazards models included baseline CD4 cell count and HIV RNA levels taken within 6 months before HAART initiation.
Viral Suppression
In this study, “viral suppressors” were defined as patients with an undetectable post-HAART viral RNA measurement < 200 HIV RNA copies/mL for the combined MACS and MHCS data sets, and with the more sensitive cutoff of <50 copies/mL in a separate analysis of the MACS. In both cohorts, measurement of HIV-1 RNA was obtained by reverse transcriptase (RT) PCR, using the Amplicore HIV Monitor Assay (Roche Diagnostics, Nutley, NJ); however, the MACS used a detection kit with a sensitivity of 50 copies/mL. Separate categoric analyses were performed on suppression to < 200 and < 50 RNA copies/mL. Time to suppression for the Cox proportional hazards model was defined as the time from HAART initiation to the first undetectable viral load for each patient. All survival analyses were adjusted for clinical AIDS before HAART in addition to baseline viral load and CD4 cell count. Patients were censored at their last viral load record date if they did not suppress.
Progression to AIDS
Clinical AIDS events were analyzed in patients without an AIDS diagnosis before initiating HAART. The relative hazard (RH) of developing AIDS was computed from the HAART start date for each patient. For those without a clinical AIDS event, data were censored at the last known AIDS-free date. A second set of analyses was performed on patients who successfully suppressed HIV RNA to < 200 copies/mL. In this latter analysis, the relative hazard of AIDS was computed from the date of the first nondetectable viral load. Categoric analyses were also performed comparing participants who developed AIDS after HAARTwith those who did not for both subsets described previously. AIDS survivorship analyses were adjusted for baseline viral load, CD4 cell count, and age at which therapy was started.
CD4 Lymphocyte Data
Data on CD4 cell count were available for the MACS cohort only. We used linear mixed-effects models (proc mixed, SAS version 9.1) to look at square root of CD4 cell counts and slope after HAART initiation. Patients were tested biannually and had an average of 9.7 (SD = 4.2) cell counts. We are aware that there is potential bias caused by possible closer monitoring of virally unsuppressed or sick patients. Intercept and time from HAART were included as random variables to allow intercepts to vary between subjects and for variability in time between visits, respectively.
RESULTS
A total of 695 European American men under surveillance in the MACS and MHCS longitudinal AIDS cohorts were examined for genetic associations of 6 genes involved in AIDS progression and the MDR1 gene with clinical, virologic, and immunologic outcomes after initiation of HAART. First, we tested for genetic influence on HIV RNA suppression. Of 692 patients from the MHCS and MACS cohorts, 498 (79.8%) suppressed to <200 HIV-1 copies/mL. Of the 548 MACS patients tested with more sensitive viral assays, 402 (74.2%) successfully suppressed to < 50 HIV-1 copies/mL. Second, we examined the influence of genotype on AIDS progression after HAART initiation in 539 patients who did not have an AIDS-defining condition before HAART initiation. Third, we examined the influence of genotype on CD4 lymphocyte slope in 499 MACS study participants after HAART initiation.
The results of the genetic association tests for HIV RNA viral suppression are presented in Table 1 and illustrated in Figure 1. Each allele was tested in categoric and survival tests using dominant, recessive, and codominant models, with the exception of RANTES, which, because of rarity, was only tested in a dominant model. All survival analyses were adjusted for clinical AIDS before HAART and for baseline viral load and CD4 cell count.
TABLE 1.
Categoric and Cox Proportional Hazards Analyses for Viral Suppression in the MACS and MHCS
| Viral Suppression (<200 HIV RNA Copies/mL) |
Viral Suppression (<50 HIV RNA Copies/mL) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Categoric (n = 607) |
Cox Proportional Hazard (n = 461) |
Categoric (n = 531) |
Cox Proportional Hazard (n = 413) |
||||||
| Marker | Model | OR | P | HR | P | OR | P | HR | P |
| CCR5-Δ32 | D* | 2.08 (1.16 to 3.93) | 0.0096 | 1.40 (1.10 to 1.78) | 0.008 | 1.52 (0.91 to 2.61) | 0.12 | 1.21 (0.94 to 1.56) | 0.15 |
