ABSTRACT
We aimed to characterize the expression pattern of UBE2C in rectal carcinoma and elucidate its fundamental involvement in rectal carcinoma biology. The relative expression of UBE2C in rectal carcinoma was determined by immunoblotting and QPCR. The cell viability was measured using CCK-8 assay. The anchorage-independent growth was evaluated with soft agar assay. Cell apoptosis was detected by Annexin V-PI staining. Invasion capacity was determined by transwell chamber. Tumor growth was monitored in xenograft mice model. We demonstrated that UBE2C was aberrantly up-regulated in rectal carcinoma. SiRNA-mediated knockdown of UBE2C significantly inhibited cell viability, proliferation, colony formation, invasion and induced apoptosis in vitro. Moreover, tumor growth in xenograft mice was markedly suppressed upon UBE2C silencing. Furthermore, we have identified that miR-381 was involved in regulation of UBE2C in rectal carcinoma. Here we demonstrated that UBE2C was over-expressed in rectal carcinoma, which was subjected to miR-381 modulation and in turn promoted cell proliferation, invasion and inhibited cell apoptosis.
KEYWORDS: apoptosis, invasion, miR-381, proliferation, rectal carcinoma, UBE2C, xenograft mice model
Introduction
Globally, it's estimated that more than 1 million people was diagnosed with colorectal cancer each year and approximately 715,000 deaths were claimed by this malignance at 2010.1 As of 2012, colorectal cancer was the second most common cancer in women and the third most common in men, and simultaneously the fourth common cause of cancer related death after lung, stomach and liver cancer.2 The majority of incidence of colorectal cancer are related to age and unhealthy lifestyle, which underlying genetic deficiency only account for a small fraction of all cases.3 The common recognized risk factors include diet, obesity, smoking and lack of physical activity.4 The chronic inflammatory bowel disease also significantly contributed to the incidence of colorectal cancer.5 The clinical diagnosis of this disease relies heavily on the medical imaging of biopsy obtained from the colon during sigmoidoscopy or colonoscopy. The clinical treatment options for this disease include some combination of surgery, radiation therapy, chemotherapy and targeted therapy based on variety determinant factors such as patient's health and stage of the tumor.6 The localized and early-diagnosed malignance may be curative with complete surgical removal. Chemotherapy is the regularly applicable while in combination with surgery for some cases dependent on the stage of the disease, especially for the metastasis to the lymph nodes and distant organs. Similarly, radiotherapy was adopted in the neoadjuvant and adjuvant setting for some stages of rectal cancer. Histologically, despite of the high similarity between rectal and colon cancers in many facets, their clinical managements are quite distinct mainly due to the complicated localization and structure of rectum, which severely impose great challenge for complete surgical excision.7 Therefore, elucidation of the novel molecular mechanisms underlying incidence and progression of rectal carcinoma is particularly critical for new therapeutics exploitation and early diagnosis.
UBE2C encodes a member of the E2 ubiquitin-conjugating enzyme family, which physiologically accepts ubiquitin from the E1 complex and catalyzes lys-11 and lys-48-specific polyubiquitination of the anaphase promoting complex/cyclosome (APC/C) substrates, and eventually leads to the degradation of APC/C substrates by the ubiquitin-proteasome system and promotes mitotic exit and cell cycle progression.8 More recently, accumulative evidences suggested that UBE2C might involve in cancer progression. In respect to colorectal cancer, the previous study detected aberrantly high expression of UBE2C in advanced colon cancer using DNA microarray and FISH technologies.9 However, the mechanistic involvement of overexpression of UBE2C in this disease is still to be elucidated. In another way, the expression pattern of UBE2C in rectal carcinoma has not been characterized so far and its potential role in tumor biology of this disease remains unclear. To address this question, here we first set out to determine the expression level of UBE2C in clinical rectal carcinoma samples, and the impact on malignant behavior of rectal carcinoma imposed by UBE2C manipulation has been fully investigated. Furthermore, we have predicted and validated that miR-381 specifically modulated UBE2C expression at the post-transcriptional level. UBE2C-silencing by exogenous introduction of either siRNAs or microRNA effectively suppressed tumor proliferation and growth.
Results
UBE2C up-regulated in rectal carcinoma
The previous study reported that the UBE2C was aberrantly up-regulated in colon cancer. Here we sought to investigate the expression pattern of UBE2C in rectal carcinoma. The relative expression of UBE2C at both protein and transcript level was measured in clinically collected tumor samples. As shown in Fig. 1A, UBE2C was dramatically higher in the rectal carcinoma tissues in comparison with adjacent benign tissues. This observation was consistent with the previous reports and further consolidated by our quantitative PCR results. The relative level of UBE2C transcript was almost three-times higher in the rectal carcinoma than normal counterparts (Fig. 1B). Our data unambiguously demonstrated that UBE2C was significantly over-expressed in rectal carcinoma, which suggested a potentially oncogenic activity of UBE2C in this disease.
