ABSTRACT
Puberty is a developmentally plastic phase. Variations in pubertal tempo have implications for the risk of later adult diseases. Influences on pubertal tempo have been widely discussed, but the underlying biological mechanisms remain unclear. Epigenetic modifications are known to regulate development processes; they could play an important role in affecting pubertal outcomes. We conducted a population-based analysis to investigate the association of peripubertal blood DNA methylation at LINE-1 and growth-related candidate genes with pubertal onset and progression in healthy adolescents. The analytic sample included 114 males and 129 females aged 10 to 18 years. DNA methylation at growth-related candidate loci IGF2, H19, HSD11B2, as well as LINE-1 repetitive elements were quantified. Cox survival and ordinal regression models were used to examine sex- and locus-specific associations of epigenetic markers with pubertal development using physician-assessed Tanner stages and self-reported menarche, adjusted for covariates. Among boys, DNA methylation at H19 was associated with later pubarche. HSD11B2 methylation was associated with earlier onset of pubic hair and genitalia development and slower pubertal progression. IGF2 was associated with later onset of genital development. Among girls, LINE-1 methylation was associated with later onset of breast development. For each percent increase of methylation at H19, there was 5% increased odds in the earlier onset of breast development. DNA methylation of IGF2 was associated with earlier onset of pubic hair. DNA methylation at genes known to influence early-life growth may also influence pubertal outcomes.
KEYWORDS: DNA methylation, pubertal onset and progression, sexual maturation, epigenetics
Introduction
Puberty is the process by which and during which sexual maturation occurs and reproductive capacity is attained [1]. The timing of puberty ranges widely. Early onset of puberty has been documented, with U.S girls exhibiting pubertal change before age 6 among African Americans and age 7 among Caucasians, age 8 for girls in other parts of the world, and age 9 for Caucasian boys [2–5]. Delayed puberty, on the other hand, is defined as the lack of pubertal onset by an age ≥2 SD above the population mean and can occur as late as age 13 in girls and age 14 in boys [6]. These variations in pubertal timing have implications for the risk of later adult diseases, including polycystic ovary syndrome (PCOS), obesity, type 2 diabetes, cardiovascular disease and reproductive tract cancers [1,6]. Moreover, they may negatively affect adult psychosocial functioning, educational achievement, height, and bone mineral density [1,6]. Over recent decades, the risk factors of earlier or later puberty, including chemical exposures, unbalanced diet, and abnormal hormone levels caused by diseases and psychological stress, have been discussed [7,8]. However, the underlying biological mechanisms that lead to early or delayed puberty remain unclear. Moreover, studies in racially and ethnically diverse groups are few and have methodologic limitations (for instance, cross-sectional study design).
Epigenetic modification is a biological mechanism that may underline pubertal timing and progression, since it is known to regulate development processes and is responsive to environmental factors. The epigenome consists of heritable, yet potentially reversible, modifications including DNA methylation, posttranslational histone tail modifications and non-coding RNA (ncRNA)-associated gene silencing which regulate gene expression but do not alter the DNA sequence [9]. Epigenetic modifications including DNA methylation are known to regulate developmental processes [10–12]. Given that sexual maturation is continual from the time it is initiated in intrauterine life through the life cycle [1], epigenetic programming may contribute to the timing of puberty. Several reports have examined the role of epigenetics in regulating changes in body composition and growth milestones, which typically occur at the same time as puberty [12,13]. Epigenetic age, an estimate of biological age based on changes in DNA methylation at particular locations along the genome [14], was associated with longitudinal changes in weight, BMI, height and fat mass during childhood and adolescence in a sample of 1018 children [15]. Other studies found the levels of cord blood DNA methylation were related to development of adiposity later in life [12,16]. However, few studies have examined the specific effect of epigenetics on pubertal onset and progression. A rat study revealed epigenetic control of Kiss1 is important for pubertal timing in females, and an epidemiological study linked repetitive element DNA hypomethylation at long interspersed nucleotide elements (LINE-1) to increased odds of menarche by age 12 among girls and lower luteinizing hormone levels at age 9 years among boys [17,18].
Based upon previous biological evidence, there are two reasons to hypothesize an association between altered DNA methylation and changes in pubertal timing. Firstly, a growing body of literature has demonstrated that increased body mass index (BMI) is associated with altered methylation at multiple genes [19–21]. Moreover, evidence from both animal and human studies suggest that pre-pubertal obesity might be causally related to earlier puberty [22,23]. Secondly, reproductive hormones, primarily testosterone and dehydroepiandrosterone (DHEA), two androgens that facilitate masculine development, and estradiol, an estrogen that facilitates feminine development [24], advance puberty. DNA methylation levels could potentially modify reproductive hormone levels or sensitivity/expression of hormone receptors [18,25–27]. These two proposed mechanisms are possibly intertwined, given that feedback from reproductive hormones fat mass might stimulate the central pulsatile gonadotrophin secretion and trigger the onset of puberty [28,29]. However, in spite of evidence to support the possible association between DNA methylation, BMI and sexual maturation outcomes, no research has examined relationships among all three in adolescent children.
To address these research gaps, this longitudinal observational study in Mexico City tests the hypothesis that peripubertal blood leukocyte DNA methylation at LINE-1 and specific genes (HSD11B2, as well as imprinted genes IGF2 and H19) will be associated with pubertal onset and progression assessed at two time periods, adjusted for BMI, age and household socioeconomic status (SES). Regions were selected for DNA methylation analysis based on demonstrated variability across age and/or by various environmental factors in previous studies [30–35]. In addition, LINE-1 repetitive elements have been associated with pubertal status and hormone levels in Mexican Americans. H19, IGF2, and HSD11B2 are linked to early life growth but their implications for adolescent health, specifically with regards to pubertal onset and tempo, have not yet been studied.
Results
The cohort included 250 subjects who attended the early-teen visit (boys: 118 (47.2%), girls: 132 (52.8%), and 222 subjects who attended both visits (boys: 108 (48.6%), girls: 114 (51.4%)). After eliminating 7 individuals with missing predictors, the analytical sample included 243 subjects (boys: 114 (46.9%), girls: 129 (53.1%)). The mean age for the early-teen visit was 10.4 years in boys and 10.3 years in girls; the mean was 13.7 years in boys and 13.5 for girls for the late-teen visit. We observed children moving to more advanced pubertal stages from the early- to late-teen visits. Among boys, 79.7% and 48.3% were at Tanner stage 1 for pubic hair and genital development in the early-teen visit; and the number dropped down to 25.0% and 6.5% in the second visit. In terms of testicular volume, the percentage of boys in the pre-pubertal stage dropped from 15.3% to 0%. Among girls, 74.2% of them had Tanner stage 1 for pubic hair and 65.9% for breast development in the early-teen visit; and the percentage dropped to 7.9% and 4.4% later (Table 1).
Table 1.
