Abstract
SHORT ABSTRACT:
Patient-derived xenograft (PDX) models more robustly recapitulate melanoma molecular and biological features, and are more predictive of therapy response compared to traditional plastic tissue culture-based assays. Here we will describe our standard operating protocol for the establishment of new PDX models, and the characterization/experimentation of existing PDX models.
LONG ABSTRACT:
Accumulating evidence suggests that molecular and biological properties differ in melanoma cells grown in traditional two-dimensional tissue culture vessels versus in vivo in human patients. This is due to bottleneck selection of clonal populations of melanoma cells that can robustly grow in vitro in the absence of physiological conditions. Further, responses to therapy in two-dimensional tissue culture overall do not faithfully reflect responses to therapy in melanoma patients, with the majority of clinical trials failing to show efficacy of therapeutic combinations shown to be effective in vitro. Although xenografting of melanoma cells into mice provides the physiological in vivo context absent from two-dimensional tissue culture assays, the melanoma cells used for engraftment have already undergone bottleneck selection for cells that could grow under two-dimensional conditions when the cell line was established. The irreversible alterations that occur as a consequence of the bottleneck include changes in growth and invasion properties, as well as loss of specific subpopulations. Therefore, models that better recapitulate the human condition in vivo may better predict therapeutic strategies that effectively increase the overall survival of patients with metastatic melanoma. The patient-derived xenograft (PDX) technique involves the direct implantation of tumor cells from the human patient to a mouse recipient. In this manner, tumor cells are consistently grown under physiological stresses in vivo and never undergo the two-dimensional bottleneck, which preserves the molecular and biological properties present when the tumor was in the human patient. Notably, PDX models derived from organ sites of metastases (i.e., brain), therapy naïve patients, and patients with acquired resistance to therapy display similar metastatic capacity and sensitivity to therapy, respectively.
Keywords: melanoma, patient-derived xenograft, therapy, resistance, in vivo models, metastasis
INTRODUCTION:
Preclinical models are critical for all aspects of translational cancer research, including disease characterization, discovery of actionable vulnerabilities unique to cancer versus normal cells, and the development of efficacious therapies that exploit these vulnerabilities to increase the overall survival of patients. In the melanoma field, tens of thousands of cell line models have been heavily utilized for drug screening, with > four thousand contributed by our group alone (WMXXX series). These cell line models were derived from melanoma patients with various forms of cutaneous melanoma (i.e., acral, uveal, superficial spreading) and diverse genotypes (i.e., BRAFV600-mutant, NRAS-mutant), which span the spectrum of disease present in the clinic (1, 2).
Unequivocally, the most successful targeted therapy strategy in the melanoma field has emerged from 1) genomic characterization of patients’ tumors identifying rapidly accelerated fibrosarcoma B (BRAF) mutations in ~50% of melanomas (3), and 2) preclinical investigation leveraging melanoma cell line models (4). The BRAF inhibitor and mitogen-activated protein/extracellular signal-regulated kinase kinase (MEK) inhibitor combination was Food and Drug Administration (FDA)-approved in 2014 for the treatment of patients whose melanomas harbor activating BRAFV600E/K mutations and boasts a >75% response rate (5). Despite this initial efficacy, resistance rapidly arises in nearly every case due to multifarious intrinsic and acquired resistance mechanisms and intra-tumoral heterogeneity. Unfortunately, cell line models do not recapitulate representative biological heterogeneity when grown in two-dimensional culture in plastic vessels, which masks their clinically predictive potential when investigators attempt to experimentally determine therapies that might be effective in patients with a specific form or genotype of melanoma (6). Understanding how to best model patient intra-tumoral heterogeneity will allow investigators to better develop therapeutic modalities that can kill therapy resistant subpopulations that drive failure to current standard of care therapies.
