Abstract
We present a framework to model microbial transformations in chemostats and retentostats under transient or quasi-steady state conditions. The model accounts for transformation-induced isotope fractionation and mass-transfer across the cell membrane. It also verifies that the isotope fractionation ϵ can be evaluated as the difference of substrate-specific isotope ratios between inflow and outflow. We explicitly considered that the dropwise feeding of substrate into the reactor at very low dilution rates leads to transient behavior of concentrations and transformation rates and use this information to validate conditions under which a quasi-steady state treatment is justified. We demonstrate the practicality of the code by modeling a chemostat experiment of atrazine degradation at low dilution/growth rates by the strain Arthrobacter aurescens TC1. Our results shed light on the interplay of processes that control biodegradation and isotope fractionation of contaminants at low (μg/L) concentration levels. With the help of the model, an estimate of the mass-transfer coefficient of atrazine through the cell membrane was achieved (0.0025s–1).
Introduction
Organic chemicals such as pesticides, pharmaceuticals, or personal-care products are ubiquitously used and have increasingly been detected in surface water and groundwater.1,2 Even though the concentrations are low (submicrograms-per-liter), levels are still high enough to be of potential concern.3 For instance, atrazine concentrations investigated in this study are, although low (20–50 μg/L), still above threshold values for drinking water worldwide (0.1 μg/L).4,5 These trace organics have received increased attention as micropollutants.6 While many micropollutants are biodegradable at high concentrations, their microbial degradation is observed to decrease at trace levels, down to a threshold at which natural attenuation appears to diminish.7 The question whether the reason is physiological adaptation of microorganisms (i.e., down-regulation of catabolic enzymes in response to substrate scarcity8), or bioavailability limitation of substrate (i.e., rate-limiting mass transfer into microbial cells when enzyme kinetics is no longer zero-order9,10) has been a long-standing debate. An answer to this question may offer a new perspective on the behavior of microorganisms at low concentrations.
Until now, it has been difficult to observe the onset of mass-transfer limitations directly. Even though the concept of bioavailability limitations is well-established,10 so far it is uncertain at which exact concentrations such a mass-transfer restriction comes into play, and how this relates to physiological adaptation. Recently, isotope fractionation has been brought forward as a new opportunity to accomplish precisely such a direct detection of mass transfer limitations.11−13 Basically, the isotope ratio of a micropollutant changes during a biochemical reaction since molecules with heavy isotopes are transformed at a slightly different rate than those with light isotopes.14−16 These changes, however, can only be observed if there is a rapid exchange of molecules within the cell interior at the enzyme level (bioavailable) with those outside the cell (bulk) where samples are taken for analysis. The exchange rate between bioavailable and bulk domains is described by a linear model in which the mass-transfer coefficient of the cell membrane is included.9,10,13 In the presence of mass transfer limitations (i.e., when mass transfer coefficients are small), the slow exchange rate of isotopologues between these domains generates pools of different isotopic ratios across the exchanging interface (i.e., the cell membrane). At the scale of a cell, this means molecules diffuse into or out of the cell at a rate much slower than the rate at which the enzymatic isotope effect occurs. The phenomenon has been usually referred to as masking of isotopic signatures meaning that the measured isotopic fractionation at bulk domain is notably different than the actual, transformation-induced isotopic fractionation occurring at bioavailable domain (i.e., the cell interior).12,13 As such, carbon and nitrogen isotope signatures provide direct evidence of mass-transfer limitations and have the potential to be used to quantify associated mass-transfer coefficients.
Previous studies examined such mass-transfer effects at relatively high concentration levels where bacteria were cultivated at sufficiently high substrate concentrations and then suddenly exposed to a low substrate concentration.11,12 The drawback was that cells could not adapt to a specific concentration in batch experiments, obstructing the interpretation of measured concentrations and isotope ratios. Thus, to assess the degree of influence that mass-transfer limitations exert at steady low concentrations, an experimental system is required that continuously maintains the contaminant concentration at a low and environmentally relevant level for a reasonably long time so that cells have enough time to adapt to low-energy conditions. This was beyond the reach of previously conducted batch experiments involving atrazine.14
A solution is offered by chemostats and retentostats that run at very low dilution rates. Here, substrate is continuously added and residual substrate and cells are continuously washed out from the bioreactor. Such chemostats are operated in a way that the essential growth rate equals the dilution rate so that biomass and residual concentrations remain constant within the reactor. While chemostat experiments have a long tradition in bioengineering,17,18 few studies have used them to study isotope fractionation.19,20 To our knowledge all preceding isotope studies in chemostat have measured isotope fractionation by taking the difference between substrate and product. This is particularly true for studies on photosynthesis which were run on nitrate limitation so that although mass transfer of carbon dioxide was addressed, bicarbonate was always present in great excess and was never the limiting substrate.21,22 In contrast, none has determined isotope fractionation by relating isotope ratios of the same substrate from the feed and the outflow of a reactor. In an experimental study submitted along with this contribution23 we therefore set out to study degradation of atrazine by the strain Arthrobacter aurescens TC1 in a chemostat at very low dilution rates (and thus low concentration levels) with the aim to pinpoint the onset of mass transfer limitation by compound-specific isotope analysis (CSIA).
Application of CSIA can unravel the underlying dynamics if validated by a chemostat model that is able to account for the mechanisms of mass transfer and transformation-based isotopic fractionation at low dilution rates. Furthermore, the model allows the delineation of the interactions between these processes in a traceable manner and thus provides a platform to critically evaluate the experimental setup, guide the experimental approach, precheck possible pitfalls, and assist in quantification of the results. The first aspect is the usual concern associated with chemostats running at very low dilution rates where a dropwise input may create discontinuities in substrate levels and result in adverse consequences. For instance, too-slow drip feeds may create “feast and famine” conditions for microorganisms preventing adaptation to a certain condition.24 As a consequence, the typical analyses of chemostats, which are based on the assumption of constant inflow conditions,25 do not accurately resolve the change of concentrations and isotopic values in waiting times between two subsequent droplets. To overcome this issue, we present a chemostat/retentostat model that considers the transient behavior under rapid changes of boundary conditions (here addressed by a periodic inlet). The model then enabled us to illustrate the extent of influence that inlet discontinuities may have on the steady-state observations. The second aspect is the new way in which degradation-associated isotope fractionation is evaluated in chemostats. Isotope fractionation has so far been calculated as a function of remaining substrate in batch experiments according to the Rayleigh equation.26,27 In chemostats, however, the substrate continuously enters and leaves the reactor, and the observed isotope fractionation must thus be derived from the difference between isotopic ratios of the same compound in inlet and outlet. This is again different from previous approaches which also considered the substrate in the inlet, but determined isotope fractionation by comparison to the product in the outlet. Using the model, we were able to confirm the validity of the experimental approach in the companion paper.23 The third aspect is the inclusion of mass transfer across the cells’ membrane (i.e., between the monitored bulk solution and the cell interior9) into the chemostat equations. It is worth noting that due to high stirring speeds in chemostat the effects of incomplete mixing in the bulk phase are negligible so that the transfer through the cell membrane remains as the only physical barrier. The model offers a platform to describe mass transfer through the cell wall and to derive tentative quantitative estimates on mass-transfer coefficients. The fourth and final aspect is related to sensitivity and error propagation analyses of the model in order to understand the relationships between the uncertainty of input parameters and model estimates. Global sensitivity analysis further contributes to our understanding of how the variation in the model estimates can be apportioned to the variation in the input parameters. The model was then applied to the experimental study of atrazine degradation by Arthrobacter aurescens TC1 at low concentrations, detailed in the companion paper.23
The overall aim of this contribution is to introduce a comprehensive modeling tool in order to quantitatively analyze the interactions between the following processes: (1) mass transfer through the cell membrane, (2) enzymatic transformation, and (3) transformation-induced compound-specific isotope fractionation in chemostats/retentostats with (4) periodic input of substrate.
