Abstract
Background
The complex preparation procedures and severe toxicities are two major obstacles facing the wide use of chimeric antigen receptor-modified T (CAR-T) cells in clinical cancer immunotherapy. The nanotechnology-based T cell temporary CAR modification may be a potential approach to solve these problems and make the CAR-T cell-based tumor therapy feasible and broadly applicable.
Methods
A series of plasmid DNA-loaded self-assembled nanoparticles (pDNA@SNPsx/y) prepared from adamantane-grafted polyamidoamine (Ad-PAMAM) dendrimers of different generations (G1 or G5) and cyclodextrin-grafted branched polyethylenimine (CD-PEI) of different molecular weights (800, 2000, or 25,000 Da) were characterized and evaluated. The detailed physicochemical properties, cellular interaction, and cytotoxicity of selected pDNA@SNPG1/800 were systematically investigated. Thereafter, the epidermal growth factor receptor variant III (EGFRvIII) CAR-expression plasmid vector (pEGFRvIII-CAR) was constructed and encapsulated into SNPG1/800. The resulting pEGFRvIII-CAR@SNPG1/800 was used for Jurkat cell transient transfection, and the EGFRvIII-CAR expressed in transfected cells was measured by flow cytometry and Western blot. Finally, the response of EGFRvIII CAR-positive Jurkat T cell to target tumor cell was evaluated.
Results
The pDNA@SNPG1/800 showed the highest efficacy in Jurkat cell gene transfection and exhibited low cytotoxicity. pEGFRvIII-CAR@SNPG1/800 can efficiently deliver pEGFRvIII-CAR into Jurkat T cells, thereby resulting in transient EGFRvIII-CAR expression in transfected cells. EGFRvIII-CAR that is present on the cell membrane enabled Jurkat T cells to recognize and bind specifically with EGFRvIII-positive tumor cells.
Conclusion
These results indicated that pEGFRvIII-CAR@SNPG1/800 can effectively achieve T-cell transient CAR modification, thereby demonstrating considerable potential in CAR-T cancer therapy.
Keywords: self-assembled nanoparticle, PAMAM dendrimer, polyethylenimine, epidermal growth factor receptor variant III, chimeric antigen receptor, T lymphocyte
Introduction
Chimeric antigen receptors (CARs) expressed on the T-cell surface can redirect them against targets on malignant cells, thereby enabling their destruction by exerting T-cell effector functions.1–4 The adoptive transfer of CAR-modified T (CAR-T) cells has emerged as a promising cancer treatment method.5–7 This strategy has demonstrated its clinical efficacy in hematologic malignancies and shown evident responses when targeting solid tumors.8,9
The clinical-scale manufacturing of CAR-T lymphocytes requires harsh and complex processes, including T lymphocyte isolation, viral vector-based genetic modification, and selective expansion of redirected T cells before infusing them back into the patient.10,11 The complex procedures and high costs involved in producing genetically modified T lymphocytes remain the major obstacles in implementing CAR-T in cancer treatment.12 CAR-T-based therapy can also elicit expected and unexpected toxicities, including cytokine release syndrome, neurologic toxicity, “on target/off tumor” recognition, and anaphylaxis.13 Thus, abrogating toxicity has also become a critical step in the successful application of this emerging technology with considerable potential. Various strategies, such as the use of true tumor targeting, combinational targeting or “tumor-sensing” CAR-T to enhance tumor specificity, site-specific insertion of CAR, inducible suicide or apoptosis as a safety switch for adoptive cell therapy, have been used to abrogate or reduce the toxicity of CAR-T-based tumor therapy.13,14 However, the effectiveness of these strategies is potentially limited. As a result, effective strategies still need to achieve ideal therapeutic efficiency without serious accompanying side effects.
A temporary approach toward CAR expression and T-cell activation may be a potential strategy to solve the problems encountered in CAR-T-based cancer immunotherapy.15 June et al constructed transient CAR-redirected T cells by using the in vitro electroporation of mRNA encoding a CD19- or Mesothelin-specific CAR, and the results showed that the adoptive transfer of these CAR-T cells is feasible, effective, and safe in xenograft animal model and patients.16,17 Although the in vitro electroporation of CAR mRNA for transient CAR expression has produced impressive results, the dismal cell viability post electroporation, instability of mRNA molecules, and the in vitro methods required to generate large numbers of tumor-specific T cells all limit the widespread application of in vitro CAR-mRNA electroporation-based cancer immunotherapy.
Nonviral vector-mediated gene delivery is relatively safer and more easy to use than in vitro electroporation. Among the potential nonviral vectors, lipid nanoparticle is one of the most common vectors used for in vitro gene delivery. Lipid-mediated transfection is extremely effective in adhesion cells, while their transfection rates in the hard-to-transfect lymphoma/leukemia cells are unsatisfactory and generally accompanied by cytotoxicity and potent interferon response.18 Chemical cationic polymers, such as linear or branched polyethylenimine (PEI), linear poly(amido-amine), polyamidoamine (PAMAM) dendrimers, and poly(β-amino ester) (PBAE), have demonstrated considerable potential for gene transfection in adhesion and suspension cells.18 Stephan et al successfully delivered leukemia-specific CAR genes into host T cells in situ by using functionalized polymeric nanocarriers that were prepared from PBAE 447 polymer-microtubule-associated nuclear localization peptide, CAR-expression plasmid DNA, polyglutamic acid, and lymphocyte-targeting ligand. PBAE nanoparticle-reprogrammed T cells can destroy tumor cells and are sufficient to induce tumor regression with efficacies that are similar to the conventional infusions of T cells transduced ex vivo with CAR-encoding viral vectors.19 These results indicated that the cationic-polymer vector-based transient T cell CAR modification may provide a feasible and broadly applicable cancer treatment in various settings due to its excellent advantages, such as simple procedures and predictable low toxicity. Therefore, new gene delivery vectors with considerable potential in gene loading and T cell genetic modification need to be developed.
PEI is the gold standard approach for cell transfection, and some PEI derivatives are currently being investigated in clinical trials.20,21 High-molecular-weight PEI or high-generation PAMAM dendrimers have high gene loading and transfection efficiency but are generally accompanied by serious cytotoxicity and cargo release difficulty.22–25 By contrast, low-molecular-weight (LMW) PEI or low-generation PAMAM dendrimers have low transfection efficiency and cytotoxicity.26–28 In our previous study, supramolecular self-assemble nanoparticles (SNPs) that were prepared from cyclodextrin-grafted branched PEI25000 (CD-PEI25000), adamantane-grafted PAMAMG1 dendrimer (Ad-PAMAMG1), and Ad-grafted poly(ethylene glycol) (Ad-PEG) were used for gene loading, and high-efficiency gene delivery was realized in pDNA@SNPs combined with Ad-grafted silicon nanowire substrates (Ad-SiNWS) gene transfer in vitro.29 Meanwhile, the application potential and ideal formulation of pDNA@SNPs in T-cell gene delivery and transient CAR modification are unclear.
