Abstract
Purpose:
To introduce a novel, deep-learning method to generate synthetic computed tomography (SCT) scans for proton treatment planning and evaluate its efficacy.
Materials and Methods:
50 Patients with base of skull tumors were divided into 2 nonoverlapping training and study cohorts. Computed tomography and magnetic resonance imaging pairs for patients in the training cohort were used for training our novel 3-dimensional generative adversarial network (cycleGAN) algorithm. Upon completion of the training phase, SCT scans for patients in the study cohort were predicted based on their magnetic resonance images only. The SCT scans obtained were compared against the corresponding original planning computed tomography scans as the ground truth, and mean absolute errors (in Hounsfield units [HU]) and normalized cross-correlations were calculated. Proton plans of 45 Gy in 25 fractions with 2 beams per plan were generated for the patients based on their planning computed tomographies and recalculated on SCT scans. Dose-volume histogram endpoints were compared. A γ-index analysis along 3 cardinal planes intercepting at the isocenter was performed. Proton distal range along each beam was calculated.
Results:
Image quality metrics show agreement between the generated SCT scans and the ground truth with mean absolute error values ranging from 38.65 to 65.12 HU and an average of 54.55 ± 6.81 HU and a normalized cross-correlation average of 0.96 ± 0.01. The dosimetric evaluation showed no statistically significant differences (p > 0.05) within planning target volumes for dose-volume histogram endpoints and other metrics studied, with the exception of the dose covering 95% of the target volume, with a relative difference of 0.47%. The γ-index analysis showed an average passing rate of 98% with a 10% threshold and 2% and 2-mm criteria. Proton ranges of 48 of 50 beams (96%) in this study were within clinical tolerance adopted by 4 institutions.
Conclusions:
This study shows our method is capable of generating SCT scans with acceptable image quality, dose distribution agreement, and proton distal range compared with the ground truth. Our results set a promising approach for magnetic resonance imaging–based proton treatment planning.
Keywords: MRI, treatment planning, synthetic CT, proton therapy
Introduction
Current dose calculation in photon and proton radiotherapy is based on computed tomography (CT) as the primary imaging modality [1, 2]. The CT provides density information on patients' anatomy [3] in terms of Hounsfield units (HU), which, in turn, can be converted to relative electron density and a relative stopping power ratio (rSPR) to water for accurate photon and proton dose calculations. However, precise target delineation cannot be achieved with CT alone. Currently, magnetic resonance imaging (MRI) is routinely used for contouring in radiation therapy. Superior soft tissue contrast obtained from MRI, compared with CT scans, enhances the delineation of the target and other critical structures [4, 5], whereas CT enables precise calculation of dose distribution [6–8] and provides reference images that are essential for patient positioning [9]. The MRI scans are CT-registered images, so that contours of the targets and organs-at-risk (OARs) can be transferred [10, 11]. However, image registration introduces systematic spatial uncertainties, which are carried downstream to all phases of treatment planning and delivery [12–16].
With the advancements from machine-learning methods, several studies have developed techniques to facilitate dose calculations based on MRI alone. These methods construct synthetic CT (SCT) images based on MRI, aiming for the complete elimination of CT imaging from the workflow [17–25]. They generally consist of 2 stages. In the training stage, pairs of CT and MRI scans from a training-group of patients are used, with the CT images as the learning-based target of their corresponding MRI scans. Next, during the synthesizing stage, only MRI scans from new patients are provided to the trained model to predict the SCT image. The constructed SCT is then compared with the original planning CT (pCT) to evaluate the performance of the model. Their results show that MRI-based dose calculation can be achieved, and reference images compatible for image-guided radiotherapy with x-ray localization techniques can be constructed. The MRI-only based radiation therapy eliminates image-registration errors, simplifies the workflow, reduces treatment workload and cost, and decreases the concomitant dose to the patient. The latter is of particular interest in pediatric patients who are more likely to receive proton therapy.
Proton therapy is ideal for treating intracranial and base-of-skull (BoS) tumors [26, 27]; for which, the sharp dose falloff beyond the target volume allows for superior sparing of healthy tissue. Studies show that the risk of developing secondary cancers is less for patients who receive proton therapy compared with conventional photon therapy, which makes it ideal for pediatric patients [28–30]. Proton dose is calculated based on CT images to extract HU values associated with each voxel within the volume of the patient. Those values are then converted to respective rSPRs with a conversion curve. Proton range along the beam path is calculated by integration of the rSPR of the tissues. The uncertainties in the calculation of the proton range alter dose distribution in the patient [31], which necessitates an evaluation of the robustness of the plan. Accurate target delineation is of particular importance for generating robust proton plans. Errors in target delineation will result in undercoverage of the planning volume or irradiation of OARs, particularly because of the sharp distal dose falloff of proton beams. During routine proton-planning workflow, MRI scans are acquired and registered to the corresponding CT images to accommodate accurate target delineation. However, image registration may introduce a 0.5 to 3.5 mm systematic error in the head region [32–34]. That error propagates downstream, resulting in a systematic error that remains throughout the course of treatment, which, in turn, may cause suboptimal tumor coverage or tissue sparing.
The main challenge in SCT generation is the lack of a 1-to-1 relationship between corresponding the CT and the MRI for 2 main reasons: (1) MRI and CT scans are 2 different modalities that work on different physical phenomena, and (2) unless an MRI is acquired at the same time as the CT scan on a combined MRI-CT machine, setup differences at the time the two images are acquired are unavoidable. Those setup differences can be reduced by applying rigid registration; however, they cannot be eliminated. In this article, we introduce our approach to SCT generation.
Methods
Patient Selection and Image Acquisition
This study was based on 50 patients who were diagnosed with BoS tumors and who originally underwent stereotactic radio surgery (SRS) in our clinic in May to October of 2018. All MRI scans selected were acquired with the same 3-dimensional (3D) sequences and with fine spatial resolutions of the entire head. Those images, along with their CT images, were extracted retrospectively under an institutional review board–approved protocol. For the brain images, standard T1-weighted MRI was captured with an Avanto 1.5T (Siemens, Munich, Germany) with a T1 gradient rephasing/inversion recovery sequence of 1-mm-thick slices and a resolution of 0.977 mm in 256 × 256 pixels. The CT images were captured with a SOMATOM Definition AS CT scanner (Siemens) with 1.0 × 1.0 × 1.0–mm3 voxel size and 120 peak kilovolt. Patients were positioned and marked for repositioning. At the time of MRI acquisition, patients were positioned with the same immobilization devices based on the tattoo marks.
Image Preprocessing and Registration
Intensity inhomogeneity of the MRI was corrected with an N4ITK MRI Bias-correction filter, implemented in 3D Slicer (version 4.8.1) software. Because CT and MRI scans had different resolutions, MRI was resampled to match the resolution of the CT images. Intrasubject, rigid registration was performed within Velocity AI 3.2.1 (Varian Medical Systems, Palo Alto, California) to register the MRI and CT scans of each patient.
