ABSTRACT
MiR-146a could stimulate tumor growth or block tumor proliferation in systemic malignancies, referring to the specific downstream targeted gene. However, its roles in breast cancer stem-like cells (BrCSCs) are barely known. To dig out its mechanistic functions, we explored the indicative roles of miR-146 in preclinical study, regardless of the hormone receptor status, and the positive correlation between miR-146 and better prognosis was proved, as its correlation to Let-7c was. To uncover the implicated mechanisms, we first identified the suppressive role of miR-146a in stem cells’ renewal, which was achieved by promoting the asymmetric division of BrCSCs. Let-7c was previously revealed with its suppressive functions in stem-like cells expansion, and miR-146 was predicated and successfully proved to bind to and degrade the 3’UTR of LIN28, a maturation blocker of Let-7 family. Results further showed that miR-146a increased the Let-7c level through degrading LIN28, and LIN28 inhibition is required for miR-146a induction of asymmetric stem cells’ division. Moreover, Let-7 controlled Wnt signaling pathway activity could be strengthened due to the miR146 inhibition of H19, later of which was often activated in stem cells group with functional existence of Wnt signaling. H19 itself in turn formed the positive feedback regulation with Let-7. Our results suggested the miR-146a/LIN28/Wnt signaling circle in restraining the symmetric cells division, which was specifically referred to the controlling of the small circle of Let-7c and H19, and together, this dual axis could help to prohibit the stem cells expansion.
KEYWORDS: Breast cancer stem-like cells, Non-coding RNAs, division manners
1. Introduction
Breast cancer stem cells (BrCSCs), also known and accepted as cancer stem-like cells or cancer initiating cells, have been well identified for their stimulation of drug resistance and long term recurrence. Accurate therapy strategy aiming at eliminating these typical cells with little and controllable side-effect has been raised out in Western world for years, and China is chasing up to implement the huge public health plans [1], [2]. Non-coding RNAs could interact with each other to perform the complicated functions [3] The interaction between non-coding RNAs have attracted our attention long time ago, and we identified the mechanisms of miRNA-miRNA regulation and miRNA-lncRNA regulation. In breast cancer, we focused on eliminating the stem cells hiding in the estrogen receptor positive cells, however, intense mission is to explore strategies that could influence breast cancer cells with any kind of hormone receptors status.
The long non-coding genes of RNAs (lnc-RNAs) were well explored, but their roles were only studied in breast cancer of mono hormone receptor status, and the lnc-RNA of H19 functions in signatures of BrCSCs have not been well explored [4,5].
The division manners was recently found to be related to the self-renewal capacity of stem cells and cancer stem cells, and the disruption of the normal procedure during the cells split could be critical for suppressing the certain division mode. How could the non-coding RNAs impact on the division process was rarely covered.
In this study, we plan to dig the roles and related mechanisms of H19 in patterns of molecule status of breast cancer, and to identify the role of non-coding RNAs in integrating the stem cells’ split, which will help to find one universal and effective regulator of the stem cells group in breast cancer, regardless of status of hormone receptor.
2. Methods and materials
2.1. Online analysis of significance of certain factors in breast cancer specimens
To investigate the expression level of Let-7 family and miR-146 in breast cancer, Kaplan-Meier analysis (http://kmplot.com/analysis/) was used to acquire the clinical data from multiple data base [6]. For the expression patterns of H19 and several Wnt signaling factors, data base of CBIOPORTAL FOR CANCER GENOMIC (http://www.cbioportal.org/) was used for statistical analysis [7,8]. No specific selection or exclusion was made when determining the subtypes of enrolled patients, and the hormone receptor status includes of every type of estrogen, progestogen and HER-2 receptor. Study of H19 and Wnt signaling factors was performed to identify the alternations induction of survival changes. EMT inductor of Snai1 and stem cell marker of ALDH1 was analyzed by using Kaplan-Meier analysis and CBIOPORTAL FOR CANCER GENOMIC respectively. The heatmap of factors of Wnt signaling were generated from CBIOPORTAL FOR CANCER GENOMIC, based on their irregular expressions.
2.2. Cell culturing, mimics/inhibitors transfection and lentiviral transduction
Three breast cell lines of T47-D, MM-231 and BT-20, and HEK-293T were originally purchased from American Type Culture Collection and maintained in either DMEM 1x (Gibco) or in RPMI 1640 Medium (Hyclone). 15% FBS (Gibco), 1% penicillin and 1% streptomycin (Hyclone) were also added. For constructing cells with H19 overexpression or H19 deletion, the genomic regions encoding H19 or targeted inhibiting H19 were PCR-amplified using PrimeSTAR® HS DNA Polymerase (TaKaRa) and subcloned into the pcDNA3.1 vector (Invitrogen), named pcDNA3.1-H19 or shRNA-H19. The pcDNA3.1 vector was used as an empty control. Breast cancer were transfected or infected with the plasmid pcDNA3.1-H19, Let-7 mimic, miR-146 mimic, Let-7c lentivial or miR-146 lentiviral by using Lipofectamine 3000, or calcium transfection, referring to the manufacturer instruction. Cells were selected with neomycin (800μg/ml) for two weeks. The stem-like cells of spheres of different groups were suspended in DMEM/F12 Medium supplementing with 20 ng/mL EGF (BD Biosciences), bFGF and 4 μg/mL insulin (Sigma), and then plating at 1000cells/ml in 6-well ultra-low attachment dishes (Corning Incorporated) for about one week. To analyze the self-renewal ability, spheres number was counted by using phase contrast microscope (Nikon).