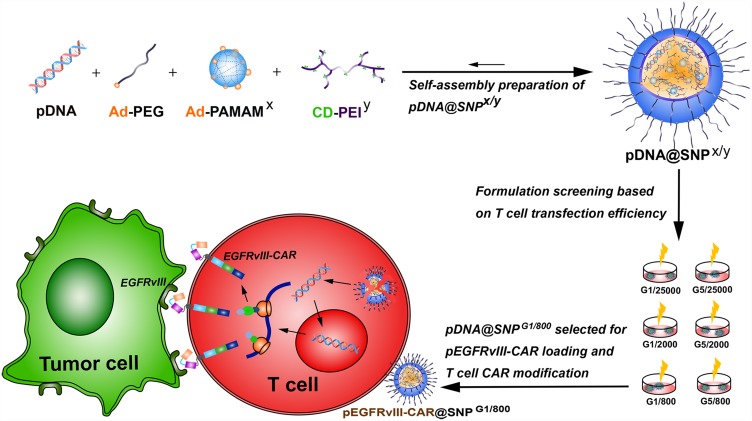
This study was conducted to evaluate the application potential of pDNA@SNPs in T lymphocyte CAR modification and explore promising formulation for T cell transfection (Figure 1). Molecular blocks, including CD-PEI800, CD-PEI2000, CD-PEI25000, Ad-PAMAMG1, Ad-PAMAMG5, and Ad-PEG5000, were synthesized for plasmid DNA-loaded self-assembled nanoparticle (pDNA@SNPx/y; x: PAMAM dendrimer generation, y: PEI molecular weight) preparation. The formulation pDNA@SNPG1/800 was screened out from pDNA@SNPx/y series on the basis of gene transfection efficiency evaluation, and its detailed physicochemical properties, cellular interaction, and cytotoxicity were systematically investigated. Therefore, the epidermal growth factor receptor variant III (EGFRvIII) CAR-expression plasmid vector (pEGFRvIII-CAR) for the intracellular expression of third-generation CAR targeting EGFR variant III was constructed and encapsulated into pDNA@SNPG1/800. The resulting pEGFRvIII-CAR@SNPG1/800 was used for Jurkat cell transient transfection, and the EGFRvIII-CAR expressed in transfected cells was measured by flow cytometry and Western blot. Finally, the response of EGFRvIII CAR-positive Jurkat T cell to target tumor cell was evaluated.
Figure 1.
Schematic illustration of the preparation and formulation screening of pDNA@SNPs for pEGFRvIII-CAR loading and T cell CAR modification.
Abbreviations: pDNA, plasmid DNA; Ad-PEG, adamantane-grafted poly(ethylene glycol); Ad-PAMAMx, adamantane-grafted polyamidoamine dendrimer (x: PAMAM dendrimer generation); CD-PEIy, cyclodextrin-grafted branched polyethylenimine (y: PEI molecular weight); SNPs, self-assembled nanoparticles; EGFRvIII, epidermal growth factor receptor variant III; CAR, chimeric antigen receptor; pEGFRvIII-CAR, epidermal growth factor receptor variant III-chimeric antigen receptor expression plasmid.
Materials and Methods
Materials and Chemicals
Branched PEI (bPEI, MW of 800, and 2000 Da) was obtained from Sigma-Aldrich (St. Louis, MO). bPEI (MW of 25 kD) was obtained from Polysciences Inc (Washington, PA). First- and fifth-generation PAMAM dendrimers in MeOH were obtained from Dendritic Nanotechnologies, Inc. (Mount Pleasant, MI). NHS-PEG5000 was obtained from Nanocs, Inc. (New York, NY). 1-Adamantanamine hydrochloride and β-cyclodextrin were obtained from TCI America (San Francisco, CA). LipofectamineTM 2000 was obtained from Invitrogen (Carlsbad, CA). Plasmid gWIZ-GFP encoding green fluorescent protein (GFP) was obtained from Gene Therapy Systems, Inc. (San Diego, CA). pGL3-control vector and Luciferase assay kits were obtained from Promega Corporation (Madison, USA). HuH7 hepatocarcinoma cell line and Jurkat cell line were purchased from the Institute of Biochemistry and Cell Biology (SIBS, CAS, China) and routinely maintained in RMPI1640 containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a humid cell incubator. YOYO-1 dye was obtained from Thermo Fisher Scientific (Madison, USA). Cy5.5 NHS ester was obtained from ApxBio (Glendale, USA). LysoTracker Red, BeyoECL Star, Carboxyfluorescein diacetate, carboxyfluorescein succinimidyl ester (CFSE), and 1,1′-Dioetadeeyl-3,3,3′,3′-tetramethylindocarboeyanine perchlorate (DiI) were obtained from Beyotime Biotechnology Co., Ltd. (Shanghai, China). Anti-CD3ζ, anti-GAPDH, and mouse IgG H&L (horseradish peroxidase, HRP) were obtained from Santa Cruz Biotechnology, Inc. (Dallas, USA). Other reagents and solvents were obtained from Sigma–Aldrich (St. Louis, MO) or Sinopharm Chemical Reagent Co., Ltd (Shanghai, China) and used as received without further purification, unless otherwise noted. 6-Mono-tosyl-β-CD (6-OTs-β-CD) was prepared by following the method reported in the literature.30 Ad-PAMAMx, CD-PEIy, and Ad-PEG were prepared according to the method previously reported by Wang et al31.
Construction of EGFRvIII-CAR Expression Plasmid
pcDNA3.1(+) plasmid containing anti-EGFRvIII scFv-CD28-4-1BB-CD3ζ expression cassette was constructed and named as pEGFRvIII-CAR. The products were transformed into DH5a competent cells, and a single clone was selected, amplified, and sequenced.
pDNA@SNPx/y Preparation (X: PAMAM Generation, Y: PEI Molecular Weight)
pDNA@SNPs were prepared by applying a self-assembly procedure.32 Briefly, 2 μL of DMSO solution containing Ad-PAMAMG1 or Ad-PAMAMG5 (2.1 mg/mL) was added into 600 μL of ddH2O mixture with 6 µg plasmid DNA and 1.69 nmol Ad-PEG. The resulting mixture was incubated at room temperature for 2 min. Then, 1.5 µL of CD-PEI800, CD-PEI2000, or CD-PEI25000 (10 mg/mL) was added slowly under vigorous stirring. The mixture was kept at room temperature for 20 min, thereby yielding pDNA@SNPsx/y by using a self-assembly procedure.