Figure 1.
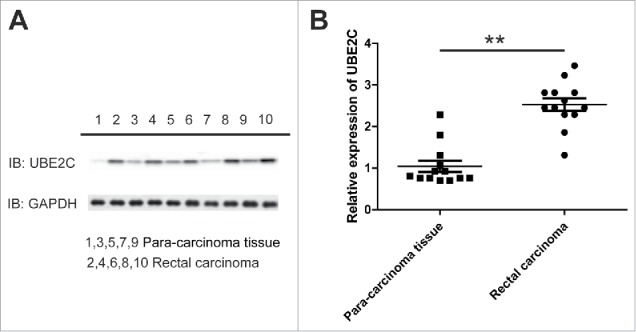
UBE2C up-regulated in rectal carcinoma. A) Western bolt showed protein levels of UBE2C in 5 rectal carcinomas compare to their para-carcinoma tissues. B) Q-PCR showed the expression level of UBE2C in another 13 rectal carcinoma tissue samples. ** indicated p<0.01.
UBE2C promoted the cell proliferation in HR-8348 cells
Next, we set out to clarify the potential involvement of UBE2C in malignant behavior of rectal carcinoma. To this purpose, we manipulated the relative expression of UBE2C in human rectal carcinoma cell line HR-8348 with exogenous introduction of siRNAs. The knockdown efficiency was first evaluated by either immunoblotting or real-time PCR. As shown in Fig. 2A and B, the UBE2C expression at both translation and transcription level was significantly silenced by two different siRNAs. Both siRNAs caused approximately 80% reduction of UBE2C in HR-8348 cells.
Figure 2.
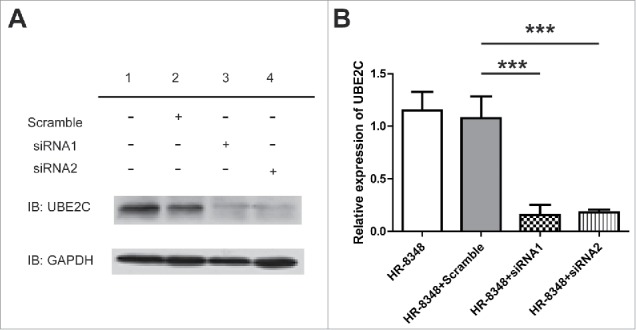
siRNA of UBE2C down-regulated the expression level of UBE2C in HR-8348 cells. A) Western blot showed the protein level of UBE2C after the treatment of siRNA. B) Q-PCR showed the expression level of UBE2C in the cells with different treatments as indicated. *** indicated p<0.001.
We then assessed the impact of UBE2C silence on HR-8348 cell proliferation. The average number of indicated cells was counted up to 6 days of consecutive culture. As shown in Fig. 3A, the cell growth was significantly slowed down by introduction of UBE2C siRNAs. The cell viability upon UBE2C silencing was further determined by CCK-8 assay. Consistent with the reduction of cell number once UBE2C knockdown, our data demonstrated similar decrease in respect to cell viability (Fig. 3B). Furthermore, the anchorage-independent growth capacity in UBE2C-silencing cells was measured by the soft agar assay. In line with the suppressive effect on cell proliferation elicited by UBE2C knockdown, our data displayed significant reduction in colony formation in UBE2C-deficient HR-8348 cells (Fig. 3C, D). Our results suggested an indispensable role of UBE2C in promoting cell proliferation in rectal carcinoma.
Figure 3.
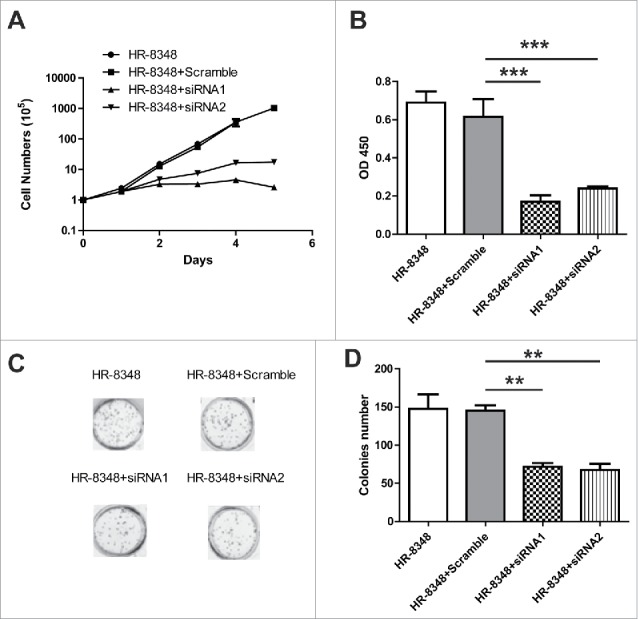
UBE2C promoted the cell proliferation in HR-8348 cells. A) Cell counting assay showed the cell proliferation with the different treatments as indicated. B) The viability of HR-8348 cells with different treatments were determined by cck-8 kit. *** indicated p<0.001. C) and D) Colonies assay showed the less colonies numbers after the RNAi of UBE2C.
