Abstract
“Non-fouling” polymer brush surfaces can greatly improve the performance of in vitro diagnostic (IVD) assays due to the reduction of non-specific protein adsorption and consequent improvement of signal-to-noise ratios. In this progress report, we briefly review the development of synthetic polymer brush architectures that suppress adventitious protein adsorption and discuss their integration into surface plasmon resonance and fluorescent sandwich immunoassay formats. We also highlight a novel, self-contained immunoassay platform (the D4 assay), that transforms time-consuming laboratory-based assays into a user-friendly and point-of-care format with a sensitivity and specificity comparable or better than standard ELISA directly from unprocessed samples. These advancements clearly demonstrate the utility of non-fouling polymer brushes as a substrate for ultra-sensitive and robust diagnostic assays that may be suitable for clinical testing, in field and laboratory settings.
Keywords: Non-fouling surfaces, polymer brushes, immunoassays, in vitro diagnostics
Graphical Abstract
The table of contents entry should be 50–60 words long, and the first phrase should be bold. The entry should be written in the present tense and impersonal style. The text should be different from the abstract text.
The development of non-fouling polymer brushes surfaces has ushered in a new wave of vitro diagnostic (IVD) tests. Polymer brushes eliminate non-specific protein adsorption to the surface and thus enable ultra-sensitive detection of proteins directly from complex biological milieu. Diagnostic platforms capable of point-of-care testing are highlighted in this article.
1. INTRODUCTION
In vitro diagnostics (IVDs) play a critical role in the clinical diagnosis, management, and monitoring of disease. IVDs are designed to* analyze biological specimens to identify analytes of interest (i.e. proteins, lipids, metabolites, etc.) that can be insightful in guiding clinical decision making. Enzyme-linked immunosorbent assay (ELISA) and “ELISA-like” sandwich immunoassays are the clinical gold standard IVD for precise, sensitive, and quantifiable protein detection [1, 2] with many tests developed for the detection of protein biomarkers, ranging from infectious diseases to cancer. Recently, much effort has been dedicated to the development of IVDs amenable for point-of-care testing, thereby democratizing access to clinical-grade diagnostics independent of clinical infrastructure.
While ELISA offers exquisite analytical sensitivity and specificity, the assay procedure is quite complex, cumbersome and time consuming, and typically requires a dedicated centralized laboratory with highly-trained personnel and specialized equipment. A typical 96-well microplate ELISA procedure requires incubation of a clinical sample such as blood or serum (which often needs to be diluted) onto a plate pre-coated with a capture reagent—most commonly an antibody (Ab) or antigen (Ag)—addition and incubation of a detection Ab followed by a secondary Ab, and several wash steps in between.[2] Even though ELISA is often automated in centralized laboratories and manufacturers supply ready to use kits, which greatly reduce the challenges related to assay fabrication and assay standardization, the procedure typically takes 4 – 5 hours, which can delay clinical decision making. To improve upon the standard 96-well plate ELISA systems, miniaturized and microfluidic-driven ELISAs have been developed, which can speed up the process workflow tremendously (< 2 hours) using small sample volumes to facilitate clinical decision making.[3–8] The analytical performance of several of these platforms is encouraging and discussed later in this progress report. However, miniature and microfluidic ELISAs often have “active” components (i.e. pumps and valves), which increases the complexity, costs, and likelihood of device malfunction, especially if the intended use is in point-of-care settings. In both traditional ELISA and many of the new configurations, several steps in the assay attempt to maximize the signal-to-noise ratio (SNR), either by amplifying the signal or by reducing non-specific binding (NSB) of non-target proteins in the sample. For example, enzymes are often used to amplify the signal and thereby the SNR, and blocking buffers are used to eliminate adventitious protein adsorption, which can lead to an elevation in background signal and loss in sensitivity of the assay.
Increasing analytical sensitivity has traditionally been accomplished by using signal enhancement techniques; however, these methods can often raise the noise threshold of the detection system and can also increase the complexity of the assay. Due to the importance of eliminating NSB for IVDs, much effort has been dedicated into further understanding protein adsorption processes with the goal of creating “non-fouling” surfaces that resist NSB of proteins below the limit-of-detection (LOD) of label-free sensitive techniques.[9–11] Rather than employing blocking buffers, synthetic polymer coatings offer an alternative approach to engineer surfaces to prevent non-specific protein adsorption. The use of non-fouling surfaces can reduce or eliminate NSB and therefore increase the SNR, improving the assay’s LOD while eliminating the need for complex signal amplification.[10, 12, 13] Increasing the sensitivity of clinical diagnostics can enable earlier and more reliable diagnosis of disease, which can lead to early intervention, better patient outcomes and lower healthcare costs.[14]
Historically, polyethylene glycol (PEG) has been the most popular coating to confer protein resistance to a surface.[15, 16] PEG’s protein resistance is attributed to its highly hydrated and random conformation. PEG surface modification can be performed by either “grafting to” or “grafting from” the surface. Physisorption [17], chemisorption [18] and covalent grafting [19, 20] are amongst the most popular “grafting to” techniques. Although these immobilization methods can create strongly bound polymer chains to the surface (especially with covalent grafting), the “grafting to” approaches usually suffer from low polymer density due to the diffusion-limited nature of the immobilization process.[21] During immobilization, mushroom-like structures are formed by the long chains of the polymer, sterically hindering the approach of other chains in their radius, ultimately preventing their attachment. Surface densities normally achieved with PEG coatings by “grafting to” methods can reduce the adsorption of proteins in the ng cm−2 regime, but have difficulty preventing non-specific binding upon exposure to complex biological milieu.[21–23]
The extensive prior work performed on PEG and the development of non-fouling self-assembled monolayers (SAMs) of oligoethylene glycol-terminated alkanethiols on gold by Prime and Whitesides [24, 25] laid the foundation for our laboratory’s interest in the rational design of non-fouling polymer coatings. We sought to combine the ease of formation of SAMs with thicker and more robust polymer architectures to engineer a protein resistant surface coating. Specifically, surface-initiated polymerization (SIP) of three-dimensional, PEG-like brushes was a promising alternative to design thicker and more robust surface coatings. With SIP, the architecture and overall properties of brushes such as the grafting density, thickness of coating and hydrophilicity of the structure can be precisely controlled, as described extensively in a separate review by Krishnamoorthy et al.[26] The development of SIP by living radical polymerization (LRP) provided the tools to synthesize high density polymer brushes with accurate control of thickness and composition. Amongst the different types of LRP, atom transfer radical polymerization (ATRP) is most commonly used for the synthesis of non-fouling surfaces for several reasons: (1) ATRP initiators can be easily synthesized and immobilized onto surfaces by different methods such as plasma grafting, covalent coupling, Langmuir-Blodgett deposition and chemical self-assembly; (2) ATRP conditions can be tuned to control the thickness and polymer graft density; and (3) ATRP can be performed in a diverse range of solvents and under mild experimental conditions [27, 28] and recently, various SI-ATRP schemes have been developed that can be performed in air under ambient conditions without the need for deoxygenation.[29–31]
Herein, we discuss the progress in designing non-fouling synthetic polymer brushes and using them as a substrate for IVDs, with a special focus on immunoassays using optical transduction methods. We highlight advances from our laboratory towards the successful translation of IVDs from a microarray format to a point-of-care test (POCT). Finally, we discuss future opportunities and directions for the field to enable the successful development and clinical implementation of novel POCTs.
2. POLYMER BRUSHES WITH NON-FOULING PROPERTIES
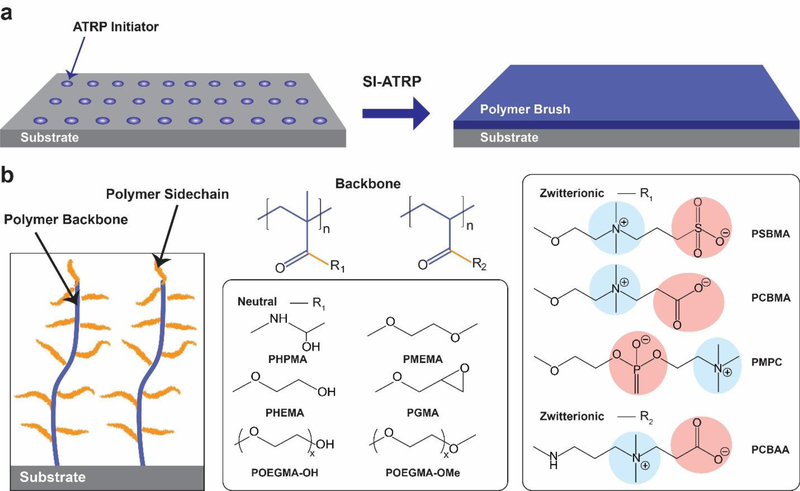
While many different polymer brushes with varying chemical structures have been synthesized, (Figure 1) [26] poly(oligo(ethylene glycol) methyl ether methacrylate), poly[N-(2-hydroxypropyl) methacrylamide], and zwitterionic brushes are most commonly used coatings to suppress NSB in complex biological milieu and for IVDs.[26] Although multiple surface coatings have been developed, comparing and ranking them is challenging as there is a lack of a standard evaluation methodology. There are multiple techniques and analytical methods to evaluate non-fouling surfaces. Surfaces can be challenged by either: (1) complex biological samples, such as human serum, plasma, or whole blood; (2) living cells; and or (3) purified protein solutions. NSB is typically determined analytically by measurement of protein adsorption by radiolabeling [32], surface plasmon resonance (SPR) spectroscopy [33], ellipsometry [34], quartz crystal microbalance [30], or fluorescence imaging [35] and of cell adhesion by optical microscopy. Because of the variability in NSB studies performed and analytical methods used, it is difficult to compare (or rank) surfaces across studies in the literature. Further, the conditions used for brush synthesis can vary depending on the study, which can lead to different brush properties and therefore additional variability in NSB determination. There have also been reports of divergent results for fouling studies from human plasma depending on the source of plasma.[36] Therefore, throughout the text, the nature of the non-fouling studies performed are explicitly stated. The synthesis and non-fouling behavior of several high-performing brushes are discussed in the following section.
Figure 1. Chemical structures of polymer brushes used for biomedical applications.
A) General strategy for polymer brush growth from a surface. An ATRP initiator is attached to a substrate from which a polymer brush is grown by surface-initiated ATRP. B) Architecture of surface tethered polymer brush (left panel) and commonly used polymer backbones and side-chains used for polymer brush growth, including polymethacrylates and polyacrylates (middle panel). A set of neutral and zwitterionic monomers are also shown (right panel).