Gel Electrophoresis Assay
The pDNA loading capacity of SNPsx/y was analyzed by gel electrophoresis. Briefly, 1 kb DNA ladder, naked pGL3, and pGL3@SNPsx/y containing 0.3 µg pGL3 were used for electrophoresis in 1% agarose gel 100 V for 30 min. Plasmid DNA was visualized under UV illumination by staining the gels with gel red at room temperature.
Hydrodynamic Size and Zeta Potential Measurements
The hydrodynamic sizes and zeta potentials of pDNA@SNPsx/y were determined via dynamic light scattering method (DLS) by using Malvern Nano-ZS90 (Malvern Instruments, Malvern Worcs, U.K.). To test the stability of pDNA@SNPG1/800, we determined the particle size distribution of pGL3@SNPG1/800 dispersions prepared with PBS by using NanoSight NS300 system (Malvern Instruments, UK) at different time points (0, 2, 4, 8, 12, and 24 h).
Transmission Electron Microscopy (TEM) Analysis
A drop of aqueous solution containing pDNA@SNPsG1/800 was placed and dried on the C-coated copper grid. The morphology of pDNA@SNPsG1/800 was analyzed by a Tecnai G2 Spirit BioTwin TEM (FEI, Netherlands).
pGL3 Transfection and Assay
Jurkat cells in 48-well plates were incubated with pGL3@SNPsx/y in an equivalent pGL3 dosage of 2 μg for 4 h and then replenished with fresh medium comprising 10% FBS and cultured for another 24 h. Luciferase gene expression was quantified by using a commercial kit and photon counting. All experiments were run in triplicate, and data were expressed as relative light units.
gWIZ-GFP Transfection and Assay
Jurkat cells in 24-well plates were incubated with pGFP/PEI800 complex (N/P 20), pGFP/LipofectamineTM 2000 complex (prepared according to the manufacturer’s instruction), or pGFP@SNPsG1/800 at an equivalent gWIZ-GFP dosage of 2 μg for 4 h and then replenished with fresh medium with 10% FBS and cultured for another 48 h. The GFP expression in transfected cells was evaluated using fluorescent microscopy.
Cellular Binding and Endosome Escape Evaluation
Dual fluorescence-labeled pGL3@SNPsG1/800 were prepared from Cy5.5-labeled CD-PEI800 and YOYO-1-labeled plasmid DNA. To analyze their cellular binding properties, we incubated the Jurkat cells with pGL3@SNPsG1/800 at 37°C for 15 or 30 min, followed by washing and fixing. The cells without pGL3@SNPsG1/800 treatment served as a control group. Then, the cells were incubated with DAPI at 37°C for 7 min. After three washes with PBS, the cells were dispersed on slides and then mounted with coverslips with antifade mounting medium. To analyze the endosome escape of pGL3@SNPsG1/800, Jurkat cells were incubated with YOYO-1-labeled pGL3@SNPsG1/800 at 37°C for 4 h. After incubation, the cells were washed three times with PBS (pH of 7.4) and then incubated with Lysotracker Red (DND 99) at 37°C for 1 h, followed with DAPI dyeing and mounting, as mentioned above. The resulting slides were visualized using a Leica TCS SP8 confocal microscope (Leica, Germany).
Cytotoxicity Assay
The cytotoxicity of pDNA@SNPsG1/800 was measured using the CCK-8 assay. Briefly, Jurkat cells in 96-well plate were incubated with fresh cultured medium containing 10% FBS (negative control group), pGL3@SNPsG1/800, or PEI800/pGL3 complexes (N/P 20) at designated plasmid DNA dosage for 4 h and then replenished with fresh medium. After 24 h, the treated cells were subjected to CCK-8 assay following the manufacturer’s protocol. All experiments were performed in triplicate.
Flow Cytometry (FACS) and Western Blot Analysis of EGFRvIII-CAR Expressed in pEGFRvIII-CAR@SNPG1/800-Transfected Jurkat Cells
EGFRvIII-CAR expression plasmid-loaded pEGFRvIII-CAR@SNPsG1/800 were prepared by the method mentioned above. Jurkat cells were incubated with pEGFRvIII-CAR@SNPsG1/800 at 37°C for 4 h, and then the medium was replaced with fresh RPMI1640 containing 10% FBS. After 24 h, the pEGFRvIII-CAR-transfected Jurkat cells were harvested and incubated with FITC-protein-L (2.5 μg/mL) at 4°C for 30 min. After being washed with ice-cold wash buffer three times, the cells were analyzed using a BD FACScan flow cytometer (BD Biosystems).
For the Western blot analysis of the EGFRvIII-CAR expressed in pEGFRvIII-CAR@SNPG1/800-transfected Jurkat cells, the Jurkat cells were harvested 24 h after transfection, followed by washing and lysis procedures. After centrifugation, the supernatant was collected, denatured under reducing condition, and electrophoresed by 12% SDS-PAGE. Then, the sample was transferred to PVDF membrane (Millipore) and immunoblotted with antihuman CD3ζ and anti-GAPDH antibody. The blot was incubated with HRP-conjugated goat anti-mouse IgG H&L and detected by ECL Western blotting analysis system.
Confocal Microscopy of Interaction of EGFRvIII-CAR-Redirected Jurkat Cells with EGFRvIII-Positive HuH7 Cells
Human hepatocarcinoma HuH7 cells that were engineered to stably overexpress EGFRvIII (Huh-7EGFRvIII+) were used to test the response of EGFRvIII-CAR-redirected Jurkat cells to EGFRvIII-expressing target cells. The adherent Huh7EGFRvIII+ were labeled with CFSE and then incubated with DiI-labeled pEGFRvIII-CAR-transfected Jurkat cells at 37°C for 1 h. After washing, cells were mounted in antifade mounting medium and visualized using a Leica TCS SP8 confocal microscope (Leica, Germany).
Statistical Analysis
All experiments were carried out at least in triplicate. Experimental data were shown as the mean ± standard deviation, unless otherwise indicated. Statistical differences were evaluated by analysis of variance (ANOVA), and p<0.05 was considered significant.