Training Phase
The MRI and pCT imaging pairs of 25 of the patients were used to construct the training cohort. Despite performing rigid registration, some local mismatches remained between the MRI and corresponding CT imaging, which is one of the major obstacles in training an MRI-to-CT transformation model. To overcome that difficulty, we introduced a novel, 3D generative adversarial network (cycleGAN), which uses inverse transformation and inverse supervision to capture the relationship between the MRI and CT image pairs. The architecture of our 3D cycleGAN was constructed based on dense blocks [35], which is different from previously published approaches, such as residual blocks used in the Wolterink et al [36] cycleGAN architecture. Rather than learning the difference between 2 image modalities, dense blocks aim to represent the image patch by both high- and low-frequency information, such as textural and structural features. In the training stage, the 3D MRI patches at the 64, 64, and 64 voxel size with overlapping 18 × 18 × 18 patches were fed to the MRI-to-CT generator to construct an SCT. Inversely, the SCTs are fed to another (CT-to-MRI) to generate synthetic MRI (cycle MRI) scans. Similarly, the training CT was fed to the CT-to-MRI generator to generate synthetic MRI and, inversely, to the MRI-to-CT generator to generate cycle CT scans. The constraints for MRI-to-CT and CT-to-MRI transformations were obtained so that (1) cycle images were similar to the original images, (2) synthetic images were similar to the original images, and (3) discriminators differentiated between synthetic and original images. The input of our generator was a 3D patch extracted by a sliding window from the MRI scans with overlap to its neighboring patches. That overlap ensures that a continuous whole-image output can be obtained and allows for increased training data for the network. We trained and tested on 2 Tesla V100 (NVIDIA, Santa Clara, California) with 32 GB of memory for each graphics processing unit. A batch size of 4.10 GB of central processing unit memory and 58 GB of graphics processing unit memory was used for each batch optimization.
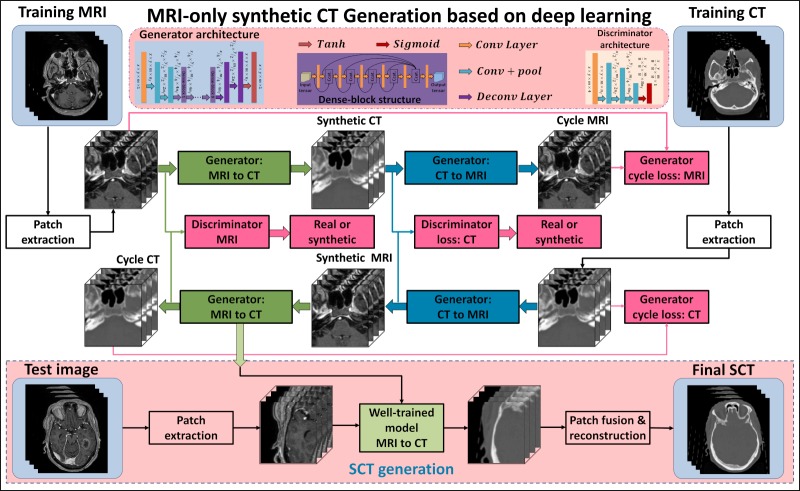
The loss function is typically constructed based on l1-norm (mean absolute distance [MAD]) or l2-norm (mean squared distance [MSD]) separation between synthetic and original images. However, MSD-based loss functions tend to generate blurry images [37], and MAD-based loss functions potentially introduce tissue-classification errors, especially for bone and air differentiation. To demonstrate the efficiency of the lp-norm (p-space = 1.5) distance (MPD) loss, we have compared it against MAD and MSD-based loss functions. The details can be found elsewhere [38]. Such errors are particularly significant for dose calculations because of the considerable differences in the electron density of air and bone [39]. Therefore, we introduced MPD as a measure of the distance between the original and synthetic images. We constructed a loss function based on the MPD and the gradient-difference loss [40]. Generator loss is defined as the sum of the compound loss between synthetic and real images, the compound loss between cycle and real images, and the adversarial loss obtained from discriminators, as recommended by Wolterink et al [36]. The discriminator loss was defined as a function of MPD between the discriminator results of input synthetic and planning CT images. To update kernels for all the hidden layers, Adam (adaptive moment estimation) gradient descent method was applied to minimize generator loss and discriminator loss. In the synthesizing stage, only MRI patches were fed into the MRI-to-CT generator to predict SCT patches, which were fused, consecutively, to construct the final SCT images. The patch fusion is performed by averaging voxel values when overlapping exists. Figure 1 outlines the workflow schematic for our proposed method. A thorough comparison of our method against recent random forest–based [41], deep learning–based [42], and cycleGAN-based [36] methods shows the advantages of our method in detail [38].
Figure 1.
Schematic flowchart of the magnetic resonance imaging-based synthetic computed tomography generation algorithm. The upper part shows the training stage of the method, which consisted of 4 generators. The bottom part shows the synthesizing stage.
Image Quality Evaluation
Mean absolute error (MAE) and normalized cross-correlation (NCC) metrics were used to evaluate absolute differences and image similarity between pCT and SCT scans, which were defined as follows:
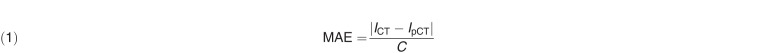 |
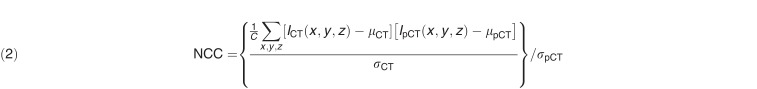 |
where ICT is the ground truth CT, IpCT is the corresponding pCT, C is the number of image voxels within the body outline, and μCT and μpCT are the means of the CT and pCT intensity values, respectively. The σCT and σpCT are the standard deviations of the CT and pCT intensity values, respectively.
Dosimetric Evaluation
The original CT scans for the study cohort, as well as the corresponding SCT scans were transferred to RayStation (version 8A, RayStation Laboratories, Stockholm, Sweden) treatment planning software. Experienced dosimetrists generated proton plans on pCT scans with pencil-beam scanning technique. Dose calculation was achieved with Monte Carlo (version 4.2) software, and default spot and energy-layer spacing were used for all the cases. Each plan consisted of 2 beams at optimized gantry angles (minimum healthy tissue irradiation and robust to setup, HU, and anatomy uncertainties). Dose was prescribed at 45 Gy in 25 fractions and normalized to 98% of the planning target volume (PTV) receiving 100% of the prescription. As the purpose of this study was to evaluate the HU accuracy from the SCT scan, no robust optimization (to compensate for setup and range uncertainty) was used. A simple PTV-based technique was used, and the dose to the PTV was inversely optimized. Dose falloff and maximum dose-volume histogram (DVH) objective functions were used as constraints for minimization of the dose outside of the clinical target volume. To validate dose calculation using SCT, each plan generated and optimized on pCT was recalculated on SCT. Several clinically relevant DVH endpoints (eg, the dose covering 10% of the target volume [D10], D50, D95, Dmean, Dmax, relative volume receiving > 10 Gy, [V10], V15, V20) were obtained for each plan based on both pCT and SCT. The distribution of deviations at each endpoint was displayed with a boxplot. To evaluate dose accuracy based on SCT, we performed a γ-index analysis with MyQA (IBA Dosimetry, Schwarzenbruck, Germany) along the coronal, transverse, and sagittal planes intersecting at the isocenter within: 3% and 3 mm; 2% and 2 mm; and 1% and 1 mm.