Paramount to the limited predictive value of cell line models rests on how cell line models are initially established. Irreversible alterations occur in the tumor clonal landscape when a single-cell suspension of a patients’ tumor is grown on two-dimensional, plastic tissue culture vessels, including changes in proliferative and invasive potential, elimination of specific subpopulations, and alteration of genetic information (7). Xenografts into mice of these melanoma cell line models represent the most frequently used in vivo platform for preclinical studies, however this strategy also suffers from the poor recapitulation of complex tumor heterogeneity observed clinically. To overcome this shortcoming, there has been a growing interest in incorporating more sophisticated preclinical models of melanoma, including the patient-derived xenograft (PDX) model. PDX models have been utilized for > 30 years with seminal studies in lung cancer patients demonstrating concordance between the patients’ response to cytotoxic agents and the response of the PDX model derived from the same patient (8). Recently, there has been a drive to utilize PDX models as the tool of choice for preclinical investigations both in industry and in academic centers. Briefly, our protocol to generate PDX models requires the subcutaneous implantation of fresh tissue from primary or metastatic melanomas (collected by biopsy or surgery) into NOD/SCID/IL2-receptor null (NSG) mice. A variety of variations in methodologic approach are used by different groups, however a fundamental core exists (9).
PROTOCOL:
The Generation of Patient-Derived Xenograft (PDX) Melanoma Models:
The following protocols follow the guidelines of The Wistar Institute’s humane ethic’s committee and animal care guidelines.
-
Collect tumors tissue (termed passage 0) from melanoma patients by one of the following surgery or biopsy methods:
Surgical excision: maintain, in transport storage media (RPMI 1640 + 0.1% fungizone+ 0.2% gentamicin) at 4°C or ice, a minimum of 1g of tissue.
Surgical biopsy: maintain in transport storage media at 4°C or ice, less than 1g of tissue (often punch biopsies of subcutaneous (s.c.) metastases and lymph node (LN) metastases).
Core biopsy: wash out, into a 15 ml conical tube containing 5 ml transport storage media at 4°C or ice, approximately 10×1 mm3 core cylinder tissue (often liver biopsies.
Fine needle aspirates (FNAs): keep in a needle and syringe, at 4°C or ice, a small amount of tissue (size of the head of a needle) taken directly from the patient.
-
Tumor tissue processing for mouse implantation:
-
Tumor tissue dissociation for surgical excision or surgical biopsy samples.
Transfer tumor tissue into sterile petri dish, separate tumor tissue from surrounding normal tissue as much as possible.
Remove necrotic tissue (usually identified as pale-whitish tissue located centrally within tumor) from the remaining tumor as much as possible.
-
Either implant tumor chunks, whereby approximately 5 equal pieces (~2 × 2 mm3) can be subdivided with a scalpel from one initial tumor (Figure 2). Or make a tumor slurry by mincing tumor tissue using a cross blade technique with two scalpel blades. Mince tumors as finely as possible to form slurry now ready for surgical mouse implantation.
**Optional: If tumor tissue is too hard for mechanical dissociation, a digestion dissociation procedure can be used to get gel-like slurry and a single cell suspension for implantation and/or injection.
Put slurry in 50 mL Falcon tube with cold HBSS−/− (w/o Ca++, Mg++), centrifuge and pellet at 220 x g for 4 minutes at 4°.
Resuspend slurry in 10 mL of warmed fresh digest media (200 U/ml Collagenase IV + 5 mM CaCl2 + 50 U/mL DNase in HBSS−/−) per 1 g of tumor tissue.
Place tube in 37° C water bath for 20 minutes, mix vigorously every 5 minutes with a disposable pipette.
Wash with up to 50 mL of HBSS−/−, centrifuge at 220 x g for 4 minutes at 4°C
Add 5 mL of prewarmed TEG (0.025% Trypsin + 40 ug/ml EGTA + 10 ug/ml PVA) per 1 g of tumor tissue, gently resuspend/shake, place tube at 37°C for 2 minutes without mixing.
Add at least 1 equal volume of cold staining media (1% BSA + 10 mM Hepes + 1x Penstrep in L15 media) to quench trypsin, centrifuge at 220 x g for 4 minutes at 4°C.
Resuspend in 10 mL of staining media per 1 g of tumor tissue and filter through a 40 μm cell strainer to get single cell suspension for mouse injection (Figure 2).
Slurry remaining on top of the cell strainer can also be collected for surgical mouse implantation.
-
Tumor tissue dissociation for core biopsy samples.
Pour the core cylinder tissue-transport tube into a 5 cm petri dish.
Remove excess liquid, scrape tissue to the edge of the petri dish, finely mince the tumor tissue, add ~100–150 ul of HBSS−/− on top of the tumor tissue, and quickly draw the HBSS/tumor tissue suspension into the 1 mL syringe.
Attach a 23 gauge (G) needle to the 1 mL syringe, pass the HBSS/tumor tissue suspension through the needle into an 1.5 mL spin tube, redraw the tumor suspension into the syringe with the needle still on, and repeat using the shearing forces of the needle to further process the tumor sample until it can pass smoothly through the needle.
