Abstract
Purpose:
Congenital microcephaly (CM) is an important birth defect with long term neurological sequelae. We aimed to perform detailed phenotypic and genomic analysis of patients with Mendelian forms of CM.
Methods:
Clinical phenotyping, targeted or exome sequencing, and autozygome analysis.
Results:
We describe 150 patients (104 families) with 56 Mendelian forms of CM. Our data show little overlap with the genetic causes of postnatal microcephaly. We also show that a broad definition of primary microcephaly —as an autosomal recessive form of nonsyndromic CM with severe postnatal deceleration of occipitofrontal circumference—is highly sensitive but has a limited specificity. In addition, we expand the overlap between primary microcephaly and microcephalic primordial dwarfism both clinically (short stature in >52% of patients with primary microcephaly) and molecularly (e.g., we report the first instance of CEP135-related microcephalic primordial dwarfism). We expand the allelic and locus heterogeneity of CM by reporting 37 novel likely disease-causing variants in 27 disease genes, confirming the candidacy of ANKLE2, YARS, FRMD4A, and THG1L, and proposing the candidacy of BPTF, MAP1B, CCNH, and PPFIBP1.
Conclusion:
Our study refines the phenotype of CM, expands its genetics heterogeneity, and informs the workup of children born with this developmental brain defect.
Keywords: autozygome, primary microcephaly, dwarfism, CNTRL
INTRODUCTION
Congenital microcephaly (CM) is defined by an occipital frontal circumference (OFC) that is >2 SD below the mean at birth, although some suggest 3 SD as the cutoff value.1–3 In the United States, it is estimated that this birth defect affects 2–12/10,000 live births.4 The small brain size usually carries poor neurodevelopmental prognosis. The etiology of CM is highly heterogeneous.2 Mendelian forms of CM are particularly interesting because they are amenable to human genetics approaches and can unravel the molecular underpinning of early brain development.
Primary microcephaly (PM) is a severe form of isolated CM that was defined on the basis of lack of syndromic features or major brain malformations. Many of the underlying genes were mapped using linkage analysis by exploiting their fully penetrant recessive inheritance pattern. However, recent advances in sequencing technology have greatly accelerated their discovery and there are currently 18 PM genes listed in OMIM. Early ventricular zone neural progenitors are stem-like cells that divide symmetrically to expand the pool of progenitor cells but also asymmetrically to allow neurons to differentiate and migrate to form the multilayered cortex in a highly orchestrated manner. Most of the PM genes identified to date encode proteins that appear to play a role in orienting the mitotic spindle or otherwise control cell division.5,6 Another aspect of PM pathogenesis that has also become apparent through the identification of novel PM genes is DNA damage repair, although the exact link between this fundemental and ubiquitous cellular process and PM remains poorly understood.2
The expanding list of biological processes that are perturbed by PM-related pathogenic variants clearly shows that multiple mechanisms can lead to isolated CM. Furthermore, the finding that several genes cause isolated CM though not classified as PM-related genes suggests a potential overlap that can be exploited to further our molecular insight into brain development. To our knowledge, however, there has not been any systematic phenotypic analysis of molecularly confirmed CM phenotypes to draw patterns that inform the clinical classification of genes that are critical for neurogenesis. In this study, we exploit the large referral base to our program to conduct such systematic clinical and molecular analysis, which sheds light on the genetic control of early brain development in humans.
MATERIALS AND METHODS
Human subjects
Individuals with OFC at birth of ≥−2 SD or those reported by history to have microcephaly at birth were eligible for the study. The SDs were calculated based on standard World Health Organization (WHO) charts taking into consideration both gender and gestational age. A written informed consent was signed by the parents (or legal guardians) of all subjects prior to enrollment (KFSHRC RAC 2080 006 and 2121 053). Blood was collected in EDTA tubes from the index and available family members, and in sodium heparin tubes for the creation of Lymphoblastoid cell lines (LCLs) when needed.
Additionally, we include two previously unpublished cases, each with a biallelic pathogenic variant in ANKLE2 identified by exome sequencing in the research setting. Informed consent was obtained for their research participation and inclusion in this study with approval from their respective institutions (Boston Children’s Hospital, Boston, MA and Institute of Human Genetics, University of California–San Francisco, San Francisco, CA).