| CCR2–64I | D | 0.84 (0.51 to 1.44) | 0.53 | 0.92 (0.69 to 1.20) | 0.53 | 0.67 (0.41 to 1.12) | 0.10 | 0.79 (0.58 to 1.08) | 0.13 |
| R | 0.82 (0.21 to 4.72) | 0.73 | 0.65 (0.27 to 1.59) | 0.31 | 0.91 (0.21 to 5.39) | 1.00 | 0.62 (0.23 to 1.05) | 0.32 | |
| C | 0.87 (0.57 to 1.32) | 0.51 | 0.90 (0.71 to 1.15) | 0.40 | 0.74 (0.49 to 1.11) | 0.14 | 0.80 (0.61 to 1.05) | 0.10 | |
| CCR5 P1 | D | 0.77 (0.50 to 1.20) | 0.24 | 0.99 (0.80 to 1.23) | 0.94 | 0.72 (0.47 to 1.11) | 0.13 | 0.88 (0.71 to 1.11) | 0.28 |
| R | 0.54 (0.28 to 1.09) | 0.07 | 0.76 (0.51 to 1.13) | 0.16 | 0.58 (0.30 to 1.16) | 0.11 | 0.65 (0.42 to 0.99) | 0.03 | |
| C | 0.76 (0.56 to 1.03) | 0.08 | 0.95 (0.11 to 0.51) | 0.51 | 0.74 (0.55 to 1.00) | 0.05 | 0.96 (0.72 to 1.02) | 0.08 | |
| SDF1–3′ A | D | 0.61 (0.40 to 0.94) | 0.02 | 0.87 (0.70 to 1.08) | 0.21 | 0.78 (0.51 to 1.18) | 0.22 | 0.86 (0.69 to 1.09) | 0.21 |
| R | 0.80 (0.34 to 2.01) | 0.53 | 0.86 (0.48 to 1.54) | 0.59 | 0.77 (0.30 to 2.09) | 0.50 | 0.74 (0.40 to 1.36) | 0.31 | |
| C | 0.69 (0.50 to 0.95) | 0.06 | 0.88 (0.73 to 1.07) | 0.20 | 0.81 (0.59 to 1.12) | 0.21 | 0.87 (0.71 to 1.06) | 0.16 | |
| IL10–5′ A | D | 0.88 (0.57 to 1.36) | 0.60 | 1.01 (0.81 to 1.26) | 0.91 | 1.07 (0.71 to 1.64) | 0.76 | 0.98 (0.78 to 1.23) | 0.87 |
| R | 0.98 (0.38 to 3.00) | 1.00 | 1.20 (0.73 to 1.96) | 0.48 | 0.98 (0.39 to 2.81) | 1.00 | 1.34 (0.81 to 2.23) | 0.28 | |
| C | 0.91 (0.65 to 1.28) | 0.60 | 1.03 (0.86 to 1.24) | 0.73 | 1.05 (0.75 to 1.46) | 0.79 | 1.02 (0.84 to 1.25) | 0.81 | |
| RANTES H1 | D | 0.30 (0.01 to 1.99) | 0.32 | 0.72 (0.41 to 1.26) | 0.27 | 0.44 (0.05 to 2.00) | 0.38 | 0.70 (0.39 to 1.25) | 0.25 |
| R | 1.13 (0.70 to 1.81) | 0.64 | 1.02 (0.81 to 1.28) | 0.87 | 0.93 (0.59 to 1.46) | 0.83 | 0.93 (0.73 to 1.12) | 0.57 | |
| C | 1.02 (0.69 to 1.52) | 0.92 | 0.98 (0.80 to 1.19) | 0.83 | 0.89 (0.61 to 1.30) | 0.54 | 0.91 (0.74 to 1.12) | 0.38 | |
| RANTES H2 | D* | 0.78 (0.40 to 1.64) | 0.48 | 0.96 (0.68 to 1.36) | 0.83 | 0.76 (0.40 to 1.49) | 0.42 | 1.03 (0.71 to 1.48) | 0.88 |
| RANTES H35 | D* | 0.94 (0.30 to 3.96) | 1.00 | 1.02 (0.58 to 1.79) | 0.94 | 0.79 (0.28 to 2.56) | 0.61 | 0.94 (0.52 to 1.73) | 0.85 |
| RANTES H4 | D* | 1.11 (0.64 to 2.01) | 0.79 | 1.04 (0.80 to 1.37) | 0.75 | 1.68 (0.94 to 3.11) | 0.07 | 1.18 (0.90 to 1.56) | 0.25 |
| RANTES H3 | D* | 0.48 (0.02 to 28.35) | 0.48 | 1.78 (0.44 to 7.22) | 0.48 | 0.17 (0 to 3.30) | 0.16 | 0.73 (0.10 to 5.24) | 0.74 |
| RANTES H5 | D* | 0.86 (0.03 to 0.86) | 1.00 | 0.94 (0.52 to 1.74) | 0.86 | 1.12 (0.34 to 4.82) | 1.00 | 0.98 (0.52 to 1.84) | 0.95 |
| MDR1-C3435T | D | 0.91 (0.55 to 1.47) | 0.73 | 0.88 (0.70 to 1.12) | 0.31 | 0.85 (0.53 to 1.34) | 0.51 | 0.93 (0.72 to 1.19) | 0.57 |
| R | 0.76 (0.46 to 1.27) | 0.25 | 0.94 (0.72 to 1.22) | 0.62 | 0.77 (0.47 to 1.28) | 0.27 | 0.93 (0.70 to 1.23) | 0.59 | |
| C | 0.87 (0.65 to 1.17) | 0.35 | 0.93 (0.79 to 1.08) | 0.34 | 0.85 (0.64 to 1.13) | 0.27 | 0.94 (0.80 to 1.11) | 0.49 | |
| CX3CR1-T280M | D | 1.08 (0.68 to 1.73) | 0.82 | 1.08 (0.86 to 1.35) | 0.53 | 1.32 (0.84 to 2.09) | 0.24 | 1.15 (0.91 to 1.46) | 0.25 |
| R | 1.21 (0.25 to 11.47) | 1.00 | 1.42 (0.67 to 3.01) | 0.39 | 1.37 (0.27 to 13.38) | 1.00 | 1.73 (0.77 to 3.90) | 0.22 | |
| C | 1.08 (0.72 to 1.61) | 0.72 | 1.09 (0.88 to 1.34) | 0.42 | 1.29 (0.87 to 1.92) | 0.21 | 1.17 (0.94 to 1.46) | 0.17 | |
| CX3CR1-V249I | D | 1.08 (0.70 to 1.66) | 0.75 | 1.13 (0.91 to 1.40) | 0.28 | 1.44 (0.95 to 2.19) | 0.07 | 1.28 (1.02 to 1.60) | 0.04 |
| R | 1.63 (0.66 to 4.84) | 0.33 | 1.29 (0.86 to 0.92) | 0.23 | 1.62 (0.68 to 4.46) | 0.34 | 1.58 (1.05 to 2.39) | 0.04 | |
| C | 1.13 (0.81 to 1.58) | 0.46 | 1.13 (0.95 to 1.34) | 0.17 | 1.38 (0.99 to 1.91) | 0.05 | 1.27 (1.06 to 1.53) | 0.01 | |
Categoric events were defined as undetectable viral RNA after HAART. Only MACS participants are included in analyses of suppression to <50 HIV RNA copies/mL. Categoric P values are from the Fisher exact test for recessive (R) and dominant (D) models, and the Mantel-Haenszel test for codominant (C) models. Cox proportional hazard P values are based on log-likelihood. Survival analyses were adjusted for clinical AIDS, baseline viral load, and baseline CD4 cell count.
Recessive and codominant models are not shown because of the few homozygotes present.