Results and Discussion
pDNA@SNPx/y Preparation and Evaluation
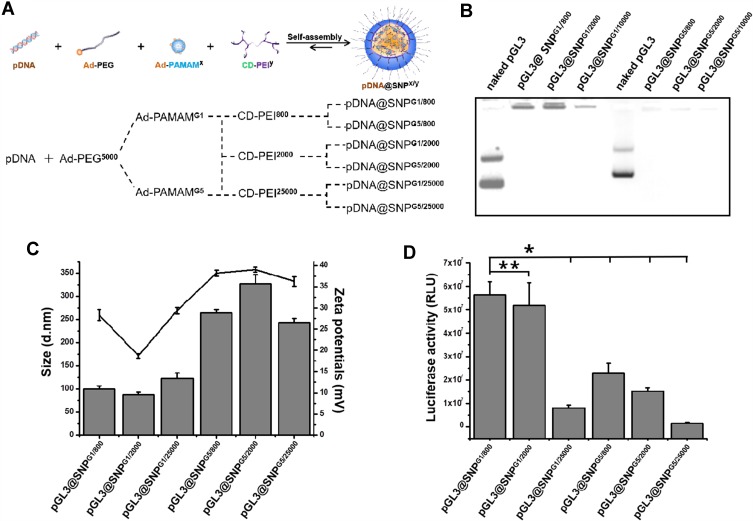
Supramolecular SNPs prepared from CD-PEI25000, Ad-PAMAMG1, and Ad-PEG2000 on the basis of the molecular recognition of Ad and CD motifs can effectively encapsulate plasmid DNA.29,32,33 To prepare pDNA@SNPsx/y with different CD-PEI molecular weights and different Ad-PAMAM generations, we synthesized, purified, and further confirmed the molecular building blocks, such as CD-PEI800, CD-PEI2000, CD-PEI25000, Ad-PAMAMG1, and Ad-PAMAMG5, by MALDI-TOF-MS and 1 H nuclear magnetic resonance spectroscopy (data not supplied). pDNA@SNPsx/y were prepared as previously reported (Figure 2A),29,32,33 and their physicochemical properties were evaluated. The agarose gel retardation assay indicated the complete retardation of the plasmid DNA formulated with all SNPs (Figure 2B). This result suggested the successful incorporation of plasmid DNA molecules into the pDNA@SNPsx/y. The mean hydrodynamic diameter of pDNA@SNPsx/y was approximately 100–300 nm, and the surface charge approximately ranged from +20 mV to 40 mV (Figure 2C).
Figure 2.
pDNA@SNPx/y preparation and evaluation. (A). Scheme of pDNA@SNPx/y preparation. (B). Gel retardation assay of pGL3@SNPsx/y. (C). Hydrodynamic size (column diagram) and zeta potentials (broken line diagram) of pGL3@SNPsx/y obtained from DLS measurement. (D). Activity of luciferase expressed by Jurkat cells transfected with pGL3@SNPsx/y (*P<0.05, **P>0.05).
Abbreviations: pDNA, plasmid DNA; Ad-PEG, adamantane-grafted poly(ethylene glycol) (molecular weight: 5000); Ad-PAMAMx, adamantane-grafted polyamidoamine dendrimer (x: PAMAM dendrimer generation); CD-PEIy, cyclodextrin-grafted branched polyethylenimine (y: PEI molecular weight); SNPs, self-assembled nanoparticles; DLS, dynamic light scattering.
We further compared the transfection efficiency of pDNA@SNPsx/y in Jurkat T cell by using pGL3 luciferase reporter vector. The results presented in Figure 2D showed that the transfection efficiency was relatively different among the six groups, and the highest luciferase expression level was found in the pGL3@SNPG1/800 group. The high PEI molecular weight or PAMAM generation used in pDNA@SNPsx/y decreased the transfection efficiency of pDNA@SNPsx/y. The activity of luciferase expressed in the pGL3@SNPG1/800 group was not significantly different from that in the pGL3@SNPG1/2000 group (P>0.05) and approximately 7-, 2.4-, 3.7-, and 40-fold higher than those of the pGL3@SNPG1/25000, pGL3@SNPG5/800, pGL3@SNPG5/2000, and pGL3@SNPG5/25000 groups (P<0.05), respectively. Compared with poor transfection efficiency of LMW PEI and low-generation PAMAM dendrimers,27,28,34 the pDNA@SNPG1/800 prepared from CD-PEI800 and Ad-PAMAMG1 had the highest transfection efficiency in addition to the effective encapsulation of plasmid DNA. This enhanced gene encapsulation efficiency of SNPs may contribute to the cationic hydrogel generated from CD-PEI800 and Ad-PAMAMG1 on the basis of the molecular recognition of CD and Ad.32 Similar to the enhanced transfection efficiency and low cytotoxicity of degradable PEI or PAMAM due to the cleavage of the disulfide linkages in the polymer,34–38 the noncovalent crosslink of CD-PEI800 and Ad-PAMAMG1 may also allow the efficient release of plasmid DNA from pDNA@SNPsG1/800 and then increase the transgene expression in transfected cells.
pDNA@SNPG1/800 Preparation and Characterization
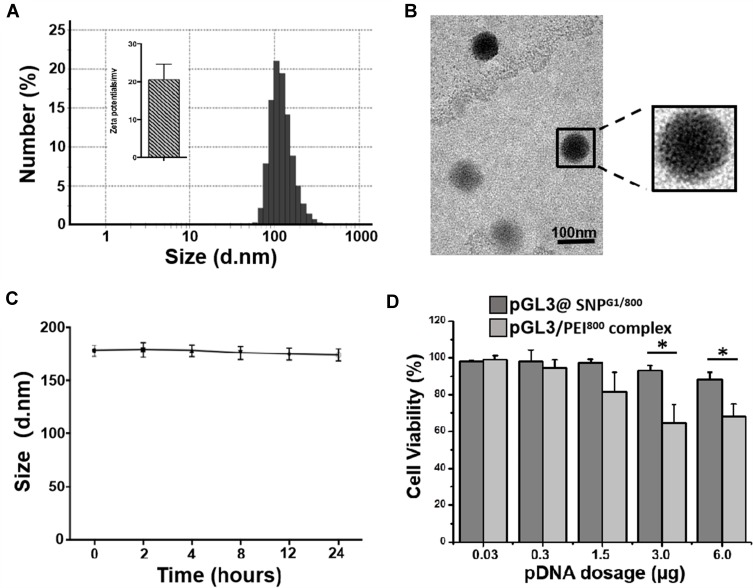
Given that pDNA@SNPG1/800 has the highest transfection efficiency in Jurkat cell among the prepared pDNA@SNPsx/y, its physicochemical properties and cytotoxicity were further evaluated. The mean hydrodynamic diameter of pDNA@SNPsG1/800 was approximately 120 nm, and the surface charge was approximately +20 mV (Figure 3A). Most pDNA@SNPsG1/800 were uniform in size and morphology, as shown by TEM in Figure 3B. To test the stability of prepared nanoparticles, we determined the size distribution of the pDNA@SNPG1/800 dispersions prepared with PBS by NanoSight at different time points, and the variation for 24 h was insignificant (Figure 3C). The stability of these nanoparticles can be attributed to CD/Ad host–guest recognition and electrostatic interactions between Ad-PAMAM/CD-PEI hydrogel and plasmid DNA.32 The difference in cell viability with the increase in pDNA@SNPG1/800 concentrations in the present dosage range was insignificant (Figure 3D). The cytotoxicity of pDNA@SNPsG1/800 was even lower than that of PEI800, a LMW PEI showed low cytotoxicity and transfection efficiency in in vitro gene delivery.26,27,37
Figure 3.