Distributions of tanner stages and other covariates among 250 ELEMENT children at the early-teen visit (Visit 1) and again at the late-teen visit (Visit 2) for 222 children who continued follow-up.
| Visit 1 (N = 118) |
Visit 2 (N = 108) |
|||
|---|---|---|---|---|
| Boys | N | % | N | % |
| Pubic Hair: Tanner Stage | ||||
| Refused Observation/Missing | 3 | 2.54 | 3 | 2.78 |
| 1 | 94 | 79.66 | 27 | 25.00 |
| 2 | 17 | 14.41 | 16 | 14.81 |
| 3 | 3 | 2.54 | 30 | 27.78 |
| 4 | 1 | 0.85 | 18 | 16.67 |
| 5 | 0 | 0.00 | 14 | 12.96 |
| Genital Development: Tanner Stage | ||||
| Refused Observation/Missing | 3 | 2.54 | 3 | 2.78 |
| 1 | 57 | 48.31 | 7 | 6.48 |
| 2 | 43 | 36.44 | 17 | 15.74 |
| 3 | 10 | 8.47 | 26 | 24.07 |
| 4 | 5 | 4.24 | 37 | 34.26 |
| 5 | 0 | 0.00 | 18 | 16.67 |
| Testicular Development (L) | ||||
| Refused Observation/Missing | 3 | 2.54 | 3 | 2.78 |
| 1–3 ml | 18 | 15.25 | 0 | 0.00 |
| 3–11 ml | 75 | 63.56 | 16 | 14.81 |
| >11 ml | 22 | 18.65 | 89 | 82.41 |
| Testicular Development (R) | ||||
| Refused Observation/Missing | 4 | 3.39 | 3 | 2.78 |
| 1–3 ml | 18 | 15.25 | 0 | 0.00 |
| 3–11 ml | 76 | 64.41 | 16 | 14.81 |
| >11 ml | 20 | 16.95 | 89 | 82.41 |
| Age | 10.35 ± 1.61 | 13.72 ± 1.75 | ||
| BMI | 19.06 ± 3.14 | 20.43 ± 3.68 | ||
| Household SES: Quartile | N = 100 | |||
| 1 | 24 | 24.00 | ||
| 2 | 27 | 27.00 | ||
| 3 | 24 | 24.00 | ||
| 4 |
25 |
25.00 |
|
|
|
Visit 1 (N = 132) |
Visit 2 (N = 114) |
|||
|
Girls |
N |
% |
N |
% |
| Pubic Hair: Tanner Stage | ||||
| Refused Observation/Missing | 0 | 0.00 | 2 | 1.75 |
| 1 | 98 | 74.24 | 9 | 7.90 |
| 2 | 22 | 16.67 | 39 | 34.21 |
| 3 | 9 | 6.82 | 29 | 25.44 |
| 4 | 2 | 1.52 | 21 | 18.42 |
| 5 | 1 | 0.76 | 14 | 12.28 |
| Breast Development: Tanner Stage | ||||
| Refused Observation/Missing | 0 | 0.00 | 2 | 1.75 |
| 1 | 87 | 65.90 | 5 | 4.39 |
| 2 | 20 | 15.15 | 12 | 10.53 |
| 3 | 18 | 13.63 | 46 | 40.35 |
| 4 | 7 | 5.30 | 31 | 27.19 |
| 5 | 0 | 0.00 | 18 | 15.79 |
| Menarche | ||||
| Refused Observation/Missing | 0 | 0.00 | 1 | 0.88 |
| Yes | 30 | 22.73 | 90 | 78.95 |
| No | 102 | 77.27 | 23 | 20.17 |
| Age | 10.30 ± 1.72 | 13.54 ± 1.75 | ||
| BMI | 19.66 ± 3.95 | 21.61 ± 4.07 | ||
| Household SES: Quartile | N = 102 | |||
| 1 | 25 | 24.51 | ||
| 2 | 29 | 28.43 | ||
| 3 | 24 | 23.53 | ||
| 4 | 24 | 23.53 | ||
LINE-1 methylation was higher among boys compared to girls, both before (Table 2) and after (Supplemental Table 1) imputation of missing values. No statistically significant sex differences of DNA methylation levels at HSD11B2, H19 or IGF2 were observed.
Table 2.
DNA Methylation at LINE-1, H19, HSD11B2 and IGF2 among all individuals and stratified by sexa.
| Entire Cohort |
Boys |
Girls |
|||||
|---|---|---|---|---|---|---|---|
| N | Mean % methylation (SD) | N | Mean % methylation (SD) | N | Mean % methylation (SD) | P valueb | |
| LINE-1 methylation | |||||||
| Site 1 | 243 | 79.88 (3.87) | 113 | 80.48 (4.13) | 130 | 79.37 (3.57) | 0.026 |
| Site 2 | 243 | 81.89 (1.99) | 113 | 82.20 (2.15) | 130 | 81.62 (1.80) | 0.022 |
| Site 3 | 243 | 78.64 (2.93) | 113 | 79.26 (2.99) | 130 | 78.10 (2.77) | 0.002 |
| Site 4 | 243 | 73.54 (2.14) | 113 | 73.96 (1.96) | 130 | 73.18 (2.22) | 0.004 |
| H19 methylation | |||||||
| Site 1 | 245 | 59.08 (8.24) | 115 | 58.46 (7.57) | 130 | 59.63 (8.79) | 0.266 |
| Site 2 | 245 | 58.23 (4.83) | 115 | 58.23 (3.32) | 130 | 58.22 (5.87) | 0.990 |
| Site 3 | 245 | 59.26 (3.78) | 115 | 59.05 (3.54) | 130 | 59.45 (3.98) | 0.412 |
| Site 4 | 245 | 56.66 (8.51) | 115 | 55.83 (7.74) | 130 | 57.39 (9.09) | 0.153 |
| HSD11B2 methylation | |||||||
| Site 1 | 246 | −1.54 (2.14) | 115 | −1.57 (2.35) | 131 | −1.51 (1.94) | 0.844 |
| Site 2 | 246 | 0.08 (0.92) | 115 | −0.04 (0.92) | 131 | 0.19 (0.92) | 0.057 |
| Site 3 | 245 | −2.20 (2.25) | 115 | −2.23 (2.33) | 130 | −2.18 (2.19) | 0.856 |
| Site 4 | 244 | −0.76 (1.75) | 115 | −0.70 (1.58) | 129 | −0.81 (1.89) | 0.601 |
| Site 5 | 229 | 0.24 (4.46) | 108 | 0.20 (4.33) | 121 | 0.28 (4.58) | 0.898 |
| IGF2 methylation | |||||||
| Site 1 | 223 | 35.15 (12.23) | 101 | 34.71 (11.98) | 122 | 35.51 (12.46) | 0.630 |
| Site 2 | 213 | 44.85 (14.06) | 94 | 42.99 (14.56) | 119 | 46.31 (13.54) | 0.087 |
| Site 3 | 228 | 53.79 (6.40) | 102 | 53.57 (6.92) | 126 | 53.97 (5.97) | 0.644 |
| Site 4 | 196 | 37.87 (4.26) | 88 | 37.69 (4.65) | 108 | 38.01 (3.94) | 0.607 |
| Site 5 | 229 | 52.94 (6.51) | 103 | 52.90 (6.21) | 126 | 52.97 (6.76) | 0.932 |
a LINE-1, HSD11B2, and H19 data exhibited batch effects and as such were standardized to controls included on experimental plates as previously described [30], while the IGF2 data exhibited no batch effect. Batch adjustment for HSD11B2 involves subtracting the value of the 0% methylated control from each experimental plate and sometimes results in negative values due to low raw methylation values for HSD11B2.
b P-value of 2-sample t test comparing boys and girls.
