Abstract
Telomeres are protein–DNA complexes that protect chromosome ends from illicit ligation and resection. Telomerase is a ribonucleoprotein enzyme that synthesizes telomeric DNA to counter telomere shortening. Human telomeres are composed of complexes between telomeric DNA and a six-protein complex known as shelterin. The shelterin proteins TRF1 and TRF2 provide the binding affinity and specificity for double-stranded telomeric DNA, while the POT1-TPP1 shelterin subcomplex coats the single-stranded telomeric G-rich overhang that is characteristic of all our chromosome ends. By capping chromosome ends, shelterin protects telomeric DNA from unwanted degradation and end-to-end fusion events. Structures of the human shelterin proteins reveal a network of constitutive and context-specific interactions. The shelterin protein–DNA structures reveal the basis for both the high affinity and DNA sequence specificity of these interactions, and explain how shelterin efficiently protects chromosome ends from genome instability. Several protein–protein interactions, many provided by the shelterin component TIN2, are critical for upholding the end-protection function of shelterin. A survey of these protein–protein interfaces within shelterin reveals a series of “domain–peptide” interactions that allow for efficient binding and adaptability towards new functions. While the modular nature of shelterin has facilitated its part-by-part structural characterization, the interdependence of subunits within telomerase has made its structural solution more challenging. However, the exploitation of several homologs in combination with recent advancements in cryo-EM capabilities has led to an exponential increase in our knowledge of the structural biology underlying telomerase function. Telomerase homologs from a wide range of eukaryotes show a typical retroviral reverse transcriptase-like protein core reinforced with elements that deliver telomerase-specific functions including recruitment to telomeres and high telomere-repeat addition processivity. In addition to providing the template for reverse transcription, the RNA component of telomerase provides a scaffold for the catalytic and accessory protein subunits, defines the limits of the telomeric repeat sequence, and plays a critical role in RNP assembly, stability, and trafficking. While a high-resolution definition of the human telomerase structure is only beginning to emerge, the quick pace of technical progress forecasts imminent breakthroughs in this area. Here, we review the structural biology surrounding telomeres and telomerase to provide a molecular description of mammalian chromosome end protection and end replication.
Keywords: Telomerase, Telomeres, Shelterin, End replication, End protection, Meiosis, DNA damage response, DNA repair, ATM, ATR
Introduction
Eukaryotic chromosomes are linear and end in nucleoprotein complexes called telomeres. Telomeric DNA is composed of a repetitive sequence (GGTTAG/CCAATC in humans) that is largely double-stranded (ds; 10–15 kb in humans), but ends in a short single-stranded (ss; 50–500 nt in humans) G-rich 3′ overhang [1]. Linear chromosomes present two major biological hurdles at chromosome ends: the end replication and end protection problems. The end-replication problem exists, because DNA polymerases are unable to fully replicate chromosome ends. During lagging strand DNA replication, the RNA primer at the extreme 5′ end is removed, leaving a gap that cannot be filled by DNA polymerase. This results in the loss of DNA at the very ends of chromosomes during each replication cycle [2]. The specialized ribonucleoprotein enzyme telomerase counteracts this DNA attrition by synthesizing new telomeric repeats at chromosome ends (Fig. 1) [3]. The protein subunit of telomerase, called telomerase reverse transcriptase (TERT), contains a catalytic reverse transcriptase domain for DNA synthesis [4–6]. The RNA subunit of telomerase called Telomerase RNA component (TERC or TR) contains, amongst other elements, the template for telomeric repeat addition (Fig. 1) [7, 8].
Fig. 1.
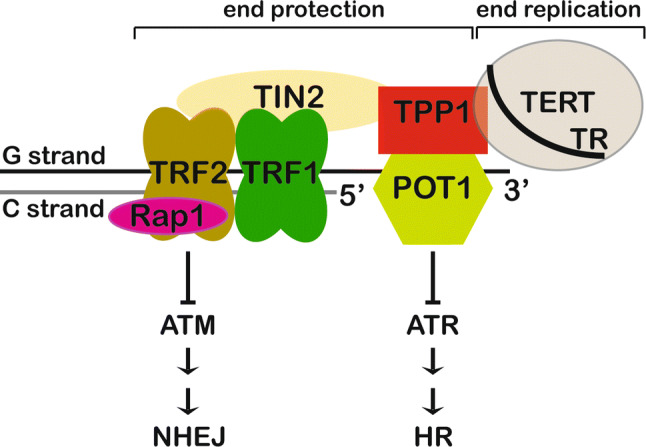
Schematic showing the composition of complexes involved in chromosome end protection and end replication. The shelterin complex (POT1, TPP1, TRF1, TRF2, TIN2, and Rap1 proteins) protects chromosome ends from ATM kinase and ATR kinase-mediated DNA damage response pathways. The telomerase RNP (TERT protein and TR RNA subunits) adds telomeric DNA de novo at the 3′ ends of chromosomes
Telomerase is expressed in both germ line and somatic stem cells to facilitate their continued cell division [9]. It is, therefore, not surprising that mutations in telomere- and telomerase-associated genes result in inherited stem cell-dysfunction diseases collectively known as “telomeropathies”, the most notable of which is dyskeratosis congenita or DC [10–12]. Normally, nondividing cells lacking telomerase enter a non-proliferative state called senescence once their telomere length falls below a certain threshold [13]. This is an anti-tumorigenic mechanism to prevent uncontrolled cell division. However, if a rare cell escapes senescence and aberrantly resumes telomerase expression to re-establish telomere length maintenance, it could attain “replicative immortality”, a hallmark of cancer [14, 15]. In fact, an overwhelming majority of cancers (~ 80%) show telomerase expression [16, 17], and thus, this enzyme is a promising target for anti-cancer drug discovery.
While telomerase solves the end-replication problem, linear chromosomes must also solve the end protection problem. The end protection problem occurs when the natural ends of linear chromosomes are misrecognized by the DNA damage response and repair machinery, as double strand breaks requiring repair [1]. The six-protein complex shelterin, consisting of proteins POT1, TPP1, TIN2, TRF1, TRF2, and Rap1, solves the end protection problem by coating telomeric DNA (Figs. 1, 2) [1, 18–28]. While TRF1 and TRF2 recognize ds telomeric DNA, POT1 binds the ss overhang with high specificity and affinity [21, 29]. By coating telomeric DNA, the shelterin complex sequesters it away from the ATM and ATR-mediated DNA damage response pathways (Fig. 1) [1, 11]. Although continuously dividing cells must protect chromosome ends from the DNA damage response machinery, they must also allow recruitment of telomerase to those ends. This is the essence of the telomerase recruitment problem, which is primarily solved by the telomere protein TPP1 [30]. Finally, telomeres also perform an essential function in meiosis. During prophase I in meiosis, homologous chromosomes pair together and undergo DNA recombination, a process central to generating genetic diversity [31]. Telomeres play an essential role in this process by attaching chromosomes to the inner nuclear membrane (INM), and facilitating homolog pairing and genetic crossover [32–36]. Telomere–INM engagement is achieved by interaction of the shelterin component TRF1 with the meiosis-specific complex TERB1–TERB2–MAJIN that localizes at the INM [37]. In this review, we will focus on the structural biology surrounding telomeres and telomerase to describe our current understanding of how the coordinated action of various protein–protein and protein–nucleic acid interactions results in the proper maintenance of chromosome ends and genome stability.
Fig. 2.
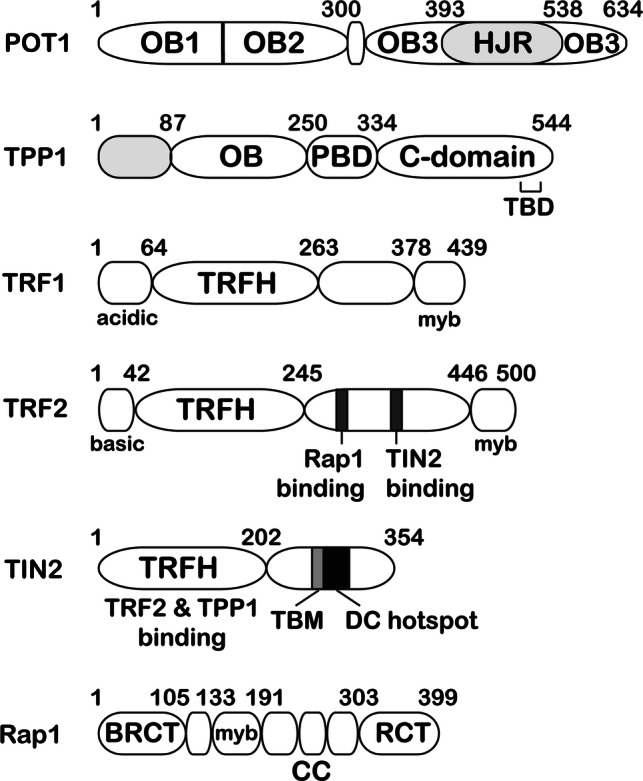
Domain diagrams of the six human shelterin proteins. DC hotspot indicates a stretch of amino acids in TIN2 that is host to mutations associated with telomeropathies such as dyskeratosis congenita (DC). TBD indicates the TIN2-binding region of TPP1. TBM of TIN2 contains the TRF1-binding F–X–L–X–P motif
Single-stranded chromosome end protection
Ciliates provide the first insights
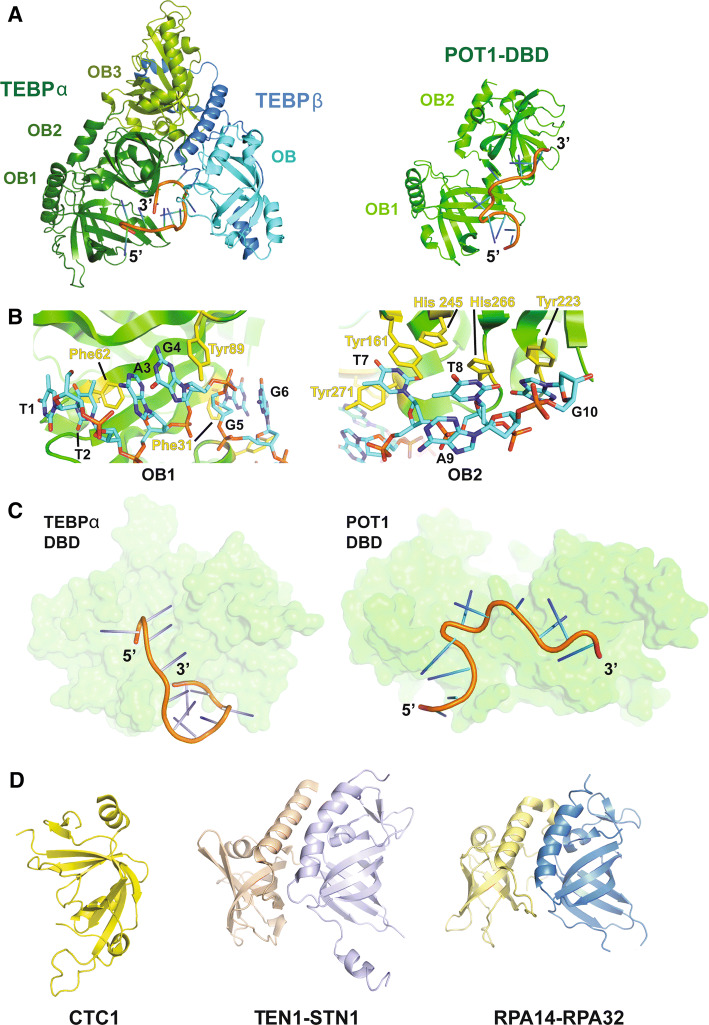
The very end of any eukaryotic chromosome is single-stranded and thus a potential substrate for illicit homology-driven recombination or repair. Thus, a major challenge in end protection involves preventing this DNA from participating in such processes. The G-rich 3′ ss overhang of telomeric DNA is also involved in end replication, as it provides the site for telomerase to bind and extend chromosome ends. Both of these functions are either directly dictated by (in case of end protection) or facilitated by (in case of end replication) proteins that bind the ss overhang. The first major structural insights into G-rich 3′ ss DNA-binding proteins came from the structure of the Sterkiella nova (S. nova; formerly Oxytricha nova) proteins TEBP-α and TEBP-β, which bind as a heterodimer to ss DNA at chromosome ends (left; Fig. 3a) [38, 39]. The TEBP-α protein is composed of three oligosaccharide/oligonucleotide binding (OB) domains. The two N-terminal OB domains collectively form a DNA-binding domain (DBD) that binds specifically to the 3′ ss overhang of the telomeric DNA. [39, 40]. The protein–DNA interface is formed between residues on both TEBP-α and β, and the bases and deoxyribose groups of the DNA. Thus, this interface is rich in hydrogen bonding, and hydrophobic and pi-stacking interactions. In fact, almost every base in the protein–DNA co-crystal structure stacks with an aromatic amino acid or another base. Following the track that the DNA takes through the structure reveals that the 3′ end interacts not only with TEBP-α, but also several residues in TEBP-β (left; Fig. 3a). The terminal 3′ nucleotide G11 folds back to stack with nucleotide T7 burying the extreme end of the chromosome within the protein–DNA complex (Fig. 3a, c). Because of these extensive interactions, the TEBP-α and TEBP-β heterodimer is able to protect the ss tail of the telomere from potential interactions with DNA damage response proteins. Although this structure provides a compelling solution to the end protection problem, it is likely that the TEBP-α-β-DNA complex undergoes a conformational change to allow telomerase to access and extend the DNA ends.