Materials and Methods
Model Equations
We consider the concentrations of light and heavy isotopologues of a substrate (lS and hS [ML–3]), and the biomass concentration (X[ML–3]) as dynamic state variables. Note that the dimensions of all variables are introduced by bracketed variables T, M, and L, respectively, referring to the units of time, mass, and length. The turnover of substrate is described by Monod kinetics28 with competitive inhibition among the isotopologues, and is coupled to the input and output of substrate through the inflow and the outflow of the reactor, respectively. Biomass growth is assumed proportional to the substrate turnover via a yield factor. This leads to the following system of ordinary differential equations:
| 1a |
| 1b |
| 1c |
where rD[T–1] is the dilution rate coefficient (flow rate divided by the reactor volume), qmax[T–1] denotes the maximum specific conversion rate, Km[ML–3] is the half-saturation constant, m[T–1] is the maintenance term, α[−] indicates the isotopic fractionation factor, Y[−] is the yield coefficient, and f[−] denotes the fraction of biomass filtered at the outflow, ranging between zero (biomass leaves the system at the reactor current concentration; chemostat) and one (complete filtration of biomass thus no biomass discharges from the outlet; perfect retentostat). The maximum specific growth rate μmax[T–1] is related to qmax by μmax = Y(qmax – m).29,30
The chemostat equations accounting for the mass-transfer through the cell membrane are modified such that the concentrations outside the cells (S) differ from the concentrations inside the cells (Sbio). Thus, S and Sbio are referred to as the substrate concentrations in the bulk and bioavailable phases, respectively.9,31,32 A linear-driving force model with the mass-transfer coefficient ktr[T–1] was assumed to control the exchange between these two phases. Including such mass-transfer limitations, eqs 1a to 1c change as follows:
| 2a |
| 2b |
| 2c |
| 2d |
| 2e |
in which the observable isotope fractionation in the bulk phase is affected by the transformations inside the cell and the mass transfer between bulk and bioavailable phases. The initial concentrations for the substrate and biomass are indicated by Sini[ML–3] and Xini[ML–3]. The isotope ratio of the heavy and the light isotopologues of the substrate is evaluated in the common δhS[‰] notation:
| 3 |
typically expressed in parts per thousand, where R is the reference isotope ratio of VPDB (Vienna Pee Dee Belemnite). The model is presented in a general form and in principle can be applied to any stable isotope element. In this study, we examined the carbon isotope effects of atrazine and thus hS and lS are respectively replaced by 13S and 12S, representing the concentrations of substrate isotopologues containing heavy (13C) and light (12C) carbon isotopes. As a result, the δ13C notation replaces δhS and represents the observed isotopic signatures of carbon.
Model Solution
We solved the above systems of ordinary differential equations, ODE, (eqs 1a to 1c and eqs 2a to 2e) with the MATLAB ODE suite (e.g., the ode15s solver).33,34 To avoid unintended numerical instabilities, the input pulses were smoothed using forth-order analytical expressions.35 For smoothing the pulses, the user can choose the time period over which the pulse is smoothed, which may be interpreted as the mixing time in the system depending on agitation, droplet size, and reactor volume. A higher numerical stability is achieved when the smoothing intervals are larger. However, the smoothing interval should be substantially smaller than the interval between the pulses in order to avoid flattening the periodicity of the incoming droplets. Increasing the smoothing intervals will negate the very purpose of examining the droplet effect, as extreme smoothing would in principle be identical to having a continues feed (averaging the droplet volume over the time period and resulting in a constant feed). The smoothing type can be chosen between the following two polynomial spike functions:
| 4a |
| 4b |
producing either a smoothed pulses with a constant area underneath (in case of eq 4a) or a pulse that is set to reach to a specific peak height (in case of eq 4b). t[T] denotes the time variable which varies between zero and the time until the next droplet, s[T] denotes the length of the smoothing interval. Although both approaches are available in the model, we used the first smoothing function eq 4a as the other expression overestimates the introduction of mass into the system. We also skipped the maintenance term in the chemostat model since its effect on isotope signatures was found to be negligible (discussed in more details in Ehrl et al.23). According to Pirt,30 μmax = Yqmax when m is small enough to be treated as zero. The forthcoming sensitivity and uncertainty analyses then considers μmax as an input parameter instead of qmax. The parameter values are taken from the companion paper of Ehrl et al.23 for degradation of atrazine by the strain Arthrobacter aurescens TC1 in chemostat, and are listed in Table 1.
Table 1. Model Solution. Model Parameter Values Taken from Ehrl et al.23.
| reactor volume (V) | 2000 mL |
| dilution rate (rD) | 0.009 hr–1 |
| average droplet size (Vd) | 0.1 mL |
| average time between droplets (td) | 20 s |
| atrazine concentration at the inlet (Sin) | 30 000 μg/L |
| maximum specific conversion rate (qmax) | 6.01 hr–1 |
| half-saturation constant (Km) | 237 μg/L |
| yield factor (Y) | 0.018 |
| isotopic fractionation factor (α) | 0.9946 |
| initial atrazine concentration in reactor (Sini) | 65 μg/L |
| initial concentration of biomass in reactor (Xini) | 550 μg/L |
| fraction of biomass retained from chemostat outflow (f) | 0 |
Model Accuracy and Stability
The model is validated by comparing the results with the experiment23 and its accuracy is evaluated through the comparison with the analytical model of Thullner et al.13 Although eqs 1 and 2 are written in a general perspective and include essential terms such as maintenance energy, additional processes can still be introduced within the existing potentials of the model. For instance, the model allows introducing degradation mechanisms other than Monod (or Michaelis–Menten) kinetics, for example, at very small concentration levels ([S] ≪ Km) using a first-order kinetics might describe the system behavior more effectively, or in cases where the concentrations of both reaction partners (electron donor and acceptor) become rate-limiting, a dual Monod kinetics can be introduced. A similar flexibility holds for changing the mechanism controlling the rate of exchange across the cell membrane, which is currently expressed by a linear term and can be substituted by more sophisticated nonlinear expressions.
Use of MATLAB ODE suite as the internal solver increased model stability on handling relatively stiff problems. However, it should be noted that the model can still turn out numerically unstable if the smoothing interval of droplet is not sufficiently large with respect to the time period between droplets. As a rule of thumb, the smoothing interval should be around 15% of the period between droplets, that is, the time between each input cycle.
