Abstract
Mitochondrial disease is a multifactorial disorder involving both nuclear and mitochondrial genomes. Over the past 20 years, great progress was achieved in the field of gene editing which raised the possibility of partial or complete elimination of mutant mtDNA that causes disease phenotypes. Each cell contains thousands of copies of mtDNA which can be either wild-type (WT) or mutant, a condition called heteroplasmy. As there are multiple copies of mtDNA inside a cell, the percentage of mutant mtDNA can vary and a directional shift in the heteroplasmy ratio towards an increase of WT mtDNA copies would have therapeutic value. Gene editing tools have been adapted to translocate to mitochondria and were able to change heteroplasmy in a predictable manner. These include mitochondrial targeted restriction endonucleases, Zinc-finger nucleases, and TAL-effector nucleases. These procedures could also be adapted to reduce the levels of mutant mtDNA in embryos, offering an option to the controversial mitochondrial replacement techniques during in vitro fertilization. The current strategies to induce heteroplasmy shift of mtDNA and its implications will be comprehensively discussed.
Keywords: Mitochondrial DNA, Heteroplasmy, Restriction Endonucleases, Mitotalens, mitoZFNs, Review
2. INTRODUCTION
Mitochondria are unique organelles that actively participate in cell metabolism and homeostasis regulation. Most of the energy production is achieved by electron transfer in the respiratory chain through oxidative phosphorylation (OXPHOS) (1, 2). Aerobic tissues rely on OXPHOS for ATP production and the overall process is highly controlled involving the integration and balance of the oxidation of fatty acids, pyruvate, ketoacids and a range of other intermediary metabolites and regulatory ions such as calcium.
The human mitochondrial DNA (mtDNA) is a small double stranded circular genome which is maternally inherited (3). Each mammalian cell contains in average one thousand copies of mtDNA and each molecule contains 37 genes, encoding ribosomal RNAs (12S and 16S), 22 transfer rRNAs (tRNAs) and 13 OXPHOS complexes subunits (4). Defects in the mtDNA, both point mutations and large scale rearrangements, have been associated with severe mitochondrial syndromes, which cause diverse phenotypes including: stroke-like episodes, cardiomyopathies, ragged red fibers, optic neuropathies, neurodegenerative disorders, among others. When pathogenic mutations occur in the mtDNA most often both mutant and wild-type copies co-exist within the same cell, a phenomenon known as heteroplasmy (5), and, in general, only when the mutation load is higher than approximately 80% symptoms manifest (6). Mitochondrial diseases have a prevalence of roughly one in 5000 live births, with only 8% of children with mitochondrial disease diagnosed before one year of age, while 28% are diagnosed by age 18 (7). Currently, there are no effective strategies to cure mitochondrial disease and, in spite of the advances in genetics and biotechnology, there are still some gaps in the understanding of mitochondrial genetics. For instance, we do not know what controls mtDNA copy number (8), and mechanisms of mtDNA replication are still controversial (9, 10). Moreover, there is an emerging need for in vivo models and studies that could be used to explore these crucial questions of mitochondrial biology.
During the past 16 years our lab and others have been focusing in the use of endonucleases to target mitochondria and induce double strand breaks (DSB) in the mtDNA. Taking advantage of the fact that mitochondria lack an established DSB repair mechanism, it has been shown that mtDNA is quickly degraded after a DSB (11). Therefore, heteroplasmy can be manipulated and the mutant genomes can be efficiently eliminated through cleavage of mutant mtDNA and repopulation of the cells with WT mtDNA. Recently, a new door has been open regarding translation of these techniques into the clinics by the development of precise DNA editing tools, which can be targeted to mitochondria to promote DSB in the mtDNA. The current gene editing tools to shift mDNA heteroplasmy, their molecular and cellular implications and possible applications will be comprehensively discussed in this review.
2.1. Mitochondrial energy production and genetics
Mitochondria are under a dual genetic control of both mitochondrial and nuclear genomes. Human mtDNA is a 16,569-bp double stranded circular genome which is located within the matrix and is packaged in protein-DNA complexes known as nucleoids. Each nucleoid contains one or two mtDNA molecules (12). Because of its proximity to the electron transport chain, mtDNA is highly exposed to oxidative damage and therefore prone to mutations (13, 14). In fact, many studies suggested that mtDNA has a higher mutation rate when compared to the nuclear genome (13) and repair is less efficient than in the nucleus (15, 16). Nonetheless, other studies contradict this concept. For instance, the biomarker 7,8-dihydro-8-oxoguanine (8-oxoG), the oxidized form of guanine, has been a pivotal marker for measuring the effect of endogenous oxidative damage to DNA and it is often associated with carcinogenesis and neurodegenerative diseases. Its highly mutagenic potential is believed to be caused by misincorporation of an adenine instead of cytosine causing G: C→T: A transversion mutations. It has been reported that repair of the damage caused by 8-oxoG in Chinese hamster ovary fibroblasts, was faster in the mitochondrial DNA than in the nuclear DNA, in a non-strand specific manner (17). Another study showed that the levels of oxidatively induced base lesions in mitochondrial DNA were not raised in livers of old wistar rats (18). The authors not only concluded that age is not playing a role in increasing the oxidative damage but also the mtDNA damage was not increased when compared to the nuclear DNA (18). The comparison of mtDNA damage with nuclear DNA, as well as the role of mitochondrial-induced oxidative damage in aging, are still topics of controversy, which will depend on the development of more accurate techniques and studies (19, 20). However, it is curious that by simply analyzing the whole tissue instead of isolated rat liver mitochondria, the levels of 8-oxoG were surprisingly lower than in isolated mitochondria and were also not higher than in the nuclear DNA (21). The authors of this study study and others concluded that it is then likely that the mitochondrial DNA oxidative damage levels have been overestimated due to technical artifacts, over the past years (19, 21).
Mitochondria are considered to be the powerhouse of the cells as they are the main energy producers. Shortly, pyruvate is channeled to enter mitochondria via pyruvate dehydrogenase (PDH) yielding acetyl CoA, NADH + H+, and CO2. Acetyl CoA enters the Krebs cycle or Tricarboxylic Acid (TCA) cycle generating NADH + H+ (22). Fatty acids are oxidized in mitochondria via β-oxidation to generate Acetyl-CoA, NADH + H+ and reduced FAD co-factor. Two electrons are then transferred from NADH to the OXPHOS complex NADH dehydrogenase (Complex I) or from FADH2-containing enzymes as the electron transfer factor dehydrogenase or succinate dehydrogenase (SDH, complex II) to reduce ubiquinone (coenzyme Q10, CoQ) to ubiquinol (CoQH2). The electrons are then transferred to complex III (bc1 complex), cytochrome c, complex IV (cytochrome c oxidase, COX), and finally to oxygen, generating H2O (23). Coupled to the electron flow down the chain is the pumping of protons across the mitochondrial inner membrane space by complexes I, III and IV, which creates a proton electrochemical gradient (Δp=ΔΨ+ΔμH+) which is acidic and positive in the intermembrane space and negative and alkaline in the matrix side. The potential energy (also called driving force) stored in the proton motive gradient (Δp) is used to aid in the import of proteins and calcium to the mitochondrial matrix, to generate heat and to synthesize ATP (2). The maximal proton motive force across the inner membrane is around 180–220 mV (24). The energy to convert ADP+Pi to ATP comes from the flow of protons through the ATP synthase (complex V) back into the matrix. The matrix ATP is also exchanged by cytosolic ADP through the ANT (adenine nucleotide translocator) located in the inner mitochondrial membrane (25). The production of ATP is dependent on the efficiency by which the protons are pumped out of the matrix by the electron transfer chain (ETC) complexes and by the efficiency of proton flux through complex V. However, this machinery is actually prone to proton leakage, leading to variations in the coupling efficiency of the respiratory chain (26). Furthermore, alterations in the coupling efficiency can influence the generation of reactive oxygen species (ROS), ultimately leading to apoptosis (27). Finally, each of the ETC complexes incorporates multiple electron carriers. Complex I and II utilize flavins and iron-sulfur (Fe-S) centers (25). The Krebs cycle enzyme aconitase also utilizes a Fe-S group, susceptible to oxidative stress. Complex III encompasses a Fe-S center plus cytochromes b and c1-Complex IV encompasses two Cu centers plus cytochromes a and a3 (25). The importance of mitochondria in the cell cannot be overemphasized thus pathogenic mutations that affect any of the mitochondrial components will disturb cellular function.
As mentioned above, the 37 genes are encoded by the mtDNA include: the large and small RNA (12S rRNA and 16S rRNA), 22 transfer rRNAs (tRNA) and 13 key respiratory chain (RC) subunits. The structural proteins encoded by mtDNA include seven subunits from Complex I (ND1-ND6 and ND4L), one subunit from Complex III (cytochrome b), three subunits from Complex IV (COXI- III), and two subunits from Complex V (ATPase 6 and 8) (28). The remaining of the mitochondrial proteins, including the ones involved in maintenance, expression, transcription and translation, dynamics, are actually nuclear encoded and synthetized in the cytosol prior to mitochondrial import. Therefore, the optimal functioning of the mitochondria is crucially dependent of these two distinct genomes and their dynamic crosstalk. The mitochondrial genetic code also differs from the nuclear. In vertebrates, codons AUA (which require 5-formylcitidine (F5C) present at the wobble position, (29)) and AUG code for the first methionine, UGA codes for tryptophan (in contrast to the nuclear genome whereas is a stop codon), and AGA and AGG were thought to read as stop codons (unlike arginine, as in the nuclear genome) (30), until recently. It has now been shown that at least in humans a −1 mitoribosome frameshift occurs at the AGA and AGG codons predicted to terminate the CO1 and ND6 ORFs, and consequently both ORFs terminate in the standard UAG codon (31). There are several other unique features of the mtDNA genome that are crucial to understand the mechanisms of mitochondrial disease, but the most important are: 1) mitochondrial DNA is maternally inherited, 2) there are multiple copies of mtDNA within each cell, and mutations can occur in all (homoplasmy) or in a proportion of the molecules (heteroplasmy); 3) in the presence of heteroplasmy, there is a threshold effect and mitotic segregation of the mtDNA, leading to different penetrance of bioenergetic defects; 4) the transmission of the heteroplasmy levels from the mother to the offspring is often random and unpredictable, determined by a “bottleneck” germline transmission. These concepts will be discussed and clarified in the next section.