Fig. 3.
Structural basis for single-stranded chromosome end protection. a Left: the structure of TEBP-α-β in complex with ss DNA (5′-d(GGGTTTTGGGG)-3′) (PDB ID: 2I0Q). The DBD and TEBP-β-binding domain of TEBP-α are shown in forest green and split pea green, respectively. The TEBP-α binding domain and OB domain of TEBP-β are shown in blue and cyan, respectively. Right: the human POT1 DNA-binding domain (DBD; PDB ID: 1XJV) shown in green in complex with telomeric DNA [5′-d(TTAGGGTTAG)-3′]. The DNA is shown in cartoon representation. b Views showing stacking interactions between the two OB domains of hPOT1 DBD and telomeric DNA. The stacking protein and DNA residues are shown in stick representation. c Views of TEBP-α DBD in complex with telomeric DNA (left; from the TEBP-α–β–DNA structure) and POT1 DBD in complex with telomeric DNA (right) to highlight the different tracks adopted by the DNA molecules in the two structures. The proteins are shown in surface representation and the DNA is rendered in cartoon representation. d Left: the central domain of human CTC1 (PDB ID: 5W2L). Center: high-resolution structure of the human STN1–TEN1 complex (PDB ID: 4JOI). STN1 is shown in wheat and TEN1 is shown in pale violet. Right: heterodimeric structure of RPA14 (yellow) and RPA 32 (blue) of the trimeric RPA complex (PDB ID: 1QUQ)
Protection of ss DNA at chromosome ends by human POT1-TPP1
Human telomeres are longer than that of S. nova and are comprised of more proteins, but they still must overcome the same biological hurdles. The protein that protects the ss overhang at human chromosome ends is called POT1 (Figs. 1, 2), a homolog of which also exists in Schizosaccharomyces pombe (S. pombe) [23]. Not surprisingly, both S. pombe and human POT1 have a similar overall structure to that of S. nova TEBP-α [29, 41]. The DNA-binding domain (DBD) of human POT1 uses two N-terminal OB domains (OB1 and OB2) to bind to the ss overhang of telomeric DNA (right; Fig. 3a). A crystal structure of this complex details how OB1 of POT1 is able to interact with the first six bases of the 10 nt telomeric sequence (1-TTAGGG-6) (left; Fig. 3b) in a fashion similar to that of the homologous S. pombe POT1pN protein fragment (not shown). Both structures detail stacking interactions between aromatic residues in POT1 and the bases of the telomeric DNA. In addition to these stacking interactions, OB1 makes several hydrogen bonds with the telomeric DNA. OB2 of human POT1 makes less extensive hydrogen bonds with telomeric DNA, but the four 3′ bases in the crystal structure (7-TTAG-10) all stack with aromatic residues present on OB2 (right; Fig. 3b). Interestingly, OB2 of S. pombe Pot1 exhibits lower sequence specificity compared to the adjacent OB1 domain and binds optimally to a 9-mer DNA sequence [42]. Although the overall structure of human POT1’s DBD is very similar to that of TEBP-α’s DBD, the track of the DNA through the two OB domains in these structures is strikingly different (Fig. 3c). In POT1, the DNA is kinked, as it passes from OB1 to OB2. This occurs, because the two OB domains in the POT1 structure are oriented differently relative to one another than they are in the S. nova structure (Fig. 3a). In addition, the 3′ end of the DNA does not curve back towards POT1, as it does in the S. nova structure (Fig. 3c). These differences may be attributed to the fact that the S. nova structure contains the full TEBP-α-β complex bound to DNA, while the POT1-DNA structure lacks both TPP1 (mammalian TEBP-β homolog) and the TPP1-binding domain of POT1. Alternatively, these structural differences may suggest distinct species-specific solutions for end protection (see discussion of human POT1–TPP1–DNA SAXS data below).
The high affinity and specificity of POT1 for ss telomeric DNA provide an elegant mechanism for protection against an ATR kinase-mediated response and subsequent homologous recombination (HR) at chromosome ends [41, 43]. An essential initial step for HR is binding of the participating ss DNA with replication protein A (RPA), a heterotrimer composed largely of OB domains (Fig. 3d) [44]. By binding ss telomeric DNA with high affinity and sequence specificity, POT1 blocks access of RPA to prevent ATR activation at chromosome ends (Fig. 1) [45]. Indeed, mutations in the DNA-binding domain of POT1 are associated with chromosome end aberrations including fusions in several chronic lymphocytic leukemias (CLL) and familial melanomas (FM), highlighting the importance of this protein in maintaining genome integrity [46, 47].
Faithful protection of ss chromosome ends not only mandates POT1 to outcompete RPA, but also requires POT1 to avoid binding to TERRA, a non-coding ss G-rich RNA that is transcribed from and localized to the telomeres [48, 49]. TERRA contains many potential POT1 binding sites and is more abundant than the ss overhang. Yet, POT1 is able to avoid this RNA decoy and selectively bind to telomeric DNA. Binding studies and a structure of the POT1 DBD complexed with dTrUd(AGGGTTAG) have revealed that the introduction of a ribonucleotide (rU) at the second dT position from the 5′ end severely weakens the interaction between POT1 and its cognate nucleic acid, providing the basis for DNA versus RNA discrimination by POT1 [50]. The preference for DNA over RNA of POT1 is further increased by binding of TPP1 [50], although the structural basis for this remains unknown.
Another protein complex that localizes to telomeres and which can bind to ss DNA is the CST (CTC1–STN1–TEN1) complex [51]. This complex was first identified in yeast and is essential to protect yeast telomeres. In humans, CST is able to act as terminator of telomerase activity [52] and is responsible for recruiting polymerase-α-primase for fill-in synthesis of the C-strand after telomerase extension of the G-strand [53]. CST has been reported to bind both POT1 and TPP1, providing a possible mechanism for how this complex is recruited to telomeres [52, 54]. The role of CST in fill-in synthesis extends beyond telomeres, as it is now known that CST is recruited to double-stranded breaks containing 53BP1, RIF1, and the shieldin complex to counteract resection [55]. Yeast and human CTC1 and STN1–TEN1 structures have been solved [56–59]. Both STN1 and TEN1 have OB folds that resemble that of the RPA complex, suggesting that the CST complex evolved from an RPA-like ancestral complex (Fig. 3d). More recently, human STN1–TEN1 has been shown to help counteract replication fork stalling, further suggesting that these proteins perform a wide array of functions that extends beyond their role at telomeres [60, 61].
POT1–TPP1 interface
The first structural insights into how the POT1–TPP1 heterodimer forms to protect chromosome ends came from the analysis of the TEBP-α–β complex. TEBP-α has a third C-terminal OB domain that makes extensive interactions with a C-terminal region of TEBP-β that is partly helical, but mostly extended in conformation (Fig. 3a). However, the C-terminus of POT1 does not share much sequence similarity with TEBP-α making it unclear if TPP1 and POT1 would interact in a similar manner. Two similar structures of the C-terminus of POT1 in complex with the POT1 binding domain (PBD) of TPP1 were solved using X-ray crystallography [62, 63]. These structures revealed that the C-terminus of POT1, like that of TEBP-α, forms an OB domain that is able to make extensive interactions with TPP1 (Fig. 4a). Unexpectedly, the authors also identified an additional TPP1-binding element within POT1: a holiday junction resolvase-like domain (HJRD) inserted within POT1’s third OB domain (OB3). In both structures, the two α-helices and a 310 helix of TPP1’s PBD lie in grooves formed by the OB3 and the HJRD domains of POT1. Somatic mutations associated with several CLL and FL malignancies are mapped to the TPP1-binding region of POT1 [62, 63]. This is not surprising, because the inability to bind TPP1 will prevent recruitment of POT1 to telomeres, thereby unleashing chromosome end deprotection (see section on Hierarchical assembly of shelterin at telomeres below) [45, 64, 65].
Fig. 4.
