Abstract
Background
Breast cancer remains the most lethal malignancy in women worldwide. Aberrant O-glycosylation is closely related to many human diseases, including breast carcinoma; however, its precise role in cancer development is insufficiently understood. Cosmc is an endoplasmic reticulum-localized chaperone that regulates the O-glycosylation of proteins. Cosmc dysfunction results in inactive T-synthase and expression of truncated O-glycans such as Tn antigen. Here we investigated the impact of Cosmc disruption-mediated aberrant O-glycosylation on breast cancer cell development through in vitro and in vivo experiments.
Materials and Methods
We deleted the Cosmc gene in two breast cancer cell lines (MCF7, T47D) using the CRISPR/Cas-9 system and then measured the expression levels of Tn antigen. The proliferation of Tn-positive cells was examined by RTCA, colony formation and in vivo experiments. The effects of Cosmc deficiency on glycoprotein CD44 and MAPK pathway were also determined.
Results
Both in vitro and in vivo studies showed that Cosmc deficiency markedly suppressed breast cancer cell growth compared with the corresponding controls. Mechanistically, Cosmc disruption impaired the protein expression of CD44 and the associated MAPK signaling pathway; the latter plays a crucial role in cell proliferation. Reconstitution of CD44 substantially reversed the observed alterations, confirming that CD44 requires normal O-glycosylation for its proper expression and activation of the related signaling pathway.
Conclusion
This study showed that Cosmc deficiency-mediated aberrant O-glycosylation suppressed breast cancer cell growth, which was likely mediated by the impairment of CD44 expression.
Keywords: breast cancer, O-glycosylation, Tn antigen, tumor growth
Introduction
Breast cancer is the leading malignancy in women across the world.1 The underlying mechanisms related to breast cancer development are yet to be defined. Mucin-type O-linked glycosylation (hereafter referred to as O-glycosylation) is one of the most common posttranslational protein modifications and has wide-ranging effects on glycoprotein stability and function.2 O-glycosylation is involved in various biological processes, such as signal transduction, cell cycle progression, transcription, and metabolism.3,4 Aberrant O-glycosylation of many transmembrane and secreted proteins can translate into altered biological behaviors involving cell proliferation, differentiation, adhesion, and immune surveillance, and has thereby been associated with multiple steps of tumor progression, including breast cancer.2,5,6
The O-glycosylation process is normally initiated by the addition of N-acetyl galactosamine (GalNAc) to a serine or threonine residue in proteins and is branched by other sugars to produce more complex O-glycans.7,8 Biosynthesis of O-glycans is solely controlled by the enzyme called T-synthase (C1galt1).9,10 In addition, an endoplasmic reticulum-localized molecular chaperone Cosmc is required for the activity and expression of T-synthase.11–13 Cosmc dysfunction results in inactive T-synthase and subsequent expression of the Tn antigen, a truncated O-linked structure that is universally regarded as a hallmark for aberrant O-glycosylation (Figure 1A).14,15 Breast cancers arise in most cases from mammary gland epithelial cells. Tn antigen is seldom detected in normal mammary epithelial cells but is prevalently expressed in almost 90% of breast cancers, suggesting a prominent occurrence of abnormal O-glycosylation during the development of breast cancer.15,16 However, the precise role of Tn antigen (representative of abnormal O-glycosylation) in tumor progression, particularly breast cancer, is largely unclear. The prevailing studies supported a tumor-promoting role for aberrant O-glycosylation in a broad range of human cancers, including pancreatic, colorectal, and gastric cancer,17–20 whereas several recent reports indicated rather an opposite effect of aberrant O-glycosylation, which was shown to delay tumor development in breast, liver, and head and neck cancers.21–23 Overall, there is much to be learned about the impact of aberrant O-glycosylation on breast cancer development.
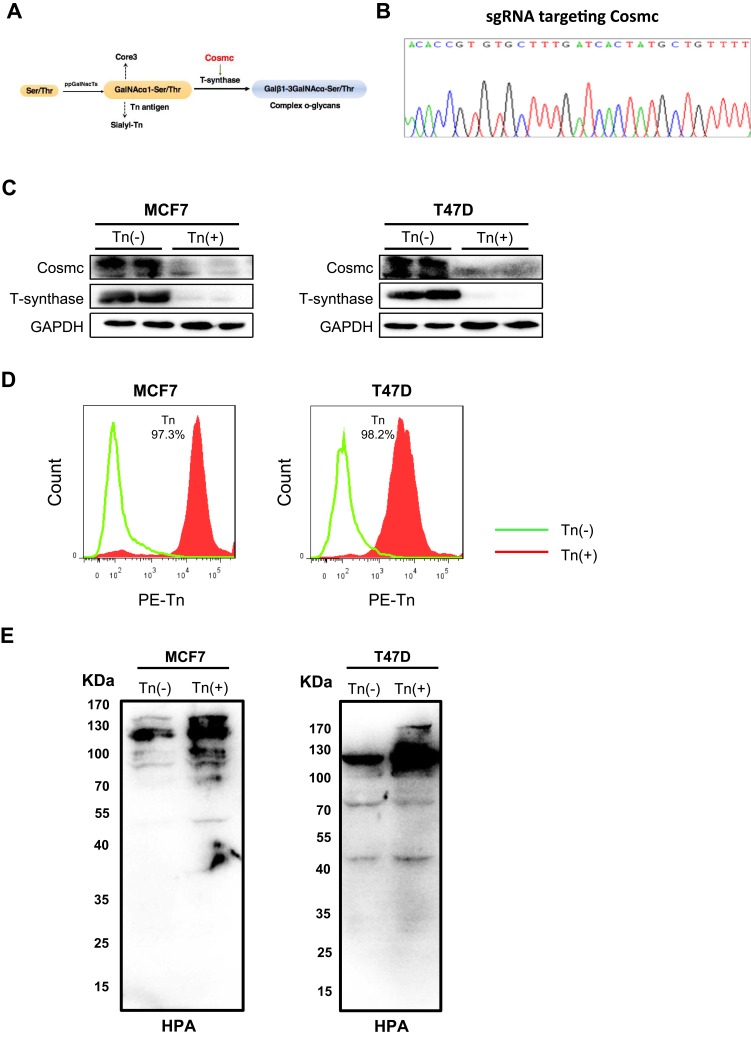
Figure 1.