Autozygome analysis and targeted and exome sequencing
The likely causal variants were identified using either targeted exome (“Mendeliome assay”) or exome sequencing as described previously.7 For the sporadic cases and those born to nonconsanguineous parents, all the rare (minor allele frequency [MAF] <0.001) heterozygous and homozygous variants were investigated for candidacy. For familial consanguineous cases, DNA samples from affected and available unaffected family members were genotyped using Axiom single-nucleotide polymorphism (SNP) chip array following the manufacturer’s instructions. This was followed by genomewide autozygome analysis using runs of homozygosity >2 Mb as surrogates of autozygosity using AutoSNPa and variants therein were prioritized for candidacy. Rare variants on X-chromosome were also investigated in male cases.
Computational structural analysis of mutants
RESULTS
Marked genetic heterogeneity of CM
Our Mendelian Genomics Program (MGP) serves as a key referral center for a wide range of birth defects, including CM. In this study, we used 2 SD below the mean at birth as the cutoff value for defining CM, OFC measurement below 35 cm within the first 3 months, or recorded microcephalic at birth. For the period 2008–2017, we enrolled 137 families with affected children who met this definition. A potentially causal variant was identified in 104, and these were considered eligible for inclusion in this study, which specifically examines the phenotype of molecularly characterized CM cases.
The variants identified in this cohort fall in four categories.
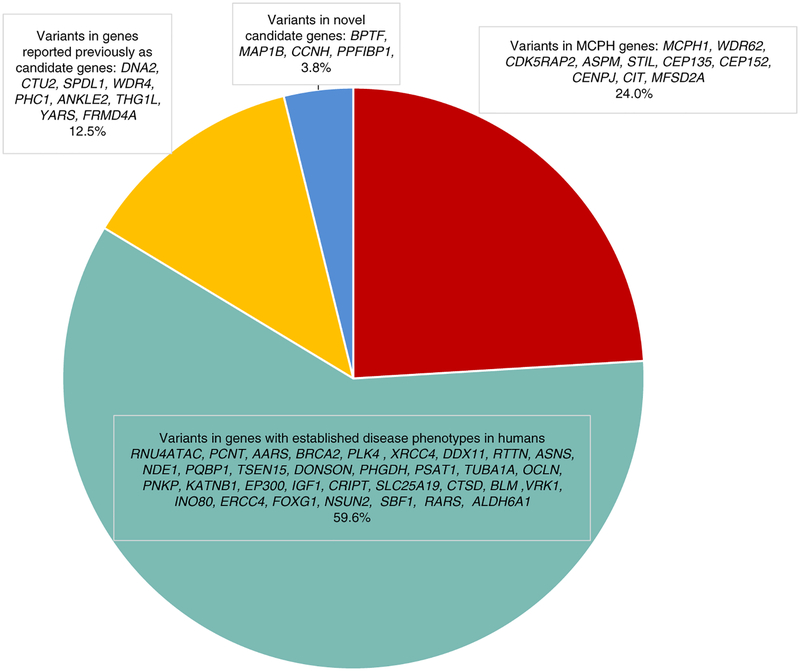
Category 1: variants in MCPH genes
According to OMIM, 18 genes are classified as microcephaly primary hereditary (MCPH) genes. Pathogenic variants were encountered in ten of these genes, including a common founder pathogenic variant in ASPM, and these accounted for 25/104 (24%) of our cohort (Fig. 1, Table S1, S2, and S3). Case 16DG0186 was found to harbor a previously reported homozygous intronic pathogenic variant in CDK5RAP2: NM_001011649.2:c.4005–15A>G confirmed to cause aberrant splicing by reverse transcription polymerase chain reaction (RT-PCR).8 Unlike the previously reported cases with this variant, our case had evidence of primordial dwarfism (PD) in addition to microcephaly. Interestingly, this case was also found to have a homozygous nonsense variant in CNTRL, a gene approximately 508 kb downstream of CDK5RAP2 and known to encode a centrosomal protein that has not been linked yet to human disease. Both genes are located within the same region of homozygosity (ROH) of ≈17 Mb. The truncated nature of the variant identified in CNTRL and the involvement of CNTRL in centrosome function indicate that this variant may also contribute together with the variant in CDK5RAP2 to the pathogenicity of the disease in our patient.