UBE2C inhibited apoptosis and promoted the tumor invasion
Next, we attempted to determine the impact of UBE2C deficiency on tumor cell apoptosis and metastasis-related invasion capacity. The spontaneous apoptosis in wild-type, scramble control and specific UBE2C knocked down HR-8348 cells was measured by Annexin V/PI double staining method. As shown in Fig. 4A and B, we only detected background apoptosis in either sham or scramble siRNA-transfected HR-8348 cells. However, the apoptosis was remarkably induced upon UBE2C-specific siRNAs introduction. Similarly, the invasion capacity of HR-8348 was significantly compromised by UBE2C silencing (Fig. 4C, D). The average number of invaded colony decreased from 81±4 to 36±5 once UBE2C was silenced by specific siRNAs. Our data suggested that in addition to pro-proliferative effect, UBE2C was crucial for the inhibitory effect on tumor cell apoptosis and invasion.
Figure 4.
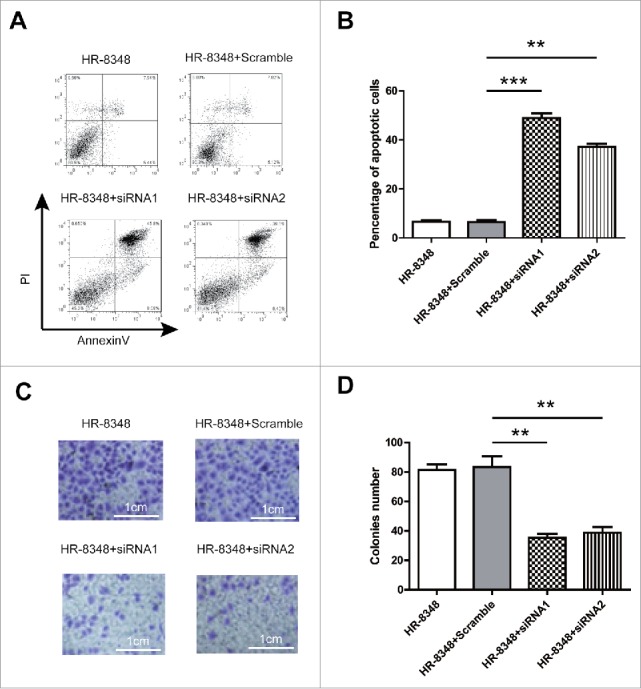
UBE2C inhibited apoptosis and promoted the tumor invasion. A) The apoptosis of the HR-8348 cells with different treatment as indicated was determined by using the Annexin-V-FITC & PI Apoptosis Kit and assessed by flow cytometry. n = 3 independent experiments and this panel presented one of these repeats. B) Statistic of percentage of the apoptosis cells performed in panel (A). Data showed the Annexin-V and PI double positive cells. Data were represented as mean ± s.d.; n = 3 independent experiments. ***indicated p < 0.001 while ** indicated p < 0.01. C) Tumor invasion was determined by transwell. D) Statistic of transferred cells performed in panel (C). ** indicated p < 0.01.
UBE2C promoted rectal carcinoma in vivo
Our previous in vitro results demonstrated an important role of UBE2C in promoting cell proliferation and invasion and inhibiting cell apoptosis. Next, we sought to consolidate these critical roles of UBE2C in vivo. To this purpose, here we employed xenograft mice with indicated cell inoculated subcutaneously. The tumor growth was monitored with respect to UBE2C expression. Our data obviously demonstrated that UBE2C-silencing significantly suppressed tumor growth in vivo (Fig. 5A). Furthermore, the total survival of tumor-bearing mice was analyzed using Kaplan-Meier regression method. As shown in Fig. 5B, the overall survival in UBE2C deficient mice (105 and 118 days) was significantly prolonged while compared to the control group (70 days post-inoculation). Our data suggested an important pro-tumor activity of UBE2C in vivo.
Figure 5.
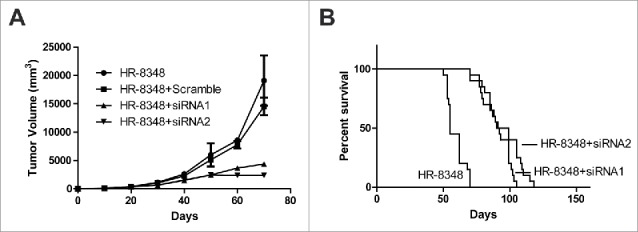
UBE2C promoted rectal carcinoma in vivo. A) Tumor generated by HR-8348 with different treatment as indicated was measured in diameters and calculated to volume according to the time point. B) Survival curve of the mice injected with different cells. Each group contains 20 mice.