Forced depletion of Cosmc results in Tn antigen expression in breast cancer cells. (A) Schematic depiction of the biosynthesis of mucin-type O-glycans showing the key regulatory role of Cosmc in the process of O-glycosylation. Deletion of Cosmc results in inactive T-synthase and expression of Tn antigen. (B) One pair of single guide RNAs (sgRNAs) was designed to specifically target the Cosmc gene. (C) The knockout of the Cosmc chaperone and the degradation of T-synthase were confirmed by Western blotting in two breast cancer cells. (D) Flow cytometry analysis of Tn antigen expression with anti-Tn antibody. The percentages of Tn-positive cells were 97.1% and 98.3% in Cosmc-deficiency MCF7 and T47D cells, respectively. (E) Western blot analysis showed a strong expression of Tn antigen in MCF7 and T47D Tn-positive cells compared to the control cells. The membrane was blotted with HPR-labeled HPA.
In the present study, we performed a precise gene editing technology (CRISPR/Cas-9) to delete Cosmc in human breast cancer cells (MCF7, T47D), which resulted in aberrant O-glycosylation (Tn antigen expression) in cells. Our results showed that Cosmc disruption-mediated aberrant O-glycosylation markedly suppressed breast cancer cell proliferation both in vitro and in vivo. Mechanistically, we found that the expression level of CD44, a heavily O-glycosylated protein, was drastically impaired in the absence of Cosmc, which may contribute to the observed biological alterations.
Materials and Methods
Cell Lines and Cell Culture
The human breast cancer cell lines (MCF7, T47D) and a human embryonic kidney cell line (HEK293T) were obtained from the Cell Bank of the Chinese Academy of Sciences (Beijing, China). MCF7 and HEK293T cells were cultured in DMEM, while T47D cells were cultured in RPMI-1640 medium. All media were supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution. All cells were maintained at 37°C in an atmosphere of 5% CO2.
The CRISPR/Cas9 System Was Used for Cosmc Depletion in Cells
To knock out Cosmc in MCF7 and T47D cells, one pair of single guide RNA (sgRNA) sequences was designed to target Cosmc. The Cosmc sgRNA sequences were: F:5′-CACCGTGTGCTTTGATCACTATGCT3′; R:5′AAACAGCATAGTGATCAAACA-CAC-3′. We cloned the sgRNA sequences into a LentiCRISPRv2 plasmid, which was co-transfected into HEK293T cells using Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA) with the psPAX2 and pMD2.G plasmids to generate lentivirus containing the CRISPR/Cas9 system targeting Cosmc. After transfection for 48 hrs, the produced viruses were harvested and used to infect MCF7 and T47D cells. Finally, the transfected cells were selected with puromycin (2 μg/mL) for 1 week.
Flow Cytometric Analysis
The cultured breast cancer cells were washed, collected and suspended in cold PBS (1 × 106/mL). Then, the cells were incubated with a mouse anti-Tn mAb24 or an isotype control (mouse IgM) for 1 hr on ice, followed by incubation with PE-labeled goat anti-mouse IgM (BD, 562033) for 1 h. The cells were finally resuspended in 500 μL PBS and analyzed on a flow cytometer (Canto II; BD Bioscience, NJ, USA).
Cell Proliferation Assay
The cell proliferation rate was assessed using a real-time cell analyzer (RTCA, ACEA Biosciences). Cells were suspended in the culture medium and 5000 cells were added into every well of the E-plate with 4 replicates. After incubation at room temperature for 15 mins, the E-plate was placed onto the RTCA Station in the incubator for continuous recording. Cell index values were recorded every 15 mins for a period of up to 120 hrs.
Colony Formation Assay
The cells were harvested and seeded into 6-well plates with 800 cells in each well. These cells were cultured under standard conditions for 9 days, and then fixed with methanol and stained with 0.1% crystal violet for 20 mins at room temperature. Finally, the cells were rinsed using deionized water and dried. Cell colonies consisting of >50 cells/plate were counted under anatomical lenses.
RNA Isolation and Quantitative RT-PCR Analysis
TRIzol reagent (Invitrogen, CA, USA) was used to extract total RNA from cells. The Prime ScriptTM RT Master Mix system (TaKaRa) was used to reverse mRNA into complementary DNA (cDNA). The relative expression level of CD44 was determined by real-time PCR analysis using SYBR Premix (Applied Biosystems, CA, USA) on an ABI 7500 Real-Time PCR system (Applied Biosystems). GAPDH was used for normalization. The amplification reaction was set as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. Primer sequences were as follows: GAPDH, forward primer 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse primer 5′-GAAGATGGTG-ATGGGATTTC-3′; CD44, forward primer 5′-ACCCCAACTCCATCTGTGC-3′, reverse primer 5′-TTCTGGACATAGCGGGTG-3′.
Nude Mouse Xenograft Experiment
All animal experiments were approved according to the guidelines of Institutional Animal Care and Use Committee at Capital Medical University (Beijing, China). Six-week-old female Balb/C nude mice were obtained from Charles River Laboratories (Beijing, China). For tumor growth experiments, 2×106 cells were subcutaneously injected into nude mice at day 0 and tumor sizes were measured every 3 days. Xenograft volume was defined as V = L (length of tumor) × W2 (width of tumor)/2. All mice were sacrificed after 21 days, and then the tumor weight and volume were measured.