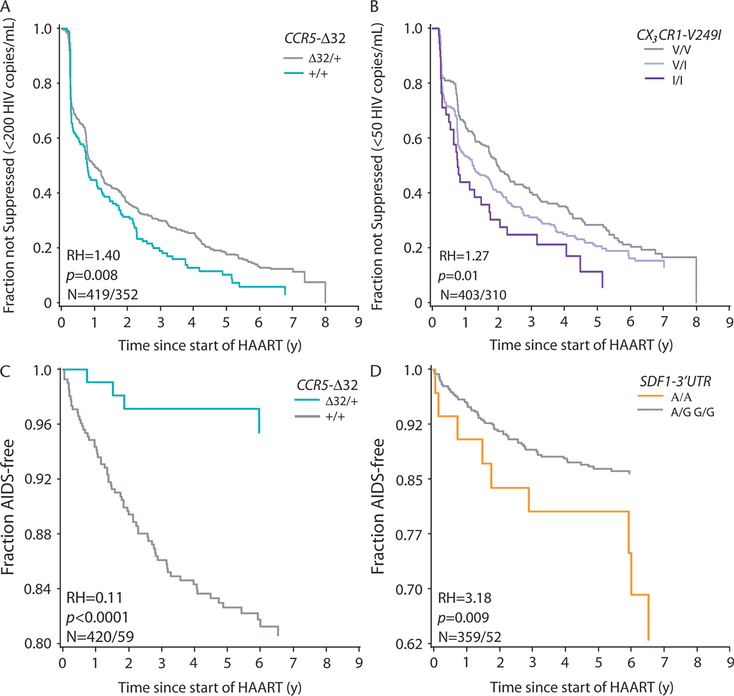
FIGURE 1.
Selected results of survival analysis of time to viral suppression (A, B) and time to AIDS after HAART (C, D). Number of patients and number of events (N), P value (P), and RH are based on Cox proportional hazards models. A, Kaplan-Meier plot of time to < 200 HIV RNA copies/mL for the CCR5-Δ32 dominant model. B, Viral suppression to < 50 HIV RNA copies/mL for the CX3CR1-V249I codominant model. C, D, Time to AIDS after HAART initiation for the CCR5-Δ 32 dominant and SDF1–3′ UTR recessive models, respectively. All hazard ratios were adjusted for baseline viral load and baseline CD4 lymphocyte count. Cox proportional hazards models for time to viral suppression were also adjusted for the occurrence of clinical AIDS before HAART initiation. Those patients with their first clinical AIDS before HAART initiation were excluded from analyses of AIDS progression. Age at HAART initiation was included as a covariate in Cox proportional hazards for AIDS progression.
CCR5-Δ32 was associated with accelerated viral suppression (< 200 RNA copies/mL, odds ratio [OR] = 2.08, P < 0.0096 for categoric analyses and RH = 1.40, P = 0.008 for Cox RH analyses; results < 50 RNA copies/mL were considered nonsignificant [ns]). The CCR5 P1 promoter allele (+.P1.+), which increases CCR5 transcription and accelerates AIDS progression, was associated with delayed viral suppression to < 50 RNA copies/mL (RH = 0.65, P = 0.03; < 200 RNA copies/mL = ns). CX3CR1-V249I allele-bearing genotypes were associated with accelerated viral suppression (< 50 RNA copies/mL: RH = 1.27, P = 0.01; < 200 RNA copies/mL = ns). In categoric models, fewer patients with SDF1–3′ A suppressed to < 200 RNA copies/mL (OR = 0.61, P = 0.02; < 50 RNA copies/mL = ns). Kaplan-Meier plots for cumulative incidence of viral suppression after HAART for CCR5-Δ32 and CX3CR1-V249I genotypes are presented in Figure 1. Other tested genotypes did not show significant associations in the same tests.
Among the HAART users with no prior AIDS diagnoses, a total of 75 developed AIDS after initiating therapy. Five ARGs (CCR5-Δ32, CCR2–64I, CCR5 P1, SDF1–3′ A, and RANTES) showed significant associations with AIDS progression (Table 2; see Fig. 1). The most dramatic influence was CCR5-Δ32, which was associated with delayed AIDS progression after HAART. This influence was detected among all AIDS-free persons who initiated HAART (ie, viral suppressors and nonsuppressors: n = 524, RH = 0.11; P ≤ 0.0001; see Table 2; see Fig. 1C). This AIDS delaying trend was also evident among viral suppressors (n = 360), although this did not achieve statistical significance. SDF-3′ A was associated with accelerated AIDS progression (RH = 3.18; P = 0.009; see Table 2; see Fig. 1D). The RANTES H1 haplotype, which lacks the RANTES-IN1.1C AIDS-accelerating enhancer sequence, and the 3′ 222C variant were strongly associated with delayed AIDS progression in patients receiving HAART, whereas RANTES H3 and RANTES H4 were associated with accelerated AIDS. RANTES H5 differs from haplotype H3 by the weakly upregulating RANTES −28G SNP.10 This haplotype did seem to be protective in our study; however, it was so rare that this requires further testing. The recessive CCR5 P1 promoter haplotype also was associated with accelerated AIDS progression after HAART (RH = 2.68; P = 0.02). Surprisingly, CCR2–64I was associated with an increased risk of AIDS in categoric analyses (OR = 1.64; P = 0.04), but these results were based on few individuals and not statistically robust. Neither MDR1 nor the other ARGs were consistently associated with post-HAART AIDS progression (see Table 2).
TABLE 2.