2.1. Poly(oligo(ethylene glycol) methyl ether methacrylate) brushes
Over the past 15 years, poly(oligo(ethylene glycol) methyl ether methacrylate) (POEGMA) —a PEG derivative with a comb-like architecture— has been investigated as a substitute for linear PEG for various biomedical applications.[10, 34, 35, 37–49] POEGMA brushes exhibit a highly branched architecture with a high density of oligoethylene glycol (EG) moieties, which are responsible for the excellent non-fouling behavior of the material.[50] The first demonstration of the non-fouling properties of POEGMA brushes in serum were reported by our laboratory nearly 15 years ago.[34, 44, 45] In these initial reports, Ma et al. used surface-initiated atom transfer radical polymerization (SI-ATRP) to grow POEGMA brushes from gold, glass, and silicon oxide surfaces using bifunctional molecules and different initiators with an appropriate functional group for SAM’s formation. We —and later others— observed that protein resistance is a function of brush thickness and density which can be tuned by varying the polymerization times and surface density of the active initiator.[34, 44, 51–53] As the chain length and brush density can be controlled independently of each other, they provide two orthogonal parameters to precisely control the architecture and —thereby— properties of the polymer brush. We also found that a relatively low initiator surface density is sufficient to create protein-resistant coatings due to the much larger footprint of the polymer brush. POEGMA brushes with five EG units in the PEG sidechain (EG5) that were greater than 10 nm thick (in the dry state) conferred resistance to lysozyme, fibronectin, bovine serum albumin (BSA), and undiluted fetal bovine serum (FBS) solutions below the detection limit of ellipsometry (1 Å of adsorbed protein). Additionally, these POEGMA brushes prevented cell adhesion, as measured by fluorescence microscopy[34, 44, 54, 55]
We also expanded the range of materials capable of supporting the formation of POEGMA coatings to make this technology broadly useful. Engineering plastics are cheap and ubiquitous but are more difficult to functionalize with an ATRP initiator as they do not support the formation of SAMs. To solve this problem, Hucknall et al. developed facile methods for installation of an ATRP initiator on their surface.[55] A POEGMA overlayer could be grown by ATRP from a spin or dip coated layer of poly(vinylbenzylchloride) (PVBC), as its benzyl chloride side-chain acts as an ATRP initiator. This work also showed that plasma polymerization of 2-chloroethylmethacrylate yielded surface functionalization with sufficiently high concentration of ATRP-initiators to permit ATRP of POEGMA on the surface. More recently, we have shown controlled POEGMA growth for brushes grafted from high-κ dielectric materials, such as TiO2, ZrO2, Al2O3 and that they show a low level of BSA adsorption. This finding may enable future development of complementary metal–oxide–semiconductor (CMOS) devices for biosensing in complex biological mediums.[35] Combined, these studies demonstrate that POEGMA brushes fabricated via SI-ATRP offer a robust approach towards coatings that greatly reduce NSB on a variety of substrates.
Despite the success of POEGMA surfaces in greatly reducing NSB, there are several drawbacks to PEG-coated surfaces. First, PEG and its derivatives can auto-oxidize when exposed to oxygen or transition-metals, which may affect the stability of the brush and the non-fouling properties.[56] Second, the antigenicity of PEG has become a major concern in recent year [57], as human anti-PEG antibodies (APAs) can bind to PEGylated drugs causing premature clearance of the drug from circulation and thereby decrease its effectiveness. APAs can also – albeit in a small minority of patients— cause an adverse immune response.[58]
Of particular relevance to human IVDs, APAs may cause interference in Ag-down assays for the detection of Abs (“serology”), such as in indirect sandwich immunoassay formats (ISIA). Although APAs may not interfere in sandwich immunoassays, serological assays are routinely performed in clinical laboratories and APA binding is thus an important consideration. In ISIA, APAs may bind to PEG-derived surfaces and thus be labelled (along with the Ab of interest) resulting in an elevation in background signal and a poorer assay sensitivity. In a recent study unrelated to clinical diagnostics, we showed that the anti-PEG antigenicity of a drug-POEGMA conjugate was significantly reduced compared to two FDA-approved PEGylated drugs (Krystexxa® and Adagen®).[59, 60] We also demonstrated that shortening the EG side-chain length of POEGMA from EG9 to EG3 eliminated nearly all APA reactivity for our drug-POEGMA conjugate.[59] Therefore, to mitigate the possible consequences of APA interference for IVDs, we investigated the effect of EG side-chain length on surface APA reactivity. We found that shortening of EG side-chain lengths increased the hydrophobicity of the polymer brush, but still provided sufficient surface coverage with EG side-chains to eliminate Cy5-labeled BSA adsorption, cell adhesion, and did not bind to a panel of APAs.[61]
While we have developed strategies to drastically reduce NSB and to alleviate the presence of interfering APAs, another solution is to use polymer brushes that are non-fouling but do not have a PEG repeat unit. Poly[N-(2-hydroxypropyl) methacrylamide] and zwitterionic polymer brushes, discussed next, offer such an alternative. In many of the studies described next, POEGMA brushes were also included, so their performance is highlighted as well.
2.2. Poly[N-(2-hydroxypropyl) methacrylamide] brushes
Although poly[N-(2-hydroxypropyl) methacrylamide] (PHPMA) has been extensively studied for use in drug delivery systems [62], Rodriguez-Emmenegger et al. were the first to investigate PHPMA brushes as a substrate for IVDs.[63] Controlled ATRP reactions are well established for methacrylate (and acrylate) monomers [64], however development of a robust system for methacrylamides has proven to be more difficult.[65–67] For polymerizations performed in bulk or solution, monomer conversion can be low and the reaction may not proceed by a living process.[67, 68] Further, control over the reaction may be poor due to nucleophilic displacement or inactivation.[63] For IVDs, precise control over the thickness and architecture of the brush are necessary to eliminate variability in assay performance, and if more complex copolymer architectures are desired.
Despite these limitations, Rodriguez-Emmenegger et al. identified polymerization conditions to graft PHPMA, POEGMA, poly(2-hydroxypropyl methacrylate) (PHPM), and poly(carboxybetaine acrylamide) (PCBAA) —a zwitterionic polymer— brushes from a gold SPR sensor using SI-ATRP.[63] All surfaces prevented NSB below the detection limit of SPR (0.3 ng cm−2) when exposed to single protein solutions of human fibrinogen, immunoglobulin G (IgG), human serum albumin (HSA), and lysozyme. When challenged with undiluted human blood plasma, PCBAA and PHPMA surfaces remained non-fouling; however, POEGMA and PHPM surfaces had plasma deposits of 22.5 ng cm−2 and 40.5 ng cm−2, respectively.[63] In a subsequent study, Vaisocherová-Lísalová et al. synthesized PHPMA brushes—along with copolymer brushes composed of HPMA and zwitterionic monomers—and assessed fouling from food samples using SPR. The PHPMA brushes exhibited undetectable fouling from milk, spinach, cucumber, hamburger, and lettuce samples, making them a suitable substrate for a biosensor for food security and safety.[69]
In another study, PHPMA brushes (as well as POEGMA and zwitterionic polymer brushes) were grafted from a polycarbonate substrate using SI-ATRP.[70] Fouling from human blood plasma was measured using an electromagnetic piezoelectric acoustic sensor. Modification with the polymer brushes resulted in a drastic reduction in NSB, consistent with previous results.[63, 71] Thrombogenicity of the surfaces was also assessed by flowing whole human blood with fluorescently labeled cells over the surface. Surface modification with a PHPMA brush led to a substantial reduction in thrombus formation and platelet aggregation as visualized by fluorescence microscopy.[70]
Collectively, recent reports have demonstrated that PHPMA coatings can drastically reduce protein adsorption to levels below the detection limit of SPR, even from undiluted human serum, plasma, and complex food samples. Although the synthesis of PHPMA brushes by ATRP is less controlled than other monomers, optimal conditions identified by some groups [63, 71] or alternative polymerization schemes [72] may provide a robust route for the synthesis of these coatings for biomedical applications.
2.3. Zwitterionic polymer brushes
As another alternative to PEG-derived coatings, zwitterionic polymer materials have been investigated extensively for the generation of non-fouling surfaces. Zwitterionic polymer materials vary widely in architecture, but their common characteristic is a side-chain that contains a cation and anion within the same side chain. Zwitterionic polymers with varied backbones, [33, 73] charged groups, [52] and distances between the positive and negative charges [74] have been studied; of these, phosphorylcholine (PC), carboxybetaine (CB), and sulfobetaine (SB) are the most widely used –through which it has been learned that their non-fouling behavior is due to the strong hydration at the solid-liquid interface of zwitterionic polymer brushes.[75, 76] It has also been shown that zwitterionic polymer materials are “invisible” to potential interfering APAs. For further details of the different types of zwitterionic polymer materials, we refer the reader to the following reviews.[77–79]
Like POEGMA brushes, zwitterionic polymers are compatible with “grafting to” and “grafting from” LRP techniques. Although a dip-coating “grafting to” procedure has also successfully been applied [80], “grafting from” approaches are most commonly used. One of the first demonstrations of the excellent non-fouling properties of zwitterionic materials came from Lad et al. where poly(carboxybetaine methacrylate) (PCBMA) and poly(sulfobetaine methacrylate) (PSBMA) brushes (as well as POEGMA) were grafted from a gold-substrate.[33] In this study, PCBMA, PSBMA, POEGMA, and oligoethylene glycol SAM surfaces were subjected to 10% human serum and plasma, and the non-fouling properties were characterized by SPR spectroscopy. All three brush surfaces exhibited superior protein resistance compared to the oligoethylene glycol SAM, with fouling levels approaching the detection limit of SPR. When undiluted serum and blood plasma were tested, PCBMA coatings slightly outperformed both PSBMA and POEGMA coatings, although the protein adsorption was below 50 ng cm−2 on all three surfaces.
In a follow-up study Yang et. al, fabricated poly(carboxybetaine acrylamide) (PCBAA) polymer brushes via SI-ATRP.[81] In this study, the polymer brush thickness was varied, and protein adsorption from undiluted human serum and plasma was measured at both 25°C and 37°C. PCBAA coatings with a dry thickness of ~20 nm exhibited undetectable levels of fouling using SPR. This result was further corroborated by Rodriguez-Emmenegger et al. who demonstrated undetectable fouling from human plasma for PCBAA coatings with a dry thickness of 17 nm.[63] In follow-up study, they showed that PCBAA and PHPMA coatings completely resisted NSB from human cerebrospinal liquid, saliva, urine, FBS, calf serum, chicken egg and whole cow milk below the detection limit of SPR (0.3 ng cm−2), suggesting these surfaces could be useful for diagnostics from a variety of sample mediums.[71] Interestingly, both PCBAA and PHPMA showed zero fouling despite having very different hydrophilicity, which suggests that surface hydrophilicity is not the primary factor that prevents fouling.