Immunohistochemical Staining of Ki67
Xenograft tumors were collected, and paraffin-embedded sections were prepared. For immunohistochemistry staining, tumor sections were deparaffinized in xylene and rehydrated in graded alcohols. Then, sections were treated with 3% hydrogen peroxide for 15 mins and incubated with 10% goat serum for 30 mins to decrease nonspecific binding. After washing three times in PBS, the sections were incubated with anti-Ki67 (1:500, ZSGB-BIO, China, ZM-0166) at 4°C overnight, followed by incubation with peroxidase labeled secondary antibody at room temperature for 30 mins. The nuclei were counterstained with hematoxylin for 5 min, dehydrated, and mounted for microscopic examination.
Western Blotting Analysis
The cells were washed with cold PBS, scraped in RIPA lysis buffer and lysed for 20 mins. Protein concentrations were determined using a BCA assay kit (Thermo Fisher, MA, USA). Electrophoresed samples were separated by SDS-PAGE, transferred onto PVDF membranes (Millipore, MA, USA) and blocked using 8% nonfat milk for 1 hr at room temperature. After washing in TBST, the membranes were incubated with primary antibodies at 4°C overnight. Then, the membranes were washed three times in TBST before the addition of HRP-labeled secondary antibodies. The signals were detected by a chemiluminescent HRP substrate (Millipore) on the Bio-Rad imaging system (Bio-Rad ChemiDoc MP). Primary antibodies against the following antigens were used: Cosmc (1:500; Santa Cruz Biotechnology), T-synthase (1:500; Santa Cruz Biotechnology), CD44 (1:1000; CST), Erk1/2 (1:1000; CST), p-Erk (1:1000; CST), JNK (1:1000; CST), p-JNK (1:1000; CST), p38 (1:1000; CST), p-p38(1:1000; CST), and GAPDH (1:1000; CST). HRP-labeled lectins Helix pomatia agglutinin (HPA, 2 μg/mL) and Arachis hypogaea agglutinin (PNA, 2 μg/mL) were purchased from Vector laboratories.
Reconstitution of CD44 in Cells
The CD44 lentivirus plasmid and control lentivirus vector were obtained from Hanbio Biotechnology (Beijing, China). The cells were seeded in 6-well plates with complete medium and transfected with the lentiviral particles with or without a CD44 gene insert using Lipofectamine 3000 (Invitrogen, CA, USA). After 48 h of transfection, the expression levels of CD44 protein were determined by Western blotting.
Statistical Analysis
All values were presented as the mean±standard deviation (SD). Student’s t-test (unpaired, 2-tailed) was used to analyze significant differences between groups and p<0.05 was considered significant.
Results
Cosmc Disruption Causes Tn Antigen Expression in Human Breast Cancer Cells
To study the biological roles of aberrant O-glycosylation, we first deleted the gene encoding Cosmc (Figure 1B), which is specifically required for the normal process of O-glycosylation,25 using the CRISPR/Cas9 system in human breast cancer cell lines MCF7 and T47D. Cosmc depletion was validated by Western blotting analysis. The results demonstrated that T-synthase was also absent in Cosmc-deficient cancer cells, which was in line with previous studies that the expression of T-synthase is dependent on Cosmc (Figure 1C).26 We then used flow cytometry to analyze aberrant O-glycosylation by measuring the expression of Tn antigen in cells. The results revealed that both Cosmc-deficient breast cancer cell lines showed abundant Tn antigen expression (Tn-positive), indicative of the occurrence of aberrant O-glycosylation. In contrast, the control cells transfected with mock plasmids had no detectable Tn antigen (Tn-negative) (Figure 1D). Additionally, HPA lectin blot analysis showed Tn antigen structures abundantly present in whole cell lysates of MCF7 and T47D Tn-positive cells (Figure 1E).
Cosmc Disruption-Mediated Aberrant O-glycosylation Inhibits Breast Cancer Cell Growth
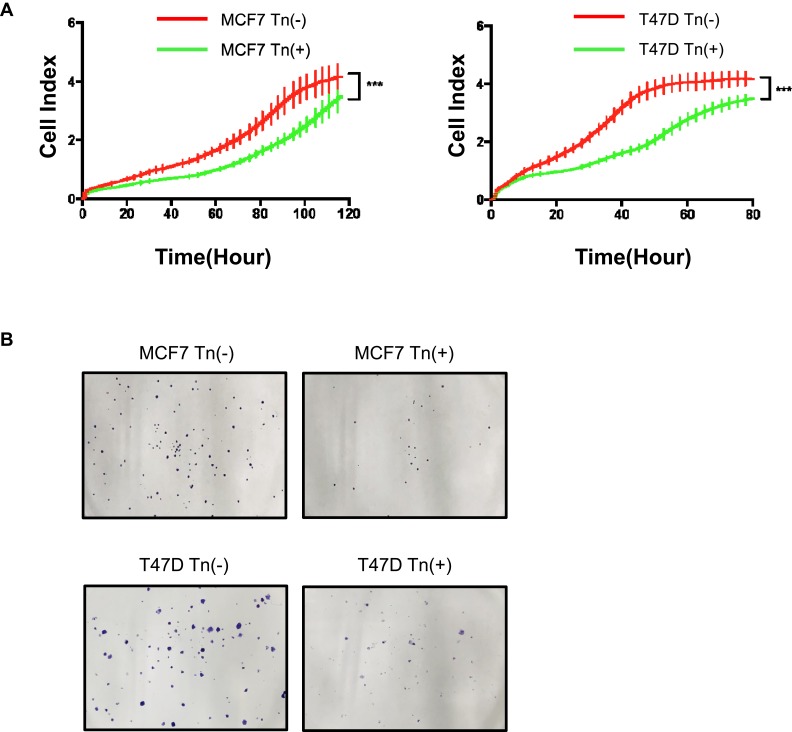
We next assessed the biological effects of aberrant O-glycosylation on breast cancer cell growth. We used RTCA to measure the cell proliferation ability and found that both Cosmc-deficient cancer cells expressing high levels of Tn antigen (Tn-positive) had decreased cell proliferation rates relative to the control cells that were Tn-negative (Figure 2A). Likewise, the results of the colony formation assay showed that both Tn-positive MCF7 and T47D cells formed fewer colonies in contrast to the control Tn-negative cells (Figure 2B).