Results and Discussion
Model Results
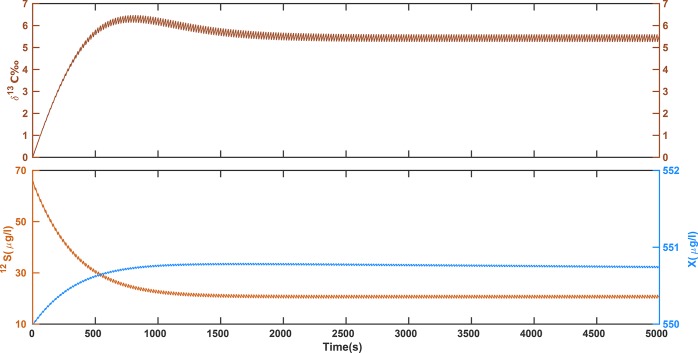
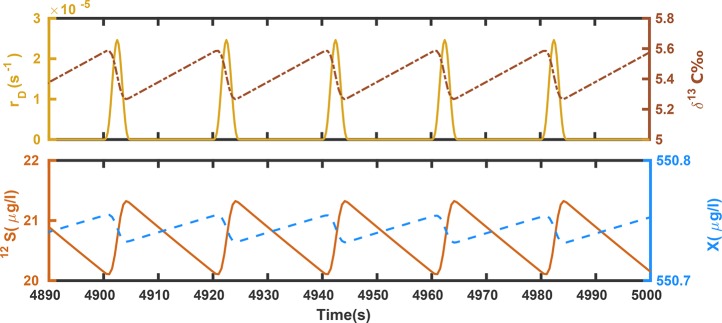
Regarding the first question—the effect of discontinuities—Figures 1 and 2 show that the model is well capable of capturing the transient behavior caused by drip-feeding of substrate (as it is perceived in the chemostats at very low dilution rates). The results confirm that the effects from a discontinuous input on concentrations and isotope compositions are small at the given dilution rate. Figure 2 displays the same data as Figure 1 over a short time period when dynamic steady state has been reached, and magnifies the recurrent fluctuations for better recognition of details. Under dynamic steady-state conditions the periodic input of droplets causes concentrations to fluctuate by 3% at most, which justifies the steady-state treatment adopted in the companion paper.23
Figure 1.
Solution of eqs 1a to 1c (in the absence of mass-transfer limitations across the cell membrane) for the following set of parameters: Sin = 30 000 μg/L, μmax = 0.11hr–1, Km = 237 μg/L, Y = 0.018, α = 0.9946, Sini = 65 μg/L, Xini = 550 μg/L, and rD = 0.009 hr–1. For better illustration of the droplet spikes, the dilution rates together with the changes of concentration, biomass, and δ13C at steady-state are shown over a short time span (100 s) in Figure 2. Although the concentrations of the substrate isotopologues decrease monotonically, the slight shift of timing between the light and heavy isotopologues cause a nonmonotonic behavior of the isotope ratios. As a result, the values of δ13C exceed slightly above the final value between times 500 and 1000 s.
Figure 2.
Solution of eqs 1a to 1c at steady-state. The figure is a close-up snapshot of the last 100 s in Figure 1 at which the system has reached steady-state. Based on size of droplet (0.1 mL), volume of chemostat (2 L), and the dilution rate (rD = 0.009 hr–1) the droplet frequency is calculated as one drop per every 20 s. The smoothing interval is assumed 5 s. For this setup, the results at steady-state are averaged as δ13C = 5.4 ± 0.2‰, 12S = 20.66 ± 0.6 μg/L, X = 550.74 ± 0.01 μg/L.
To address the second aspect,
the evaluation of isotope fractionation
from the inlet and the outlet of chemostat: the model was provided
with the actual, enzymatic, intrinsic isotopic fractionation for degradation
of atrazine by strain Arthrobacter aurescens TC1
ϵ13C = α–1 = −5.4‰
as input parameter (see Table 1). This value had been determined in batch experiments with
bacterial cultures degrading atrazine at high (mg/L) concentrations14,31 and with pure enzyme in the absence of bacterial cells.16 In all of these cases, mass-transfer limitations
are either absent or insignificant. Therefore, in the absence of a
mass-transfer term (solving eqs 1a to 1c), the model should predict
that the carbon isotope signatures δ13C inside the chemostat differs from that in the inflow by almost the
same enrichment factor ϵ13C of batch
studies (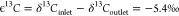 ). Figure 1 shows the simulated time series of concentrations
and δ-values for this case where the concentration inside the
cells equals the concentration in the bulk solution (eqs 1a to 1c).
As shown, the obtained δ13C values
at steady-state eventually approach the actual fractionation coefficient
reported from the batch experiments (δ13C = 5.4‰),14,16 validating the method of calculating
the evaluation of ϵ13C between the
inlet and the outlet of chemostat experiments. ϵ13C has been traditionally determined as the difference
between isotope values of an infinitely large reservoir of bicarbonate
in the chemostat and the biomass formed.21,22 The approach clearly does not work for our experiments for the following
reasons. In previous studies, bicarbonate was present in excess and
nitrate was the limiting source for growth whereas in our experiments
the carbon-containing substrate (atrazine) is the limiting source
and required to be depleted in order to mimic the environmentally
related conditions. Hence, the only way to determine epsilon is to
measure it as the difference between atrazine in inflow and outflow
(as theoretically derived by Hayes36).
In addition, the flow-through rate in a chemostat must be reasonably
slower than the rate of degradation in order to be able to identify
and measure the substrate decay, and to prevent overwriting the enzymatic
isotope fractionation by isotope ratios of the inflow. Solving eqs 1a and 1b at steady-state and assuming that
). Figure 1 shows the simulated time series of concentrations
and δ-values for this case where the concentration inside the
cells equals the concentration in the bulk solution (eqs 1a to 1c).
As shown, the obtained δ13C values
at steady-state eventually approach the actual fractionation coefficient
reported from the batch experiments (δ13C = 5.4‰),14,16 validating the method of calculating
the evaluation of ϵ13C between the
inlet and the outlet of chemostat experiments. ϵ13C has been traditionally determined as the difference
between isotope values of an infinitely large reservoir of bicarbonate
in the chemostat and the biomass formed.21,22 The approach clearly does not work for our experiments for the following
reasons. In previous studies, bicarbonate was present in excess and
nitrate was the limiting source for growth whereas in our experiments
the carbon-containing substrate (atrazine) is the limiting source
and required to be depleted in order to mimic the environmentally
related conditions. Hence, the only way to determine epsilon is to
measure it as the difference between atrazine in inflow and outflow
(as theoretically derived by Hayes36).
In addition, the flow-through rate in a chemostat must be reasonably
slower than the rate of degradation in order to be able to identify
and measure the substrate decay, and to prevent overwriting the enzymatic
isotope fractionation by isotope ratios of the inflow. Solving eqs 1a and 1b at steady-state and assuming that 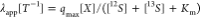 is the apparent first-order decay coefficient,
the following equation can be derived:
is the apparent first-order decay coefficient,
the following equation can be derived:
| 5 |
which is analogous to eq (8) in Farquhar et al.37 (see also the derivation in “Materials and Methods” of Ehrl et al.23). Thus, the difference between inflow and outflow would be expected to approach ϵ13C under realistic, sufficiently small dilution rates as it is also confirmed by the model.