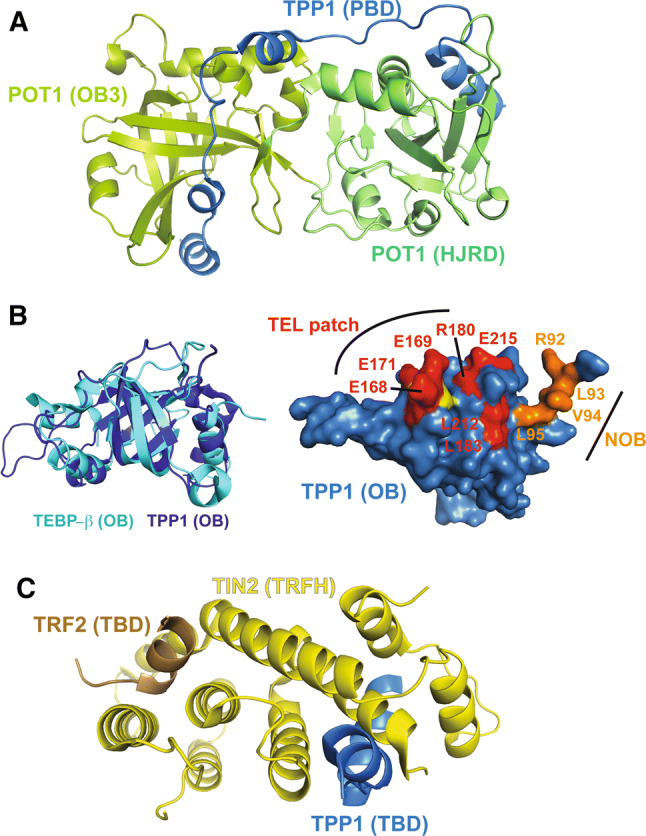
Crystal structures of TPP1 domains and related protein–protein interfaces. a Crystal structure of the C-terminal domains of POT1 (green) in complex with the PBD of TPP1 (blue) (PDB ID: 5UN7 and 5H65). b Left: overlay of the human TPP1 OB domain (dark blue; PDB ID: 2I46) and the OB domain of TEBP-β (cyan; PDB ID: 2I0Q). Right: surface view of the TPP1 OB domain. TEL patch residues are shown in red and NOB residues are shown in orange. K170, deletion of which is implicated in telomeropathies such as DC, is shown in yellow. c High-resolution structure of TIN2’s TRFH domain (brown) bound to TPP1 (blue) and TRF2 (brown) (PDB ID: 5XYF)
Although there is no high-resolution structure for the human POT1–TPP1–DNA complex, small-angle X-ray scattering (SAXS) analysis of the TPP1-N–POT1–DNA complex has revealed an elongated V-shaped envelope. In the model generated with these data, TPP1 OB is proposed to be distal to both the DBD of POT1 and the telomeric DNA [62]. This is in stark contrast to the overall heart-shaped structure of TEBP-α–β–DNA that brings both subunits in close proximity and allows them to interact simultaneously with ss DNA. A high-resolution human POT1–TPP1–DNA structure will be instrumental to address the apparent differences in how ss DNA ends are protected in S. nova versus human.
To protect chromosome ends or to allow telomerase access?
One major question that still eludes the field is how telomerase accesses a chromosome end that is so efficiently protected by POT1–TPP1. POT1–TPP1 plays a dual role at telomeres by stimulating telomerase activity and processivity while also protecting telomeric DNA with the help of POT1 [66]. POT1 tethers TPP1 close to the 3′ end, where TPP1 is able to directly recruit telomerase to the telomere using its N-terminal OB domain [66–68]. The crystal structure of the TPP1 OB domain illustrates a five-stranded β-barrel that resembles the OB domain found in TEBP-β (Fig. 4b) [66]. Several independent studies led to the discovery of the TEL patch [TPP1 glutamate (E) and leucine (L)-rich patch] in TPP1 OB as a critical surface element for binding telomerase [69–71]. Three critical glutamate residues in the TEL patch (E168, E169, and E171) lie in a long flexible loop that protrudes from the β-barrel and ends in a short α-helix (Fig. 4b). Indeed, deletion of residue K170, which is flanked by these three glutamate residues, is associated with telomeropathies in two unrelated families, further highlighting the role of this patch in telomerase function (Fig. 4b) [72–74]. In the asymmetric unit of the TPP1 OB crystal structure, TPP1 forms a non-physiological dimer using its largely hydrophobic N-terminal tail. This region, termed the NOB (N-terminus of the OB domain), is also involved in recruiting telomerase (Fig. 4b) [75]. Although much has been uncovered about the surface of TPP1 that recruits telomerase, far less is known about the surface of telomerase that interacts with TPP1. To date, only one direct salt-bridging interaction is firmly mapped between human TPP1 and telomerase [76]. While structural information is lacking for the human TPP1–telomerase interaction, provocative insights into this interface can be obtained from the existing structures of Tetrahymena thermophila telomerase holoenzyme (discussed later). Future structural analysis of POT1–TPP1–DNA in the presence and absence of telomerase is necessary to understand how telomeres switch between the end-protection state provided by POT1–TPP1 and the more open end-replication state that allows for telomerase action.
Double-stranded chromosome end protection
Shelterin and telomere-associated proteins
While sequestration of the ss G-rich overhang is an essential part of end protection, telomeric DNA is mostly double-stranded in nature [1]. Therefore, a major task of shelterin is to coat and protect ds telomeric DNA. TRF1 and TRF2 constitute the ds DNA-binding proteins of shelterin [21, 77]. Both proteins have a similar domain layout, with each having a dimerization domain and a C-terminal DNA-binding myb domain (Fig. 2) [1]. At the N-terminus, TRF1 contains a region rich in acidic residues, while TRF2 contains a highly basic stretch of amino acids known as the basic domain or GAR (Gly/Arg-rich) region. TRF1 and TRF2 assemble the remaining shelterin proteins (TIN2, TPP1, POT1, and Rap1) qualifying these as critical determinants of chromosome end protection [78]. Given their importance in maintaining genome stability, it is not surprising that knock out of either TRF1 or TRF2 results in mouse embryonic lethality [79, 80]. TRF1 aids DNA replication through the G-rich repetitive telomeric DNA sequences. As a result, TRF1 deficiency leads to the characteristic “fragile telomere” phenotype [81]. TRF2 plays a pivotal role in end protection by preventing activation of the ATM kinase DNA damage response (Fig. 1) [82–84]. TRF2 also facilitates formation of telomeric structures known as t-loops, which bury the ss telomeric DNA end presumably to help prevent its recognition by the DNA damage response machinery [85–87]. In addition to their direct roles in maintaining chromosome integrity, TRF1 and TRF2 serve as docking sites for transiently recruiting accessory factors in a biological context-specific manner (discussed later) [88].
Double-stranded telomeric DNA binding by TRF1 and TRF2: TRF1 and TRF2 are able to specifically bind ds telomeric DNA via a C-terminal DNA-binding domain (Fig. 5a) related to the DNA-binding region of the transcriptional activator protein c-myb [21]. In both structures, the Myb-related domain adopts a similar three-helix conformation, where the third helix specifically contacts the major groove, binding to the double-stranded sequence 5′ AGGGTT 3′ [89–92]. Although in vivo dimerization of TRF1 and TRF2 enhances their DNA-binding abilities, there seems to be no constraint on the distance between bound myb domains, allowing these TRF proteins to induce loops and higher order structures in telomeric DNA [89]. This is facilitated by the presence of a long-linker region between the TRFH and myb domains of both TRF1 and TRF2 (Fig. 2).
Fig. 5.
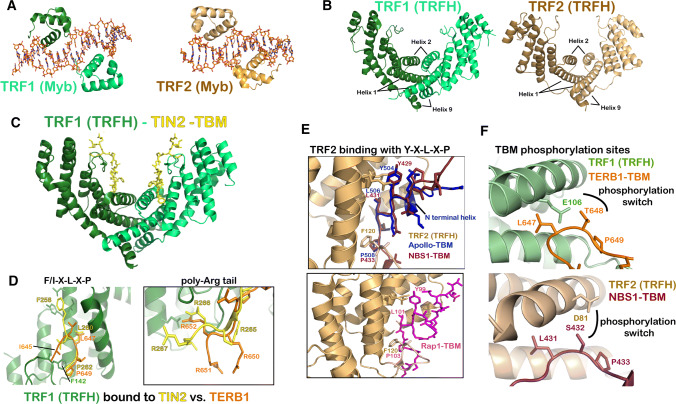
Structural biology of TRF1, TRF2, and binding partners. a Myb domain of TRF1 (left; PDB ID: 1W0T) and TRF2 (right; PDB ID: 1W0U) bound to ds telomeric DNA. The protein is shown in cartoon representation and the DNA is rendered in stick representation. b Structure of the TRFH dimerization domains of TRF1 (Left; PDB ID: 1H6O) and TRF2 (Right; PDB ID: 1H6P). c Overall heart-shaped structure of heterodimeric TRFH domain of TRF1 bound to two TIN2 peptides (PDB ID: 3BQO). TRFH is shown as a cartoon in green and the TIN2–TBM is shown as sticks in yellow. d Overlay of TIN2-bound (yellow; PDB ID: 3BQO) and TERB1-bound (orange; PDB ID 5WIR and 5XUP) TRF1 TRFH structures. e Top: overlay of Apollo-bound (blue; PDB ID: 3BUA) and NBS1-bound (ruby; PDB ID: 5WQD) TRF2 TRFH structures. Bottom: structure of TRF2 TRFH bound to a Y–X–L–X–P containing peptide in the BRCT domain of Rap1 (PDB ID: 4RQI). f Structural basis for phosphorylation regulated dissociation of TRF1-TERB1 (top; PDB ID: 5WIR and 5XUP) and TRF2-NBS1 (PDB ID: 5WQD; bottom) interactions
Homodimerization of TRF1 and TRF2: The TRF homology domain (TRFH) not only accomplishes homodimerization of TRF1 and TRF2 proteins, but also prevents their heterodimerization [93]. Although the TRFH domains of TRF1 and TRF2 have a relatively low sequence identity (27%), their overall structures are almost indistinguishable [93]. Nine alpha-helices make up one monomer, with a symmetrical horseshoe-shaped dimer being formed from two antiparallel monomers (Fig. 5b). The TRFH structures show that a large homodimeric interface is formed by helices 1, 2, and 9 from each monomer. Helices 1 and 2 from one monomer stack against the corresponding helices from the second monomer forming an antiparallel helical bundle. Helix 9 from each monomer packs perpendicular to its respective helix 1, helping to stabilize this bundle. A large hydrophobic core formed between the helices at the interface explains what drives TRF1 and TRF2 to exist as stable homodimers in vitro and in vivo [93]. The TRFH structures also explain why these two proteins with similar properties, domain architecture, and overall structures do not heterodimerize. A combination of differences in helix length and interacting side chains would result in charge and steric clashes between approaching TRF1 and TRF2 monomers.
TRF1 and TRF2 form docking sites for telomere accessory proteins
In addition to facilitating homodimerization and preventing heterodimerization, sequence differences between the TRFH domains of TRF1 and TRF2 enable them to recruit distinct sets of client proteins to telomeres. It has been shown that TRF1 has an affinity for proteins which contain an F–X–L–X–P motif (where X can be any amino acid), the most notable of which is fellow shelterin protein TIN2, while TRF2 has a preference for binding proteins containing a Y–X–L–X–P motif, examples of which include Apollo/SNM1B and NBS1 (Fig. 5c–e) [88].
