Abstract
Mitochondrial energy metabolism depends upon high-flux and low-flux electron transfer pathways. The former provide the energy to support chemiosmotic coupling for oxidative phosphorylation. The latter provide mechanisms for signaling and control of mitochondrial functions. Few practical methods are available to measure rates of individual mitochondrial electron transfer reactions; however, a number of approaches are available to measure steady-state redox potentials (Eh) of donor/acceptor couples, and these can be used to gain insight into rate controlling reactions as well as mitochondrial bioenergetics. Redox changes within the respiratory electron transfer pathway are quantified by optical spectroscopy and measurement of changes in autofluorescence. Low-flux pathways involving thiol/disulfide redox couples are measured by redox Western blot and mass spectrometry-based redox proteomics. Together, the approaches provide the opportunity to develop integrated systems biology descriptions of mitochondrial redox signaling and control mechanisms.
Keywords: NADH, NADPH, NADH dehydrogenase, Ubiquinone, Cytochromes, Hydrogen peroxide, Glutathione, Thioredoxin-2, Redox Western blot, Redox proteomics, Peroxiredoxin
1. Introduction
Mitochondrial function depends upon oxidation-reduction (redox) processes with three areas especially relevant to contemporary mitochondrial research. The first is the use of energy from oxidation to support electrochemical coupling of oxidative phosphorylation [1]. The second involves a small fraction of the O2 consumed by mitochondria which is converted to the so-called reactive oxygen species (ROS), superoxide anion radical (O2−), and H2O2 [2], with mitochondria being selectively vulnerable to oxidative damage [3, 4]. The third encompasses low-flux redox reactions which function in cell signaling and control [5]. Another chapter in this volume (Wieckowski) addresses mitochondrial ROS generation, so this will not be included in the present article.
The first section addresses measurement of steady-state redox potentials of the high-flux redox systems of the mitochondrial respiratory chain. These methods were developed over 50 years ago and currently provide a convenient but underutilized approach to improve understanding of mitochondrial bioenergetics and respiratory control. As described by Britton Chance in 1957 [6], the mitochondrial electron transfer chain undergoes a “cushioning effect” in which the steady-state reduction of components changes with substrate conditions to maintain function of oxidative phosphorylation. Because the system functions in vivo under a non-equilibrium steady state, the steady-state redox potentials of the components within the chain are always relevant to accurate descriptions of respiratory function.
A second section describes use of redox Western blot methods to measure components of low-flux thiol/disulfide systems. These methods are relatively newly developed and provide the capability to selectively study mitochondrial function in intact cells and tissues due to the redox-sensitivities of specific mitochondrially localized proteins. Critically important enzymes within mitochondria are subject to redox regulation. Examples include NADH dehydrogenase which undergoes S-glutathionylation [7], apoptosis signal-regulating kinase-1 which is regulated by the redox state of thioredoxin-2 [8], and glutaredoxin-2 which is present as an inactive iron-sulfur complex until oxidatively activated [9].
A subsequent section describes a mass spectrometry-based method to measure fractional reduction of cysteines in specific proteins [10]. This is a highly versatile approach to profile redox-sensitive proteins which relies upon rapidly advancing technology. A final section provides some general comments concerning future needs for mitochondrial redox biology and briefly discusses related methods to study redox regulation by S-glutathionylation and S-nitrosylation. Combinations of immunoassay, mass spectrometry, and molecular manipulations provide means to define and discriminate these covalent modifications from other redox mechanisms in signaling and control.
1.1. Methodologic Limitations Due to Complexity of Mitochondria
Accurate quantification is essential to progress in redox biochemistry. However, several features of mitochondria have hindered accurate assessment of key reaction rates under relevant physiologic conditions. A first problem is that the tissue-specific regulation of mitochondria which occurs in vivo cannot readily be replicated in vitro. Because of the inability in in vitro studies to completely mimic pO2, pCO2, substrate supply, workload, and physiologic signaling as found in vivo, there is a possibility that key observations are misinterpreted. Furthermore, even if these conditions can be controlled, one is faced with the in vitro adaptations of cells and limitations concerning function of individual mitochondria within populations of mitochondria and cells. Mitochondria are not uniform in biochemical characteristics, and there is likelihood that spatial constraints within the cell further impact redox reactions at the individual mitochondrial level. Thus, the complexity of mitochondrial structure, function, and regulation requires that in vitro findings be validated in vivo and that “bottom-up” models of mitochondrial function be mirrored with “top-down” investigation.
Despite the limitations of in vitro studies, measurements of mitochondrial function at the cellular level can be more reflective of physiologic rates than studies of isolated mitochondria. Fractionation of cells and isolation of mitochondria result in perturbation of rate-control characteristics [11]. Isolated mitochondria invariably have some extent of physical damage, and even if this is considered minimal, there is an uncertainty about how to create a synthetic aqueous cytoplasm which appropriately reflects the intracellular environment. On the other hand, mitochondria which have been isolated from normal tissues contain proteins which are present at the correct in vivo level. Most tumor-derived cell lines have aberrant mitochondria, and mitochondrial characteristics often change dramatically when normal cells are grown in vitro. Thus, there is a need to study mitochondrial functions at multiple levels of organization. Importantly, the methods described in the present article can be applied at all levels of organization. Thus, an advantage of available quantitative redox biology methods is that they can provide a foundation for a comprehensive understanding of mitochondrial respiratory control.
1.2. Steady-State Reduction Potentials as Quantitative Descriptors in Redox Biology
As described elsewhere in this volume, quantitative information for specific biochemical processes can be obtained using isolated mitochondria and submitochondrial fractions. These preparations provide mechanistic detail that cannot be obtained with more intact systems. There is a critical need to integrate the information obtained from mitochondria and submitochondrial fractions with data obtained from cellular and in vivo models. A particular challenge lies in the difficulty of knowing whether kinetic principles or thermodynamic principles govern specific redox processes. Measurement of steady-state redox potential (Eh) values provides means to characterize operational characteristics of mitochondria without specific knowledge of reaction rates. Eh values are reduction potentials for electron acceptor/donor pairs relative to a standard hydrogen electrode [12]. By definition, the reduction potentials are given for an acceptor/donor pair, e.g., pyruvate/lactate. The biological and medical research literature more commonly uses the inverted expression of the donor/acceptor pair, e.g., lactate/pyruvate; while technically incorrect, common use requires that, regardless of how the couple is ordered, Eh expressions be interpreted as reduction potentials, unless explicitly stated otherwise. In the present article, we use donor/acceptor to refer to a redox couple and the term Eh with the acceptor to define the couple, e.g., for the GSH/GSSG couple, EhGSSG refers to the reduction potential for the reaction GSSG +2e−+ 2H+ ↔ 2 GSH. For proteins, we use the protein abbreviation with the redox-sensitive thiols implied, e.g., for thioredoxin-2, EhTrx2 refers to the reduction potential for the active site dithiol/disulfide couple.
In biological systems, Eh values are calculated using the Nernst equation, Eh = Eo + RT/nF ln ([acceptor]/[donor]), where Eo is the standard potential for the couple, R is the gas constant, T is the absolute temperature, n is the number of electrons transferred, and F is Faraday’s constant. The difference in Eh values between two couples, ΔEh, is related to Gibbs free energy for the electron transfer according to the equation ΔG = nFΔEh.
Changes in Δψ and ΔpH depend upon the ΔEh for electron transfer, yet countless studies of Δψ, proton leak, ROS production, etc. have been performed and interpreted without knowledge of Eh values for donors or ΔEh for donor and acceptor couples under the conditions of assay. Because the validity of interpretation depends upon the energetics determined by the redox potentials of the respective upstream and downstream couples in the electron transfer pathway, there is a need to incorporate measurements redox potentials into bioenergetic descriptions.
1.3. Steady-State Redox Potentials of Mitochondrial Electron Transfer Components
In vivo, the mitochondrial electron transfer pathway operates under non-equilibrium steady-state conditions. The principles for measurement are derived from the historically important discovery of the mitochondrial cytochrome chain by David Keilin, in which oxidation-reduction reactions were characterized in terms of spectral changes in the cytochromes (Fig. 1) [13, 14]. While the rates of electron transfer through the entire pathway are conveniently measured in terms of O2 consumption, measurement of specific donor pathways is more difficult. However, changes in steady-state potentials can be readily measured for many of the components because of their inherent spectral and fluorescent properties (Fig. 2) [15].
Fig. 1.
Changes in steady-state redox levels of mitochondrial components are based upon absorbance characteristic as shown by this original study of yeast [13]. The reduced pyridine nucleotides, NADH and NADPH, absorb light at 320 nm. Oxidized flavoproteins absorb light at 455 in the 450–500 nm range. Cytochromes have distinct absorbance in the reduced forms. Reproduced with permission of the publisher
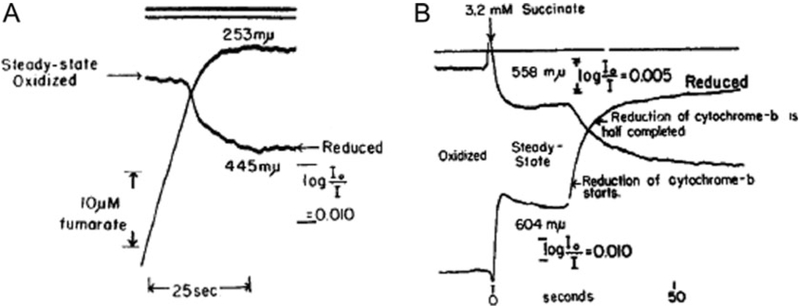
Fig. 2.