Fig. 1. Pie chart showing the grouping and distribution of the variants identified in this cohort into four categories: variants in MCPH genes, variants in genes with established disease phenotypes in humans, variants in genes reported previously as candidate genes, and variants in genes with no established disease phenotypes in humans.
MCPH, microcephaly primary hereditary.
Category 2: variants in genes with established disease phenotypes in humans other than MCPH
This category accounts for 60% of our cohort (Fig. 1). The majority involved genes where CM was in the published phenotype and these include RNU4ATAC, PCNT, AARS, BRCA2, PLK4, XRCC4, DDX11, RTTN, ASNS, NDE1, PQBP1, TSEN15, DONSON, PHGDH, PSAT1, TUBA1A, OCLN, PNKP, KATNB1, EP300, IGF1, CRIPT, SLC25A19, CTSD, BLM, INO80, ERCC4, FOXG1, NSUN2, and VRK1. There is one report of a prenatally diagnosed microcephaly in the setting of a VRK1 pathogenic variant.9 Two families in our cohort were found to have a pathogenic variant in VRK1 with extremely progressive microcephaly of −4.3 SD at age 2 months in one family and −11.1 SD at the age 11 years in the second one (Fig. 2). Although ERCC4 pathogenic variants typically cause postnatal microcephaly, our data corroborate those of Kashiyama et al., who reported the only patient with documented ERCC4-related prenatal onset microcephaly.10 Furthermore, microcephaly is also a common feature of NSUN2-related phenotype but the association of congenital microcephaly was only reported once by Martinez FJ, 2012 and we are confirming in this paper that congenital microcephaly can be part of the phenotype. Reassuringly, the contribution of genes linked only to postnatal and not congenital microcephaly in our cohort was minimal and was limited to RARS, SBF1, and ALDH6A1. To our knowledge only five missense pathogenic variants were reported in ALDH6A1 as disease-causing variants.11–13 Thus, the truncating nature of the variant in ALDH6A1 identified in our patient may have allowed us to observe the broader spectrum of ALDH6A1-related microcephaly. RARS-related microcephaly was only reported once in a 2-month-old patient who was compound heterozygous for a startloss and a missense pathogenic variants (patient 4 in Wolf et al.),14 but we show here that RARS pathogenic variants can also cause congenital microcephaly. Similarly, we previously reported the only family with SBF1-related microcephaly caused by a missense variant but had no records of the birth OFC.15,16 The patient we describe here with a homozygous, presumably more severe pathogenic variant clearly has a congenital microcephaly phenotype.
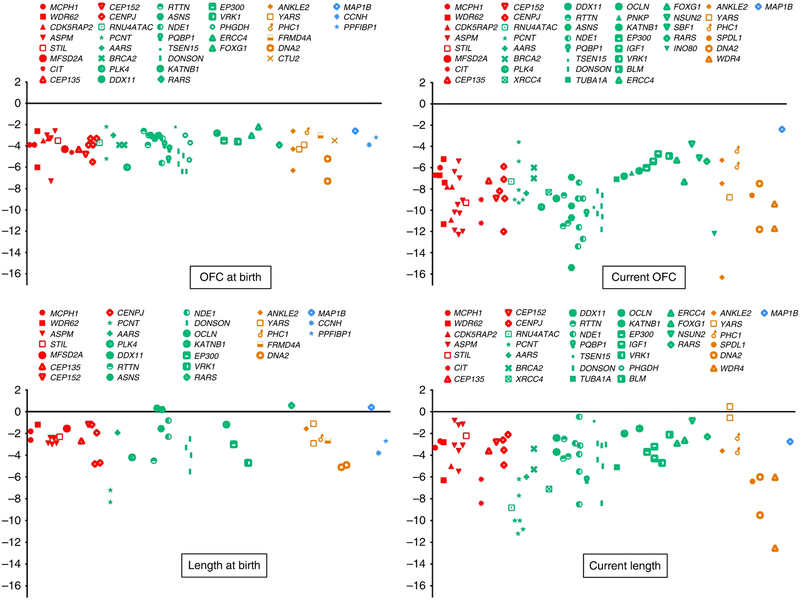
Fig. 2. Graph plot showing the available OFC (upper panel) and height/length (lower panel) data points at birth and on the last clinical evaluation that we collected for the cohort.