Figure 2.
Cosmc disruption-mediated aberrant O-glycosylation inhibits cellular growth in vitro. (A) A RTCA assay was performed to determine the cell growth curves. MCF7 and T47D cell numbers were analyzed by RTCA at different time points up to 160 hrs. Data were presented as the means±SD from four replicated wells. (*** P < 0.001). Both Tn-positive cells had decreased cell proliferation rate relative to the control Tn-negative cells. (B) Colony formation assay was performed in MCF7 and T47D cells. The results showed that both Tn-positive MCF7 and T47D cells formed fewer colonies than the control Tn-negative cells.
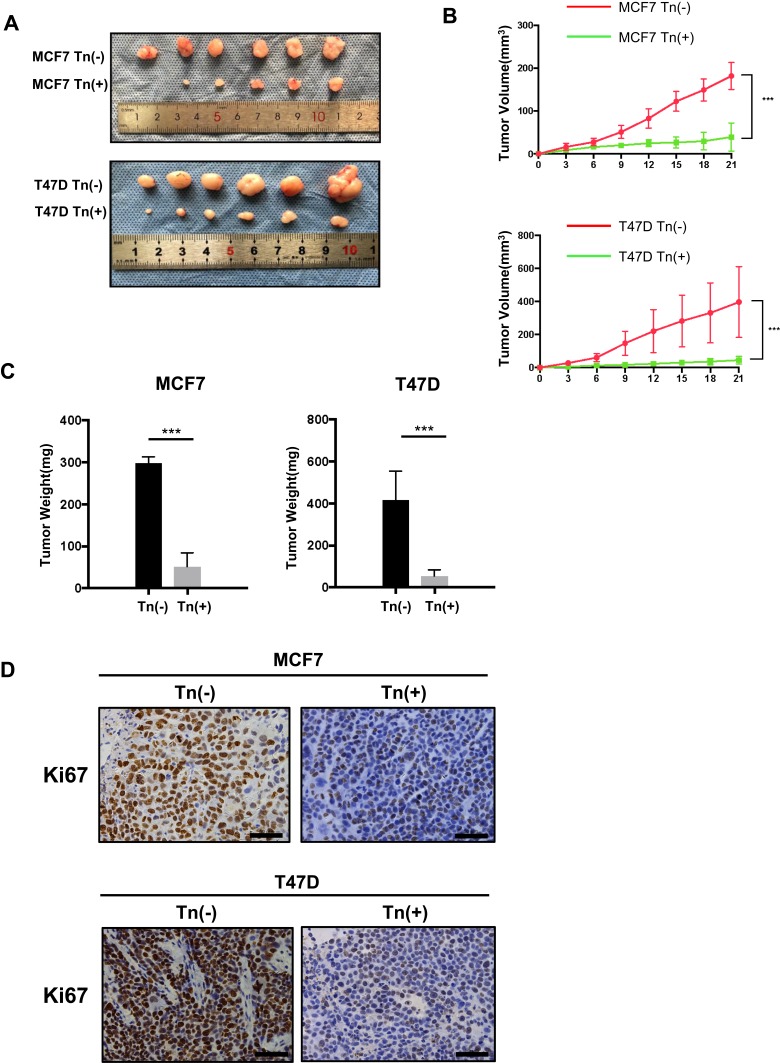
We further performed in vivo experiments to confirm these in vitro results. Here, we used both Tn-positive MCF7 and T47D cells along with the corresponding Tn-negative control cells to establish a subcutaneous tumor formation murine model. After 3 weeks of inoculation, we found that the size and weight of Tn-positive xenotransplants were much smaller than those in the control (Tn-negative) mice (Figure 3A–C). We further used immunohistochemical staining of Ki67 to examine the rate of breast cancer cell proliferation in mice. The results showed that the number of Ki67-positive cancer cells was markedly reduced in mice treated with Tn-positive cells compared with mice treated with Tn-negative cancer cells (Figure 3D). Altogether, these results suggested that Cosmc disruption-mediated aberrant O-glycosylation inhibited breast cancer cell growth both in vitro and in vivo.
Figure 3.
Cosmc disruption delays tumor growth in vivo. (A) Effects of Cosmc disruption on tumor growth and proliferation in a Balb/c nude mouse model. Tn-positive MCF7 and T47D cells as well as the corresponding Tn-negative cells were subcutaneously injected into mice. Six mice were used for each group. Images of the excised tumors were shown. (B) The volume of tumors was measured at different time points in 21 days (*** P < 0.001). (C) Tumor weight of Tn-positive MCF7 and T47D cell tumors in nude mice compared with control cell tumors (*** P < 0.001). (D) Cell proliferation of tumors was evaluated by immunohistochemical staining of Ki67, and representative images were shown. The results showed that the size and weight of Tn-positive xenotransplants were reduced compared with Tn-negative xenotransplants. Besides, the number of Ki67-positive cells were fewer in mice treated with Tn-positive cells than in mice treated with Tn-negative cells.
Cosmc Deletion Impairs CD44 Expression and the Associated MAPK Signaling Pathway
We next aimed to explore the molecular mechanisms underlying decreased breast cancer development in the absence of Cosmc. CD44 is a non-kinase transmembrane glycoprotein and is closely implicated in intracellular signaling involved in cell proliferation and motility.27 CD44 is also an abundantly O-glycosylated protein in the mammary gland. Altered expression of CD44 has been correlated with breast cancer development and poor prognosis.28 Thus far, it is uncertain whether aberrant O-glycosylation impacts the function and/or expression levels of CD44.