Proton Range Evaluation
Proton distal range was calculated with an IronPython script within the RayStation treatment planning software and was defined at the 80% distal dose falloff along each beam direction:
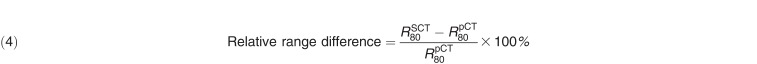 |
The 3 uncertainty criteria defined by 4 institutions—Massachusetts General Hospital (MGH, Boston), MD Anderson Cancer Center (Houston, Texas), and the University of Florida, respectively—are as described below:
The range differences were compared against all 3 criteria.
To further investigate the range differences, a new plan based on the SCT image was created for each case, using the same beam configurations and optimization objectives as the pCT plan. The proximal and distal energies used to cover the target were recorded for both plans, and the energy differences between the 2 plans were analyzed.
Results
Image Quality Evaluation
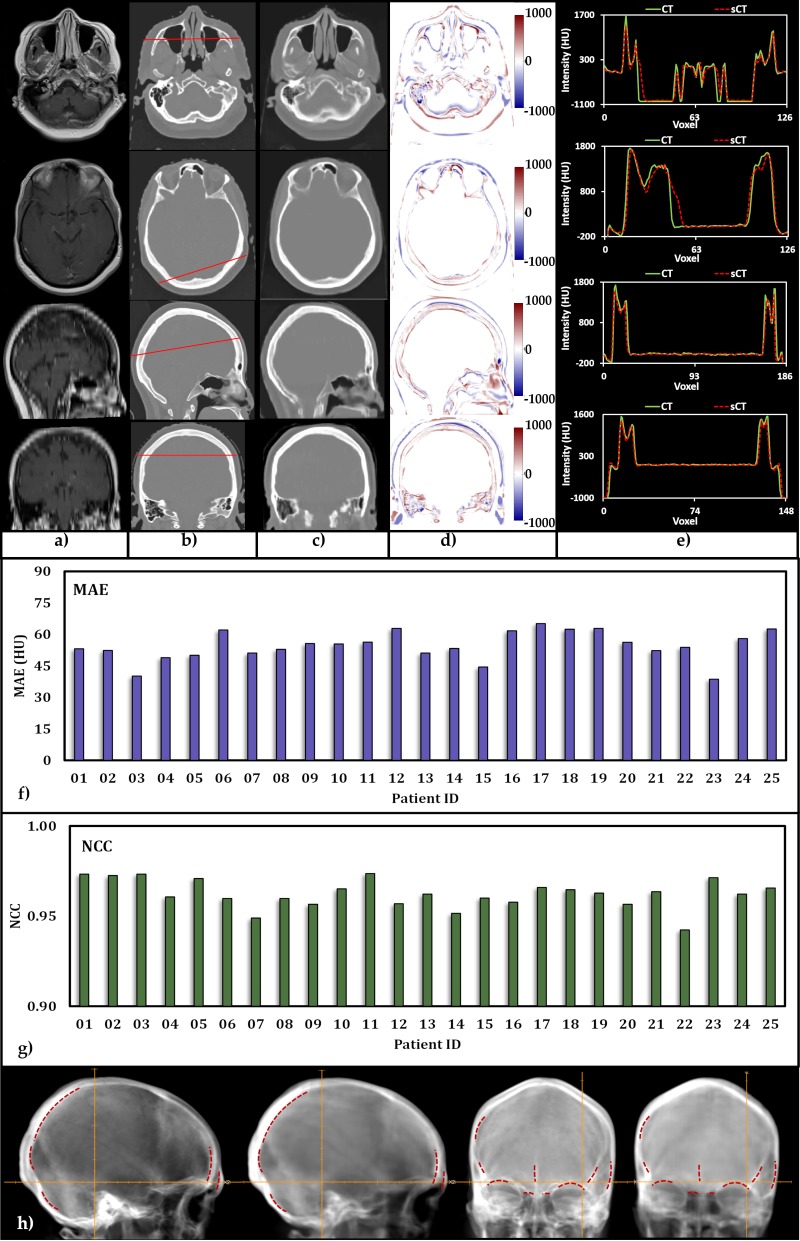
Figure 2 shows 2 transverse, a sagittal, and a coronal plane view of an MRI (Figure 2a) and a pCT (Figure 2b), which was acquired pretreatment as the routine workflow of treatment for one of the patients, and the column shows the, respectively, generated SCT scans (Figure 2c). As can be seen, the SCT scans contain anatomical details similar to the pCT scans at each slice. There are few structural discrepancies between the 2, particularly in the detection of air and bone. These discrepancies may affect proton range if they appear along the beam path. However, traditionally, beam paths that transverse cavities are avoided because of range uncertainties that are brought about by the extreme inhomogeneity. The difference in HU value between the pCT and SCT scans at each plane is presented in Figure 2d. As expected, the discrepancies appear at bone, as well as air, interfaces. Profiles of pCT and SCT scans along the red line are plotted in Figure 2e.
Figure 2.
Image quality: (a) magnetic resonance imaging, (b) planning computed tomography (pCT), and (c) synthetic computed tomography (SCT) for 1 of the 25 patients in the study cohort. (d) Hounsfield unit (HU) differences between the SCT and pCT pairs. (e) Comparison of HU profiles along the red line shown in b. (f and g) Histograms of calculated mean absolute error and normalized cross-correlation. (h) Lateral and anterior digitally reconstructed radiographs obtained from the pCT (left) and SCT, respectively. Red dotted lines show the delineated structures obtained from the SCT, copied to the pCT.
Figure 2f and 2g provides MAEs and NCCs for each individual patient in the study cohort, respectively. The MAEs of the SCT scans range from 38.65 to 65.12 HU, with a mean of 54.55 ± 6.81 HU, which is considerably lower than the reported values of 84.8 [43], 92.5 [40], 97 [44], and 149 HU [45, 46]. The NCC is a metric to quantify image similarity between the SCT and the original pCT scans, and the closer the value is to 1, the more similarity between the 2 images is indicated. Our results show that the SCT scans for the study cohort ranged from 0.94 to 0.97 with an average of 0.96 ± 0.01, indicating very high similarity. Overall, our machine-learning approach has provided very promising results in terms of image quality.
Lateral and anterior projections of the SCT and pCT were generated within RayStation and are presented in Figure 2h. The SCT-based digitally reconstructed radiographs (DRRs) appear to be more blurred compared with their pCT counterparts. However, structures such as superior orbital margin, ethmoid sinus, nasal septum, and crista galli are visible in the SCT-based DRRs with relatively good details. With the SCT-based DRRs, several bony structures relevant for patient positioning are delineated and marked with dashed red lines. Once those marks are copied and transferred intact to the pCT-based DRRs, they match.