2.3. Luciferase reporter assay
For analysis of luciferase activity, 2 × 105 cells of different groups were seeded into 24-wells plate filling with Opti-MEM (GIBCO) 24 hours before transfection. Pretreated cells were co-transfected with either 300ng luciferase reporter vectors (H19-3’UTR-wt or H19-3’UTR-mt), 30nM Let-7c mimic, or 30nM miR-146 mimic using Lipofectamine 3000 (Invitrogen). Renilla luciferase plasmid (100ng/well, Promega) was used as an internal control. The promoter reporter plasmid pGL3-TCF4-wt (LEF/TCF reporter, TOP) or pGL3-TCF4-mut (LEF/TCF reporter, FOP) was co-transfected with Let-7c or miR-146 mimic into different cells. For detection of HEK-293T cells with certain treatment, cells were co-transfected with 200ng TOP or FOP plasmid, 25nm mimics or mutant and empty vector, 20ng pRL-TK Renilla luciferase vector (Promega) were set as normalization control. 16 hours after transfection, cells were harvested and lysed for standard luciferase assay. The positive control of using Wnt3a (mouse Wnt3a protein, 400 ng/ml) and LiCl (10mM) were added to cells at the 24th hour after transfection, and the cells were cultured for another 18 hours. Dual Luciferase assay was performed 42 hours after transfection, and promoter activity were indicated as arbitrary units using a Renilla reporter for internal normalization. Experiments were done in triplicates, and the standard deviation is indicated.
2.4. Immunofluorescence of EDU staining and western blotting
For immunofluorescence detection of EDU staining, cells of different groups were seeded into chamber slides with RS glass (Nunc, Thermo Scientific) after treatments. EdU intensity was detected after cells being incubated with 10uM EdU for 24 hours. Detection was performed with using Click-iT Plus EdU Alexa imaging Kit (MP 10,637, Life Technologies). The fluorescence images were obtained using an Olympus microscope. For western blotting, the protein from cell extracts were separated by 10% SDS-PAGE electrophoresis, and was later transferred onto PVDF membrane. Membranes were incubated with Snai1 (1:5000, C15D3, #3879, Cell Signaling Technology), TCF-4 (1:1000, sc-166,699, Santa Cruz Biotechnology), WNT1 (1:2000, ab15251, Abcam), C-myc (1:2000, ab32072, Abcam), ALDH1 (1:1000, sc-50,385, Santa Cruz Biotechnology), CTNNB1 (1:200, ab16051, Abcam), and Vinculin (1:10,000, #4650, Cell Signaling Technology).
2.5. Statistical analysis
Numerical data were presented as mean ± standard deviation (SD), of three to six independent experiments. Statistical analysis was carried out by using Graph Pad Prism 6 and Microsoft Excel software. Differences between groups were assessed by using Student’s t test or ANOVA individually. P < 0.05 was statistically significant.
3. Results
3.1. Correlation between Let-7 family of miRNAs and prognosis of patients with breast cancer
The overall survival ratios were acquired as months lasting, and results were generated from both METABRIC database and TCGA database. Results of TCGA were shown in Figure S1, and no candidate Let-7 family member was chosen. Data from METABRIC data base was analyzed, and results were displayed in Figure 1. Higher levels of Let-7a, Let-7b, Let-7c, Let-7e, Let-7f and Let-7g were proven to be related with longer survival time. Based on previous results, Let-7b and Let-7c were chosen as candidates for their strong suppressive functions in cancer generation and development, especially for their roles in cancer stem cells [9,10].
Figure 2.
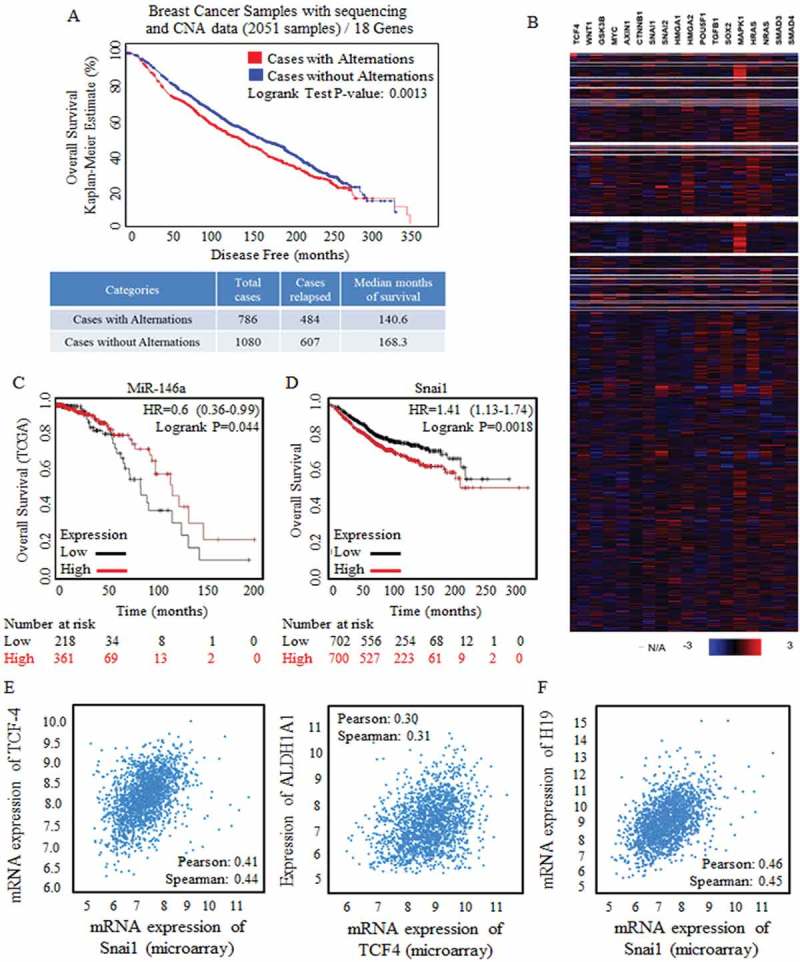
Wnt signaling activation were strongly correlated with poorer prognosis.