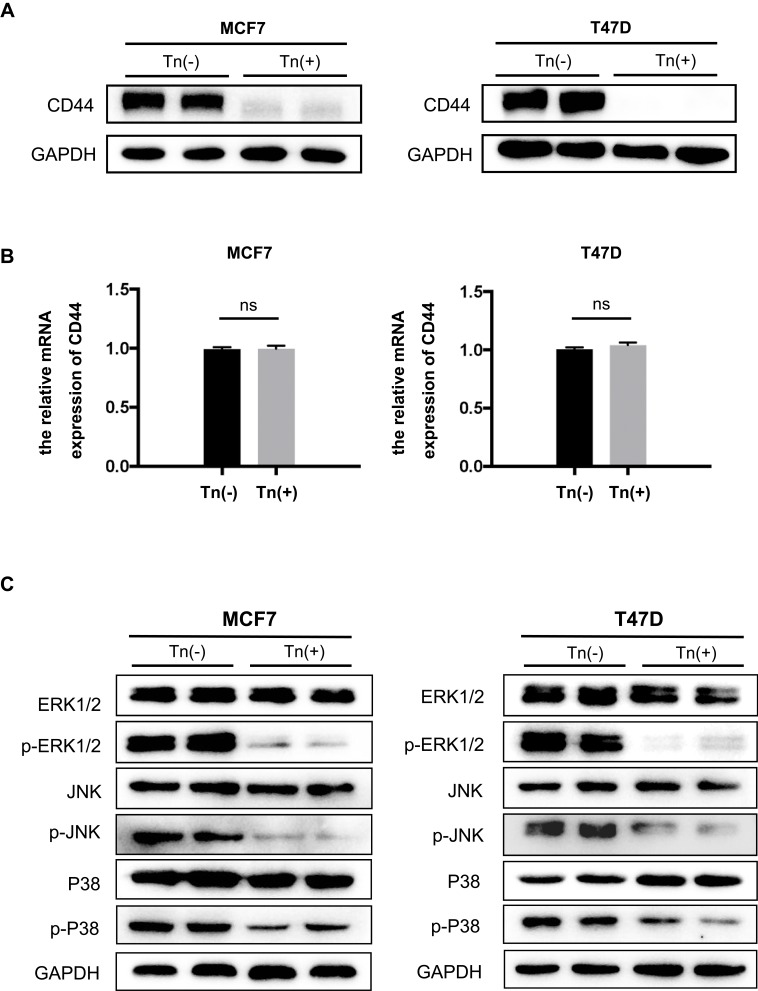
Here, we observed that in both Tn-positive breast cancer cell lines, CD44 expression was remarkably downregulated at the protein levels compared with that in Tn-negative cells (Figure 4A). Of note, there were no appreciable changes in the mRNA levels of CD44 between Tn-positive and negative cancer cells (Figure 4B). It is theoretically assumed that O-glycosylation is a posttranslational modification and may not directly affect substrate glycoproteins at the mRNA levels. Our results hence suggest that correct O-glycosylation is critical for the expression of CD44.
Figure 4.
CD44 expression and the associated MAPK signaling pathway were impaired in Tn-positive cancer cells. (A) The expression of CD44 at its protein levels was determined by Western blot analysis. It showed that Cosmc deficiency drastically reduced the expression of CD44 in Tn-positive MCF7 and T47D cells compared with the corresponding Tn-negative control cells. (B) Quantitative RT-PCR analysis showed that the mRNA levels of CD44 were not different between the Tn-positive and Tn-negative MCF7 and T47D cells. ns indicates no significant difference. (C) Cosmc disruption inhibited the phosphorylation of the MAPK signaling pathway. The expression levels of ERK1/2, p-ERK1/2, JNK, p-JNK, p38 and p-p38 were determined by Western blot analysis.
Accumulating evidence has demonstrated that CD44 interacts with a wide range of signaling pathways to mediate its roles.29 Of these, the MAPK pathway is primarily associated with cell growth in various human cancers.30 We conducted Western blot analysis to determine the phosphorylation of MAPK signaling molecules. The results demonstrated that the phosphorylation of ERK1/2, JNK and P-38 MAPK in both Tn-positive cancer cells was appreciably reduced when compared with that in the control cells (Tn-negative) (Figure 4C). These observations suggested that Cosmc disruption-induced aberrant O-glycosylation alters the expression of glycoproteins such as CD44, which further inhibits cell growth by impairing the MAPK signaling pathway.