Regarding the third aspect—in order to assess how observable isotope fractionation is influenced by mass-transfer limitations—we applied the model to the experimental data obtained in chemostat experiments of our companion paper.23 At high dilution rates (>0.018 hr–1) and at high bulk concentrations (>100 μg/L), the measured difference between isotopic ratios in the inlet and the outlet perfectly matched the isotope fractionation from batch experiments, similar to our model predictions in the absence of the mass transfer limiting term (see above). In contrast, Ehrl et al.23 observed lower isotope fractionation with decreasing chemostat dilution rates. At a dilution rate of 0.009 hr–1 an isotopic fractionation of ϵ13C = −2.2‰ was measured which was noticeably smaller in magnitude than the previously reported values for this reaction. This revealed the importance of mass transfer through the cell membrane under low-energy conditions. To reproduce a dilution rate of 0.009hr–1 in our model, a periodic input of every 20 s was assumed with droplets of approximately 0.1 mL into a chemostat with 2 L volume. Figure 3 shows the concentration and isotope time-series for this case (solution of eqs 2a to 2e). By solving eqs 2a to 2e, in which mass-transfer mechanisms are taken into account, the model was able to reproduce smaller δ13C values in the outlet (and, hence, smaller apparent isotope fractionation ϵ13C) when the exchange rate through the cell membrane was slowed by assigning low values of the mass-transfer coefficient ktr. In order to determine the value of ktr in the experiment, we used a trial and error fitting procedure. In this procedure, the value of ktr is constrained such that the late-time δ13C-values (at steady-state) equal the value observed in the experiment. At the dilution rate of 0.009 hr–1 using ktr value of 0.0025 s–1, we achieved an apparent isotopic enrichment value of ϵ13C = −2.2‰ which corresponds well to the reported value in Ehrl et al.23Figure 3 shows the concentration and isotope time-series for this case (solution of eqs 2a to 2e). Here, the simulated concentrations inside the cell Sbio were found to be only about 40% of the concentrations S outside the cell. Boosting the exchange rate between bulk and bioavailable domains through gradually increasing the value of the mass-transfer coefficient ktr in the model increased the late-time δ13C-values and eventually reached the value of the actual, transformation-induced, intrinsic isotopic fractionation coefficient ϵ13C = −5.4‰ (identical to the late-time δ13C-value in Figure 1).
Figure 3.
Solution of eqs 2a to 2e (in the presence of mass-transfer limitations across the cell membrane) for the set of parameter values in Figure 1 and ktr = 0.0025s–1. Note that due to mass-transfer limitations the observed δ13C = 2.2‰ at steady-state notably reduced from 5.4% in Figure 1. It is worth mentioning that inside cells (i.e., at the bioavailable domain) the δ13C is equal to the expected value of 5.4‰ (data not shown).
The evaluation of the forth aspect—sensitivity of model estimates to the input parameters (Table 1)—is detailed as follows.
Sensitivity and Uncertainty Analyses
Uncertainty Propagation Analyses
A Monte Carlo simulation was used to propagate the uncertainty originating from experimental and analytical variability of the parameters ktr, Km, μmax, and Sin onto concentrations and isotopic signatures. In order to reduce the total runtime of the Monte Carlo simulations, we reduced the walltime needed for simulating a single scenario to 7.5 s on a quad-cores Intel Core i5–4590 CPU at 3.30 GHz with 16GB RAM by optimizing the code and performing parallel computations.
Eqs 2a to 2e were solved for 50 000 randomly generated sets of parameters, which took about 105 h walltime. In each realization, the parameters of eqs 2a to 2e were perturbed at random, scaled to the experimentally obtained standard error. Mean values and standard deviations were calculated from repeated replicates (237 ± 57 μg/L for Km, 0.11 ± 0.02 hr–1 for μmax, and 30 000 ± 600 μg/L for Sin). In case of ktr, since the value is not experimentally determined, a relative standard error of 20% was presumed (0.0025 ± 0.0005s–1). All parameters were drawn from normal distributions and no correlation was assumed between the input parameters.
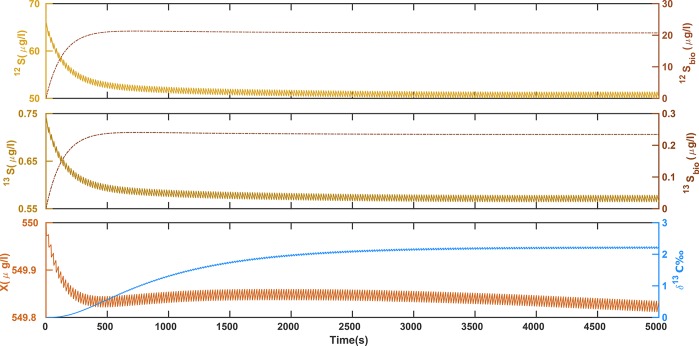
The Monte Carlo simulations showed probability distributions of the model outputs (δ13C, 12S, 13S, 12Sbio, 13Sbio, and X) as the result of the input parameter variabilities. Figure 4 shows the 16–84% probability range of model outcomes which corresponds to ±1 standard deviation of a normal distribution. Table 2 lists the average and standard deviation of all model predictions at late time. There is a small offset between the mean output of the ensemble calculation and a single run using the mean input parameter values which can be attributed to the nonlinear dependence of model outputs on the parameters. Figure 4 shows that the parameter uncertainty translates into a large uncertainty of model predictions, with coefficients of variation (also known as relative standard deviations) between 20% and 33% for solute concentrations and δ-values. Among all model predictions, biomass (X) was clearly the least affected by uncertainties.
Figure 4.
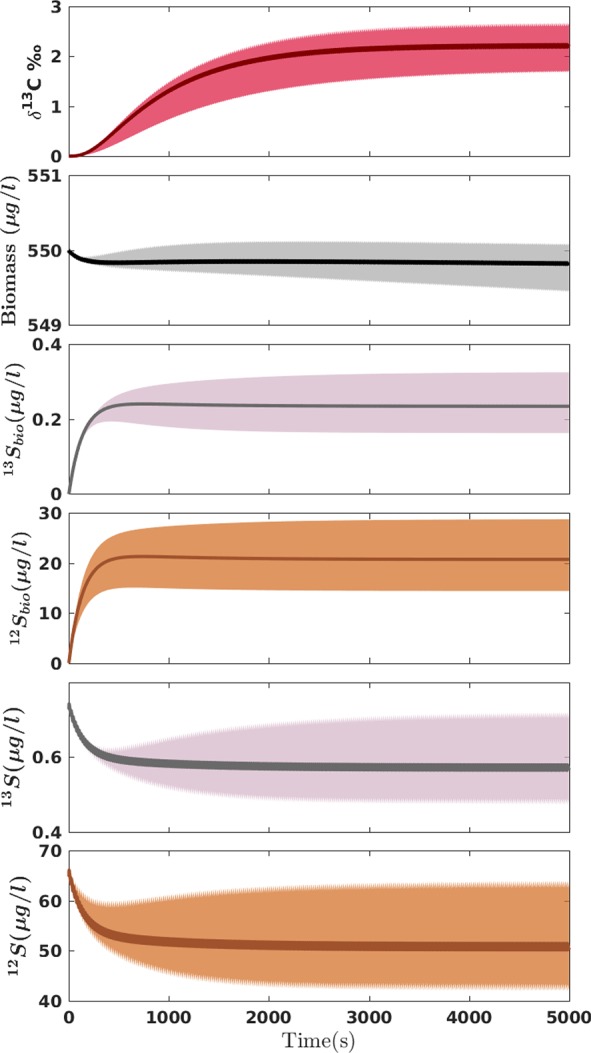
Uncertainty analysis using a Monte Carlo simulation. The 68% confidence intervals are shown for all the output parameters, from top to bottom, isotopic signature, biomass concentration, bioavailable, and bulk substrate concentrations (for both heavy and light isotopologues respectively). Note that the perturbations resulting from the periodic inlet are more visible at the profiles for bulk concentrations and δ13C-values.