Categoric and Cox Proportional Hazards Models for AIDS in All Post-HAART Patients and in the Subset Who Suppressed Virus to <200 HIV RNA Copies/mL
| AIDS Progression in All Patients Receiving HAART |
AIDS Progression in Patients With Viral Suppression |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Categoric (n = 524) |
Cox Proportional Hazard (n = 420) |
Categoric (n = 376) |
Cox Proportional Hazard (n = 319) |
||||||
| Marker | Model | OR (95% CI) | P | HR (95% CI) | P | OR (95% CI) | P | HR (95% CI) | P |
| CCR5-Δ32 | D | 0.19 (0.05 to 0.52) | 0.0002 | 0.11 (0.03 to 0.45) | <0.0001 | 0.37 (0.04 to 1.61) | 0.26 | 0.40 (0.09 to 1.76) | 0.18 |
| CCR2–64I | D | 1.66 (0.91 to 2.94) | 0.09 | 1.23 (0.67 to 2.38) | 0.48 | 2.51 (0.80 to 7.21) | 0.07 | 2.11 (0.71 to 6.26) | 0.20 |
| R | 3.04 (0.48 to 14.58) | 0.13 | 2.93 (0.90 to 9.53) | 0.12 | 4.65 (0.09 to 49.91) | 0.24 | 5.19 (0.59 to 45.33) | 0.21 | |
| C | 1.64 (1.02 to 2.63) | 0.04 | 1.37 (0.82 to 2.29) | 0.25 | 2.34 (1.04 to 5.27) | 0.03 | 2.12 (0.87 to 5.20) | 0.13 | |
| CCR5 P1 | D | 1.00 (0.59 to 1.71) | 1.00 | 1.32 (0.78 to 2.25) | 0.30 | 0.69 (0.22 to 2.00) | 0.48 | 0.74 (0.26 to 2.07) | 0.56 |
| R | 1.43 (0.58 to 3.21) | 0.38 | 2.68 (1.30 to 5.52) | 0.02 | 0.65 (0.02 to 4.51) | 1.00 | 0.91 (0.12 to 6.95) | 0.93 | |
| C | 1.08 (0.74 to 1.58) | 0.68 | 1.48 (1.00 to 2.20) | 0.06 | 0.73 (0.33 to 1.63) | 0.44 | 0.81 (0.34 to 1.89) | 0.61 | |
| SDF1–3′ A | D | 1.05 (0.61 to 1.79) | 0.90 | 1.07 (0.62 to 1.86) | 0.80 | 0.59 (0.16 to 1.83) | 0.46 | 0.65 (0.20 to 2.09) | 0.46 |
| R | 2.75 (1.06 to 6.54) | 0.03 | 3.18 (1.49 to 6.77) | 0.009 | 2.36 (0.24 to 11.40) | 0.25 | 4.49 (0.99 to 20.32) | 0.10 | |
| C | 1.26 (0.85 to 1.86) | 0.26 | 1.35 (0.89 to 2.07) | 0.17 | 0.85 (0.37 to 1.96) | 0.70 | 1.04 (0.41 to 2.64) | 0.93 | |
| IL10–5′ A | D | 1.10 (0.64 to 1.86) | 0.70 | 0.95 (0.55 to 1.64) | 0.85 | 0.90 (0.29 to 2.57) | 1.00 | 0.67 (0.23 to 1.97) | 0.45 |
| R | 0.26 (0.01 to 1.65) | 0.23 | 0.38 (0.05 to 2.75) | 0.26 | 0 | 0.61 | 0 | 0.19 | |
| C | 0.96 (0.63 to 1.47) | 0.85 | 0.88 (0.55 to 1.41) | 0.60 | 0.78 (0.34 to 1.80) | 0.57 | 0.62 (0.23 to 0.31) | 0.31 | |
| MDR1-C3435T | D | 0.83 (0.47 to 1.49) | 0.48 | 0.01 (0.56 to 1.81) | 0.97 | 0.41 (0.14 to 1.18) | 0.07 | 0.60 (0.21 to 1.68) | 0.34 |
| R | 1.09 (0.58 to 2.00) | 0.77 | 1.26 (0.68 to 2.36) | 0.47 | 1.24 (0.34 to 3.80) | 0.78 | 1.51 (0.47 to 4.81) | 0.50 | |
| C | 0.96 (0.68 to 1.35) | 0.80 | 1.09 (0.74 to 1.59) | 0.66 | 0.72 (0.37 to 1.40) | 0.33 | 0.91 (0.43 to 1.91) | 0.80 | |
| CX3CR1-T280M | D | 0.70 (0.38 to 1.25) | 0.23 | 0.72 (0.39 to 1.33) | 0.28 | 0.94 (0.28 to 2.74) | 1.00 | 0.67 (0.21 to 2.12) | 0.49 |
| R | 1.27 (0.13 to 6.32) | 0.67 | 1.51 (0.21 to 11.16) | 0.70 | 2.95 (0.06 to 26.34) | 0.33 | 4.02 (0.51 to 31.57) | 0.27 | |
| C | 0.77 (0.47 to 1.27) | 0.30 | 0.77 (0.43 to 1.37) | 0.36 | 1.08 (0.45 to 2.59) | 0.87 | 0.87 (0.32 to 2.41) | 0.79 | |
| CX3CR1-V249I | D | 0.68 (0.40 to 1.16) | 0.17 | 0.91 (0.53 to 1.55) | 0.72 | 1.59 (0.56 to 4.89) | 0.36 | 1.76 (0.60 to 5.17) | 0.29 |
| R | 1.08 (0.39 to 2.60) | 0.83 | 1.43 (0.56 to 3.61) | 0.47 | 2.93 (0.66 to 10.07) | 0.08 | 3.05 (0.84 to 11.04) | 0.13 | |
| C | 0.80 (0.54 to 1.19) | 0.28 | 1.00 (0.65 to 1.54) | 0.99 | 1.71 (0.87 to 3.37) | 0.12 | 1.84 (0.86 to 3.92) | 0.12 | |
| RANTES H1 | D | 0.51 (0.12 to 2.99) | 0.40 | 0.78 (0.24 to 2.53) | 0.69 | 0.12 (0.03 to 0.80) | 0.014 | 0.20 (0.06 to 0.75) | 0.04 |
| R | 0.84 (0.48 to 1.48) | 0.50 | 0.78 (0.45 to 1.37) | 0.39 | 0.51 (0.17 to 1.58) | 0.19 | 0.39 (0.13 to 1.14) | 0.08 | |
| C | 0.81 (0.51 to 1.29) | 0.37 | 0.82 (0.52 to 1.29) | 0.40 | 0.43 (0.20 to 0.91) | 0.02 | 0.41 (0.20 to 0.86) | 0.02 | |
| RANTES H2 | D | 0.85 (0.31 to 2.00) | 0.84 | 0.68 (0.27 to 1.74) | 0.40 | 1.11 (0.12 to 5.09) | 0.70 | 1.01 (0.22 to 4.64) | 0.99 |
| RANTES H35 | D | 0.89 (0.09 to 4.05) | 1.00 | 0.90 (0.21 to 3.78) | 0.88 | 3.70 (0.36 to 19.28) | 0.14 | 4.00 (0.80 to 19.99) | 0.14 |
| RANTES H4 | D | 1.41 (0.73 to 2.60) | 0.26 | 1.65 (0.89 to 3.03) | 0.13 | 1.81 (0.48 to 5.79) | 0.34 | 2.33 (0.77 to 7.00) | 0.15 |
| R | 11.88 (0.61 to 702.66) | 0.06 | 4.59 (1.05 to 20.04) | 0.09 | 42 (2.00 to 2484.38) | 0.007 | 23.11 (4.00 to 133.55) | 0.005 | |
| C | 1.54 (0.89 to 2.66) | 0.12 | 1.71 (1.00 to 2.94) | 0.06 | 2.53 (1.03 to 6.21 | 0.04 | 3.10 (1.25 to 7.68) | 0.02 | |
| RANTES H3 | D | ∞ | 0.02 | 9.82 (2.24 to 43.12) | 0.02 | ∞ | 0.003 | 39.46 (6.78 to 229.70) | 0.002 |
| RANTES H5 | D | 0 (0 to 1.47) | 0.23 | 0 | 0.04 | 0 (0 to 6.00) | 1.00 | 0 | 0.26 |
Categoric events are the occurrence of first AIDS after HAART. Categoric P values are from the Fisher exact test for recessive (R) and dominant (D) models and by the Mantel-Haenszel test for the codominant (C) model. All P values for the Cox proportional hazard models are from log-likelihood. Cox proportional hazards models were adjusted for age at HAART initiation as well as baseline viral load and CD4 cell count. ORs and hazard ratios of infinity (∞) and 0 have undefined confidence ratios. In these cases, no people with the genotype had an event or all people with the genotype did, respectively.