Among boys, we observed associations between early-teen DNA methylation and pubertal outcomes cross-sectionally at the early teen visit as well as prospectively at the late teen visit, and with the progression between the two. In the cross-sectional adjusted analysis (Table 3), we found for each percent increase of DNA methylation at H19 CpG sites 2 and 3 (equivalent to 0.31 and 0.29 SD increase of DNA methylation, respectively; they were obtained as 1/SD in % (sex-specific and site-specific) from Supplemental Table 1), there were 36% and 26% increased odds of later pubarche (p = 0.025 and 0.039). However, the associations did not remain statistically significant in the prospective analysis (Table 3). We also found for each percent methylation increase of HSD11B2 site 4 (0.62 SD), there was a 60% increased odds of earlier pubarche (p = 0.030), and a 67% increased odds in earlier onset of genital development (p = 0.003), in cross-sectional analysis. Prospectively, for each percent methylation increase of HSD11B2 site 4, there was 20% increased odds of earlier pubarche (p = 0.034). HSD11B2 site 4 was also associated with 17% increased odds of slower genital development progression (p = 0.016, Table 4). For each percent methylation increase of IGF2 site 3 (0.13 SD), there was 7% increased odds of later onset of genital development, in both cross-sectional and prospective analyses (p = 0.010 and 0.005). IGF2 methylation was also associated with faster genital development progression (p = 0.036) (Table 4). Among girls, we found for each percent increase in methylation of LINE-1 CpG sites 3 and 4 (0.36 and 0.44 SD equivalents, respectively), there were 11% and 17% increased odds of later onset of breast development (p = 0.008 and <0.001) in the cross sectional analysis. For each percent increase in methylation of H19 sites 1 and 4 (0.11 SD of each), there were 5% increased odds in the earlier onset of breast development (both p values <0.001). However, the associations mentioned above were not statistically significant in prospective analyses. In terms of HSD11B2, we found for each percent increase in methylation of site 1 and 3 (0.51 and 0.45 SD), there were 20% and 13% increased odds of later onset of breast development (p < 0.001 and 0.02); for each percent increase in methylation of site 4 (0.53 SD), there were 25% increased odds in the expected earlier onset of breast development (p < 0.001). In addition, with one percent increase methylation of IGF2 site 5 (0.18 SD), there was 7% increased odds of earlier pubarche (p = 0.020) (Table 3). DNA methylation was not found to be associated with pubertal progression among girls (Table 4).
Table 3.
Associations between site-specific visit 1 (early-teen) DNA methylation and visit 1 (early-teen) or visit 2 (late-teen) pubertal onset, in adjusted Cox survival modela,b. CpG sites with at least one significant relationship (p < 0.05) are shown. See Supplemental Table 2 for full results.
| Pubic Hair |
Genital Development |
Testicular Volume (L) |
Testicular Volume (R) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Hazard Ratio(CI) |
Hazard Ratio (CI) |
Hazard Ratio (CI) |
Hazard Ratio (CI) |
||||||
| Boys (N = 114) |
Visit 1 |
Visit 2 |
Visit 1 |
Visit2 |
Visit 1 |
Visit 2 |
Visit 1 |
Visit 2 |
|
| Site | Adjusted | Adjusted | Adjusted | Adjusted | Adjusted | Adjusted | Adjusted | Adjusted | |
| H19 | 2 |
0.64 (0.43, 0.94) |
0.96 (0.87, 1.05) |
0.93 (0.84, 1.03) |
0.96 (0.90, 1.03) |
1.08 (0.97, 1.21) |
1.08 (0.97, 1.20) |
1.07 (0.97, 1.18) |
1.07 (0.97, 1.18) |
| 3 |
0.74 (0.55, 0.99) |
0.97 (0.88, 1.06) |
0.93 (0.84, 1.03) |
0.95 (0.89, 1.02) |
1.10 (0.98, 1.24) |
1.10 (0.98, 1.24) |
1.08 (0.97, 1.19) |
1.08 (0.98, 1.19) |
|
| HSD11B2 | 4 |
1.60 (1.05, 2.44) |
1.20 (1.01, 1.43) |
1.67 * (1.19, 2.33) |
1.01 (0.88, 1.16) |
0.99 (0.78, 1.27) |
1.01 (0.78, 1.30) |
1.03 (0.83, 1.29) |
1.05 (0.84, 1.31) |
|
IGF2 |
3 |
0.95 (0.89, 1.01) |
0.96 (0.92, 1.01) |
0.93 (0.88, 0.99) |
0.93 (0.89, 0.98) |
1.03 (0.97, 1.10) |
1.03 (0.96, 1.10) |
1.01 (0.96, 1.07) |
1.01 (0.95, 1.07) |
|
Pubic Hair |
Breast Development |
Menarche (Y/N) |
Menarche Age |
||||||
|
Girls (N = 129) |
Hazard Ratio (CI) |
Hazard Ratio (CI) |
Hazard Ratio (CI) |
Hazard Ratio (CI) |
|||||
| |
Site |
Adjusted |
Adjusted |
Adjusted |
Adjusted |
Adjusted |
Adjusted |
Adjusted |
Adjusted |
| LINE1 | 3 | 1.07 (0.86, 1.34) |
0.95 (0.84, 1.08) |
0.89 (0.82, 0.97) |
0.97 (0.87, 1.09) |
0.83 (0.67, 1.03) |
0.98 (0.89, 1.08) |
0.87 (0.72, 1.06) |
0.94 (0.92, 1.10) |
| 4 | 1.01 (0.78, 1.31) |
0.92 (0.79, 1.07) |
0.83 * (0.75, 0.93) |
0.88 (0.76, 1.02) |
0.85 (0.66, 1.09) |
0.88 (0.77, 1.01) |
0.89 (0.74, 1.06) |
1.01 (0.84, 1.04) |
|
| H19 | 1 | 1.03 (0.95, 1.12) |
1.02 (0.98, 1.07) |
1.05 * (1.02, 1.07) |
1.02 (0.99, 1.06) |
0.99 (0.94, 1.06) |
1.02 (0.99, 1.06) |
1.01 (0.95, 1.06) |
1.01 (0.99, 1.04) |
| 2 | 1.21 (1.00, 1.47) |
1.02 (0.96, 1.07) |