When sample goes smoothly through the needle, finally draw it back into the syringe and detach the 23 G needle.
Attach a 27 G needle and draw an equal volume (~100–150 ul) of Matrigel into the syringe.
Administer the equal volume of Matrigel slowly into the 1.5 mL spin tube with the tumor/HBSS suspension, carefully avoiding the formation of bubbles.
Draw one last time back into the syringe and the core biopsy tumor tissue is now ready for mouse injection.
-
Tumor tissue dissociation for FNA samples.
Place the syringe containing the FNA samples (trapped in the needle) on ice.
Separate the needle (containing FNA tissue) from the syringe and remove the plunger from the syringe, add ~150–200 ul of HBSS−/− into the top of the syringe.
Re-insert the plunger into the syringe and the needle (containing FNA tissue), and push out the HBSS−/− through the needle into an 1.5 mL spin tube. This step removes the FNA tumor from the needle.
Repeat step 3 with the HBSS/FNA suspension twice using the same syringe to maximize the retrieval of tumor tissue from the needle.
Add an equal volume (~100–150 ul) of Matrigel to the 1.5 mL spin tube containing the HBSS/FNA suspension, adequately mix by slowly drawing up the Matrigel/HBSS/FNA suspension back into the 23 G needle and repeating twice.
Draw the entire Matrigel/HBSS/FNA volume back into the syringe, replace the 23 G needle with a 27 G needle, and the FNA sample is now ready for mouse injection.
-
-
Tumor implantation and injection in mice:
-
Implantation of surgical excision or surgical biopsy samples.
Shave hair from the lower back of NOD/SCID/IL2-receptor null (NSG) mice leaving an approximately 1.5 × 3 cm2 area with no hair, anesthetize mice by using isoflurane.
Place individual mice in the nose cone of anesthesia machine, douse shaved area with 70% ethanol and allow to evaporate.
Divide tumor slurry in petri dish into individual mounds for surgical implantation (i.e., into three equal mounds if to be implanted into 3 mice).
Using the scalpel blade, make an incision of approximately 1 cm long on the center of the back of the mouse, take one pair of forceps and lift up skin on the side of the incision opposite of operator.
Take the scissors into the other hand and separate the skin from the muscle layer by gently cutting the fascial membrane with small scissor cuts, thereby creating a “pocket” for the tumor tissue
Pick up one individual mound of tumor tissue with the scalpel blade and gently place tissue into the created pocket.
Administer 100 ul of Matrigel on the tumor tissue mound in the pocket.
Using two pairs of forceps, pull up the incision on both ends so that the wound edges come close together, close the wound by applying one or two wound clips, take the mouse out of the nose cone and place it back into its original cage, observe mouse while waking up.
Remove wound clips after approximately 7 days. If healing isn’t complete after 7 days, leave wound clip in for an additional one or two days.
-
Injection of FNA or core biopsy samples.
Hold an NSG mouse firmly by the tail and allow it to hold on to a grid with its front legs.
Insert the needle slowly into the skin of the mouse, slightly pulling up to avoid injecting into the muscle layer underneath the skin.
Slowly and steadily inject the contents of the syringe under the skin of the mouse. Before removal of the needle from the skin, pinch the skin with two fingers to avoid forming a channel in which the sample could leak out
Pull out needle and place mouse back into its cage.
-
-
Monitor tumor growth:
Monitor mice once weekly to check for palpable tumors.
Once tumors are at a measurable size (approximately 50 mm3), use a caliper to record tumor dimensions. We use the formula (width2 x length) / 2 to calculate tumor volumes.
Harvest tumor once the tumor volume reaches around 1.5 cm3 (takes approximately 4–10 weeks). The tumor is now called mouse passage 1 (MP1).
-
Harvest tumor from mice for banking tissue, reimplantation and experiment/characterization:
Place the euthanized mouse in Virkon for 30 seconds in the bio-cabinet.
Use curved scissors and surgical forceps to lift skin adjacent to the tumor and make a horizontal cut. Using blunt separation technique, mobilize the skin on both sides of the tumor and over the tumor, exposing the tumor.
Using scissors or a scalpel blade, separate the tumor from the fascia.
-
Resect the tumor and transfer the tumor to a sterile Petri dish, cut the tumor into small pieces and remove necrotic tissue from the tumor.