A cluster of Wnt signaling factors were selected and applied for survival risk analysis by using CBIOPORTAL FOR CANCER GENOMIC (http://www.cbioportal.org/). (a) Alternation (Red line, 786 cases) was defined with the amplification only mode, and the amplification of Wnt signaling factors were detected and referred to shorter survival periods significantly, p = 0.0013. (b) Heatmap results of factors of Wnt signaling clusters (18 genes) from data base of Breast Cancer Samples with sequencing and CNA data (2051 samples) were showed, and multiple Wnt signaling factors were universally highly overexpressed. Kaplan-Meier data analysis was also made, and results revealed the tumor suppressive miR-146 (c), and the oncogenic Snai1 (d) when predicating breast cancer prognosis. Enrolled samples number was indicated below the diagram. (e) Wnt signaling effector of TCF-4 was strongly correlated to the stem cells’ effector (left) and the ratio of stem cells with ALDH1A1 phenotype (right). (f) The predicated Wnt signaling activator of non-coding H19 was strongly correlated with Snai1 overexpression.
3.2. Wnt activation indicated poorer survival outcomes of patients with breast cancer
Wnt signaling factors were analyzed for their potential correlation with overall survival at CBIOPORTAL FOR CANCER GENOMIC (http://www.cbioportal.org/). Wnt signaling was proven to be related with multiple malignancy progression [3,11,12], and was identified with activation status in breast cancer with phenotype of positive estrogen receptor status [9]. In patients with breast cancer of any kind of hormone receptor status, the activation and overexpression of Wnt signaling factors were correlated to short survival months (Figure 2(a)). Multiple Wnt signaling factors were applied for expression detection in patients with breast cancer, and the heatmap panel revealed their expression status in breast cancer specimens (Figure 2(b)).
Figure 1.
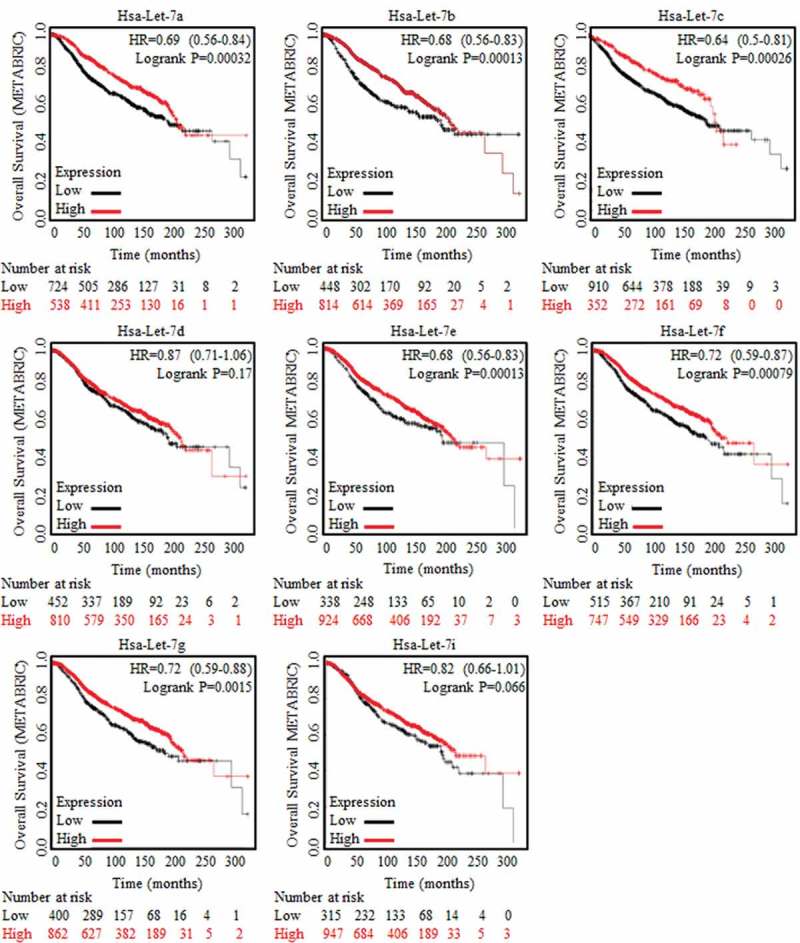
Results of Let-7 family generated based on METABRIC database analysis.
Clinical analysis of Let-7 family of miRNAs were conducted and concluded relying on Kaplan-Meier analysis (http://kmplot.com/analysis/), where the huge amount of data and trends of clinical specimens in analyzing survival prognosis could be found. Enrolled samples number was indicated below each diagram. Let-7a, Let-7b, Let-7c, Let-7d, Let-7e, Let-7f, Let-7g and Let-7i were generated and captured from METABRIC data base, as were indicated in the figures. Results of Let-7a, Let-7b, Let-7c, Let-7e, Let-7f and Let-7g showed their significant correlation to better prognosis.
MiR-146 was predicated with tumor suppressive roles for its indication of better survival outcomes (Figure 2(c)), the miR-146 stimulator of Snai1 [13] was correlated to shorter survival time (Figure 2(d)). Snai1 is the crucial promoter of pluripotency capacity, and ALDH1A1(usually known as ALDH1) was one of the typical and reliable marker for stem cells identification and sorting, and we confirmed the positive correlation between either Snai1 or ALDH1A1 (Figure 2(e), left) and TCF-4 (Figure 2(e), right), one core factor to perform Wnt signaling function, by analyzing the validated data from CBIOPORTAL FOR CANCER GENOMIC (http://www.cbioportal.org/) [7,8].