1.02 (0.96, 1.09) |
0.93 (0.88, 0.99) |
0.98 (0.88, 1.09) |
0.99 (0.94, 1.04) |
0.96 (0.85, 1.09) |
1.00 (0.96, 1.04) |
|
| 3 |
1.22 (1.01, 1.49) |
1.07 (0.99, 1.17) |
1.06 (0.99, 1.13) |
0.99 (0.91, 1.07) |
0.99 (0.87, 1.13) |
1.01 (0.93, 1.10) |
0.97 (0.86, 1.09) |
1.01 (0.95, 1.08) |
|
| 4 | 1.03 (0.95, 1.13) |
1.03 (0.98, 1.08) |
1.05 (1.03, 1.08) |
1.03 (0.99, 1.06) |
1.00 (0.94, 1.06) |
1.03 (1.00, 1.06) |
1.01 (0.95, 1.06) |
1.01 (0.99, 1.04) |
|
| HSD11B2 | 1 | 0.98 (0.79, 1.22) |
0.97 (0.84, 1.11) |
0.80 * (0.71, 0.91) |
0.86 (0.72, 1.02) |
0.93 (0.76, 1.14) |
0.95 (0.80, 1.12) |
0.92 (0.77, 1.10) |
0.98 (0.88, 1.09) |
| 3 | 1.07 (0.87, 1.33) |
1.04 (0.90, 1.20) |
0.87 (0.77, 0.98) |
0.92 (0.79, 1.07) |
0.89 (0.72, 1.11) |
0.91 (0.79, 1.06) |
0.91 (0.77, 1.08) |
0.96 (0.87, 1.06) |
|
| 4 | 1.32 (0.91, 1.90) |
1.06 (0.93, 1.20) |
1.25 * (1.13, 1.38) |
1.08 (0.95, 1.24) |
1.18 (0.87, 1.60) |
0.93 (0.76, 1.13) |
1.21 (0.94, 1.57) |
0.90 (0.76, 1.07) |
|
| IGF2 | 3 | 0.96 (0.88, 1.04) |
0.98 (0.90, 1.07) |
1.00 (0.95, 1.05) |
0.97 (0.91, 1.04) |
1.12 (1.02, 1.22) |
0.99 (0.94, 1.04) |
1.03 (0.98, 1.09) |
0.98 (0.94, 1.01) |
| 5 | 1.05 (0.94, 1.17) |
1.07 (1.01, 1.14) |
1.01 (0.96, 1.07) |
1.02 (0.96, 1.09) |
1.12 (1.00, 1.26) |
1.00 (0.95, 1.06) |
1.02 (0.96, 1.09) |
0.99 (0.95, 1.03) |
|
a Adjusted for age, BMI and SES status at visit 1 (early-teen).
b Bolded value indicates the association is significant with a P value < 0.05.
* Association is significant after multiple testing with a P value < 0.0028.
Table 4.
Associations between site-specific visit 1 (early-teen) DNA methylation and pubertal progression from visit 1 (early-teen) to visit 2 (late-teen), in adjusted multivariate regression model a,b,c. CpG sites with at least one significant relationship (p < 0.05) are shown (see Supplemental Table 3 for full results). Odds Ratios are shown from adjusted models for the main effect of DNA methylation along with the interaction between DNA methylation and time between visits.
| Pubic Hair |
Genital Development |
Testicular Volume (L) |
Testicular Volume (R) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Boys (N = 114) |
Odds Ratio (CI) |
Odds Ratio (CI) |
Odds Ratio (CI) |
Odds Ratio (CI) |
|||||
| Site | Main Effect | Site by Time | Main Effect | Site by Time | Main Effect | Site by Time | Main Effect | Site by Time | |
| HSD11B2 | 1 | 0.84 (0.42, 1.68) |
1.04 (0.90, 1.20) |
0.77 (0.53, 1.12) |
1.12 (1.01, 1.24) |
0.94 (0.60, 1.50) |
1.03 (0.92, 1.15) |
0.96 (0.64, 1.45) |
1.03 (0.93, 1.14) |
| 4 | 1.36 (0.47, 3.96) |
0.97 (0.76, 1.23) |
1.59 (0.92, 2.74) |
0.83 (0.72, 0.97) |
1.08 (0.49, 2.40) |
1.01 (0.85, 1.19) |
1.14 (0.55, 2.38) |
1.00 (0.85, 1.18) |
|
| IGF2 | 4 | 0.95 (0.78, 1.15) |
1.02 (0.98, 1.06) |
0.93 (0.82, 1.05) |
1.04 (1.00, 1.07) |
1.04 (0.85, 1.27) |
1.01 (0.96, 1.06) |
1.04 (0.87, 1.25) |
1.01 (0.96, 1.06) |
a Adjusted for age, BMI and SES status at visit 1 (early-teen).
b Bolded value indicates the association is significant with a P value < 0.05.
c Model: logit (Y ij) = β0 + β1*age + β2*time_difference + β3*CpG_Site + β4*CpG_Site*time_difference +β5*age*time_difference
After corrections for multiple testing, associations of LINE-1, H19 and HSD11B2 with breast onset among girls, as well as the association of HSD11B2 with genital onset among boys remained significant with p values < 0.0028 (Table 3). None of the pubertal progression results maintained statistical significance, however, after correction for multiple testing (Table 4).
Discussion
There have been a limited number of population-based longitudinal studies examining the potential association between epigenetics and puberty. The goal of this sex-specific analysis was to investigate the potential effects of peri-pubertal DNA methylation on pubertal status and progression. We found that DNA methylation of H19, IGF2, and HSD11B2 were associated with pubic hair and genital onset in boys, while methylation of LINE-1, H19, IGF2 and HSD11B2 were associated with pubic hair and breast development among girls. These findings suggest that DNA methylation at genes known to influence early-life growth and development may also influence pubertal outcomes, though the mechanism (direct or indirect) remains to be elucidated.
While it is well established that pubertal timing and progression is controlled by many genes [36–38], the epigenetic regulation involved in this process is less understood. One study found rats treated with 5-azacytidine (Aza), a DNA methylation inhibitor had delayed vaginal opening, failed to reach puberty, as assessed by the lack of ovulation, and showed no estrous cyclicity [17]. In our gene-specific analysis, we found decreased DNA methylation levels in H19 and IGF2 were linked to later onset of breast development and menarche in girls. Among boys, decreased DNA methylation levels in HSD11B2 were associated with delayed pubic hair onset and genitalia onset.