2.2. Maternal inheritance of the mtDNA
There are multiple copies of mtDNA in both somatic and germ cells. As it was previously mentioned, the inheritance of mtDNA is exclusively maternal, although a rare exception has been reported (32). The clinical importance of this phenomenon is that pathogenic mutations in an affected mother are transmitted to all her offspring, but only females will transmit the mutation to their children. A disease that affects both genders but that is not transmitted by the paternal line is highly suggestive of a mtDNA deficiency (33, 34). There are two main processes that may explain why the mtDNA present in sperm is not transmitted to the next generation; first, there is a significant downregulation of the mtDNA copy number during spermatogenesis, the so-called “dilution effect”. Second, sperm mitochondria undergoes ubiquitination, after fertilization. Consequently, the mtDNA content of the zygote was thought to be determined exclusively by the previously unfertilized egg (35, 36). Recently, a mathematical model suggested that the selection against heteroplasmy may explain the uniparental inheritance of mitochondria (37).
2.3. Heteroplasmy of mtDNA and mitotic segregation
Pathogenic mutant mtDNA often co-exists with wild-type mtDNA within the same cell (mtDNA heteroplasmy, (33)). The percentage of mutant mtDNA can vary between 0 and 100%. A minimum number of mutant mitochondrial genomes is required for the expression of a mitochondrial dysfunction or disease, and this is called the “threshold effect” (38). This effect is relative, as the percentage of mutated genomes necessary to impair mitochondrial function varies according to the metabolic requirements of the cell or tissue at any given time, the particular base pair/gene involved and, the nuclear genetic background (38–40). In general, the higher the levels of mutant mtDNA the more severe the disease state is, although occasional exceptions exist (41). Furthermore, mutations in the mtDNA may contribute to common age-related disease phenotypes including blindness, deafness, cardiovascular and neurological disease, renal and endocrine dysfunction (33). It is generally assumed that age-associated mtDNA mutations are caused by the accumulation of damage during aging (25); but, recent studies support the hypothesis that somatic mtDNA mutations created during embryogenesis may contribute to aging phenotypes in adult life (42, 43).
Mitochondria replicate and divide at any time during the cell cycle and the proportion of mutant mtDNA passed onto the daughter cell at cell division may not be identical to that of the parental cell (44). Besides, in response to metabolic demands of the whole cell, WT and mutant mtDNA may replicate at varying degrees during cell cycle. Therefore, the percentage of mutant mtDNA may change quickly between the parent and the daughter cells due to a random drift (a phenomenon known as mitotic segregation) (40). This phenomenon of mitotic segregation of heteroplasmic mtDNA mutations in somatic tissues, and, the principles for maternal transmission of mtDNA are cornerstones of mammalian mitochondrial genetics (33). Unfortunately, the mechanisms of these events are not clearly understood at the cell or molecular levels despite their fundamental importance.
2.4. The mtDNA genetic bottleneck hypothesis and germline transmission
It has been reported that a phenotypically healthy mother harboring 50% of mutant mtDNA can produce both healthy and severely affected offspring (45). The reason behind this generational shift is designated as the “bottleneck effect” (46). A restriction in the number of mtDNA molecules or groups of molecules arranged in segregating units allows oocytes to inherit a sub-set of the mother’s mtDNA pool. As a consequence, the heteroplasmy levels in children of an affected mother can be remarkably variable with respect to the maternal heteroplasmy, while the average heteroplasmy across the offspring is often comparable to that of the mother (46). Rapid intergenerational shifts in the level of mtDNA heteroplasmy levels were first observed in Holstein cows (47), and subsequently documented in human pedigrees transmitting pathogenic mtDNA mutations (35, 48). Observations in mice also supported this phenomenon of selective transmission (bottleneck effect), since it was observed that female mtDNA genomes repopulating the germline was restricted to homogeneous haplotypes (49, 50).
Random genetic drift mechanisms acting in all heteroplasmic mtDNA mutations were revealed by limited data analysis (35). Two laboratories have measured a dramatic reduction in the amount of mtDNA present in the first discernible germ cells (the primordial germ cells) that appear shortly after implantation in the female embryo (49, 50). By crossing heteroplasmic mice with others expressing fluorescent germ-cell markers, it was shown that the majority of heteroplasmy variation and migration, and the levels within individual oocytes, are actually determined before birth (51). These data are compatible with a preferential replication of a subpopulation of molecules either before or after birth, or some form of compartmentalization of the mtDNA, speeding up the genetic drift.
More recently, it has been hypothesized that there is a purifying selection mechanism in the maternal germ line that decreases the inheritance of mutant mtDNA (52, 53). In humans, females with high levels of mtDNA mutations in the germline have been associated with decreased fertility. Recently, the largest in silico analysis to date was performed in 577 mother-child pairs transmitting different mtDNA mutations. The study revealed that different mtDNA mutations segregate at different rates in human pedigrees (54). However, they found no evidence of selection during transmission. Certain pedigrees such as m.8993T>G/C segregated faster than the others analyzed in this study. These findings can help us understand the inheritance risks of heteroplasmic mtDNA diseases, and their prevention using conventional techniques such as prenatal and pre-implantation diagnosis. Furthermore, strategies that aim to reduce the mtDNA content could also lead to a faster segregation of heteroplasmic alleles increasing the probability of generating germ cells with very low mutant mtDNA levels or very high as well, which may not be viable at an early stage pregnancy (54). Interestingly, a very recent study corroborates this theory showing that gene editing strategies such us mitochondrial targeted-restriction endonucleases and mitoTALENs were able to prevent germline transmission of specific mtDNA haplotypes, by shifting the mtDNA ratio towards the wild-type mtDNA molecules, by the elimination of mutated mtDNA in oocytes and embryos (55). Moreover, this study provided a new road in the search for an effective treatment for individual mitochondrial diseases, and it will be discussed below in section 3.
3. MITOCHONDRIAL DISEASE AND HETEROPLASMY
Currently, more than 200 point mutations and large scale rearrangements in the human mtDNA are known to be associated with a large diversity of clinical symptoms, ranging from progressive muscle weakness to fatal infantile disorders (56). The first molecular evidence that heteroplasmic mtDNA mutations (deletions) could cause disease was demonstrated in 1988 by Holt and others (57). In addition, Wallace and others (58) also reported that a homoplasmic missense mutation in the ND4 gene (m.11778G>A) was maternally inherited and responsible for the Leber’s hereditary optic neuropathy (LHON). It was also discovered that myoclonic epilepsy and ragged red fiber (MERRF) disease was caused by an heteroplasmic A>G transition mutation in position m.8344 in the tRNALys gene (59, 60). These interesting findings set the stage for the research of mitochondrial familial and age-related diseases. With the advance of molecular techniques, the following decade saw the description of several hundred different point mutations and deletions in patients with a wide range of clinical phenotypes (12). By contrast to what was thoughtmutations in the mtDNA are relatively common. It was reported that the incidence of clinical mitochondrial diseases is about one in 5,000, including the most common pathogenic mutations (60, 61). Furthermore, it was also revealed that one in 200 newborns harbored some level of one of 10 common pathogenic mutations (62, 63). Because of the high rate of mutations in the mtDNA, new pathogenic mutations are recurrently introduced into the human population. When a mutation arises in the mtDNA, there is an intracellular mixture of mutant and wild-type mtDNA molecules (as it was discussed previously), in some manner the initial mutant mtDNA becomes enriched within certain cells, and predominates influencing the cellular patient phenotype. In general, organs with very high energy demand require a very efficient ATP production, consequently are the first ones to reveal the consequences of partial mitochondrial bioenergetic deficits due to pathogenic mutations. The most affected organs by these mutations are the brain followed by the heart, muscle, kidney, and endocrine systems (64). Therefore, a mitochondrial disease or syndrome can be defined as a chronic loss of cellular energy that results in a clinical phenotype. MtDNA point mutations are usually maternally inherited, with multiple individuals of the same family being affected, while mtDNA deletions are rarely inherited and are never homoplasmic. Actually, homoplasmic mtDNA point mutations cause in general a mild biochemical defect that typically affects only one organ or tissue (with some exceptions, (65, 66)). In contrast, heteroplasmic mutations affect multiple organs and the levels of heteroplasmy usually correlate with degree of the clinical phenotype, which is severe in the affected tissues. Recently, next-generation sequencing technology has been used to identify and quantify mtDNA mutations (67). However, these techniques have a high intrinsic error rate when applied to detection of low-level heteroplasmy. The data can be contaminated with pseudogene sequences present in the nuclear DNA which are derived from mitochondrial DNA, designated as nuclear mitochondrial DNA sequences (NUMTs), and because of this, it can be particularly challenging to accurately filter the data (68).
Despite all the technological difficulties, it is believed that mtDNA heteroplasmy exists in almost every healthy individual studied, even though at very low levels (69). These heteroplasmic variants can also be passed down the maternal lineage, raising the possibility that some presumably somatic mutations measured late in life are actually low-level heteroplasmies that have been inherited and somehow clonally expanded (e.g. in tumors).
Mitochondrial DNA polymerase gamma (POLG) is a heterotrimeric enzyme containing a Pol I-like catalytic core (PolgA) and an accessory subunit (70). A provocative report in 2013 showed that heterozygous mice for the mitochondrial DNA mutator allele (PolgA) have reduced fertility due to maternally transmitted mtDNA mutations (43). The phenotype is aggravated in PolgA (mut/mut) showing signs of premature ageing. The authors concluded that a pre-existing mutation load increases clonal expansion of mtDNA mutations, in a similar manner maternally transmitted mutations exacerbate the aspects of normal human aging (43).
3.1. Point mutations
Inherited point mutations primarily occur in protein coding tRNA genes and result in a reduction of cellular energy, via reduction in the activity of the electron transport chain respiratory complexes or due to an impairment of mitochondrial protein synthesis (71). Point mutations are usually rare and are often heteroplasmic. The threshold levels for the disease onset are usually between 80–90%. Many classical syndromes have been comprehensively described over the last few decades. A few examples of different syndromes and their associated-point mutations will be shortly described.
3.1.1. Mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS)
Mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS) is typically manifested in childhood and the symptoms range from tonic-clonic seizures, recurrent headaches, and anorexia with vomiting and postlingual hearing loss but can also be associated with motor, vision and mental impairment due to the accumulation of stroke-like episodes (72, 73). The mitochondrial 3243 A>G in MT-TL1 accounts for 80% of the cases (74), but is also associated with variants in ND5 (75), ND1 complex I subunits and MT-CYB. Therefore, defects in complex I and IV activities have been described (76, 77).
3.1.2. Myoclonic epilepsy with ragged red fibers (MERRF)
Myoclonic epilepsy with ragged red fibers (MERRF) is a neuromuscular disorder associated with m.8344 A>G mt-tRNALys (MTTK) gene mutation (60, 78). The clinical manifestations are myoclonus, epilepsy, muscle weakness, cerebellar ataxia and dementia, and neurological symptoms can evolve with age (59, 60). High heteroplasmic levels of this mutation have been associated with complex I and IV deficiencies (45, 59, 78). This point mutation has also been associated with Leigh’s syndrome (79, 80).