TRF1 and TRF1-binding proteins: TIN2 is able to bind ds DNA-binding proteins TRF1 and TRF2, as well as TPP1, thereby bridging the ds and ss DNA-binding proteins in the shelterin complex [1]. It is, therefore, not surprising that removing TIN2 has a negative effect on shelterin assembly and stability. In mice, TRF1, but not TRF2, is required to recruit TIN2 to telomeres [94]. It has been shown that TIN2 utilizes an F–X–L–X–P-binding motif (258FNLAP262) to interact with a docking site formed by alpha-helices 3 and 4, and loop 34 of the dimerization domain (TRFH) of TRF1 (Fig. 5d) [88]. The structure shows that two TIN2 TRF-binding motifs (TBMs) can bind to a single TRF1 dimer and this stoichiometry has also been suggested by biochemical analysis of the full-length protein (Fig. 5c) [88, 95].
Although TIN2 is the primary TRF1-binding protein in the cell, the TBM docking site of TRF1 is also exploited by other proteins that require temporary access to telomeres in particular biological contexts. TERB1 (telomere-repeat-binding bouquet formation protein 1) is one such protein, which is expressed specifically in meiotic prophase I during which it binds TRF1 [96]. Along with two other meiotic proteins, TERB2 and MAJIN, TERB1 initiates the formation of a meiotic complex that connects telomeres to the inner nuclear membrane (INM) [37, 96–98]. This allows chromosomes to be tethered to the cytoskeletal motors, via proteins that span the nuclear membrane. Rapid chromosome movements that result from these cytoskeletal forces facilitate homologous chromosome pairing during meiosis [36]. The telomere–INM link results from direct binding of TERB1 to the TRFH domain of TRF1 [96]. Two recently solved structures of TRF1TRFH bound to its binding partner site in TERB1 (called TERB1TBM) demonstrated that TERB1 utilizes an I–X–L–X–P motif (645 ILLTPRRR 652) to bind TRF1, a slight variation on that of the TIN2TBM (258 FNLAPLGRRR 267), with both TBMs containing a downstream arginine tail for making additional interactions with TRF1 (Fig. 5d) [95, 99]. The structures show that the TERB1 isoleucine 645 is excluded from the hydrophobic pocket that the corresponding phenylalanine 258 of TIN2 binds, explaining the weaker affinity of TERB1TBM for TRF1 compared to TIN2TBM. Mutations in the TRF1-TERB1 interface resulted in a decrease in telomere–INM tethering in mouse spermatocytes [95, 99]. The physiological importance of the TRF1-TERB1 interface is further highlighted by the finding that interface mutants slow down spermatogenesis, allowing autosomes but not X–Y chromosomes to pair [99]. This results in male-specific infertility in mice and may be explained by the limited region of pairing between X–Y chromosomes that would result in a greater dependence on intact TRF1–TERB1 binding.
During meiotic prophase I, after telomere attachment to the INM, phosphorylation of TERB1 triggers a phenomenon known as telomere cap exchange [37]. During this process, the shelterin complex appears to be temporarily displaced from the telomeric DNA, and replaced by meiotic proteins TERB1, TERB2, and MAJIN. Cap exchange suggests that these meiotic proteins can somehow fulfill the function of protecting chromosome ends even in the absence of shelterin bound to the DNA, although a molecular/structural basis for this mechanism is currently lacking. The TRF1TRFH–TERB1TBM structures show that phosphorylation of T648 in TERB1 would cause an electrostatic clash with negatively charged E106 of TRF1 providing a structural basis for how TERB1 post-translational modification facilitates cap exchange (top; Fig. 5f) [95, 99]. It has also been shown that a gain of function I645F mutation in TERB1 (that reinstates the F–X–L–X–P motif found in TIN2) perturbs TRF1–TERB1 dissociation in the late pachytene stage of prophase I [95]. These observations suggest that the TERB1–TRF1 interaction is optimally tuned, such that it is strong enough to facilitate initial telomere–INM tethering, but can subsequently be severed later in meiosis via phosphorylation of TERB1. The recent discovery of meiosis-specific proteins TERB1, TERB2, and MAJIN provides a fertile ground for future research on the structural biology of telomere–INM tethering.
TRF2 and TRF2-binding proteins: Like TRF1, TRF2 also utilizes its dimerization domain to recruit accessory proteins to telomeres. However, the F–X–L–X–P motif is non-optimal for an interaction with TRF2, as TRF2 binds TIN2TBM with much less affinity than TRF1 [88]. TRF2TRFH binds strongly to at least two other telomere-associated proteins that are important for TRF2’s role in ATM repression: exonuclease Apollo-SNM1B, and NBS1, a member of the double strand break sensing complex MRN (MRE11–RAD50–NBS1) [100]. Both Apollo-SNM1B and NBS1 utilize a Y–X–L–X–P motif to bind TRF2, and crystallographic studies have shown that these TRF2 TBMs bind to TRF2TRFH in a highly similar fashion (top; Fig. 5e) [88, 100]. NBS1’s decreased affinity for TRF2 compared to that of Apollo is explained by differences in the N-terminal regions of the two TBMs. The ApolloTBM N-terminal residues form short helices which are able to pack on helices 2 and 3 of the TRF2TRFH domain, whereas the N-terminus of the NBS1TBM forms either a pseudo-helix or is largely unstructured (in the two protomers that bind to the two TRF2 monomers), neither of which makes substantial contacts with TRF2. It is interesting to note that while additional interactions C-terminal to the F–X–L–X–P motif give rise to stronger interactions with TRF1, regions N-terminal to the Y–X–L–X–P accomplish the same goal with TRF2.
NBS1 contains a CDK2 phosphorylation site which regulates its binding to TRF2 [100]. In stage G1 of the cell cycle, NBS1 is maintained in a dephosphorylated state, promoting its interaction with TRF2. In S/G2 phase, CDK2-mediated phosphorylation of NBS1 releases NBS1 from TRF2. This allows Apollo-SNM1B to bind TRF2 and protect leading strand telomeres from NHEJ-mediated repair. Phosphorylated NBS1 is able to promote ATM signaling at telomeres, where TRF2 and Rap1 are absent. Conversely dephosphorylated NBS1 is necessary to promote ATR signaling at telomeres devoid of POT1 and TPP1 [100]. This model shows that TRF2 and its interacting proteins are key in managing DNA repair pathways and end protection [100]. Both TRF2–NBS1 and TRF1–TERB1 interactions seem weaker than with canonical ligands [88], and are disrupted by CDK-mediated phosphorylation, suggesting a common theme of post-translational modification-mediated regulation of TRF1/2 binding by client proteins (Fig. 5f).
TRF2 and Rap1: Another critical feature of TRF2 is its ability to recruit shelterin protein Rap1 to telomeres. Unlike S. cerevisiae Rap1, mammalian Rap1 has no telomeric DNA-binding function and thus depends entirely on TRF2 for recruitment [27]. Although Rap1 does not help TRF2 repress ATM signaling, Rap1 is important in repressing homology directed repair (HDR), which can alter telomere length and cause chromosome fusions [101, 102]. The structure of the mammalian TRF2RBM (Rap1-binding motif) in complex with Rap1RCT (Rap1 C-terminal domain) has been solved [103]. The Rap1RCT contains six alpha-helices divided into two three-helix bundles. The TRF2RBM contains a helix–turn–helix motif that stacks against two of the Rap1 helices forming a bundle that brings the two proteins together. Interestingly, there is a secondary interaction between TRF2 and Rap1. The C-terminus of the BRCT domain of Rap1 contains a Y–X–L–X–P motif that is able to bind to the dimerization domain of TRF2 (bottom; Fig. 5e) [104]. This raises the possibility that the TBM-binding site of TRF2 is normally blocked by Rap1, probably to prevent binding of non-cognate TRF1TRFH-binding proteins to this site. Indeed, the measurable (non-physiological) binding between TERB1TBM− and TRF2 in vitro is lost when Rap1 protein is also present [95].
The TRF2–Rap1 duo plays an important role in preventing unnecessary homology directed repair (HDR) at telomeres. Rap1 cooperates with the basic domain of TRF2 to prevent telomere resection and chromosome fusions [101]. The basic domain of TRF2 binds to the holiday junction (HJ) formed during t-loop formation and strand invasion, which prevents PARP localization to telomeres [105]. This is important for end protection, because PARP1 recruits to telomeres the protein SLX4 that is capable of resolving t-loops and exposing telomeric DNA ends [106, 107]. Rap1 plays an important role in this process insofar as chromosome fusions in the context of the basic domain deletion of TRF2 were only observed when Rap1 was also absent [101].
Hierarchical assembly of shelterin at telomeres
POT1 is required for telomere maintenance, yet it cannot arrive at the chromosome end without being recruited by its binding partner TPP1 (Figs. 1, 4a) [64]. Before recruiting POT1, TPP1 must itself localize to telomeres with the help of shelterin protein TIN2 (Fig. 1) [68, 108]. TIN2 plays a central role at telomeres, as it is able to bind TRF1, TRF2, and TPP1 (Fig. 1). TIN2 recruitment to telomeres requires binding to TRF1 [94], the structural basis of which is already discussed above. Although binding to TRF2 is not mandatory for TIN2 recruitment to telomeres, it is important for stabilizing TRF2 protein for proper repression of ATM-mediated damage response [109]. Due to the hierarchical nature of shelterin assembly, defects in TIN2 recruitment or expression negatively impact TRF2, POT1, and TPP1 function. Indeed depletion of TIN2 results in the activation of both ATM and ATR DNA damage response pathways [94]. A new structure consisting of a TRF2–TIN2–TPP1 interface provides insights into how TIN2 binds cooperatively to both TRF2 and TPP1 to help bridge the ss and ds DNA-binding shelterin components [110]. Although the F–X–L–X–P motif of TIN2 can also bind TRF2TRFH, it does so with low affinity [88]. Instead, a different region, TIN22−202, contains the primary site for binding both TRF2 and TPP1 (Fig. 2). Interestingly, it adopts a helical structure related to that of the TRFH domains of TRF1 and TRF2, suggesting that TRF1, TRF2, and TIN2 may have structurally (and functionally) diverged from a common ancestral telomeric protein with TRFH topology (Fig. 4c) [110]. The first seven helices of the TIN2 structure align with helices 3-9 of the TRF1/2 TRFH domains. In addition, the TRFH-like domain of TIN2, like its counterparts in TRF1 and TRF2, serves as a protein–protein interaction domain. TIN22−202 forms two helical bundles that pack against each other, with one bundle interacting with TRF2 and the other contacting TPP1. Helix 7 of TIN22−202 contacts both TPP1 and TRF2 to give rise to cooperativity of binding, such that binding of TPP1 to TIN2 enhances binding of TIN2 to TRF2. This structure suggests a TRF2–TIN2–TPP1–POT1 connection that is critical for end protection. In vitro studies showing the proficiency of this complex to stimulate telomeric DNA end extension suggests that it might play a similarly important role in end replication too [111]. Further structural and functional studies will be required to understand how specific the roles of TRF1 and TRF2 are in recruiting the remaining shelterin components and dictating their biological functions.