Our findings build on prior studies in other important ways, with both consistent and contrasting results. For example, in a longitudinal study, Huen et al examined the relationship of Alu and LINE-1 repetitive element DNA methylation measured in umbilical cord and 9-year old child blood samples with puberty status in Mexican-American participants of the Center for Health Assessment of Mothers and Children of Salinas (CHAMACOS) cohort. They found no association between child LINE-1 methylation and odds of genital or pubic hair development in boys, but found a significant association with later onset of menarche in girls [18]. We also did not observe any association between peri-pubertal LINE-1 methylation and male sexual characteristics in either cross-sectional or longitudinal analyses. However, cross-sectional analysis showed that elevated LINE-1 methylation was suggestively associated with later breast onset, but not menarche in girls. The findings were consistent in terms of directionality. As both breast development and menarche are advanced by estrogens [39], the different findings may be due to age differences between the two cohorts. Moreover, based on observational studies, the estimated mean age at menarche is different in Mexico City (11.40 years [40]), versus among Mexican Americans (12.25 years [41]), and timing differed between the two studies as children were only followed through age 12 in the CHAMACOS study.
To our knowledge, although no population-based studies have examined the associations between H19 and IGF2 methylation and pubertal outcomes specifically, several articles support the effect of methylation on imprinted genes with growth, growth-related hormone concentrations, adiposity and birth weight. For instance, Huang et al found greater peripheral blood H19/IGF2 methylation was associate with elevated subcutaneous fat measures in 315 young adults [42]. In addition, Deodati et al observed that elevated IGF2 methylation levels from blood lymphocytes were associated with higher levels of triglycerides, triglyceride/HDL-cholesterol ratio and C-peptide concentrations among overweight and obese adolescents [43]. Future studies are needed to explore the sexually differentiated mechanisms behind DNA methylation and pubertal status.
In the post-hoc analysis examining the pubertal progression pattern, our findings that may provide statistical evidence to support the ‘catch-up growth’ and ‘compensatory growth’ theory in pubertal development. Based on previous studies [44], we had hypothesized that individuals with early pubertal onset would have faster pubertal progression, and those with later onset would have slower pubertal progression. However, we found older age of onset of puberty was associated with shorter duration (tempo) of puberty, and vice versa [45]. IGF2 site 3 was associated with later onset of genital development in both cross-sectional and prospective analyses (Table 3). Correspondingly, we observed 20% increased odds of faster genital development progression with IGF2 methylation, though the association was not statistically significant. Similarly, HSD11B2 site 4 was associated with earlier onset of genital development but slower tempo among boys. Much research on catch-up growth has been published since 1963 [46–48]. Most studies observed that among infants and children whose growth had been slowed by illness or starvation, there was a rapid and longer phase of growth until the children reached their pre-illness growth curve [46,47]. However, catch-up in pubertal characteristics during adolescence is less well-described. Based on our results, pubertal timing and tempo adjustments may exist among Mexican boys. Considering pre-adult periods of adaptive plasticity from juvenility to adolescence establishes longevity and the age of reproduction and fecundity [49], our results indicated modified DNA methylation levels may affect this timing and progression. Some significant associations seen at pubertal onset were attenuated in the progression model, which may be due in underpowered statistics part to less sensitivity of the ordinal regression model compared to the Cox proportional hazard model [50].
This analysis had some limitations, including a moderate sample size with approximately 20% missing rates of IGF2 DNA methylation at some of the CpG sites. Though we used multiple regression imputation to increase the number of predictors, this method can underestimate standard error, which might result in inflated p-values [51]. While pubertal status was based on a highly trained physicians’ observation, age of menarche was self-reported and may not be accurate due to recall bias. Our sample comes from a mixed ancestry population but we did not genotype this population in order to estimate ancestry. As such, it is possible that ancestry differences could influence the relationships we are observing between DNA methylation and pubertal timing. Since the epigenome and transcriptome vary by cell and tissue type, analyzing DNA methylation in blood leukocytes which consist of multiple cell types is a limitation, though recent studies have identified blood leukocyte differentially methylated genes associated with BMI or adiposity in adults that replicated in adipose or skeletal muscle, biologically relevant tissues [19,52,53]. The number of genes studied was also a limitation, and we recommend epigenome-wide studies in this area to identify key genes regulating puberty. In addition, we performed both unadjusted and adjusted model analyses of onset and progression and observed that including BMI did not significantly attenuate the associations. Nevertheless, we cannot rule out the possibility that DNA methylation is a mediator between BMI and sexual maturation, if BMI had been measured at an earlier time point.
In conclusion, this is the first study to evaluate the effect of DNA methylation of H19, IGF2, and HSD11B2 in combination with LINE-1 on pubertal onset and progression among boys and girls. Unlike previous puberty- related studies that primarily used menarche as the only pubertal indicator [54,55], we applied multiple secondary sexual characteristics (pubic hair, genital development and testicular volumes in boys, as well as pubic hair, breast development and menarche in girls). We found suggestive evidence of associations of DNA methylation with pubertal onset among adolescents. By using ordinal regression models and repeated measurements of Tanner stages, we observed the effect of DNA methylation on pubertal progression in boys only. The findings also raise the possibility of influencing pubertal timing by regulating DNA methylation levels. Future work in this field should consider epigenetic regulation in a larger panel of genes that may directly or indirectly influence puberty.
Methods
Study population
The study population comprised a subset of participants from the Early Life Exposure in Mexico to ENvironmental Toxicants (ELEMENT) project, a longitudinal epidemiological study consisting of three sequentially-enrolled birth cohorts. As originally designed, ELEMENT focused primarily on lead exposure and its impact on cognitive performance, and analysis of other metals, chemicals and epigenetics have been incorporated over time [56,57]. Participants were recruited at three maternity hospitals representing low- to moderate-income populations (Mexican Social Security Institute, Manuel Gea Gonzalez Hospital, and the National Institute of Perinatology) in Mexico City from 1994 to 2005. Mothers provided written consent upon enrollment in the study, and children also provided assent at peri-adolescent study visits. The research protocol was approved by the Human Subjects Committee of the National Institute of Public Health of Mexico, participant hospitals, and the Internal Review Board at all participating institutions including the University of Michigan. The subjects in this project were a subset of mother-child pairs from the second and third birth cohorts (n = 1079 pairs at baseline). At the clinic visit after the child was born, mothers provided household and demographic information, including age, education, and previous numbers of pregnancies. Their offspring were followed from birth until 4 years of age. Starting in 2011, we re-contacted a subset of the offspring (n = 250; henceforth referred to as the early-teen visit, see Figure 1) based on availability of prenatal and neonatal biospecimens [56]. One more peri-pubertal visit (late-teen visit) was completed approximately five years later (n = 549, with 223 having also participated in the early-teen visit). Fasting blood, pubertal status and anthropometry were collected at both teen visits [58] (Figure 1).
Figure 1.