3.1.3. Leber’s hereditary optic neuropathy (LHON)
Leber’s hereditary optic neuropathy (LHON) is a common cause of inherited blindness presenting bilateral, painless, sub-acute visual failure in young adult males (81). This was the first maternally inherited disease to be associated with mtDNA point mutations (58, 82, 83). There are three common point mutations associated with this syndrome that can be diagnosed by molecular genetic analysis, such as m.3460 G>A, m. 1178 G>A and m. 14484 T>C (84). These mutations are associated with complex I defects and LHON mutations are typically homoplasmic (85).
3.2. Large scale rearrangements and deletions
In contrast to point mutations, primary mitochondrial rearrangements of mtDNA are not inheritable, they are sporadic. Large-scale deletions are typically heteroplasmic and result in disease. To date, roughly 120 different mtDNA deletions have been found in patients with mitochondrial disease. In this case, the heteroplasmic threshold is reported to be lower than the one for point mutations, the patients manifest the disease symptoms with as low as 50–60% heteroplasmic mtDNA levels (86). Two different models arise to explain deletions in the mtDNA, while one points to replication errors, the other one points to poor and inefficient mtDNA repair mechanisms. MtDNA deletions are associated with three main clinical phenotypes: Kearns-Sayre syndrome (KSS) (87), sporadic progressive external ophthalmoplegia (PEO) (88) and Pearson’s syndrome (89). Mitochondrial dysfunction manifests usually with ragged red fibers (RRFs), which is the accumulation of dysfunctional mitochondrial in the sub-sarcolemmal region detected by Gomori trichrome staining (88). Person’s bone marrow pancreas syndrome is also a rare infantile disorder characterized by sideroblastic anemia including severe exocrine pancreatic insufficiency (89). Pearson’s patients have high levels of mtDNA deletions in affected organs, and might develop KSS later in life (90).
Finally, many mitochondrial disorders have been associated with reduction of the mtDNA copy number (91). Alpers syndrome, has been associated with mutations in POLG, which can impair mtDNA replication (92, 93). In addition, there is a plethora of mutations in nuclear genes coding for proteins associated with nucleotide metabolism and availability which affect mtDNA integrity. Recessive mutations in thymidine phosphorylase (TP) cause neurogastrointesinal encephalopathy with mtDNA depletion, multiple deletions and point mutations (94, 95). Mitochondrial DNA depletion has also been found in early onset of hypotonia with myopathy and hepatic involvement, caused by mutations in thymidine kinase 2 (TK2) or deoxyguanosine kinase (DGUOK), which reduced replication efficiency (96–99).
4. CURRENT STRATEGIES TO SHIFT MTDNA HETEROPLASMY: THE BRIDGE TOWARDS CLINICS
The concept of shifting the balance between healthy and mutated mtDNA as a treatment for heteroplasmic mtDNA disease has been under investigation over the past 20 years. Many publications demonstrated that it is possible to manipulate the mtDNA and shift heteroplasmy, either in vitro or in vivo. By simply reducing the levels of the mutant allele below a certain threshold, an improvement in pathology is achieved (11, 100–102).
Metabolic strategies to shift mtDNA heteroplasmy have been tested and evaluated throughout the last decades. It was shown that treating cybrids harboring a large-scale partial deletion with ketogenic supplements (deprived of glucose) was enough to shift mtDNA heteroplasmy towards wild-type mtDNA, rescuing metabolic function. This improvement in function though was not associated with any obvious decrease in the mutation levels (103). A possible explanation lies in the fact that the ketogenic stress diluted the deleterious effect of the mutation, because cells contained more mitochondria. Another example of heteroplasmy changes through metabolic intervention was trough exercise. Activation of satellite cells proliferation is known to occur in response to resistance exercise and muscle fiber injury (104, 105). In 1999, a report from a patient that started a program of resistance exercise showed an increase in the proportion of wild-type mtDNA molecules after the training (106). A larger study was performed years later by Murphy et al., showing that after 12 weeks of resistance training, many positive physiological changes occurred in the patients, including improved strength and oxidative capacity in the trained muscles; however no decrease in the proportion of mutated mtDNA was detected (107). Other studies followed these and the same conclusion was reached, no change in the heteroplasmy and if so, there was an increase in the mutant mtDNA instead of wild-type. In a similar manner, promoting mitochondrial biogenesis through overexpression of PGC-1α/β improved partially the OXPHOS in cells with a pathogenic mtDNA mutation (108). Again, this improvement was not associated with a heteroplasmic shift. Despite these concerns, a clinical trial was advanced investigating exercise versus inactivity in a cohort of patients with mitochondrial myopathy (109).
The idea of shifting the mtDNA heteroplasmy was finally efficiently demonstrated by the use of mitochondrial-targeted restriction endonucleases both in mouse and human-derived cell lines. This strategy has heralded a new era for mtDNA gene editing and heteroplasmy shift.
4.1. Mitochondrial-targeted restriction endonucleases (REs)
As it was previously discussed, mitochondria do not have an established/accepted double strand break repair mechanism and the little evidence that it may occur comes mainly from work from our lab and others using mitochondrial targeted restriction endonucleases (REs). These are recombinant REs with mitochondrial localization signals (MLSs) that are imported to the mitochondrial matrix, accessing mtDNA and creating double strand-breaks. A hybrid cell line containing both rat and mouse mtDNA was used to test if mitochondrial PstI could alter mtDNA heteroplasmy. Because the mouse mtDNA has two PstI restriction sites, and the rat none, upon expression of the endonuclease a significant shift in heteroplasmy was observed (110). These results suggested that cleavage of mtDNA leads to the degradation of the targeted mtDNA molecules and expansion of the residual mtDNA species (Figure 1). This line of research was applied to the pathogenic m.8993 T>G mutation in the mitochondrial ATP6 gene, which is known to cause Leigh’s syndrome or neurogenic muscle weakness, ataxia and NARP (neuropathy, ataxia, and retinitis pigmentosa). This particular mutation creates a unique SmaI restriction site that could be targeted by a mitochondrially-targeted SmaI enzyme in heteroplasmic cells harboring m.8993 T>G mutation. This strategy resulted in the depletion of the mutated mtDNA, accompanied by an increase of WT mtDNA molecules (111). Later, XmaI (SmaI isoschizomer) was used to target the same mutation in human cybrids and in this study an adenovirus was used. At a viral multiplicity of infection of 50, the point mutation was effectively eliminated from cells with high mutation load, leading to the restoration of OXPHOS. However, it took five cycles of transfection to completely eliminate the mutated mtDNA (112).
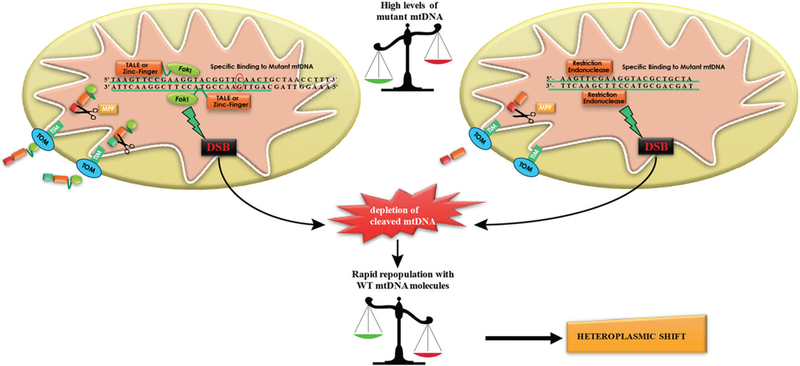
Figure 1.
Schematic representation of the action mode of mitochondrial-targeted restriction endonucleases and designer endonucleases such as mitoZinc-fingers and mitoTALENs. First, the cytosolic synthesized unfolded proteins are imported into the mitochondrial matrix through the outer and inner mitochondrial import machinery (TOM/TIM complexes). When inside the matrix they are folded by molecular chaperones. The mitochondrial localization signal/sequence (MLS) is cleaved by a mitochondrial peptidase (MPP) and the mature protein is then ready to bind a specific mtDNA haplotype. When the two monomers are close enough, FokI dimerizes and either the mitochondrial targeted-Zinc finger or TALEN cleave the specific mtDNA by inducing double-strand breaks. A similar pathway occurs with a mitochondrial targeted restriction endonuclease which will bind to the respective restriction site in the mtDNA. Since mitochondria do not have an effective double-strand break repair mechanism, most of the targeted mtDNA is degraded leading to a repopulation of the cell with the residual mtDNA (mostly WT), inducing a shift in heteroplasmy towards the WT.
The use of mitochondrial-targeted REs was also shown to be effective in vivo. Our group further advanced the potential of this concept and proceeded with in vivo studies utilizing the NZB/BALB heteroplasmic mouse model (11). This mouse model was created by cytoplasmic fusion in single cell embryos (113). The BALB mtDNA variant harbors a unique ApaLI site that is absent in the NZB mtDNA mouse variant. Inducible expression of mito-ApaLI in cells derived from these mice showed a rapid, directional and complete shift in mtDNA heteroplasmy, resulting in transient depletion. The initial depletion correlated with the initial levels of the target BALB mtDNA. By using recombinant viral vectors expressing the mito-ApaLI it was observed a significant shift in the mtDNA heteroplasmy, reflected by a decrease of BALB mtDNA levels in the muscle and brain transduced with recombinant viruses. It is also important to notice that in this study a shift in heteroplasmy seen in the brain was achieved with the use of AAV1,2-based viral vector (11). In addition, recombinant adenovirus (rAd5-Mito-ApaLI) transduction was very effective in the muscle fibers of 5-day-old heteroplasmic mice. This study showed for the first time that this strategy could be used in vivo. Nevertheless, this concept was limited to mutations that create unique restriction sites, such as NARP, which are unfortunately rare. Our group attempted to extend the approach to a multiple “cleavage site model”, by the use of a different restriction endonuclease- ScaI, which recognizes three sites in the BALB mtDNA and five in the NZB mtDNA (114). In a similar manner to the previous study, a “mammalianized” mito-ScaI enzyme construct was designed and Ad5 was injected in the jugular vein, to target the liver and local injection was used to target the muscle. A shift towards the BALB mtDNA that has fewer ScaI restriction sites was observed in both tissues. However, this approach resulted in a lower magnitude of the shift when compared with a single differential cleavage of mito-ApaLI. Moreover, it was more prone to trigger severe mtDNA depletion in treated cells. NZB/BALB mtDNA heteroplasmy could also be manipulated in heart, using a cardiotropic adeno-associated virus type-6 (AAV6) and in liver, using the hepatotropic adenovirus type-5 (Ad5) (115). Another study showed that mitoApaLI expressed from AAV9 particles when delivered by intraperitoneal or intravenous injections in neonates, promoted dramatically shifts in heteroplasmy in all striated muscles, including heart (116).