Chromosome end replication by telomerase
Although telomerase does not play a direct role in end protection, it does prevent the severe erosion of telomeres (in continuously dividing cells) that could ultimately result in genome instability. The core telomerase enzyme is made of an RNA subunit termed telomerase RNA (TR), and a protein subunit termed telomerase reverse transcriptase (TERT) [4, 6–8]. This section will focus on our current understanding of the structural organization of TERT, TR, and the RNP core of the telomerase holoenzyme.
Telomerase RNA
Although the length and sequence of telomerase RNA can vary widely between species, the RNA often contains three structurally conserved domains: the template/pseudoknot, the CR4/5 (STE) domain, and the H/ACA box [112, 113]. The secondary structure conservation of these motifs highlights the essential role they play in telomere maintenance. The 5′ end of TR folds into a template-containing pseudoknot. The template is flanked by a 5′ template boundary element (TBE) and a 3′ template recognition element (TRE) [114–118]. The ss TRE helps TERT recognize and position the template in the active site of the enzyme to facilitate repeat addition processivity. The template boundary element (TBE) prevents the addition of nucleotides outside of the defined telomeric repeat. Telomerase is a processive enzyme, adding multiple telomeric repeats per replication cycle, which is a property unique to telomerase and absent in any other known DNA/RNA polymerase. It is still unclear how the template is translocated to the end of the nascent terminal repeat to facilitate continued addition, although a few models have been proposed. Single-molecule FRET and biochemical experiments suggest an accordion model. In this model, the TBE and template recognition element (TRE) expand and contract to allow movement of the template during the catalytic cycle [119]. Another hypothesis is inspired by the mode of action of translesion DNA polymerase v. It suggests that after synthesis of a telomeric repeat, the GT-rich newly added DNA loops out into a non-canonical hairpin, while the template translocates to pair with the AG at the 3′ end. Ultimately, an incoming dGTP allows the DNA to realign, so another step of synthesis can proceed [120]. Further biophysical and structural studies will be necessary to gain a better understanding of telomerase repeat addition processivity. Outside of the template, the pseudoknot is formed by a series of helices and loops for which several structures from ciliates, yeast, and vertebrates exist. We point the readers to two articles that describe these structures in great detail [121, 122].
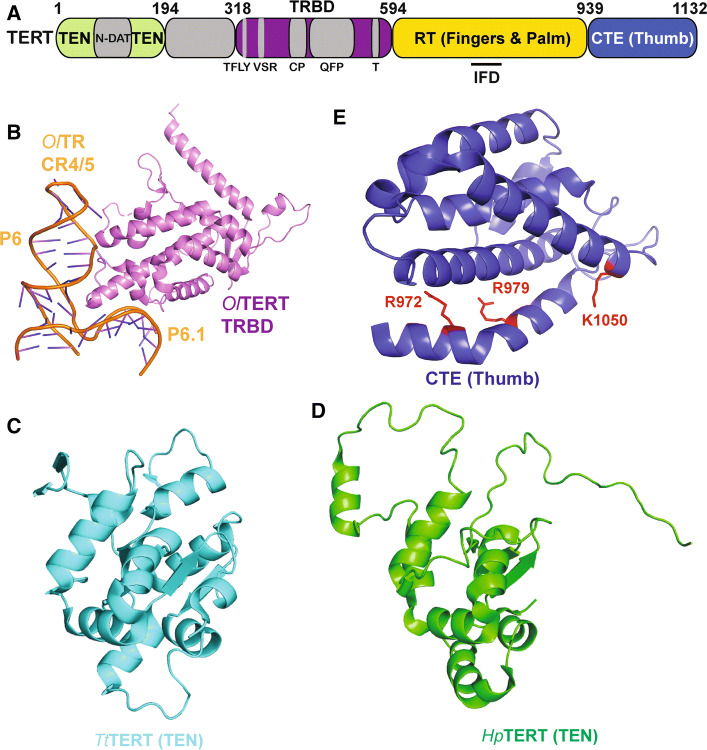
Following the pseudoknot is the CR4/5 domain (SL4 region in Tetrahymena thermophila), which is the major TERT-binding domain within telomerase RNA. Crosslinking studies [123] combined with the solved crystal structure of the CR4/5 domain bound to the TRBD (telomerase RNA-binding domain) from the fish Oryzias latipes (medaka) [124] have provided atomic details of the interface between the core telomerase RNA and protein subunits that depends both on sequence and conformation of the RNA. The CR4/5 domain forms a three-way junction that takes on an L-shape. Two stems, P6 and P6.1, of the CR4/5 domain directly interact with TERT by forming hydrogen bonds between their RNA backbone and residues on two separate TERT surfaces (Fig. 6b) [124]. When comparing this structure to that of CR4/5 alone [125], it can be seen that upon TRBD binding, there is a change in the spatial orientation of stem P6.1 relative to P6. While the structure of the human CR4/5 domain remains unsolved, there is an NMR structure of human stem P6.1 that is structurally similar to that of Oryzias latipes (medaka), suggesting that the human CR4/5 may interact with TERT in a similar manner [126].
Fig. 6.
Crystal structures of telomerase domains and related interfaces. a Domain diagram of hTERT. bOryzias latipes TERT RNA-binding domain (TRBD) bound to the CR4/5 domain of cognate TR (PDB ID: 4O26). cTetrahymena thermophila telomerase TEN domain (PDB ID: 2B2A). d NMR structure of the Hansenula polymorpha TEN domain (PDB ID: 5LGF). e Atomic resolution structure of the CTE (thumb) domain of human TERT (PDB ID: 5UGW). Dyskeratosis congenita (DC) associated mutations are highlighted in red
The TRBD forms the protein half of the interface between TERT and the CR4/5 of TR. Sequence conservation suggests that TERT TRBDs from many organisms contain three motifs (CP, QFP, and T motifs) that are important for binding TR (Fig. 6a). Vertebrate TRBDs have an additional vertebrate-specific RNA-binding motif (VSR) found on the N-terminal portion of the TRBD. All four of these motifs have been shown biochemically to take part in TR binding at some capacity [123, 124, 127]. Atomic details of this interface have been elucidated with the help of TRBD structures from Tetrahymena thermophila, medaka, and Tribolium castaneum. Each of these structures has been solved in complex with a TR fragment, while the structure of the TRBD from Takifugu rubripes (Japanese puffer fish) has been solved in apo form [124, 127–131]. In all the cases, the TRBD is mostly helical with the differences in the structures residing mainly in the N-terminal linker region. In medaka, a portion of the QFP motif along with N-terminal residues of the α2 helix of TRBD is responsible for interacting with CR4/5 in a sequence and conformation specific manner (Fig. 6a) [124]. Alanine-scanning mutagenesis of surface exposed residues in Takifugu rubripes TRBD found a TFLY motif in a pocket formed by the conserved T-CP domains that is important for binding to the TBE and orienting the template in the active site (Fig. 6a). In fact, loss of binding to TBE in this region results in a loss of telomerase activity and processivity [127]. The Tribolium castaneum TERT structure also places this region close to the T-CP cleft [130]. The solved crystal structure of Tetrahymena thermophila TERT TRBD in complex with its cognate TR further depicts TERT’s ability to bind and orient the TBE in a manner that prevents nucleotides outside of the telomeric repeat from being aberrantly incorporated. Ciliate TRBDs have a conserved CP2 motif that is structurally analogous to the highly conserved TFLY motif found in higher eukaryotic homologs. The structure revealed that, along with the CP2 motif, the CP and T motifs all take part in making polar contacts with the TBE. These extensive protein–RNA interactions help prevent nucleotides outside of the template from entering the active site of TERT (for the context of TBE in the holoenzyme structure, see Fig. 7c) [131].
Fig. 7.
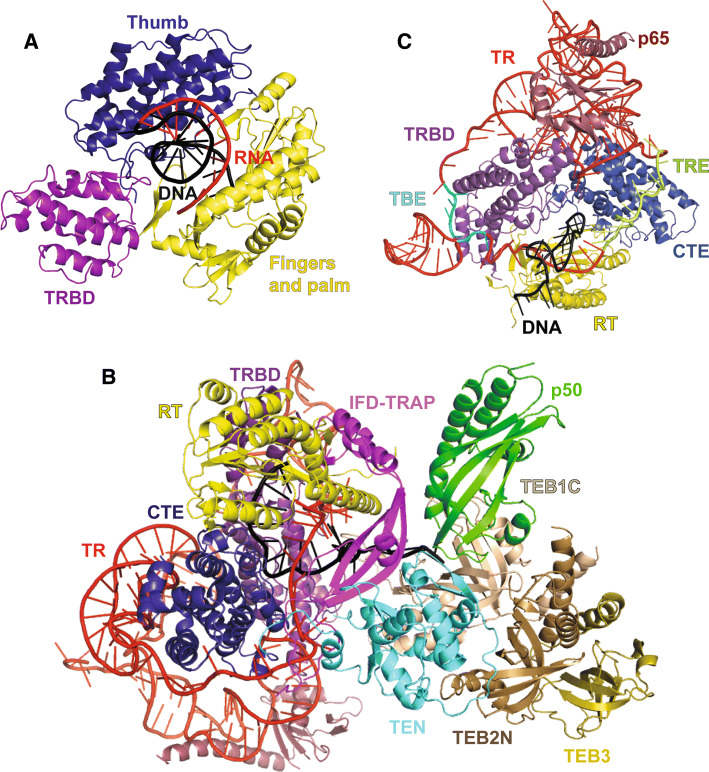
Structures of telomerase holoenzyme. aTribolium castaneum TERT ring bound to an RNA–DNA hairpin (PDB ID: 3KYL). Nucleic acid is rendered in cartoon representation with the RNA in red and DNA in black. The TRBD is shown in mauve, the fingers and palm are shown in yellow, and the thumb is shown in blue. b Atomic model of Tetrahymena thermophila telomerase (PDB ID: 6D6V). The RNA is depicted in red and the DNA in black. The TRBD is shown in mauve, the CTE is shown in blue, the RT is shown in yellow, the TEN domain is shown in cyan, the IFD-TRAP is shown in purple, and p50 is shown in green. TEB1C and TEB2 N are shown in different shades of brown and TEB3 is shown in gold. c TERT-TR-p65 core of the Tetrahymena thermophila telomerase holoenzyme structure highlighting components of the catalytic core of the RNP including the TBE (template boundary element) and TRE (template recognition element) of TR (PDB ID: 6D6V)
At the far 3′ end of the mature, RNA lies the H/ACA domain. The H/ACA domain ensures TR stability by acting as a scaffold for proteins such as Dyskerin, GAR1, NHP2, and NOP10 [132–134]. One such element within the H/ACA domain is the CAB box, which is responsible for binding the telomerase accessory protein TCAB1 and essential for Cajal body localization of telomerase [135–137]. TCAB1 stimulates telomerase activity through an interaction between its WD40 repeat domain and the CAB box of TR. This interaction is proposed to stabilize the CR4/5 domain allowing it to interact with the TRBD of TERT and generate an active conformation of telomerase [138]. Structures of the H/ACA domain bound to its partner proteins have been solved in certain archaea and budding yeast species [139, 140]. Along with these structures the recent cryo-EM structure of human telomerase provides abundant structural insights into how the H/ACA domain at the 3′ end of TR interacts with and is protected by Dyskerin, GAR1, NHP2, NOP10, and TCAB1 (discussed further below) [141].