Red color: Cases with variants in microcephaly primary hereditary (MCPH) genes. Green: Cases with variants in genes with established disease phenotypes in humans with congenital microcephaly (CM). Orange: Cases with variants in genes reported previously as candidate genes. Blue: Cases with variants in genes with no established disease phenotypes in humans. OFC, occipital frontal circumference.
Category 3: variants in genes reported previously as candidate genes
These genes include those with only tentative links based on single families. One particularly interesting gene in this category is ANKLE2, which was reported in a single Mexican family comprising two affected children who were compound heterozygous for a missense and nonsense allele.17 In our cohort, we identified one patient with a homozygous likely deleterious variant (Table S1). Through an international collaboration, we were able to identify two further families who share a homozygous likely deleterious variant (Table S3). These families strongly support the designation of ANKLE2 as MCPH16. Another interesting finding is the identification of a likely deleterious homozygous variant in YARS, a gene only known to cause an autosomal dominant form of Charcot–Marie–Tooth disease. This variant in YARS was found in family 89 with two affected siblings (14DG0613 and 14DG0614) born to consanguineous parents (Table S1, Fig. 2). Nowaczyk et al. recently reported two siblings who presented with a multisystem disease and both were found to harbor biallelic likely deleterious variants within the YARS gene.18 Thus, our homozygous variant likely confirms YARS as a bona fide congenital microcephaly gene in the recessive form and adds to the list of genes with markedly distinct dominant and recessive phenotypes.19,20 The third candidate gene that our results appear to confirm is THG1L. The index (14DG0824) in family 91 was born with intrauterine growth restriction (IUGR) and diffuse cerebral and cerebellar atrophy (Table S3). Exome sequencing revealed a novel missense variant in THG1L that is predicted to be pathogenic (it likely destabilizes a short helix resulting in lack of anchorage to the core of the protein, see Supplemental Computational Biology Material). The phenotype is similar to that reported recently by Edvardson et al.,21 which adds to the growing list of human neurodevelopmental diseases linked to abnormal transfer RNA (tRNA) modification.22–24 We also highlight FRMD4A, as the fourth previously reported candidate (one presumably pathogenic variant was reported by Fine et al.25) that we confirm in this cohort based on case 15DG0781 (Tables S2 and S3).
Category 4: variants in genes with no established disease phenotypes in humans
These are genes we propose as novel candidates for congenital microcephaly in humans and include BPTF, MAP1B, CCNH, and PPFIBP1. The justification for their listing as candidates is provided in Supplemental Computational Biology Material.
Distinct patterns of OFC in CM patients
We collected extensive data points of OFC for our cohort and this allowed us to observe two distinct patterns (Fig. 2). Pattern A is characterized by progressive and severe microcephaly (>−5 SD), whereas pattern B shows a much milder form of microcephaly that remains largely stable with age. As expected, all patients with MCPH pathogenic variants belonged to pattern A. However, we note that pathogenic variants in the following genes also followed a similar pattern: RNU4ATAC, PCNT, XRCC4, PLK4, RTTN, DDX11, BRCA2, AARS, TUBA1A, NDE1, KATNB1, IGF1, OCLN, EP300, PQBP1, ASNS, DONSON, TSEN15, PNKP, FOXG1, VRK1, and SBF1.
Phenotypic overlap between CM and microcephalic primordial dwarfism
We first reported the overlap between PM and microcephalic primordial dwarfism (MOPD) when we showed that CENPJ, a strictly PM gene, can also underlie MOPD.26 Similarly, our cohort revealed that CEP135, a gene only reported in the context of severe PM, can be mutated in MOPD (11DG0561, see Table S2 and S3).27,28 This genetic overlap prompted us to further explore the role of CM genes in somatic growth. Therefore, we plotted the height/length data points (Fig. 2) that we collected for our cohort and noticed an interesting pattern of short stature (>−3 SD, not necessarily congenital) in 11/21 (52%). Further analysis of this trend revealed that it is strongest in PM genes as well as most of the genes that follow pattern A of OFC growth, e.g., ASNS.