In our own work, we have confirmed the non-fouling nature of zwitterionic polymer brushes. PSBMA and POEGMA surfaces were exposed to FITC-BSA in PBS, washed, and then fluorescently imaged. We found that the protein resistance of both surfaces were comparable.[82] Zwitterionic polymer brushes are highly water wettable with a water contact angle close to zero which contributes to their excellent non-fouling properties. However, unlike POEGMA brushes that have an intermediate surface wettability, they do not allow inkjet printing of reagents on to their surface.[82] To enable non-covalent inkjet printing functionalization, we engineered hybrid zwitterionic–cationic polymer coatings where by controlling the surface density of the zwitterionic and cationic side chains, we could minimize NSB but also tune the surface wettability to allow inkjet printing Abs, thus allowing for simple fabrication without the need for covalent chemistry to biofunctionalize the surfaces, as described in further detail in the next section.[82]
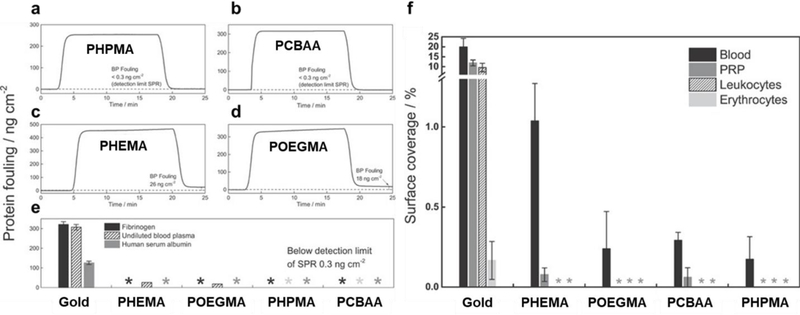
Overall, several polymer brushes have been investigated to suppress NSB from complex biological samples. Surman and coworkers reported that PHPMA and PCBAA were the most successful non-fouling surfaces (compared to POEGMA and PHEMA coatings), when challenged with undiluted serum or plasma samples as they suppressed NSB below the analytical sensitivity of SPR (0.3 ng cm−2) (Figure 2A–E). Further, these surfaces drastically reduced cell adhesion (Figure 2F).[83] These non-fouling surfaces and others have been utilized for the development of many sensitive IVDs, several of which are discussed in the next section.
Figure 2. Protein fouling and cell adhesion for various polymer brushes/.
SPR sensorgrams of protein adsorption from undiluted blood plasma for (A) PHPMA, (B) PCBAA, (C) PHEMA, and (D) POEGMA polymer brushes. (E) Protein fouling for surfaces when challenged by from fibrinogen (1 mg mL−1), human serum albumin (5 mg mL−1) and undiluted blood plasma. (F) Surface coverage (%) of blood, platelet rich plasma (PRP), leukocytes, and erythrocytes incubated on gold, PHEMA, POEGMA, PCBAA, and PHPMA polymer brush coated gold. Adapted with permission.[83] Copyright 2015, John Wiley and Sons.
3. NON-FOULING SURFACES FOR IN VITRO DIAGNOSITCS
3.1. Surface biofunctionalization
For protein-resistant surfaces to be relevant to clinical diagnostics, their surface needs to be functionalized with biomolecules that enable the detection of analytes from a complex milieu. The challenge of devising a surface that is non-fouling when exposed to complex media (i.e. serum, plasma, or whole blood), but also allows substrate functionalization with biomolecules, has been approached in several different ways. For a thorough discussion, we refer the reader to a review by Badoux et al.[84]
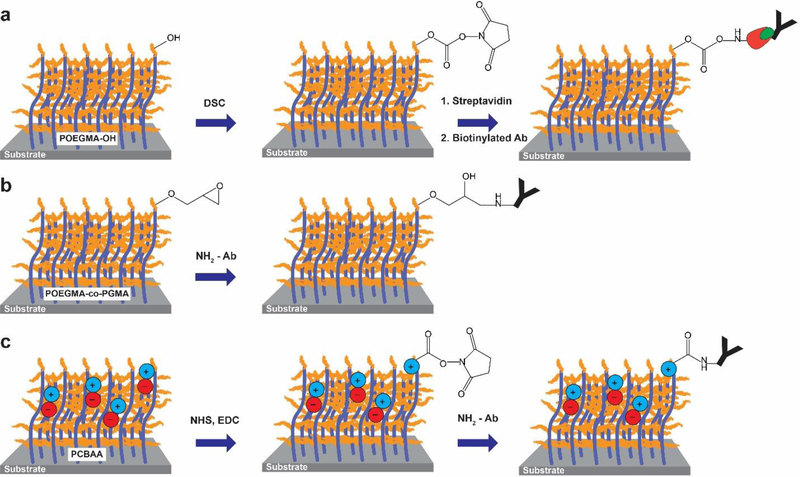
Covalent attachment is the most widely used method for immobilization of biomolecules on polymer brushes. One method for covalent attachment is to change the monomer composition of the brush to include a monomer with a reactive chemical handle that can be used for conjugation. For example, Trmcic-Cvitas et al. used hydroxy-terminated POEGMA, which contains hydroxy moieties for immobilization, but still retains non-fouling properties.[85] Streptavidin was attached through N,N-disuccinimidyl carbonate (DSC) coupling to the terminal hydroxyl group so that biotinylated Abs could be immobilized onto the surface (Figure 3A).[85] Hu et al. –and later others—employed poly[oligo (ethylene glycol) methacrylate-b-glycidyl methacrylate] (POEGMA-co-PGMA) co-polymerization to include both non-fouling properties and epoxide groups for functionalization via reaction with reactive nucleophilic groups on Abs under mild conditions (Figure 3B).[86–88] Another common surface functionalization technique, popular with zwitterionic polymer brushes, is surface activation with 1‐ethyl‐3‐(3‐dimethylaminopropyl)‐carbodiimide and N‐hydroxysuccinimide (EDC/NHS) coupling chemistry. Yang et al. functionalized their PCBAA surface with Abs via EDC/NHS coupling and subsequent deactivation of unreacted NHS ester groups by hydrolysis under mild conditions (Figure 3C).[81] Importantly, no further blocking was required after surface functionalization, as the PCBAA surface in the deactivated state provided sufficient reduction of NSB. However, some studies have found contradictory results in that surface functionalization with Abs using EDC/NHS-activated covalent chemistries can lead to an increase in fouling from plasma compared to non-functionalized surfaces.[89] Therefore, it is important to test how surface functionalization impacts the non-fouling properties of brushes during IVD development.
Figure 3. Covalent attachment techniques for surface bio-functionalization.
A) Hydroxyl-terminated POEGMA brushes are activated with DSC. The brushes are then functionalized with streptavidin (red symbol) and then a biotinylated (green symbol) Ab for biosensing. B) Abs are contact printed onto a POEGMA-co-PGMA block copolymer brush, resulting in covalent attachment of the Abs to the epoxide groups under mild conditions. Subsequently, the remaining epoxide groups are inactivated. C) Carboxylic acid groups from PCBAA brushes are reacted with NHS/EDC to yield a reactive NHS-ester, followed by functionalization with an Ab via the formation of an amide bond. Subsequently, the surface is inactivated and reverts to a non-fouling state.
Despite the success of covalent coupling methods, biofunctionalization of a surface with multiple different biological recognition elements requires multiple sequential steps and thus increases the complexity of fabrication of the sensing interface. Previously, we devised a simple and direct method of fabricating Ab-based microarrays via non-covalent immobilization of Abs on POEGMA using noncontact inkjet printing followed by mild desiccation.[10, 48] Upon rehydration of the surface, the printed Ab spots do not bleed, even upon overnight exposure to biological fluids, showing that the printed Abs remain stably immobilized, even upon rehydration of the brush.
3.2. Fluorescent antibody microarray immunoassays
In a previous review [10], we reported on Ab microarrays for IL-1β, IL-6, IL-8, tumor necrosis factor alpha (TNF-α) and osteoprotegerin (OPG) that were fabricated on POEGMA brushes and were used in a sandwich immunoassay format. Analysis of the dose-response curves for OPG in buffer and whole serum revealed that the dynamic range and LODs were virtually identical in PBS and serum and that the LOD of an IL-6 assay was 100-fold lower compared to the same assay on nitrocellulose.[10, 48] This result was intriguing as complex biological matrices typically present LODs that are orders of magnitude greater than those in analyte-spiked buffer due to background noise caused by NSB. This simple yet novel approach addressed several key limitations of protein microarrays: (1) inkjet printing of Abs onto a dried POEGMA brush is a simple method to immobilize the Ab on the surface and obviates the need for complicated surface chemistry or activation/inactivation steps. (2) When hydrated, the tendency of proteins in the sample milieu to adsorb non-specifically was dramatically reduced due to the non-fouling properties of the surface coating. (3) The need for separate blocking steps after capture reagent immobilization becomes unnecessary due to the dramatically reduced NSB. We have recently transformed this microarray platform into a POCT, which is highlighted later in this progress report.
Since our initial demonstration, other groups have developed similar technologies for protein microarray fabrication and Ag detection, although they use covalent chemistry for surface biofunctionalization. For example, Liu et al. developed a flow-through microarray immunoassay utilizing a polyGMA-co-poly(ethylene glycol) methacrylate] (P(GMA-co-PEGMA)) brush surface that could be functionalized with Abs via covalent coupling of the GMA moiety with amine groups on Abs using a contact printing method.[87] In the flow-through configuration, samples were injected through a microfluidic network over the microarray, followed by fluorescently-labeled Abs, enabling the detection of carcinoembryonic antigen (CEA) with a detection limit of 10 pg mL−1 in 10% human serum.
Lei et al. synthesized P(GMA-HEMA) brushes as a substrate for protein and peptide microarrays for the detection of rabbit antihuman IgG Abs and matrix metalloproteinases (MMP) activity.[90] While detection of rabbit anti-human IgG Abs was used as a proof-of-concept study for the platform, MMP activity is related to several pathological processes [91] and is thus potentially important clinically. MMP activity was assessed by immobilizing biotinylated peptide sequences (subsequentially labeled with avidin-Cy3) that can be cleaved by specific MMP enzymes. Therefore, as the concentration of MMP increases, the fluorescence signal decreases, due to the biotin-avidin-Cy3 complex being liberated from the surface. When combined with an RGD anchoring motif, this platform was used to determine MMP-2 and MMP-9 activity secreted from cells cultured on-chip. In this study, while the P(GMA-HEMA) brush presumably decreased NSB significantly, the fouling studies were carried out using only single protein solutions and the chips were further blocked with BSA, suggesting that additional blocking was required to sterically hide the unbound and reactive GMA groups.
Hu et al. developed a sensitive competitive immunoassay built upon a POEGMA-co-GMA polymer brush modified glass slide for the detection of mycotoxins.[92] The copolymer brush was grown via SI-ATRP, followed by spotting of aflatoxin B1-, ochratoxin A-, and zearalenone-BSA conjugates. In the competition format, mycotoxins in a sample compete with the mycotoxins immobilized to the brush to bind a monoclonal Ab introduced to the system, which is then labeled by a secondary Ab. Therefore, a weaker signal is observed when higher concentrations of mycotoxin are present. Using mycotoxin-spiked PBS, the LOD for aflatoxin B1, ochratoxin A and zearalenone were 4, 4 and 3 pg mL−1, respectively.[92] Further, spiked peanut samples were tested and the recovery rate was 85.8 – 109.2%, suggesting good agreement with the PBS calibration curve. Overall, the performance of the test was far superior to that of an immunoassay carried out on a glycidoxypropyl trimethoxy silane-based microarray blocked with BSA, which is attributed to the high protein loading capacity of the GMA moiety and non-fouling characteristics provided by the POEGMA brush.