Reconstitution of CD44 Restores Defective Cellular Growth
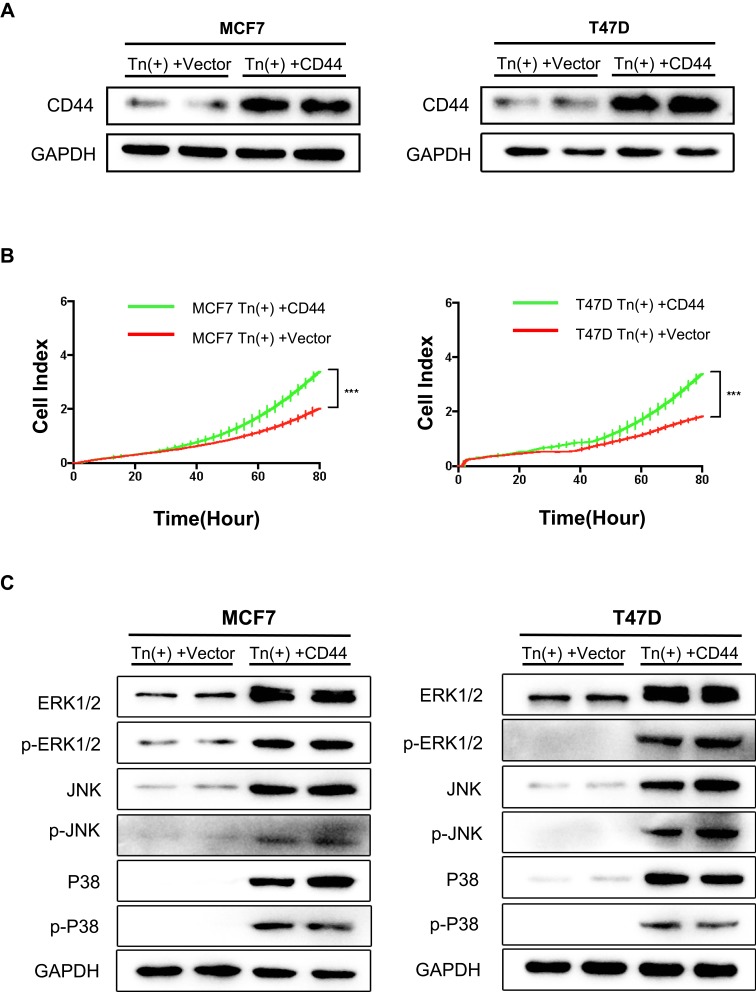
To further confirm the above findings, we transiently transfected human CD44 into these Tn-positive breast cancer cells caused by Cosmc depletion. Western blot confirmed the re-expression of CD44 in these cells (Figure 5A). As a consequence, we observed that CD44 reconstitution significantly enhanced the cell proliferation rate when compared with the control cells transfected with empty vectors (Figure 5B). Accordingly, CD44 re-expression in both Tn-positive cancer cells activated the MAPK signaling pathway, as evidenced by an enhancement of ERK1/2, JNK and P-38 phosphorylation (Figure 5C).
Figure 5.
Re-expression of CD44 in Tn-positive cells reversed the inhibitory effects. (A) Re-expression of CD44 in Tn-positive MCF7 cells and T47D cells was analyzed by Western blot. (B) RTCA was performed to measure the cell proliferation rate. Data were represented as the means±SD from four replicated wells. (*** P < 0.001). (C) Expression of ERK1/2, p-ERK1/2, JNK, p-JNK, p38 and p-p38 was determined by Western blot analysis in Tn-positive cells transfected with CD44.
Our findings indicated that loss of Cosmc led to aberrant O-glycosylation (characterized by Tn antigen expression), which further inhibited the expression of key glycoproteins such as CD44 and the associated signaling pathway related to cell proliferation in breast cancer development.
Discussion
O-glycosylation occurs on many transmembrane and secreted glycoproteins and is critical for protein expression and/or function.31,32 Aberrant O-glycosylation (characterized by the expression of Tn antigen) has been frequently observed in many human cancers, including breast cancer.5,33,34 There are a few proposed mechanisms that may influence the O-glycosylation process and lead to Tn antigen expression.35 Of these, dysfunction of T-synthase and/or its specific molecular chaperone-Cosmc constitute the prevailing causes.36,37 T-synthase and Cosmc deficiency equally impair the biosynthesis of O-glycans and induce Tn antigen expression.14 Aberrant O-glycosylation has been considered to play a causative role in tumor development and progression.38,39 However, some contradicting reports on the biological role of aberrant O-glycosylation in many cancers were recently published.40 Most studies support the notion that aberrant O-glycosylation promotes tumor development and metastasis in various human cancers. For example, it has been reported that disruption of either Cosmc or T-synthase equally induced aberrant O-glycosylation and significantly promoted malignant behaviors in pancreatic cancer.18,19 Liu et al also showed that Cosmc deficiency in human colorectal cancer cells markedly enhanced cellular growth and metastasis.41 Conversely, several recent studies unexpectedly found that loss of T-synthase, but not Cosmc, resulted in Tn antigen expression and subsequently delayed tumor development including breast, head and neck, and hepatocellular carcinoma.21–23 It appears that aberrant O-glycosylation may exert opposite effects in different types of cancers, which requires more investigations.
In the present study, we studied the functional impact of Cosmc deprivation-mediated aberrant O-glycosylation on breast cancer development through in vitro and in vivo investigations. In two human breast cancer cell lines (MCF7, T47D), we found that Cosmc loss-of-function and subsequent expression of Tn antigen reproducibly suppressed breast cancer cell growth. Accordingly, the results from in vivo experiments demonstrated that mice bearing Tn-positive cancer cells exhibited an obvious reduction in tumor size and weight in comparison with mice treated with Tn-negative cells. In addition, significantly decreased cell proliferation was confirmed by the staining of Ki67, a canonical marker for cell proliferation,42 in mice bearing Tn-positive cancer cells. Our findings are in accordance with a recent report showing that T-synthase deficiency-mediated aberrant O-glycosylation delayed mouse breast cancer development.21 This suggests that either Cosmc or T-synthase disruption-mediated aberrant O-glycosylation has similarly inhibitory effects on breast cancer development.
We next explored the mechanisms by which Cosmc disruption-mediated aberrant O-glycosylation inhibited breast cancer development. It is known that loss of Cosmc produces global effects on protein O-glycosylation,43 which may influence the expression and/or function of various O-glycoproteins that modulate mammary gland proliferation. Of particular interest, CD44 is a highly O-glycosylated glycoprotein that plays an essential role in multiple biological processes.27 CD44 also has a functional role in maintaining the stemness of cancer stem cells.44 Overexpression of CD44 is frequently observed in breast cancer and correlates with cancer progression and poor outcomes.45,46 CD44 has been reported to activate many signaling pathways including Rho GTPases, PI3K/AKT, and MAPK, to mediate its roles.29,47 Because MAPK is primarily associated with cellular proliferation, we determined the changes in CD44 and the associated MAPK signaling pathway in Cosmc-deficient breast cancer cells. We found that loss of Cosmc significantly impaired the expression of CD44 at its protein levels but not its mRNA levels, implying that O-glycosylation may affect posttranslational protein stability or degradation. Previous studies also revealed that O-glycans can protect glycoproteins from proteolytic attack.48 In addition, we observed that the MAPK signaling pathway, which is known to be one of the downstream pathways for CD44 and is essential for cell proliferation, was hampered in the absence of Cosmc. The reconstitution of CD44 could substantially reverse these changes, further confirming that CD44 and its related MAPK signaling may account for the aberrant O-glycosylation-mediated biological consequences in breast cancer. However, it is noticeable that breast cancer cells may have many other O-glycan substrates and identifying more key glycoproteins and related downstream signaling molecules for a comprehensive understanding of the role of aberrant O-glycosylation is highly warranted.