Measurement of changes in steady-state levels of oxidation in the mitochondrial cytochrome chain. Changes in steady-state oxidation of (a) cytochrome a3 and (b) cytochrome b are illustrated in this original study of Chance [15]. In a, the response of heart-muscle mitochondrial preparation to metabolism of added fumarate is shown. In b, changes in steady-state oxidation are observed in response to added succinate, with the cytochromes approaching complete reduction as O2 becomes exhausted. Reproduced with permission of the publisher
The materials needed to perform experiments include appropriate spectrophotometry or fluorometry instrumentation, appropriate means to calibrate the system for complete oxidation and reduction, and means to control O2 and substrate availability. Spectral changes are measured as changes in light absorbance by a chemical at a specific wavelength defined by Beer’s law, A = εCl, where A is the absorbance, ε is the extinction coefficient, C is the concentration, and l is the path length. ε is usually given as the mM extinction coefficient, and the path length is 1 cm in standard spectrophotometers. Studies of steady-state levels of oxidation of respiratory chain components in isolated mitochondria and cell suspensions can be obtained in 1 cm cuvettes or in small beakers [16] with appropriate positioning of light source and detector. With appropriate instrumentation, absorbance changes can be measured in perfused organs and adherent cells [17]. In all cases, light scatter can be limiting so that dual-wavelength spectrometry is used to minimize this problem [13, 17].
Stock reagents needed for total oxidation are either 100 mM potassium ferricyanide (added at 5 μL/mL incubation) or 1 mM FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone) in ethanol (added at 1 μL/mL incubation). Reagent needed for total reduction is sodium dithionite. The latter must be maintained dry so that only a few grains can be added; if a solution is used, this is prepared fresh as a 1 M solution so that only 1 μL/mL incubation is needed to effect complete reduction. It should be noted that reduction by dithionite is slower with larger amounts of dithionite due to altered chemical reactions.
1.4. NADH/NAD+ Measurement
The NADH/NAD+ couple provides a central electron carrier connecting oxidation of many food-derived metabolites and energy production. Enzyme-coupled and HPLC assays are available to measure NADH and NAD+ concentrations [18, 19], and with selective solubilization of cell membranes, measurements of mitochondrial contents can be obtained [20]. However, a substantial fraction of the total NADH + NAD+ pool is protein bound, so the measurements of the total amounts of NADH and NAD+ in the mitochondria do not allow accurate estimate of EhNAD+. Studies of different dehydrogenase reactions in rat liver and hepatocytes showed that the reaction catalyzed by β-hydroxybutyrate dehydrogenase is near equilibrium so that the β-hydroxybutyrate/acetoacetate ratio can be used to calculate the matrix EhNAD+, which is about 318 mV [20]. Because Eo for NADH/NAD+ is −337 mV, this means that free NAD+ is present at a considerably higher concentration than free NADH. This characteristic means that the energetics of Complex I can be altered by the rates of the NAD+-linked dehydrogenases.
Assay of β-hydroxybutyrate and acetoacetate can be performed with several standard approaches, including on a fluorescence plate reader using enzyme-coupled assays [19], by 1H-NMR spectroscopy [21] or gas chromatography-mass spectrometry [22]. EhNAD+ is then estimated using the Nernst equation, the β-hydroxybutyrate and acetoacetate concentrations, an Eo value of −297 mV [20], and the assumption that the β-hydroxybutyrate dehydrogenase reaction is at equilibrium. It must be pointed out that it is not clear whether this reaction is at equilibrium in all mitochondria so this assumption will require validation for extension to other cell types and tissues.
Measures of relative changes in the NADH/NAD+ couple can be readily obtained by absorbance and fluorescence methods. NADH has an absorbance at 340 nm (extinction coefficient, 6.2/mM/cm) which is not present in NAD+. Although not directly useful for Eh determination, measurement with dual-wavelength spectroscopy can provide a sensitive means to measure relative changes. In this approach, absorbance at 375 nm is used as a reference to control for light scatter so that absorbance change can be used to provide a measure of the absolute change in NADH concentration in intact cells [23].
More sensitive detection of changes in NADH redox state is obtained by measuring fluorescence of NADH with excitation at 366 nm and emission at 450 nm [24, 25]. Protein-binding results in enhanced NADH fluorescence efficiency, and fractionation studies have shown that most of the NADH fluorescence is associated with mitochondria. Consequently, relative changes in fluorescence of NADH provide means to measure changes in redox state of this couple in intact cells and tissues. This approach has been used effectively to study redox control in cell culture, often described as measurement of “autofluorescence” because it does not require addition of fluorescence probes.
The flow of electrons from substrates to NAD+ is regulated at many levels to balance utilization of carbohydrate, fat, and amino acid-derived precursors. Some of the key dehydrogenases are activated by Ca2+, and several of the monocarboxylate, dicarboxylate and tricarboxylate substrates are exchanged by antiporters. Multiple fatty acid transporters control utilization of different chain lengths, and variations in amino acid transporters also contribute to rates of utilization of these substrates for mitochondrial respiration. The relatively limited range of substrates and conditions used for most studies of isolated mitochondria raises the possibility that key regulatory mechanisms related to supply of reducing equivalents to maintain EhNAD+ in different tissues remain to be discovered. To date, relatively limited efforts have been made to mimic physiologically relevant metabolic conditions in studies with isolated mitochondria.
1.5. Coenzyme Q and Complex III
Electron transfer from NADH to ubiquinone (CoQ) occurs through Complex I, a redox-driven proton pump. This is a large protein complex containing a covalently bound, redox-active flavin, as well as multiple iron-sulfur centers. The ΔEh for the overall reaction is determined by the EhCoQ relative to EhNAD+. CoQ exists as three redox forms, ubiquinone, ubiquinol, and ubisemiquinone. The redox characteristics of quinones have recently been reviewed [26]; CoQ is hydrophobic and mostly membranal; estimates of EhCoQ are obtained using the Nernst equation with relative amounts of ubiquinol and ubiquinone obtained by HPLC [27]. Because CoQ is an intermediate between Complex I and Complex III and also substrate for Complex II and a number of other substrate-linked dehydrogenases (e.g., acyl CoA dehydrogenases), studies of Complex I function are subject to variable energetics due to steady-state reduction of the CoQ pool. In this way, changes in ΔEh can alter the energetics for maintenance of Δψ independently of leak currents. Thus, there is a need to include ΔEh measurements along with Δψ and O2 consumption measurements.
Absorbance and fluorescence of the flavin in complex III change upon reduction, and these changes have also been used to measure redox in cells and intact tissues [28]. In contrast to the increases in absorbance and fluorescence of NADH upon reduction, flavoproteins have a decrease in absorbance and fluorescence upon reduction. Fluorescence is measured with excitation at 436 nm and emission at 570 nm, largely a measure of mitochondrial flavoproteins [29]. Because there are multiple flavoproteins which can contribute to the signals, there is a limitation to the mechanistic information which can be derived. On the other hand, ratiometric studies of the NADH fluorescence and flavoprotein fluorescence provide very sensitive indicators of tissue anoxia which can be used to visualize regional differences in oxidation due to pathophysiologic changes in vivo [24, 25]. With modern instrumentation, these approaches could provide improved capabilities for in vivo study of mitochondrial redox reactions.
1.6. Cytochromes
Complex III and Complex IV contain hemoproteins termed “cytochromes” which are readily detected by spectrophotometry. These were used as early as 1952 to measure mitochondrial redox changes in cells [15]. The measurements of the redox states of cytochromes depend upon the absorbance characteristics of the hemes in the cytochromes, which have more intense absorbance in the reduced forms (Fig. 1) [13]. In the visible range, the absorbance bands are termed α, β, and γ (Soret) absorbance bands. While the Soret band has the greatest extinction coefficients, it occurs at the shortest wavelength where there is the greatest light scatter in preparations containing mitochondria; consequently, measurements in this region can be more prone to error due to this light scatter. In addition, there is considerable overlap of signals from hemes b and c, and these can be further obscured by signals from myoglobin or hemoglobin [30]. Consequently, measurements in the Soret region are usually limited to cytochromes a3. Cytochromes a and a3 contain heme a, which is structurally distinct from heme b, present in hemoglobin, myoglobin, catalase, and b- and c-type cytochromes. Cytochromes a and a3 also absorb light in the near-infrared region, and this has provided a means to measure oxidation in vivo [17]. Considerable spectral overlap occurs with the β bands, so most measurements are obtained with the α bands. Cytochromes b and c contain heme b (Fe-complexed protoporphyrin IX) but differ in absorbance because the heme is covalently bound in cytochromes c and c1 but not in bH or bL. Other minor differences in absorbance maxima occur due to the interactions with amino acids in the vicinity of the heme. Addition of ligands like KCN, which bind to the O2 site, can be used to discriminate cytochromes a and a3. However, this is not useful for steady-state redox measurement because it blocks electron flow.
Complex III accepts two electrons from CoQ and transfers one to cytochrome c1 and the other to the cytochrome bH and bL pair. The electron from cytochrome c1 is then transferred to cytochrome c, while the electron from cytochrome b is transferred back to CoQ. The system is a proton pump, with energetics of proton pumping probably defined by steady-state potential of the donor ubiquinol/ubisemiquinone couple and cytochrome c1. As above, changes in the steady-state Eh values determine the energetics available for proton pumping. Consequently, interpretations of Complex III function in generation of ΔμH+, ROS generation or support of ATP production, are limited by changes in steady-state Eh values which determine the available ΔEh.
Steady-state reduction of cytochromes b is measured by dual-wavelength measurement with 575 as an isosbestic point. Absorbance maxima for the two forms are at 561 and 566 nm, but the signals overlap considerably so that redox changes of the mixture are most readily obtained by measuring absorbance at 562 nm relative to 575 nm. The extinction coefficient value for 562 nm minus 575 nm is 23/mM/cm. Fractional oxidation under steady-state conditions is obtained by use of an uncoupler to approximate 100% oxidation followed by addition of sodium dithionite to obtain 100% reduction. Because dithionite breaks down rapidly in solution, complete reduction is usually achieved by addition of a few grains of solid sodium dithionite. FCCP and CCCP (carbonyl cyanide m-chlorophenylhydrazone) are commonly used as uncouplers to obtain maximal oxidation, but this approach assumes that the only limitation to oxidation is the existence of the proton gradient. Because this assumption may not be valid, an alternative means to obtain maximal oxidation is to use a chemical oxidant such as 0.5 mM potassium ferricyanide [31]. In intact organs and some cell preparations, the fully reduced state can be approximated by removal of O2 supply or addition of potassium cyanide [6, 15]; however, these treatments may not result in complete reduction due to endogenous respiratory control characteristics.