95% CI indicates 95% confidence interval.
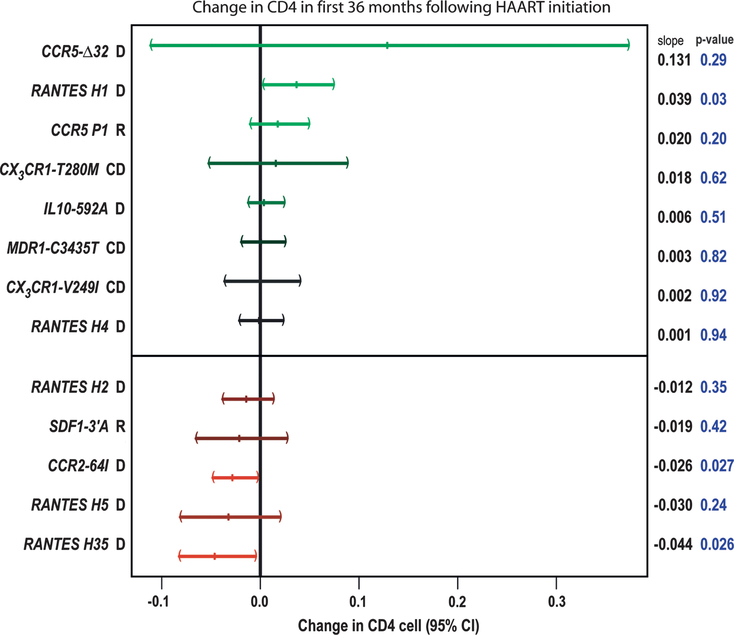
The long-term CD4 cell slope was examined using a mixed-effects model to detect genetic influence on CD4 lymphocyte recovery after HAART initiation. During an average 4.5 years of follow-up, RANTES was demonstrated to have moderate effects (H1: increase of 0.039 √CD4 cells/μL, P = 0.03; H35: lower CD4 cell slope = −0.044 √CD4 cells/μL, P = 0.026) as was CCR2–64I (lower slope = −0.03 √CD4 cells/μL, P = 0.027) (Fig. 2), both of which were consistent with the AIDS progression associations noted previously.
FIGURE 2.
Change in CD4 lymphocyte slope after HAART initiation for each genotype relative to other genotypes. Data shown are from the MACS cohort only. Patients were followed biannually for an average of 9.7 (SD = 4.2) visits after HAART.
DISCUSSION
Significant attention has been focused on understanding the role of genetics on AIDS progression. Several ARG SNPs have been identified and found to influence HIV-1 infection and AIDS progression. We sought to explore how these genes and the drug metabolism gene MDR1 affect viral suppression, AIDS progression, and CD4-cell trajectory in European-American MACS and MHCS patients undergoing HAART therapy. Although many of the ARGs did not seem to influence HAART outcomes in this study, significant associations were found in SNPs from CCR5, CCR2, CX3CR1, SDF1, and RANTES that extend their role to a post-HAART influence.
Some of the strongest positive associations were observed with the RANTES H1 haplotype and CCR5-Δ32, whereas strong deleterious associations were observed for RANTES haplotypes bearing In1.1C and 3′ 222C, the CCR5 P1 promoter haplotype (+.P1.+), and SDF1–3′ A. The significance of different SNPs in the 3 hypotheses suggests that certain SNPs play a role early in HAART treatment (ie, during viral suppression), whereas RANTES and others show strong associations with outcomes of AIDS progression and long-term CD4 lymphocyte recovery, suggesting different mechanisms. We do recognize that not all these results would remain statistically significant with corrections for multiple tests; however, it can be said that there is a priori evidence for most of the associations.
CCR5-Δ32, CCR5 P1 promotes haplotype, SFD1–3′ A, and CX3CR1-V249I were associated with different degrees of viral suppression. CCR5 is the main coreceptor used by R5 HIV to gain entry into the cell. In our study of HAART, CCR5-Δ32 was associated with accelerated viral suppression to < 200 RNA copies/mL and with delayed AIDS progression. Our viral suppression data support earlier results observed for better response4,20,49–52 and delayed disease progression28 after HAART. CCR5-Δ32 did not play a significant role in CD4 lymphocyte slope (see Fig. 2); however, it had a significant positive effect on CD4 cell counts at baseline in the mixed model analysis (data not shown), Further, it has been observed that there is a stronger association between HIV-1 RNA level and AIDS progression than between CD4 cell count slope and progression53; therefore, these results provide consistent evidence that CCR5-Δ32 is protective against disease progression after HAART.