4.2. Mitochondrial-targeted Zinc-fingers
One way to solve the limitation of REs is to develop sequence-specific nucleases that could be designed to target any sequence of the mtDNA. Zinc-fingers technology allows the engineering of zinc-finger proteins that can bind any predetermined sequence. Dr Minczuk’s group was a pioneer in this field and in 2006 the first paper was published showing that it was possible to deliver zinc-finger peptides (ZFPs) to mitochondria (117). They showed that when targeting ZFPs fused to the catalytic domain of a human methyltransferase it was possible to induce a site-specific alteration in the mtDNA, in cells harboring the m.8993 T>G mutation, responsible for NARP and maternally inherited Leigh’s syndrome (MILS) (117). However, this approach revealed some limitations in terms of toxicity.
By fusing a particular zinc-finger to a nuclease domain, a zinc-finger nuclease can be created. The nuclease domain is FokI, a type II restriction enzyme, which requires dimerization to induce a double strand break in the DNA (fig.1). In order to achieve dimerization, two zinc-finger endonuclease (FokI) monomers need to be used, binding to adjacent sites on complementary DNA strands containing the target sequence. This technology has been successfully applied to nuclear DNA and site-specific gene correction and addition via homologous recombination was achieved by the induction of double-strand breaks (118, 119). In a similar manner, ZFNs could be targeted to mitochondria and selectively induce the depletion of mutated mtDNA molecules, shifting the heteroplasmy towards the intended direction. Engineered zinc finger nucleases (ZFNs) are chimeric enzymes utilizing the modular Cys2His2 zinc finger DNA-binding moiety conjugated to the C-terminal catalytic subunit of FokI (120, 121). In 2008, it was described for the first time the application of ZFNs to mitochondria, targeting the same point mutation, m.8993 T>G. Initially, the authors used conventional pairs of heterodimeric ZFNs, however they were not able to find suitable pairs for that particular mtDNA sequence. Therefore, they have created a novel approach by designing a single-chain ZFN that recognizes 12 bp of the mtDNA target sequence, by conjugating two FokI nuclease domains, connected to a flexible linker (of 35 aminoacids), to a ZFP with an N-term mitochondrial targeting sequence. This strategy proved to be effective in heteroplasmic cybrids through selectively cleaving the mutated mtDNA molecules (122) and increasing the ratio towards the wild-type mtDNA. However, this approach raised some cytotoxic concerns. To minimize nuclear-related cytotoxicity the authors used a nuclear export signal (NES) linked to the mito-ZFNs but this still resulted in cytotoxicity. In 2014, advances in the architecture of the dimeric mito-ZFNs were made by placing MTS-epitope tag-NES at the N-terminus. In contrast to the previous designs, the Myc tag of one of the monomers was replaced with a FLAG tag. Modified FokI domains, which are paired as a heterodimer (ELD/KKR), were also incorporated in this new study (123). These FokI variants exhibit efficiency comparable to the wild-type architecture, but with a greater than 40-fold reduction in homodimer activity (124–126). In addition, this approach not only showed to be effective to target the m.8993 T>G but also the common deletion (m.8483_13459del4977). With several combinations of mtZFNs capable of specifically degrading targeted mtDNA variants, these data anticipates its clinical application to patients with primary mitochondrial diseases.
4.3. Mitochondrial-targeted TALENs
Since the discovery of a naturally occurring family of proteins secreted by plant pathogens from the Xanthomonas species, another breakthrough was achieved for engineering programmable DNA-binding proteins. Transcription activator-like effectors or TALEs contain DNA-binding domains composed of a series of 33–35-amino-acid repeat domains, each of which recognizes a single base-pair in a DNA strand (127). This nucleotide specificity of each repeat domain is determined by the two amino acids at positions 12 and 13 designated as the repeat variable di-residues (RVDs). Four different RVD modules - namely Asn-Asn (NN), Asn-Ile (NI), His-Asp (HD) and Asn-Gly (NG) are most widely used to recognize guanine or adenine, adenine, cytosine and thymine, respectively. Using this code, targets of new TALEs have been correctly predicted, and functional targets of TALEs composed of randomly assembled repeats have been generated (128–130). Encouraged by the success of ZFNs architecture, a combination of TALEs fused to a non-specific nuclease (FokI) lead to the creation of TAL effector nucleases (TALENs). TALENs can be designed to target almost any given DNA sequences posing only one constrain, the requirement for a thymine at the 5’ end of the target sequence, which is recognized by two amino-terminal cryptic repeat folds. Recently, new TALE variants that can recognize other bases at the 5’ end have broaden the range of targetable sites by TALENs (131). TALE-like proteins from Ralstonia spp. (which is another phytopathogenic bacteria) can also be engineered to bind to predetermined DNA sequences (132). Furthermore, and as it was previously mentioned, FokI works as a dimer, so TALENs require two monomers to bind DNA and produce double strand breaks (fig.1). Even though they are potentially more powerful than ZFNs, their bulky size may limit their use with adeno-associated viral vectors (AAVs) in vivo. In 2013, our laboratory reported for the first time the use of mitochondrial-targeted TALENs- MitoTALENs (100). The constructs were designed to target the mtDNA “common deletion” (m.8483_13459del4977) and a point mutation m. 14459G>A in the MT-ND6 known to cause Leber’s hereditary optic neuropathy plus dystonia (100, 133). The mitoTALENs were coded by the basic principles shown by Cermak and others; the DNA binding domain was kept in between 10–16 repeats and whenever possible, the critical discriminating base pairs were at position 0 or 1 (134). A typical TALEN monomer contains nuclear localization signals and in order to build mitoTALENs, these signals were replaced by mitochondrial localization sequences, that differ between the two monomers used. It was found that some TALE domains lead to poor mitochondrial import for reasons that are still unknown. Thus, in this study ATP5B, COX8A and SOD2 were selected as they could efficiently direct the TALEN monomers to mitochondria. Immuno-tags were also included in this design in the N-terminus of each TAL domain, either –HA or FLAG. Fluorescent markers to select for expression, co-localization and sorting were also placed in the constructs, either eGFP in one monomer and mCherry in the other monomer. A T2A’ (picornaviral 2A-like sequence) was also added between the mitoTALENs and the fluorescent markers, to allow their simultaneous expression under the same promoter (135). At last, FokI heterodimeric domains were placed in all constructs (124, 126). Next, the constructs were first tested in either HeLa cells or COS-7 for expression and mitochondrial localization. To test if mitoTALENs were capable of shifting mtDNA heteroplasmy, trans-mitochondrial cybrids were produced by fusing human osteosarcoma 143B (TK-) cells with the patient fibroblasts, generating cells with different levels of heteroplasmy. These heteroplasmic cells were transfected with both monomers and cell sorting was performed in order to separate the untransfected from the double-transfected cells. This was possible because of presence of eGFP or m-cherry tags in the two plasmids. The DNA from the cells was extracted, a PCR using mtDNA primers followed by a “last-cycle hot PCR” were performed, to avoid interference from heteroduplexes formed during the final PCR cycles. Restriction-digestion fragment length polymorphism (RFLP) was performed by using an appropriate restriction enzyme which enabled to distinguish between the wild-type and mutant mtDNA. It is also important to mention that when the dimers of FokI are formed, the mitoTALENs cleaves the mtDNA haplotype which is subsequently degraded. To target the mutant mtDNA which differs by a single-base pair in the case of the point mutation, one of the TALEN monomers must be able to bind only to the mutated sequence and not to the wild-type. This goal was achieved efficiently in this study, demonstrating as well that the shift in heteroplasmy persisted 2 weeks after growing the sorted cells, and when the constructs were no longer detected by western-blot (100).
Another study from our group showed that it was possible to efficiently target two relatively common pathogenic mtDNA point mutations, the m.8344A>G tRNALys gene mutation associated with myoclonic epilepsy with ragged red fibers (MERRF) and the m.13513G>A ND5 mutation associated with MELAS/Leigh syndrome (136). MitoTALENs efficiently reduced the levels of the targeted pathogenic mtDNAs in the respective cells and improved their OXPHOS activity. To ameliorate the design in the context of the low complexity of mtDNA, the authors designed shorter versions of the mitoTALEN specific for the MERRF m.8344A>G mutation. These shorter mitoTALENs also eliminated the mutant mtDNA. This anticipates that reductions in size will improve the ability to package these large sequences into viral vectors, bringing the use of these genetic tools closer to the clinics (136). Recently, a breakthrough strategy to prevent germline transmission of mitochondrial diseases by inducing mtDNA heteroplasmy shift through the selective elimination of mutated mtDNA with mito nucleases has been demonstrated (55). The authors of this study took advantage of the NZB/BALB heteroplasmic mouse model and selectively prevented the germline transmission of one of the haplotypes, either by using endonucleases (mito-ApaLI) or TALENs, preventing their transmission to the next generation. Furthermore, they were also able to successfully reduce the mutation levels of human LHON and NARP- associated point mutations, in mammalian oocytes by the use of mitoTALENs (55). These results demonstrate the potential of germline heteroplasmy shift as a clinical strategy to prevent the transgenerational transmission of mitochondrial genomes.
The most recent addition to this group of gene editing endonucleases is the Cas9 nuclease. Cas9 is targeted using a very small short RNA sequence- clustered regularly interspaced short palindromic repeats (CRISPR) (137). CRISPRs are much easier to design in comparison to ZFNs or TALENs because the process requires only making a short RNA sequence. As an example of its potential role for nuclear mutations, Cas9/CRISPR has been shown to restore the weight loss phenotype of a mouse model of a Fah mutation in hepatocytes in a mouse model of the human disease hereditary tyrosinemia (138). However, its high off-target effects were a concern. There has been a report that FLAG-Cas9 can localize to mitochondria to edit mitochondrial DNA with sgRNAs targeting specific loci of the mitochondrial genome. Expression of FLAG-Cas9 together with gRNA targeting Cox1 and Cox3 led to the cleavage of the specific mtDNA loci. In addition, the authors observed disruption of mitochondrial protein homeostasis following mtDNA truncation or cleavage by CRISPR/Cas9. To overcome FLAG-Cas9 unspecificity they have created a mitochondrial-targeted Cas9 (mitoCas9) that showed specific cleavage of mtDNA together with expression of gRNA. MitoCas9-induced reduction of mtDNA and mitochondrial membrane potential disruption, as well as cell growth inhibition (139). However, this study did not show direct evidence that the Cas9 system was cleaving mtDNA, but rather the copy number of regions targeted to be cleaved (Cox1 and Cox3). Because mtDNA behaves as a genetic unit, it is difficult to envision how the levels of a specific mtDNA region would be different from other regions, unless there is a deletion, which would be observable by Southern blot. Such experiment was not presented. In our lab, we have tried to use Cas9/CRISPR for mtDNA heteroplasmy changes but failed. Although we were able to deliver Cas9 to mitochondria with a targeting sequence, we suspect that the guide RNA was not efficiently imported, even though we used different RNA extensions to promote RNA import (140). The advantages and limitations of the described techniques are summarized in Table 1.