Cytochrome c has an absorbance maximum at 550 nm in the reduced form, while cytochrome c1 has a maximum at 554 nm. The two are not easily resolved spectrally in cells or tissues; therefore, the two are measured together as cytochrome c + c1. For these measures, dual-wavelength spectroscopy is used by measuring 550 nm minus 540 nm as an isosbestic point. The extinction coefficient is 19/mM/cm. This measurement is usually expressed relative to maximal oxidation and reduction as performed for cytochromes b.
Steady-state reduction of cytochromes a and a3 is also frequently measured together as cytochrome a + a3, using the wavelength pair 605 nm and 630 nm [32]. The extinction coefficient is 13.1/mM/cm.
1.7. Oxygen Consumption Rate
Changes in redox state of cytochromes can be used to measure O2 consumption rate in closed systems where known amounts of O2 are added [15, 23]. Typically, the system is allowed to consume all O2 and become anoxic. Rapid addition of a known amount of O2 then results in oxidation of the cytochromes, and the time required to consume the O2 and become reduced allows calculation of O2 consumption rate. Because the rate of O2 consumption decreases when O2 concentration reaches the low micromolar range, measurements are calculated from the difference in time to consume different amounts of added O2. This approach has been directly validated relative to measurements with a Clark-type electrode [23]. With this approach, the duration of anoxia prior to addition of O2 pulses affects the O2 consumption rate [33], necessitating care in the timing of the measurements. A variation on this approach has also been used to measure rates of catalase activity in terms of the amount of O2 produced and cytochrome oxidized following addition of H2O2 under anaerobic conditions [34].
1.8. NADPH/NADP+ Measurement
As indicated above, mitochondria contain low-flux redox systems that are qualitatively different from those which support oxidative phosphorylation and energy metabolism. Although originally described in terms of their role in protection against oxidative stress, accumulating evidence shows that these systems function in regulation of the redox states of cysteine (Cys) residues of proteins, controlling their structures and activities. Consequently, assays to measure fractional reduction of specific Cys residues of proteins are of considerable importance to understand mitochondrial signaling and control.
Important conceptual advances in understanding redox regulation of cell functions have come from redox proteomics methods, some of which are especially useful to measure mitochondrial redox systems. These systems are oxidized by the endogenous mitochondrial generation of O2− and H2O2. An early estimate that H2O2 production by mitochondria is about 1% of the total O2 consumption rate [2] is frequently repeated, but accurate values for mitochondria in intact systems are not available. Measurements obtained with isolated mitochondrial preparations are likely to provide exaggerated values due to the presence of damage mitochondria and the analysis under supraphysiologic conditions of oxidizable substrates and pO2. On the other hand, because there is also evidence for electron transfer bypass, in which NADH dehydrogenase generates O2−, the O2− is transported into the inter-membrane space according to the electrochemical gradient, and the O2− donates the electron to cytochrome c [35]. The rate of this pathway is not known, so that there is a possibility that the estimated rate of 1% could also be an underestimate. Available methods to measure oxidant production by mitochondria under relevant in vivo conditions are difficult to calibrate so that better approaches are needed. Nonetheless, the data are accurate enough to allow the conclusion that only a small fraction of the total O2 consumed by cells is converted by mitochondria to oxidants. Because thiol-dependent regulatory mechanisms are dependent upon these oxidants for oxidation, this low rate of oxidant production defines redox regulatory systems as “low-flux” systems when compared to mitochondrial respiration. The practical meaning is that minor disruption of normal high-flux electron transfer by the respiratory chain can produce oxidants at rates which are comparable to or higher than the rates which normally function in regulation. These issues have been previously reviewed and provide the basis for a definition of oxidative stress which includes the disruption of redox signaling and control pathways [36, 37].
While rates of electron transfer through the low-flux redox signaling and control pathways are not known, information about their function can be obtained from measurements of steady-state redox potentials. The thiol-dependent systems are maintained by electron transfer from NADPH. The NADPH/NADP+ couple in mitochondria is maintained at relatively negative Eh value, about −415 mV in the liver [20]. As with the NADH/NAD+ couple, direct measurement of total concentrations does not give a reliable measure of the potential because of extensive protein binding. Consequently, better estimates are obtained in the liver by measuring the glutamate, ammonia, and 2-oxoglutarate concentrations and calculating the Eh with the assumption that the reaction is at equilibrium. Alternatively, isocitrate, 2-oxoglutarate, and CO2 can be used [20]. Little information is available concerning whether these reactions are at equilibrium in different tissues, so some extent of validation would be important in use of this approach.
2. Materials
2.1. GSH/GSSG Sample Buffer and Derivatization Reagents
10% PCA/BA (10% perchloric acid/0.2 M boric acid) is prepared by adding 71 mL of 70% perchloric acid, 6.2 g boric acid (0.2 M) and adjusting total volume to 500 mL.
10% PCA/BA with internal standard is made by adding 1.38 mg γ-glutamylglutamate (10 μM) to the solution above.
20 mg/mL dansyl chloride solution prepared in 100% acetone.
9.3 mg/mL iodoacetic acid solution prepared in water.
1% digitonin (w/v) solution in water.
Chloroform.
Acetone.
2.2. Mobile Phase Buffers for GSH/GSSG HPLC Determination
Solvent A: 80% methanol in water.
Solvent B: 64% methanol, 4 M sodium acetate buffer, pH 4.6.
2.3. SDS-Polyacrylamide Gel Electrophoresis
Running buffer (10×): 250 mM Tris–HCl, 1.92 M glycine, 1% SDS.
30% acrylamide/bis solution 37.5:1 (2.6% C) (BioRad, Hercules, CA) and N,N,N,N′-tetramethylethylenediamine (TEMED) (Sigma).
Ammonium persulfate (APS): Prepare a 10% stock solution and store at −20 °C.
Precision plus protein standards (BioRad).
Gel casting stock solutions: 1.5 M Tris–HCl (pH 8.8) to be used for casting the resolving gel; 1.0 M Tris–HCl (pH 6.8) to be used in casting the stacking gel; 10% SDS.
Mini-Protean III cell (BioRad) and Power Pac 200 power source (BioRad).
2.4. Redox Western Blotting (Trx2, Prx3, TrxR2)
Towbin’s transfer buffer: 25 mM Tris–HCl, 192 mM glycine, 20% methanol. Store at 4 °C.
Nitrocellulose membrane (BioRad) and extra thick filter paper (7.5 × 10 cm) (BioRad).
Trans-blot SD semidry transfer apparatus (BioRad) and Power Pac 200 power source (BioRad).
PBS with Tween-20 (PBS-T)/wash buffer: add 2 mL of Tween-20 (Sigma) to 1 L of 1 × PBS.
Blocking buffer/antibody dilution buffer: 1:1 mixture of PBS-T and Li-Cor Blocking Buffer (Li-Cor, Lincoln, NE).
Primary antibody: rabbit antisera against Trx2; Prx3 mouse monoclonal antibody (Abcam, Cambridge, MA); TrxR2 rabbit polyclonal antibody (AbFrontier, Korea).
Secondary antibody: goat anti-rabbit or goat anti-mouse Alex-afluor680 antibody (Invitrogen, Eugene, OR).
Li-Cor Odyssey infrared imager (Li-Cor).
2.5. Cell Lysis and Protein Alkylation: Trx2 Redox Western Blot
10% trichloroacetic acid solution (Sigma).
100% acetone (Sigma).
Phosphate-buffered saline (PBS) (Mediatech).
Disposable cell scraper (Fisher).
Alkylation buffer: 50 mM Tris–HCl (pH 8), 0.1% SDS, 15 mM 4-acetoamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) (Invitrogen).
Nonreducing sample loading buffer (5×): 300 mM Tris–HCl (pH 6.8), 50% glycerol, 1% SDS, 0.05% bromophenol blue.
2.6. Cell Lysis and Protein Alkylation: Prx3 Redox Western Blot
DMEM/F-12 (1:1) (with L-glutamine) cell culture medium (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Atlanta, GA) and 1% penicillin/streptomycin (Hyclone, Logan, UT).
Paraquat (Sigma, St. Louis, MO) 10 mM stock solution in water. Prepare stock solution and use within 15 min of preparation.
Phosphate-buffered saline (Mediatech, Manassas, VA).
Alkylation buffer: 40 mM Hepes, 50 mM NaCl, 1 mM EGTA, protease inhibitor cocktail (Sigma), 100 mM N-ethylmaleimide (NEM). Add NEM to buffer immediately prior to use.
25% CHAPS stock solution for cell lysis.
Disposable cell scraper (Fisher).
Nonreducing sample loading buffer (5×): 300 mM Tris–HCl (pH 6.8), 50% glycerol, 1% SDS, 0.05% bromophenol blue.
2.7. BIAM Modification and Pulldown: TrxR2 Redox Western Blot
Biotin iodoacetamide [N-(biotinoyl)-N′-(iodoacetyl)ethylenediamine, BIAM, Molecular Probes]: Dissolved in 50 mM Tris–HCl buffer (pH 6.8) to make a stock solution at 0.1 M concentration.
Iodoacetamide (IAM, Sigma-Aldrich Co.): Dissolve in Tris–HCl buffer at 5 mM concentration.
Lysis buffer A [50 mM Bis-Tris–HCl (pH 6.5), 0.5% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, leupeptin, aprotinin, and 0.1 mM PMSF] containing 10 μM BIAM.
Protein G Sepharose (Sigma-Aldrich, St. Louis, MO).
2.8. Cell Lysis and ICAT Reagents: Redox Proteomics
10% TCA.
Denaturation buffer: 50 mM Tris–HCl, pH 8.5, 0.1% SDS.
ICAT assay kit (Applied Biosystems, Foster City, CA).
2.9. Mass Spectrometry-Based Redox Proteomics
Ultimate 3000 nanoHPLC system (Dionex) with a nanobore column (0.075 × 150 mm PepMap C18 100 Å, 3 μm, Dionex).
QSTAR XL Q-TOF mass spectrometer (Proxeon Biosystems).
ProteinPilot software (Applied Biosystems).