In our study, individuals homozygous for the CCR5 P1 promoter (+.P1.+) haplotype had delayed viral RNA suppression and accelerated AIDS after HAART, which agrees with the worse prognosis found with this genotype in other posttherapy studies.21
RANTES was found to be strongly associated with HAART efficacy in our study. RANTES binds CCR5 and inhibits CCR5-mediated entry of R5 strains of HIV.32,33 In suppressors, there was significant correlation between wild-type RANTES haplotype H1, which is devoid of known deleterious SNPs in AIDS natural history studies, and CD4 lymphocyte recovery (see Fig. 2). Contrary to this, haplotype H3, which has RANTES-In1.1C, and haplotype H4, which has RANTES-In1.1C and RANTES-3′ 222C, had lower CD4 cell count recovery. These results, together with the results from CCR5-Δ32, suggest that control of HIV R5 strains is important for HAART efficacy. Although RANTES seems to be upregulated early in treatment,54 the SNPs with significant deleterious associations in our study are believed to downregulate RANTES production, and therefore may increase the number of R5 receptors for HIV, impeding control of HIV replication by HAART.
We found SDF1–3′ A to be associated with lower viral suppression and accelerated progression to AIDS. The role of SDF1 in AIDS progression has been inconsistent in the presence and absence of therapy, suggesting that the interaction between SDF1 and HIV is complicated. Although a trend (P = 0.073) for a higher proportion of non–wild-type SDF1 in HAART responders has been reported,23 in our study and that reported by Lathey et al,28 there was a negative association between SDF1–3′ A and treatment response. One reason for this inconsistency may be the involvement of SDF1 in the transition from HIV R5 to the more pathogenic HIV X4 strains, as has been suggested for pretherapy AIDS.29 Perhaps patients with SDF1–3′ A who were receiving HAART were more likely to harbor HIV X4 strains when they initiated therapy than those with the wild-type SDF1 genotype. If so, SDF1–3′ A would seem to be detrimental, although the exact mechanism of the association remains unclear.
CCR2–64I was associated with accelerated AIDS progression and lower CD4 lymphocyte recovery after HAART in our study. CCR2–64I is associated with delayed progression in untreated HIV-positive patients but seems to be insignificant in most HAART studies,4,21,23,25 with the exception of a study of 149 patients that found more rapid suppression in 30 months.26 In natural history studies, CCR2–64I has a time-restricted effect on survival. Early in infection, CCR4–64I is protective; however, studies have suggested that CCR2–64I carriers develop X4 strains more rapidly than do noncarriers, after which protection against AIDS conferred by CCR2–64I is lost.55,56 In our study, most of the long-term HAART users started HAART long after HIV infection. This could suggest that CCR2–64I would not be beneficial and may even encourage higher levels of X4 strains in HAART users.
CX3CR1 has been identified as a coreceptor of HIV.57 In our patients receiving HAART, CX3CR1-V249I was associated with earlier viral suppression. Brumme et al27 found that CX3CR1-V249I was associated with earlier immunologic failure, although we found no association with CD4 lymphocyte long-term recovery. CX3CR1-T280M is reported to be rare in cohorts that contain patients who have been infected for a long time, purportedly because of its deleterious effect; thus, inconsistencies between prior natural history studies may be spurious.58,59 Indeed, in our study, the T280M/T280M genotype was only present in only ∼1% of the population and we saw no association between this SNP and efficacy.
Despite suggestive data that MDR1-C3435T alters gene activity and intestinal absorption of substances such as PIs38 and that homozygosity for the MDR1-C3435T allele was associated with increased CD4 cell count response in a study of Swiss patients receiving HAART,26 the findings of our study agree with those of Nasi et al41 and Haas et al60 in that there does not seem to be an association between MDR1-C3435T and HAART efficacy. IL10–5′ A did not seem to have an effect on viral suppression, AIDS progression, or CD4-cell trajectory after HAART.
In conclusion, our study examined a range of genes that were predicted to influence HAART through various interactions with the HIV life cycle and drug metabolism. Although the changes at the viral and cellular levels involved in HAART efficacy are likely quite complex, our data may suggest that genes involved with the R5 viral pathway are important to therapy success. Finally, the concurrence of ARG influence after HAART supports the notion that residual HIV-1 replication and spread continue even in patients successfully suppressing detectable viral RNA. That conclusion emphasizes the urgency to discover better therapies, which unlike HAART, clear HIV effectively.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Holli Hutcheson, Cheryl Winkler, Louise McKenzie, and An Ping and 2 anonymous reviewers for useful discussion of the manuscript. Bailey Kessing, Janet Schollenberger, and Francis Yellin assisted with data acquisition.
Supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The Multicenter AIDS Cohort Study is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute. National Cancer Institute contracts include: UO1-AI-35042, 5-MO1-RR-00722 (General Clinical Research Centers), UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613, UO1-AI-35041, and N01-CO-12400.