Table 2. Uncertainty Analysisa.
| δ13C‰ | 12S (μg/L) | 13S (μg/L) | 12Sbio (μg/L) | 13Sbio (μg/L) | X (μg/L) | |
|---|---|---|---|---|---|---|
| model run with mean input parameters | 2.21 | 50.72 | 0.57 | 20.75 | 0.23 | 549.82 |
| Monte Carlo simulations | 2.17 ± 0.47 | 52.89 ± 10.25 | 0.59 ± 0.12 | 21.60 ± 7.18 | 0.24 ± 0.08 | 549.77 ± 0.31 |
The estimated average and standard error of output parameters calculated from Monte Carlo analyses of 50 000 randomly generated sample scenarios based on the error variability of input parameters (Km = 237 ± 57 μg/L, μmax = 0.11 ± 0.02hr–1, Sin = 30 000 ± 600 μg/L, and ktr = 0.0025 ± 0.0005s–1).
The 95% confidence interval of δ13C ≈ 2.17 ± 0.92‰ does not cover the value of δ13C = 5.4‰ expected from the isotope fractionation of the reaction.14,16 This clearly illustrates the ability of the model to pinpoint the limitations of mass transfer across the cell membrane as the origin of masked isotope fractionation in chemostats at low dilution rates. As a result, the observed isotopic signatures (δ13C) are noticeably smaller than the expected transformation-induced isotopic signatures. Sources of uncertainty exist that are not addressed by the Monte Carlo simulations, for example, the error in measuring the dilution rate or the uncertainties associated with the size of droplets. The error propagation of these factors is assumed to be insignificant and is partly lumped into the uncertainty of the inlet concentration (Sin).
Local Sensitivity Analysis
A tornado diagram is used here to depict the local sensitivity of the simulated δ13C-value at steady state with respect to the changes in the input parameters: ktr, Km, μmax, Sin, and the time between droplets 1/rD. To compare the relative importance of the above input parameters, we varied the value of one input parameter at a time by 20% while keeping all the other input parameters at their base values. As expected, the results (depicted in Figure 5) show a strong sensitivity toward the mass-transfer coefficient ktr in the chemostat model accounting for mass-transfer limitations eqs 2a to 2e. The modeled isotope signatures show a similar but weaker sensitivity to Sin and Km whereas variations of μmax and 1/rD inversely influence the values of δ13C noting the absolute sensitivity to μmax is on par with that to ktr. The results clearly indicate that the impact of physiological parameters (Km and μmax) are as significant as that of the physically motivated parameter (ktr).
Figure 5.
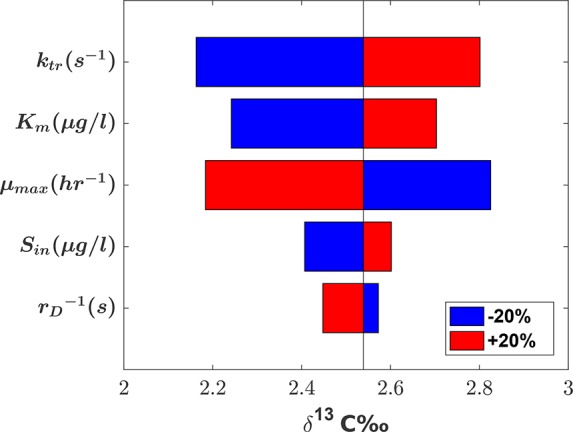
Local sensitivity analysis. Tornado plot showing the sensitivity of the observed δ13C values to the input variables ktr, Km, μmax, Sin, and the inlet periodic time (1/rD) when mass-transfer limitations across the cell membrane are present eqs 2a to 2e.
A similar sensitivity analysis was performed with the model neglecting mass-transfer limitations, eqs 1a to 1c. Unlike the previous model, the simulated late-time δ13C-values showed no sensitivity to the changes of the input parameters Km, μmax, Sin and 1/rD (data not shown). This implies that in the presence of mass transfer limitations, the sensitivity of the observed δ13C-values even to other input parameters (e.g., Km and μmax) is affected by the magnitude of the mass-transfer coefficient ktr.
Global Sensitivity Analysis
We used the variance-based analysis of Sobol38 for global sensitivity analysis (GSA). The benefit of a global over a local sensitivity analysis is that it accounts for the entire range of all parameter values rather than focusing on one parameter value at a time. As such, GSA offers a more robust solution in elucidating the impact of an individual parameter considering that all other parameters are also uncertain. To this end, a quasi Monte Carlo method (here, a Latin hypercube sequencing sampler) was employed to generate 60 000 sample scenarios that uniformly covered the space of input parameters. The first-order index (FOi) and the total-order index (TOi) were then calculated similar to Pianosi et al.39 and Sobol and Levitan.40 FOi indicates the effect of an individual parameter variation alone on an output variable, whereas TOi includes also the effects caused by the interactions of that parameter with all other parameters.
The pie charts in Figure 6 demonstrate the sensitivity of output variables: δ13C-values, biomass (X), bioavailable (Sbio) and bulk concentrations (S) to the input parameters Sin, μmax, Km, and ktr. The GSA confirms the relatively equal sensitivity of the δ13C-values to Km, μmax and ktr as previously estimated from the local sensitivity analysis (Figure 5). Bulk concentration showed a relatively high sensitivity of about 50% to the ktr values which is in the range of the combined sensitivity to all other input parameters. Among the model predictions, bulk concentrations are affected the most by mass transfer followed by δ13C-values at the second place. To our surprise, the bioavailable concentrations showed no sensitivity to mass-transfer effects. The variation of Km showed a predominant effect on the variation of all predicted quantities except biomass. In fact, biomass showed no sensitivity to variation of any input parameter. This might be due to the reason that in all scenarios the biomass concentration hardly changed with time (see Figure 4).
Figure 6.
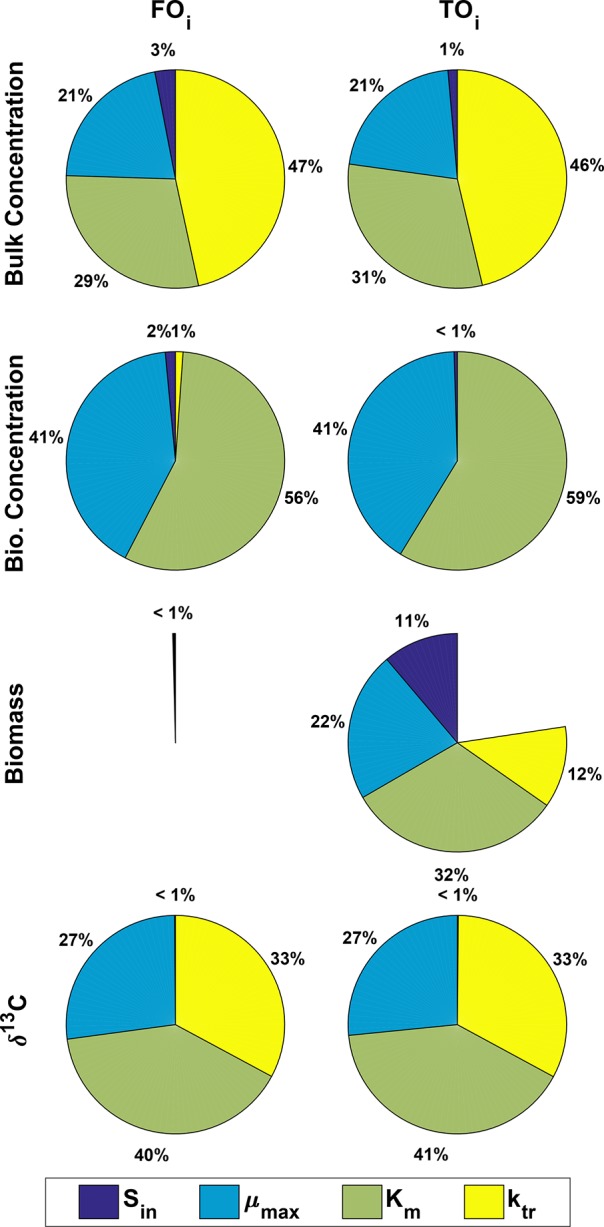
Global sensitivity analysis. The pie charts show the contributions of variability in the parameters ktr, Km, μmax, and Sin to the steady-state δ13C-values, X, Sbio, and S (Table 3) for the case where mass transfer is limited across the cell membrane eqs 2a to 2e.