Table 1.
Currently described mitochondrial-targeted gene editing tools
| Gene editing tool | Nuclease | Target | Limitations | Advantages | References |
|---|---|---|---|---|---|
| Mitochondrial-targeted restriction endonucleases | PstI, SmaI, ApaLI, XmaI, ScaI, among others | DNA | Limited to mutations that create unique restriction sites | Compact size Effective shift in heteroplasmy in vivo |
11, 110–112, 114–116 |
| Mitochondrial-targeted Zinc Fingers | Heterodimeric FokI | DNA | Toxicity Large size (requires two monomers or quasi-dimeric architecture) |
Virtually targets any mitochondrial DNA sequence | 122, 123, 125 |
| Mitochondrial-targeted TALENs | Heterodimeric FokI | DNA | Large size (requires two monomers) T0 binding requirement (in general) |
Virtually targets any mitochondrial DNA sequence | 100, 152 |
| Mitochondrial CAS91/CRISPR2 | Cas9 | RNA | Problems with gRNA import to mitochondria. Potential off target effects | Compact and easy to design, it only requires the design of the short RNA | 139, 152 |
CRISPR associated protein 9
Clustered regularly interspaced short palindromic repeats
5. CLINICAL AVENUES AND PERSPECTIVES
There have been great advances for avoiding the transmission of mtDNA mutations. Pre-implantation genetic diagnosis (PGD) can assist females which carry mtDNA mutations, by preventing the transmission of deleterious mtDNA. After in vitro fertilization embryos are analyzed and only those with very low mutant levels are transferred to the uterus, but for woman that carry intermediate-levels of heteroplasmic mtDNA mutations, this technique may not be effective since there is uncertainty regarding the clinical mutation threshold remains (141, 142). Techniques such as “pro-nuclear transfer” involve the transfer of nuclear DNA from a donor zygote to an enucleated recipient zygote via fusion (143). The new “reconstructed zygote” retains the nuclear DNA from the mother, but the mtDNA from a donor. This approach has made significant steps towards treating mitochondrial disease at mtDNA level. Similarly, “spindle transfer” of nDNA to an enucleated donor was also successful in animal models and human embryos (144). Unlike pro-nuclear transfer, the nDNA isolation occurs pre-fertilization, meaning that it can be integrated into established in vitro fertilization techniques. Mitochondrial replacement techniques involve a series of complex technical manipulations of nuclear genome between patient and donor oocytes that result in the generation of embryos carrying genetic material from three different origins and for these reasons, these techniques raised different ethical and medical concerns. In spite of all the polemics, mitochondrial replacement was approved in the UK, early 2015 (145). Mitochondrial nucleases can be a powerful alternative for these techniques, as it does not require any mtDNA donor. Using a single mRNA or adenovirus injection into patient oocytes or embryos (respectively) expressing a mitochondrial nuclease is technically simpler and less traumatic compared to mitochondrial replacement techniques and it also does not require a third donor, circumventing some of the ethical issues associated with this matter.
There are currently at least two strategies for applying gene therapy to patients with mtDNA diseases: 1) Allotopic expression of mitochondrial genes; 2) Manipulation of mtDNA heteroplasmy. Allotopic expression of mitochondrial genes which consists in the synthesis of a wild-type version of the mutated protein in the nuclear-cytosolic compartment followed by its import into mitochondria has been a controversial approach because of the high hydrophobicity of mtDNA-encoded proteins and the competition with endogenous counterparts (146–149). Nonetheless, clinical trials for Leber’s optic neuropathy are currently ongoing (150, 151). The use of mitochondrial endonucleases is still in its infancy, but hopefully will move into the clinics in the next few years.
To conclude, the manipulation of mtDNA heteroplasmy either by using mito restriction endonucleases, mito zinc-fingers or mitoTALENs could facilitate delivery and increase specificity of mtDNA editing, having the potential to eliminate mutant mitochondrial genomes from germline treated affected patients.
6. REFERENCES
- 1.Chance B and Williams GR: The respiratory chain and oxidative phosphorylation. Adv Enzymol Relat Subj Biochem, 17, 65–134 (1956) DOI: 10.1002/9780470122624.ch2 [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P: Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism. Nature, 191, 144–8 (1961) DOI: 10.1038/191144a0 [DOI] [PubMed] [Google Scholar]
- 3.Giles RE, Blanc H, Cann HM and Wallace DC: Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A, 77(11), 6715–9 (1980) DOI: 10.1073/pnas.77.11.6715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace DC: Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem, 76, 781–821 (2007) DOI: 10.1146/annurev.biochem.76.081205.150955 [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC and Chalkia D: Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol, 5(11), a021220 (2013) DOI: 10.1101/cshperspect.a021220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuppen HA, Blakely EL, Turnbull DM and Taylor RW: Mitochondrial DNA mutations and human disease. Biochim Biophys Acta, 1797(2), 113–28 (2010) DOI: 10.1016/j.bbabio.2009.09.005 [DOI] [PubMed] [Google Scholar]
- 7.Sanderson S, Green A, Preece MA and Burton H: The incidence of inherited metabolic disorders in the West Midlands, UK. Arch Dis Child, 91(11), 896–9 (2006) DOI: 10.1136/adc.2005.091637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clay Montier LL, Deng JJ and Bai Y: Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics, 36(3), 125–31 (2009) DOI: 10.1016/S1673-8527(08)60099-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton DA: Mitochondrial DNA replication: what we know. IUBMB Life, 55(4–5), 213–7 (2003) DOI: 10.1080/1521654031000134824 [DOI] [PubMed] [Google Scholar]
- 10.Holt IJ, Lorimer HE and Jacobs HT: Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell, 100(5), 515–24 (2000) DOI: 10.1016/S0092-8674(00)80688-1 [DOI] [PubMed] [Google Scholar]
- 11.Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA and Moraes CT: Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc Natl Acad Sci U S A, 102(40), 14392–7 (2005) DOI: 10.1073/pnas.0502896102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart JB and Chinnery PF: The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet, 16(9), 530–42 (2015) DOI: 10.1038/nrg3966 [DOI] [PubMed] [Google Scholar]
- 13.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL and Alexeyev MF: Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res, 37(8), 2539–48 (2009) DOI: 10.1093/nar/gkp100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandy B and Davison AJ: Mitochondrial mutations may increase oxidative stress: implications for carcinogenesis and aging? Free Radic Biol Med, 8(6), 523–39 (1990) DOI: 10.1016/0891-5849(90)90152-9 [DOI] [PubMed] [Google Scholar]
- 15.Alexeyev M, Shokolenko I, Wilson G and LeDoux S: The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harb Perspect Biol, 5(5), a012641 (2013) DOI: 10.1101/cshperspect.a012641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen NB, Rasmussen M and Rasmussen LJ: Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion, 5(2), 89–108 (2005) DOI: 10.1016/j.mito.2005.02.002 [DOI] [PubMed] [Google Scholar]
- 17.Thorslund T, Sunesen M, Bohr VA and Stevnsner T: Repair of 8-oxoG is slower in endogenous nuclear genes than in mitochondrial DNA and is without strand bias. DNA Repair (Amst), 1(4), 261–73 (2002) DOI: 10.1016/S1568-7864(02)00003-4 [DOI] [PubMed] [Google Scholar]
- 18.Anson RM, Senturker S, Dizdaroglu M and Bohr VA: Measurement of oxidatively induced base lesions in liver from Wistar rats of different ages. Free Radic Biol Med, 27(3–4), 456–62 (1999) DOI: 10.1016/S0891-5849(99)00091-X [DOI] [PubMed] [Google Scholar]
- 19.Lim KS, Jeyaseelan K, Whiteman M, Jenner A and Halliwell B: Oxidative damage in mitochondrial DNA is not extensive. Ann N Y Acad Sci, 1042, 210–20 (2005) DOI: 10.1196/annals.1338.023 [DOI] [PubMed] [Google Scholar]
- 20.Kauppila JH and Stewart JB: Mitochondrial DNA: Radically free of free-radical driven mutations. Biochim Biophys Acta, 1847(11), 1354–61 (2015) DOI: 10.1016/j.bbabio.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 21.Anson RM, Hudson E and Bohr VA: Mitochondrial endogenous oxidative damage has been overestimated. FASEB J, 14(2), 355–60 (2000) [DOI] [PubMed] [Google Scholar]
- 22.Rich PR: The molecular machinery of Keilin’s respiratory chain. Biochem Soc Trans, 31(Pt 6), 1095–105 (2003) DOI: 10.1042/bst0311095 [DOI] [PubMed] [Google Scholar]
- 23.Wallace DC: The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene, 354, 169–80 (2005) DOI: 10.1016/j.gene.2005.05.001 [DOI] [PubMed] [Google Scholar]
- 24.Mitchell P and Moyle J: Estimation of membrane potential and pH difference across the cristae membrane of rat liver mitochondria. Eur J Biochem, 7(4), 471–84 (1969) DOI: 10.1111/j.1432-1033.1969.tb19633.x [DOI] [PubMed] [Google Scholar]
- 25.Halestrap AP and Brenner C: The adenine nucleotide translocase: a central component of the mitochondrial permeability transition pore and key player in cell death. Curr Med Chem, 10(16), 1507–25 (2003) DOI: 10.2174/0929867033457278 [DOI] [PubMed] [Google Scholar]
- 26.Jastroch M, Divakaruni AS, Mookerjee S, Treberg JR and Brand MD: Mitochondrial proton and electron leaks. Essays Biochem, 47, 53–67 (2010) DOI: 10.1042/bse0470053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orrenius S, Zhivotovsky B and Nicotera P: Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol, 4(7), 552–65 (2003) DOI: 10.1038/nrm1150 [DOI] [PubMed] [Google Scholar]