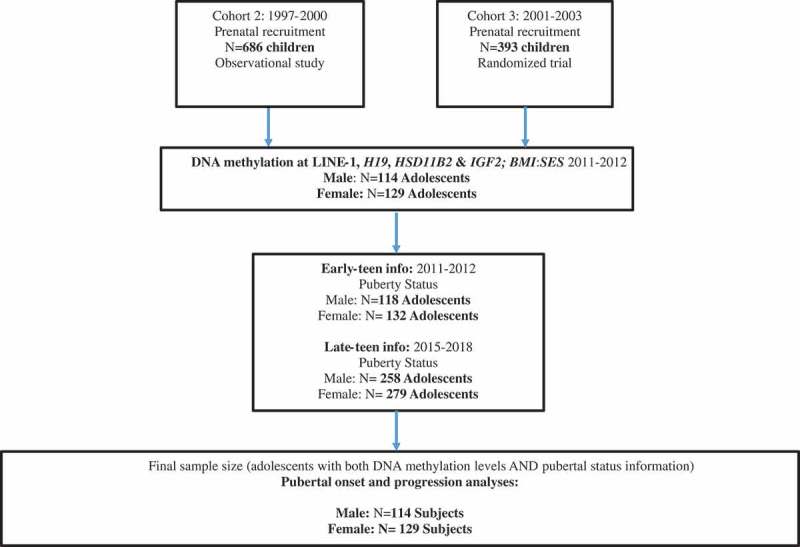
Selection of ELEMENT subjects for the study1.
1Offspring from enrollment cohorts 2 and 3 were re-contacted and re-enrolled based on availability of prenatal and neonatal biospecimens. We did not re-contact Cohort 1 participants, originally recruited in 1994–96, because the majority had advanced stages or had completed pubertal development.
Laboratory measurements and outcomes
DNA Methylation
Blood samples were obtained at the early teen visit and collected in PAXGene tubes by trained staff following standard protocols. High-molecular-weight DNA was extracted from blood leukocytes with the PAXgene Blood DNA kit (PreAnalytix, Switzerland). DNA samples were treated with sodium bisulfite using kits from Zymo or Qiagen [59]. Percent of methylated cells was then quantitatively analyzed in well-characterized differentially methylated regions (DMRs) of two imprinted genes (the DMR upstream of the H19 paternally imprinted, maternally expressed transcript (non-coding). H19, which is within the imprinting control region [60]; and the DMR within exon 3 of the IGF2AS transcript of the maternally imprinted, paternally expressed insulin-like growth factor, IGF2 [61], the promoter region of a non-imprinted gene (hydroxysteroid (11-beta) dehydrogenase 2, HSD11B2), and a conserved sequence found in the promoter region of LINE-1 repetitive elements of all subfamilies (sequence: 5ʹ- CTCGTGGTGCGCCGTTTCTTAAGCCG). DNA methylation was quantified via pyrosequencing at 4 (H19, LINE-1) or 5 (HSD11B2) CpG sites. The Sequenom EpiTYPER was used to quantify DNA methylation at 5 units of IGF2 representing a total of 7 CpG sites due to the resolution capabilities of EpiTYPER. Quality control includes running >10% of samples in duplicate and including human DNA controls of known methylation status (i.e. 0%, 100%) on each batch. Full details on primers, quality control, and analysis methods have been previously published [30]. Primer sequences and loci of CpG sites can be found in Supplemental Table 4. LINE-1, HSD11B2, and H19 data exhibited batch effects and as such were standardized to controls included on experimental plates as previously described [30]. For example, the value of 0% methylation controls on each plate of samples amplified and pyrosequenced together (a batch) for HSD11B2 was subtracted from the raw DNA methylation values generated for each sample in the same batch.
Pubertal Outcomes
Pubertal outcomes were obtained at both early-teen and late-teen visits. Tanner stages of breast and pubic hair growth in girls as well as Tanner stages of genitalia and pubic hair growth in boys were examined and collected by trained physicians [62]. Outcomes were recorded with a range from stage 1 indicating pre-puberty to stage 5 indicating full maturation [63]. Testicular volumes were measured by trained physicians using orchidometers (range from 1 to 25 ml). Occurrence and age of menarche were gathered from a self-reported questionnaire [62,64].
Covariates
Based on a priori knowledge and preliminary correlation tests between predictors and potential confounders, covariates included in the final model were SES and BMI of the child, obtained at the early-teen visit. The socioeconomic status (SES) information was collected, using a validated questionnaire consisting of thirteen questions on housing quality, services, material goods and education of the head of household by AMAI (Asociación Mexicana de Agencias de Investigación de Mercados y Opinión Pública, version 13 × 6). With the use of fourteen hierarchical trees this scale classified households into six SES categories (A/B, C+, C, D+, D, E; with A/B being the highest category) [8,65]. This scale was validated using the results of the National Survey of Household Income and Expenditure 2005, Mexico (ENIGH, Encuesta Nacional de Ingresos y Gastos de los Hogares 2004), classifying households into seven SES categories (A/B, C+, C, C-, D+, D, E; with A/B being the highest category) using a point based system [8,65].
[8,65]. Weight and height of the child were measured by trained nurses, following standardized protocols we have previously described [66]; BMI was calculated as weight over height squared (kg/m2) [66]. Children’s age was recorded at each visit.
Statistical methods
We examined the distribution of Tanner stages among individuals who attended only the early-teen visit and among those who attended both early- and late teen visits, and compared the distributions across categories of background characteristics using χ2 tests.
Some participants had missing CpG site values for HSD11B2 (n of missing = 14 at site 5), IGF2 (n of missing = 20 at site 1, 30 at site 2, 15 at site 3, 47 at site 4, 14 at site 5), SES (n of missing = 48). Thus, we performed multiple imputation [67] including all covariates; five imputed datasets were obtained. In order to test the assumption that the gene variables were missing at random, we examined the DNA methylation distribution (means ± SD) of CpG sites in all genes before and after imputation. The final sample size for the DNA methylation dataset was 114 boys and 129 girls. Values were not imputed for 7 subjects that were missing 3 out of 4 genes.
We used interval censored regression models to analyze time to pubertal onset at each visit separately. We created binomial Tanner stage outcomes using > 1 as the pubertal onset cut-off point [45,63] for pubic hair, genital development and breast development characteristics. Testicular volume ≤ 3 mL indicated pre-pubertal stage; testicular volume > 3mL but ≤ 11mL indicates pubertal onset; and > 11mL indicated sexual maturity [45,63]. Age from early-teen visit was used as the time to follow-up here.
We used the following ordinal regression model:
to analyze pubertal progression between early- and late-teen visits in boys and girls separately where methylation is the percent methylation at a given CpG site in one of the four genes, age is the age at the early teen visit, and time difference is the time between the early and late teen visits.
We selected confounders based on a priori knowledge and variables that were significantly associated with DNA methylation and pubertal stage. We included BMI and SES in adjusted interval censored regression models. BMI, SES and age from early-teen visit as well as the time difference between these two visits in the adjusted ordinal regression model. We used a cutoff value of p < 0.05 to define statistical significance. However, since we conducted a large number of tests, we also considered significance after adjusting for multiple testing. A Bonferroni correction for multiple testing would be overly conservative given that correlations among the 18 CpG sites can be large (e.g. several CpG sites within LINE-1 (Pearson r > 0.7) or within H19 (r > 0.85) are highly correlated within assay). Moreover, pubertal outcomes are also correlated. Thus, we corrected for multiple testing by using a cutoff point of 0.0028, obtained as 0.05/18, which considers that for each outcome we tested 18 CpG sites. All analyses were conducted using SAS software version 9.4 (SAS Institute Inc., Cary, NC, USA).