Dosimetric Evaluation
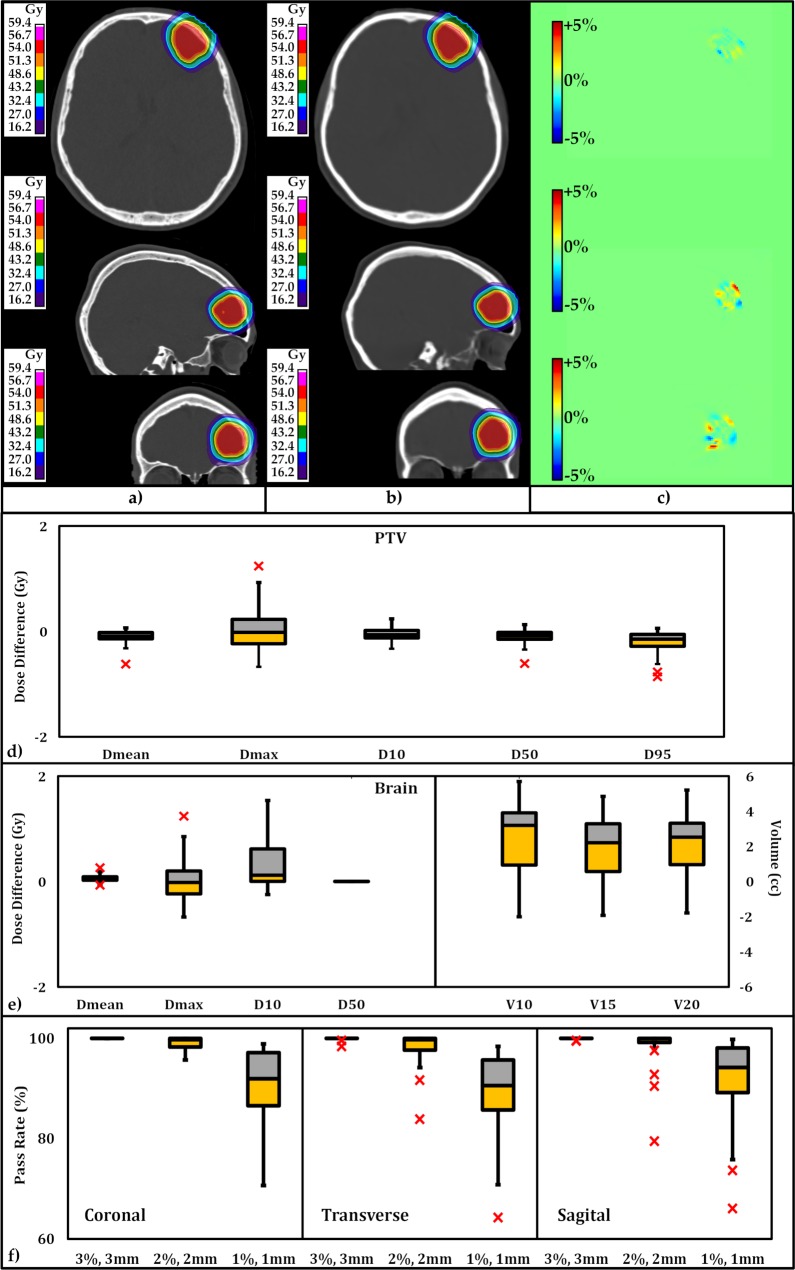
Figure 3a and 3b provide dose distributions based on pCT and SCT scans, at identical transverse, sagittal, and coronal planes intercrossing at the isocenter. Figure 3c presents the dose difference between the 2 at each plane. As can be seen in Figure 3c, the dose difference is small. To quantitatively analyze the difference, we performed γ-index analysis at 3D global γ criteria: 2% and 2 mm, with 10% threshold for comparison with previously published data, and we find them to be 98% and higher, presenting good agreement between dose distributions obtained from pCT and SCT images. Differences in PTV and OAR (whole brain) DVH metrics for all 25 plans calculated for 25 patients are presented with boxplots in Figure 3d and 3e. The central line in each box shows the median, and the yellow and gray bars present 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points, excluding the outliers, which are shown individually with the × symbol. Differences in DVH metrics for all 25 plans are summarized in Table 1. Standard deviations and the P values calculated with the null hypothesis of the zero dose difference between SCT and pCT scans are presented. For all PTV metrics, the average of the differences for Dmean, Dmax, D10, D50, and D95 among the 25 plans were small. The P values for all aforementioned metrics (with the exception of D95) showed no statistical difference. The relative differences for D95 were < 0.47%, which is very small. Within the OAR (whole brain), the metrics and DVH endpoints are in good agreement, as depicted in Figure 3e.
Figure 3.
Transverse, sagittal, and coronal views of (a) dose distribution based on planning computed tomography, (b) dose distribution based on synthetic computed tomography, and (c) dose distribution difference. The statistical distribution of absolute dose distribution difference at dose-volume histogram endpoints for (d) planning target volumes and (e) organs-at-risk (brain) for all 25 plans In the study cohort. The central mark indicates the median. The yellow and gray bars indicate the second and third quartile. (f) The statistical distribution of the pass-rate results of the γ-index analyses along the coronal, transverse, and sagittal planes are within 3 distinct criteria (3% and 3 mm; 2% and 2 mm; and 1% and 1 mm).
Table 1.
Standard deviation and P values calculated with the null hypothesis of the zero planning target volume dose difference between the synthetic computed tomography and the planning computed tomography.
| Value |
Dmean (Gy) |
Dmax (Gy) |
D10 (Gy) |
D50 (Gy) |
D95 (Gy) |
| Mean | −0.130 | −0.028 | 0.046 | 0.130 | 0.259 |
| Standard deviation | 0.172 | 0.357 | 0.119 | 0.178 | 0.321 |
| P | .181 | .872 | .728 | .229 | .003 |
A γ-index analysis along the coronal, transverse, and sagittal planes intercepting at isocenter was performed. The statistics of the pass-rate results are summarized in Table 2, and their statistical distributions are depicted as boxplots in Figure 3f. As can be seen, all plans pass within the 3% and 3 mm criteria. At the tighter criteria of 2% and 2 mm, the γ-index pass rate shows an average of > 98%. At the tightest criteria within this study (1% and 1 mm), the γ-index pass rate averages are 89.2, 91.5, and 91.7, within transverse, coronal, and sagittal plane, respectively.
Table 2.