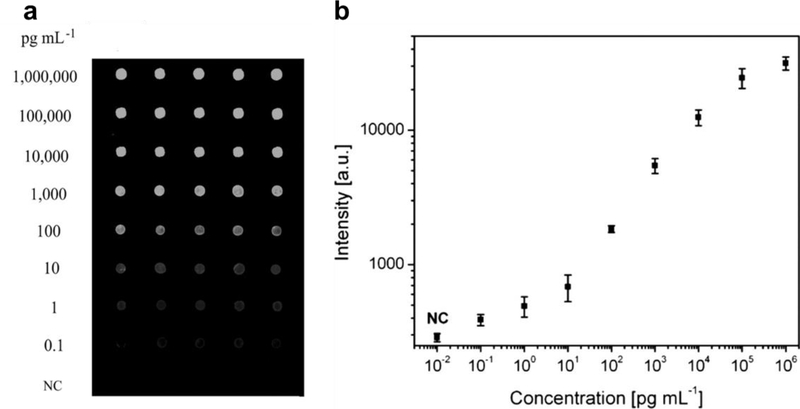
A particularly successful demonstration of an Ab microarray built upon a protein resistant surface was also developed by Hu et al.[93] Here, randomly-oriented zinc oxide (ZnO) nanorods were grown from a glass slide, followed by SI-ATRP of POEGMA-co-GMA copolymer brushes. The ZnO nanorods were used to amplify the fluorescence intensity of nearby fluorophores and the POEGMA-co-GMA brush was functionalized with Abs at a high-loading density and used to suppress NSB, thereby greatly enhancing the performance of the assay. In a sandwich immunoassay format, CEA was detected from undiluted human serum with an LOD of 100 fg mL−1 (Figure 4), which is 100 times more sensitive than the same assay performed without ZnO nanorods on a POEGMA-co-GMA surface. This work clearly demonstrated how signal amplification and suppression of background signal can be integrated together to yield ultra-sensitive detection schemes.
Figure 4. Antibody microarray on POEGMA‐co‐GMA brushes “grafted from” ZnO nanorods.
A) Fluorescent images, with concentration of CEA and B) plotted dose response curve for detection of CEA spiked into human serum, with a LOD of 100 fg mL-1. Adapted with permission.[93] Copyright 2014, John Wiley and Sons.
While the work described in this section utilized fluorescent transduction, non-fouling polymer brushes are of broad utility across all transduction modalities in clinical diagnostics. We discuss in the next section, their use in a label-free detection system —SPR— that is in even more acute need for reduction of NSB, because the lack of a label does not provide any intrinsic mechanism to discriminate between the signal originating from receptor-analyte interactions and noise that originates from NSB.
3.3. Surface plasmon resonance immunoassays
SPR is a label-free optical biosensing modality that measures the change in refractive index when a specific biorecognition event occurs (i.e. Ab binding Ag) or a non-specific event occurs (i.e. adventitious protein adsorption) at or in close proximity to a noble metal surface. SPR can measure the binding kinetics of analytes to a surface bound receptor and records the binding event as resonance units versus time in a sensorgram. Sensorgrams can be fit to a kinetic model to determine the association rate constant or on-rate (kon) and the dissociation rate constant or off-rate (koff) and hence the equilibrium dissociation constant (KD = kon koff−1).[94] SPR can also be used for the detection of a target protein using a surface-coupled biorecognition element (usually an Ab) [95, 96], among many other applications.[97–99] As SPR detects binding events at a surface, the performance of SPR immunoassays is extremely sensitive to NSB. Commercial SPR sensors are often coated with a dextran-based coating which greatly reduces NSB from single protein solutions or biofluids diluted 10-times; however, significant fouling occurs (> 800 ng cm−2) when exposed to undiluted serum.[96, 100, 101] Therefore, integration with a high-performing non-fouling surface is vital for detection from complex biological milieu.[102] Although not intended to be a comprehensive review of SPR biosensors, several examples are summarized below. The topic of SPR biosensors and their application for label-free sensing from complex biological milieu has been more thoroughly reviewed in the following reviews.[22, 103]
One of the first demonstrations of an SPR immunoassay functionalized with a non-fouling surface was conducted by Jiang’s group at the University of Washington. In this study, Zhang et al. grafted a PCBMA polymer brush upon a gold-coated SPR sensor and functionalized the surface with an anti-human chorionic gonadotropin monoclonal Ab.[104] They showed that the sensor exhibited minimal non-specific adsorption, even when exposed to high concentrations of lysozyme and fibrinogen. In a follow up study, Vaisocherová et al. grafted PCBAA from a gold-coated SPR chip and further functionalized with Abs specific for activated leukocyte cell adhesion molecule (ALCAM), a potential biomarker for various types of cancer.[105] Remarkably, they were able to detect ALCAM with a 10-fold greater sensitivity using the PCBAA coated surface as compared to traditional −COOH/OH oligo(ethylene glycol) (OEG) surfaces that had also been blocked using standard procedures.
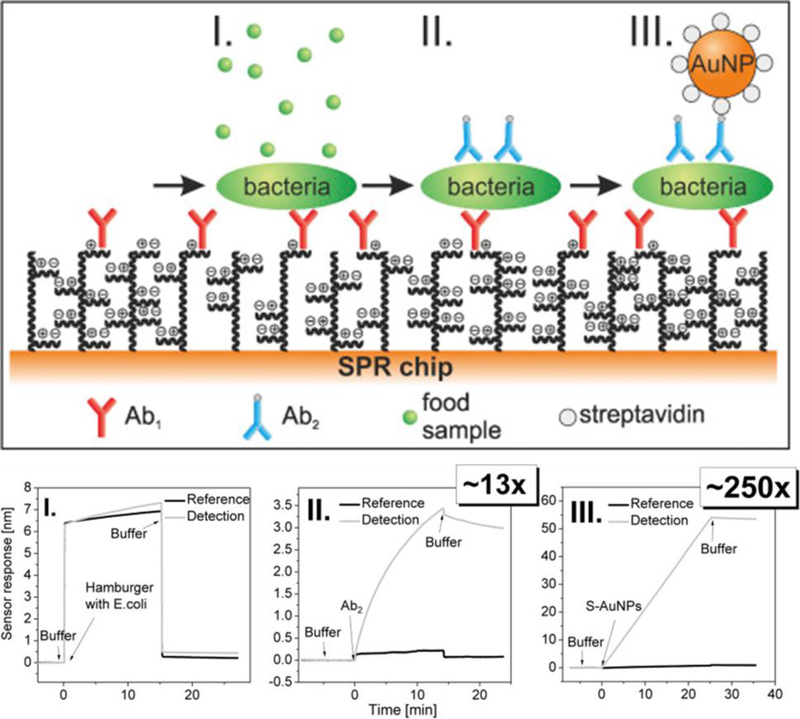
More recently, an SPR biosensor with a non-fouling PCBAA coating was used for the detection of Escherichia coli (E. coli) and Salmonella sp. contamination in food samples.[106] In this study, several signal amplification steps were incorporated. Bacteria were captured with a primary anti-E. coli or anti-Salmonella Abs immobilized to the surface, followed by labeling with a biotinylated secondary Ab, followed by streptavidin-coated spherical gold nanoparticles. With the additional labeling steps, the sensor response enhancement was 250-fold compared to the label-free format, which resulted in detection limits below 60 cfu mL−1 for E. coli and 11.7×103 for Salmonella sp, even from crude food samples (Figure 5). Another SPR sensor, developed by the same group, utilized a P(CBMAA-co-HPMA) coating for the detection of E. coli from hamburger and cucumber samples.[69] The LOD was 2.1×104 CFU mL−1 for direct detection and 81 CFU mL−1 when secondary labeling was incorporated.
Figure 5. Detection of E. coli from hamburger samples using SPR.
Scheme of the three-step assay is shown, with corresponding SPR sensorgrams and enhancement in LOD. (I) direct detection using primary Ab, (II) flowing a biotinylated Ab over the sensor, and (III) flowing a streptavidin-coated spherical gold nanoparticle over the sensor. Adapted with permission.[106] Copyright 2016, Elsevier.
In another study, Riedel et al. developed a hydroxy-functional POEGMA-coated SPR sensor for the detection of Epstein–Barr virus (EBV) Abs from clinical serum samples.[107] Here, the brush surface was functionalized with several EBV Ags using a covalently bound streptavidin and biotinylated-Ag system. Using SPR, they were able to stage EBV infection by monitoring the presence of Abs in clinical samples. Riedel et al. later developed an SPR sensor built upon a P(HPMA-co-CBMAA) polymer brush for the detection of hepatitis B Abs from diluted (10%) clinical serum samples.[108] Hepatitis B Ags were attached to the brush using EDC/NHS chemistry. The copolymer brush completely resisted NSB (fouling below the detection limit of SPR) when challenged with undiluted serum both before and after functionalization with the hepatitis B Ags. The sensor was capable of clearly discriminating between positive and negative hepatitis B samples, suggesting it could be a viable platform for clinical testing.
Zhang et al. engineered a methoxy-PEG-coated SPR sensor surface which served a dual purpose: (1) the surface was non-fouling and greatly reduced NSB in diluted serum or plasma, and (2) the methoxy-PEG modified surfaces were reactive towards APAs.[109] Because the surface was “invisible” to serum proteins, but presented many binding sites for APAs, it was used as an SPR sensor to quantitatively detect APAs in serum. Other surfaces were investigated, such as EG9 POEGMA; however, methoxy-PEG proved to be the most sensitive. Importantly, this method produced a linear response with concentration of APA, and APA isotypes could be discriminated by binding of an isotype-specific secondary Ab.
Overall, several groups have demonstrated the successful integration of non-fouling surfaces for SPR-based diagnostics. SPR immunoassays have the benefit of label-free detection, and binding of a secondary Ab improves the sensitivity and specificity of the assay, albeit at the cost of a longer workflow. As the performance of these assays is extremely sensitive to NSB, non-fouling surfaces play a crucial role in eliminating non-specific adsorption from blood and serum samples. Despite the examples outlined above, there have been very few demonstrations of SPR systems capable of detecting protein biomarkers at sufficiently low levels from complex biological samples at the point-of-care for successful translation to clinical practice.
4. TRANSITION TOWARDS POINT-OF-CARE IMMUNOASSAY DIAGNOSTICS
IVDs must be easy to use with minimal user intervention, yet provide clinically acceptable performance. While clinical ELISA can be implemented in abundant resource settings, it is not amenable for point-of-care testing nor in limited-resource settings. A particularly successful example of an IVD platform that can be performed at the point-of-care with no ancillary instrumentation is the lateral flow assay (LFA). LFAs require minimal user intervention and usually provide a visual qualitative readout in less than 30 minutes. They have become the most widely used POCT because of their simplicity and reliability.[110, 111] However, LFAs relying on colorimetric readout are typically not as sensitive as their clinical laboratory equivalent (i.e. ELISA), thus limiting their utility for detection of low abundance disease biomarkers.[112, 113] Addressing this limitation requires incorporating an amplification scheme into the LFA to increase sensitivity, which can significantly improve the performance,[114, 115] but the added complexity then dilutes the most attractive attribute of LFAs— their simplicity.
An ideal POCT should provide a low-cost, single-step procedure where the detection and capture reagents can be contained in the same device, while retaining the exquisite sensitivity and quantitative readout of clinical ELISAs. The World Health Organization (WHO) has developed the ASSURED —affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free and deliverable to end users— criteria for evaluation of POCTs.[116] Successful implementation of the ASSURED criteria in point-of-care diagnostics can have a tremendous impact in improving global health in resource-limited settings but also in reducing health care costs in the developed world. Our groups’ contribution toward this goal is highlighted in the next section.