In conclusion, we revealed that Cosmc disruption can cause abnormal O-glycosylation in breast cancer cells, which further delays tumor cell proliferation in vitro and in vivo possibly through impairment of the CD44 glycoprotein. This study provides novel insights into the functional role of Cosmc, and further investigations are considered in the future.
Funding Statement
This work was supported by National Natural Science Foundation of China (81672338 to Tao Wen).
Abbreviations
Cosmc, Core 1 β3-galactosyltransferase Specific Molecular Chaperone; MAPK, Mitogen-activated protein kinase; RTCA, Real-Time cell analyzer; T-synthase, C1GalT1 or Core 1 β3-galactosyltransferase.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicting interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14(4–5):525–546. doi: 10.1002/pmic.v14.4-5 [DOI] [PubMed] [Google Scholar]
- 3.Tian E, Ten Hagen KG. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj J. 2009;26(3):325–334. doi: 10.1007/s10719-008-9162-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kudelka MR, Antonopoulos A, Wang Y, et al. Cellular O-Glycome Reporter/Amplification to explore O-glycans of living cells. Nat Methods. 2016;13(1):81–86. doi: 10.1038/nmeth.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 2006;7(6):599–604. doi: 10.1038/sj.embor.7400705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burchell JM, Beatson R, Graham R, Taylor-Papadimitriou J, Tajadura-Ortega V. O-linked mucin-type glycosylation in breast cancer. Biochem Soc Trans. 2018;46(4):779–788. doi: 10.1042/BST20170483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim Biophys Acta. 1999;1473(1):67–95. doi: 10.1016/S0304-4165(99)00170-1 [DOI] [PubMed] [Google Scholar]
- 8.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. doi: 10.1038/nrc3982 [DOI] [PubMed] [Google Scholar]
- 9.Xia L, McEver RP. Targeted disruption of the gene encoding core 1 beta1-3-galactosyltransferase (T-synthase) causes embryonic lethality and defective angiogenesis in mice. Methods Enzymol. 2006;416:314–331. [DOI] [PubMed] [Google Scholar]
- 10.Ju T, Xia B, Aryal RP, et al. A novel fluorescent assay for T-synthase activity. Glycobiology. 2011;21(3):352–362. doi: 10.1093/glycob/cwq168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Ju T, Ding X, et al. Cosmc is an essential chaperone for correct protein O-glycosylation. Proc Natl Acad Sci U S A. 2010;107(20):9228–9233. doi: 10.1073/pnas.0914004107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju T, Lanneau GS, Gautam T, et al. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008;68(6):1636–1646. doi: 10.1158/0008-5472.CAN-07-2345 [DOI] [PubMed] [Google Scholar]
- 13.Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci U S A. 2002;99(26):16613–16618. doi: 10.1073/pnas.262438199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju T, Otto VI, Cummings RD. The Tn antigen-structural simplicity and biological complexity. Angew Chem Int Ed Engl. 2011;50(8):1770–1791. doi: 10.1002/anie.v50.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju T, Wang Y, Aryal RP, et al. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteomics Clin Appl. 2013;7(9–10):618–631. doi: 10.1002/prca.201300024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konska G, Guerry M, Caldefie-Chezet F, De Latour M, Guillot J. Study of the expression of Tn antigen in different types of human breast cancer cells using VVA-B4 lectin. Oncol Rep. 2006;15(2):305–310. doi: 10.3892/or.15.2.305 [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y, Liu Z, Xu F, et al. Aberrant O-glycosylation contributes to tumorigenesis in human colorectal cancer. J Cell Mol Med. 2018;22(10):4875–4885. doi: 10.1111/jcmm.13752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann BT, Schluter L, Lange P, et al. COSMC knockdown mediated aberrant O-glycosylation promotes oncogenic properties in pancreatic cancer. Mol Cancer. 2015;14:109. doi: 10.1186/s12943-015-0386-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chugh S, Barkeer S, Rachagani S, et al. Disruption of C1galt1 Gene Promotes Development and Metastasis of Pancreatic Adenocarcinomas in Mice. Gastroenterology. 2018;155(5):1608–1624. doi: 10.1053/j.gastro.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua D, Shen L, Xu L, et al. Polypeptide N-acetylgalactosaminyltransferase 2 regulates cellular metastasis-associated behavior in gastric cancer. Int J Mol Med. 2012;30(6):1267–1274. doi: 10.3892/ijmm.2012.1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song K, Herzog BH, Fu J, et al. Loss of Core 1-derived O-Glycans Decreases Breast Cancer Development in Mice. J Biol Chem. 2015;290(33):20159–20166. doi: 10.1074/jbc.M115.654483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin MC, Chien PH, Wu HY, et al. C1GALT1 predicts poor prognosis and is a potential therapeutic target in head and neck cancer. Oncogene. 2018;37(43):5780–5793. doi: 10.1038/s41388-018-0375-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YM, Liu CH, Huang MJ, et al. C1GALT1 enhances proliferation of hepatocellular carcinoma cells via modulating MET glycosylation and dimerization. Cancer Res. 2013;73(17):5580–5590. doi: 10.1158/0008-5472.CAN-13-0869 [DOI] [PubMed] [Google Scholar]
- 24.Avichezer D, Springer GF, Schechter B, Arnon R. Immunoreactivities of polyclonal and monoclonal anti-T and anti-Tn antibodies with human carcinoma cells, grown in vitro and in a xenograft model. Int J Cancer. 1997;72(1):119–127. doi: 10.1002/(ISSN)1097-0215 [DOI] [PubMed] [Google Scholar]