Banking tumor tissue for future implantation: take small tumor pieces and mince them, transfer minced tissue to a 2 mL cryogenic vial, add 1 mL of freezing media (10% DMSO + 90% FBS), mix well and place cryogenic vials into a pre-cooled container of dry ice. Store container in −80° C freezer overnight and finally transfer cryogenic vials to LN2.
Snap frozen tissue for downstream assays (i.e., RNASeq, WES etc.): place tumor tissue pieces (~3×3 mm3) into cryogenic vial and put cryogenic vial in LN2 immediately. Store in −80° C freezer.
-
PDX therapy trials:
-
First need to grow sufficient tumor tissue to implant the necessary number of mice for the therapy trial in two expansion phases
First implant 5 NSG mice with banked tissue from 1 cryogenic vial.
Once tumors reach ~600–800mm3, harvest 1–2 tumors and process tumor tissue per the above protocol for Tumor tissue dissociation for surgical excision or surgical biopsy samples.
Use mechanical dissociation or enzymatic dissociation with collagenase iv for 30 minutes on a shaker at room temperature.
After 3 washes with HBSS−/−, pass tumor suspension through a 40 uM cell strainer to get single cell suspension.
Pellet cells and resuspend in 6 ml HBSS/Matrigel (1 to 1 ratio), inject 50–60 NSG mice, with 100 uL cell mixture per mouse.
Measure tumor size biweekly.
When tumors reach ~100mm3, start treating mice.
When tumor size reaches your maximum IACUC-approved volume, stop treatment and harvest tumor.
-
Regarding animal treatment, animals are euthanized by CO2 chamber and confirmed by checking for vital signs. Anytime mice are under anesthesia, we do not leave them unattended and we use vet ointment on their eyes to prevent dryness. We gently squeeze the foot to test for responsiveness. All mice that have undergone surgical implantation of PDX are not returned to a cage until fully recovered.
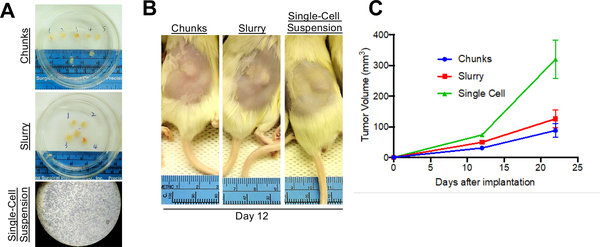
Figure 2:
Alternative Implantation Methods. (A) Tumors can be processed into either chunks, a slurry suspension, or as a single-cell suspension. (B) All three methods will allow for growth of tumors in vivo. Shown here are mice subcutaneously implanted with tumor and imaged 12 days after implantation. (C) Shown are tumor growth curves for the mice injected with one of the three implantation methods. N=5 per arm.
REPRESENTATIVE RESULTS:
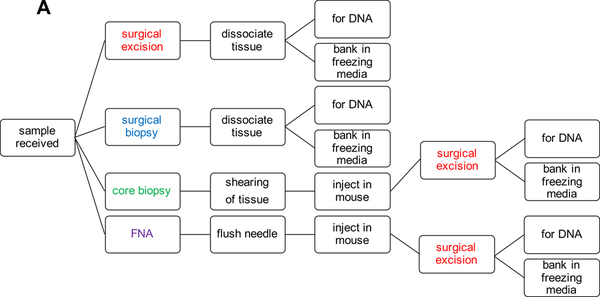
Tumor tissue for melanoma PDX models can come from a variety of different sources and can also be processed per the growth dynamics of individual models and the desired use of the PDX tissue. The priority when establishing a PDX model is to have sufficient material to bank for future use and DNA for characterization (Fig. 1A).
Figure 1:
PDX Model Generation Work Flow.