4.1. The D4 point-of-care immunoassay
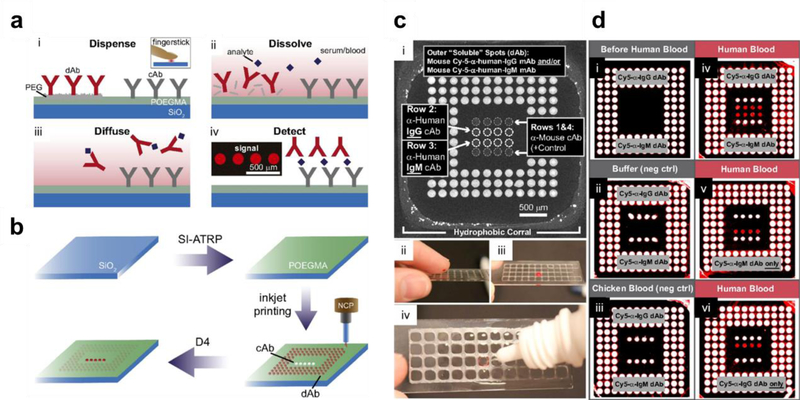
Recently, we set out to re-design and simplify the sandwich immunoassay to turn it into a POCT.[49] Towards this goal, Ab microarrays were fabricated on POEGMA brushes by inkjet printing of capture Abs (cAb), as outlined in our previous work.[10, 48] However, in contrast to our previous work, fluorescently labeled detection Abs (dAb) were also printed —mixed with an excipient such as PEG or trehalose— directly onto the POEGMA brushes in close proximity to the cAbs. Printing the dAb with an excipient allows the dAb to solubilize upon exposure to a liquid sample. Therefore, with this architecture, all the necessary reagents to complete a sandwich immunoassay are contained on-chip, thus avoiding multiple separate incubation and or wash steps that are required for ELISA and other protein microarrays.
This platform is named the “D4” assay for the series of events that occur during the assay (Figure 6A): (1) blood is Dispensed onto the array, (2) the dAb Dissolve from the POEGMA brush into solution, (3) both the dAb and target Ag in the sample Diffuse towards the immobilized cAb, and (4) Detection of the binding event with a fluorescent detector, where the fluorescent intensity scales with Ag concentration. In addition to being user-friendly, assay fabrication is also simple and easily scalable (Figure 6B). Glass slides are functionalized with a ~30 nm thick POEGMA brush by SI-ATRP (dry thickness), followed by inkjet printing of the cAb and dAb. Upon printing the Abs onto the brush, they are stabilized and retain their tertiary structure in the brush even upon prolonged storage at room temperature without the need for refrigeration or hydration because of the interfacial hydration provided by the polymer brush.[48, 117] Our fabrication strategy also consumes a minimum amount —pico to nano-liters— of Ab reagents to fabricate a single chip, which reduces the cost of assay fabrication and makes it more amenable for point-of-care applications, especially in low-resource settings. The non-fouling POEGMA interface is the core technology of the D4, which enables simple assay fabrication, a long shelf-life of printed chips, and a simple assay with high analytical performance. As an example, the qualitative detection of human IgG and human IgM from a fingerstick of blood is shown in Figure 6C–D.
Figure 6. D4 immunoassay on POEGMA brushes.
A) A schematic of the steps in the D4 assay. The cAb and dAb are inkjet printed onto the POEGMA brush. (i) A drop of blood or serum is dispensed directly onto the chip. (ii) dAb dissolves and binds to Ag in the sample. (iii) Ab-Ag complexes diffuse and bind to their respective cAb. (iv) a quantifiable fluorescent signal is detected using a fluorescent detector. B) D4 chip fabrication. A POEGMA brush is grown on glass chips by SI-ATRP. The cAb and dAb are inkjet printed onto the surface. After desiccation, the D4 chips are ready for use. C) D4 detection of human IgG and IgM from fingerstick blood. (i) D4 chip layout with each of the components labeled. (ii) Addition of a drop of blood to the D4 array. (iii) Incubation with blood, followed by (iv) rinse with a wash buffer. D) D4 array scanned: (i) before the addition of blood, (ii) after incubation in buffer (negative control), (iii) after addition of chicken blood (negative control), (iv) after addition of human blood with both anti-IgG and anti-IgM dAbs printed on the chip, (v) after addition of human blood with only anti-IgM dAb printed on the chip, and (vi) after addition of human blood with only anti-IgG dAb printed on the chip. Adapted with permission.[49]
We also demonstrated the ability of the D4 assay to quantify the concentration of analytes from complex matrices such as blood and serum in a multiplexed format. Multiplexed detection is a highly desirable attribute, especially for diagnosis of infectious diseases, as many infectious diseases share common symptoms and are endemic in the same region.[118] Multiplexed screening is also important for the diagnosis of various types of cancer where single biomarkers have poor clinical sensitivity and specificity.[119] To demonstrate a multiplexed D4 assay, we developed a 2-analyte assay for IL-6 and TNF-α and compared it to the respective single-plex assays. For multiplexed detection of multiple target analytes, we print the cAbs at spatially separate addresses on the chip and print microspots of a “cocktail” of fluorescently-labeled dAb conjugates targeting each analyte. Because the cAb spots for each analyte are spatially discrete and defined, the same fluorophore can be used to label each dAb, which greatly simplifies assay readout.
The resulting assays yielded sub-picomolar LODs and showed minimal cross-reactivity between assays. In addition, the dynamic range of each assay was roughly 3-log in concentration or greater. Importantly, we demonstrated the assays could be run with incubation times as low as 15 min in unmodified serum or whole blood while retaining acceptable sensitivity.[49] More recently, we have extended multiplexed detection to six analytes for the diagnosis of hepatocellular carcinoma (unpublished).
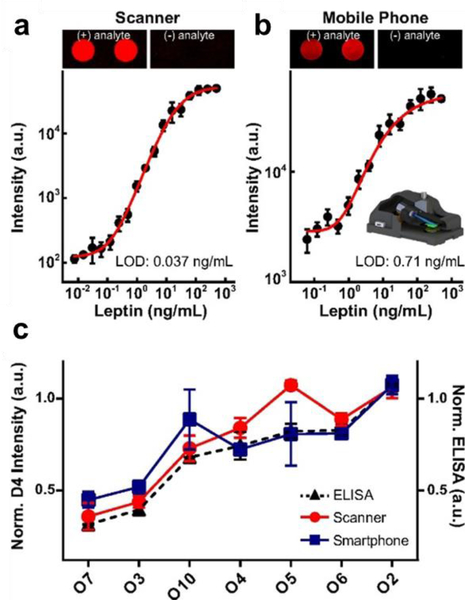
To ascertain the feasibility of the D4 assay in a point-of-care setting, we deployed the D4 in a pilot clinical study. We used the D4 assay to quantify serum levels of leptin –recently identified as a predictive marker of mortality due to severe malnutrition [120]—and compared these results to an ELISA performed in a clinical laboratory. The data showed a strong correlation between D4 and ELISA measurements, suggesting that the D4 can replace conventional ELISA in clinical settings. We also demonstrated that the D4 assay is compatible with an inexpensive mobile phone-based fluorescence detector (Figure 7). Although the sensitivity on the mobile platform was inferior to a tabletop fluorescence scanner, we are actively developing a next generation, low-cost detector, which we anticipate will match the performance of traditional tabletop scanners.
Figure 7. Mobile phone-based imaging of D4 arrays and concordance with ELISA.
Dose–response curves in calf serum spiked with leptin Ag acquired by a (A) benchtop scanner and (B) a mobile phone-based fluorescence microscope, with representative D4 microspot images. C) Patient sera tested on the D4 assay using a benchtop scanner (red trace) versus the mobile phone microscope (blue trace), and comparison with ELISA results (dashed black trace). Reproduced with permission.[49]
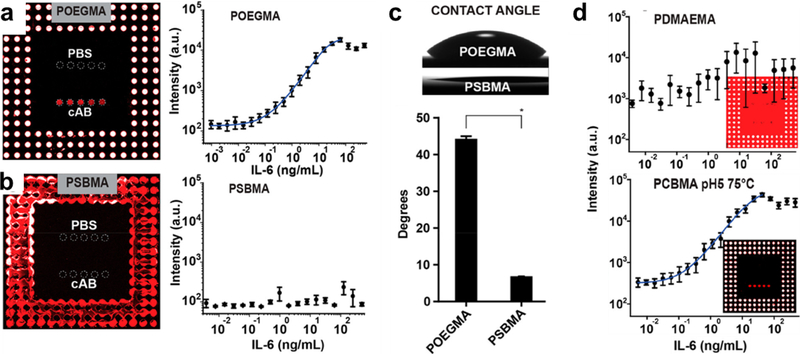
The design of the D4 assay is not limited to POEGMA as the interface; we have also explored a hybrid zwitterionic polymer brush surface as the biointerface, motivated by their lack of protein adsorption.[82] We found that the D4 assay on a PSBMA brush failed (Figure 8A–B), as the surface is far too hydrophilic for inkjet printing of Abs due to its extremely low water contact angle (Figure 8C). To solve this problem, we synthesized a poly-2-(dimethylamino) ethyl methacrylate (PDMAEMA) brush on glass and derivatized a fraction of the amine groups to create a hybrid zwitterionic-cationic surface. By controlling the degree of derivatization, we identified coatings that are suitable for inkjet printing of Abs, but which also exhibit low protein adsorption.[82] The LOD and dynamic range of the D4 assay on these surfaces was similar to the same assay fabricated on a POEGMA brush and much better than assays fabricated directly on PDMAEMA brushes (Figure 8D).[82]
Figure 8. D4 immunoassay performance on zwitterionic polymer brush surfaces.
D4 assay dose-response curves for IL-6 Ag on (A) POEGMA and (B) PSBMA polymer brush surfaces. The PSBMA surface is too hydrophilic to allow inkjet printing immobilization of Ab reagents, resulting in no visible dose-response behavior in panel B. C) Optical images and quantification of contact angles for POEGMA and PSBMA brushes, showing the significantly lower water contact angle on the zwitterionic PSBMA surface. D) D4 assay dose-respose curves for IL-6 Ag on a poly-2-(dimethylamino)ethyl methacrylate (PDMAEMA) brush and a PDMAEMA polymer brush surface that was partially derivatized to PCBMA at 75 °C, pH 5. The assay metrics for the PCBMA surface are similar to the same assay on a POEGMA brush surface. Adapted with permission.[82] Copyright 2018, American Chemical Society.
The D4 technology satisfies most or all of the WHO-defined ASSURED criteria.[49, 116] It is affordable, as the consumption —and hence cost— of Ab reagents per chip is minimal. It has exquisite sensitivity and specificity due to the high SNR afforded by the non-fouling interface. The platform is extremely user-friendly, as only a fingerstick of blood and one wash step is required in the current format and further automation to eliminate the manual wash step is being developed. The test is rapid and robust, with results possible in 15 minutes and requires no sample pre-processing even with whole blood. Although not entirely equipment free, the D4 can be imaged and analyzed using smartphones that are ubiquitous in both the developed and developing world. Finally, it is deliverable to end users, as the D4 chip has a long shelf life without the needs for cold-chain —the need for hydration and refrigeration— as the POEGMA brush preserves Ab activity. The flexible design of the D4 suggests that it can be rapidly developed —on the order of weeks— to target any protein biomarker for which Ab pairs are available. When coupled with advancements in Ab generation and selection, where Abs can be developed against new targets quickly [121–124] the D4 platform is ideally suited for the on-demand development of the next generation of diagnostic tests.