TERT protein
The second major ingredient of the telomerase RNP is the reverse transcriptase TERT. TERT is largely made up of four conserved domains: the telomerase essential N-terminal domain (TEN), the TERT RNA-binding domain (TRBD), the reverse transcriptase domain (RT), and the thumb domain also called C-terminal extension (CTE) (Fig. 6a) [142]. Though the high-resolution structure of human TERT remains unsolved, many studies have helped elucidate structural aspects of TERT in ciliates, yeast, and insects among other species.
One major focus of structural studies has been the essential TEN domain, which is a TERT-specific element that is absent in other known reverse transcriptase families. Structural studies in Hansenula polymorpha and Tetrahymena thermophila suggest that although the TEN domain sequence varies widely between species, the core structure is well conserved (Fig. 6c, d) [143, 144]. Although structural data for the human TEN domain are unavailable, functional data confirm that it is essential for recruiting telomerase to telomeres. Mutations in the DAT (dissociates activities of telomerase) region of the TEN domain render the enzyme unable to function in vivo, yet it retains catalytic activity in vitro (Fig. 6a) [145]. Indeed, in vivo telomerase function is rescued by linking the mutant telomerase to either POT1 or TRF2, further suggesting a role for the DAT region in telomerase recruitment to telomeres [146]. Three mutants in the DAT region (K78E, G100V, and R132E) of human telomerase have been implicated in reducing TPP1’s ability to stimulate processivity [76, 147, 148]. Charge-swap experiments suggest a direct interaction between residue K78 in the DAT region of the TEN domain and E215 in the TEL patch of TPP1 [76]. Although establishing this direct interaction was paramount towards building an understanding of telomerase recruitment, much more remains to be learned regarding the remaining residues of the TEN domain and other regions of telomerase that interact with TPP1. TPP1’s OB domain, specifically the TEL patch and NOB regions, are essential for telomerase recruitment to telomeres (right; Fig. 4b) [69, 71, 75]. Together, these data strongly suggest a direct interaction between the TPP1 TEL patch or NOB region and the DAT region in the TEN domain of telomerase. However, the possibility of a bridging protein cannot be ruled out as a protein called Ccq1 has been implicated in bridging the interaction between telomerase and the TPP1 homolog in S. pombe [149].
The TEN domain has also been implicated in binding nucleic acids. HSQC NMR experiments with Hansenula polymorpha TEN domain have revealed that it interacts more specifically with DNA/RNA hybrids than DNA or RNA alone [144]. This supports the fact that the TEN domain helps facilitate recognition of the DNA–RNA template hybrid in the active site [150]. In addition, crosslinking data suggest that the TEN domain binds DNA in a fashion that would allow it to act as an anchor site 5′ to the template/DNA duplex [151, 152]. The interaction between the TEN domain and DNA/RNA could help facilitate telomerase processivity by helping orient the primer/template hybrid for proper catalysis.
The three subsequent domains make up the TERT ring (Fig. 6a). The first of these is the TRBD, which is responsible for interacting with the CR4/5 domain of TR forming the major interface between TERT and TR (discussed above). High-resolution structural information about the rest of the TERT catalytic subunit comes largely from crystal structures of the T.castaneum catalytic subunit (TcTERT; Fig. 7) and the human CTE domain (Fig. 6e) [129, 130, 153]. Structures exist for both non-canonical RNA–DNA hairpin-bound and apo TcTERT [129, 130]. Both structures suggest that the catalytic core of the telomerase protein subunit is similar to that of retroviral reverse transcriptases and viral RNA polymerases [129]. The structure is composed of an RT domain and a CTE, which together form a hand-like structure composed of fingers and palm domains in the RT, and a thumb domain represented by the CTE. Together, the TRBD, fingers, palm, and thumb form a ring with a positively charged cavity, where the RNA–DNA hairpin resides. This TERT ring interacts with the RNA–DNA hairpin in a manner that orients the RNA template in the active site, generating a conformation that allows for the synthesis of additional nucleotides. To facilitate this, the 5′ end of the RNA interacts with the fingers and palm regions, while the thumb interacts with the minor groove formed by the RNA–DNA heteroduplex. A rigid loop in the thumb domain forms the primer grip region that directs the 3′ end of the DNA towards the active site of the protein. The thumb domain is also in close contact with the TRBD and has been shown to interact with CR4/5 through an FVYL motif [154]. The FVYL motif was further shown to be important in both structural and biochemical studies of the human CTE, where DC-associated mutations have been found [153]. The fingers and CTE also contain residues that have been implicated in recruiting telomerase [71]. The CTE contains a second DAT region called C-DAT that when mutated does not affect telomerase activity in vitro, but stops telomerase from maintaining telomere length in vivo [155]. Unlike the N-DAT, it is less clear if the C-DAT plays a direct role in telomerase recruitment, as it was not tested if the in vivo telomere shortening phenotype is rescued by linking the mutants to POT1 or TRF2 [155]. Finally, TERT is unique among reverse transcriptases in that it has an insertion in fingers domain (IFD), residues in which have also been implicated in TPP1 binding [156–158]. Specifically, an IFD mutant, hTERT-V791Y, showed defective telomerase recruitment and was not stimulated by TPP1 (and POT1) overexpression, suggesting that this mutation interferes with TERT–TPP1 binding. Although the TcTERT structures have yielded unprecedented insight into telomerase structure and function, it should be noted that the T. castaneum telomerase RNA subunit has not been identified. Moreover, the TcTERT protein lacks a TEN domain and contains a relatively inconspicuous IFD domain. Thus, it remains possible that mammalian telomerase has acquired new functionalities relative to its flour beetle counterpart or that these orthologs perform some functions using divergent structural mechanisms.
Electron microscopy structures of the telomerase holoenzyme
Electron microscopy (EM) has been instrumental in providing initial insights into telomerase holoenzyme structure [159–161]. The negative stain EM structures of the human and Tetrahymena thermophila holoenzymes provided the overall shape and structural layout of the full RNP. The low-resolution EM map for human telomerase showed two globular lobes connected by a flexible linker region suggesting telomerase may form a dimer. This dimer was hypothesized to be mediated by the H/ACA domain of TR [159]. This quaternary structure remains controversial; some reports suggest human telomerase acts as a monomer, while others suggest that dimeric telomerase is the functional enzyme [159, 162–164]. Recent major breakthroughs in telomerase structural biology have come in the form of two Tetrahymena thermophila telomerase and one human telomerase cryo-EM structures solved to ~ 9 Å, 4.8 Å, and ~ 8 Å resolution, respectively [141, 160, 165]. The human telomerase holoenzyme structure shows an overall bilobal architecture consistent with the previously reported negative stain EM map. However, the cryo-EM reconstruction of human telomerase shows that one of these lobes represents the catalytic core of TERT and TR, while the other contains the TR H/ACA domain bound to the biogenesis protein core composed of one TCAB1, two Dyskerin, two NOP10, and two GAR1 protein subunits [141]. The bilobal nature of human telomerase places TCAB1 on the opposite end of TR relative to CR4/5 [140] failing to explain how TCAB1 might allosterically activate the CR4/5–TERT interaction [137]. It is possible that the TCAB1-mediated “switch” is not captured by the conformation characterized in the cryo-EM structure. Characterizing a greater fraction of the holoenzyme particles in the active chromatographic fraction will likely lead to the identification of additional conformational states. The cryo-EM characterization of human telomerase also reveals the presence of a small population of (bilobal) telomerase that is dimeric, but the biological importance of this, if any, remains unknown [141]. Although the current cryo-EM data are insufficient to build a complete atomic model for human telomerase, they are clearly consistent with the presence of a typical reverse transcriptase topology for TERT, known structures of TR fragments, and predicted structures of human telomerase biogenesis factors such as TCAB1.
Tetrahymena thermophila, the model system that led to the discovery of telomeric DNA repeats and telomerase activity, has provided the deepest structural insights into the telomerase holoenzyme. The 8.9 Å structure of Tetrahymena thermophila telomerase published in 2015 allowed for the identification of secondary structure elements and the building of a pseudo-atomic model with the help of structurally solved protein and RNA subunits/fragments (or their close homologs) [160]. This structure, which confirmed the monomeric status of Tetrahymena thermophila telomerase RNP, afforded the first opportunity to place the TEN domain with respect to the catalytic ring structure of TERT. The Tetrahymena thermophila TEN domain was modeled above the CTE on the front (active) side of the TERT ring. This is also obvious in the most recent structure of the holoenzyme described below (see Fig. 7b). While the TEN domain is in the vicinity of the template–DNA duplex in the cryo-EM structure, further structural elucidation will be required to confirm the direct interaction alluded by functional studies. The authors were also able to identify an RPA-like heterotrimeric TEB1–TEB2–TEB3 complex and a CST-like P75–P45–P19 complex highlighting how a combination of cryo-EM and mass spectrometry could serve as a powerful approach to discovering new subunits in a purified holoenzyme. For a full description of the RPA-like complexes contained in this telomerase holoenzyme structure, we point the readers to the following references [122, 160].
The recently published 4.8 Å resolution structure of Tetrahymena thermophila telomerase reveals the structural details of this complex interaction network by providing the first structural information about both p50 and a new structural element within TERT called IFD-TRAP (Fig. 7b). The IFD-TRAP forms an L-shape with two interacting terminal helices forming the short arm of the L, and an extended β-sheet region termed as the TRAP forming the long arm (Fig. 7b). The IFD-TRAP is able to interact with the TEN domain, RBD, and RT to form a ring that helps trap the pseudoknot of TR onto the ring formed by the catalytic core domains of TERT [165]. The TEN domain and IFD interact to form a surface on TERT that is in close proximity to p50, TEB1, and TEB2. p50 is made up of a six-stranded β-barrel that forms an OB fold like that of TPP1 (Fig. 7b) [165]. Thus, this structure provides the first three-dimensional insights into how telomerase may be recruited to human telomeres and supports previous data that implicate the TEN and IFD domains in recruiting human telomerase to TPP1 at telomeres. Finally, by determining the relative positioning of various important motifs of TR (e.g., TBE, TRE; Fig. 7c) in the context of the holoenzyme, the Tetrahymena thermophila telomerase cryo-EM structures provide important structural insights into the role of TR in facilitating telomere repeat definition and processive synthesis.