3. Methods
3.1. GSH/GSSG Measurement in Cultured Cells
Mitochondria contain two central systems controlling protein thiol/disulfide states, one dependent upon GSH and the other dependent upon a mitochondria-specific thioredoxin, thioredoxin-2 (Trx2). These systems are parallel, nonredundant systems [38] which are both reduced by NADPH-dependent mechanisms and oxidized by H2O2-dependent mechanisms. The GSH/GSSG couple is maintained by a splice variant of GSSG reductase which is targeted to mitochondria. Oxidation of GSH can occur by many enzymes, most notably glutathione peroxidase-1 (Gpx1) and glutathione peroxidase-4 (Gpx4) and a number of GSH transferases [39]. Glutaredoxin-2 also oxidizes GSH to GSSG in its reaction to remove GSH from glutathionylated proteins.
3.1.1. Sample Derivatization
Remove treatment media from cells and wash three times using cold PBS.
Add approximately 500 μL of 1% digitonin (w/v) to each plate/well of cells, and incubate for 5 min on ice to permeabilize.
Aspirate the digitonin solution from the cells and carefully wash the cells three times with cold PBS.
Apply approximately 500 μL of 10% PCA/BA directly to the permeabilized, adherent cells, scrape each well, and collect cell extracts in a 1.5 mL tube.
Samples are spun for 2 min in a microcentrifuge at approximately 14,000 × g to pellet protein.
An aliquot (300 μL) of each supernatant is transferred to a fresh microcentrifuge tube.
9.3 mg/mL Iodoacetic acid solution (60 μL) is added to each tube and vortex to mix.
The pH is adjusted to 9.0 ± 0.2 with the KOH/tetraborate solution (approximately 220–250 μL).
After about 3 min to allow complete precipitation of potassium perchlorate, the pH of at least some of the samples should be checked to verify that they are in the correct range.
Incubate samples for 20 min at room temperature.
300 μL of dansyl chloride (20 mg/mL acetone) is added, and the samples are mixed and placed in the dark at room temperature for 16–26 h. Dansylation of GSH is complete by 8 h, but GSSG has 2 amino groups that must be modified. The rate of the second dansylation is slower than for the first with the result that mono-dansyl derivatives of GSSG (eluting between the N-dansyl-S-carboxymethyl-GSH and bis-dansyl-GSSG) will be present if dansylation is incomplete.
After derivatization, chloroform (500 μL) is added to each tube to extract the unreacted dansyl chloride.
Samples are stored at 0–4° in the dark in the presence of both the perchlorate precipitate and the chloroform layer until assay by HPLC. Stability tests show that samples can be stored under these conditions for 12 months with little change in the amounts of GSH and GSSG derivatives.
3.1.2. HPLC Analysis of GSH/GSSG Redox
Samples are centrifuged for 2 min in a microcentrifuge prior to transfer of an aliquot of the upper (aqueous) layer to an autosampler.
Typical injection volume is 25 μL.
Separation is achieved on 3-aminopropyl columns (5 μm; 4.6 mm × 25 cm; Custom LC, Houston; or Supelcosil LC-NH2, Supelco, Bellefonte, PA).
Initial solvent conditions are 80% A, 20% B run at 1 mL/min for 10 min.
A linear gradient to 20% A, 80% B is run over the period from 10 to 30 min.
From 30 to 46 min, the conditions are maintained at 20% A, 80% B and returned to 80% A, 20% B from 46 to 48 min.
Equilibration time for the next run is 12 min.
Detection is obtained by fluorescence monitoring with band-pass filters, 305–395 nm excitation and 510–650 nm emission (Gilson Medical Electronics, Middleton, WI).
Quantification is obtained by integration relative to the internal standard, γ-glutamylglutamate.
3.2. Trx2 Redox Western Blot Analysis in Cultured Cells: Sample Preparation
Human Trx2 contains two Cys residues in the active site; the dithiol and disulfide forms are separated by nonreducing PAGE following modification of the protein with AMS, a reagent which increases the mass from 12 kD to 13 kD (1 kD/2 Cys) (Fig. 3). To obtain EhTrx2, a redox Western blot method is used [40]. This approach has an advantage over the GSH/GSSG method in that study of proteins which are present only in mitochondria provides a compartment-specific assay without subcellular fractionation. Redox Western blots have been developed which resolve reduced and oxidized forms of proteins based upon both changes in molecular mass and changes in charge. For the former, a thiol reagent with relatively large mass, such as AMS (4-acetoamido-4′-maleimidylstilbene-2,2′-disulfonic acid), is used to increase the mass of the thiol-containing form of the protein. This allows separation by electrophoresis using SDS-PAGE and is described below.
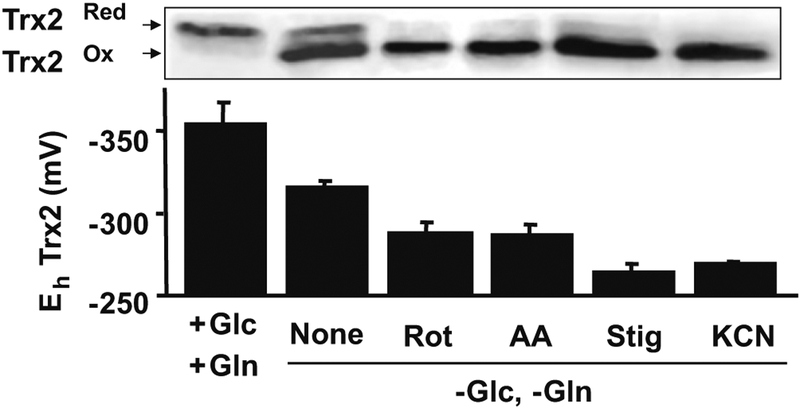
Fig. 3.
Trx2 redox state in HT29 cells exposed to mitochondrial respiratory inhibitors in control media and glucose (Glc)-, glutamine (Gln)-free media. Cells were grown to 80% confluency and then cultured for 24 h with media as indicated. After 24 h, inhibitors were added at 5 μM (Rot, AA, Stig) or 0.5 mM (KCN) for 30 min prior to extraction and analysis by redox Western blotting. Separation was provided by reaction of samples with AMS, a thiol reagent which adds approximately 500 Da per thiol, thereby slowing mobility sufficiently to separate the reduced form from the disulfide form. All incubations in Glc,—Gln-free media were significantly different from respective +Glc,+Gln controls. N = 5. Rot rotenone, AA antimycin A, Stig stigmatellin
3.2.1. Cell Lysis and Protein Alkylation
Assays are performed with cells grown in a 35-mm or 6-well culture plate (1–2 × 106 cells).
After experimental treatment, cells are washed with ice-cold PBS.
The cells are then treated with 1 mL of ice-cold TCA (10%), scraped, transferred to microcentrifuge tubes, incubated on ice for 30 min, and centrifuged at 12,000 × g for 10 min.
The supernatant is removed and the protein pellet is saved.
One milliliter of 100% acetone is added to the pellet. The tube is mixed, incubated on ice for 30 min, and centrifuged at 12,000 × g for 10 min.
The acetone is removed and the pellet (30–50 μg protein) is used for analysis.
After addition of 100 μL of lysis/derivatization buffer (50 mM Tris–HCl, pH 8.0, 0.1% SDS, 15 mM AMS), pellets are resuspended by sonication and incubated at room temperature in the dark for 3 h. The samples can be used for redox Western blotting at this step or be saved at −20 °C for subsequent analysis.
3.3. Prx3 Redox Western Blot Analysis in Cultured Cells: Sample Preparation
Peroxiredoxins (Prx) are a class of thiol-dependent antioxidant proteins that exhibit peroxidase activity. These proteins utilize redox-active Cys residues in their active site to reduce hydrogen peroxide and other organic peroxides. Prx are classified into two distinct groups, 1-Cys and 2-Cys, based on the number of Cys residues involved in the reduction of peroxides. Mammals possess six isoforms, five of which are considered 2-Cys Prx (Prx1–5). Prx 6 is the sole member of the 1-Cys class. The reaction mechanism for Prx occurs in two steps, the first of which is common for both 1-Cys and 2-Cys Prx. First, peroxide reacts with the “peroxidatic” Cys residue in the active site resulting in the formation of a cysteine sulfenic acid. This newly formed sulfenic acid then reacts with the “resolving” Cys, resulting in the formation of a disulfide linkage. It should be noted that Prx are present as domain-swapped homodimers, where the “peroxidatic” Cys is located on one subunit and “resolving” Cys on the opposite subunit. Therefore, when a 2-Cys Prx becomes oxidized, the enzyme is locked in a dimer due to the newly formed disulfide bonds (Fig. 4). 2-Cys Prx utilize the thioredoxin/thioredoxin reductase system to reduce this disulfide and effectively recycle the enzyme back to its active form [41].
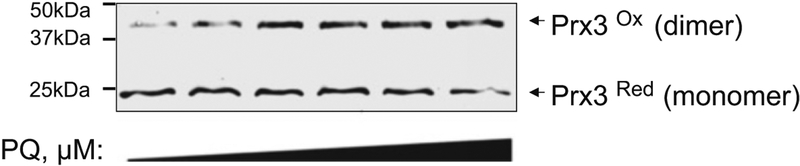
Fig. 4.
Measurement of oxidation of Prx3 by redox Western blot analysis. SH-SY5Y human neuroblastoma cells were treated 24 h with increasing amounts of paraquat (0, 10, 25, 50, 75, 100 μM). Following extraction and treatment with NEM to prevent further oxidation, separation by SDS-PAGE followed by Western blotting reveals the oxidized (disulfide) form increases in response to PQ compared to the reduced, NEM-modified form (monomer)
Prx3 and Prx5 are mitochondrial isoforms; however, Prx5 is an “atypical” 2-Cys Prx that does not form an intermolecular disulfide upon reaction with a peroxide because it functions as a monomer. Based on the fact that Prx form stable disulfides resulting in a “locked” dimerized state, Cox et al. have exploited this concept in order to measure redox changes in Prx3 [42]. This method involves alkylation of cellular proteins with N-ethylmaleimide (NEM) followed by SDS-PAGE and standard immunoblotting techniques.
3.3.1. Cell Lysis and Protein Alkylation
Approximately 1–2 × 106 SH-SY5Y neuroblastoma cells are plated into each well of a 6-well plate containing 2 mL of media and allowed to adhere overnight.
Medium is aspirated from each well and replaced with treatment media containing desired concentrations of paraquat, and cells are incubated for 24 h.