miR-381 regulated the expression level of UBE2C
Our previous data demonstrated that UBE2C was aberrantly up-regulated in rectal carcinoma, which in turn promoted tumor cell proliferation, invasion and suppressed cell apoptosis, and eventually contributed to the its oncogenic property. However, the regulatory mechanism underlying the UBE2C overexpression was still elusive. To address this issue, here we concentrated on potential microRNAs which might involve in UBE2C regulation. We first closely inspected the 3’UTR region of UBE2C and attempted to identify the candidate miRs with the aid of bioinformatics online tool (http://www.microrna.org/). As shown in Fig. 6A, we have identified the target sequence in the 3’UTR of UBE2C which perfectly matched with the seed region of miR-381, which suggested that miR-381 might play a crucial role in regulation of UBE2C in rectal carcinoma. Next, we sought to experimentally validate this possibility by exogenously introducing miR-381 mimic. As shown in Fig. 6B, the forced expression of miR-381 in HR-8348 cells dramatically inhibited UBE2C expression at the protein level, which clearly revealed that miR-381 post-transcriptionally modulated UBE2C in rectal carcinoma. Consistent with previous observation that siRNA mediated silencing of UBE2C led to cell viability reduction, here we demonstrated that ectopic expression of miR-381 significantly compromised cell proliferation as well (Fig. 6C). Our data indicated that miR-381 might be a crucial player in regulating the UBE2C expression in rectal carcinoma.
Figure 6.
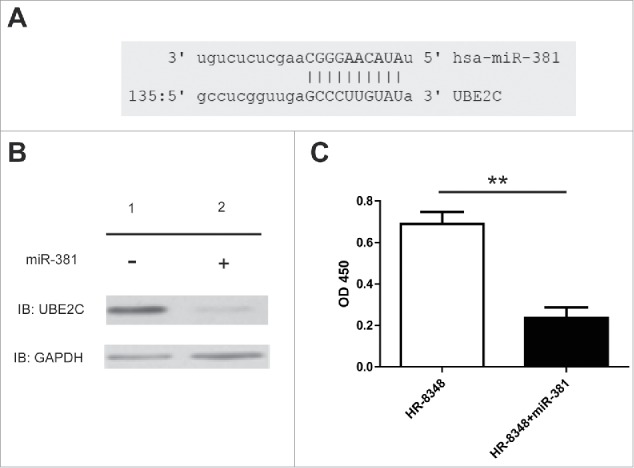
miR-381 regulated the expression level of UBE2C. A) Schema showed the complimentary domain between miR-381 and UBE2C. B) Western blot showed the protein level of UBE2C after the treatment of miRNA. C) The viability of HR-8348 cells with different treatments were determined by cck-8 kit. ** indicated p < 0.01.
miR-381 directly inhibited the growth of rectal carcinoma both in vitro and in vivo
Based on the bioinformatics prediction, we have demonstrated that miR-381 specifically modulated UBE2C expression in human rectal carcinoma cell line HR-8348. Here we sought to investigate the impact of miR-381 expression on the malignant behavior of rectal carcinoma cells. In line with previous results, the anchorage-independent growth of HR-8348 was remarkably suppressed by exogenous introduction of miR-381 (Fig. 7A). Similarly, the invasion capacity interrogated by the transwell assay demonstrated that ectopic expression of miR-381 significantly inhibited the invasion into Matrigel (Fig. 7B). We further confirmed this inhibitory effect in xenograft mice model. As shown in Fig. 7C, the survival analysis showed remarkable survival extension upon introduction of miR-381 into UBE2C cells (the median survival time from 51 days in control group to 90 days in miR-381 group). Our data suggested a tumor suppressor role of miR-381 in rectal carcinoma via down-regulating UBE2C expression.
Figure 7.
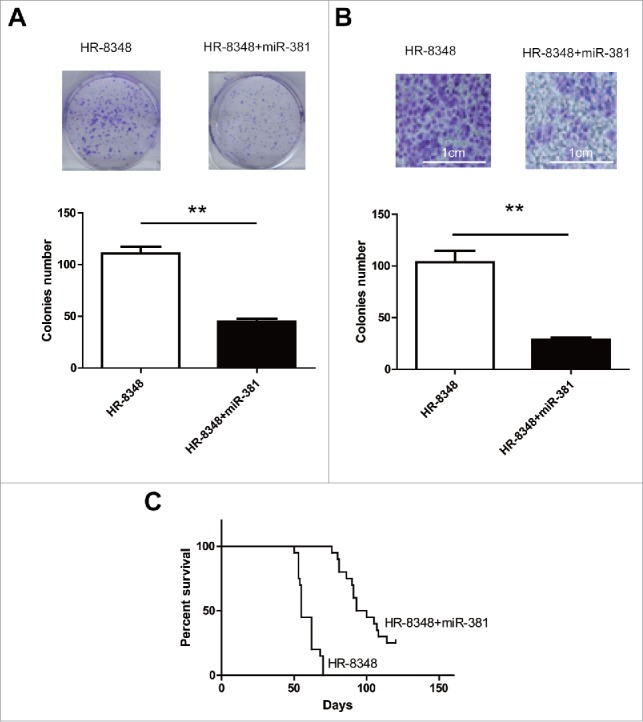
miR-381 directly inhibited the growth of rectal carcinoma both in vitro and in vivo. A) Colonies assay showed the colonies numbers after the transfection of miR-381. ** indicated p < 0.01. B) Tumor invasion was determined by transwell. ** indicated p<0.01. C) Survival curve of the mice injected with different cells. Each group contains 20 mice.