Concluding remarks and perspectives
Structural data acquired over almost two decades have led to the construction of a detailed, although incomplete, structural framework for mammalian end protection and replication. A strong theme emerges from the study of various protein–protein interactions involving shelterin. Most of these interfaces involve a “domain–peptide” interaction mechanism [110]. The “domains” involved in these interfaces (e.g., TRFH, OB) were likely retained in evolution due to their superior structural stability. However, the specificity and affinity of these interactions originated and improved via changes to the structurally unrestrained “peptide” partners. Indeed a mere F or Y choice in the F/Y–X–L–X–P peptide dictates preference of a client protein for TRF1 versus TRF2. Thus, this domain-peptide combination offers a binding platform that is structurally robust yet readily tunable for acquiring new or improved functions at telomeres. Major unresolved areas in shelterin structural biology include the structural analysis of partial/full complexes of shelterin (beyond just peptides and domains), the rules guiding shelterin assembly and disassembly, and allosteric effects within shelterin complexes that unravel the full potential of individual components. Although a high-resolution structure of human telomerase is still lacking, the enormous amount of structural and functional data from various telomerase components and complexes has led to a detailed understanding of human telomerase function. These studies help rationalize how acquisition of telomerase-specific structural elements such as TEN, IFD-TRAP, and TR allowed for conversion of a basic HIV-like reverse transcriptase scaffold to an extraordinary enzyme that can add multiple telomeric repeats de novo at chromosome ends. However, aggressive efforts to solve the human telomerase structure will continue as success in these endeavors will provide an unparalleled structural scaffold for designing drugs against telomerase, which remains a prime target for anti-cancer drug design.
Acknowledgements
We thank the entire telomere and telomerase biology community for the groundbreaking research conducted over three decades that has led to our current understanding of chromosome end protection and replication. Although we strived to discuss and cite all publications that are relevant to this topic, we sincerely apologize if we may have overlooked any important contributions. We thank the entire Nandakumar laboratory for critical feedback and proofreading of the manuscript. Author salary/stipend and research in the lab during the writing of this review were funded by R01GM120094 (to J.N.), R01AG050509 (to J.N.; co-investigator), NIH Biology of Aging Training Grant (T32AG000114) awarded to the University of Michigan Geriatrics Center from the National Institute on Aging (fellowship to E.M.S.), and the American Cancer Society Research Scholar grant RSG-17-037-01-DMC (to J.N.).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J Mol Biol. 1992;225(4):951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 3.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 4.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276(5312):561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 5.Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94(17):9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90(4):785–795. doi: 10.1016/S0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 7.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51(6):887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 8.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337(6205):331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 9.Shay JW, Wright WE. Telomeres and telomerase in normal and cancer stem cells. FEBS Lett. 2010;584(17):3819–3825. doi: 10.1016/j.febslet.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dokal I. Dyskeratosis congenita. Hematol Am Soc Hematol Educ Program. 2011;2011:480–486. doi: 10.1182/asheducation-2011.1.480. [DOI] [PubMed] [Google Scholar]
- 11.Jones M, Bisht K, Savage SA, Nandakumar J, Keegan CE, Maillard I. The shelterin complex and hematopoiesis. J Clin Invest. 2016;126(5):1621–1629. doi: 10.1172/JCI84547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage SA. Human telomeres and telomere biology disorders. Prog Mol Biol Transl Sci. 2014;125:41–66. doi: 10.1016/B978-0-12-397898-1.00002-5. [DOI] [PubMed] [Google Scholar]
- 13.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 14.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 15.Wright WE, Pereira-Smith OM, Shay JW. Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol Cell Biol. 1989;9(7):3088–3092. doi: 10.1128/MCB.9.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart SA, Weinberg RA. Telomeres: cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–557. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 17.Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21(6):349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16(7):1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broccoli D, Chong L, Oelmann S, Fernald AA, Marziliano N, van Steensel B, Kipling D, Le Beau MM, de Lange T. Comparison of the human and mouse genes encoding the telomeric protein, TRF1: chromosomal localization, expression and conserved protein domains. Hum Mol Genet. 1997;6(1):69–76. doi: 10.1093/hmg/6.1.69. [DOI] [PubMed] [Google Scholar]
- 20.Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17(2):236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 21.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17(2):231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 22.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6(5):801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 23.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292(5519):1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6(7):673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 25.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18(14):1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14(18):1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 27.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101(5):471–483. doi: 10.1016/S0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 28.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23(4):405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11(12):1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 30.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14(2):69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhalla N, Dernburg AF. Prelude to a division. Annu Rev Cell Dev Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17(5):598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 33.Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5(12):983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 34.Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2(8):621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 35.Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134(5):1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe Y. Geometry and force behind kinetochore orientation: lessons from meiosis. Nat Rev Mol Cell Biol. 2012;13(6):370–382. doi: 10.1038/nrm3349. [DOI] [PubMed] [Google Scholar]
- 37.Shibuya H, Hernandez-Hernandez A, Morimoto A, Negishi L, Hoog C, Watanabe Y. MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell. 2015;163(5):1252–1266. doi: 10.1016/j.cell.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Gray JT, Celander DW, Price CM, Cech TR. Cloning and expression of genes for the Oxytricha telomere-binding protein: specific subunit interactions in the telomeric complex. Cell. 1991;67(4):807–814. doi: 10.1016/0092-8674(91)90075-A. [DOI] [PubMed] [Google Scholar]
- 39.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95(7):963–974. doi: 10.1016/S0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 40.Buczek P, Horvath MP. Structural reorganization and the cooperative binding of single-stranded telomere DNA in Sterkiella nova. J Biol Chem. 2006;281(52):40124–40134. doi: 10.1074/jbc.M607749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426(6963):198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 42.Dickey TH, McKercher MA, Wuttke DS. Nonspecific recognition is achieved in Pot1pC through the use of multiple binding modes. Structure. 2013;21(1):121–132. doi: 10.1016/j.str.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448(7157):1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 44.Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26(20):2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell. 2011;44(4):647–659. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu P, Wang Y, Bisht KK, Wu L, Kukova L, Smith EM, Xiao Y, Bailey SM, Lei M, Nandakumar J, Chang S. Pot1 OB-fold mutations unleash telomere instability to initiate tumorigenesis. Oncogene. 2017;36(14):1939–1951. doi: 10.1038/onc.2016.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinzaru AM, Hom RA, Beal A, Phillips AF, Ni E, Cardozo T, Nair N, Choi J, Wuttke DS, Sfeir A, Denchi EL. Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep. 2016;15(10):2170–2184. doi: 10.1016/j.celrep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25(1):29–36. doi: 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 50.Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci USA. 2010;107(2):651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice C, Skordalakes E. Structure and function of the telomeric CST complex. Comput Struct Biotechnol J. 2016;14:161–167. doi: 10.1016/j.csbj.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen LY, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488(7412):540–544. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 53.Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284(9):5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu P, Takai H, de Lange T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012;150(1):39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirman Z, Lottersberger F, Takai H, Kibe T, Gong Y, Takai K, Bianchi A, Zimmermann M, Durocher D, de Lange T. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature. 2018;560(7716):112–116. doi: 10.1038/s41586-018-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shastrula PK, Rice CT, Wang Z, Lieberman PM, Skordalakes E. Structural and functional analysis of an OB-fold in human Ctc1 implicated in telomere maintenance and bone marrow syndromes. Nucleic Acids Res. 2018;46(2):972–984. doi: 10.1093/nar/gkx1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bryan C, Rice C, Harkisheimer M, Schultz DC, Skordalakes E. Structure of the human telomeric Stn1-Ten1 capping complex. PLoS One. 2013;8(6):e66756. doi: 10.1371/journal.pone.0066756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gelinas AD, Paschini M, Reyes FE, Heroux A, Batey RT, Lundblad V, Wuttke DS. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc Natl Acad Sci USA. 2009;106(46):19298–19303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23(24):2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chastain M, Zhou Q, Shiva O, Fadri-Moskwik M, Whitmore L, Jia P, Dai X, Huang C, Ye P, Chai W. Human CST facilitates genome-wide RAD51 recruitment to GC-rich repetitive sequences in response to replication stress. Cell Rep. 2016;16(5):1300–1314. doi: 10.1016/j.celrep.2016.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain PD, 2nd, Wright WE, Price CM. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012;31(17):3537–3549. doi: 10.1038/emboj.2012.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C, Gu P, Wu J, Chen X, Niu S, Sun H, Wu L, Li N, Peng J, Shi S, Fan C, Huang M, Wong CC, Gong Q, Kumar-Sinha C, Zhang R, Pusztai L, Rai R, Chang S, Lei M. Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer. Nat Commun. 2017;8:14929. doi: 10.1038/ncomms14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rice C, Shastrula PK, Kossenkov AV, Hills R, Baird DM, Showe LC, Doukov T, Janicki S, Skordalakes E. Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat Commun. 2017;8:14928. doi: 10.1038/ncomms14928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14(8):754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 65.Gong Y, de Lange T. A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol Cell. 2010;40(3):377–387. doi: 10.1016/j.molcel.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445(7127):506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 67.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O’Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445(7127):559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 68.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30(12):2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492(7428):285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sexton AN, Youmans DT, Collins K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem. 2012;287(41):34455–34464. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150(3):481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bisht K, Smith EM, Tesmer VM, Nandakumar J. Structural and functional consequences of a disease mutation in the telomere protein TPP1. Proc Natl Acad Sci USA. 2016;113(46):13021–13026. doi: 10.1073/pnas.1605685113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo Y, Kartawinata M, Li J, Pickett HA, Teo J, Kilo T, Barbaro PM, Keating B, Chen Y, Tian L, Al-Odaib A, Reddel RR, Christodoulou J, Xu X, Hakonarson H, Bryan TM. Inherited bone marrow failure associated with germline mutation of ACD, the gene encoding telomere protein TPP1. Blood. 2014;124(18):2767–2774. doi: 10.1182/blood-2014-08-596445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kocak H, Ballew BJ, Bisht K, Eggebeen R, Hicks BD, Suman S, O’Neil A, Giri N, Laboratory NDCGR. Group NDCSW. Maillard I, Alter BP, Keegan CE, Nandakumar J, Savage SA. Hoyeraal-hreidarsson syndrome caused by a germline mutation in the TEL patch of the telomere protein TPP1. Genes Dev. 2014;28(19):2090–2102. doi: 10.1101/gad.248567.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grill S, Tesmer VM, Nandakumar J. The N terminus of the OB domain of telomere protein TPP1 Is critical for telomerase action. Cell Rep. 2018;22(5):1132–1140. doi: 10.1016/j.celrep.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmidt JC, Dalby AB, Cech TR. Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife. 2014 doi: 10.7554/eLife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270(5242):1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 78.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336(6081):593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7(7):712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 80.Karlseder J, Kachatrian L, Takai H, Mercer K, Hingorani S, Jacks T, de Lange T. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol Cell Biol. 2003;23(18):6533–6541. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283(5406):1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 83.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2(8):E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13(17):1549–1556. doi: 10.1016/S0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 85.Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, Lototska L, Kaur P, Bauwens S, Djerbi N, Latrick CM, Fraisier V, Pei B, Gay A, Jaune E, Foucher K, Cherfils-Vicini J, Aeby E, Miron S, Londono-Vallejo A, Ye J, Le Du MH, Wang H, Gilson E, Giraud-Panis MJ. TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol Cell. 2016;61(2):274–286. doi: 10.1016/j.molcel.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155(2):345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–514. doi: 10.1016/S0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 88.Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319(5866):1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 89.Hanaoka S, Nagadoi A, Nishimura Y. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 2005;14(1):119–130. doi: 10.1110/ps.04983705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nishikawa T, Okamura H, Nagadoi A, Koig P, Rhodes D, Nishimura Y. Structure of the DNA-binding domain of human telomeric protein, TRF1 and its interaction with telomeric DNA. Nucleic Acids Res Suppl. 2001;1(1):273–274. doi: 10.1093/nass/1.1.273. [DOI] [PubMed] [Google Scholar]