4.2. Microfluidic devices
Microfluidic integration is important for the design of next generation point-of-care diagnostics as it can minimize user intervention. An illustrative example is a fully microfluidic POCT for the simultaneous detection of HIV and syphilis developed by Sia and coworkers that brought liquid-based assays traditionally performed in 96-well plate format (i.e. ELISA) into a field-ready format with data upload capabilities.[4, 7] This work utilized HIV and syphilis Ags immobilized in different locations of a microchannel network manufactured by injection molding in polystyrene. Samples and reagents were delivered by a bubble-based method where 14 reagents were sequentially passed through the microchannels in the assay (reagents consisting of labeled Abs, washing solution and signal-enhancing solutions) (Figure 9). This assay interfaced with a smartphone dongle for rapid quantification of the assay and data transmission. The performance of the POCT was similar to laboratory-based ELISA and was capable of detecting HIV and syphilis with reasonably high sensitivity and specificity from fingerprick and venous whole blood; however, signal amplification techniques were required, which increased the complexity of the design.[4, 7]
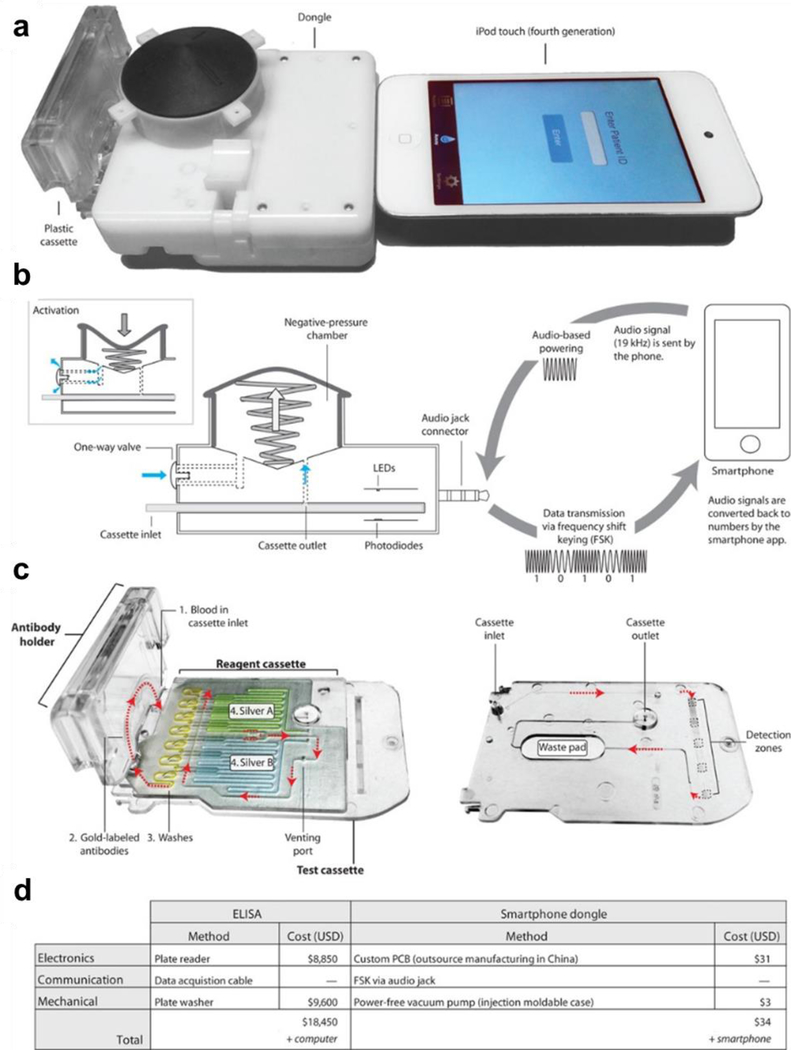
Figure 9. Smartphone-based point-of-care diagnosis for human immunodeficiency virus and syphilis.
A) Smartphone dongle with a microfluidic cassette for the detection of HIV and syphilis, connected to an iPod touch (or smartphone). B) The dongle uses the audio jack connector as the power source to generate vacuum and to transmit the data to a smartphone. C) A reagent cassette that contains the wash reagents in yellow, the silver enhancing reagents in blue and green and the detection zones and waste pad in the bottom layer. Reagents are numbered in the order they flow through the test cassette. First, blood is collected in the cassette via capillary action, allowing for HIV or syphilis Abs in the sample to bind to Ags immobilized in the detection zones. Next, resolubilized gold-labeled Abs flow through the channels and bind to the captured Abs followed by a water rinse. Once the venting port is closed, the A and B silver reagents mix and flow through the channel and enhance the signal of the colloidal gold-Ab complexes captured in the detection zones. D) Comparison of costs for ELISA versus the smartphone dongle POCT. Reproduced with permission.[7] Copyright 2015, American Association for the Advancement of Science.
Tokel et al. developed a point-of-care microfluidic chip for the detection of pathogenic E. coli. They immobilized an anti-lipopolysaccharide Ab by capture on a protein G functionalized gold substrate and used a portable SPR as the detection modality (Figure 10).[8] The microfluidic chip design was optimized to maximize bacteria capture based on modifications to the location of inlet and outlet ports. Calibration samples, clinical samples, and wash buffers were introduced into the system using a syringe pump. The LOD for E. coli was approximately 105 CFU mL−1, in both PBS and peritoneal dialysis fluid. While this level of analytical sensitivity may be sufficient for some applications and is on par with other POCTs [125, 126], it is several orders of magnitude below the sensitivity of a research-grade Biacore SPR instrument.[8] The sensitivity could presumably be increased by using secondary labeling steps, but this would involve further user intervention and increase the complexity of the test.
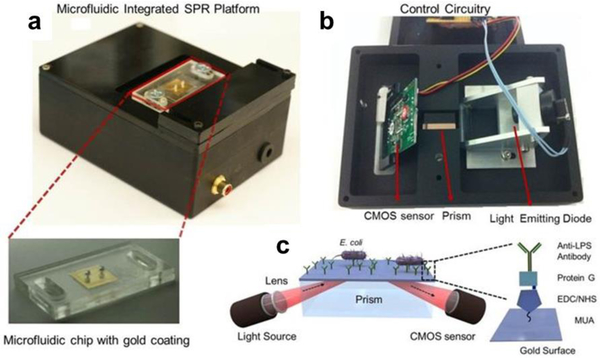
Figure 10. Portable microfluidic plasmonic platform for pathogen detection.
A) The microfluidic chips are mounted on top of the device. The chip with the inlet and outlet ports, and the 50 nm thick gold coated glass substrate is shown below. B) The light emitting diode illuminates a cylindrical lens, which collimates the light onto a rectangular prism. The reflected light is captured by a CMOS sensor and the image is transferred to a computer by the control circuitry. The chip is placed on the rectangular prism, with a refractive index matching oil in between. C) Integrated SPR platform. The bacteria are captured by Abs immobilized via surface bound protein G in the microchannel, and the capture event induces a change in the local refractive index. This change is captured by the sensor and transferred to a computer for analysis. Reproduced with permission.[8] Copyright 2015, Nature.
The challenges illustrated by these microfluidic assays is that they can automate an assay, but they come at the cost of introducing significant technological complexity that can drive up the manufacturing cost of the assay and reduce its reliability, especially when many sequential steps are involved. Further, many microfluidic platforms utilize BSA blocking to suppress NSB, which decreases surface fouling, but not as well compared to polymer brush surfaces, especially when challenged by undiluted serum or plasma.[105] Therefore, sensitivity could be presumably increased with the incorporation of a non-fouling surface, such as POEGMA, PHPMA, or PCBAA. We believe that microfluidics has an important role to play in POCTs, but that their design must be governed by the constraints of ensuring minimal technological complexity and low manufacturing cost for them to have a broad impact on POCTs.
We are pursuing this goal by developing a low-cost, passive microfluidic chip design for the D4 assay. The POEGMA interface of the D4 assay minimizes NSB and cell adhesion to the surface and the passive, capillary flow driven microfluidics eliminates the need for an external power source and moving parts. These modifications will transform the D4 assay into a POCT that requires no user intervention beyond the addition of a drop of liquid containing the analyte of interest to a sample port and another drop of wash buffer into a rinse port.
5. CONCLUSION
IVDs are crucial for effective clinical practice. Over the past decade, advancements in surface-based polymerization have spurred the development of novel non-fouling polymer brush surfaces. Polymer brushes are ideal substrates for testing complex biological milieu because they can be designed to significantly reduce NSB, which greatly improves the SNR and thereby analytical sensitivity of the assay. Here, we reported on the D4 POCT, a novel sandwich immunoassay with all the necessary reagents contained on a polymer brush coated chip, enabling the sensitive detection of low abundance protein biomarkers from complex mixtures, such as whole blood.
The convergence of a number of innovative approaches in the design of IVDs has created an exciting time in clinical diagnostics. Currently, much effort is being devoted to the miniaturization, automation, and integration of assay components into self-contained platforms that are amenable for testing at the point-of-care. While these advancements have begun to give rise to versatile platforms that are low-cost and “field-ready” but also retain the performance, sensitivity, specificity and reproducibility of laboratory-based assays, we caution that for these developments to realize their full potential, they must be simple in design, cheap to manufacture and easy-to-use. If these requirements are met, we anticipate a future of low-cost, reliable, easy-to-use, and sensitive POCTs that will empower individuals to track their health and thereby democratize healthcare.
Acknowledgments
J.T.H. is the lead author, and wrote the first draft and revised the manuscript. C.M.F., D.Y.J., A.M.H., and A.C. wrote parts of, and edited the manuscript. A.M.H. and A.C. are co-senior authors who oversaw revisions to the manuscript. The authors gratefully acknowledge funding support from National Cancer Institute Grant 1UG3CA211232‐01, Department of Defense United States Special Operations Command Grant W81XWH‐16‐C‐0219 and Combat Casualty Care Research Program (JPC‐6) Grant W81XWH‐17‐2‐0045.
Biography
 Jacob T. Heggestad received his Bachelor’s degree in chemical engineering from Northwestern University in 2017. He is currently a doctoral student in the Chilkoti lab at Duke University, where he is conducting research on point-of-care diagnostics.
Jacob T. Heggestad received his Bachelor’s degree in chemical engineering from Northwestern University in 2017. He is currently a doctoral student in the Chilkoti lab at Duke University, where he is conducting research on point-of-care diagnostics.
 Ashutosh Chilkoti is the Alan L. Kaganov Professor and the Chair of the Department of Biomedical Engineering at Duke University. His areas of research include genetically encoded materials and biointerface science. He has worked extensively on developing non-fouling polymer brush surface for point-of-care diagnositcs. He is a member of the National Academy of Inventors. He is also the founder of five start‐up companies.