Footnotes
None of the authors in this manuscript have commercial or other associations that pose a conflict of interest. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
REFERENCES
- 1.Palella FJ Jr, Delaney KM, Moorman AC, et al. Declining morbidity andmortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338: 853–860. [DOI] [PubMed] [Google Scholar]
- 2.Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirmthe stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. [DOI] [PubMed] [Google Scholar]
- 3.Ledergerber B, Egger M, Opravil M, et al. Clinical progression and virological failure on highly active antiretroviral therapy in HIV-1 patients: a prospective cohort study. Swiss HIV Cohort Study. Lancet. 1999;353:863–868. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita TE, Phair JP, Munoz A, et al. Immunologic and virologicresponse to highly active antiretroviral therapy in the Multicenter AIDS Cohort Study. AIDS. 2001;15:735–746. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett JA, DeMasi R, Quinn J, et al. Overview of the effectiveness oftriple combination therapy in antiretroviral-naive HIV-1 infected adults. AIDS. 2001;15:1369–1377. [DOI] [PubMed] [Google Scholar]
- 6.Cozzi LA, Sabin CA, Staszewski S, et al. Resistance profiles in patientswith viral rebound on potent antiretroviral therapy. J Infect Dis. 2000;181: 1143–1147. [DOI] [PubMed] [Google Scholar]
- 7.Wensing AM, Boucher CAN. Worldwide transmission of drug-resistantHIV. AIDS Rev. 2003;5:140–155. [PubMed] [Google Scholar]
- 8.May MT, Sterne JA, Costagliola D, et al. HIV treatment response andprognosis in Europe and North America in the first decade of highly active antiretroviral therapy: a collaborative analysis. Lancet. 2006;368:451–458. [DOI] [PubMed] [Google Scholar]
- 9.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. [DOI] [PubMed] [Google Scholar]
- 10.An P, Nelson GW, Wang L, et al. Modulating influence on HIV/AIDS byinteracting RANTES gene variants. Proc Natl Acad Sci USA. 2002;99: 10002–10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien SJ, Moore JP. The effect of genetic variation in chemokines andtheir receptors on HIV transmission and progression to AIDS. Immunol Rev. 2000;177:99–111. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004; 36:565–574. [DOI] [PubMed] [Google Scholar]
- 13.Javanbakht H, An P, Gold B, et al. Effects of human TRIM5alphapolymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354:15–27. [DOI] [PubMed] [Google Scholar]
- 14.Martin MP, Lederman MM, Hutcheson HB, et al. Association of DCSIGN promoter polymorphism with increased risk for parenteral, but not mucosal, acquisition of human immunodeficiency virus type 1 infection. J Virol. 2004;78:14053–14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An P, Duggal P, Wang LH, et al. Polymorphisms of CUL5 are associatedwith CD4+ T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bashirova AA, Bleiber G, Qi Y, et al. Consistent effects of TSG101 genetic variability on multiple outcomes of exposure to human immunodeficiency virus type 1. J Virol. 2006;80:6757–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrington M, O’Brien SJN. The influence of HLA genotype on AIDS.Annu Rev Med. 2003;54:535–551. [DOI] [PubMed] [Google Scholar]
- 18.Mallal S, Nolan D, Witt C, et al. Association between presence of HLAB*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. [DOI] [PubMed] [Google Scholar]
- 19.Martin AM, Nolan D, Gaudieri S, et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc Natl Acad Sci USA. 2004;101:4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasten S, Goldwich A, Schmitt M, et al. Positive influence of theDelta32CCR5 allele on response to highly active antiretroviral therapy (HAART) in HIV-1 infected patients. Eur J Med Res. 2000;5:323–328. [PubMed] [Google Scholar]
- 21.O’Brien TR, McDermott DH, Ioannidis JP, et al. Effect of chemokinereceptor gene polymorphisms on the response to potent antiretroviral therapy. AIDS. 2000;14:821–826. [DOI] [PubMed] [Google Scholar]
- 22.Tang J, Kaslow RA. The impact of host genetics on HIV infection anddisease progression in the era of highly active antiretroviral therapy. AIDS. 2003;17(Suppl 4):S51–S60. [DOI] [PubMed] [Google Scholar]
- 23.Bogner JR, Lutz B, Klein HG, et al. Association of highly active antiretroviral therapy failure with chemokine receptor 5 wild type. HIV Med. 2004;5:264–272. [DOI] [PubMed] [Google Scholar]
- 24.Puissant B, Roubinet F, Massip P, et al. Analysis of CCR5, CCR2,CX3CR1, and SDF1 polymorphisms in HIV-positive treated patients: impact on response to HAART and on peripheral T lymphocyte counts. AIDS Res Hum Retroviruses. 2006;22:153–162. [DOI] [PubMed] [Google Scholar]
- 25.Wit FW, van Rij RP, Weverling GJ, et al. CC chemokine receptor 5 delta32 and CC chemokine receptor 2 64I polymorphisms do not influence the virologic and immunologic response to antiretroviral combination therapy in human immunodeficiency virus type 1-infected patients. J Infect Dis. 2002;186:1726–1732. [DOI] [PubMed] [Google Scholar]
- 26.Passam AM, Zafiropoulos A, Miyakis S, et al. CCR2–64I and CXCL12 3#A alleles confer a favorable prognosis to AIDS patients undergoing HAART therapy. J Clin Virol. 2005;34:302–309. [DOI] [PubMed] [Google Scholar]
- 27.Brumme ZL, Dong WW, Chan KJ, et al. Influence of polymorphisms within the CX3CR1 and MDR-1 genes on initial antiretroviral therapy response. AIDS. 2003;17:201–208. [DOI] [PubMed] [Google Scholar]
- 28.Lathey JL, Tierney C, Chang SY, et al. Associations of CCR5, CCR2, and stromal cell-derived factor 1 genotypes with human immunodeficiency virus disease progression in patients receiving nucleoside therapy. J Infect Dis. 2001;184:1402–1411. [DOI] [PubMed] [Google Scholar]
- 29.Daar ES, Lynn HS, Donfield SM, et al. Stromal cell-derived factor-1 genotype, coreceptor tropism, and HIV type 1 disease progression. J Infect Dis. 2005;192:1597–1605. [DOI] [PubMed] [Google Scholar]
- 30.Shin HD, Winkler C, Stephens JC, et al. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci USA. 2000;97:14467–14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An P, Vlahov D, Margolick JB, et al. A tumor necrosis factor-alphainducible promoter variant of interferon-gamma accelerates CD4+ T cell depletion in human immunodeficiency virus-1-infected individuals. J Infect Dis. 2003;188:228–231. [DOI] [PubMed] [Google Scholar]