- 91.Nishikawa T, Okamura H, Nagadoi A, Konig P, Rhodes D, Nishimura Y. Solution structure of a telomeric DNA complex of human TRF1. Structure. 2001;9(12):1237–1251. doi: 10.1016/S0969-2126(01)00688-8. [DOI] [PubMed] [Google Scholar]
- 92.Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6(1):39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001;8(2):351–361. doi: 10.1016/S1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- 94.Frescas D, de Lange T. TRF2-tethered TIN2 can mediate telomere protection by TPP1/POT1. Mol Cell Biol. 2014;34(7):1349–1362. doi: 10.1128/MCB.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pendlebury DF, Fujiwara Y, Tesmer VM, Smith EM, Shibuya H, Watanabe Y, Nandakumar J. Dissecting the telomere-inner nuclear membrane interface formed in meiosis. Nat Struct Mol Biol. 2017;24(12):1064–1072. doi: 10.1038/nsmb.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shibuya H, Ishiguro K, Watanabe Y. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat Cell Biol. 2014;16(2):145–156. doi: 10.1038/ncb2896. [DOI] [PubMed] [Google Scholar]
- 97.Daniel K, Trankner D, Wojtasz L, Shibuya H, Watanabe Y, Alsheimer M, Toth A. Mouse CCDC79 (TERB1) is a meiosis-specific telomere associated protein. BMC Cell Biol. 2014;15:17. doi: 10.1186/1471-2121-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Tu Z, Watanabe Y, Shibuya H. Distinct TERB1 domains regulate different protein interactions in meiotic telomere movement. Cell Rep. 2017;21(7):1715–1726. doi: 10.1016/j.celrep.2017.10.061. [DOI] [PubMed] [Google Scholar]
- 99.Long J, Huang C, Chen Y, Zhang Y, Shi S, Wu L, Liu Y, Liu C, Wu J, Lei M. Telomeric TERB1-TRF1 interaction is crucial for male meiosis. Nat Struct Mol Biol. 2017;24(12):1073–1080. doi: 10.1038/nsmb.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rai R, Hu C, Broton C, Chen Y, Lei M, Chang S. NBS1 phosphorylation status dictates repair choice of dysfunctional telomeres. Mol Cell. 2017;65(5):801–817. doi: 10.1016/j.molcel.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rai R, Chen Y, Lei M, Chang S. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat Commun. 2016;7:10881. doi: 10.1038/ncomms10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327(5973):1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, Hiraoka Y, Shore D, Hu HY, Chang S, Lei M. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol. 2011;18(2):213–221. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gaullier G, Miron S, Pisano S, Buisson R, Le Bihan YV, Tellier-Lebegue C, Messaoud W, Roblin P, Guimaraes BG, Thai R, Giraud-Panis MJ, Gilson E, Le Du MH. A higher-order entity formed by the flexible assembly of RAP1 with TRF2. Nucleic Acids Res. 2016;44(4):1962–1976. doi: 10.1093/nar/gkv1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schmutz I, Timashev L, Xie W, Patel DJ, de Lange T. TRF2 binds branched DNA to safeguard telomere integrity. Nat Struct Mol Biol. 2017;24(9):734–742. doi: 10.1038/nsmb.3451. [DOI] [PubMed] [Google Scholar]
- 106.Gonzalez-Prieto R, Cuijpers SA, Luijsterburg MS, van Attikum H, Vertegaal AC. SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. EMBO Rep. 2015;16(4):512–519. doi: 10.15252/embr.201440017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149(4):795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 108.O’Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci USA. 2006;103(32):11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004;279(42):43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 110.Hu C, Rai R, Huang C, Broton C, Long J, Xu Y, Xue J, Lei M, Chang S, Chen Y. Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex. Cell Res. 2017;27(12):1485–1502. doi: 10.1038/cr.2017.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lim CJ, Zaug AJ, Kim HJ, Cech TR. Reconstitution of human shelterin complexes reveals unexpected stoichiometry and dual pathways to enhance telomerase processivity. Nat Commun. 2017;8(1):1075. doi: 10.1038/s41467-017-01313-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang Q, Kim NK, Feigon J. Architecture of human telomerase RNA. Proc Natl Acad Sci USA. 2011;108(51):20325–20332. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100(5):503–514. doi: 10.1016/S0092-8674(00)80687-X. [DOI] [PubMed] [Google Scholar]
- 114.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17(22):2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288(5467):863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- 116.Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P. A flexible template boundary element in the RNA subunit of fission yeast telomerase. J Biol Chem. 2008;283(35):24224–24233. doi: 10.1074/jbc.M802043200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci USA. 2002;99(10):6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell. 2003;11(6):1673–1683. doi: 10.1016/S1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol. 2011;18(12):1371–1375. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang W, Lee YS. A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat Struct Mol Biol. 2015;22(11):844–847. doi: 10.1038/nsmb.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Y, Yesselman JD, Zhang Q, Kang M, Feigon J. Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. Proc Natl Acad Sci USA. 2016;113(35):E5125–E5134. doi: 10.1073/pnas.1607411113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chan H, Wang Y, Feigon J. Progress in human and tetrahymena telomerase structure determination. Annu Rev Biophys. 2017;46:199–225. doi: 10.1146/annurev-biophys-062215-011140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bley CJ, Qi X, Rand DP, Borges CR, Nelson RW, Chen JJ. RNA-protein binding interface in the telomerase ribonucleoprotein. Proc Natl Acad Sci USA. 2011;108(51):20333–20338. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang J, Brown AF, Wu J, Xue J, Bley CJ, Rand DP, Wu L, Zhang R, Chen JJ, Lei M. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol. 2014;21(6):507–512. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim NK, Zhang Q, Feigon J. Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function. Nucleic Acids Res. 2014;42(5):3395–3408. doi: 10.1093/nar/gkt1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leeper T, Leulliot N, Varani G. The solution structure of an essential stem-loop of human telomerase RNA. Nucleic Acids Res. 2003;31(10):2614–2621. doi: 10.1093/nar/gkg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure. 2013;21(10):1870–1878. doi: 10.1016/j.str.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15(11):1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 129.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455(7213):633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 130.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17(4):513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 131.Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol. 2015;22(11):883–888. doi: 10.1038/nsmb.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18(10):1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402(6761):551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 134.Kiss T, Fayet-Lebaron E, Jady BE. Box H/ACA small ribonucleoproteins. Mol Cell. 2010;37(5):597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 135.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323(5914):644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34(1):47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Venteicher AS, Artandi SE. TCAB1: driving telomerase to Cajal bodies. Cell Cycle. 2009;8(9):1329–1331. doi: 10.4161/cc.8.9.8288. [DOI] [PubMed] [Google Scholar]
- 138.Chen L, Roake CM, Freund A, Batista PJ, Tian S, Yin YA, Gajera CR, Lin S, Lee B, Pech MF, Venteicher AS, Das R, Chang HY, Artandi SE. An activity switch in human telomerase based on RNA conformation and shaped by TCAB1. Cell. 2018 doi: 10.1016/j.cell.2018.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443(7109):302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 140.Li S, Duan J, Li D, Yang B, Dong M, Ye K. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev. 2011;25(22):2409–2421. doi: 10.1101/gad.175299.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E, Collins K. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature. 2018;557(7704):190–195. doi: 10.1038/s41586-018-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Podlevsky JD, Chen JJ. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutat Res. 2012;730(1–2):3–11. doi: 10.1016/j.mrfmmm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13(3):218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 144.Petrova OA, Mantsyzov AB, Rodina EV, Efimov SV, Hackenberg C, Hakanpaa J, Klochkov VV, Lebedev AA, Chugunova AA, Malyavko AN, Zatsepin TS, Mishin AV, Zvereva MI, Lamzin VS, Dontsova OA, Polshakov VI. Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol. 2001;21(22):7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Armbruster BN, Linardic CM, Veldman T, Bansal NP, Downie DL, Counter CM. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol Cell Biol. 2004;24(8):3552–3561. doi: 10.1128/MCB.24.8.3552-3561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24(6):613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sexton AN, Regalado SG, Lai CS, Cost GJ, O’Neil CM, Urnov FD, Gregory PD, Jaenisch R, Collins K, Hockemeyer D. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28(17):1885–1899. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320(5881):1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 150.Wu RA, Collins K. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 2014;33(8):921–935. doi: 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lue NF. A physical and functional constituent of telomerase anchor site. J Biol Chem. 2005;280(28):26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15(8):870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hoffman H, Rice C, Skordalakes E. Structural analysis reveals the deleterious effects of telomerase mutations in bone marrow failure syndromes. J Biol Chem. 2017;292(11):4593–4601. doi: 10.1074/jbc.M116.771204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bryan C, Rice C, Hoffman H, Harkisheimer M, Sweeney M, Skordalakes E. structural basis of telomerase inhibition by the highly specific BIBR1532. Structure. 2015;23(10):1934–1942. doi: 10.1016/j.str.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Banik SS, Guo C, Smith AC, Margolis SS, Richardson DA, Tirado CA, Counter CM. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol Cell Biol. 2002;22(17):6234–6246. doi: 10.1128/MCB.22.17.6234-6246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lue NF, Lin YC, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol. 2003;23(23):8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chu TW, D’Souza Y, Autexier C. The insertion in fingers domain in human telomerase can mediate enzyme processivity and telomerase recruitment to telomeres in a TPP1-dependent manner. Mol Cell Biol. 2016;36(1):210–222. doi: 10.1128/MCB.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chu TW, MacNeil DE, Autexier C. Multiple mechanisms contribute to the cell growth defects imparted by human telomerase insertion in fingers domain mutations associated with premature aging diseases. J Biol Chem. 2016;291(16):8374–8386. doi: 10.1074/jbc.M116.714782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20(4):454–460. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O’Brien Johnson R, Collins K, Loo JA, Zhou ZH, Feigon J. Structure of tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350(6260):aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496(7444):187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alves D, Li H, Codrington R, Orte A, Ren X, Klenerman D, Balasubramanian S. Single-molecule analysis of human telomerase monomer. Nat Chem Biol. 2008;4(5):287–289. doi: 10.1038/nchembio.82. [DOI] [PubMed] [Google Scholar]
- 163.Egan ED, Collins K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol. 2010;30(11):2775–2786. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20(13):3526–3534. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jiang J, Wang Y, Susac L, Chan H, Basu R, Zhou ZH, Feigon J. Structure of telomerase with telomeric DNA. Cell. 2018;173(5):1179–1190. doi: 10.1016/j.cell.2018.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]