Mean, standard deviation, and range of the pass rate from the γ-index analyses for 3 different criteria (3% and 3 mm; 2% and 2 mm; and 1% and 1 mm).
| Criteria |
Transverse (%) |
Coronal (%) |
Sagittal (%) |
Total (%) |
| γ-Analysis, 1% and 1 mm | ||||
| Mean | 89.2 | 91.5 | 91.7 | 90.8 |
| Standard deviation | 8.0 | 6.4 | 8.5 | 7.8 |
| Range | 64.3–98.4 | 72.2–98.9 | 66.1–99.8 | 66.1–99.8 |
| γ-Analysis, 2% and 2 mm | ||||
| Mean | 98.17 | 98.97 | 98.13 | 98.42 |
| Standard deviation | 3.56 | 1.62 | 4.62 | 3.51 |
| Range | 83.9–100 | 93.7–100 | 79.5–100 | 79.5–100 |
| γ-Analysis, 3% and 3 mm | ||||
| Mean | 99.92 | 100.00 | 99.96 | 99.96 |
| Standard deviation | 0.33 | 0.00 | 0.13 | 0.21 |
| Range | 98.4–100 | 100–100 | 99.5–100 | 98.4–100 |
Proton range evaluation
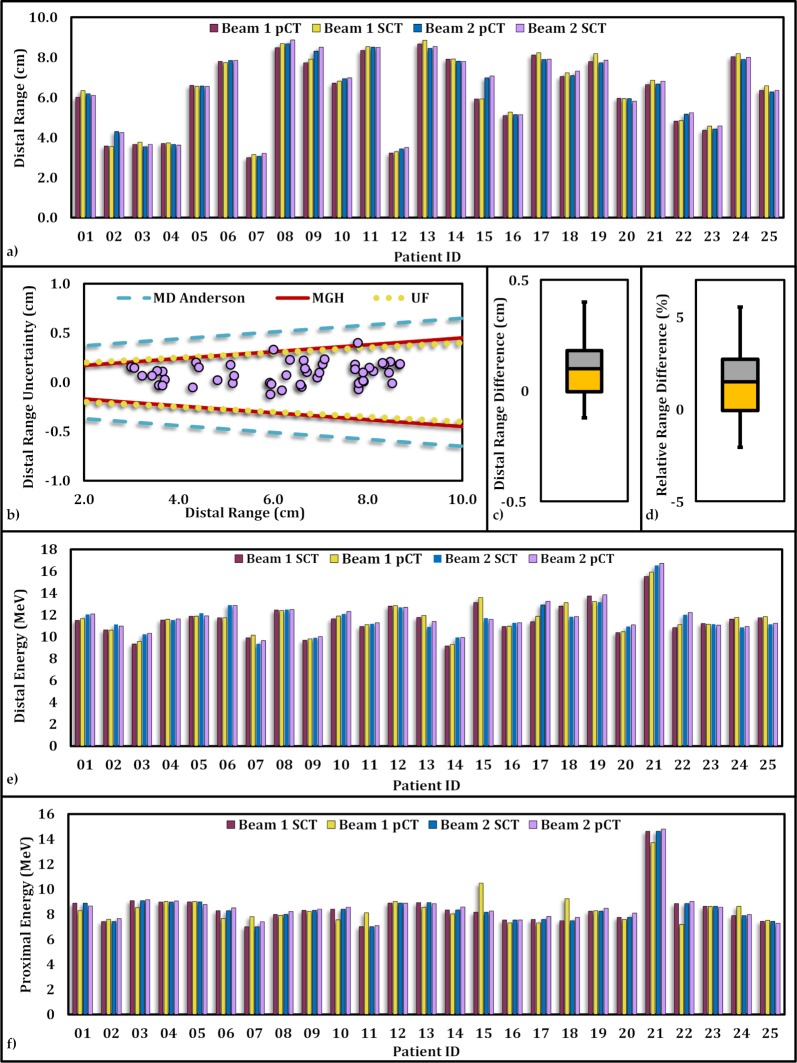
Figure 4a presents the proton distal range calculated along each beam for each plan in the study cohort based on the pCT as well as the SCT. The ranges were retrieved from dose grids in the along-beam direction intercepting the isocenter. The absolute (and relative) distal range differences vary from 0.0 to 4.0 mm (0.0% to 5%) with a mean of 1.1 ± 0.9 mm (1.9 ± 1.5 %). Figure 4b shows the distal range differences between pCT and SCT compared with clinically acceptable criteria adopted by MGH, the University of Florida, and MD Anderson Cancer Center and the University of Pennsylvania. All data points fall within the acceptable region for uncertainty, with the exception of 2 points (1 beam for patient 1, and 1 beam for patient 19), which fell inside the criteria for MD Anderson Cancer Center and the University of Pennsylvania, but slightly outside the border for MGH and the University of Florida criteria. Distal and proximal energies obtained from the SCT plan and the pCT plan for each patient are displayed in Figure 4e and 4f, respectively. The maximum absolute (and relative) proximal energy difference was 4.0 MeV (5.4%), with an average of 1.43 ± 1.0 MeV (1.7% ± 1.2%). The maximum absolute (and relative) distal energy difference is 6.7 MeV (4.8%), with an average of 1.69 ± 1.5 MeV (1.4% ± 1.2%).
Figure 4.
Proton distal range comparison along the direction of each beam calculated based on planning computed tomography (pCT) and synthetic computed tomography (SCT) for the study cohort. (a) The beam range corresponding to each beam for each patient in the study cohort based on pCT and SCT. (b) Proton distal range differences along each beam are depicted in purple circles. Solid red, dotted yellow, and dashed blue represent the criteria adopted from Massachusetts General Hospital, the University of Florida, and MD Anderson and the University of Pennsylvania, respectively. (c) The statistical distribution of differences in proton distal range calculated based on pCT and SCT along each beam. (d) The statistical distribution of relative distal range differences. Distal (e) and proximal (f) energies obtained from SCT and pCT along each beam for each patient.
Discussion
Recent works [44] have studied the feasibility of using MRI-only treatment planning so that a given, planned distribution yields dose distributions that are in close agreement to those based on CT image sets. Because the main challenge in SCT generation is correct detection of bone and air and given the sensitivity of proton beam dose distribution to inhomogeneities, we have established a novel approach for SCT generation with a 3D cycleGAN model. We used CT and MRI scans of 25 patients to train the algorithm. Then, the trained model predicted SCT images based on only MRI of another 25 patients in the study cohort. The results were evaluated based on image quality, dosimetric analysis, and proton distal range.
A side-by-side comparison of SCT and pCT scans shows qualitative agreement. The MAE values obtained for all plans in the study cohort showed a median of 53.82 HU with a maximum value of 65.12 HU. This is considerably smaller than previously reported values (125–129 HU) [22, 45, 47, 48], and the maximum MAE value for our SCT scans us almost half of those previously described. Koivula et al [44] reported an MAE value of 34, which is smaller than our values. It is worth noting that their MAE was based on voxels that excluded the air cavity, whereas our MAE results included the air cavity as well. Given that the detection of air and bone is the most challenging obstacle in SCT generation, air cavities comprise the most errors. As can be seen in Figure 2d, the differences between the SCT and pCT pair arise mainly from detection of air and bone around air cavities. Therefore, our MAE results, which already include such error-prone regions, are indeed promising and show close agreement between SCT and pCT scans. Our results are even better than the recent report [49] of and MAE of 67 HU (ranging from 51 to 115 HU). The profiles along different directions show close agreement in HU values for the 2 as illustrated in Figure 2e. The image similarity is further demonstrated with NCC values close to 1, which show close to perfect agreement between the SCT and pCT pairs. The SCT-based DRRs provide several detailed structures that are clinically relevant for patient positioning, suggesting the possible competency of such DRRs for clinical use. Further study of such DRRs is needed to estimate associated positioning error compared with that of pCT-based DRRs.