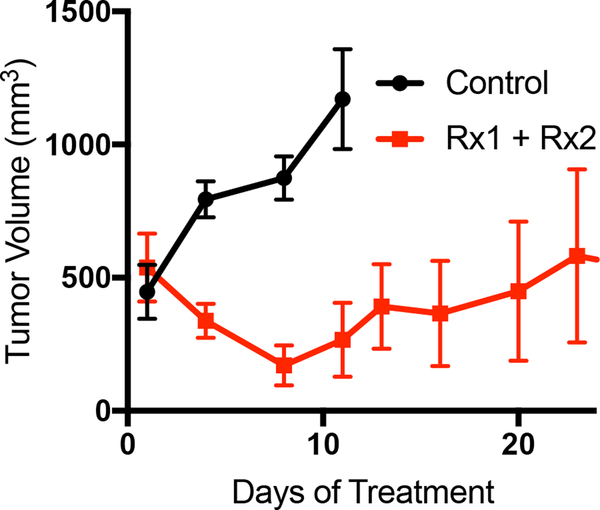
Once sufficient material is banked, tumor tissue can be expanded in one of three main methods to grow enough tumor to perform a formal therapy study (Fig. 2A). Each of the methods described herein will allow for the expansion of tumor from PDXs (Fig. 2B). It is our experience that creating a single-cell suspension of tumor cells with the use of enzymatic digestion (collagenase iv) can allow for more rapid tumor growth, and can allow one initial tumor to be expanded into 10 – 20 mice, whereas the tumor chunk and slurry method can only be expanded into 5 – 10 mice (Fig. 2C). As has been previously demonstrated in other tumor types, melanoma PDX models often reflect the drug sensitivity the patient displayed when on therapy. Shown here is a representative therapy curve from a melanoma patient with BRAFV600E mutant melanoma who initially responded a BRAF inhibitor but ultimately relapsed. The PDX derived from this patient also displayed initial sensitivity to BRAF inhibition (Rx1) plus an additional inhibitor (Rx2), however the tumors ultimately relapsed (Fig. 3A).
Figure 3:
Representative data for a PDX therapy trial. (A) Mice were implanted with PDX tumors and treated with either vehicle control or a two-inhibitor combination. N=6
DISCUSSION:
We have herein described generating PDX models of melanoma with patient tissue derived from primary and metastatic tumors, core biopsies, and FNAs. When directly engrafted into NSG mice, tumors present similar morphologic, genomic, and biologic properties to those observed in the patient. In the case when only a small quantity of tissue is available to investigators, as often occurs with FNAs, the PDX technique allows for the expansion of the tumor tissue for DNA, RNA, and protein characterization, as well as for therapy trials to allow preclinical drug development.
Importantly, response to therapy of PDX melanoma models recapitulates the sensitivity of the donor patient, allowing for robust preclinical investigations to develop improved therapeutic strategies to combat therapy resistance and improve the durability of response. Most metastatic melanoma patients do not experience cures with existing standard of care (SOC) therapies (5). Our PDX melanoma collection contains more than 500 distinct models, including those derived from patients who relapsed on targeted therapy and immunotherapy (1, 2). This resource will be critical for the development of therapeutic modalities that overcome resistance to current SOC. High throughput approaches leveraging PDX models in drug screens have been developed and allow the strength of the PDX approach to be utilized as efficiently and cost-effectively as possible (10).
In summary, PDX models allow for preclinical investigations of melanoma cells that better recapitulate the tumor heterogeneity and melanoma aggressiveness observed in the clinic (versus other two-dimensional and standard xenograft approaches). PDX models allow for a deeper understanding of which genes are involved in therapy resistance and provide a model from which more effective therapies can be developed to increase the overall survival of patients with metastatic melanoma.
ACKNOWLEDGMENTS:
We thank the Wistar Institute Animal Facility, Microscopy Facility, Histotechnology Facility and Research Supply Center. This study was funded in part by grants from the U54 (CA224070–01), SPORE (CA174523), P01 (CA114046–07), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the Melanoma Research Foundation.