Seeking a definition of primary microcephaly
We exploited the extensive phenotypic (including imaging) and molecular data available in our cohort to test the commonly used definition of primary microcephaly, i.e., congenital nonsyndromic microcephaly that progresses with age with largely normal brain magnetic resonance imaging (MRI). Because there is no consensus on the different elements of this definition, we opted to test different combinations of the following cutoff values:
Congenital microcephaly: The cutoff value of OFC at birth that we used was >–2 SD but even a more restrictive cutoff value of >−2.5 SD would have been met by all our molecularly confirmed MCPH cases.
Progressive microcephaly: although we did not have long term OFC data on all the cases, our cohort suggests that all MCPH cases have an OFC >−5SD after 6 months of age.
Nonsyndromic: defining syndromic association can be very problematic because we have shown a strong overlap with primordial dwarfism. In addition, microcephaly-associated facial dysmorphism is common, as well as epilepsy (16/25 [64%]). Therefore, we opted to define “syndromic association” as evidence of a major birth defect in a non–central nervous system organ other than short stature. Such definition will again be met by all of our molecularly confirmed MCPH cases.
Lack of major brain malformation: our analysis shows that this is perhaps the most difficult aspect of the definition because we found abnormal neuronal migration (lissencephaly/pachygyria and variation thereof in 11/22), and, to a lesser extent abnormal postmigrational development (3/22) to be common with 64% of the molecularly confirmed MCPH cases having at least one element of these defects (Table S2 and Fig. 3). These accompanying major defects were particularly common in those with pathogenic variants in ASPM, WDR62, CEP152, MCPH1, CIT, CENPJ, and STIL. Because the nondiscriminate use of this criterion will greatly reduce the sensitivity of the common primary microcephaly definition, we suggest that the presence of lissencephaly and pachygyria should be tolerated. Accordingly, we think that RTTN and NDE1 should be classified as primary microcephaly genes. On the other hand, we note the conspicuous absence of “atrophy” in MCPH cases. Indeed, replacing “lack of major brain defects” with “lack of atrophy” would be met by virtually all of our MCPH cases.
Fig. 3. Representative magnetic resonance images (MRI) for the cases with pathogenic variant in microcephaly primary hereditary (MCPH) genes and other genes highlighted in this study.
(a) Case 13DG0605 with pathogenic variant in ASPM showing pachygyria. (b) Case 15DG1001 with pathogenic variant in MFSD2A showing hydrocephaly. (c) Case 17DG0679 with a pathogenic variant in STIL showing the partial agenesis of the corpus callosum and pachygyria. (d, e) Case 17DG0680 with a pathogenic variant in CEP152 showing polymicrogyria, severe callosal hypogenesis with inter-hemispheric cyst at left aspect of the falx compressing the left cerebral hemisphere continuous with the third ventricle, and mild dilatation of the lateral ventricles. (f) Case 15DG0077 with a pathogenic variant in BRCA2 showing hypoplastic corpus callosum. (g) Case 13DG0152 with a pathogenic variant in DDX11 showing hypoplastic corpus callosum. (h) Case 12DG1528 with a pathogenic variant in SPDL1 showing virtually no brain in computed tomography (CT) scan. (i) Case with a pathogenic variant in ANKLE2 showing sloping of the forehead along with simplified gyration, partial agenesis of the corpus callosum, and hypoplastic cerebellum.
DISCUSSION
Congenital microcephaly is a birth defect that offers a window into the early human brain development, and deciphering its genetics has greatly added to our understanding of the molecular mechanisms that control this developmental process. Despite the strong interest in this line of research, prior studies have focused on individual novel disease gene identification. When approached as a collective entity, studies have typically only focused on primary microcephaly as a subgroup.29–31 Although our study with its unbiased inclusion of all forms of microcephaly supports a distinct clinical and molecular profile of primary microcephaly, we note that a strict definition that requires lack of syndromic and other associated brain malformations has important limitations. First, we note that several genes not classified as primary microcephaly genes can result in a very similar profile, namely RTTN and NDE1. Second, we note the remarkable phenotypic and molecular overlap between primary microcephaly and microcephalic primordial dwarfism, which suggests that the definition should not exclude primordial dwarfism as an acceptable syndromic association. Third, major brain malformations are not uncommon in our molecularly confirmed MCPH cases. Thus, while we support the continued use of the term “primary microcephaly,” it should be emphasized that it has limited sensitivity and specificity.