Discussion
The rectal carcinoma is one of the most common human cancer and imposes great challenge for clinical management owing to its complicated localization and histologic structure in comparison with colon cancer.10 The mainstay of treatment against rectal carcinoma heavily relies on conventional chemotherapy and radiotherapy. Target therapeutics widely used in other human malignance are still very limited in this setting. Therefore, identification of novel molecular target and hereby development of corresponding therapeutics is crucial for this disease.
UBE2C plays critical role in cell cycle regulation via catalyzing polyubiquitination-mediated degradation of APC/C substrates.11 Noteworthily, the assembling evidences demonstrated that UBE2C was aberrantly expressed in variety human malignances and proposed crucial roles in incidence and progression of tumors. For instance, Jiang et al. reported that knockdown of UBE2C by RNA interference inhibited glioma cell proliferation and enhanced cell apoptosis in vitro.12 Leta et al. have identified overexpressed genes in hepatocellular carcinoma with special reference to UBE2C.13 Takahashi et al. detected aberrant expression of UBE2C in advanced colon cancer with liver metastasis by DNA microarray and two-color FISH.9 Yang et al. demonstrated that UBE2C promoted gastric cancer growth and served as a potential biomarker for gastric cancer diagnosis.14 Li et al. demonstrated that UBE2C overexpression increased carcinogenesis and blocked ALLN susceptibility in colorectal cancer.15 Furthermore, UBE2C has been suggested that regulated apoptosis-dependent tumor progression of non-small cell lung cancer via ERK pathway.16 Regarding to the regulatory mechanism, Han et al. reported the miR-196a post-transcriptionally up-regulated the UBE2C proto-oncogene and promoted cell proliferation in breast cancer.17 And inhibition of microRNA-17/20a suppressed cell proliferation in gastric cancer by modulating UBE2C.18 Intriguingly, high expression of UBE2C has also been suggested that associated with drug resistance. For instance, Wang et al. showed that knockdown of UBE2C enhanced the chemo-sensitivity of dual drug resistant breast cancer cells to epirubicin and docetaxel.19 Rawat et al. reported that inhibition of UBE2C reduced proliferation and sensitized breast cancer cells to radiation, doxorubicin, tamoxifen and letrozole.20 Bose et al. suggested that dominant negative UBE2C sensitized cervical cancer cells to radiation.21 The potential prognostic value of UBE2C has been implicated in several investigations. For example, Cacciola et al. suggested the prognostic value of UBE2C expression and correlation with the sensitivity to the antineoplastic treatment for colorectal cancer patients.22 UBE2C exhibited additive effect with AZGP1, PIP, S100A8 as biomarkers improved outcome prediction in breast carcinoma.23 Morikawa et al. suggested that UBE2C was a marker of unfavorable prognosis in bladder cancer after radical cystectomy.24 A multicenter based investigation validated that cyclin D1, MCM7, TRIM29 and UBE2C as prognostic protein markers in non-muscle-invasive bladder cancer.25 In lung cancer, Zhao proposed that UBE2C expression provided a useful tool for the prognosis and treatment of non-small cell lung cancer.26 The prognostic significance of UBE2C mRNA expression also was confirmed in high-risk early breast cancer patients.27 In line with all abovementioned observations, here we presented evidences that UBE2C was aberrantly over-expressed in rectal carcinoma, which promoted proliferation, invasion and inhibited apoptosis, and convergently contributed to its oncogenic activity in this disease.
Noteworthily, with the aid of bioinformatics tool, here we predicted and experimentally validated that miR-381 actively involved in UBE2C regulation via post-transcriptional mechanism. Accumulation of data indicated a tumor suppressor role of miR-318 in diverse human cancers. For instance, Xue et al. demonstrated that miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4.28 Xia et al. reported that miR-381 inhibited epithelial ovarian cancer malignancy via YY1 suppression.29 Ming et al. showed that miR-381 suppressed C/EBPI+-dependent Cx43 expression in breast cancer.30 In metastatic prostate cancer, miR-381 along with other microRNAs including miR-154, miR-299, miR-376, miR-377, miR-487, miR-485, miR-495 and miR-654 regulated proliferation, apoptosis, migration and invasion.31 In hepatocellular carcinoma, miR-381 suppressed cell growth and invasion by targeting the liver receptor homolog-1.32 In agreement with this notion, here we demonstrated that miR-381 functioned as a tumor suppressor microRNA in rectal carcinoma through specific inhibition of UBE2C expression, which indicated the diversity in the mode of action of this microRNA. Although we have unambiguously showed that miR-381 negatively modulated UBE2C expression, how miR-381 expression was suppressed in this disease was still elusive and warranted further investigations.