The TOi pie charts provide a measure on the importance of interactions (of any order) between the input parameters. As shown in Table 3, the total order indices TOi and the first-order indices FOi were almost identical, indicating that the interactions between parameters did not impose any significant effect on variability of the model predictions except for biomass (X). We extended our GSA for another 60 000 sample scenarios to the total amount of 120 000 scenarios to check the consistency of the results and to see whether the sensitivity indices can be improved. Similar indices as those listed in Table 3 were calculated for all model outputs except for the biomass (data not shown). The inconsistency between biomass indices (obtained from 60 000 and 120 000 sample scenarios) indicates that the calculated sensitivity indices for biomass are possibly incorrect. This might have been caused by numerical errors originating mainly from the negligible change of biomass with time.
Table 3. Global Sensitivity Analysisa.
| FOi |
TOi |
|||||||
|---|---|---|---|---|---|---|---|---|
| δ13C | X | Sbio | S | δ13C | X | Sbio | S | |
| Sin | 0.0014 | 0 | 0.0152 | 0.0377 | 0 | 0.01119 | 0.0049 | 0.0165 |
| μmax | 0.2706 | 0 | 0.4153 | 0.2663 | 0.2658 | 0.2217 | 0.4108 | 0.2559 |
| Km | 0.4010 | 0 | 0.5737 | 0.3584 | 0.4052 | 0.3194 | 0.5914 | 0.3690 |
| ktr | 0.3294 | 0 | 0.0114 | 0.5780 | 0.3276 | 0.1208 | 0 | 0.5531 |
The first-order index (FOi) and the total-order index (TOi) of the output parameters (δ13C-values, X, Sbio, and S) in respect to the input parameters (Sin, μmax, Km, and ktr). The higher the value, the more impact the input variability exerts on the variance of the output parameter. Note that both heavy and light isotopologues showed a similar sensitivity trend in bulk and bioavailable domains.
Temporal Dynamics of Biomass Growth
The model accounts for the temporal dynamics of biomass growth and washout in the chemostat system eqs 1c and 2e. We assumed standard Monod kinetics28 in which biomass growth is proportional to the turnover rate. Growth depends only on the concentration of a single substrate, indicating that all other compounds required for growth are available in excess. The only removal term is described by washout via outflow. This is a reasonable assumption for a chemostat system, in which the loss due to washout is considerably greater than the biomass death rate. Maintenance terms are also not considered since the energy demand for maintenance is constant under quasi steady-state conditions. Hence, the maintenance effect is conveniently assumed to be subsumed in the yield factor (for an explicit treatment of maintenance energy see the Supporting Information of Ehrl et al.23). Furthermore, we did not consider a prescribed carrying capacity, or maximum biomass concentration, since the simulated biomass concentration remained fairly low as a result of limited supply of substrate and continuous washout of cells. Such an assumption is not valid for a model of a perfect retentostat where washout of biomass is prohibited and as a result biomass growth must be balanced by the maintenance energy requirement, biomass decay, or reaching the maximum carrying capacity.
In Figure 3, the biomass decreases at late times while substrate concentrations reach steady state. This can be explained by the initial biomass concentration being higher than the steady-state biomass which is controlled by the balance between bacterial growth and dilution rate. Here, a high initial biomass concentration mimics the conditions of an inoculum at high concentration levels.
Comparison with the Analytical Model of Thullner et al. (2008)
We compared our model to the analytical model of Thullner et al.13 which estimates the observed isotopic fractionation factor α under steady-state conditions in relation to the intrinsic isotropic fractionation of the enzymatic reaction α°,
| 6 |
where T = (a/ktr – S/Km – 1) is a dimensionless term and a = μmax/Km is the specific affinity of the microorganism promoting the enzymatic reaction. For an arbitrary case of ktr = 0.002s–1, Km = 50 μg/L, μmax = 0.027 hr–1, Y = 0.036, Sini = 65 μg/L, Xini = 1000 μg/L, α° = 0.994 and rD = 2.5× 10–6s–1, the observed δ13C at steady-state was calculated by our model to about 2.64‰. Using eq 5, the apparent fractionation factor α was calculated as 0.99738 which yields the observed δ13C = 2.62‰. This means that the two models estimated similar observed vs expected isotopic signatures. It is worth noting that unlike the analytical model,13 the presented numerical model can determine the observed isotopic signatures also under transient conditions.
Implications for Natural Systems
The model validated the approach of isotope fractionation measurements between the outflow and the inflow of a chemostat where a steady, low, and environmentally related concentration of a micropollutant is maintained for a time long enough to allow the adaptation of bacterial cultures. The model elucidates the role of mass-transfer limitations across the cell membrane in regulating the observed vs expected compound-specific isotopic signatures in chemostats. In addition, our results confirm that slow mass transfer across the cell membrane can mask the true isotope fractionation of a chemical transformation. So far the differences between observed isotopic signatures from laboratory and field were attributed to other factors, such as leakage from other contaminant sources or hydrologically driven mechanisms (e.g., by transverse dispersion at plume fringes41). As shown here, such differences in isotope fractionation can also stem from bioavailability limitations and may even originate from mass-transfer limitations across the cell membrane. The effect from bioavailability limitations is much more pronounced at low concentrations, and therefore is of high relevance for many micropollutants of which concentrations typically do not exceed micrograms-per-liter. Recognition and understanding of the interplay of bioavailability limitations with other existing processes thus enhance the overall interpretation of isotope signatures under field conditions.
Under the influence of other processes the isotopic signatures show no dependency on enzymatic reaction rates. Thus, one way to identify the masking of isotope signatures as the result of mass-transfer through a cell membrane is to focus on the fact that isotopic signatures are highly sensitive to enzymatic transformation rates in the presence of mass-transfer limitations (see the sensitivity of δ13C to μmax in the presence of ktr). Therefore, two strains with different metabolic activities when feeding on a single substrate must exhibit different isotopic signatures under mass-transfer limitations, assuming both have an identical isotopic fractionation factor and similar cell membrane characteristics.
Potential Model Applications
The presented model improves the mechanistic understanding of contaminant degradation in microbial ecosystems. While the model in its current form is only applied to fully mixed reactors, it can be easily coupled to solute transport equations42−44 contributing to the development of models that more realistically describe fixed-bed reactors and natural subsurface systems.