At the completion of the treatment period, the medium is aspirated from each well, and wells are gently washed three times with 2 mL of cold PBS.
After washing approximately 100 μL of alkylation buffer containing 100 mM NEM is added to each well and allowed to incubate 10–15 min at room temperature.
Add approximately 4 μL of 25% CHAPS solution to each well (1% final concentration) to lyse cells.
Using a cell scraper, scrape cells and debris from each well, and collect cell extracts in 1.5 mL tubes and place on ice.
Using a benchtop sonicator, sonicate each tube briefly to ensure complete cell lysis and protein extraction. Keep cells on ice.
Pellet debris and insoluble protein by centrifugation at 17,000 × g for 5 min on a benchtop microcentrifuge. Place samples back on ice.
Assay protein content of each sample using BCA protein assay (Thermo Scientific, Rockford, IL).
Add 5× nonreducing sample buffer to approximately 20–25 μg of protein. Place samples on a heating block at 95 °C for 5 min.
3.4. TrxR2 Redox Western Blot Analysis: Sample Preparation
Trx system is composed of Trx, thioredoxin reductase (TrxR), NADPH, and Trx peroxidases/peroxiredoxins. Trx reduces protein disulfides directly and serves as a reductant for the peroxiredoxins [43]. Oxidized form of Trx is reduced by catalytic activity of TrxR using an electron from reduced NADPH [44]. TrxR2 is an isoform of TrxRs including E. coli TrxR and human TrxR1. The catalytic site of TrxR is -Cys-Val-Asn-Val-Gly-Cys- and located in the FAD domain of enzymes [45]. Furthermore, TrxR2 has a C-terminal selenocysteine residue that is required for catalytic activity but is not part of the conserved active site [46]. Since TrxRs are known to reduce oxidized Trx, alterations in TrxR activity may regulate Trx activity. TrxR2 is localized in mitochondria, while TrxR1 is predominantly found in cytosol and nucleus [47]. In this chapter, we describe an assay for measuring mitochondrial TrxR2 redox state using BIAM-labeling technique (Fig. 5). The thiol-reactive biotin iodoacetamide and biotin maleimide derivatives can be used for BIAM-labeling technique described in this chapter. This method is based on the procedure of Kim et al. [48], with modifications to measurement of redox states of proteins containing thiols in catalytic site such as TrxR and redox factor-1 (Ref-1). The procedures are described below.
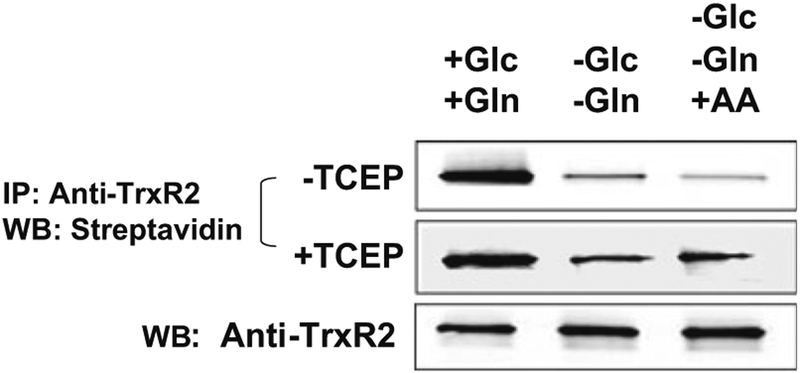
Fig. 5.
Semiquantitative analysis of fractional reduction of thioredoxin reductase-2 (TrxR2) by BIAM-blot. Cells were incubated 24 h with +Glc,+Gln, −Glc, −Gln, or −Glc, −Gln and antimycin A (30 min with 5 μM; +AA). Aliquots of cell lysates were treated with the biotinylated iodoacetamide reagent, BIAM, and parallel aliquots were reduced with TCEP and then reacted with BIAM. Following immunoprecipitation with anti-TrxR2, samples were separated by SDS-PAGE, blotted and probed with fluorescently labeled streptavidin. Controls for recovery following immunoprecipitation were performed by Western blotting with anti-TrxR2 and showed similar recovery. Although these methods are reproducible, the limiting conditions of BIAM labeling selected to maximize detection of reactive thiols in the presence of less reactive thiols do not allow strict quantification
3.4.1. Cell Culture and Affinity Purification
Cells (1–2 × 107 in 10 cm plate) after treatment [e.g., glucose (Glc)- and glutamine (Gln)-deficient media] were washed with cold PBS and fractionated, and then nuclear fraction was lysed with 1 mL of lysis buffer A [50 mM Bis-Tris-HCl (pH 6.5), 0.5% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, leupeptin, aprotinin, and 0.1 mM PMSF] containing 10 μM BIAM. As a control, cells that were not stimulated with treatment (e.g., Glc- and Gln-deficient media) were lysed and labeled with BIAM.
After incubation for 10 min at 37 °C in the dark, the labeling reaction was stopped by adding IAM to 5 mM. TrxR2 in the reaction mixtures was precipitated with the use of rabbit antibodies to TrxR2 and Protein G Sepharose (40 μL per sample). (Optional: Streptavidin-agarose can be used instead of TrxR2 antibody to immunoprecipitate all BIAM-labeled proteins.)
Immunocomplex of BIAM-labeled TrxR2 (use of TrxR2 antibody) or other BIAM-labeled proteins (use of streptavidin-agarose, 40 μL) will be washed with 1 mL of ice-cold lysis buffer A three times.
Add 40 μL of 2× gel loading buffer to each sample; heat at 95 °C for 10 min.
3.5. SDS-PAGE: Trx2, Prx3, and TrxR2 Redox Western Analysis
These instructions assume the use of a BioRad Mini-Protean II or III gel electrophoresis system. Clean each glass plate first with water then 100% methanol prior to gel casting.
For Trx2 and Prx3: prepare a 1.5 mm thick, 15% resolving gel by mixing 2.3 mL water, 5 mL 30% acrylamide solution, 2.5 mL 1.5 M Tris–HCl (pH 8.8), 100 μL 10% SDS, 100 μL APS, and 4 μL TEMED in a 15 mL tube. Invert tube to mix, pour gel solution between glass plates, and allow gel to polymerize for approximately 45–60 min at room temperature. Note: be sure to leave sufficient room for a stacking gel. Also, fill the remainder of the glass plate with water to ensure that the gel polymerizes with a clean, straight edge.
For TrxR2: prepare a 1.5 mm thick, 10% resolving gel by mixing 4 mL water, 3.3 mL 30% acrylamide solution, 2.5 mL 1.5 M Tris–HCl (pH 8.8), 100 μL 10% SDS, 100 μL APS, and 4 μL TEMED in a 15 mL tube. Invert tube to mix, pour gel solution between glass plates, and allow gel to polymerize for approximately 45–60 min at room temperature. Note: be sure to leave sufficient room for a stacking gel. Also, fill the remainder of the glass plate with water to ensure that the gel polymerizes with a clean, straight edge.
After polymerization of resolving gel, prepare and pour the 5% stacking gel by mixing the following in a 15 mL tube: 2.7 mL water, 0.67 mL 30% acrylamide solution, 0.5 mL 1.0 M Tris–HCl (pH 6.8), 40 μL 10% SDS, 40 μL APS, and 4 μL TEMED. Pour off the water layer on the resolving gel and add stacking gel to the top of the glass plates. Place in desired comb (10 or 15 well) and allow to polymerize for 15–30 min.
Assemble the gel apparatus.
Prepare 500 mL of running buffer by adding 50 mL 10× running buffer to 450 mL water. Then fill inner gel chamber and add remainder of buffer to outer chamber. Allow the assembly to sit for a few min to ensure that there are now leaks. If no leaks are discovered, then samples may be loaded.
Load samples and the precision plus protein standards to individual wells.
Place lid on gel apparatus and connect to the Power Pac 200. Run the gel at 150 V for approximately 60–80 min or until the dye front runs off the gel.
3.6. Western Blotting: Trx2, Prx3, and TrxR2 Redox Western Analysis
After proteins are separated on the SDS-PAGE gel, proteins need to be transferred to a nitrocellulose (or PVDF) membrane for immunoblotting. The procedure described here assumes the use of a BioRad semidry transfer apparatus. A “wet” transfer can also be conducted; however, this procedure will not be described here.
Preincubate two filter papers and an appropriately cut nitrocellulose membrane in cold transfer buffer for approximately 10–15 min.
After SDS-PAGE, discard the running buffer and carefully pry apart the glass plates to free the gel.
Place a filter paper saturated with transfer buffer on the semidry transfer apparatus, and use a serological pipette to roll out any trapped air bubbles. Next place the membrane on the filter paper and roll out any air bubbles. Place the gel on top of the membrane and orient it to your liking. Again, carefully roll out any air bubbles. Place the last filter paper on top the gel and roll out any air bubbles. Note: it is important to not introduce any air bubbles in the filter paper-membrane-gel-filter paper sandwich. These bubbles will prevent proper protein transfer and produce poor quality blots.
After the filter paper sandwich is made, assemble the transfer apparatus, and connect it to the Power Pac 200. Run the transfer at 20 V for 60 min. Note: if you cast a thinner gel, run the transfer for slightly shorter time period, i.e., 0.75 mm gels for 45 min at 20 V.
After the transfer is complete, place the membrane in 10–20 mL of blocking buffer, and incubate on a shaker table for at least 30 min at room temperature.
Prepare primary antibody solution by adding 4 μL of peroxiredoxin 3 antibody to 10 mL of fresh blocking buffer (1:2500 dilution); 10 μL of Trx2 or TrxR2 antibody to 10 mL of fresh blocking buffer (1:1000 dilution).
Discard blocking solution and add the primary antibody solution to the membrane and incubate on a shaker table overnight at 4 °C.
After primary antibody incubation, pour used antibody solution into a clean 15 mL tube and save for another use.
Wash membrane 3 times for 15 min each in 10–20 mL of PBS-T. Discard wash buffer after every wash.
For Trx2 and TrxR2: Prepare secondary antibody solution immediately prior to use by adding 2 μL of goat anti-rabbit Alexa Fluor 680 secondary antibody to 10 mL of blocking buffer (1:5000 dilution). Protect this solution from light to avoid photobleaching of the fluorophore.