3.3. Non-coding gene of H19 was possibly correlated with Wnt activation and stem cells’ potency of regeneration
The long non-coding gene of H19 correlates with higher risk of cancer death in multiple cancers [14,15], and selected key members of Wnt signaling were analyzed for their co-occurrence with H19 amplification, such as Wnt1 (p<0.05), CTNNB1 (p<0.05), Snai1 (p<0.001). HMGA2 (p<0.05), SOX4 (p<0.01), LIN28A (p<0.05), data not shown [7,8]. Additionally, based on the data base of microarray results, results showed the positive correlation between H19 and the strong pluripotency stimulator of Snai1 (Figure 2(f))
3.4. Wnt activation is critical for maintaining the symmetric division of BrCSCs
Wnt activation in stem cells of breast cancer with positive hormone receptor status functioned through sustaining the self-renewal ability, and we explored the effects of Wnt signaling in manipulation of division modes [9]. Three cell lines of BT-20, MDA-MB-231 and T47-D were selected for studying the universal role of Wnt activation, regardless of the hormone status [16]. Axitinib induced Wnt signaling repression was used as the positive control, and Let-7c exerted the most effective functions in promoting asymmetric division of stem cells of breast cancer (Figure 3(a), above). Symmetric division was inhibited significantly, comparing to control group (Figure 3(a), below). The representative images of BdU staining of DNA was showed (Fig, 3B, 40X). The manipulation of division mode sequentially altered the self-renewal ability of breast cancer stem cells, and Let-7c overexpression deducted the ratio of spheres (Figure 3(c), left), as were shown in Figure 3(c) (right).
Figure 3.
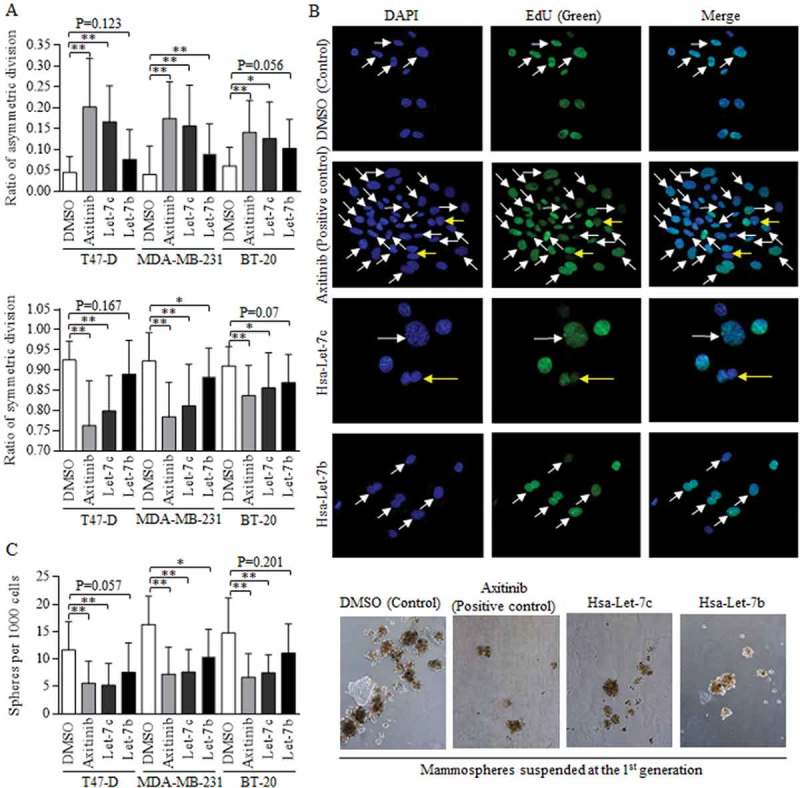
Let-7c and Wnt signaling inhibitor blocks the symmetric division.
Wnt activation inhibitor of Axitinib was used as positive control. (a) Both Axitinib and Let-7c effectively promoted the asymmetric division (above), suppressed the symmetric division (below). The differences of ACD ratios among groups of Let-7c, Let-7b, and Axitinib in T47-D (F = 0.223, p = 0.801), MDA-MB-231 (F = 2.066, p = 0.136) and BT-20 (F = 0.526, p = 0.594) are not significant, and similarly, the differences of SCD ratios among groups of Let-7c, Let-7b, and Axitinib in T47-D (F = 0.584, p = 0.561), MDA-MB-231 (F = 2.159, p = 0.126) and BT-20 (F = 0.253, p = 0.778) are not significant, processing by ANOVA analysis. (b) Images of fluorescence results of EdU dye were shown, and the dividing cells were marked with either the white arrow (SCD) or the yellow arrow (ACD). (c) Axitinib and Let-7c decreased number of the forming spheres significantly (left), which was directly attributed to the self-renewal ability of stem cells, with representative images shown in right. ACD, asymmetric division, SCD, symmetric division. Results were acquired and calculated from six independent experiments.
3.5. MiR-146a promoted Let-7c expression to inactivate Wnt signaling in a LIN28 depression dependent way
Either miR146a or Let-7c overexpression could increase the ACD of BrCSCs (Figure 4(a), left), and decrease the SCD significantly (Figure 4(a), right). The combination of both Let-7c and miR146 overexpression did not further promote the ACD ratio (Figure 4(a)), as were exhibited in Figure S2. Their effects on division modes consequently affected the self-renewal ability of stem cells, and both miR146 and Let-7c overexpression decreased the number of spheres acquired from the 1st generation, inhibiting the self-renewal (Figure 4(a)).
Figure 4.