- 28.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R and Young IG: Sequence and organization of the human mitochondrial genome. Nature, 290(5806), 457–65 (1981) DOI: 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- 29.Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T and Watanabe K: Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res, 37(5), 1616–27 (2009) DOI: 10.1093/nar/gkp001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osawa S, Jukes TH, Watanabe K and Muto A: Recent evidence for evolution of the genetic code. Microbiol Rev, 56(1), 229–64 (1992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Temperley R, Richter R, Dennerlein S, Lightowlers RN and Chrzanowska-Lightowlers ZM: Hungry codons promote frameshifting in human mitochondrial ribosomes. Science, 327(5963), 301 (2010) DOI: 10.1126/science.1180674 [DOI] [PubMed] [Google Scholar]
- 32.Schwartz M and Vissing J: New patterns of inheritance in mitochondrial disease. Biochem Biophys Res Commun, 310(2), 247–51 (2003) DOI: 10.1016/j.bbrc.2003.09.037 [DOI] [PubMed] [Google Scholar]
- 33.Wallace DC and Fan W: Energetics, epigenetics, mitochondrial genetics. Mitochondrion, 10(1), 12–31 (2010) DOI: 10.1016/j.mito.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park CB and Larsson NG: Mitochondrial DNA mutations in disease and aging. J Cell Biol, 193(5), 809–18 (2011) DOI: 10.1083/jcb.201010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chinnery PF, Thorburn DR, Samuels DC, White SL, Dahl HM, Turnbull DM, Lightowlers RN and Howell N: The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet, 16(11), 500–5 (2000) DOI: 10.1016/S0168-9525(00)02120-X [DOI] [PubMed] [Google Scholar]
- 36.Sutovsky P: Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: killing three birds with one stone. Microsc Res Tech, 61(1), 88–102 (2003) DOI: 10.1002/jemt.10319 [DOI] [PubMed] [Google Scholar]
- 37.Christie JR, Schaerf TM and Beekman M: Selection against heteroplasmy explains the evolution of uniparental inheritance of mitochondria. PLoS Genet, 11(4), e1005112 (2015) DOI: 10.1371/journal.pgen.1005112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DiMauro S and Moraes CT: Mitochondrial encephalomyopathies. Arch Neurol, 50(11), 1197–208 (1993) DOI: 10.1001/archneur.1993.00540110075008 [DOI] [PubMed] [Google Scholar]
- 39.Davis RL and Sue CM: The genetics of mitochondrial disease. Semin Neurol, 31(5), 519–30 (2011) DOI: 10.1055/s-0031-1299790 [DOI] [PubMed] [Google Scholar]
- 40.Battersby BJ, Loredo-Osti JC and Shoubridge EA: Nuclear genetic control of mitochondrial DNA segregation. Nat Genet, 33(2), 183–6 (2003) DOI: 10.1038/ng1073 [DOI] [PubMed] [Google Scholar]
- 41.Sue CM, Bruno C, Andreu AL, Cargan A, Mendell JR, Tsao CY, Luquette M, Paolicchi J, Shanske S, DiMauro S and De Vivo DC: Infantile encephalopathy associated with the MELAS A3243G mutation. J Pediatr, 134(6), 696–700 (1999) DOI: 10.1016/S0022-3476(99)70283-0 [DOI] [PubMed] [Google Scholar]
- 42.Larsson NG: Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem, 79, 683–706 (2010) DOI: 10.1146/annurev-biochem-060408-093701 [DOI] [PubMed] [Google Scholar]
- 43.Ross JM, Stewart JB, Hagstrom E, Brene S, Mourier A, Coppotelli G, Freyer C, Lagouge M, Hoffer BJ, Olson L and Larsson NG: Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature, 501(7467), 412–5 (2013) DOI: 10.1038/nature12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogenhagen D and Clayton DA: Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell, 11(4), 719–27 (1977) DOI: 10.1016/0092-8674(77)90286-0 [DOI] [PubMed] [Google Scholar]
- 45.Larsson NG, Tulinius MH, Holme E, Oldfors A, Andersen O, Wahlstrom J and Aasly J: Segregation and manifestations of the mtDNA tRNA(Lys) A-->G(8344) mutation of myoclonus epilepsy and ragged-red fibers (MERRF) syndrome. Am J Hum Genet, 51(6), 1201–12 (1992) [PMC free article] [PubMed] [Google Scholar]
- 46.Jenuth JP, Peterson AC, Fu K and Shoubridge EA: Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet, 14(2), 146–51 (1996) DOI: 10.1038/ng1096-146 [DOI] [PubMed] [Google Scholar]
- 47.Olivo PD, Van de Walle MJ, Laipis PJ and Hauswirth WW: Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature, 306(5941), 400–2 (1983) DOI: 10.1038/306400a0 [DOI] [PubMed] [Google Scholar]
- 48.Howell N, Halvorson S, Kubacka I, McCullough DA, Bindoff LA and Turnbull DM: Mitochondrial gene segregation in mammals: is the bottleneck always narrow? Hum Genet, 90(1–2), 117–20 (1992) DOI: 10.1007/BF00210753 [DOI] [PubMed] [Google Scholar]
- 49.Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HH and Chinnery PF: A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet, 40(2), 249–54 (2008) DOI: 10.1038/ng.2007.63 [DOI] [PubMed] [Google Scholar]
- 50.Wai T, Teoli D and Shoubridge EA: The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet, 40(12), 1484–8 (2008) DOI: 10.1038/ng.258 [DOI] [PubMed] [Google Scholar]
- 51.Freyer C, Cree LM, Mourier A, Stewart JB, Koolmeister C, Milenkovic D, Wai T, Floros VI, Hagstrom E, Chatzidaki EE, Wiesner RJ, Samuels DC, Larsson NG and Chinnery PF: Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat Genet, 44(11), 1282–5 (2012) DOI: 10.1038/ng.2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A and Larsson NG: Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol, 6(1), e10 (2008) DOI: 10.1371/journal.pbio.0060010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR and Wallace DC: A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science, 319(5865), 958–62 (2008) DOI: 10.1126/science.1147786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson IJ, Carling PJ, Alston CL, Floros VI, Pyle A, Hudson G, Sallevelt SC, Lamperti C, Carelli V, Bindoff LA, Samuels DC, Wonnapinij P, Zeviani M, Taylor RW, Smeets HJ, Horvath R and Chinnery PF: Mitochondrial DNA sequence characteristics modulate the size of the genetic bottleneck. Hum Mol Genet, 25(5), 1031–41 (2016) DOI: 10.1093/hmg/ddv626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy P, Ocampo A, Suzuki K, Luo J, Bacman SR, Williams SL, Sugawara A, Okamura D, Tsunekawa Y, Wu J, Lam D, Xiong X, Montserrat N, Esteban CR, Liu GH, Sancho-Martinez I, Manau D, Civico S, Cardellach F, Del Mar O’Callaghan M, Campistol J, Zhao H, Campistol JM, Moraes CT and Izpisua Belmonte JC: Selective elimination of mitochondrial mutations in the germline by genome editing. Cell, 161(3), 459–69 (2015) DOI: 10.1016/j.cell.2015.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schon EA, DiMauro S and Hirano M: Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet, 13(12), 878–90 (2012) DOI: 10.1038/nrg3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holt IJ, Harding AE and Morgan-Hughes JA: Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature, 331(6158), 717–9 (1988) DOI: 10.1038/331717a0 [DOI] [PubMed] [Google Scholar]
- 58.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ 2nd and Nikoskelainen EK: Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science, 242(4884), 1427–30 (1988) DOI: 10.1126/science.3201231 [DOI] [PubMed] [Google Scholar]
- 59.Wallace DC, Zheng XX, Lott MT, Shoffner JM, Hodge JA, Kelley RI, Epstein CM and Hopkins LC: Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell, 55(4), 601–10 (1988) DOI: 10.1016/0092-8674(88)90218-8 [DOI] [PubMed] [Google Scholar]
- 60.Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW and Wallace DC: Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell, 61(6), 931–7 (1990) DOI: 10.1016/0092-8674(90)90059-N [DOI] [PubMed] [Google Scholar]
- 61.Shoffner JM, Lott MT, Voljavec AS, Soueidan SA, Costigan DA and Wallace DC: Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc Natl Acad Sci U S A, 86(20), 7952–6 (1989) DOI: 10.1073/pnas.86.20.7952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elliott HR, Samuels DC, Eden JA, Relton CL and Chinnery PF: Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet, 83(2), 254–60 (2008) DOI: 10.1016/j.ajhg.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chinnery PF, Elliott HR, Hudson G, Samuels DC and Relton CL: Epigenetics, epidemiology and mitochondrial DNA diseases. Int J Epidemiol, 41(1), 177–87 (2012) DOI: 10.1093/ije/dyr232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wallace DC: A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet, 39, 359–407 (2005) DOI: 10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McFarland R, Clark KM, Morris AA, Taylor RW, Macphail S, Lightowlers RN and Turnbull DM: Multiple neonatal deaths due to a homoplasmic mitochondrial DNA mutation. Nat Genet, 30(2), 145–6 (2002) DOI: 10.1038/ng819 [DOI] [PubMed] [Google Scholar]
- 66.Taylor RW, Giordano C, Davidson MM, d’Amati G, Bain H, Hayes CM, Leonard H, Barron MJ, Casali C, Santorelli FM, Hirano M, Lightowlers RN, DiMauro S and Turnbull DM: A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J Am Coll Cardiol, 41(10), 1786–96 (2003) DOI: 10.1016/S0735-1097(03)00300-0 [DOI] [PubMed] [Google Scholar]
- 67.Huang T: Next generation sequencing to characterize mitochondrial genomic DNA heteroplasmy. Curr Protoc Hum Genet, Chapter 19, Unit19 8 (2011) DOI: 10.1002/0471142905.hg1908s71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Just RS, Irwin JA and Parson W: Mitochondrial DNA heteroplasmy in the emerging field of massively parallel sequencing. Forensic Sci Int Genet, 18, 131–9 (2015) DOI: 10.1016/j.fsigen.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Payne BA, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M and Chinnery PF: Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet, 22(2), 384–90 (2013) DOI: 10.1093/hmg/dds435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C and Bogenhagen DF: Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J Biol Chem, 281(1), 374–82 (2006) DOI: 10.1074/jbc.M509730200 [DOI] [PubMed] [Google Scholar]