In summary, here we characterized aberrant overexpression of UBE2C in rectal carcinoma, which was subjected to miR-381 post-transcriptional modulation. Our both in vivo and in vitro data implicated that either siRNA or miR introduction effectively suppressed tumor growth, which highlighted the potential that UBE2C as therapeutic target for drug development in the future.
Materials and methods
Animal
The B-NSG nude mice were purchased from Beijing Biocytogen and housed in the pathogen-free environment. All the animal experiments were performed in strict accordance with protocol approved by the Institutional Committee of Animal Care and Use of The Forth Affiliated Hospital of Harbin Medical University.
Patient sample
The paired benign and rectal carcinoma tissue samples were collected from patients whose written informed consent was obtained in advance in the The Forth Affiliated Hospital of Harbin Medical University. All the experimental protocol complied strictly with the guideline from the Medical Ethics Committee of The Forth Affiliated Hospital of Harbin Medical University.
Western blotting
The protein from tissue samples were prepared by ultrasonication, and cell lysates were prepared by lysis in RIPA buffer on ice. The cell debris was discarded by refrigerated centrifugation. The equal amount of protein samples was resolved by SDS-PAGE gel and transferred to PVDF membrane. After brief blocking with 5% skim milk in TBST buffer (TBS plus 0.5% Tween-20), the membrane was hybridized with indicated primary antibody (UBE2C, CST#14234, 1:1,000; GAPDH, CST#5174, 1:1,000) at 4°C overnight. Next day, the PVDF membrane was washed with TBST and subjected to secondary antibody (anti-rabbit, CST#7074, 1:5,000) incubation for one hour at room temperature. The bands were visualized using the enhanced chemiluminescence (ECL, Millipore, Billerica, MA, USA). The housekeeping gene GAPDH was used as loading control.
Real-time PCR
The total RNA was extracted from either tissue samples or cell lines using Trizol reagent (Invitrogen) in accordance with the manufacturer's instruction. The quantity and quality of RNA was determined by the BioAnalyzer 2100 (Agilent, Santa Clara, CA) prior to further processing. Each 1 μg RNA was reversely transcribed into cDNA using the PrimeScript 1st strand cDNA Synthesis Kit (Clontech). Real-time PCR was performed with TaqMan Assay (UBE2C Assay ID: Hs00964100_g1; GAPDH Assay ID: Hs02786624_g1, ThermoFisher, Waltham, MA, USA). The relative expression of UBE2C was calculated using 2−ΔΔCt method and normalized to GAPDH.
Cell culture
The human rectal carcinoma cell line HR-8348 was obtained and authenticated by the American Type Culture Collection (ATCC). The cells were cultured in PRMI-1640 medium supplemented with 1% PSG (penicillin-streptavidin-glutamine) and maintained in the humidified incubator at 37°C with 5% CO2 supply. The exponentially growing cells were harvested for any further analysis.
Transfection
The log phase cells were seeded into 6-well plate and allowed for 24-hour culture. The transfection was performed with Lipofectamine 2000 following the manufacturer's instruction. The transfection efficiency was evaluated by the parallel transfection of GFP plasmid.
Cell proliferation assay
The relative cell viability was measured using the commercial CCK-8 Kit (Dojindo, Kumamoto, Japan) in accordance with the manufacturer's guide. Briefly, the indicated cells were seeded into 96-well plate in triplicate. After 24 consecutive hours of culture, 10 μl of CCK-8 solution was added into each well and incubated for one hour at 37°C. The absorption at 450 nm was recorded by the Microplate Reader (Molecular Devices, Sunnyvale, CA, USA) and cell number was counted.
Soft agar assay
The exponentially growing cells were harvested and prepared into single-cell suspension in 0.35% low melting point agar at concentration of 104/ml. The mixture was cautiously lay on the base layer (0.6% agar) and chilled at 4°C for 15 min. After incubation for 14 days, the colony was stained with 0.25% crystal violet and counted in 5 random field under light microscope.
Apoptosis assay
The cell apoptosis was analyzed by the Annexin V-Propidium Iodide (PI) double staining method. Briefly, the indicated cells were prepared into single-cell suspension in HEPES buffer and stained with Annexin V and PI for 15 min in the dark simultaneously. The apoptotic cells were measured by the BD flow cytometry and analyzed with the Flowjo software.
Transwell assay
The cell invasion capacity was measured with the transwell chamber (BD, Franklin Lakes, NJ, USA). The indicated cells at log phase were trypsinized and prepared into single-cell suspension in the serum-free medium (Gibco, Grand Island, NY, USA) and cautiously lay on the top of polycarbonate transwell membrane pre-coated with Basement Membrane Extract (BD, USA). The lower compartment in the chamber was supplemented with complete culture medium containing 15% fetal bovine serum. The uninvaded cells were completely removed with cotton swab from the top of insert after 24 hours consecutive culture. The invaded cells in BME were stained with crystal violet and counted under light microscope.