A practical aspect of our model is its capacity to calculate the membrane permeability of a specific cell in conjugation with chemostat/batch experiments. The differences between the observed isotopic signatures (δ13C) in batch and chemostat experiments are linked to mass-transfer limitations through the cell membrane which is widely referred to as membrane permeability. The formulation on how to obtain the value of membrane permeability Papp[LT–1] and the diffusion coefficient through the membrane Dmem[L2T–1] from the mass-transfer limiting coefficient ktr[T–1] is presented and discussed by Ehrl et al.23 According to the model results, atrazine permeation through the cell wall of Arthrobacter aurescens TC1 was approximated as Papp = 3.5 × 10–5ms–1 and Dmem = 1.9 × 10–16m2s–1, which are close to the values reported for a typical range of small organic molecules.45−47 While different techniques are used in pharmaceutical studies to determine the membrane permeability, the present model provides an alternative way of estimating it.
Sensitivity analysis of the model enables users to inspect the influence of different physical and physiological parameters on the observable isotopic signature before performing the experiments. The results provide clarity into the specific features influencing isotopic signatures in chemo- and retentostats. The modeling framework used in this study allows for a delineation of features such as (i) biodegradation dynamics of a contaminant, (ii) metabolic activity of the microbial degrader, (iii) the role of bioavailability limitations and typical mass-transfer restrictions through a cell’s membrane, and (iv) whether the interplay between these mechanisms is responsible for observing uncommon isotopic signatures at low concentration levels. As shown above, these results have relevant implications for both theory building and practical application.
Acknowledgments
This research has received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7-IDEAS-ERC)/ERC Grant Agreement No. 616861 (MicroDegrade).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.8b02498.
The authors declare no competing financial interest.
Supplementary Material
References
- Daughton C.International Encyclopedia of Public Health; Heggenhougen H. K. K., Ed.; Academic Press: Oxford, 2008; pp 66–102. [Google Scholar]
- Ohe T.; Watanabe T.; Wakabayashi K. Mutagens in surface waters: a review. Mutat. Res., Rev. Mutat. Res. 2004, 567, 109–149. 10.1016/j.mrrev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Daughton C. G.; Ternes T. A. Pharmaceuticals and personal care products in the environment: agents of subtle change?. Environ. Health Perspect. 1999, 107 (Suppl 6), 907–938. 10.1289/ehp.99107s6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council Directive 98/83/EC on the quality of water intended for human consumption. Official Journal of the European Communities, 1998; adopted on 3 November 1998, L 330/32.
- Villanueva C. M.; Durand G.; Coutté M.-B.; Chevrier C.; Cordier S. Atrazine in municipal drinking water and risk of low birth weight, preterm delivery, and small-for-gestational-age status. Occup. Environ. Med. 2005, 62, 400–405. 10.1136/oem.2004.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach R. P.; Escher B. I.; Fenner K.; Hofstetter T. B.; Johnson C. A.; von Gunten U.; Wehrli B. The Challenge of Micropollutants in Aquatic Systems. Science 2006, 313, 1072–1077. 10.1126/science.1127291. [DOI] [PubMed] [Google Scholar]
- Egli T. How to live at very low substrate concentration. Water Res. 2010, 44, 4826–4837. 10.1016/j.watres.2010.07.023. [DOI] [PubMed] [Google Scholar]
- Wick A.; Wagner M.; Ternes T. A. Elucidation of the Transformation Pathway of the Opium Alkaloid Codeine in Biological Wastewater Treatment. Environ. Sci. Technol. 2011, 45, 3374–3385. 10.1021/es103489x. [DOI] [PubMed] [Google Scholar]
- Best J. The inference of intracellular enzymatic properties from kinetic data obtained on living cells: Some kinetic considerations regarding an enzyme enclosed by a diffusion barrier. J. Cell. Comp. Physiol. 1955, 46, 1–27. 10.1002/jcp.1030460102. [DOI] [PubMed] [Google Scholar]
- Bosma T. N. P.; Middeldorp P. J. M.; Schraa G.; Zehnder A. J. B. Mass Transfer Limitation of Biotransformation: Quantifying Bioavailability. Environ. Sci. Technol. 1997, 31, 248–252. 10.1021/es960383u. [DOI] [Google Scholar]
- Aeppli C.; Berg M.; Cirpka O. A.; Holliger C.; Schwarzenbach R. P.; Hofstetter T. B. Influence of Mass-Transfer Limitations on Carbon Isotope Fractionation during Microbial Dechlorination of Trichloroethene. Environ. Sci. Technol. 2009, 43, 8813–8820. 10.1021/es901481b. [DOI] [PubMed] [Google Scholar]
- Kampara M.; Thullner M.; Richnow H. H.; Harms H.; Wick L. Y. Impact of Bioavailability Restrictions on Microbially Induced Stable Isotope Fractionation. 2. Experimental Evidence. Environ. Sci. Technol. 2008, 42, 6552–58. 10.1021/es702781x. [DOI] [PubMed] [Google Scholar]
- Thullner M.; Kampara M.; Richnow H. H.; Harms H.; Wick L. Y. Impact of Bioavailability Restrictions on Microbially Induced Stable Isotope Fractionation. 1. Theoretical Calculation. Environ. Sci. Technol. 2008, 42, 6544–51. 10.1021/es702782c. [DOI] [PubMed] [Google Scholar]
- Meyer A. H.; Penning H.; Elsner M. C and N Isotope Fractionation Suggests Similar Mechanisms of Microbial Atrazine Transformation Despite Involvement of Different Enzymes (AtzA and TrzN). Environ. Sci. Technol. 2009, 43, 8079–8085. 10.1021/es9013618. [DOI] [PubMed] [Google Scholar]
- Meyer A.; Dybala-Defratyka A.; Alaimo P.; Geronimo I.; Sanchez A.; Cramer C.; Elsner M. Cytochrome P450-catalyzed dealkylation of atrazine by Rhodococcus sp. strain NI86/21 involves hydrogen atom transfer rather than single electron transfer. Dalton Transactions 2014, 43, 12175–86. 10.1039/C4DT00891J. [DOI] [PubMed] [Google Scholar]
- Schürner H. K. V.; Seffernick J. L.; Grzybkowska A.; Dybala-Defratyka A.; Wackett L. P.; Elsner M. Characteristic Isotope Fractionation Patterns in s-Triazine Degradation Have Their Origin in Multiple Protonation Options in the s-Triazine Hydrolase TrzN. Environ. Sci. Technol. 2015, 49, 3490–3498. 10.1021/es5055385. [DOI] [PubMed] [Google Scholar]
- Hartmann R.; Hany R.; Witholt B.; Zinn M. Simultaneous Biosynthesis of Two Copolymers in Pseudomonas putida GPo1 Using a Two-Stage Continuous Culture System. Biomacromolecules 2010, 11, 1488–1493. 10.1021/bm100118t. [DOI] [PubMed] [Google Scholar]
- Khunjar W. O.; Mackintosh S. A.; Skotnicka-Pitak J.; Baik S.; Aga D. S.; Love N. G. Elucidating the Relative Roles of Ammonia Oxidizing and Heterotrophic Bacteria during the Biotransformation of 17α-Ethinylestradiol and Trimethoprim. Environ. Sci. Technol. 2011, 45, 3605–12. 10.1021/es1037035. [DOI] [PubMed] [Google Scholar]