It is crucial to evaluate the accuracy of SCT-based dose distribution against those obtained from the original pCT scans. For tumors of the BoS, accurate detection of bone structure, cartilage, and air cavities is imperative for accurate dose calculation. Figure 3a–3c shows dose distribution maps for plans obtained based on pCT and SCT imaging along 3 planes for 1 of the 25 patients under study. The dosimetric evaluation of the plans obtained based on SCT and pCT scans reveals no statistically significant differences between among clinically relevant DVH endpoints (Dmean, Dmax, D10, and D50), with the exception of D95, which shows difference with a mean of 0.259 Gy. The relative dose difference in this case was 0.47%, which is small. This is further demonstrated in Figure 3d. With the exception of 2 outlying points, the D95 for all plans falls close to zero. Figure 3e illustrates the statistical distribution of a few clinically relevant metrics. The differences are mainly for Dmax and D10. The V10, V15, and V20 within the whole brain show very small variations. The similarity in dose calculations based on the SCT and pCT pairs is further supported by γ-index analysis of the plans along 3 planes coinciding with the isocenter. All plans pass along 3 planes, with a total average of 99.96% within the clinical criteria of 3% and 3 mm. For the tighter criteria of 2% and 2 mm, the total pass rate averaged 98.62%. For the tightest criteria of 1% and 1 mm, the average pass rate was 90.8%, which is lower than the 95% for intensity-modulated proton therapy reported by Koivula et al [44].
Proton ranges along each beam of all plans for the 25 patients in the study cohort were calculated and are summarized in a boxplot in Figure 4. The median value for the difference in distal range was 0.99 mm, with a maximum value of 4 mm. The median is higher than the median value of 0.5 mm reported by Pileggi et al [50]. However, our maximum falls slightly lower than the maximum value of 4.4 mm in their report. Relative distal range differences from all beams within the accepted range for the 3 institutions, with 2 beams (1 beam for patient 1 and 1 beam for patient 19), falling marginally outside the accepted range. In both cases, beams in question travel through the thick part of bone (in the case of patient 1 because of a very oblige beam angle, and in the case of patient 19 because of a combination of beam angle and the thickness of the posterior aspect of the skull).
The sources that comprise the discrepancies in proton range are errors in detection of air and bone, imperfect image registration, and MRI distortion. The latter is addressed as one major challenge for highly conformal plans [39]. Many manufacturers have provided solutions to rectify such distortions, and a standard guideline is under development (American Association of Physicists in Medicine [Alexandria, Virginia] Task Group No. 117: Use of MRI Data in Treatment Planning and Stereotactic Procedures—Spatial Accuracy and Quality Control Procedures).
We would like to emphasize that we have performed this novel study based on images of patients originally treated by the SRS technique, and thus immobilized accordingly. However, because proton beams are more sensitive to the variations along the beam path, immobilization of patients with BoS tumors is performed in a more efficient method. Patients are positioned on a BoS frame to which their masks are attached. Then, the frame is indexed to the robotic couch. This method allows for a more-precise patient positioning with fewer patient-repositioning errors. Had these patients been treated with protons, they would have been immobilized with a proton-specific mask and table top for both CT and MRI simulations. This superior immobilization technique reduces patient movement during, and in between, CT and MRI scans, as well as for every fraction of treatment. One desirable aspect of this approach is the minimized differences in patient setup for CT and MRI scans, leading to fewer image-registration errors, which, in turn, make the training of the model an easier task. Even though our results are promising as they are, we expect better performance with images obtained from proton-specific immobilization. Another interesting objective is to investigate the performance of our method after it has been used to train for cases in which an MRI with contrast has been acquired.
Another issue with SCT is metal surgical implants because MRI does not have signal for them (only some artifacts around them). In a pCT scan, the implants are typically contoured with a predefined HU threshold, and their materials and physical densities (or stopping power) are overridden with known values. Similar strategies can be used for the SCT scan. The implants could be outlined, and their physical properties overridden with correct numbers for proton planning. In many cases, the geometry and physical properties are available from the surgery log book, which can be used to double check their contours. In addition, studies have shown that thin titanium surgical mesh and plate (< 1 mm thick) have a minimal effect on the dose distribution and proton range [51]. We also noticed, in our clinics, that small titanium screws (< 5 mm long) have little effect on the dose distribution when they are overridden with the same materials as the surrounding tissue.
Conclusions
We introduced a novel learning-based approach for an MRI-only method of generating SCT scans for proton therapy of BoS tumors with a 3D cycleGAN. We investigated the efficacy of our method for image quality, dosimetric performance, and proton distal range. The nonoverlapping training and study cohorts were composed of 50 patients. Results obtained for the study cohort show that our method is capable of predicting images with close to 1 NCC values and an MAE of 54.55, including regions of air cavities. The dosimetric analysis based on 50 beams from plans for 25 patients in the study cohort showed promising results for MRI-only–based proton treatment planning. The DVH endpoints and other metrics show no statistically significant differences, with the exception of D95 which shows a P value of .003 and comprises a 0.47% relative dose difference within PTV. The γ-index analysis showed an average passing rate of 90% at 1% and 1 mm. The proton distal range for 48 of the 50 beams (96%) fell below the criteria of 3 institutions. We have demonstrated, through several quantitative analyses, that our method is capable of predicting SCT images with desirable image quality, appropriate dose distribution (even in regions in proximity to air cavities), and acceptable proton distal range along each beam. These results highlight the efficacy of our method because the model has overcome the additional and unwarranted challenges from SRS-specific immobilization and will improve further if repeated with patients who are treated with proton therapy and immobilized with a more-effective BoS frame and mask.
ADDITIONAL INFORMATION AND DECLARATIONS
Conflicts of Interest: The authors have no conflicts of interest to disclose.
Acknowledgments: This research was supported, in part, by the National Cancer Institute of the National Institutes of Health, under award R01CA215718, and an Emory Winship Cancer Institute pilot grant.
Ethical Approval: All patient data were collected under an internal review board–approved protocol.