pGL3@SNPG1/800 preparation and evaluation. (A). Hydrodynamic size distribution and zeta potential of pGL3@SNPsG1/800. (B). TEM image of pGL3@SNPsG1/800. (C). Hydrodynamic size of pGL3@SNPG1/800 in PBS buffer (pH of 7.4) at different time points. (D). Viability of Jurkat cells treated with pGL3@SNPsG1/800 or pGL3/PEI800 complexes at varying doses of pGL3 (*P<0.05).
Abbreviations: pDNA, plasmid DNA; SNPs, self-assembled nanoparticles; PEI800, polyethylenimine (molecular weight: 800); TEM, transmission electron microscope.
Cellular Binding and Endosome Escape of pDNA@SNPG1/800
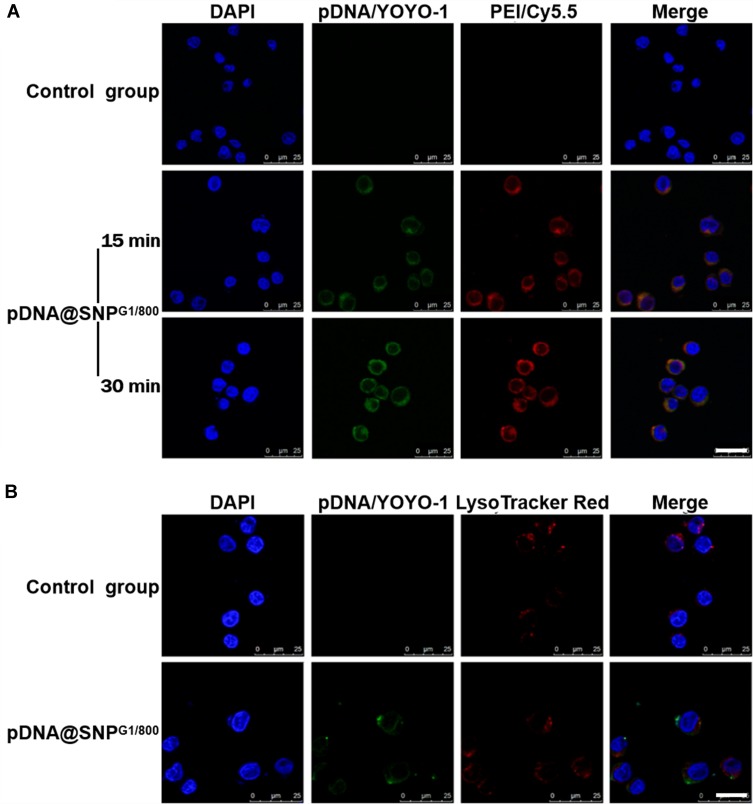
Cellular binding and internalization are the prerequisites for pDNA@SNPG1/800 to achieve high transfection efficiency. Here, pDNA@SNPsG1/800 containing cy5.5-labeled PEI800 and YOYO-1-labeled plasmid DNA were first prepared, and their binding ability to Jurkat cells was evaluated by confocal laser scanning microscopy. The results showed evident overlapping green and red fluorescence on the Jurkat cell surface after incubation with dual fluorescence-labeled pDNA@SNPG1/800 for 15 min, and the fluorescent signal increased with increase in incubation time (Figure 4A). After cellular binding and consequential internalization, the effective endosome escape of pDNA@SNPsG1/800 was also involved in achieving efficient gene delivery and transgene expression. The results of intracellular trafficking experiments presented in Figure 4B showed that the fluorescence of YOYO-1-labeled pDNA and LysoTracker Red indicated endosomes was separated 4 h after incubation. This observation can be interpreted as follows: after pDNA@SNPsG1/800 are internalized into endosome, the proton sponge effect of CD-PEI and Ad-PAMAM blocks in pDNA@SNPsG1/800 enhances the escape of gene cargo from the endosome to cytoplasm.20,26
Figure 4.
Cellular binding and endosome escape of pDNA@SNPsG1/800. (A). Confocal microscopy images of Jurkat cells incubated with dual-fluorescence-labeled pDNA@SNPsG1/800. (B). Confocal microscopy images of endosome escape of pDNA@SNPsG1/800. Scale bars, 20 μm.
Abbreviations: DAPI, 4ʹ,6-diamidino-2-phenylindole; pDNA, plasmid DNA; YOYO-1, YOYO™-1 Iodide; PEI, polyethylenimine; cy5.5, Cyanine5.5; SNP, self-assembled nanoparticle.
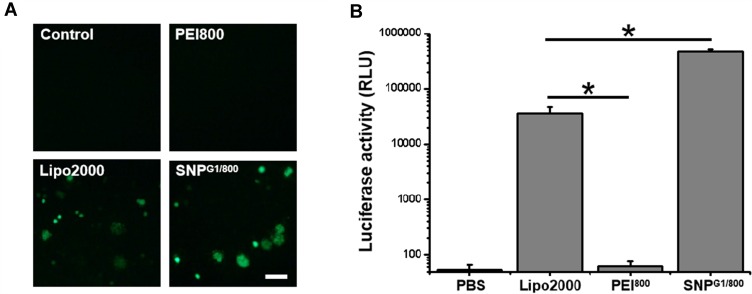
Transgenic Expression in pDNA@SNPG1/800-Transfected Jurkat Cells
After confirming the cellular binding and endosome escape of pDNA@SNPsG1/800, we further evaluated the transgenic expression of the reporter gene in Jurkat cells treated with GFP or luciferase encoding plasmid DNA-loaded pDNA@SNPsG1/800. Fluorescence microscopy images presented in Figure 5A indicated the high-level GFP transgenic expression in pGFP@SNPG1/800 or pGFP/LipofectamineTM 2000 complex-transfected Jurkat cells, whereas no GFP fluorescence was observed in pGFP/PEI800 complex group. Consistent with these results, the luciferase activities in pGL3/PEI800 complex group were extremely low, while it was approximately 600-fold higher in pGL3/LipofectamineTM 2000 complex group. The luciferase activity in the pGL3@SNPG1/800 group was even 10-fold higher than that in pGL3/LipofectamineTM 2000 complex group (Figure 5B). The high transfection efficiency of pDNA@SNPG1/800 should contribute to its effective cellular binding, endosome escape, plasmid DNA release, and low cytotoxicity. These experiments revealed that pDNA@SNPG1/800 significantly enhanced transfection efficiency while maintaining or even reducing cytotoxicity compared with LMW PEI.