- 71.Rotig A: Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta, 1807(9), 1198–205 (2011) DOI: 10.1016/j.bbabio.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 72.Pavlakis SG, Phillips PC, DiMauro S, De Vivo DC and Rowland LP: Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Ann Neurol, 16(4), 481–8 (1984) DOI: 10.1002/ana.410160409 [DOI] [PubMed] [Google Scholar]
- 73.Hirano M, Ricci E, Koenigsberger MR, Defendini R, Pavlakis SG, DeVivo DC, DiMauro S and Rowland LP: Melas: an original case and clinical criteria for diagnosis. Neuromuscul Disord, 2(2), 125–35 (1992) DOI: 10.1016/0960-8966(92)90045-8 [DOI] [PubMed] [Google Scholar]
- 74.Goto Y, Nonaka I and Horai S: A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature, 348(6302), 651–3 (1990) DOI: 10.1038/348651a0 [DOI] [PubMed] [Google Scholar]
- 75.Santorelli FM, Tanji K, Kulikova R, Shanske S, Vilarinho L, Hays AP and DiMauro S: Identification of a novel mutation in the mtDNA ND5 gene associated with MELAS. Biochem Biophys Res Commun, 238(2), 326–8 (1997) DOI: 10.1006/bbrc.1997.7167 [DOI] [PubMed] [Google Scholar]
- 76.Kirby DM, McFarland R, Ohtake A, Dunning C, Ryan MT, Wilson C, Ketteridge D, Turnbull DM, Thorburn DR and Taylor RW: Mutations of the mitochondrial ND1 gene as a cause of MELAS. J Med Genet, 41(10), 784–9 (2004) DOI: 10.1136/jmg.2004.020537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petruzzella V, Moraes CT, Sano MC, Bonilla E, DiMauro S and Schon EA: Extremely high levels of mutant mtDNAs co-localize with cytochrome c oxidase-negative ragged-red fibers in patients harboring a point mutation at nt 3243. Hum Mol Genet, 3(3), 449–54 (1994) DOI: 10.1093/hmg/3.3.449 [DOI] [PubMed] [Google Scholar]
- 78.Blakely EL, Alston CL, Lecky B, Chakrabarti B, Falkous G, Turnbull DM, Taylor RW and Gorman GS: Distal weakness with respiratory insufficiency caused by the m.8344A > G “MERRF” mutation. Neuromuscul Disord, 24(6), 533–6 (2014) DOI: 10.1016/j.nmd.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hammans SR, Sweeney MG, Brockington M, Lennox GG, Lawton NF, Kennedy CR, Morgan-Hughes JA and Harding AE: The mitochondrial DNA transfer RNA(Lys)A-->G(8344) mutation and the syndrome of myoclonic epilepsy with ragged red fibres (MERRF). Relationship of clinical phenotype to proportion of mutant mitochondrial DNA. Brain, 116 (Pt 3), 617–32 (1993) DOI: 10.1093/brain/116.3.617 [DOI] [PubMed] [Google Scholar]
- 80.Silvestri G, Ciafaloni E, Santorelli FM, Shanske S, Servidei S, Graf WD, Sumi M and DiMauro S: Clinical features associated with the A-->G transition at nucleotide 8344 of mtDNA (“MERRF mutation”). Neurology, 43(6), 1200–6 (1993) DOI: 10.1212/WNL.43.6.1200 [DOI] [PubMed] [Google Scholar]
- 81.Kerrison JB and Newman NJ: Clinical spectrum of Leber’s hereditary optic neuropathy. Clin Neurosci, 4(5), 295–301 (1997) [PubMed] [Google Scholar]
- 82.Huoponen K: Leber hereditary optic neuropathy: clinical and molecular genetic findings. Neurogenetics, 3(3), 119–25 (2001) DOI: 10.1007/s100480100115 [DOI] [PubMed] [Google Scholar]
- 83.Newman NJ, Lott MT and Wallace DC: The clinical characteristics of pedigrees of Leber’s hereditary optic neuropathy with the 11778 mutation. Am J Ophthalmol, 111(6), 750–62 (1991) DOI: 10.1016/S0002-9394(14)76784-4 [DOI] [PubMed] [Google Scholar]
- 84.Harding AE, Sweeney MG, Miller DH, Mumford CJ, Kellar-Wood H, Menard D, McDonald WI and Compston DA: Occurrence of a multiple sclerosis-like illness in women who have a Leber’s hereditary optic neuropathy mitochondrial DNA mutation. Brain, 115 (Pt 4), 979–89 (1992) DOI: 10.1093/brain/115.4.979 [DOI] [PubMed] [Google Scholar]
- 85.De Vries DD, Went LN, Bruyn GW, Scholte HR, Hofstra RM, Bolhuis PA and van Oost BA: Genetic and biochemical impairment of mitochondrial complex I activity in a family with Leber hereditary optic neuropathy and hereditary spastic dystonia. Am J Hum Genet, 58(4), 703–11 (1996) [PMC free article] [PubMed] [Google Scholar]
- 86.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP and Letellier T: Mitochondrial threshold effects. Biochem J, 370(Pt 3), 751–62 (2003) DOI: 10.1042/bj20021594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeviani M, Moraes CT, DiMauro S, Nakase H, Bonilla E, Schon EA and Rowland LP: Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology, 38(9):1339–46 (1988) DOI: 10.1212/WNL.38.9.1339 [DOI] [PubMed] [Google Scholar]
- 88.Moraes CT, DiMauro S, Zeviani M, Lombes A, Shanske S, Miranda AF, Nakase H, Bonilla E, Werneck LC, Servidei S and et al. : Mitochondrial DNA deletions in progressive external ophthalmoplegia and Kearns-Sayre syndrome. N Engl J Med, 320(20), 1293–9 (1989) DOI: 10.1056/NEJM198905183202001 [DOI] [PubMed] [Google Scholar]
- 89.Rotig A, Cormier V, Blanche S, Bonnefont JP, Ledeist F, Romero N, Schmitz J, Rustin P, Fischer A, Saudubray JM and et al. : Pearson’s marrow-pancreas syndrome. A multisystem mitochondrial disorder in infancy. J Clin Invest, 86(5), 1601–8 (1990) DOI: 10.1172/JCI114881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sadikovic B, Wang J, El-Hattab A, Landsverk M, Douglas G, Brundage EK, Craigen WJ, Schmitt ES and Wong LJ: Sequence homology at the breakpoint and clinical phenotype of mitochondrial DNA deletion syndromes. PLoS One, 5(12), e15687 (2010) DOI: 10.1371/journal.pone.0015687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Copeland WC: Defects in mitochondrial DNA replication and human disease. Crit Rev Biochem Mol Biol, 47(1), 64–74 (2012) DOI: 10.3109/10409238.2011.632763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kollberg G, Moslemi AR, Darin N, Nennesmo I, Bjarnadottir I, Uvebrant P, Holme E, Melberg A, Tulinius M and Oldfors A: POLG1 mutations associated with progressive encephalopathy in childhood. J Neuropathol Exp Neurol, 65(8), 758–68 (2006) DOI: 10.1097/01.jnen.0000229987.17548.6e [DOI] [PubMed] [Google Scholar]
- 93.Naviaux RK and Nguyen KV: POLG mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann Neurol, 55(5), 706–12 (2004) DOI: 10.1002/ana.20079 [DOI] [PubMed] [Google Scholar]
- 94.Nishino I, Spinazzola A and Hirano M: Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science, 283(5402), 689–92 (1999) DOI: 10.1126/science.283.5402.689 [DOI] [PubMed] [Google Scholar]
- 95.Hirano M, Lagier-Tourenne C, Valentino ML, Marti R and Nishigaki Y: Thymidine phosphorylase mutations cause instability of mitochondrial DNA. Gene, 354, 152–6 (2005) DOI: 10.1016/j.gene.2005.04.041 [DOI] [PubMed] [Google Scholar]
- 96.Saada A, Shaag A and Elpeleg O: mtDNA depletion myopathy: elucidation of the tissue specificity in the mitochondrial thymidine kinase (TK2) deficiency. Mol Genet Metab, 79(1), 1–5 (2003) DOI: 10.1016/S1096-7192(03)00063-5 [DOI] [PubMed] [Google Scholar]
- 97.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S and Elpeleg O: Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet, 29(3), 342–4 (2001) DOI: 10.1038/ng751 [DOI] [PubMed] [Google Scholar]
- 98.Mandel H, Szargel R, Labay V, Elpeleg O, Saada A, Shalata A, Anbinder Y, Berkowitz D, Hartman C, Barak M, Eriksson S and Cohen N: The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat Genet, 29(3), 337–41 (2001) DOI: 10.1038/ng746 [DOI] [PubMed] [Google Scholar]
- 99.Salviati L, Sacconi S, Mancuso M, Otaegui D, Camano P, Marina A, Rabinowitz S, Shiffman R, Thompson K, Wilson CM, Feigenbaum A, Naini AB, Hirano M, Bonilla E, DiMauro S and Vu TH: Mitochondrial DNA depletion and dGK gene mutations. Ann Neurol, 52(3), 311–7 (2002) DOI: 10.1002/ana.10284 [DOI] [PubMed] [Google Scholar]
- 100.Bacman SR, Williams SL, Pinto M, Peralta S and Moraes CT: Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med, 19(9), 1111–3 (2013) DOI: 10.1038/nm.3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y and Nonaka I: Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc Natl Acad Sci U S A, 88(23), 10614–8 (1991) DOI: 10.1073/pnas.88.23.10614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith PM and Lightowlers RN: Altering the balance between healthy and mutated mitochondrial DNA. J Inherit Metab Dis, 34(2), 309–13 (2011) DOI: 10.1007/s10545-010-9122-6 [DOI] [PubMed] [Google Scholar]
- 103.Santra S, Gilkerson RW, Davidson M and Schon EA: Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann Neurol, 56(5), 662–9 (2004) DOI: 10.1002/ana.20240 [DOI] [PubMed] [Google Scholar]
- 104.Shoubridge EA, Johns T and Karpati G: Complete restoration of a wild-type mtDNA genotype in regenerating muscle fibres in a patient with a tRNA point mutation and mitochondrial encephalomyopathy. Hum Mol Genet, 6(13), 2239–42 (1997) DOI: 10.1093/hmg/6.13.2239 [DOI] [PubMed] [Google Scholar]
- 105.Clark KM, Bindoff LA, Lightowlers RN, Andrews RM, Griffiths PG, Johnson MA, Brierley EJ and Turnbull DM: Reversal of a mitochondrial DNA defect in human skeletal muscle. Nat Genet, 16(3), 222–4 (1997) DOI: 10.1038/ng0797-222 [DOI] [PubMed] [Google Scholar]
- 106.Taivassalo T, Fu K, Johns T, Arnold D, Karpati G and Shoubridge EA: Gene shifting: a novel therapy for mitochondrial myopathy. Hum Mol Genet, 8(6), 1047–52 (1999) DOI: 10.1093/hmg/8.6.1047 [DOI] [PubMed] [Google Scholar]
- 107.Murphy JL, Blakely EL, Schaefer AM, He L, Wyrick P, Haller RG, Taylor RW, Turnbull DM and Taivassalo T: Resistance training in patients with single, large-scale deletions of mitochondrial DNA. Brain, 131(Pt 11), 2832–40 (2008) DOI: 10.1093/brain/awn252 [DOI] [PubMed] [Google Scholar]
- 108.Srivastava S, Barrett JN and Moraes CT: PGC-1alpha/beta upregulation is associated with improved oxidative phosphorylation in cells harboring nonsense mtDNA mutations. Hum Mol Genet, 16(8), 993–1005 (2007) DOI: 10.1093/hmg/ddm045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. https://clinicaltrials.gov/ct2/show/NCT00457314.