References
- 1.Chernak ES, Rodriguez-Antunez A, Jelden GL, Dhaliwal RS, Lavik PS. The use of computed tomography for radiation therapy treatment planning. Radiology. 1975;117(pt 1):613–4. doi: 10.1148/117.3.613. [DOI] [PubMed] [Google Scholar]
- 2.Pereira GC, Traughber M, Muzic RF., Jr The role of imaging in radiation therapy planning: past, present, and future. Biomed Res Int. 2014;2014:231090. doi: 10.1155/2014/231090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Expert Panel on Radiation Oncology-Prostate. Zaorsky NG, Showalter TN, Ezzell GA, Nguyen PL, Assimos DG, D'Amico AV, Gottschalk AR, Gustafson GS, Keole SR, Liauw SL, Lloyd S, McLaughlin PW, Movsas B, Prestidge BR, Taira AV, Vapiwala N, Davis BJ. ACR Appropriateness Criteria® external beam radiation therapy treatment planning for clinically localized prostate cancer, part I of II. Adv Radiat Oncol. 2017;2:62–84. doi: 10.1016/j.adro.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debois M, yen R, Maes F, Verswijvel G, Gatti G, Bosmans H, Feron M, Bellon E, Kutcher G, Van Poppel H, Vanuytsel L. The contribution of magnetic resonance imaging to the three-dimensional treatment planning of localized prostate cancer. Int J Radiat Oncol Biol Phys. 1999;45:857–65. doi: 10.1016/s0360-3016(99)00288-6. [DOI] [PubMed] [Google Scholar]
- 5.Dirix P, Haustermans K, Vandecaveye V. The value of magnetic resonance imaging for radiotherapy planning. Semin Radiat Oncol. 2014;24:151–9. doi: 10.1016/j.semradonc.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Seco J, Evans PM. Assessing the effect of electron density in photon dose calculations. Med Phys. 2006;33:540–52. doi: 10.1118/1.2161407. [published correction appears in Med Phys. 2007;34:3121] [DOI] [PubMed] [Google Scholar]
- 7.Skrzyński W, Zielińska-Dabrowska S, Wachowicz M, Slusarczyk-Kacprzyk W, Kukołowicz PF, Bulski W. Computed tomography as a source of electron density information for radiation treatment planning. Strahlenther Onkol. 2010;186:327–33. doi: 10.1007/s00066-010-2086-5. [DOI] [PubMed] [Google Scholar]
- 8.Brock KK, Dawson LA. Point: principles of magnetic resonance imaging integration in a computed tomography-based radiotherapy workflow. Semin Radiat Oncol. 2014;24:169–74. doi: 10.1016/j.semradonc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Van den Berge DL, De Ridder M, Storme GA. Imaging in radiotherapy. Eur J Radiol. 2000;36:41–8. doi: 10.1016/s0720-048x(99)00182-5. [DOI] [PubMed] [Google Scholar]
- 10.Khoo VS, Joon DL. New developments in MRI for target volume delineation in radiotherapy. Br J Radiol. 2006;79:S2–15. doi: 10.1259/bjr/41321492. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt MA, Payne GS. Radiotherapy planning using MRI. Phys Med Biol. 2015;60:R323–61. doi: 10.1088/0031-9155/60/22/R323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlsson M1, Karlsson MG, Nyholm T, Amies C, Zackrisson B. Dedicated magnetic resonance imaging in the radiotherapy clinic. Int J Radiat Oncol Biol Phys. 2009;74:644–51. doi: 10.1016/j.ijrobp.2009.01.065. [DOI] [PubMed] [Google Scholar]
- 13.Nyholm T, Nyberg M, Karlsson MG, Karlsson M. Systematisation of spatial uncertainties for comparison between a MR and a CT-based radiotherapy workflow for prostate treatments. Radiat Oncol. 2009;4:54. doi: 10.1186/1748-717X-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraass BA, McShan DL, Diaz RF, Ten Haken RK, Aisen A, Gebarski S, Glazer G, Lichter AS. Integration of magnetic resonance imaging into radiation therapy treatment planning: I. technical considerations. Int J Radiat Oncol Biol Phys. 1987;13:1897–908. doi: 10.1016/0360-3016(87)90358-0. [DOI] [PubMed] [Google Scholar]
- 15.Lee YK, Bollet M, Charles-Edwards G, Flower MA, Leach MO, McNair H, Moore E, Rowbottom C, Webb S. Radiotherapy treatment planning of prostate cancer using magnetic resonance imaging alone. Radiother Oncol. 2003;66:203–16. doi: 10.1016/s0167-8140(02)00440-1. [DOI] [PubMed] [Google Scholar]
- 16.Edmund JM, Nyholm T. A review of substitute CT generation for MRI-only radiation therapy. Radiat Oncol. 2017;12:28. doi: 10.1186/s13014-016-0747-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korhonen J, Kapanen M, Keyriläinen J, Seppälä T, Tenhunen M. A dual model HU conversion from MRI intensity values within and outside of bone segment for MRI-based radiotherapy treatment planning of prostate cancer. Med Phys. 2014;41:011704. doi: 10.1118/1.4842575. [DOI] [PubMed] [Google Scholar]
- 18.Korhonen J, Kapanen M, Keyriläinen J, Seppälä T, Tuomikoski L, Tenhunen M. Influence of MRI-based bone outline definition errors on external radiotherapy dose calculation accuracy in heterogeneous pseudo-CT images of prostate cancer patients. Acta Oncol. 2014;53:1100–6. doi: 10.3109/0284186X.2014.929737. [DOI] [PubMed] [Google Scholar]
- 19.Kapanen M, Tenhunen M. T1/T2*-weighted MRI provides clinically relevant pseudo-CT density data for the pelvic bones in MRI-only based radiotherapy treatment planning. Acta Oncol. 2013;52:612–8. doi: 10.3109/0284186X.2012.692883. [DOI] [PubMed] [Google Scholar]
- 20.Kapanen M, Collan J, Beule A, Seppälä T, Saarilahti K, Tenhunen M. Commissioning of MRI-only based treatment planning procedure for external beam radiotherapy of prostate. Magn Reson Med. 2013;70:127–35. doi: 10.1002/mrm.24459. [DOI] [PubMed] [Google Scholar]
- 21.Dowling JA, Sun J, Pichler P, Rivest-Hénault D, Ghose S, Richardson H, Wratten C, Martin J, Arm J, Best L, Chandra SS, Fripp J, Menk FW, Greer PB. Automatic substitute computed tomography generation and contouring for magnetic resonance imaging (MRI)-alone external beam radiation therapy from standard MRI sequences. Int J Radiat Oncol Biol Phys. 2015;93:1144–53. doi: 10.1016/j.ijrobp.2015.08.045. [DOI] [PubMed] [Google Scholar]
- 22.Johansson A, Karlsson M, Nyholm T. CT substitute derived from MRI sequences with ultrashort echo time. Med Phys. 2011;38:2708–14. doi: 10.1118/1.3578928. [DOI] [PubMed] [Google Scholar]
- 23.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology. 2018;286:676–84. doi: 10.1148/radiol.2017170700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siversson C, Nordström F, Nilsson T, Nyholm T, Jonsson J, Gunnlaugsson A, Olsson LE. Technical note: MRI only prostate radiotherapy planning using the statistical decomposition algorithm. Med Phys. 2015;42:6090–97. doi: 10.1118/1.4931417. [DOI] [PubMed] [Google Scholar]
- 26.Munzenrider JE, Liebsch NJ. Proton therapy for tumors of the skull base. Strahlenther Onkol. 1999;175(suppl 2):57–63. doi: 10.1007/BF03038890. [DOI] [PubMed] [Google Scholar]