Ashutosh Chilkoti is the Alan L. Kaganov Professor and the Chair of the Department of Biomedical Engineering at Duke University. His areas of research include genetically encoded materials and biointerface science. He has worked extensively on developing non-fouling polymer brush surface for point-of-care diagnositcs. He is a member of the National Academy of Inventors. He is also the founder of five start‐up companies.
Footnotes
Conflict of Interest: The underlying technology of the D4 assay was developed by A.M.H. and A.C. and acquired by Immucor Inc. In 2014.
6. REFERENCES
- [1].Gosling JP, Clinical Chemistry 1990, 36. [PubMed] [Google Scholar]
- [2].Crowther JR, The ELISA Guidebook, Humana Press, 2001. [Google Scholar]
- [3].Campbell J, Pollock N, Sharon A, Sauer-Budge AF, Anal Methods 2015, 7, 8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK, Nat Med 2011, 17, 1015. [DOI] [PubMed] [Google Scholar]
- [5].Tan X, Khaing Oo MK, Gong Y, Li Y, Zhu H, Fan X, Analyst 2017, 142, 2378. [DOI] [PubMed] [Google Scholar]
- [6].Wang T, Zhang M, Dreher DD, Zeng Y, Lab Chip 2013, 13, 4190. [DOI] [PubMed] [Google Scholar]
- [7].Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK, Sci Transl Med 2015, 7, 273re1. [DOI] [PubMed] [Google Scholar]
- [8].Tokel O, Yildiz UH, Inci F, Durmus NG, Ekiz OO, Turker B, Cetin C, Rao S, Sridhar K, Natarajan N, Shafiee H, Dana A, Demirci U, Sci Rep 2015, 5, 9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bhat RR, Chaney BN, Rowley J, Liebmann-Vinson A, Genzer J, Adv Mater 2005, 17, 2802. [Google Scholar]
- [10].Hucknall A, Rangarajan S, Chilkoti A, Adv Mater 2009, 21, 2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Perevozchikova T, Nanda H, Nesta DP, Roberts CJ, Journal of pharmaceutical sciences 2015, 104, 1946. [DOI] [PubMed] [Google Scholar]
- [12].Duangkaew P, Tapaneeyakorn S, Apiwat C, Dharakul T, Laiwejpithaya S, Kanatharana P, Laocharoensuk R, Biosensors & bioelectronics 2015, 74, 673. [DOI] [PubMed] [Google Scholar]
- [13].Hu P, Zhu C, Jin L, Dong S, Biosensors & bioelectronics 2012, 34, 83. [DOI] [PubMed] [Google Scholar]
- [14].Rajkomar A, Dhaliwal G, The Permanente Journal 2011, 15, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kingshott P, Griesser HJ, Current Opinion in Solid State and Materials Science 1999, 4, 403. [Google Scholar]
- [16].Zhang M, Desai T, Ferrari M, Biomaterials 1998, 19, 953. [DOI] [PubMed] [Google Scholar]
- [17].Lee JH, Kopecek J, Andrade JD, Journal of biomedical materials research 1989, 23, 351. [DOI] [PubMed] [Google Scholar]
- [18].Xia N, Hu Y, Grainger DW, Castner DG, Langmuir 2002, 18, 3255. [Google Scholar]
- [19].Amiji M, Park K, Journal of biomaterials science. Polymer edition 1993, 4, 217. [DOI] [PubMed] [Google Scholar]
- [20].Llanos GR, Sefton MV, Journal of biomaterials science. Polymer edition 1993, 4, 381. [DOI] [PubMed] [Google Scholar]
- [21].Currie EPK, Norde W, Cohen Stuart MA, Advances in Colloid and Interface Science 2003, 100–102, 205. [DOI] [PubMed] [Google Scholar]
- [22].Vaisocherova H, Brynda E, Homola J, Anal Bioanal Chem 2015, 407, 3927. [DOI] [PubMed] [Google Scholar]
- [23].Rodriguez Emmenegger C, Brynda E, Riedel T, Sedlakova Z, Houska M, Alles AB, Langmuir 2009, 25, 6328. [DOI] [PubMed] [Google Scholar]
- [24].Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM, Chemical reviews 2005, 105, 1103. [DOI] [PubMed] [Google Scholar]
- [25].Prime KL, Whitesides GM, Journal of the American Chemical Society 1993, 115, 10714. [Google Scholar]
- [26].Krishnamoorthy M, Hakobyan S, Ramstedt M, Gautrot JE, Chemical reviews 2014, 114, 10976. [DOI] [PubMed] [Google Scholar]
- [27].Jakubowski W, Matyjaszewski K, Angew Chem Int Ed Engl 2006, 45, 4482. [DOI] [PubMed] [Google Scholar]
- [28].Matyjaszewski K, Macromolecules 2012, 45, 4015. [Google Scholar]
- [29].Hong D, Hung HC, Wu K, Lin X, Sun F, Zhang P, Liu S, Cook KE, Jiang S, ACS Appl Mater Interfaces 2017, 9, 9255. [DOI] [PubMed] [Google Scholar]
- [30].Navarro LA, Enciso AE, Matyjaszewski K, Zauscher S, Journal of the American Chemical Society 2019, 141, 3100. [DOI] [PubMed] [Google Scholar]
- [31].Simakova A, Averick SE, Konkolewicz D, Matyjaszewski K, Macromolecules 2012, 45, 6371. [Google Scholar]
- [32].Du YJ, Cornelius RM, Brash JL, Colloids and Surfaces B: Biointerfaces 2000, 17, 59. [Google Scholar]
- [33].Ladd J, Zhang Z, Chen S, Hower JC, Jiang S, Biomacromolecules 2008, 9, 1357. [DOI] [PubMed] [Google Scholar]
- [34].Ma H, Li D, Sheng X, Zhao B, Chilkoti A, Langmuir 2006, 22, 3751. [DOI] [PubMed] [Google Scholar]
- [35].Joh DY, McGuire F, Abedini-Nassab R, Andrews JB, Achar RK, Zimmers Z, Mozhdehi D, Blair R, Albarghouthi F, Oles W, Richter J, Fontes CM, Hucknall AM, Yellen BB, Franklin AD, Chilkoti A, ACS Appl Mater Interfaces 2017, 9, 5522. [DOI] [PubMed] [Google Scholar]
- [36].Pereira A. d. l. S, Rodriguez-Emmenegger C, Surman F, Riedel T, Alles AB, Brynda E, RSC Advances 2014, 4, 2318. [Google Scholar]
- [37].Ahmad SA, Leggett GJ, Hucknall A, Chilkoti A, Biointerphases 2011, 6, 8. [DOI] [PubMed] [Google Scholar]
- [38].Harrison RH, Steele JA, Chapman R, Gormley AJ, Chow LW, Mahat MM, Podhorska L, Palgrave RG, Payne DJ, Hettiaratchy SP, Dunlop IE, Stevens MM, Adv Funct Mater 2015, 25, 5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lee PW, Isarov SA, Wallat JD, Molugu SK, Shukla S, Sun JE, Zhang J, Zheng Y, Lucius Dougherty M, Konkolewicz D, Stewart PL, Steinmetz NF, Hore MJ, Pokorski JK, Journal of the American Chemical Society 2017, 139, 3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hill RT, Kozek KM, Hucknall A, Smith DR, Chilkoti A, ACS Photonics 2014, 1, 974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tjong V, Yu H, Hucknall A, Chilkoti A, Anal Chem 2013, 85, 426. [DOI] [PubMed] [Google Scholar]
- [42].Hill RT, Mock JJ, Hucknall A, Wolter SD, Jokerst NM, Smith DR, Chilkoti A, ACS Nano 2012, 6, 9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tjong V, Yu H, Hucknall A, Rangarajan S, Chilkoti A, Anal Chem 2011, 83, 5153. [DOI] [PubMed] [Google Scholar]
- [44].Ma H, Hyun J, Stiller P, Chilkoti A, Adv Mater 2004, 16, 338. [Google Scholar]
- [45].Ma H, Hyun J, Zhang Z, Beebe TP, Chilkoti A, Advanced Functional Materials 2005, 15, 529. [Google Scholar]
- [46].Ma H, Wells M, Beebe TP, Chilkoti A, Advanced Functional Materials 2006, 16, 640. [Google Scholar]
- [47].Ma J, Luan S, Song L, Jin J, Yuan S, Yan S, Yang H, Shi H, Yin J, ACS Applied Materials & Interfaces 2014, 6, 1971. [DOI] [PubMed] [Google Scholar]
- [48].Hucknall A, Kim D-H, Rangarajan S, Hill RT, Reichert WM, Chilkoti A, Adv Mater 2009, 21, 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Joh DY, Hucknall AM, Wei Q, Mason KA, Lund ML, Fontes CM, Hill RT, Blair R, Zimmers Z, Achar RK, Tseng D, Gordan R, Freemark M, Ozcan A, Chilkoti A, Proc Natl Acad Sci U S A 2017, 114, E7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hower JC, Bernards MT, Chen S, Tsao HK, Sheng YJ, Jiang S, J Phys Chem B 2009, 113, 197. [DOI] [PubMed] [Google Scholar]
- [51].Bao Z, Bruening ML, Baker GL, Macromolecules 2006, 39, 5251. [Google Scholar]
- [52].Zhang Z, Chao T, Chen S, Jiang S, Langmuir 2006, 22, 10072. [DOI] [PubMed] [Google Scholar]
- [53].Dehghani ES, Du Y, Zhang T, Ramakrishna SN, Spencer ND, Jordan R, Benetti EM, Macromolecules 2017, 50, 2436. [Google Scholar]
- [54].Fan X, Lin L, Messersmith PB, Biomacromolecules 2006, 7, 2443. [DOI] [PubMed] [Google Scholar]
- [55].Hucknall A, Simnick AJ, Hill RT, Chilkoti A, Garcia A, Johannes MS, Clark RL, Zauscher S, Ratner BD, Biointerphases 2009, 4, FA50. [DOI] [PubMed] [Google Scholar]
- [56].Lyu S, Untereker D, Int J Mol Sci 2009, 10, 4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].(Ed: F. a. D. A. U.S. Department of Health and Human Services), 2014. [DOI] [PubMed]
- [58].Verhoef JJ, Carpenter JF, Anchordoquy TJ, Schellekens H, Drug Discov Today 2014, 19, 1945. [DOI] [PubMed] [Google Scholar]
- [59].Qi Y, Simakova A, Ganson NJ, Li X, Luginbuhl KM, Ozer I, Liu W, Hershfield MS, Matyjaszewski K, Chilkoti A, Nat Biomed Eng 2016, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lee KC, Lee S, Nature Biomedical Engineering 2017, 1, 0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Joh DY, Zimmers Z, Avlani M, Heggestad JT, Aydin HB, Ganson N, Kumar S, Fontes CM, Achar RK, Hershfield MS, Hucknall AM, Chilkoti A, Adv Healthc Mater 2019, 8, e1801177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kopecek J, Kopeckova P, Adv Drug Deliv Rev 2010, 62, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rodriguez-Emmenegger C, Brynda E, Riedel T, Houska M, Subr V, Alles AB, Hasan E, Gautrot JE, Huck WT, Macromol Rapid Commun 2011, 32, 952. [DOI] [PubMed] [Google Scholar]