- Penning H.; Plugge C. M.; Galand P. E.; Conrad R. Variation of carbon isotope fractionation in hydrogenotrophic methanogenic microbial cultures and environmental samples at different energy status. Global Change Biol. 2005, 11, 2103–13. 10.1111/j.1365-2486.2005.01076.x. [DOI] [PubMed] [Google Scholar]
- Trautwein K.; Lahme S.; Wöhlbrand L.; Feenders C.; Mangelsdorf K.; Harder J.; Steinbüchel A.; Blasius B.; Reinhardt R.; Rabus R. Physiological and Proteomic Adaptation of Aromatoleum aromaticum EbN1 to Low Growth Rates in Benzoate-Limited, Anoxic Chemostats. J. Bacteriol. 2012, 194, 2165–80. 10.1128/JB.06519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws E. A.; Popp B. N.; Bidigare R. R.; Kennicutt M. C.; Macko S. A. Dependence of phytoplankton carbon isotopic composition on growth rate and (CO2)aq: Theoretical considerations and experimental results. Geochim. Cosmochim. Acta 1995, 59, 1131–1138. 10.1016/0016-7037(95)00030-4. [DOI] [Google Scholar]
- Wilkes E. B.; Carter S. J.; Pearson A. CO2-dependent carbon isotope fractionation in the dinoflagellate Alexandrium tamarense. Geochim. Cosmochim. Acta 2017, 212, 48–61. 10.1016/j.gca.2017.05.037. [DOI] [Google Scholar]
- Ehrl B. N.; Kundu K.; Gharasoo M.; Marozava S.; Elsner M.. Rate-Limiting Mass Transfer in Micropollutant Degradation Revealed by Isotope Fractionation in Chemostat. Environ. Sci. Technol. 2018, 10.1021/acs.est.8b05175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Verseveld H. W.; Chesbro W. R.; Braster M.; Stouthamer A. H. Eubacteria have 3 growth modes keyed to nutrient flow. Arch. Microbiol. 1984, 137, 176–184. 10.1007/BF00414463. [DOI] [PubMed] [Google Scholar]
- Fritsch C.; Harmand J.; Campillo F. A modeling approach of the chemostat. Ecol. Modell. 2015, 299, 1–13. 10.1016/j.ecolmodel.2014.11.021. [DOI] [Google Scholar]
- Abe Y.; Hunkeler D. Does the Rayleigh Equation Apply to Evaluate Field Isotope Data in Contaminant Hydrogeology?. Environ. Sci. Technol. 2006, 40, 1588–96. 10.1021/es051128p. [DOI] [PubMed] [Google Scholar]
- Kopinke F.-D.; Georgi A.; Voskamp M.; Richnow H. H. Carbon Isotope Fractionation of Organic Contaminants Due to Retardation on Humic Substances: Implications for Natural Attenuation Studies in Aquifers. Environ. Sci. Technol. 2005, 39, 6052–62. 10.1021/es040096n. [DOI] [PubMed] [Google Scholar]
- Monod J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. 10.1146/annurev.mi.03.100149.002103. [DOI] [Google Scholar]
- Heijnen J.Encyclopedia of Bioprocess Technology; American Cancer Society, 2002; pp 286–290. [Google Scholar]
- Pirt S. Maintenance energy: a general model for energy-limited and energy-sufficient growth. Arch. Microbiol. 1982, 133, 300–302. 10.1007/BF00521294. [DOI] [PubMed] [Google Scholar]
- Ehrl B. N.; Gharasoo M.; Elsner M. Isotope fractionation pinpoints membrane permeability as barrier to atrazine biodegradation in gram negative Polaromonas sp. Nea-C. Environ. Sci. Technol. 2018, 52, 4137–44. 10.1021/acs.est.7b06599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharasoo M.; Centler F.; Van Cappellen P.; Wick L. Y.; Thullner M. Kinetics of Substrate Biodegradation under the Cumulative Effects of Bioavailability and Self-Inhibition. Environ. Sci. Technol. 2015, 49, 5529–5537. 10.1021/es505837v. [DOI] [PubMed] [Google Scholar]
- Gharasoo M.; Thullner M.; Elsner M. Introduction of a new platform for parameter estimation of kinetically complex environmental systems. Environ. Model Softw 2017, 98, 12–20. 10.1016/j.envsoft.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampine L. F.; Reichelt M. W. The MATLAB ODE Suite. SIAM J. Sci. Comput 1997, 18, 1–22. 10.1137/S1064827594276424. [DOI] [Google Scholar]
- Weckesser W.A Matlab function that creates periodic pulses with a given width and amplitude one. 2007; www.warrenweckesser.net/software/matlab/.
- Hayes J. M. Fractionation of Carbon and Hydrogen Isotopes in Biosynthetic Processes. Rev. Mineral. Geochem. 2001, 43, 225–277. 10.2138/gsrmg.43.1.225. [DOI] [Google Scholar]
- Farquhar G. D.; O’Leary M. H.; Berry J. A. On the relationship between carbon isotope discrimination and the intercellular carbon dioxide concentration in leaves. Aust. J. Plant Physiol. 1982, 9, 121–137. 10.1071/PP9820121. [DOI] [Google Scholar]
- Sobol I. Global sensitivity indices for nonlinear mathematical models and their Monte Carlo estimates. Math Comput. Simul 2001, 55, 271–280. 10.1016/S0378-4754(00)00270-6. [DOI] [Google Scholar]
- Pianosi F.; Sarrazin F.; Wagener T. A Matlab toolbox for Global Sensitivity Analysis. Environ. Model Softw 2015, 70, 80–85. 10.1016/j.envsoft.2015.04.009. [DOI] [Google Scholar]
- Sobol I.; Levitan Y. On the use of variance reducing multipliers in Monte Carlo computations of a global sensitivity index. Comput. Phys. Commun. 1999, 117, 52–61. 10.1016/S0010-4655(98)00156-8. [DOI] [Google Scholar]
- Rolle M.; Chiogna G.; Bauer R.; Griebler C.; Grathwohl P. Isotopic Fractionation by Transverse Dispersion: Flow-through Microcosms and Reactive Transport Modeling Study. Environ. Sci. Technol. 2010, 44, 6167–6173. 10.1021/es101179f. [DOI] [PubMed] [Google Scholar]
- Cirpka O. A.; Frind E. O.; Helmig R. Numerical methods for reactive transport on rectangular and streamline-oriented grids. Adv. Water Resour. 1999, 22, 711–728. 10.1016/S0309-1708(98)00051-7. [DOI] [Google Scholar]
- Gharasoo M.; Centler F.; Regnier P.; Harms H.; Thullner M. A reactive transport modeling approach to simulate biogeochemical processes in pore structures with pore-scale heterogeneities. Environ. Model Softw 2012, 30, 102–114. 10.1016/j.envsoft.2011.10.010. [DOI] [Google Scholar]
- Gharasoo M.; Centler F.; Fetzer I.; Thullner M. How the chemotactic characteristics of bacteria can determine their population patterns. Soil Biol. Biochem. 2014, 69, 346–358. 10.1016/j.soilbio.2013.11.019. [DOI] [Google Scholar]
- Males R.; Herring F. A 1H-NMR study of the permeation of glycolic acid through phospholipid membranes. Biochim. Biophys. Acta, Biomembr. 1999, 1416, 333–338. 10.1016/S0005-2736(98)00236-3. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y.; Yuasa H.; Iga T.; Hanano M. Determination of the membrane permeability coefficient and the reflection coefficient by the two-dimensional laminar flow model for intestinal perfusion experiments. Biochim. Biophys. Acta, Biomembr. 1986, 854, 191–197. 10.1016/0005-2736(86)90110-0. [DOI] [PubMed] [Google Scholar]
- van Meer G.; Voelker D.; Feigenson G. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.