The contemporary expanded use of genomic tests makes it less critical to narrow the differential diagnosis prior to ordering gene sequencing. Nonetheless, knowledge of which genes can result in congenital microcephaly can be very helpful in variant interpretation. For example, our demonstration that certain disease genes not previously linked to congenital microcephaly can result in this phenotype makes it easier to consider variants in these genes as candidate causal variants in this context. More importantly, the depletion we show in our cohort for genes linked to postnatal microcephaly strongly supports the notion that these two classes of microcephaly are etiologically distinct, which is also important in variant interpretation.
We have previously shown that a gene strictly linked to primary microcephaly (CENPJ) can also cause microcephalic primordial dwarfism, a phenomenon that was later observed for CEP152. Here, we expand the overlap between these two disease entities by showing that pathogenic variants in CEP135, which has only been linked to microcephaly 8, primary, can result in microcephalic primordial dwarfism. Together with the overlapping ontology of the genes for the two conditions and the identical pattern of postnatal head growth, we conclude that primary microcephaly and microcephalic primordial dwarfism are a phenotypic continuum although the genotype/phenotype correlation remains unclear at this point.
Given the recognized extreme complexity of brain development, it is perhaps not surprising that our analysis of genes associated with congenital microcephaly revealed a remarkable diversity of biological functions. Nonetheless, the rather unique phenotype associated with the subset of congenital microcephaly genes that cause MCPH appears to correlate with a rather restricted diversity. Interestingly, our results suggest that genes not listed as MCPH, but which we argue meet the definition (see above), e.g., RTTN and NDE1, encode centriolar proteins and have a remarkably similar gene ontology, which lends further support to their classification as MCPH genes.
The novel candidate genes that we propose in this study deserve a special mention. BPTF encodes bromodomain PHD finger transcription factor, which functions in protein modeling and resembles the largest subunit of NURF (nucleosome remodeling factor) in mammals. Knockout of the ortholog gene in zebrafish (bptf) using CRISPR-Cas9 genome editing causes reduction in the head of size of the mutant larvae compared to controls.32 Furthermore, knockout mice for Bptf display embryonic lethality with failure to establish a functional distal visceral endoderm.33 Recently, pathogenic variants in BPTF were found to cause an autosomal dominant neurodevelopmental disorder with postnatal microcephaly and dysmorphic features.32 It remains to be seen if recessive BPTF variants are a bona fide cause of autosomal recessive CM, as we suggest in this study, through the identification of future patients. The available data on the function of MAP1B make it another compelling CM candidate gene. MAP1B is a neuron-specific microtubule-associated protein that is expressed in different portions of the brain and has a role in brain development. Heterozygous knockout of MAP1B in mouse leads to abnormalities in the shape and size of the cerebellum while homozygotes are embryonic lethal.34 The mouse model therefore seems compatible with MAP1B-related recessive CM as we suggest here. The third gene that we are proposing in this study as a novel candidate gene for CM, CCNH, encodes a cdk-activating kinase involved in the regulation of the cell cycle by activating the phosphorylation of several cyclin-dependent kinases.35 There is no available model for deficiency of the mouse ortholog; however, a study in zebrafish involving a dominant-negative form of cyclin H showed a delay in the onset of zygotic transcription in the early embryo, resulting in apoptosis at 5 h postfertilization and later abnormal body shape, eyes, and brain.36 Finally, we note that PPFIBP1 (liprin-β−1) belongs to the leprin family of proteins, which form complex structures that serve as scaffolds for the recruitment of other proteins.37 Although liprin-α has an established role in synapse morphogenesis, such role has not yet been demonstrated for leprin-β.38 However, the truncating nature to the pathogenic variants we identified in PPFIBP1 predicting loss of nearly 70% of the protein including the three main domains SAM (1, 2, and 3) and the segregation of these variants in three affected siblings with similar phenotype support our proposal that this gene is a candidate gene for syndromic CM.