Xenograft assay
The indicated cells were harvested and prepared into single-cell suspension in HEPES buffer. The equal volume of cell solution and Matrigel (BD, USA) was mixed on ice. The mixture was then subcutaneously inoculated into the right flank of the recipient nude mice. Tumor growth was regularly monitored twice a week. The tumor volume was calculated following the formula: TV (mm3) = length × width2 × 0.5. All mice were sacrificed at indicated time, tumor was stripped and measured.
Statistics
All results expressed in this study were obtained from at least three independent repeats. Data were analyzed with SPSS 23.0, and presented as Mean ± standard deviation (SD). The statistical significances between data sets were analyzed by one or two-way ANOVA analysis, and were expressed as p values, and p < 0.05 was considered statistically different.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Abbreviation
- APC/C
anaphase promoting complex/cyclosome
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–17. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 4.Movahedi M, Bishop DT, Macrae F, Mecklin JP, Moeslein G, Olschwang S, Eccles D, Evans DG, Maher ER, Bertario L, et al.. Obesity, Aspirin, and Risk of Colorectal Cancer in Carriers of Hereditary Colorectal Cancer: A Prospective Investigation in the CAPP2 Study. J Clin Oncol. 2015;33:3591–7. doi: 10.1200/JCO.2014.58.9952. [DOI] [PubMed] [Google Scholar]
- 5.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol. 2013;35:229–44. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Rosa M Pace U, Rega D, Costabile V, Duraturo F, Izzo P, Delrio P. Genetics, diagnosis and management of colorectal cancer (Review). Oncol Rep. 2015;34:1087–96. doi: 10.3892/or.2015.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamas K, Walenkamp AM, de Vries EG, van Vugt MA, Beets-Tan RG, van Etten B, de Groot DJ, Hospers GA. Rectal and colon cancer: Not just a different anatomic site. Cancer Treat Rev. 2015;41:671–9. doi: 10.1016/j.ctrv.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 8.David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285:8595–604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi Y, Ishii Y, Nishida Y, Ikarashi M, Nagata T, Nakamura T, Yamamori S, Asai S. Detection of aberrations of ubiquitin-conjugating enzyme E2C gene (UBE2C) in advanced colon cancer with liver metastases by DNA microarray and two-color FISH. Cancer Genet Cytogenet. 2006;168:30–5. doi: 10.1016/j.cancergencyto.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ping Z, Lim R, Bashir T, Pagano M, Guardavaccaro D. APC/C (Cdh1) controls the proteasome-mediated degradation of E2F3 during cell cycle exit. Cell Cycle. 2012;11:1999–2005. doi: 10.4161/cc.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Bao Y, Luo C, Hu G, Huang C, Ding X, Sun K, Lu Y. Knockdown of ubiquitin-conjugating enzyme E2C/UbcH10 expression by RNA interference inhibits glioma cell proliferation and enhances cell apoptosis in vitro. J Cancer Res Clin Oncol. 2010;136:211–7. doi: 10.1007/s00432-009-0651-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ieta K, Ojima E, Tanaka F, Nakamura Y, Haraguchi N, Mimori K, Inoue H, Kuwano H, Mori M. Identification of overexpressed genes in hepatocellular carcinoma, with special reference to ubiquitin-conjugating enzyme E2C gene expression. Int J Cancer. 2007;121:33–8. doi: 10.1002/ijc.22605. [DOI] [PubMed] [Google Scholar]
- 14.Yang M, Qu Y, Shi R, Wu X, Su C, Hu Z, Chang Q, Liu S, Pan G, Lei M, et al.. Ubiquitin-conjugating enzyme UbcH10 promotes gastric cancer growth and is a potential biomarker for gastric cancer. Oncol Rep. 2016;36:779–86. doi: 10.3892/or.2016.4906. [DOI] [PubMed] [Google Scholar]
- 15.Li SZ, Song Y, Zhang HH, Jin BX, Liu Y, Liu WB, Zhang XD, Du RL. UbcH10 overexpression increases carcinogenesis and blocks ALLN susceptibility in colorectal cancer. Sci Rep. 2014;4:6910. doi: 10.1038/srep06910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Liu P, Wang J, Gong T, Zhang F, Ma J, Han N. Ubiquitin-conjugating enzyme E2C regulates apoptosis-dependent tumor progression of non-small cell lung cancer via ERK pathway. Med Oncol. 2015;32:149. doi: 10.1007/s12032-015-0609-8. [DOI] [PubMed] [Google Scholar]