For Prx3: Prepare secondary antibody solution immediately prior to use by adding 2 μL of goat anti-mouse Alexa Fluor 680 secondary antibody to 10 mL of blocking buffer (1:5000 dilution). Protect this solution from light to avoid photobleaching of the fluorophore.
Add secondary antibody solution to the membrane, and incubate 45 min in the dark at room temperature on a shaker table.
Wash membrane three times for 15 min each in 10–20 mL of PBS-T in the dark. Discard wash buffer after every wash.
After the final washes, carefully place the membrane between two Kimwipes, and allow the membrane to dry in the dark.
Once the membrane is dry, scan it on a Li-Cor Odyssey infrared scanner at 700 nm. Adjust the intensity, contrast, and brightness of the scan to optimize your result.
3.7. Mass Spectrometry-Based Redox Proteomics
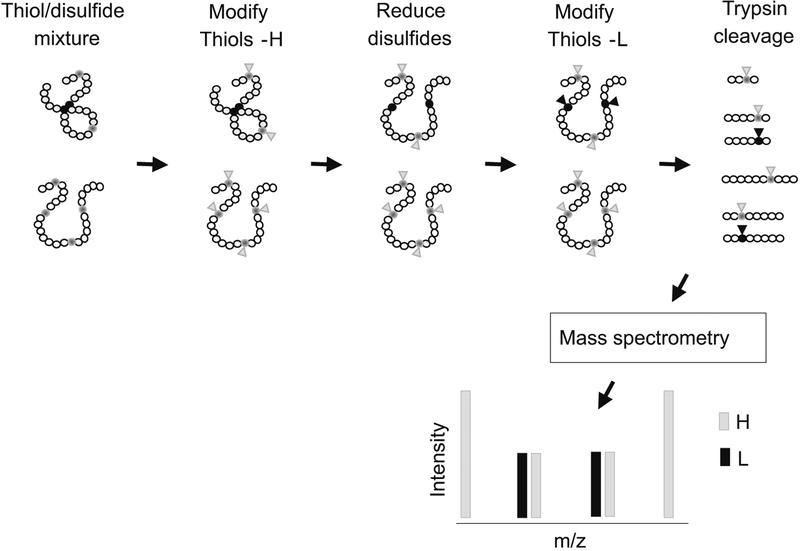
To measure the fractional reduction of mitochondrial proteins, we have used the isotope-coded affinity tag reagent (Applied Biosystems, CA) with a sequential treatment procedure designed to yield the ratio of reduced/oxidized forms of specific tryptic peptides, as measured by tandem mass spectrometry (Fig. 6) [49]. Data from this approach show that many proteins have Cys residues which are partially oxidized under steady-state conditions. Consequently, application of the mass spectrometry-based redox proteomics method can be expected to considerable improve understanding of redox signaling and control. Importantly, this approach allows detection of mitochondrial proteins even without mitochondrial isolation so that the oxidation of mitochondrial proteins can be determined within the context of other cellular compartments.
Fig. 6.
Mass spectrometry-based analysis of fractional reduction of protein using ICAT reagents. Proteins are extracted and treated with the heavy ICAT reagent (H) to label thiols. Following removal of excess reagent, samples are treated with TCEP to reduce disulfides. The newly formed thiols are modified by treatment with the light reagent (L). Following tryptic digestion, analysis by LC-MS/MS allows calculation of fractional reduction from the H:L ratio
The methods used for redox proteomic analyses of mitochondrial proteins are based upon those used for nuclear proteins [10]. Analyses can be performed in three different ways, by studying isolated mitochondria, by using digitonin to permeabilize cells and remove contaminating cytoplasm, or by directly analyzing cells and selecting mitochondrial proteins from the larger list of cytoplasmic proteins and proteins from other subcellular compartments.
3.7.1. Cell Lysis and Protein Collection
Cells [mouse aortic endothelial cells (MAEC), 1–2 106 in 3.5 cm plate] after treatment [e.g., reduced (−150 mV) or oxidized (0 mV) extracellular EhCySS] were washed with cold PBS.
Add 1 mL 10% TCA to cells, scrape, and collect lysates in a clean 1.5 mL tube. Precipitation of white protein pellets will be observed.
Place on ice for 30 min and spin for 10 min at 16,100 × g (4 °C).
Remove supernatant, add 1 mL acetone to protein pellet, vortex, and spin again at 16,100 × g for 10 min (4 °C).
Remove supernatant and air-dry pellet for 1–2 min.
Add 100 μL denaturation buffer (50 mM Tris–HCl, pH 8.5, 0.1% SDS) to pellet, and resuspend pellet by sonication for 2 s on ice.
Perform protein assay.
Transfer 150 μg protein in denaturation buffer, and add more denaturation buffer if needed to make 80 μL as total sample volume.
3.7.2. ICAT Labeling
Add 150 μg protein in 80 μL denaturation buffer prepared above to 20 μL of heavy ICAT reagent (H).
Vortex and snap-centrifuge.
Incubate for 60 min at 37 °C.
Add 10 μL 100% TCA to sample and vortex.
Add additional 400 μL of 10% TCA to “4” and transfer all to a clean 1.5 mL tube.
Place on ice for 30 min.
Centrifuge at 16,100 × g for 10 min (4 °C).
Remove supernatant.
Wash protein pellet with 500 μL, 100% acetone.
Centrifuge at 16,100 × g for 10 min.
Remove supernatant.
Add 80 μL denaturation buffer to pellet.
Add 2 μL TCEP to sample and sonicate for 2 s on ice.
Incubate for 20 min at 37 °C.
Add 20 μL light ICAT reagent (L) reagent to samples.
Vortex and snap-centrifuge.
Incubate for 60 min at 37 °C.
3.7.3. Trypsinization of ICAT-Labeled Proteins
Dissolve trypsin included in ICAT assay kit in 200 μL of MilliQ-H2O.
Add 200 μL of trypsin to a sample labeled with H- and L-ICAT reagents above.
Vortex and snap-centrifuge.
Incubate 12–16 h at 37 °C.
Vortex and snap-centrifuge.
3.7.4. Cation Exchange
Place sample in 3 mL tube.
Add 2 mL of Cation Exchange Buffer (CEB)-Load to sample (1 drop/s).
Vortex and snap-centrifuge.
Check pH; it should be around 2.5–3.3—if not add more CEB-Load until pH is correct.
Assemble cartridge holder with cation exchange cartridge.
Equilibrate cartridge by injecting 2 mL CEB-Load (1 drop/s).
Inject sample onto cartridge slowly (1 drop/s).
Wash with 1 mL CEB-Load (1 drop/s).
Elute peptides with 500 μL CEB-Elute (1 drop/s).
Collect flow-through.
3.7.5. Cleaning and Storing Cation Exchange Cartridge
Inject 1 mL CEB to clean cartridge (1 drop/s).
Inject 2 mL CEB before storing cartridge at 4 °C.
3.7.6. Purifying Peptides and Cleaving Biotin
Insert avidin cartridge into cartridge holder.
Equilibrate cartridge by injecting 2 mL of Affinity Buffer (AB)-Elute (1 drop/s) and discard waste.
Inject 2 mL AB-Load (1 drop/s) and discard waste.
Neutralize samples by adding 500 μL of AB, vortex, and snap-centrifuge.
Check pH; pH should be at 7—if not add more AB until it is correct.
Vortex and snap-centrifuge.
Slowly inject sample onto cartridge (1 drop/s).
Collect flow-through; this fraction contains unlabeled fragments and can be analyzed at a later time if necessary.
Inject an additional 500 μL of AB-Load and collect in the same tube (or a fresh tube) as step 8.
Inject 1 mL of AB (1 drop/s)-first wash and discard flow-through.
Inject 1 mL of AB (1 drop/s)-second wash and collect first 500 μL but discard the latter 500 μL.
Inject 1 mL of MilliQ-H2O and discard flow-through.
3.7.7. Eluting ICAT-Labeled Samples
Inject 800 μL AB-Elute (1 drop/s).
Discard initial 50 μL.
Collect the remaining 750 μL in tube.
Vortex and snap-centrifuge.
3.7.8. Cleaning and Storing Avidin Cartridge
Inject 2 mL AB (1 drop/s) to clean avidin cartridge.
Inject 2 mL AB and store at 4 °C.
3.7.9. Cleaving Biotin
Evaporate samples in speed vacuum.
In another tube mix cleaving agents A (95 μL) and B (5 μL) in a ratio of 95:5.
Add 95 μL of cleaving reagent solution to each sample.
Vortex and snap-centrifuge.
Incubate for 2 h at 37 °C.
Vortex and snap-centrifuge.
Evaporate in speed vacuum.
Send off for mass spectrometry analysis.
3.7.10. Mass Spectrometry Analysis of Redox ICAT
An Ultimate 3000 nanoHPLC system (Dionex) with a nanobore column (0.075 × 150 mm PepMap C18 100 Å, 3 μm, Dionex) is used with the LC eluent being directly sprayed into a QSTAR XL system using a nanospray source from Proxeon Biosystems. The data from each salt cut is combined and processed by ProteinPilot software (Applied Biosystems). All quantification is performed by the ProteinPilot V2.0.1 software using the Swiss-Prot database. Quantification for proteins of interest is manually validated by examination of the raw data.
3.7.11. Calculation of Protein Redox State
Table 1 shows an example of measuring protein reduction/oxidation state affected by changes in extracellular redox conditions [49]. The ratios of reduced (H) to oxidized (L) thiols analyzed by mass spectrometry enable to calculate protein redox state, e.g., % oxidation = [L/(H + L)] 100. Thirty mitochondrial proteins were sorted out from the original data [49] to calculate redox states of mitochondrial proteins using this approach.
Table 1.