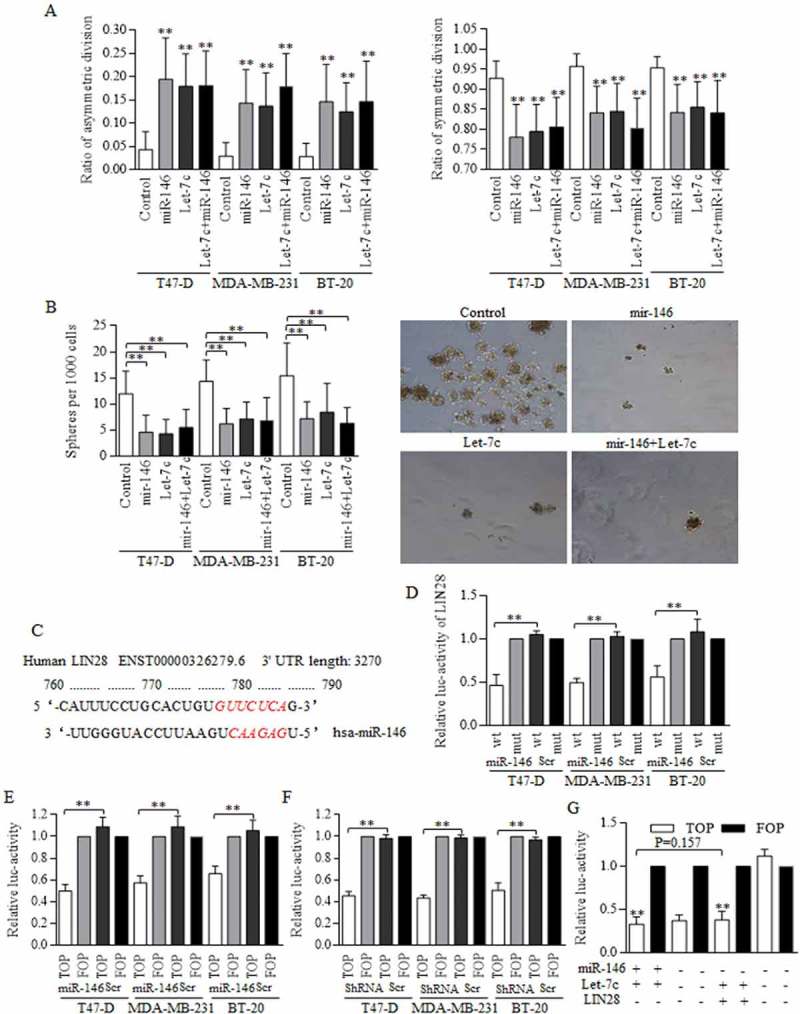
MiR-146 inhibited the Wnt activation through a Let-7c dependent way.
(a) The overexpressed miR-146 and Let-7c both exerted strong inhibition on symmetric division, and promoted the asymmetric division, ** p<0.01. The differences among groups of Let-7c, miR-146, and their combination in T47-D (F = 0.503, p = 0.610), MDA-MB-231 (F = 0.209, p = 0.813) and BT-20 (F = 0.835, p = 0.443) are not significant as written, processing by ANOVA analysis. (b) Enforcement of miR-146, Let-7c, and the combined overexpression all decreased the number of spheres acquired from the 1st generation culturing (left), as were presented (right). (c) Bioinformatic analysis of the potential binding site between miR-146 and the 3’UTR of LIN28 was analyzed and labeled in RED, as indicated. (d) Luciferase assay was conducted with detecting Renilla luciferase activity, and miR-146 decreased the luc-activity of 3’UTR of LIN28 with wild-type sequence effectively, which could be found in breast cancer of three cell lines. The functional status of Wnt signaling was tested by detecting the TCF-4 activity, and both miR-146 overexpression (e) and LIN28 inhibition (f) decreased the luc-activity of 3’UTR of TCF-4 with wild-type sequence (TOP), comparing to the controlled group of 3’UTR of TCF-4 with certain mutant sequence at the targeted region (FOP). G. The TCF-4 activity in groups of enforced miR-146 and Let-7 is similar to that of enforced Let-7 and inhibited LIN28, we hypothesized that miR-146 functioned through the LIN28/Let-7 axis, given the established interaction between LIN28 and Let-7. Results were acquired and calculated from six independent experiments.
MiR-146 was predicated to target and degrade LIN28 in a post-transcriptional way (Figure 4(c)), which was proved by luciferase assay (Figure 4(d)), and results showed that miR-146 overexpression inhibited the luc-activity of LIN28 effectively. We further identified the suppressions of either miR-146 enforcement (Figure 4(e)) or LIN28 inhibition (Figure 4(f)) on Wnt signaling activity by detecting the TCF-4 promoter activity. Let-7c was proved to inhibit the Wnt signaling activity directly, and itself could be precisely controlled by LIN28. We therefore turned to study the possible mechanism of miR-146 in Wnt signaling controlling. The combined effect of enforcing miR-146 and Let-7c is similar to that of inhibiting LIN28 in cells harboring Let-7c overexpression (Figure 4(g)), suggesting that miR-146 functioned through LIN28/Let-7c axis.
3.6. H19 suppression of Let-7c functions was achieved via the mechanistic regulation as sponge molecular
To identify the functional method of H19 regulated Let-7c effects, we constructed MS2 associated H19-Let-7c binding site to perform the RNA immunoprecipitation, which could pull down H19 and MS2 referred to Let-7c binding site. H19 and Let-7c specific MS2 were effectively grouped in groups of three cell lines (Figure 5(a)). Although firmly combined, H19 (Figure 5(b)) inhibition did not consequently restore Let-7c level (Figure 5(c)). Alternatively, Let-7c exerted no significant repression on H19 (Figure 5(d)), however, degraded the H19 binding site of wild-type effectively, further proving the mechanistic regulation of competitive sponge molecular (Figure 5(e-f)).