Funding Statement
This work was supported by U.S. Environmental Protection Agency (US EPA) grants RD834800 and RD83543601 and National Institute for Environmental Health Sciences (NIEHS) grants P20 ES018171, P01 ES02284401, R01 ES007821, and P30 ES017885. This study was also supported and partially funded by the National Institute of Public Health/Ministry of Health of Mexico. The contents of this publication are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA or the NIH. Further, the US EPA does not endorse the purchase of any commercial products or services mentioned in the publication.
Implications and contribution
Our results show that peripubertal blood DNA methylation at growth-related candidate loci IGF2, H19, HSD11B2, as well as LINE-1 repetitive elements may influence pubertal onset and progression. We believe the findings will help to understand and develop future scientific knowledge and potential intervention strategies accommodating the health needs of adolescents.
Acknowledgment
The authors acknowledge the research staff at participating hospitals and the American British Cowdray Hospital in Mexico City for providing research facilities. We thank the mothers and children for participating in the study
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- 1.Golub MS, Collman GW, Foster PMD, et al. Public health implications of altered puberty timing. Pediatrics. 2008;121(Supplement):S218–S230. [DOI] [PubMed] [Google Scholar]
- 2.Niculescu D. Puberty: ontogeny, neuroendocrinology, physiology, and disorders In: Williams textbook of endocrinology. Vol. 4 Elsevier; 2008. p. 127. [Google Scholar]
- 3.Herman-Giddens ME, Slora EJ, Wasserman RC, et al. Secondary sexual characteristics and menses in young girls seen in office practice: a study from the pediatric research in office settings network. Pediatrics. 1997;99(4):505–512. [DOI] [PubMed] [Google Scholar]
- 4.Herman-Giddens ME, Steffes J, Harris D, et al. Secondary sexual characteristics in boys: data from the pediatric research in office settings network. Pediatrics. 2012;130(5):e1058–e1068. [DOI] [PubMed] [Google Scholar]
- 5.Kaplowitz PB, Oberfield SE. reexamination of the age limit for defining when puberty is precocious in girls in the United Staes: implication for evaluation and treatment. Int J Confl Manage. 1999;10(2):130–153. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J, Chan Y-M. Adult consequences of self-limited delayed puberty. Pediatrics. 2017;139(6):e20163177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesario SK, Hughes LA. Precocious puberty: a comprehensive review of literature. J Obstet Gynecol Neonatal Nurs. 2007;36(3):263–274. [DOI] [PubMed] [Google Scholar]
- 8.Jansen EC, Miller AL, Lumeng JC, et al. Externalizing behavior is prospectively associated with intake of added sugar and sodium among low socioeconomic status preschoolers in a sex-specific manner. Int J Behav Nutr Phys Act. 2017;14(1). DOI: 10.1186/s12966-017-0591-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rozek LS, Dolinoy DC, Sartor MA, et al. Epigenetics: relevance and implications for public health. Annu Rev Public Health. 2014;35(1):105–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turan N, Ghalwash MF, Katari S, et al. DNA methylation differences at growth related genes correlate with birth weight: A molecular signature linked to developmental origins of adult disease? BMC Med Genomics. 2012;5 DOI: 10.1186/1755-8794-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banister CE, Koestler DC, Maccani MA, et al. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics. 2011;6(7):920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Relton CL, Groom A, St. Pourcain B, et al. DNA methylation patterns in cord blood DNA and body size in childhood. PLoS One. 2012;7(3). DOI: 10.1371/journal.pone.0031821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8(9):1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PA. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. [DOI] [PubMed] [Google Scholar]
- 15.Simpkin AJ, Howe LD, Tilling K, et al. The epigenetic clock and physical development during childhood and adolescence: longitudinal analysis from a UK birth cohort. Int J Epidemiol. 2017. DOI: 10.1093/ije/dyw307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perng W, Mora-Plazas M, Marín C, et al. A prospective study of LINE-1DNA methylation and development of adiposity in school-age children. PLoS One. 2013;8(4). DOI: 10.1371/journal.pone.0062587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lomniczi A, Loche A, Castellano JM, et al. Epigenetic control of female puberty. Nat Neurosci. 2013;16(3):281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huen K, Harley K, Kogut K, et al. DNA methylation of LINE-1 and Alu repetitive elements in relation to sex hormones and pubertal timing in Mexican-American children. Pediatr Res. 2016;79(6):855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: A genome-wide analysis. Lancet. 2014;383(9933):1990–1998. [DOI] [PubMed] [Google Scholar]
- 20.Milagro FI, Gómez-Abellán P, Campión J, et al. PER2 and BMAL1 DNA methylation: association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chronobiol Int. 2012;29(9):1180–1194. [DOI] [PubMed] [Google Scholar]
- 21.Soubry A, Schildkraut JM, Murtha A, et al. Paternal obesity is associated with IGF2 hypomethylation in newborns: results from a Newborn Epigenetics Study (NEST) cohort. BMC Med. 2013;11(1). DOI: 10.1186/1741-7015-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman DS, Khan LK, Serdula MK, et al. Relation of age at menarche to race, time period, and anthropometric dimensions: the bogalusa heart study. Pediatrics. 2002;110(4):e43. [DOI] [PubMed] [Google Scholar]
- 23.Kaplowitz PB. Link between body fat and the timing of puberty. Pediatrics. 2008;121(Supplement 3):S208–S217. [DOI] [PubMed] [Google Scholar]
- 24.Shirtcliff EA, Dahl RE, Pollak SD. Pubertal development: correspondence between hormonal and physical development. Child Dev. 2009;80(2):327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulrich CM, Toriola AT, Koepl LM, et al. Metabolic, hormonal and immunological associations with global DNA methylation among postmenopausal women. Epigenetics. 2012;7(9):1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi S, Inaguma S, Sakakibara M, et al. DNA methylation in the androgen receptor gene promoter region in rat prostate cancers. Prostate. 2002;52:82–88. [DOI] [PubMed] [Google Scholar]
- 27.Kumar RC, Thakur MK. Androgen receptor mRNA is inversely regulated by testosterone and estradiol in adult mouse brain. Neurobiol Aging. 2004;25(7):925–933. [DOI] [PubMed] [Google Scholar]
- 28.Ahima RS, Dushay J, Flier SN, et al. Leptin accelerates the onset of puberty in normal female mice. J Clin Invest. 1997;99(3):391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chehab FF, Lim ME, Lu R. Correction of the sterility defect in homozygous obese female mice by treatment with the human recombinant leptin. Nat Genet. 1996;12(3):318–320. [DOI] [PubMed] [Google Scholar]