- 32.Cocchi F, DeVico AL, Garzino-Demo A, et al. Identification of RANTES,MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. [DOI] [PubMed] [Google Scholar]
- 33.Trkola A, Gordon C, Matthews J, et al. The CC-chemokine RANTES increases the attachment of human immunodeficiency virus type 1 to target cells via glycosaminoglycans and also activates a signal transduction pathway that enhances viral infectivity. J Virol. 1999;73:6370–6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez E, Dhanda R, Bamshad M, et al. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci USA. 2001; 98:5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDermott DH, Beecroft MJ, Kleeberger CA, et al. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS. 2000;14:2671–2678. [DOI] [PubMed] [Google Scholar]
- 36.Ye P, Kourtis AP, Kirschner DEN. Reconstitution of thymic function in HIV-1 patients treated with highly active antiretroviral therapy. Clin Immunol. 2003;106:95–105. [DOI] [PubMed] [Google Scholar]
- 37.Back D, Gatti G, Fletcher C, et al. Therapeutic drug monitoring in HIV infection: current status and future directions. AIDS. 2002;16(Suppl 1): S5–S37. [DOI] [PubMed] [Google Scholar]
- 38.Srinivas RV, Middlemas D, Flynn P, et al. Human immunodeficiency virus protease inhibitors serve as substrates for multidrug transporter proteins MDR1 and MRP1 but retain antiviral efficacy in cell lines expressing these transporters. Antimicrob Agents Chemother. 1998;42: 3157–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marzolini C, Telenti A, Decosterd LA, et al. Efavirenz plasma levels canpredict treatment failure and central nervous system side effects in HIV-1infected patients. AIDS. 2001;15:71–75. [DOI] [PubMed] [Google Scholar]
- 40.Burger DM, Hoetelmans RM, Hugen PW, et al. Low plasma concentrations of indinavir are related to virological treatment failure in HIV-1-infected patients on indinavir-containing triple therapy. Antivir Ther. 1998;3:215–220. [PubMed] [Google Scholar]
- 41.Nasi M, Borghi V, Pinti M, et al. MDR1 C3435T genetic polymorphism does not influence the response to antiretroviral therapy in drug-naive HIV-positive patients. AIDS. 2003;17:1696–1698. [DOI] [PubMed] [Google Scholar]
- 42.Fellay J, Marzolini C, Meaden ER, et al. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet. 2002;359:30–36. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh A, Singh KK, Powell CA, et al. An MDR1–3435 variant is associated with higher plasma nelfinavir levels and more rapid virologic response in HIV-1 infected children. AIDS. 2005;19:371–380. [DOI] [PubMed] [Google Scholar]
- 44.Verstuyft C, Marcellin F, Morand-Joubert L, et al. Absence of association between MDR1 genetic polymorphisms, indinavir pharmacokinetics and response to highly active antiretroviral therapy. AIDS. 2005;19:2127–2131. [DOI] [PubMed] [Google Scholar]
- 45.Detels R Recent scientific contributions to understanding HIV/AIDS from the Multicenter AIDS Cohort. J Epidemiol. 1992;2:511–519. [Google Scholar]
- 46.Goedert JJ, Kessler CM, Aledort LM, et al. A prospective study ofhuman immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N Engl J Med. 1989;321:1141–1148. [DOI] [PubMed] [Google Scholar]
- 47.Department of Health and Human Services/Henry J. Kaiser FamilyFoundation on Clinical Practices for the Treatment of HIV Infection. Guidelines for the Use of Antiretroviral Agents in HIV-Infected Adults and Adolescents. Washington, DC; February 2002. revision. [Google Scholar]
- 48.Martin MP, Dean M, Smith MW, et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. [DOI] [PubMed] [Google Scholar]
- 49.Valdez H, Purvis SF, Lederman MM, et al. Association of the CCR5delta32 mutation with improved response to antiretroviral therapy. JAMA. 1999;282:734. [DOI] [PubMed] [Google Scholar]
- 50.Guerin S, Meyer L, Theodorou I, et al. CCR5 delta32 deletion and response to highly active antiretroviral therapy in HIV-1-infected patients. AIDS. 2000;14:2788–2790. [DOI] [PubMed] [Google Scholar]
- 51.Laurichesse JJ, Persoz A, Theodorou I, et al. Improved virological response to highly active antiretroviral therapy in HIV-1-infected patients carrying the CCR5 Delta32 deletion. HIV Med. 2007;8:213–219. [DOI] [PubMed] [Google Scholar]
- 52.Brumme ZL, Henrick BM, Brumme CJ, et al. Short communication. Association of the CCR5delta32 mutation with clinical response and >5-year survival following initiation of first triple antiretroviral regimen. Antivir Ther. 2005;10:849–853. [PubMed] [Google Scholar]
- 53.Mellors JW, Rinaldo CR Jr, Gupta P, et al. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272: 1167–1170. [DOI] [PubMed] [Google Scholar]
- 54.Ye P, Kazanjian P, Kunkel SL, et al. Lack of good correlation of serum CC-chemokine levels with human immunodeficiency virus-1 disease stage and response to treatment. J Lab Clin Med. 2004;143:310–319. [DOI] [PubMed] [Google Scholar]
- 55.van Rij RP, de Roda Husman AM, Brouwer M, et al. Role of CCR2 genotype in the clinical course of syncytium-inducing (SI) or non-SI human immunodeficiency virus type 1 infection and in the time to conversion to SI virus variants. J Infect Dis. 1998;178:1806–1811. [DOI] [PubMed] [Google Scholar]
- 56.Mulherin SA, O’Brien TR, Ioannidis JP, et al. Effects of CCR5-Delta32 and CCR2–64I alleles on HIV-1 disease progression: the protection varies with duration of infection. AIDS. 2003;17:377–387. [DOI] [PubMed] [Google Scholar]
- 57.Reeves JD, McKnight A, Potempa S, et al. CD4-independent infection byHIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231:130–134. [DOI] [PubMed] [Google Scholar]
- 58.Faure S, Meyer L, Genin E, et al. Deleterious genetic influence ofCX3CR1 genotypes on HIV-1 disease progression. J Acquir Immune Defic Syndr. 2003; 32:335–337. [DOI] [PubMed] [Google Scholar]
- 59.McDermott DH, Fong AM, Yang Q, et al. Chemokine receptor mutantCX3CR1-M280 has impaired adhesive function and correlates with protection from cardiovascular disease in humans.J Clin Invest. 2003;111:1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haas DW, Wu H, Li H, et al. MDR1 gene polymorphisms and phase 1 viral decay during HIV-1 infection: an adult AIDS Clinical Trials Group Study. J Acquir Immune Defic Syndr. 2003;34:295–298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.