Figure 5.
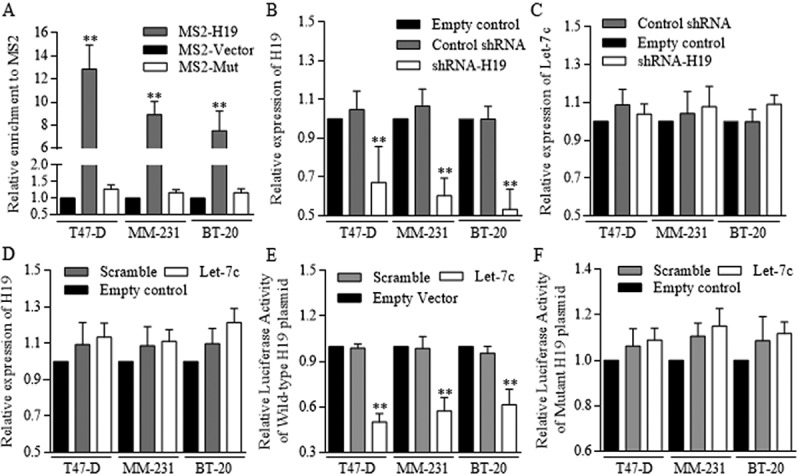
H19 inhibited the Let-7c functions as its competitive sponge molecular.
RNA immunoprecipitation was performed to identify the potential binding site between H19 and Let-7c. (a) Let-7c specific MS2 at the H19 targeted region was effectively enriched in groups of three cell lines, comparing to empty MS2 and the MS2 with mutant H19 region. ShRNA conducted H19 inhibition was successfully achieved (b) however, the inhibition did not influence Let-7c level effectively (c) (d) Let-7c exerted no significant repression on H19 expression level. The potential binding site between H19 and Let-7 was explored and synthesized, and Let-7c degraded the H19 binding site of wild-type effectively (e), and no significant results of Let-7c was detected in groups of H19 of mutant binding region (f). Results were acquired and calculated from three independent experiments.
3.7. A positive feedback loop of Wnt signaling and Let-7c was connected through interacting with H19
Wnt signaling was reported to be naturally activated in cancer stem-like cells, and we found the naturally overexpressed H19 of BrCSCs (Figure 6(a)), accompanying with Wnt activation of stem cells group (Figure 6(b)). We testified that when Wnt signaling was activated with using either 5mM LiCl or 400 ng/ml of recombinant mouse Wnt3a protein (Wnt3a) (Figure 6(c), H19 level went up significantly (Figure 6(d)), and the blocking of Wnt cascade with addition of Axitinib decreased the H19 expression significantly (Figure 6(d). H19 inhibition promotes the ACD of BrCSCs greatly, decreasing the SCD (Figure 6). Axitinib induction of Wnt signaling repression also increased the ratio of ACD significantly (Figure 6(e)). The division modes images were selected and shown in Fig. S3. H19 inhibition decreased the expression levels of multiple Wnt signaling factors (Figure 6(g)), as western blotting showed. H19 inhibition decreased the TCF-4 promoter activity in 293T cells significantly, however, no obvious changes were statistically significant when comparing groups of either combined Let-7c overexpression and H19 inhibition, or combined LIN28 inhibition and H19 inhibition (Figure 6(h)), strongly suggesting that H19 functioned through manipulating LIN28/Let-7c axis.
Figure 6.
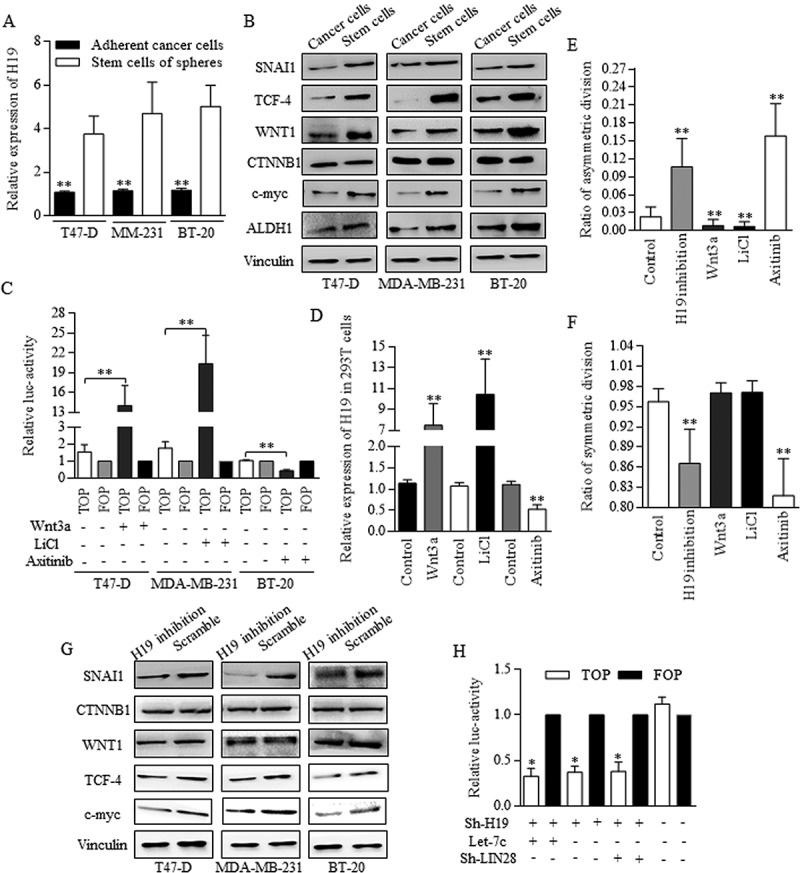
Wnt activation sustaining of H19 contributed to Let-7 disablement.