- 27.Kjellberg RN, Hanamura T, Davis KR, Lyons SL, Adams RD. Bragg-peak proton-beam therapy for arteriovenous malformations of the brain. N Engl J Med. 1983;309:269–74. doi: 10.1056/NEJM198308043090503. [DOI] [PubMed] [Google Scholar]
- 28.Miralbell R, Lomax A, Cella L, Schneider U. Potential reduction of the incidence of radiation-induced second cancers by using proton beams in the treatment of pediatric tumors. Int J Radiat Oncol Biol Phys. 2002;54:824–9. doi: 10.1016/s0360-3016(02)02982-6. [DOI] [PubMed] [Google Scholar]
- 29.Moteabbed M, Yock TI, Paganetti H. The risk of radiation-induced second cancers in the high to medium dose region: a comparison between passive and scanned proton therapy, IMRT and VMAT for pediatric patients with brain tumors. Phys Med Biol. 2014;59:2883–99. doi: 10.1088/0031-9155/59/12/2883. [DOI] [PubMed] [Google Scholar]
- 30.Allen AM, Pawlicki T, Dong L, Fourkal E, Buyyounouski M, Cengel K, Plastaras J, Bucci MK, Yock TI, Bonilla L, Price R, Harris EE, Konski AA. An evidence based review of proton beam therapy: the report of ASTRO's emerging technology committee. Radiother Oncol. 2012;103:8–11. doi: 10.1016/j.radonc.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Schaffner B, Pedroni E. The precision of proton range calculations in proton radiotherapy treatment planning: experimental verification of the relation between CT-HU and proton stopping power. Phys Med Biol. 1998;43:1579–92. doi: 10.1088/0031-9155/43/6/016. [DOI] [PubMed] [Google Scholar]
- 32.Demol B, Boydev C, Korhonen J, Reynaert N. Dosimetric characterization of MRI-only treatment planning for brain tumors in atlas-based pseudo-CT images generated from standard T1-weighted MR images. Med Phys. 2016;43:6557. doi: 10.1118/1.4967480. [DOI] [PubMed] [Google Scholar]
- 33.Andreasen D, Van Leemput K, Hansen RH, Andersen JA, Edmund JM. Patch-based generation of a pseudo CT from conventional MRI sequences for MRI-only radiotherapy of the brain. Med Phys. 2015;42:1596–605. doi: 10.1118/1.4914158. [DOI] [PubMed] [Google Scholar]
- 34.Ulin K, Urie MM, Cherlow JM. Results of a multi-institutional benchmark test for cranial CT/MR image registration. Int J Radiat Oncol Biol Phys. 2010;77:1584–9. doi: 10.1016/j.ijrobp.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen LL, Wu Y, DSouza AM, Abidin AZ, Wismüller A, Xu C. MRI tumor segmentation with densely connected 3D CNN. In: Angelini ED, Landman BA, editors. Medical Imaging 2018: Image Processing—Proceedings of SPIE. Vol. 10574. Bellingham, WA: SPIE; 2018. Vol. [Google Scholar]
- 36.Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, van den Berg CAT, Išgum I. Deep MR to CT synthesis using unpaired data. In: Tsaftaris S, Gooya A, Frangi A, Prince J, editors. Simulation and Synthesis in Medical Imaging: SASHIMI 2017—Lecture Notes in Computer Science. Vol. 10557. Cham, Switzerland: Springer; 2017. pp. 14–23. Vol. [Google Scholar]
- 37.Mathieu M, Couprie C, LeCun Y. Deep multi-scale video prediction beyond mean square error. 2016. Paper presented at. International Conference on Learning Representations; May 4, San Juan, Puerto Rico.
- 38.Lei Y, Harms J, Wang T, Liu Y, Shu HK, Jani AB, Curran WJ, Mao H, Liu T, Yang X. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med Phys. 2019;46:3565–81. doi: 10.1002/mp.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T, Manohar N, Lei Y, Dhabaan A, Shu HK, Liu T, Curran WJ, Yang X. MRI-based treatment planning for brain stereotactic radiosurgery: dosimetric validation of a learning-based pseudo-CT generation method. Med Dosim. 2018;44:199–204. doi: 10.1016/j.meddos.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, Wang Q, Shen D. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng. 2018;65:2720–30. doi: 10.1109/TBME.2018.2814538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei Y, Jeong JJ, Wang T, Shu HK, Patel P, Tian S, Liu T, Shim H, Mao H, Jani AB, Curran WJ, Yang X. MRI-based pseudo CT synthesis using anatomical signature and alternating random forest with iterative refinement model. J Med Imaging (Bellingham) 2018;5:043504. doi: 10.1117/1.JMI.5.4.043504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, Shen D. Medical image synthesis with context-aware generative adversarial networks. Med Image Comput Comput Assist Interv. 2017;10435:417–25. doi: 10.1007/978-3-319-66179-7_48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44:1408–19. doi: 10.1002/mp.12155. [DOI] [PubMed] [Google Scholar]
- 44.Koivula L, Wee L, Korhonen J. Feasibility of MRI-only treatment planning for proton therapy in brain and prostate cancers: dose calculation accuracy in substitute CT images. Med Phys. 2016;43:4634. doi: 10.1118/1.4958677. [DOI] [PubMed] [Google Scholar]
- 45.Rank CM, Hünemohr N, Nagel AM, Röthke MC, Jäkel O, Greilich S. MRI-based simulation of treatment plans for ion radiotherapy in the brain region. Radiother Oncol. 2013;109:414–8. doi: 10.1016/j.radonc.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 46.Rank CM1, Tremmel C, Hünemohr N, Nagel AM, Jäkel O, Greilich S. MRI-based treatment plan simulation and adaptation for ion radiotherapy using a classification-based approach. Radiat Oncol. 2013;8:51. doi: 10.1186/1748-717X-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edmund JM, Kjer HM, Van Leemput K, Hansen RH, Andersen JA, Andreasen D. A voxel-based investigation for MRI-only radiotherapy of the brain using ultra short echo times. Phys Med Biol. 2014;59:7501–19. doi: 10.1088/0031-9155/59/23/7501. [DOI] [PubMed] [Google Scholar]
- 48.Gudur MS, Hara W, Le QT, Wang L, Xing L, Li R. A unifying probabilistic Bayesian approach to derive electron density from MRI for radiation therapy treatment planning. Phys Med Biol. 2014;59:6595–606. doi: 10.1088/0031-9155/59/21/6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dinkla AM, Wolterink JM, Maspero M, Savenije MHF, Verhoeff JJC, Seravalli E, Išgum I, Seevinck PR, van den Berg CAT. MR-only brain radiation therapy: dosimetric evaluation of synthetic CTs generated by a dilated convolutional neural network. Int J Radiat Oncol Biol Phys. 2018;102:801–12. doi: 10.1016/j.ijrobp.2018.05.058. [DOI] [PubMed] [Google Scholar]
- 50.Pileggi G, Speier C, Sharp GC, Izquierdo Garcia D, Catana C, Pursley J, Amato F, Seco J, Spadea MF. Proton range shift analysis on brain pseudo-CT generated from T1 and T2 MR. Acta Oncol. 2018;57:1521–31. doi: 10.1080/0284186X.2018.1477257. [DOI] [PubMed] [Google Scholar]
- 51.Lin H, Ding X, Yin L, Zhai H, Liu H, Kassaee A, Hill-Kayser C, Lustig RA, McDonough J, Both S. The effects of titanium mesh on passive-scattering proton dose. Phys Med Biol. 2014;59:N81–9. doi: 10.1088/0031-9155/59/10/N81. [DOI] [PubMed] [Google Scholar]