In conclusion, we present a large cohort of molecularly characterized cases of congenital microcephaly. Our results refine the phenotypic subgrouping of congenital microcephaly, expand its genetic heterogeneity, and provide the rationale for reconsidering some historical boundaries in the phenotypic and molecular classification of this group of disorders.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the King Salman Center for Disability Research (F.S.A.), King Abdulaziz City for Science and Technology (13-BIO1113-20, F.S.A.), and the Saudi Human Genome Program (F.S.A.). M.A.S. was supported by the Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia through the research group project number RGP-VPP-301. The research by S.T.A. and A.A.M. reported in this publication was supported by funding from King Abdullah University of Science and Technology (KAUST). C.A.W. was supported by the Manton Center for Orphan Disease Research and a grant from the National Institute of Neurological Disorders and Stroke (R01 NS035129) and is an Investigator of the Howard Hughes Medical Institute. We thank the study families for their enthusiastic participation and the Sequencing and Genotyping Core Facilities at King Faisal Specialist Hospital and Research Centre (KFSHRC) for their technical help.
Footnotes
ELECTRONIC SUPPLEMENTARY MATERIAL
The online version of this article (https://doi.org/10.1038/s41436-018-0140-3) contains supplementary material, which is available to authorized users.
DISCLOSURE
The authors declare no conflicts of interest.
REFERENCES
- 1.Ashwal S, Michelson D, Plawner L, Dobyns WB. Practice parameter: evaluation of the child with microcephaly (an evidence-based review): report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology 2009;73:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcantara D and O’driscoll M, 2014, June. Congenital microcephaly. In American Journal of Medical Genetics Part C: Seminars in Medical Genetics 2014;166:124–139. [DOI] [PubMed] [Google Scholar]
- 3.Krauss MJ, Morrissey AE, Winn HN, Amon E, Leet TL. Microcephaly: an epidemiologic analysis. Am J Obstet Gynecol 2003;188: 1484–1490. [DOI] [PubMed] [Google Scholar]
- 4.Network NBDP. Major birth defects data from population-based birth defects surveillance programs in the United States, 2006–10. Birth Defects Res A Clin Mol Teratol 2013;97:S1–S172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wollnik B A common mechanism for microcephaly. Nat Genet 2010;42:923–924. [DOI] [PubMed] [Google Scholar]
- 6.Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends Genet 2009;25:501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saudi Mendeliome Group Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol 2015;16:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond J, Roberts E, Springell K, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 2005;37:353. [DOI] [PubMed] [Google Scholar]
- 9.Renbaum P, Kellerman E, Jaron R, et al. Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the VRK1 gene. Am J Human Genet 2009;85:281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kashiyama K, Nakazawa Y, Pilz DT, et al. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Human Genet 2013;92:807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcadier JL, Smith AM, Pohl D, et al. Mutations in ALDH6A1 encoding methylmalonate semialdehyde dehydrogenase are associated with dysmyelination and transient methylmalonic aciduria. Orphanet J Rare Dis 2013;8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sass JO, Walter M, Shield JP, et al. 3-Hydroxyisobutyrate aciduria and mutations in the ALDH6A1 gene coding for methylmalonate semialdehyde dehydrogenase. J Inherit Metab Dis 2012;35:437–442. [DOI] [PubMed] [Google Scholar]
- 13.Chambliss K, Gray R, Rylance G, Pollitt R, Gibson K. Molecular characterization of methylmalonate semialdehyde dehydrogenase deficiency. J Inherit Metab Dis 2000;23:497–504. [DOI] [PubMed] [Google Scholar]
- 14.Wolf NI, Salomons GS, Rodenburg RJ, et al. Mutations in RARS cause hypomyelination. Ann Neurol 2014;76:134–139. [DOI] [PubMed] [Google Scholar]
- 15.Alazami AM, Alzahrani F, Bohlega S, Alkuraya FS. SET binding factor 1 (SBF1) mutation causes Charcot-Marie-Tooth disease type 4B3. Neurology 2014;82:1665–1666. [DOI] [PubMed] [Google Scholar]