To study the possible connection among H19, Let-7, and Wnt signaling in BrCSCs, H19 inhibition was used as the key deregulation process when determining Wnt activity. A. H19 was naturally highly expressed in groups of stem cells in breast cancer. B. Wnt activation were detected in Breast cancer stem cells by using Western blot referred to the key signaling members. C. Wnt signaling could be activated with using either 5mM LiCl or 400 ng/ml of Wnt3a, and the activation status was determined by TCF-4 activity of FOP. D. H19 level went up significantly when Wnt signaling was activated in 293T cells, and the blocking of Wnt cascade with using Axitinib decreased the H19 expression significantly. E. Both H19 inhibition and Axitinib addition stimulated the asymmetric division, and the Wnt signaling activation decreased the ratio of asymmetric division, ** p<0.01 (Student’s t-test). F. Both H19 inhibition and Axitinib addition repressed the symmetric division ** p<0.01 (Student’s t-test). No significant difference was detected when comparing the addition of either Wnt3a (p = 0.0604) or LiCl (p = 0.0455) to control group. G. H19 inhibition decreased the expression levels of multiple Wnt signaling factors. H. H19 inhibition functioned the same way as either combined H19 inhibition and Let-7c overexpression, or combined H19 inhibition and LIN28 inhibition did. In these three groups, TCF-4 activity was much lower than that of controlled group. Results were acquired and calculated from three independent experiments.
3.8. Snai1/Wnt signaling promotes miR-146 transcription to form the miR-146/Wnt regulatory feedback circle
Snai1 stimulates the miR-146 expression in indirect way, which was mediated by the CCNT1/TCF-4 existence [13]. No binding site of Snai1 toward to the miR-146 promoter were detected (data not shown), and we speculated that in BrCSCs, snai1 may function in miR-146 expression deregulation in a transcription independent way. We first identified that snai1 was critical for sustaining the activation of Wnt signaling (Figure 7(a)), and snai1 exerted the similar function in regulating miR-146 expression level, as the TCF-4 did (Figure 7(b)), suggesting the snai1 manipulation of Wnt signaling is universally pivotal for sustaining of miR-146 expression. In every kind of breast cancer cell line, snai1 all acts as the crucial part in promoting miR146 expression level (Figure 7(c)). The Snai1 stimulation of Wnt activity promotes the miR-146a maturation, which repressed Wnt signaling activation through participating in Let-7/Wnt/H19 feedback loop (Figure 7(d)). The circle of miR-146/Wnt and Let-7/Wnt contributed to form the complicated feedback loop of miR-146/Let-7/Wnt cascade (Figure 7(d)).
Figure 7.
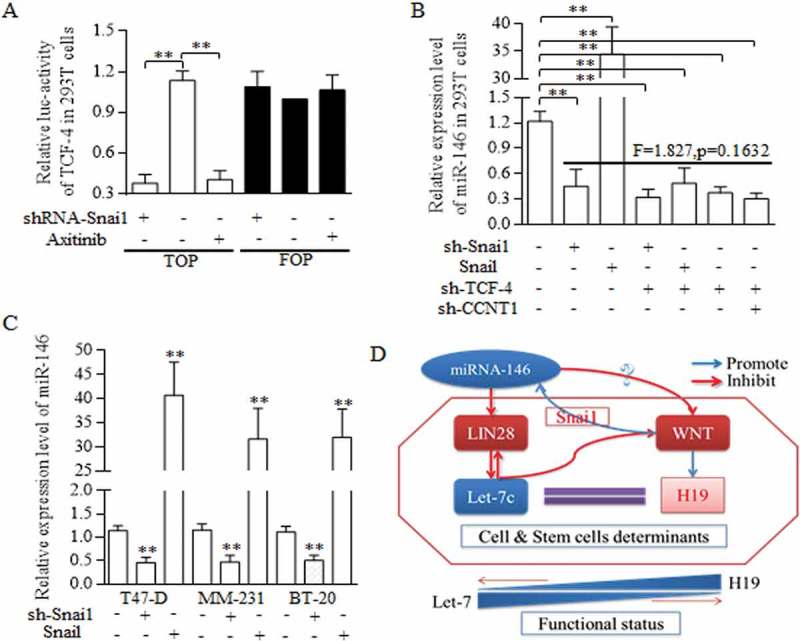
Regulatory loop of miR-146 and Wnt signaling was dependent on the fine regulating conducted by Let-7/Wnt/H19 feedback circle.
A reciprocal regulation between miR-146a and Wnt signaling pathway was dominated and conducted by Snai1. A. TOP/FOP reporter assay. Both Snai1 inhibition and Axitinib addition decreased the TCF-4 activity effectively in 293T cells, comparing to negative and control group, ** p<0.01 (Student’s t-test). B. In 293T cells, snai1 inhibition repressed the miR146 expression level greatly, and Snail transfectants promoted the miR-146 level significantly. What’s more, no significant difference was detected when analyzing the functions of either snai1 inhibition, TCF-4 inhibition, or their combined manipulation in regulations of miR-146 expression. C. Similar results were confirmed in T47-D, MM-231, and BT-20 of breast cancer cell lines, proving the pivotal role of miR-146 in controlling miR-146 level. D. The schematic illustration of the proposed regulative mechanisms of miR-146 in breast cancer stem cells. Two crucial circles formed based on the dual regulation of miR-146/Wnt and Let-7/Wnt loop, which was connected by LIN28. Data represent mean ± s.d, n = 3 independent experiments. Results were acquired and calculated from three independent experiments.