Redox ICAT results of mitochondrial proteins in mouse aortic endothelial cells treated with extracellular EhCySS of −150 mV (reduced) or 0 mV (oxidized)
| Accession | Name | −150 mV H:L |
0 mV H:L |
−150 mV % oxidation |
0 mV % oxidation |
Δ% oxidation |
|---|---|---|---|---|---|---|
| P63038 | 60 kDa heat shock protein, mitochondrial precursor | 1.9 | 1.1 | 34.4 | 46.9 | 12.6 |
| Q9CR21 | Acyl carrier protein, mitochondrial precursor | 3.9 | 2.9 | 20.4 | 25.7 | 5.3 |
| P40124 | Adenylyl cyclase-associated protein 1 | 2.3 | 1.3 | 30.0 | 42.9 | 12.9 |
| P48962 | ADP/ATP translocase 1 | 4.7 | 3.5 | 17.5 | 22.3 | 4.8 |
| P51881 | ADP/ATP translocase 2 | 2.5 | 2.3 | 28.5 | 30.5 | 2.0 |
| P62331 | ADP-ribosylation factor 6 | 1.5 | 1.2 | 40.6 | 45.5 | 4.8 |
| P47738 | Aldehyde dehydrogenase, mitochondrial precursor | 4.9 | 3.1 | 16.8 | 24.4 | 7.6 |
| P05202 | Aspartate aminotransferase, mitochondrial precursor | 2.9 | 2.2 | 25.6 | 30.9 | 5.3 |
| Q9DCX2 | ATP synthase D chain, mitochondrial | 4.1 | 2.7 | 19.6 | 26.7 | 7.1 |
| Q03265 | ATP synthase subunit alpha, mitochondrial precursor | 3.8 | 4.0 | 20.9 | 20.1 | 0.8 |
| Q61753 | D-3-phosphoglycerate dehydrogenase | 2.9 | 2.2 | 25.7 | 31.5 | 5.8 |
| Q9EQ06 | Dehydrogenase/reductase SDR family member 8 precursor | 2.9 | 1.8 | 25.7 | 35.1 | 9.4 |
| O08749 | Dihydrolipoyl dehydrogenase, mitochondrial precursor | 2.0 | 1.5 | 33.3 | 39.7 | 6.4 |
| Q91YQ5 | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 67 kDa subunit precursor | 3.7 | 0.8 | 21.3 | 54.1 | 32.8 |
| Q99LC5 | Electron transfer flavoprotein subunit alpha, mitochondrial precursor | 1.8 | 1.7 | 35.2 | 37.0 | 1.9 |
| P26443 | Glutamate dehydrogenase 1, mitochondrial precursor | 2.5 | 1.8 | 28.5 | 36.1 | 7.6 |
| P54071 | Isocitrate dehydrogenase [NADP], mitochondrial precursor | 3.5 | 1.2 | 22.4 | 44.7 | 22.2 |
| P08249 | Malate dehydrogenase, mitochondrial precursor | 3.2 | 2.3 | 23.8 | 30.1 | 6.3 |
| Q791V5 | Mitochondrial carrier homolog 2 | 2.3 | 3.1 | 30.1 | 24.4 | 5.7 |
| P52503 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial precursor | 2.5 | 1.9 | 28.7 | 34.5 | 5.8 |
| Q9DCN2 | NADH-cytochrome b5 reductase 3 | 2.8 | 2.1 | 26.5 | 32.1 | 5.5 |
| Q922Q4 | Pyrroline-5-carboxylate reductase 2 | 2.4 | 2.0 | 29.4 | 33.5 | 4.1 |
| P52480 | Pyruvate kinase isozymes M1/M2 | 3.6 | 2.0 | 21.6 | 33.8 | 12.2 |
| Q8K2B3 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor | 3.2 | 2.3 | 23.8 | 30.7 | 6.9 |
| Q9R112 | Sulfide:quinone oxidoreductase, mitochondrial precursor | 2.7 | 1.9 | 27.0 | 35.0 | 8.1 |
| Q9CZ13 | Ubiquinol-cytochrome-c reductase complex core protein 1, mitochondrial precursor | 4.5 | 3.3 | 18.3 | 23.4 | 5.1 |
| Q9Z1Q9 | Valyl-tRNA synthetase | 1.2 | 0.9 | 45.2 | 51.7 | 6.5 |
| Q60932 | Voltage-dependent anion-selective channel protein 1 | 3.9 | 2.8 | 20.5 | 26.4 | 5.9 |
| Q60930 | Voltage-dependent anion-selective channel protein 2 | 4.1 | 1.2 | 19.7 | 45.2 | 25.6 |
| Q60931 | Voltage-dependent anion-selective channel protein 3 | 2.6 | 1.1 | 27.6 | 47.3 | 19.6 |
4. Notes Regarding Redox Analyses
4.1. GSH/GSSG HPLC Analysis
To facilitate simultaneous measurement of CySS (typically >40 μM) and GSSG (typically <200 nM), two detectors with different sensitivity settings are used in series. Fluorometric detectors with monochromators set at 335 nm for excitation and 515 nm emission can also be used with equivalent results, but sensitivity is substantially less because the narrower band-widths limit the intensity of both excitation and emission light.
To calculate the EhGSSG, use the Nernst equation as follows: Eh = Eo + RT/nF ln ([GSSG]/[GSH] [2]), where Eo for this equation is equal to −276 mV.
4.2. Trx2 Redox Western Blot
An alternative redox western approach is adapted from the original method of Holmgren and Fagerstedt [50] for E. coli Trx in which iodoacetate is used to add negative charges to thiols so that separation can be obtained under native gel conditions.
Trx2 resolves at approximately 10–15 kDa, and care should be taken to prevent these lower mass proteins from running off of the gel.
Care must be taken to avoid loading too much protein because overloading interferes with separation of reduced and oxidized Trx2.
4.3. Prx3 Redox Western Blot
The amount of protein that you load for each blot may be different depending on the cell type that is used. It is wise to optimize this condition to achieve the best result.
The mouse monoclonal antibody from Abcam is used in this current protocol; however, other antibodies against peroxiredoxin 3 exist and can be used if blotting conditions are optimized.
It is very important to make sure that there are no air bubbles present in the filter paper-gel sandwich during the protein transfer step. These bubbles can obscure bands of interest in ruin an entire experiment.
If one chooses to utilize PVDF membrane instead of nitrocellulose, remember to activate the PVDF by wetting with 100% methanol prior to placing in transfer buffer. Also, do not wet nitrocellulose with 100% methanol prior to use.
The semidry transfer technique has been chosen because it is far less cumbersome and easier to perform compared to the wet transfer technique. Wet transfer can be done if the investigator so wishes.
As a negative control for peroxiredoxin 3 oxidation, simply take an aliquot of your alkylated cell extract, and add reducing sample buffer (containing DTT or β-mercaptoethanol). This will result in only one band (approx. 25 kDa) on the blot corresponding to a completely reduced protein.
5. Comments and Perspectives
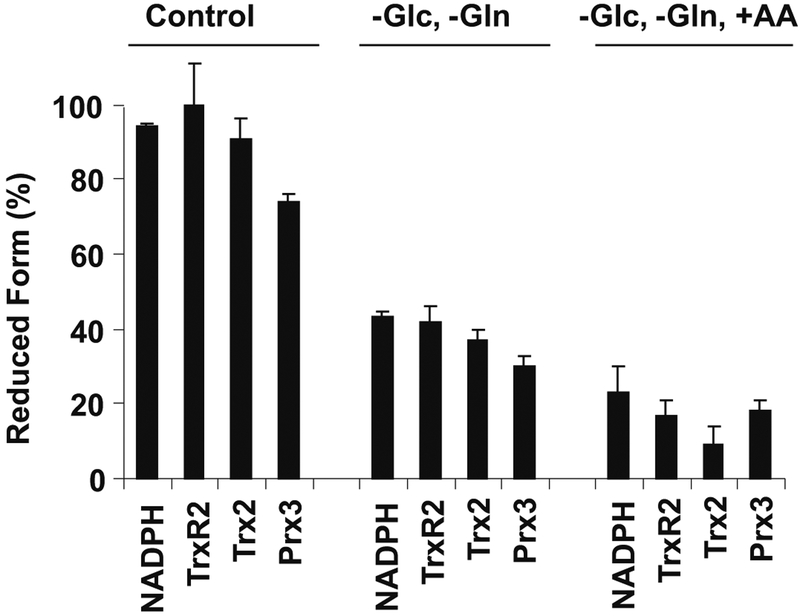
Electron transfer reactions between some of the components within the respiratory chain are very rapid, presenting a false impression that redox processes within the mitochondrion occur under near equilibrium conditions. In reality, the overall process is a non-equilibrium state, and partial rate control occurs at many steps. Similarly, proteins are synthesized with the thiol of Cys residues in the reduced state, presenting the impression that thiols are maintained in that form. However, the reality is that a fraction of the Cys residues are readily oxidized, and the steady-state generation of oxidants within mitochondria is sufficient to maintain many proteins in a partially oxidized state. An example is shown in Fig. 7 in which steady-state reduction of proteins in the pathway from NADH through TrxR2, Trx2, and Prx3 was examined under conditions which caused oxidation of NADPH. Results show that the entire pathway functions in a non-equilibrium steady state. Of critical importance, the reactions which are not at equilibrium (within the electron transfer chain and among the protein thiols) are of most interest in terms of control of mitochondrial functions, and these are the most difficult to study because of the methodological challenges to accurately trap the steady-state values.
Fig. 7.
Measurement of steady-state reduction of Trx2 by redox Western blotting and TrxR2 and Prx3 by BIAM blotting shows that mitochondrial thiol redox systems exist under non-equilibrium conditions in cells. The reduced fraction of TrxR2, Trx2, and Prx3 was decreased both by −Glc, −Gln media and by addition of respiratory substrates. Total cell NADPH was measured by HPLC and is shown for comparison. Results are representative of four experiments
Finally, an unresolved complexity lies in the relatively large number of modifications which thiols undergo under relevant biologic conditions. Irreversible modification occurs by reaction with the lipid oxidation product, 4-hydroxynonenal [51], and other reactive electrophiles. In addition, subsets of proteins are physiologically regulated by glutathionylation and nitrosylation [52]. The assays described above do not discriminate the contributions of these other modifications. Consequently, accurate descriptions of mitochondrial function will require more global approaches which capture the contributions of each rate controlling step.
Acknowledgments
Supported by NIH grants ES009047, ES011195, and ES012870.