- [64].Grimaud T, Matyjaszewski K, Macromolecules 1997, 30, 2216. [Google Scholar]
- [65].Teodorescu M, Matyjaszewski K*, Macromolecular Rapid Communications 2000, 21, 190. [Google Scholar]
- [66].Jones GR, Li Z, Anastasaki A, Lloyd DJ, Wilson P, Zhang Q, Haddleton DM, Macromolecules 2016, 49, 483. [Google Scholar]
- [67].Teodorescu M, Matyjaszewski K, Macromolecules 1999, 32, 4826. [Google Scholar]
- [68].Rademacher JT, Baum M, Pallack ME, Brittain WJ, Simonsick WJ, Macromolecules 2000, 33, 284. [Google Scholar]
- [69].Vaisocherova-Lisalova H, Surman F, Visova I, Vala M, Springer T, Ermini ML, Sipova H, Sedivak P, Houska M, Riedel T, Pop-Georgievski O, Brynda E, Homola J, Anal Chem 2016, 88, 10533. [DOI] [PubMed] [Google Scholar]
- [70].de los Santos Pereira A, Sheikh S, Blaszykowski C, Pop-Georgievski O, Fedorov K, Thompson M, Rodriguez-Emmenegger C, Biomacromolecules 2016, 17, 1179. [DOI] [PubMed] [Google Scholar]
- [71].Rodriguez-Emmenegger C, Houska M, Alles AB, Brynda E, Macromol Biosci 2012, 12, 1413. [DOI] [PubMed] [Google Scholar]
- [72].Vorobii M, De A Los Santos Pereira, Pop-Georgievski O, Kostina NY, Rodriguez-Emmenegger C, Percec V, Polymer Chemistry 2015, 6, 4210. [Google Scholar]
- [73].Colak S, Tew GN, Langmuir 2012, 28, 666. [DOI] [PubMed] [Google Scholar]
- [74].Zhang Z, Vaisocherova H, Cheng G, Yang W, Xue H, Jiang S, Biomacromolecules 2008, 9, 2686. [DOI] [PubMed] [Google Scholar]
- [75].Chen S, Zheng J, Li L, Jiang S, Journal of the American Chemical Society 2005, 127, 14473. [DOI] [PubMed] [Google Scholar]
- [76].He Y, Hower J, Chen S, Bernards MT, Chang Y, Jiang S, Langmuir 2008, 24, 10358. [DOI] [PubMed] [Google Scholar]
- [77].Schlenoff JB, Langmuir 2014, 30, 9625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Blackman LD, Gunatillake PA, Cass P, Locock KES, Chem Soc Rev 2019, 48, 757. [DOI] [PubMed] [Google Scholar]
- [79].Jiang S, Cao Z, Adv Mater 2010, 22, 920. [DOI] [PubMed] [Google Scholar]
- [80].Sun F, Wu K, Hung HC, Zhang P, Che X, Smith J, Lin X, Li B, Jain P, Yu Q, Jiang S, Anal Chem 2017, 89, 10999. [DOI] [PubMed] [Google Scholar]
- [81].Yang W, Xue H, Li W, Zhang J, Jiang S, Langmuir 2009, 25, 11911. [DOI] [PubMed] [Google Scholar]
- [82].Fontes CM, Achar RK, Joh DY, Ozer I, Bhattacharjee S, Hucknall A, Chilkoti A, Langmuir 2018, 35, 1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Surman F, Riedel T, Bruns M, Kostina NY, Sedláková Z, Rodriguez-Emmenegger C, Macromolecular Bioscience 2015, 15, 636. [DOI] [PubMed] [Google Scholar]
- [84].Badoux M, Billing M, Klok H-A, Polymer Chemistry 2019, 10, 2925. [Google Scholar]
- [85].Trmcic-Cvitas J, Hasan E, Ramstedt M, Li X, Cooper MA, Abell C, Huck WT, Gautrot JE, Biomacromolecules 2009, 10, 2885. [DOI] [PubMed] [Google Scholar]
- [86].Hu W, Liu Y, Lu Z, Li CM, Adv Funct Mater 2010, 20, 3497. [Google Scholar]
- [87].Liu Y, Wang W, Hu W, Lu Z, Zhou X, Li CM, Biomedical Microdevices 2011, 13, 769. [DOI] [PubMed] [Google Scholar]
- [88].Zhou W, Yang M, Zhao Z, Li S, Cheng Z, Zhu J, Applied Surface Science 2018, 450, 236. [Google Scholar]
- [89].Lisalova H, Brynda E, Houska M, Visova I, Mrkvova K, Song XC, Gedeonova E, Surman F, Riedel T, Pop-Georgievski O, Homola J, Anal Chem 2017, 89, 3524. [DOI] [PubMed] [Google Scholar]
- [90].Lei Z, Gao J, Liu X, Liu D, Wang Z, ACS Appl Mater Interfaces 2016, 8, 10174. [DOI] [PubMed] [Google Scholar]
- [91].Mittal R, Patel AP, Debs LH, Nguyen D, Patel K, Grati M, Mittal J, Yan D, Chapagain P, Liu XZ, J Cell Physiol 2016, 231, 2599. [DOI] [PubMed] [Google Scholar]
- [92].Hu W, Li X, He G, Zhang Z, Zheng X, Li P, Li CM, Biosensors & bioelectronics 2013, 50, 338. [DOI] [PubMed] [Google Scholar]
- [93].Hu W, Liu Y, Chen T, Liu Y, Li CM, Adv Mater 2015, 27, 181. [DOI] [PubMed] [Google Scholar]
- [94].Karlsson R, Michaelsson A, Mattsson L, J Immunol Methods 1991, 145, 229. [DOI] [PubMed] [Google Scholar]
- [95].Mullett WM, Lai EP, Yeung JM, Methods 2000, 22, 77. [DOI] [PubMed] [Google Scholar]
- [96].Masson JF, ACS Sens 2017, 2, 16. [DOI] [PubMed] [Google Scholar]
- [97].Lipschultz CA, Li Y, Smith-Gill S, Methods 2000, 20, 310. [DOI] [PubMed] [Google Scholar]
- [98].Malmqvist M, Curr Opin Immunol 1993, 5, 282. [DOI] [PubMed] [Google Scholar]
- [99].Johne B, Gadnell M, Hansen K, J Immunol Methods 1993, 160, 191. [DOI] [PubMed] [Google Scholar]
- [100].Bolduc OR, Pelletier JN, Masson JF, Anal Chem 2010, 82, 3699. [DOI] [PubMed] [Google Scholar]
- [101].Masson JF, Battaglia TM, Davidson MJ, Kim YC, Prakash AM, Beaudoin S, Booksh KS, Talanta 2005, 67, 918. [DOI] [PubMed] [Google Scholar]
- [102].Liu B, Liu X, Shi S, Huang R, Su R, Qi W, He Z, Acta Biomater 2016, 40, 100. [DOI] [PubMed] [Google Scholar]
- [103].Homola J, Chemical reviews 2008, 108, 462. [DOI] [PubMed] [Google Scholar]
- [104].Zhang Z, Chen S, Jiang S, Biomacromolecules 2006, 7, 3311. [DOI] [PubMed] [Google Scholar]
- [105].Vaisocherová H, Yang W, Zhang Z, Cao Z, Cheng G, Piliarik M, Homola J. i., Jiang S, Analytical Chemistry 2008, 80, 7894. [DOI] [PubMed] [Google Scholar]
- [106].Vaisocherova-Lisalova H, Visova I, Ermini ML, Springer T, Song XC, Mrazek J, Lamacova J, Scott Lynn N Jr., Sedivak P, Homola J, Biosensors & bioelectronics 2016, 80, 84. [DOI] [PubMed] [Google Scholar]
- [107].Riedel T, Rodriguez-Emmenegger C, de los Santos Pereira A, Bědajánková A, Jinoch P, Boltovets PM, Brynda E, Biosensors and Bioelectronics 2014, 55, 278. [DOI] [PubMed] [Google Scholar]
- [108].Riedel T, Surman F, Hageneder S, Pop-Georgievski O, Noehammer C, Hofner M, Brynda E, Rodriguez-Emmenegger C, Dostálek J, Biosensors and Bioelectronics 2016, 85, 272. [DOI] [PubMed] [Google Scholar]
- [109].Zhang P, Sun F, Hung H-C, Jain P, Leger KJ, Jiang S, Analytical Chemistry 2017, 89, 8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Bell D, Wongsrichanalai C, Barnwell JW, Nature reviews. Microbiology 2006, 4, 682. [DOI] [PubMed] [Google Scholar]
- [111].Posthuma-Trumpie G, Korf J, van Amerongen A, Analytical and Bioanalytical Chemistry 2009, 393, 569. [DOI] [PubMed] [Google Scholar]
- [112].Giljohann DA, Mirkin CA, Nature 2009, 462, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Vashist SK, Luppa PB, Yeo LY, Ozcan A, Luong JHT, Trends Biotechnol 2015, 33, 692. [DOI] [PubMed] [Google Scholar]
- [114].Hu J, Wang L, Li F, Han YL, Lin M, Lu TJ, Xu F, Lab Chip 2013, 13, 4352. [DOI] [PubMed] [Google Scholar]
- [115].Parolo C, de la Escosura-Muniz A, Merkoci A, Biosensors & bioelectronics 2013, 40, 412. [DOI] [PubMed] [Google Scholar]
- [116].Mabey D, Peeling RW, Ustianowski A, Perkins MD, Nature reviews. Microbiology 2004, 2, 231. [DOI] [PubMed] [Google Scholar]
- [117].Michel R, Pasche S, Textor M, Castner DG, Langmuir 2005, 21, 12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Engin ED, J Immunoassay Immunochem 2019, 40, 109. [DOI] [PubMed] [Google Scholar]
- [119].Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA Jr., Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N, Science 2018, 359, 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Bartz S, Mody A, Hornik C, Bain J, Muehlbauer M, Kiyimba T, Kiboneka E, Stevens R, Bartlett J, St Peter JV, Newgard CB, Freemark M, J Clin Endocrinol Metab 2014, 99, 2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Clackson T, Hoogenboom HR, Griffiths AD, Winter G, Nature 1991, 352, 624. [DOI] [PubMed] [Google Scholar]
- [122].Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR, Annu Rev Immunol 1994, 12, 433. [DOI] [PubMed] [Google Scholar]
- [123].Tabata N, Sakuma Y, Honda Y, Doi N, Takashima H, Miyamoto-Sato E, Yanagawa H, Nucleic Acids Res 2009, 37, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Zanganeh S, Rouhani Nejad H, Mehrabadi JF, Hosseini R, Shahi B, Tavassoli Z, Aramvash A, Appl Biochem Biotechnol 2019, 187, 493. [DOI] [PubMed] [Google Scholar]
- [125].Huang H, Zhao G, Dou W, Biosensors & bioelectronics 2018, 107, 266. [DOI] [PubMed] [Google Scholar]
- [126].Varshney M, Li Y, Biosensors & bioelectronics 2007, 22, 2408. [DOI] [PubMed] [Google Scholar]