4. Discussion
The stem cells helped to maintain the tumor family as the seed, generating the therapy resistance and could survive through systemic delivery of anti-cancer medicine and agents [17,18]. Precise strategy that could target at these subtype enemies could specifically help to get over the long-term recurrence and improve the therapy response [19,20].
Non-coding genes were involved in the stem cells’ potency regulations, and were proved to be of critical importance in disorders of cancer stem-like cells’ renewal [21,22]. The non-coding miRNAs were first found to affect the downstream mRNAs in a post-transcriptional manner, and numerous miRNAs have been detected and identified through bioinformatic and bench work, and even more, tentative study of clinical application were also carried out [23]. Long noncoding RNAs partially functioned the same as miRNAs did, however, one crucial difference is the way of its regulation of the targeted miRNAs. Different division status resulted in diverse outcomes of cancer stem cells group after splitting once, and the interference during the dividing procedure was crucial for controlling the stem cells’ renewal. Based on studies of stem cells formation, we further turn to exploring the asymmetric division prior to spheres number counting. Division modes could be defined with several staining assays, and the EdU dye staining was proved practicability and reliability when defining the status. To identify the possible role of miR-146 in universal stem cells of breast cancer, we selected three cell lines of breast cancer. After screening of the clinical significance of miR-146 and Let-7 family, Let-7c was selected as the candidate, and Let-7c inhibited the self-renewal ability of BrCSCs most efficiently. Lots of common sense were found when comparing their function in regulating the stem cells ‘renewal, and both miR-146 and Let-7c inhibited the activity of Wnt signaling effectively, stimulated the asymmetric division.
Wnt signaling activation, which we hypothesized to be the potential functional cascadein miR-146 regulated stem cells’ potency, were correlated to be with poorer therapy outcomes after clinical analysis, and were proved to be the most crucial node to realize the miR-146 functions. Let-7c were controlled by miR-146 through a LIN28 dependent way, and Let-7c itself formed the positive feedback loop with H19 to strengthen the Wnt signaling suppression. The molecular sponge of H19 bound to and arrested the Let-7c to stop its functioning of downstream genes deregulation. At last, miR-146 was involved in the dual circles of Let-7/Wnt signaling and Snai1/LIN28 pathways.
In conclusion, our study was trying to detected the functions of non-coding RNAs in dividing and renewing of BrCSCs, and the connected network of every in volved factor was important to implement the final functions, when aiming to controll the division manners and to restrain the stem cells’ expansion. Stem cells was obstacle when conquering the cancer empire, and symmetric division suppression could help to realizing the curing of cancer.
Funding Statement
This work was supported by the National Natural Science Foundation of China [81602597].
Acknowledgments
The authors acknowledge assistants in the Center for Translational Medicine of The First Affiliated Hospital of Xi’an Jiaotong University, for their technical assistance. This experiment was supported by National Science Foundation for Young Scientists of China, grant No. 81602597 (Referred to Xin Sun).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Xiao G, Meng J, Zhang J, et al. Clinical application of detecting 21-gene recurrence score in predicating prognosis and therapy response of patients with breast cancer from two medical centers. Cancer Invest. 2017;35:639–646. [DOI] [PubMed] [Google Scholar]
- [2].Xu C, Xiao G, Zhang B, et al. CCAT1 stimulation of the symmetric division of NSCLC stem cells through activation of the Wnt signalling cascade. Gene Ther. 2018;25:4–12. [DOI] [PubMed] [Google Scholar]
- [3].Sun X, Liu J, Xu C, et al. The insights of Let‐7 miRNAs in oncogenesis and stem cell potency. J Cell Mol Med. 2016;20:1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shima H, Kida K, Adachi S, et al. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res Treat. 2018;170:507–516. [DOI] [PubMed] [Google Scholar]
- [5].Chen S, Zhu J, Wang F, et al. LncRNAs and their role in cancer stem cells. Oncotarget. 2017;8:110685–110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lanczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. [DOI] [PubMed] [Google Scholar]
- [7].Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sun X, Xu C, Tang SC, et al. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther. 2016;23:83–89. [DOI] [PubMed] [Google Scholar]
- [10].Sun X, Tang S-C, Xu C, et al. DICER1 regulated let-7 expression levels in p53-induced cancer repression requires cyclin D1. J Cell Mol Med. 2015;19:1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sun X, Jiao X, Pestell TG, et al. MicroRNAs and cancer stem cells: the sword and the shield. Oncogene. 2014;33:4967–4977. [DOI] [PubMed] [Google Scholar]
- [12].Xiao G, Zhang B, Meng J, et al. miR-367 stimulates Wnt cascade activation through degrading FBXW7 in NSCLC stem cells. Cell Cycle. 2017;16:2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hwang WL, Jiang JK, Yang SH, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268–280. [DOI] [PubMed] [Google Scholar]
- [14].Peng F, Li -T-T, Wang K-L, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8:e2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kallen AN, Zhou X-B, Xu J, et al. The imprinted H19 LncRNA antagonizes Let-7 microRNAs. Mol Cell. 2013;52:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nassar D, Blanpain C.. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76. [DOI] [PubMed] [Google Scholar]
- [18].Bandhavkar S. Cancer stem cells: a metastasizing menace! Cancer Med. 2016;5:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jayachandran A, Dhungel B, Steel JC. Epithelial-to-mesenchymal plasticity of cancer stem cells: therapeutic targets in hepatocellular carcinoma. J Hematol Oncol. 2016;9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kim SH, Ezhilarasan R, Phillips E, et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-kappaB-dependent manner. Cancer Cell. 2016;29:201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1:93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tordonato C, Fiore PP D, Nicassio F. The role of non-coding RNAs in the regulation of stem cells and progenitors in the normal mammary gland and in breast tumors. Front Genet. 2015;6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Flynn RA, Chang HY. Long noncoding RNAs in cell fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