- 30.Goodrich JM, Dolinoy DC, Sánchez BN, et al. Adolescent epigenetic profiles and environmental exposures from early life through peri-adolescence. Environ Epigenetics. 2016;2(3). DOI: 10.1093/eep/dvw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodrich JM, Sánchez BN, Dolinoy DC, et al. Quality control and statistical modeling for environmental epigenetics: A study on in Utero lead exposure and DNA methylation at birth. Epigenetics. 2015. DOI: 10.4161/15592294.2014.989077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talens RP, Christensen K, Putter H, et al. Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell. 2012. DOI: 10.1111/j.1474-9726.2012.00835.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Xie C, Murphy SK, et al. Lead exposure during early human development and DNA methylation of imprinted gene regulatory elements in adulthood. Environ Health Perspect. 2016. DOI: 10.1289/ehp.1408577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King K, Murphy S, Hoyo C. Epigenetic regulation of newborns’ imprinted genes related to gestational growth: patterning by parental race/ethnicity and maternal socioeconomic status. J Epidemiol Community Health. 2015. DOI: 10.1136/jech-2014-204781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perkins E, Murphy SK, Murtha AP, et al. Insulin-like growth factor 2/H19 methylation at birth and risk of overweight and obesity in children. J Pediatr. 2012. DOI: 10.1016/j.jpeds.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seminara SB, Messager S, Chatzidaki EE, et al. The GPR54 Gene as a Regulator of Puberty. Obstet Gynecol Surv. 2004;59(5):351–353. [Google Scholar]
- 37.Abreu AP, Dauber A, Macedo DB, et al. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013;368:2467–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silveira LFG, Trarbach EB, Latronico AC. Genetics basis for GnRH-dependent pubertal disorders in humans. Mol Cell Endocrinol. 2010;324(1–2):30–38. [DOI] [PubMed] [Google Scholar]
- 39.Karapanou O, Papadimitriou A. Determinants of menarche. Reprod Biol Endocrinol. 2010;8 DOI: 10.1186/1477-7827-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marván ML, Catillo-López RL, Alcalá-Herrera V, et al. The decreasing age at menarche in Mexico. J Pediatr Adolesc Gynecol. 2016;29(5):454–457. [DOI] [PubMed] [Google Scholar]
- 41.Chumlea WC, Schubert CM, Roche AF, et al. Age at menarche and racial comparisons in US girls. Pediatrics. 2003;111(1):110–113. [DOI] [PubMed] [Google Scholar]
- 42.Gallou-Kabani C, Gabory A, Tost J, et al. Sex- and diet-specific changes of imprinted gene expression and dna methylation in mouse placenta under a high-fat diet. PLoS One. 2010;5(12). DOI: 10.1371/journal.pone.0014398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deodati A, Inzaghi E, Liguori A, et al. IGF2 methylation is associated with lipid profile in obese children. Horm Res Paediatr. 2013;79(6):361–367. [DOI] [PubMed] [Google Scholar]
- 44.Apter D, Vihko R. PREMENARCHEAL ENDOCRINE CHANGES IN RELATION TO AGE AT MENARCHE. Clin Endocrinol (Oxf). 1985;22(6):753–760. [DOI] [PubMed] [Google Scholar]
- 45.Marceau K, Ram N, Houts RM, et al. Individual differences in boys’ and girls’ timing and tempo of puberty: modeling development with nonlinear growth models. Dev Psychol. 2011;47(5):1389–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashworth A, Millward DJ. Catch‐up growth in children. Nutr Rev. 1986;44(5):157–163. [DOI] [PubMed] [Google Scholar]
- 47.Wi J-M, Boersma B. Catch-up growth: definition, mechanisms, and models. J Pediatr Endocrinol Metab. 2002;15(Suppl 5):1229–1241. [PubMed] [Google Scholar]
- 48.Wilmott RW. Patterns of catch-up growth. J Pediatr. 2013;162(2):220. [DOI] [PubMed] [Google Scholar]
- 49.Hochberg Z, Feil R, Constancia M, et al. Child health, developmental plasticity, and epigenetic programming. Endocr Rev. 2011;32(2):159–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon J. The strengths and limitations of the statistical modeling of complex social phenomenon: focusing on SEM, path analysis, or multiple regression models. Int J Soc Behav Educ Econ Bus Ind Eng. 2015;9(5):1604–1612. [Google Scholar]
- 51.Soley-Bori M. Dealing with missing data: key assumptions and methods for applied analysis. PM931 Dir Study Heal Policy Manag. 2013;4:20. [Google Scholar]
- 52.Day SE, Coletta RL, Kim JY, et al. Potential epigenetic biomarkers of obesity-related insulin resistance in human whole-blood. Epigenetics. 2017;12(4):254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demerath EW, Guan W, Grove ML, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24(15):4464–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demetriou CA, Chen J, Polidoro S, et al. Methylome analysis and epigenetic changes associated with menarcheal age. PLoS One. 2013;8(11). DOI: 10.1371/journal.pone.0079391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He C, Kraft P, Chen C, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41(6):724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cantoral A, Téllez-Rojo MM, Levy TS, et al. Differential association of lead on length by zinc status in two-year old Mexican children. Environ Heal A Glob Access Sci Source. 2015;14(1). DOI: 10.1186/s12940-015-0086-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watkins DJ, Sánchez BN, Téllez-Rojo MM, et al. Impact of phthalate and BPA exposure during in utero windows of susceptibility on reproductive hormones and sexual maturation in peripubertal males. Environ Heal A Glob Access Sci Source. 2017;16(1). DOI: 10.1186/s12940-017-0278-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perng W, Hector EC, Song PXK, et al. Metabolomic determinants of metabolic risk in mexican adolescents. Obesity. 2017;25(9):1594–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Tollefsbol TO. DNA methylation detection: bisulfite genomic sequencing analysis. Methods Mol Biol. 2011;791:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoyo C, Murtha AP, Schildkraut JM, et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6(7):928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heijmans BT, Kremer D, Tobi EW, et al. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet. 2007;16(5):547–554. [DOI] [PubMed] [Google Scholar]
- 62.Chavarro JE, Watkins DJ, Afeiche MC, et al. Validity of self-assessed sexual maturation against physician assessments and hormone levels. J Pediatr. 2017;186(172–178.e3). DOI: 10.1016/j.jpeds.2017.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marshall WA, Tanner JM. Variations in the pattern of pubertal changes in boys. Arch Dis Child. 1970;45(239):13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cooper R, Blell M, Hardy R, et al. Validity of age at menarche self-reported in adulthood. J Epidemiol Community Health. 2006;60(11):993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantoral A, Téllez-Rojo MM, Ettinger AS, et al. Early introduction and cumulative consumption of sugar-sweetened beverages during the pre-school period and risk of obesity at 8-14 years of age. Pediatr Obes. 2016. DOI: 10.1111/ijpo.12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nuttall FQ. Body mass index: obesity, BMI, and health: A critical review. Nutr Today. 2015;50(3):117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Buuren S. Multiple imputation of discrete and continuous data by fully conditional specification. Stat Methods Med Res. 2007. doi: 10.1177/0962280206074463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