References
- 1.Mitchell P (1979) Keilin’s respiratory chain concept and its chemiosmotic consequences. Science 206:1148–1159 [DOI] [PubMed] [Google Scholar]
- 2.Chance B, Sies H, Boveris A (1979) Hydroperoxide metabolism in mammalian organs. Physiol Rev 59:527–605 [DOI] [PubMed] [Google Scholar]
- 3.Meredith MJ, Reed DJ (1982) Status of the mitochondrial pool of glutathione in the isolated hepatocyte. J Biol Chem 257:3747–3753 [PubMed] [Google Scholar]
- 4.Wallace DC (1999) Mitochondrial diseases in man and mouse. Science 283:1482–1488 [DOI] [PubMed] [Google Scholar]
- 5.Jones DP (2006) Disruption of mitochondrial redox circuitry in oxidative stress. Chem Biol Interact 163:38–53 [DOI] [PubMed] [Google Scholar]
- 6.Chance B (1957) Cellular oxygen requirements. Fed Proc 16:671–680 [PubMed] [Google Scholar]
- 7.Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP (2003) Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J Biol Chem 278:19603–19610 [DOI] [PubMed] [Google Scholar]
- 8.Zhang R, Al-Lamki R, Bai L et al. (2004) Thioredoxin-2 inhibits mitochondria-located ASK1-mediated apoptosis in a JNK-independent manner. Circ Res 94:1483–1491 [DOI] [PubMed] [Google Scholar]
- 9.Lillig CH, Berndt C, Vergnolle O et al. (2005) Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc Natl Acad Sci U S A 102:8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Go YM, Pohl J, Jones DP (2009) Quantification of redox conditions in the nucleus. Methods Mol Biol 464:303–317 [DOI] [PubMed] [Google Scholar]
- 11.Jones DP (1984) Effect of mitochondrial clustering on O2 supply in hepatocytes. Am J Phys 247:C83–C89 [DOI] [PubMed] [Google Scholar]
- 12.Schafer FQ, Buettner GR (2001) Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med 30:1191–1212 [DOI] [PubMed] [Google Scholar]
- 13.Chance B (1954) Spectrophotometry of intracellular respiratory pigments. Science 120:767–775 [DOI] [PubMed] [Google Scholar]
- 14.Keilin D (1966) The history of cell respiration and cytochrome Cambridge University Press, Cambridge [Google Scholar]
- 15.Chance B (1952) Spectra and reaction kinetics of respiratory pigments of homogenized and intact cells. Nature 169:215–221 [DOI] [PubMed] [Google Scholar]
- 16.Jones DP, Thor H, Andersson B, Orrenius S (1978) Detoxification reactions in isolated hepatocytes. Role of glutathione peroxidase, catalase, and formaldehyde dehydrogenase in reactions relating to N-demethylation by the cytochrome P-450 system. J Biol Chem 253:6031–6037 [PubMed] [Google Scholar]
- 17.Tamura M, Hazeki O, Nioka S, Chance B (1989) In vivo study of tissue oxygen metabolism using optical and nuclear magnetic resonance spectroscopies. Annu Rev Physiol 51:813–834 [DOI] [PubMed] [Google Scholar]
- 18.Jones DP (1981) Determination of pyridine dinucleotides in cell extracts by high-performance liquid chromatography. J Chromatogr 225:446–449 [DOI] [PubMed] [Google Scholar]
- 19.Williamson JR, Corkey BE (1969) Assays of intermediates of the citric acid cycle and related components by fluorometric enzyme methods. Methods Enzymol 13:434–513 [Google Scholar]
- 20.Sies H (1982) Nicotinamide nucleotide compartmentation. In: Sies H (ed) Metabolic compartmentation. Academic Press, London, pp 205–231 [Google Scholar]
- 21.Kirwan GM, Coffey VG, Niere JO, Hawley JA, Adams MJ (2009) Spectroscopic correlation analysis of NMR-based metabonomics in exercise science. Anal Chim Acta 652:173–179 [DOI] [PubMed] [Google Scholar]
- 22.Beylot M, Beaufrère B, Normand S, Riou JP, Cohen R, Momex R (1986) Determination of human ketone body kinetics using stableisotope labelled tracers. Diabetologia 29:90–96 [DOI] [PubMed] [Google Scholar]
- 23.Jones DP, Kennedy FG (1982) Intracellular oxygen supply during hypoxia. Am J Phys 243:C247–C253 [DOI] [PubMed] [Google Scholar]
- 24.Chance B, Schoener B (1962) Correlation of oxidation-reduction changes of intracellular reduced pyridine nucleotide and changes in electroencephalogram of the rat in anoxia. Nature 195:956–958 [DOI] [PubMed] [Google Scholar]
- 25.Chance B, Cohen P, Jobsis F, Schoener B (1962) Intracellular oxidation-reduction states in vivo. Science 137:499–508 [DOI] [PubMed] [Google Scholar]
- 26.Song Y, Buettner GR (2010) Thermodynamic and kinetic considerations for the reaction of semiquinone radicals to form superoxide and hydrogen peroxide. Free Radic Biol Med 49 (6):919–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto Y, Ubiquinol YS (2002) Ubiquinone ratio as a marker of oxidative stress. Methods Mol Biol 186:241–246 [DOI] [PubMed] [Google Scholar]
- 28.Matsubara M, Ranji M, Leshnower BG et al. (2010) In vivo fluorometric assessment of cyclosporine on mitochondrial function during myocardial ischemia and reperfusion. Ann Thorac Surg 89:1532–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholz R, Thurman RG, Williamson JR, Chance B, Bucher T (1969) Flavin and pyridine nucleotide oxidation-reduction changes in perfused rat liver. I. Anoxia and subcellular localization of fluorescent flavoproteins. J Biol Chem 244:2317–2324 [PubMed] [Google Scholar]
- 30.Tamura M, Oshino N, Chance B, Silver IA (1978) Optical measurements of intracellular oxygen concentration of rat heart in vitro. Arch Biochem Biophys 191:8–22 [DOI] [PubMed] [Google Scholar]
- 31.Estabrook RW (1961) Studies of oxidative phosphorylation with potassium ferricyanide as electron acceptor. J Biol Chem 236:3051–3057 [PubMed] [Google Scholar]
- 32.Jones DP, Orrenius S, Mason HS (1979) Hemoprotein quantitation in isolated hepatocytes. Biochim Biophys Acta 576:17–29 [DOI] [PubMed] [Google Scholar]
- 33.Aw TY, Andersson BS, Jones DP (1987) Suppression of mitochondrial respiratory function after short-term anoxia. Am J Phys 252: C362–C368 [DOI] [PubMed] [Google Scholar]
- 34.Jones DP (1982) Intracellular catalase function: analysis of the catalatic activity by product formation in isolated liver cells. Arch Biochem Biophys 214:806–814 [DOI] [PubMed] [Google Scholar]
- 35.Guidot DM, Repine JE, Kitlowski AD et al. (1995) Mitochondrial respiration scavenges extramitochondrial superoxide anion via a nonenzymatic mechanism. J Clin Invest 96:1131–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DP (2006) Redefining oxidative stress. Antioxid Redox Signal 8:1865–1879 [DOI] [PubMed] [Google Scholar]
- 37.Sies H, Jones DP (2007) Oxidative stress In: Fink G (ed) Encyclopedia of stress, 2nd edn Elsevier, Amsterdam, pp 45–48 [Google Scholar]
- 38.Zhang H, Go YM, Jones DP (2007) Mitochondrial thioredoxin-2/peroxiredoxin-3 system functions in parallel with mitochondrial GSH system in protection against oxidative stress. Arch Biochem Biophys 465:119–126 [DOI] [PubMed] [Google Scholar]
- 39.Jones DP (2008) Radical-free biology of oxidative stress. Am J Physiol Cell Physiol 295: C849–C868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halvey PJ, Watson WH, Hansen JM, Go YM, Samali A, Jones DP (2005) Compartmental oxidation of thiol-disulphide redox couples during epidermal growth factor signalling. Biochem J 386:215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood ZA, Schroder E, Robin Harris J, Poole LB (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28:32–40 [DOI] [PubMed] [Google Scholar]
- 42.Cox AG, Winterbourn CC, Hampton MB (2010) Measuring the redox state of cellular peroxiredoxins by immunoblotting. Methods Enzymol 474:51–66 [DOI] [PubMed] [Google Scholar]
- 43.Padgett CM, Whorton AR (1995) S-nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am J Phys 269: C739–C749 [DOI] [PubMed] [Google Scholar]
- 44.Stadtman TC (2002) Discoveries of vitamin B12 and selenium enzymes. Annu Rev Biochem 71:1–16 [DOI] [PubMed] [Google Scholar]
- 45.Gasdaska PY, Gasdaska JR, Cochran S, Powis G (1995) Cloning and sequencing of a human thioredoxin reductase. FEBS Lett 373:5–9 [DOI] [PubMed] [Google Scholar]
- 46.Mustacich D, Powis G (2000) Thioredoxin reductase. Biochem J 346(Pt 1):1–8 [PMC free article] [PubMed] [Google Scholar]
- 47.Soini Y, Kahlos K, Napankangas U et al. (2001) Widespread expression of thioredoxin and thioredoxin reductase in non-small cell lung carcinoma. Clin Cancer Res 7:1750–1757 [PubMed] [Google Scholar]
- 48.Kim JR, Lee SM, Cho SH et al. (2004) Oxidation of thioredoxin reductase in HeLa cells stimulated with tumor necrosis factor-alpha. FEBS Lett 567:189–196 [DOI] [PubMed] [Google Scholar]
- 49.Go YM, Park H, Koval M et al. (2010) A key role for mitochondria in endothelial signaling by plasma cysteine/cystine redox potential. Free Radic Biol Med 48:275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holmgren A, Fagerstedt M (1982) The in vivo distribution of oxidized and reduced thioredoxin in Escherichia coli. J Biol Chem 257:6926–6930 [PubMed] [Google Scholar]
- 51.Roede JR, Jones DP (2010) Reactive species and mitochondrial dysfunction: mechanistic significance of 4-hydroxynonenal. Environ Mol Mutagen 51:380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Requejo R, Chouchani ET, Hurd TR, Menger KE, Hampton MB, Murphy MP (2010) Measuring mitochondrial protein thiol redox state. Methods Enzymol 474:123–147 [DOI] [PubMed] [Google Scholar]