- 25.Ju T, Aryal RP, Stowell CJ, Cummings RD. Regulation of protein O-glycosylation by the endoplasmic reticulum-localized molecular chaperone Cosmc. J Cell Biol. 2008;182(3):531–542. doi: 10.1083/jcb.200711151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aryal RP, Ju T, Cummings RD. The endoplasmic reticulum chaperone Cosmc directly promotes in vitro folding of T-synthase. J Biol Chem. 2010;285(4):2456–2462. doi: 10.1074/jbc.M109.065169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Zhao S, Karnad A, Freeman JW. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol. 2018;11(1):64. doi: 10.1186/s13045-018-0605-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senbanjo LT, Chellaiah MA. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front Cell Dev Biol. 2017;5:18. doi: 10.3389/fcell.2017.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouhtit A, Rizeq B, Saleh HA, Rahman MM, Zayed H. Novel CD44-downstream signaling pathways mediating breast tumor invasion. Int J Biol Sci. 2018;14(13):1782–1790. doi: 10.7150/ijbs.23586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35(6):600–604. doi: 10.3109/10799893.2015.1030412 [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann M, Marx K, Reichl U, Wuhrer M, Rapp E. Site-specific O-Glycosylation Analysis of Human Blood Plasma Proteins. Mol Cell Proteomics. 2016;15(2):624–641. doi: 10.1074/mcp.M115.053546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Liu B, Zeng R, Cai Y, Li Y. Predicting O-glycosylation sites in mammalian proteins by using SVMs. Comput Biol Chem. 2006;30(3):203–208. doi: 10.1016/j.compbiolchem.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 33.Deng B, Tarhan YE, Ueda K, et al. Critical Role of Estrogen Receptor Alpha O-Glycosylation by N-Acetylgalactosaminyltransferase 6 (GALNT6) in Its Nuclear Localization in Breast Cancer Cells. Neoplasia. 2018;20(10):1038–1044. doi: 10.1016/j.neo.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cazet A, Julien S, Bobowski M, Burchell J, Delannoy P. Tumour-associated carbohydrate antigens in breast cancer. Breast Cancer Res. 2010;12(3):204. doi: 10.1186/bcr2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bard F, Chia J. Cracking the Glycome Encoder: signaling, Trafficking, and Glycosylation. Trends Cell Biol. 2016;26(5):379–388. doi: 10.1016/j.tcb.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 36.Ju T, Aryal RP, Kudelka MR, Wang Y, Cummings RD. The Cosmc connection to the Tn antigen in cancer. Cancer Biomark. 2014;14(1):63–81. doi: 10.3233/CBM-130375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mi R, Song L, Wang Y, et al. Epigenetic silencing of the chaperone Cosmc in human leukocytes expressing tn antigen. J Biol Chem. 2012;287(49):41523–41533. doi: 10.1074/jbc.M112.371989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radhakrishnan P, Dabelsteen S, Madsen FB, et al. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc Natl Acad Sci U S A. 2014;111(39):E4066–E4075. doi: 10.1073/pnas.1406619111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J, Che MI, Lin NY, et al. The molecular chaperone Cosmc enhances malignant behaviors of colon cancer cells via activation of Akt and ERK. Mol Carcinog. 2014;53(Suppl 1):E62–E71. doi: 10.1002/mc.22011 [DOI] [PubMed] [Google Scholar]
- 40.Chou CH, Huang MJ, Chen CH, et al. Up-regulation of C1GALT1 promotes breast cancer cell growth through MUC1-C signaling pathway. Oncotarget. 2015;6(8):6123–6135. doi: 10.18632/oncotarget.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Liu J, Dong X, et al. Tn antigen promotes human colorectal cancer metastasis via H-Ras mediated epithelial-mesenchymal transition activation. J Cell Mol Med. 2019;23(3):2083–2092. doi: 10.1111/jcmm.2019.23.issue-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11(2):174–183. doi: 10.1016/S1470-2045(09)70262-1 [DOI] [PubMed] [Google Scholar]
- 43.Campos D, Freitas D, Gomes J, et al. Probing the O-glycoproteome of gastric cancer cell lines for biomarker discovery. Mol Cell Proteomics. 2015;14(6):1616–1629. doi: 10.1074/mcp.M114.046862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mallard BW, Tiralongo J. Cancer stem cell marker glycosylation: nature, function and significance. Glycoconj J. 2017;34(4):441–452. doi: 10.1007/s10719-017-9780-9 [DOI] [PubMed] [Google Scholar]
- 45.Menon MM, Thakur R, Simha MR, Kurkure AP, Kenkare UW, Doctor VM. Expression of cell surface glycoprotein CD44 and integrins in breast cancers among Indian women. Tumori J. 1998;84(5):589–594. doi: 10.1177/030089169808400515 [DOI] [PubMed] [Google Scholar]
- 46.Wang JS, Wang HJ, Qian HL. Biological effects of radiation on cancer cells. Mil Med Res. 2018;5(1):20. doi: 10.1186/s40779-018-0167-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bourguignon LY. CD44-mediated oncogenic signaling and cytoskeleton activation during mammary tumor progression. J Mammary Gland Biol Neoplasia. 2001;6(3):287–297. doi: 10.1023/A:1011371523994 [DOI] [PubMed] [Google Scholar]
- 48.Reddy P, Caras I, Krieger M. Effects of O-linked glycosylation on the cell surface expression and stability of decay-accelerating factor, a glycophospholipid-anchored membrane protein. J Biol Chem. 1989;264(29):17329–17336. [PubMed] [Google Scholar]