- 110.Srivastava S and Moraes CT: Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum Mol Genet, 10(26), 3093–9 (2001) DOI: 10.1093/hmg/10.26.3093 [DOI] [PubMed] [Google Scholar]
- 111.Tanaka M, Borgeld HJ, Zhang J, Muramatsu S, Gong JS, Yoneda M, Maruyama W, Naoi M, Ibi T, Sahashi K, Shamoto M, Fuku N, Kurata M, Yamada Y, Nishizawa K, Akao Y, Ohishi N, Miyabayashi S, Umemoto H, Muramatsu T, Furukawa K, Kikuchi A, Nakano I, Ozawa K and Yagi K: Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J Biomed Sci, 9(6 Pt 1), 534–41 (2002) [DOI] [PubMed] [Google Scholar]
- 112.Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G and Wilson GL: Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther, 15(7), 516–23 (2008) DOI: 10.1038/gt.2008.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jenuth JP, Peterson AC and Shoubridge EA: Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet, 16(1), 93–5 (1997) DOI: 10.1038/ng0597-93 [DOI] [PubMed] [Google Scholar]
- 114.Bacman SR, Williams SL, Hernandez D and Moraes CT: Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a ‘differential multiple cleavage-site’ model. Gene Ther, 14(18), 1309–18 (2007) DOI: 10.1038/sj.gt.3302981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bacman SR, Williams SL, Garcia S and Moraes CT: Organ-specific shifts in mtDNA heteroplasmy following systemic delivery of a mitochondria-targeted restriction endonuclease. Gene Ther, 17(6), 713–20 (2010) DOI: 10.1038/gt.2010.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bacman SR, Williams SL, Duan D and Moraes CT: Manipulation of mtDNA heteroplasmy in all striated muscles of newborn mice by AAV9-mediated delivery of a mitochondria-targeted restriction endonuclease. Gene Ther, 19(11), 1101–6 (2012) DOI: 10.1038/gt.2011.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Minczuk M, Papworth MA, Kolasinska P, Murphy MP and Klug A: Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc Natl Acad Sci U S A, 103(52), 19689–94 (2006) DOI: 10.1073/pnas.0609502103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pearson H: Protein engineering: The fate of fingers. Nature, 455(7210), 160–4 (2008) DOI: 10.1038/455160a [DOI] [PubMed] [Google Scholar]
- 119.Papworth M, Kolasinska P and Minczuk M: Designer zinc-finger proteins and their applications. Gene, 366(1), 27–38 (2006) DOI: 10.1016/j.gene.2005.09.011 [DOI] [PubMed] [Google Scholar]
- 120.Kim CA and Berg JM: A 2.2. A resolution crystal structure of a designed zinc finger protein bound to DNA. Nat Struct Biol, 3(11), 940–5 (1996) DOI: 10.1038/nsb1196-940 [DOI] [PubMed] [Google Scholar]
- 121.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S and Carroll D: Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res, 28(17), 3361–9 (2000) DOI: 10.1093/nar/28.17.3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Minczuk M, Papworth MA, Miller JC, Murphy MP and Klug A: Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res, 36(12), 3926–38 (2008) DOI: 10.1093/nar/gkn313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gammage PA, Rorbach J, Vincent AI, Rebar EJ and Minczuk M: Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol Med, 6(4), 458–66 (2014) DOI: 10.1002/emmm.201303672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO and Rebar EJ: An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol, 25(7), 778–85 (2007) DOI: 10.1038/nbt1319 [DOI] [PubMed] [Google Scholar]
- 125.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ and Cathomen T: Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol, 25(7), 786–93 (2007) DOI: 10.1038/nbt1317 [DOI] [PubMed] [Google Scholar]
- 126.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD and Holmes MC: Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods, 8(1), 74–9 (2011) DOI: 10.1038/nmeth.1539 [DOI] [PubMed] [Google Scholar]
- 127.Kim H and Kim JS: A guide to genome engineering with programmable nucleases. Nat Rev Genet, 15(5), 321–34 (2014) DOI: 10.1038/nrg3686 [DOI] [PubMed] [Google Scholar]
- 128.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A and Bonas U: Breaking the code of DNA binding specificity of TAL-type III effectors. Science, 326(5959), 1509–12 (2009) DOI: 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- 129.Moscou MJ and Bogdanove AJ: A simple cipher governs DNA recognition by TAL effectors. Science, 326(5959), 1501 (2009) DOI: 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- 130.Romer P, Recht S, Strauss T, Elsaesser J, Schornack S, Boch J, Wang S and Lahaye T: Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol, 187(4), 1048–57 (2010) DOI: 10.1111/j.1469-8137.2010.03217.x [DOI] [PubMed] [Google Scholar]
- 131.Lamb BM, Mercer AC and Barbas CF 3rd: Directed evolution of the TALE N-terminal domain for recognition of all 5’ bases. Nucleic Acids Res, 41(21), 9779–85 (2013) DOI: 10.1093/nar/gkt754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.de Lange O, Schreiber T, Schandry N, Radeck J, Braun KH, Koszinowski J, Heuer H, Strauss A and Lahaye T: Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol, 199(3), 773–86 (2013) DOI: 10.1111/nph.12324 [DOI] [PubMed] [Google Scholar]
- 133.Jun AS, Trounce IA, Brown MD, Shoffner JM and Wallace DC: Use of transmitochondrial cybrids to assign a complex I defect to the mitochondrial DNA-encoded NADH dehydrogenase subunit 6 gene mutation at nucleotide pair 14459 that causes Leber hereditary optic neuropathy and dystonia. Mol Cell Biol, 16(3), 771–7 (1996) DOI: 10.1128/MCB.16.3.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ and Voytas DF: Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res, 39(12), e82 (2011) DOI: 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF and Vignali DA: Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol, 22(5), 589–94 (2004) DOI: 10.1038/nbt957 [DOI] [PubMed] [Google Scholar]
- 136.Coggan AR, Spina RJ, King DS, Rogers MA, Brown M, Nemeth PM and Holloszy JO: Histochemical and enzymatic comparison of the gastrocnemius muscle of young and elderly men and women. J Gerontol, 47(3), B71–6 (1992) DOI: 10.1093/geronj/47.3.B71 [DOI] [PubMed] [Google Scholar]
- 137.Hsu PD, Lander ES and Zhang F: Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157(6), 1262–78 (2014) DOI: 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T and Anderson DG: Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol, 32(6), 551–3 (2014) DOI: 10.1038/nbt.2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jo A, Ham S, Lee GH, Lee YI, Kim S, Lee YS, Shin JH and Lee Y: Efficient Mitochondrial Genome Editing by CRISPR/Cas9. Biomed Res Int, 2015, 305716 (2015) DOI: 10.1155/2015/305716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang G, Shimada E, Zhang J, Hong JS, Smith GM, Teitell MA and Koehler CM: Correcting human mitochondrial mutations with targeted RNA import. Proc Natl Acad Sci U S A, 109(13), 4840–5 (2012) DOI: 10.1073/pnas.1116792109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sallevelt SC, Dreesen JC, Drusedau M, Spierts S, Coonen E, van Tienen FH, van Golde RJ, de Coo IF, Geraedts JP, de Die-Smulders CE and Smeets HJ: Preimplantation genetic diagnosis in mitochondrial DNA disorders: challenge and success. J Med Genet, 50(2), 125–32 (2013) DOI: 10.1136/jmedgenet-2012-101172 [DOI] [PubMed] [Google Scholar]
- 142.Hellebrekers DM, Wolfe R, Hendrickx AT, de Coo IF, de Die CE, Geraedts JP, Chinnery PF and Smeets HJ: PGD and heteroplasmic mitochondrial DNA point mutations: a systematic review estimating the chance of healthy offspring. Hum Reprod Update, 18(4), 341–9 (2012) DOI: 10.1093/humupd/dms008 [DOI] [PubMed] [Google Scholar]
- 143.Craven L, Tuppen HA, Greggains GD, Harbottle SJ, Murphy JL, Cree LM, Murdoch AP, Chinnery PF, Taylor RW, Lightowlers RN, Herbert M and Turnbull DM: Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature, 465(7294), 82–5 (2010) DOI: 10.1038/nature08958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tachibana M, Amato P, Sparman M, Woodward J, Sanchis DM, Ma H, Gutierrez NM, Tippner-Hedges R, Kang E, Lee HS, Ramsey C, Masterson K, Battaglia D, Lee D, Wu D, Jensen J, Patton P, Gokhale S, Stouffer R and Mitalipov S: Towards germline gene therapy of inherited mitochondrial diseases. Nature, 493(7434), 627–31 (2013) DOI: 10.1038/nature11647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. http://www.hfea.gov.uk/6896.html.
- 146.Perales-Clemente E, Fernandez-Silva P, Acin-Perez R, Perez-Martos A and Enriquez JA: Allotopic expression of mitochondrial-encoded genes in mammals: achieved goal, undemonstrated mechanism or impossible task? Nucleic Acids Res, 39(1), 225–34 (2011) DOI: 10.1093/nar/gkq769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Manfredi G, Fu J, Ojaimi J, Sadlock JE, Kwong JQ, Guy J and Schon EA: Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat Genet, 30(4), 394–9 (2002) DOI: 10.1038/ng851 [DOI] [PubMed] [Google Scholar]
- 148.Oca-Cossio J, Kenyon L, Hao H and Moraes CT: Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics, 165(2), 707–20 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Claros MG: MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci, 11(4), 441–7 (1995) DOI: 10.1093/bioinformatics/11.4.441 [DOI] [PubMed] [Google Scholar]
- 150.Guy J, Qi X, Pallotti F, Schon EA, Manfredi G, Carelli V, Martinuzzi A, Hauswirth WW and Lewin AS: Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann Neurol, 52(5), 534–42 (2002) DOI: 10.1002/ana.10354 [DOI] [PubMed] [Google Scholar]
- 151. https://clinicaltrials.gov/ct2/show/NCT02064569.
- 152.Hashimoto M, Bacman SR, Peralta S, Falk MJ, Chomyn A, Chan DC, Williams SL and Moraes CT: MitoTALEN: A General Approach to Reduce Mutant mtDNA Loads and Restore Oxidative Phosphorylation Function in Mitochondrial Diseases. Mol Ther, 23(10), 1592–9 (2015) DOI: 10.1038/mt.2015.126 [DOI] [PMC free article] [PubMed] [Google Scholar]