Figure 5.
Transfection efficiency of Jurkat cells with pDNA@SNPsG1/800. (A). Fluorescence microscope image of GFP expressed in pGFP@SNPsG1/800-transfected Jurkat cells. Scale bars, 50 μm. (B). Activity of luciferase expressed by Jurkat cells transfected with pGL3/LipofectamineTM 2000, pGL3/PEI800 complexes, or pGL3@SNPsG1/800 (*P<0.05).
Abbreviations: PEI, polyethylenimine; Lipo2000, LipofectamineTM 2000; SNP, self-assembled nanoparticle; GFP, green fluorescent protein.
EGFRvIII-CAR Expression in pEGFRvIII-CAR@SNPG1/800-Transfected Jurkat Cells
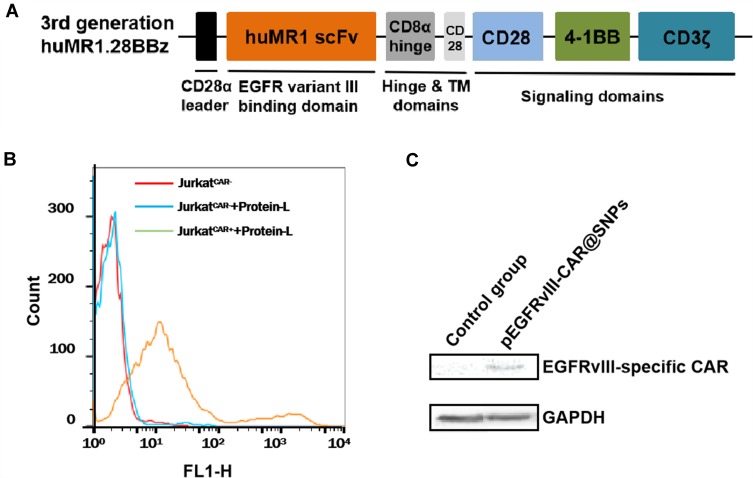
The EGFRvIII is a consistent tumor-specific mutation that is absent from normal tissues but widely expressed in various human epithelial tumors.39 Thus, EGFRvIII represents an excellent candidate for cancer target therapy, including immunotherapy. Various preclinical reports on cancer adoptive immunotherapy by using EGFRvIII CAR-redirected T cells prepared by virus infection are available.40–42 Here, a human-derived, third-generation EGFRvIII-specific CAR (EGFRvIII-CAR) was designed by fusing humanized EGFRvIII scFv43 to the hinge region of the human CD8α chain and transmembrane and cytoplasmic regions of the human CD28, 4–1BB, and CD3ζ. The structure of EGFRvIII-CAR is presented in Figure 6A, and its corresponding intracellular expression plasmid vector pEGFRvIII-CAR was constructed. pEGFRvIII-CAR was encapsulated into SNPsG1/800 according to the procedure above, and the resulting pEGFRvIII-CAR@SNPG1/800 was used for the potential evaluation of T-cell transient CAR modification. At 24 h after pEGFRvIII-CAR@SNPG1/800 treatment, Jurkat cells were stained with FITC-labeled protein-L, which was a protein that can bind to single-chain antibody fragments specifically and has been confirmed as a general reagent for CAR expression detection on transduced lymphocytes by flow cytometry.44 The FACS analysis results presented in Figure 6B showed no difference in fluorescence intensity between FITC-protein-L-treated Jurkat cells and non-FITC-protein-L-treated Jurkat cells, thereby indicating no nonspecific binding of protein L by Jurkat cells. Compared with no fluorescence change in untransfected Jurkart cell after FITC-Protein-L incubation, evident fluorescence intensity shift was found in Jurkat cells treated with pEGFRvIII-CAR@SNPsG1/800, thereby confirming the existence of EGFRvIII-CAR on the surface of the transfected Jurkat cells. The density of the CAR that is present on the cell surface is related to the transfection dosage of pEGFRvIII-CAR (data not shown). The Western blot analysis further confirmed EGFRvIII-CAR expression in pEGFRvIII-CAR@SNPG1/800-transfected Jurkat cells (Figure 6C).
Figure 6.
Flow cytometry and Western blot analysis of EGFRvIII-CAR expressed in pEGFRvIII-CAR@SNPG1/800-transfected Jurkat cells. (A). Structural diagram of EGFRvIII-CAR. (B). Protein L-based flow cytometry analysis of Jurkat cells transfected w/or w/o pEGFRvIII-CAR@SNPsG1/800. (C). Western blot analysis of EGFRvIII-CAR expressed in pEGFRvIII-CAR@SNPsG1/800-transfected Jurkat cells.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; EGFRvIII, epidermal growth factor receptor variant III; CAR, chimeric antigen receptor; pEGFRvIII-CAR, epidermal growth factor receptor variant III-chimeric antigen receptor expression plasmid; SNPs, self-assembled nanoparticles.