- 17.Han Q, Zhou C, Liu F, Xu G, Zheng R, Zhang X. MicroRNA-196a post-transcriptionally upregulates the UBE2C proto-oncogene and promotes cell proliferation in breast cancer. Oncol Rep. 2015;34:877–83. doi: 10.3892/or.2015.4049. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Han T, Wei G, Wang Y. Inhibition of microRNA-17/20a suppresses cell proliferation in gastric cancer by modulating UBE2C expression. Oncol Rep. 2015;33:2529–36. doi: 10.3892/or.2015.3835. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Pan YH, Shan M, Xu M, Bao JL, Zhao LM. Knockdown of UbcH10 enhances the chemosensitivity of dual drug resistant breast cancer cells to epirubicin and docetaxel. Int J Mol Sci. 2015;16:4698–712. doi: 10.3390/ijms16034698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rawat A, Gopal G, Selvaluxmy G, Rajkumar T. Inhibition of ubiquitin conjugating enzyme UBE2C reduces proliferation and sensitizes breast cancer cells to radiation, doxorubicin, tamoxifen and letrozole. Cell Oncol (Dordr). 2013;36:459–67. doi: 10.1007/s13402-013-0150-8. [DOI] [PubMed] [Google Scholar]
- 21.Bose MV, Gopisetty G, Selvaluxmy G, Rajkumar T. Dominant negative Ubiquitin-conjugating enzyme E2C sensitizes cervical cancer cells to radiation. Int J Radiat Biol. 2012;88:629–34. doi: 10.3109/09553002.2012.702299. [DOI] [PubMed] [Google Scholar]
- 22.Cacciola NA, Calabrese C, Malapelle U, Pellino G, De Stefano A Sepe R, Sgariglia R, Quintavalle C, Federico A, Bianco A, et al.. UbcH10 expression can predict prognosis and sensitivity to the antineoplastic treatment for colorectal cancer patients. Mol Carcinog. 2016;55:793–807. doi: 10.1002/mc.22322. [DOI] [PubMed] [Google Scholar]
- 23.Parris TZ, Kovacs A, Aziz L, Hajizadeh S, Nemes S, Semaan M, Forssell-Aronsson E, Karlsson P, Helou K. Additive effect of the AZGP1, PIP, S100A8 and UBE2C molecular biomarkers improves outcome prediction in breast carcinoma. Int J Cancer. 2014;134:1617–29. doi: 10.1002/ijc.28497. [DOI] [PubMed] [Google Scholar]
- 24.Morikawa T, Kawai T, Abe H, Kume H, Homma Y, Fukayama M. UBE2C is a marker of unfavorable prognosis in bladder cancer after radical cystectomy. Int J Clin Exp Pathol. 2013;6:1367–74. [PMC free article] [PubMed] [Google Scholar]
- 25.Fristrup N, Birkenkamp-Demtroder K, Reinert T, Sanchez-Carbayo M, Segersten U, Malmstrom PU, Palou J, Alvarez-Mugica M, Pan CC, Ulhoi BP, et al.. Multicenter validation of cyclin D1, MCM7, TRIM29, and UBE2C as prognostic protein markers in non-muscle-invasive bladder cancer. Am J Pathol. 2013;182:339–49. doi: 10.1016/j.ajpath.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 26.Zhao L, Jiang L, Wang L, He J, Yu H, Sun G, Chen J, Xiu Q, Li B. UbcH10 expression provides a useful tool for the prognosis and treatment of non-small cell lung cancer. J Cancer Res Clin Oncol. 2012;138:1951–61. doi: 10.1007/s00432-012-1275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Psyrri A, Kalogeras KT, Kronenwett R, Wirtz RM, Batistatou A, Bournakis E, Timotheadou E, Gogas H, Aravantinos G, Christodoulou C, et al.. Prognostic significance of UBE2C mRNA expression in high-risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) Study. Ann Oncol. 2012;23:1422–7. doi: 10.1093/annonc/mdr527. [DOI] [PubMed] [Google Scholar]
- 28.Xue Y, Xu W, Zhao W, Wang W, Zhang D, Wu P. miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed Pharmacother. 2017;86:426–33. doi: 10.1016/j.biopha.2016.12.051. [DOI] [PubMed] [Google Scholar]
- 29.Xia B, Li H, Yang S, Liu T, Lou G. MiR-381 inhibits epithelial ovarian cancer malignancy via YY1 suppression. Tumour Biol. 2016;37:9157–67. doi: 10.1007/s13277-016-4805-8. [DOI] [PubMed] [Google Scholar]
- 30.Ming J, Zhou Y, Du J, Fan S, Pan B, Wang Y, Fan L, Jiang J. miR-381 suppresses C/EBPalpha-dependent Cx43 expression in breast cancer cells. Biosci Rep. 2015;35. doi: 10.1042/BSR20150167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G, Candi E. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33:5173–82. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Zhao S, Pang X, Chi B. MicroRNA-381 suppresses cell growth and invasion by targeting the liver receptor homolog-1 in hepatocellular carcinoma. Oncol Rep. 2016;35:1831–40. doi: 10.3892/or.2015.4491. [DOI] [PubMed] [Google Scholar]