- 16.Bohlega S, Alazami A, Cupler E, Al‐Hindi H, Ibrahim E, Alkuraya F. A novel syndromic form of sensory-motor polyneuropathy is linked to chromosome 22q13. 31-q13. 33. Clin Genet 2011;79:193–195. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto S, Jaiswal M, Charng W-L, et al. A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 2014;159:200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowaczyk MJ, Huang L, Tarnopolsky M, et al. A novel multisystem disease associated with recessive mutations in the tyrosyl‐tRNA synthetase (YARS) gene. Am J Med Genet A 2017;173:126–134. [DOI] [PubMed] [Google Scholar]
- 19.Monies D, Abouelhoda M, AlSayed M, et al. The landscape of genetic diseases in Saudi Arabia based on the first 1000 diagnostic panels and exomes. Hum Genet 2017;136:921–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monies D, Maddirevula S, Kurdi W, et al. Autozygosity reveals recessive mutations and novel mechanisms in dominant genes: implications in variant interpretation. Genet Med 2017;19:1144–1150. [DOI] [PubMed] [Google Scholar]
- 21.Edvardson S, Elbaz-Alon Y, Jalas C, et al. A mutation in the THG1L gene in a family with cerebellar ataxia and developmental delay. Neurogenetics 2016;17:219–225. [DOI] [PubMed] [Google Scholar]
- 22.Alazami AM, Hijazi H, Al-Dosari MS, et al. Mutation in ADAT3, encoding adenosine deaminase acting on transfer RNA, causes intellectual disability and strabismus. J Med Genet 2013;50:425–430. [DOI] [PubMed] [Google Scholar]
- 23.Shaheen R, Han L, Faqeih E, et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet 2016;135:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaheen R, Abdel-Salam GM, Guy MP, et al. Mutation in WDR4 impairs tRNA m 7 G 46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol 2015;16:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fine D, Flusser H, Markus B, et al. A syndrome of congenital microcephaly, intellectual disability and dysmorphism with a homozygous mutation in FRMD4A. Eur J Hum Genet 2015;23:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS. Novel CENPJ mutation causes Seckel syndrome. J Med Genet 2010;47:411–414. [DOI] [PubMed] [Google Scholar]
- 27.Hussain MS, Baig SM, Neumann S, et al. A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am J Human Genet 2012;90:871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farooq M, Fatima A, Mang Y, et al. A novel splice site mutation in CEP135 is associated with primary microcephaly in a Pakistani family. J Hum Genet 2016;61:271. [DOI] [PubMed] [Google Scholar]
- 29.Ahmad I, Baig S, Abdulkareem A, et al. Genetic heterogeneity in Pakistani microcephaly families revisited. Clin Genet 2017;92:62–68. [DOI] [PubMed] [Google Scholar]
- 30.Wang R, Khan A, Han S, Zhang X. Molecular analysis of 23 Pakistani families with autosomal recessive primary microcephaly using targeted next-generation sequencing. J Hum Genet 2017;62: 299–304. [DOI] [PubMed] [Google Scholar]
- 31.Barbelanne M, Tsang WY. Molecular and cellular basis of autosomal recessive primary microcephaly. Biomed Res Int 2014;2014: 547986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stankiewicz P, Khan TN, Szafranski P, et al. Haploinsufficiency of the chromatin remodeler BPTF causes syndromic developmental and speech delay, postnatal microcephaly, and dysmorphic features. Am J Human Genet 2017;101:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landry J, Sharov AA, Piao Y, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet 2008;4:e1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edelmann W, Zervas M, Costello P, et al. Neuronal abnormalities in microtubule-associated protein 1B mutant mice. Proc Natl Acad Sci U S A 1996;93:1270–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lolli G, Johnson LN. CAK—cyclin-dependent activating kinase: a key kinase in cell cycle control and a target for drugs? Cell Cycle 2005;4:565–570. [PubMed] [Google Scholar]
- 36.Liu QY, Wu ZL, Lv WJ, Yan YC, Li YP. Developmental expression of Cyclin H and Cdk7 in zebrafish: the essential role of Cyclin H during early embryo development. Cell Res 2007;17:163. [DOI] [PubMed] [Google Scholar]
- 37.Serra-Pagès C,Medley QG,Tang M,Hart A,Streuli M, Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem 1998;273:15611–15620. [DOI] [PubMed] [Google Scholar]
- 38.Kaufmann N, DeProto J, Ranjan R, Wan H, Van Vactor D. Drosophila liprin-α and the receptor phosphatase Dlar control synapse morphogenesis. Neuron 2002;34:27–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.