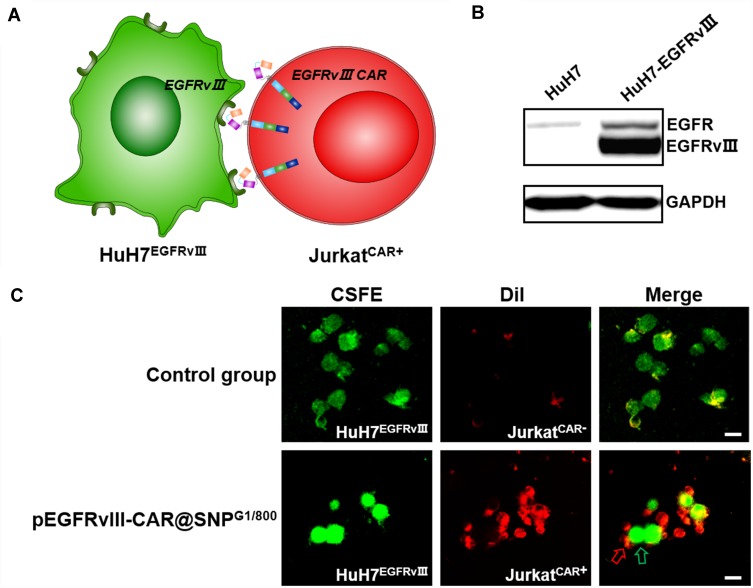
Interaction of EGFRvIII-CAR-Positive Jurkat T Cell with EGFRvIII-Positive HuH7 Cells
Whether the transient modification of CAR provides T cells with tumor cell-specific response is a prerequisite for CAR-T cell to exert cytotoxicity, thereby inducing antitumor immunity against malignancies (Figure 7A). Thus, after confirming the EGFRvIII CAR expression on the surface of pEGFRvIII-CAR@SNPG1/800-transfected Jurkat cells, we further tested the response of these EGFRvIII-CAR-redirected Jurkat cells (JurkatCAR+) to EGFRvIII-positive tumor cells. Human hepatocarcinoma HuH7 cells that were engineered to stably overexpress EGFRvIII (Huh7EGFRvIII+) were used as target tumor cells,45 and their EGFRvIII expression was validated using Western blot first (Figure 7B). DiI-labeled non-EGFRvIII CAR-engineered Jurkat T cells (JurkatCAR−) or JurkatCAR+ were cocultured with CFSE-labeled Huh7EGFRvIII+ for 1 h and then visualized using confocal microscope after washing. The results presented in Figure 7C showed that Huh7EGFRvIII+ cells were specifically recognized and bound by JurkatCAR+ but JurkatCAR−, thereby indicating that EGFRvIII-CAR molecules that are present on Jurkat cells were the critical factors for T cells’ response to target tumor cells. These results showed that Jurkat T cells can be efficiently engineered to transiently express CARs that recognize EGFRvIII by using pEGFRvIII-CAR@SNPG1/800, and the EGFRvIII-CAR-redirected Jurkat T cells exhibit specific reactivity to target tumor cells.
Figure 7.
Interaction of EGFRvIII-CAR-positive Jurkat T cells with EGFRvIII-positive HuH7 cells. (A). Scheme of the interaction of EGFRvIII-CAR-positive Jurkat T cell with EGFRvIII-positive HuH7 cell. (B). Western blot analysis of EGFRvIII expressed in HuH7 engineered to stably overexpress EGFRvIII. (C). Confocal microscopy-based studies on the interaction between EGFRvIII-positive HuH7 cells (green) and Jurkat T cells (red) transfected w/or w/o pEGFRvIII-CAR@SNPsG1/800. Scale bars, 20 µm.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; EGFR, epidermal growth factor receptor; EGFRvIII, epidermal growth factor receptor variant III; CSFE, carboxyfluorescein succinimidyl ester; Dil, 1,1′-Dioetadeeyl-3,3,3′,3′-tetramethylindocarboeyanine perchlorate.
Conclusions
A series of pDNA@SNPsx/y prepared from Ad-PAMAM dendrimers of different generations (G1 or G5) and CD-PEI of different molecular weights (800, 2000, or 25000 Da) were characterized and evaluated. pDNA@SNPG1/800 was selected for CAR-expressing plasmid DNA loading and Jurkat cell transfection due to its best transfection efficiency and low cytotoxicity. pEGFRvIII-CAR@SNPsG1/800 can efficiently deliver pEGFRvIII-CAR into Jurkat T cells, thereby resulting EGFRvIII-CAR expression on cell surface enabled Jurkat T cells to respond to EGFRvIII-positive tumor cells specifically. These results indicated that pDNA@SNPG1/800 exhibits high efficacy in Jurkat cell gene transfection with low cytotoxicity and has considerable potential in T cell transient CAR modification. Considering that the CD4+/CD8- property of Jurkat cells, future studies will be devoted to pEGFRvIII-CAR@SNPsG1/800-based ex vivo and in vivo primary T cells transient CAR modification and antitumor efficiency evaluation.
Acknowledgments
The authors thank the National Natural Science Foundation of China (Grant No.81571787; Grant No.81690262); Natural Science Foundation of Shanghai Municipal (Grant No.15ZR1422700); and the Medical-Engineering Joint Fund of Shanghai Jiao Tong University (Grant No.MS2015MS54; Grant No.MS2017MS56). The authors appreciate Prof. Hsian-Rong Tseng at University of California Los Angeles for generous technical advice with pDNA@SNPs preparation.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Eshhar Z, Waks T, Gross G, Schindler DG. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc Natl Acad Sci U S A. 1993;90:720–724. doi: 10.1073/pnas.90.2.720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brentjens RJ, Latouche J-B, Santos E, et al. Eradication of systemic B-cell tumors by genetically targeted human T lymphocytes co-stimulated by CD80 and interleukin-15. Nat Med. 2003;9:279–286. doi: 10.1038/nm827 [DOI] [PubMed] [Google Scholar]
- 3.Kochenderfer JN, Yu Z, Frasheri D, Restifo NP, Rosenberg SA. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood. 2010;116:3875–3886. doi: 10.1182/blood-2010-01-265041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–733. doi: 10.1186/s13045-018-0676-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill S, Maus MV, Porter DL. Chimeric antigen receptor T cell therapy: 25years in the making. Blood Rev. 2016;30:157–167. doi: 10.1016/j.blre.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 7.Fesnak AD, June CH, Levine BL. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat Rev Cancer. 2016;16:566–581. doi: 10.1038/nrc.2016.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maude SL, Frey N, Shaw PA, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newick K, O’Brien S, Moon E, Albelda SM. CAR T Cell Therapy for Solid Tumors. Annu Rev Med. 2017;68:139–152. doi: 10.1146/annurev-med-062315-120245 [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Riviere I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol Ther Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine BL, Miskin J, Wonnacott K, Keir C. Global Manufacturing of CAR T Cell Therapy. Mol Ther Methods Clin Dev. 2016;4:92–101. doi: 10.1016/j.omtm.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plumridge H. New costly cancer treatments face hurdles getting to patients. Wall Street J. 2014;6. [Google Scholar]
- 13.Bonifant CL, Jackson HJ, Brentjens RJ, Curran KJ. Toxicity and management in CAR T-cell therapy. Mol Ther Oncolytics. 2016;3:16011. doi: 10.1038/mto.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalaitsidou M, Kueberuwa G, Schutt A, Gilham DE. CAR T-cell therapy: toxicity and the relevance of preclinical models. Immunotherapy. 2015;7:487–497. doi: 10.2217/imt.14.123 [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Mooney DJ. Biomaterial-assisted targeted modulation of immune cells in cancer treatment. Nat Mater. 2018;17:761–772. doi: 10.1038/S41563-018-0147-9 [DOI] [PubMed] [Google Scholar]
- 16.Barrett DM, Zhao Y, Liu X, et al. Treatment of advanced leukemia in mice with mRNA engineered T cells. Hum Gene Ther. 2011;22:1575–1586. doi: 10.1089/hum.2011.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beatty GL, Haas AR, Maus MV, et al. Mesothelin-specific chimeric antigen receptor mRNA-engineered T cells induce anti-tumor activity in solid malignancies. Cancer Immunol Res. 2014;2:112–120. doi: 10.1158/2326-6066.CIR-13-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao N, Qi J, Zeng Z, et al. Transfecting the hard-to-transfect lymphoma/leukemia cells using a simple cationic polymer nanocomplex. J Control Release. 2012;159:104–110. doi: 10.1016/j.jconrel.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith TT, Stephan SB, Moffett HF, et al. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat Nanotechnol. 2017;12:813–820. doi: 10.1038/nnano.2017.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalil H, Chen T, Riffon R, Wang R, Wang Z. Synergy between polyethylenimine and different families of antibiotics against a resistant clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52:1635–1641. doi: 10.1128/AAC.01071-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godbey WT, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45:268–275. doi: [DOI] [PubMed] [Google Scholar]
- 23.Abdallah B, Hassan A, Benoist C, et al. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Hum Gene Ther. 1996;7:1947–1954. doi: 10.1089/hum.1996.7.16-1947 [DOI] [PubMed] [Google Scholar]
- 24.Zinselmeyer BH, Mackay SP, Schatzlein AG, Uchegbu IF. The lower-generation polypropylenimine dendrimers are effective gene-transfer agents. Pharm Res. 2002;19:960–967. doi: 10.1023/a:1016458104359 [DOI] [PubMed] [Google Scholar]
- 25.Malik N, Wiwattanapatapee R, Klopsch R, et al. Dendrimers: relationship between structure and biocompatibility in vitro, and preliminary studies on the biodistribution of 125I-labelled polyamidoamine dendrimers in vivo. J Control Release. 2000;65:133–148. doi: 10.1016/S0168-3659(99)00246-1 [DOI] [PubMed] [Google Scholar]
- 26.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60:149–160. doi: 10.1016/S0168-3659(99)00090-5 [DOI] [PubMed] [Google Scholar]
- 27.Kunath K, von Harpe A, Fischer D, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89:113–125. doi: 10.1016/S0168-3659(03)00076-2 [DOI] [PubMed] [Google Scholar]
- 28.Shah N, Steptoe RJ, Parekh HS. Low-generation asymmetric dendrimers exhibit minimal toxicity and effectively complex DNA. J Pept Sci. 2011;17:470–478. doi: 10.1002/psc.1347 [DOI] [PubMed] [Google Scholar]
- 29.Peng J, Garcia MA, Choi J-S, et al. Molecular recognition enables nanosubstrate-mediated delivery of gene-encapsulated nanoparticles with high efficiency. ACS Nano. 2014;8:4621–4629. doi: 10.1021/nn5003024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petter RC, Salek JS, Sikorski CT, Kumaravel G, Lin FT. Cooperative binding by aggregated mono-6-(alkylamino)-beta-cyclodextrins. J Am Chem Soc. 1990;112:3860–3868. doi: 10.1021/ja00166a021 [DOI] [Google Scholar]
- 31.Wang H, Wang S, Su H, et al. A supramolecular approach for preparation of size-controlled nanoparticles. Angew Chem Int Ed Engl. 2009;48:4344–4348. doi: 10.1002/anie.200900063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Chen K-J, Wang S, et al. A small library of DNA-encapsulated supramolecular nanoparticles for targeted gene delivery. Chem Commun (Camb). 2010;46:1851–1853. doi: 10.1039/b923711a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Liu K, Chen K-J, et al. A rapid pathway toward a superb gene delivery system: programming structural and functional diversity into a supramolecular nanoparticle library. ACS Nano. 2010;4:6235–6243. doi: 10.1021/nn101908e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Wang H, Yang W, Cheng Y. Disulfide cross-linked low generation dendrimers with high gene transfection efficacy, low cytotoxicity, and low cost. J Am Chem Soc. 2012;134:17680–17687. doi: 10.1021/ja307290j [DOI] [PubMed] [Google Scholar]
- 35.Gosselin MA, Guo WJ, Lee RJ. Efficient gene transfer using reversibly crosslinked low molecular weight polyethylenimine. Bioconjug Chem. 2001;12:989–994. doi: 10.1021/bc0100455 [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Vinogradov SV. Short biodegradable polyamines for gene delivery and transfection of brain capillary endothelial cells. J Control Release. 2010;143:359–366. doi: 10.1016/j.jconrel.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam K, Jung S, Nam JP, Kim SW. Poly(ethylenimine) conjugated bioreducible dendrimer for efficient gene delivery. J Control Release. 2015;220:447–455. doi: 10.1016/j.jconrel.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam HY, Nam K, Hahn HJ, et al. Biodegradable PAMAM ester for enhanced transfection efficiency with low cytotoxicity. Biomaterials. 2009;30:665–673. doi: 10.1016/j.biomaterials.2008.10.013 [DOI] [PubMed] [Google Scholar]
- 39.Wikstrand CJ, Hale LP, Batra SK, et al. Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res. 1995;55:3140–3148. doi: 10.1016/0165-4608(95)00039-R [DOI] [PubMed] [Google Scholar]
- 40.Morgan RA, Johnson LA, Davis JL, et al. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Hum Gene Ther. 2012;23:1043–1053. doi: 10.1089/hum.2012.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sampson JH, Choi BD, Sanchez-Perez L, et al. EGFRvIII mCAR-modified T-cell therapy cures mice with established intracerebral glioma and generates host immunity against tumor-antigen loss. Clin Cancer Res. 2015;20:972–984. doi: 10.1158/1078-0432.CCR-13-0709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson LA, Scholler J, Ohkuri T, et al. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci Transl Med. 2015;7:275ra222. doi: 10.1126/scitranslmed.aaa4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safdari Y, Farajnia S, Asgharzadeh M, Omidfar K, Khalili M. humMR1, a highly specific humanized single chain antibody for targeting EGFRvIII. Int Immunopharmacol. 2014;18:304–310. doi: 10.1016/j.intimp.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 44.Zheng Z, Chinnasamy N, Morgan RA. Protein L: a novel reagent for the detection of chimeric antigen receptor (CAR) expression by flow cytometry. J Transl Med. 2012;10:29. doi: 10.1186/1479-5876-10-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang H, Wang H, Tan Z, et al. Growth suppression of human hepatocellular carcinoma xenografts by a monoclonal antibody CH12 directed to epidermal growth factor receptor variant III. J Biol Chem. 2011;286:5913–5920. doi: 10.1074/jbc.M110.192252. [DOI] [PMC free article] [PubMed] [Google Scholar]