Footnotes
DISCLOSURES:
None
REFERENCES:
- 1.Garman B, Anastopoulos IN, Krepler C, Brafford P, Sproesser K, Jiang Y, Wubbenhorst B, Amaravadi R, Bennett J, Beqiri M, Elder D, Flaherty KT, Frederick DT, Gangadhar TC, Guarino M, Hoon D, Karakousis G, Liu Q, Mitra N, Petrelli NJ, Schuchter L, Shannan B, Shields CL, Wargo J, Wenz B, Wilson MA, Xiao M, Xu W, Xu X, Yin X, Zhang NR, Davies MA, Herlyn M, Nathanson KL. Genetic and Genomic Characterization of 462 Melanoma Patient-Derived Xenografts, Tumor Biopsies, and Cell Lines. Cell reports. 2017;21(7):1936–52. Epub 2017/11/16. doi: 10.1016/j.celrep.2017.10.052. PubMed PMID: 29141224; PMCID: PMC5709812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krepler C, Sproesser K, Brafford P, Beqiri M, Garman B, Xiao M, Shannan B, Watters A, Perego M, Zhang G, Vultur A, Yin X, Liu Q, Anastopoulos IN, Wubbenhorst B, Wilson MA, Xu W, Karakousis G, Feldman M, Xu X, Amaravadi R, Gangadhar TC, Elder DE, Haydu LE, Wargo JA, Davies MA, Lu Y, Mills GB, Frederick DT, Barzily-Rokni M, Flaherty KT, Hoon DS, Guarino M, Bennett JJ, Ryan RW, Petrelli NJ, Shields CL, Terai M, Sato T, Aplin AE, Roesch A, Darr D, Angus S, Kumar R, Halilovic E, Caponigro G, Jeay S, Wuerthner J, Walter A, Ocker M, Boxer MB, Schuchter L, Nathanson KL, Herlyn M. A Comprehensive Patient-Derived Xenograft Collection Representing the Heterogeneity of Melanoma. Cell reports. 2017;21(7):1953–67. Epub 2017/11/16. doi: 10.1016/j.celrep.2017.10.021. PubMed PMID: 29141225; PMCID: PMC5726788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. Epub 2002/06/18. doi: 10.1038/nature00766. PubMed PMID: 12068308. [DOI] [PubMed] [Google Scholar]
- 4.Paraiso KH, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, Messina JL, Flaherty KT, Smalley KS. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. British journal of cancer. 2010;102(12):1724–30. Epub 2010/06/10. doi: 10.1038/sj.bjc.6605714. PubMed PMID: 20531415; PMCID: PMC2883709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long GV, Eroglu Z, Infante J, Patel S, Daud A, Johnson DB, Gonzalez R, Kefford R, Hamid O, Schuchter L, Cebon J, Sharfman W, McWilliams R, Sznol M, Redhu S, Gasal E, Mookerjee B, Weber J, Flaherty KT. Long-Term Outcomes in Patients With BRAF V600-Mutant Metastatic Melanoma Who Received Dabrafenib Combined With Trametinib. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2018;36(7):667–73. Epub 2017/10/11. doi: 10.1200/jco.2017.74.1025. PubMed PMID: 28991513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM, Roman-Roman S, Seoane J, Trusolino L, Villanueva A. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer discovery. 2014;4(9):998–1013. Epub 2014/09/04. doi: 10.1158/2159-8290.Cd-14-0001. PubMed PMID: 25185190; PMCID: PMC4167608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hausser HJ, Brenner RE. Phenotypic instability of Saos-2 cells in long-term culture. Biochemical and biophysical research communications. 2005;333(1):216–22. Epub 2005/06/09. doi: 10.1016/j.bbrc.2005.05.097. PubMed PMID: 15939397. [DOI] [PubMed] [Google Scholar]
- 8.Fiebig HH, Neumann HA, Henss H, Koch H, Kaiser D, Arnold H. Development of three human small cell lung cancer models in nude mice. Recent results in cancer research Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 1985;97:77–86. Epub 1985/01/01. PubMed PMID: 2986247. [DOI] [PubMed] [Google Scholar]
- 9.Meehan TF, Conte N, Goldstein T, Inghirami G, Murakami MA, Brabetz S, Gu Z, Wiser JA, Dunn P, Begley DA, Krupke DM, Bertotti A, Bruna A, Brush MH, Byrne AT, Caldas C, Christie AL, Clark DA, Dowst H, Dry JR, Doroshow JH, Duchamp O, Evrard YA, Ferretti S, Frese KK, Goodwin NC, Greenawalt D, Haendel MA, Hermans E, Houghton PJ, Jonkers J, Kemper K, Khor TO, Lewis MT, Lloyd KCK, Mason J, Medico E, Neuhauser SB, Olson JM, Peeper DS, Rueda OM, Seong JK, Trusolino L, Vinolo E, Wechsler-Reya RJ, Weinstock DM, Welm A, Weroha SJ, Amant F, Pfister SM, Kool M, Parkinson H, Butte AJ, Bult CJ. PDX-MI: Minimal Information for Patient-Derived Tumor Xenograft Models. Cancer research. 2017;77(21):e62–e6. Epub 2017/11/03. doi: 10.1158/0008-5472.Can-17-0582. PubMed PMID: 29092942; PMCID: PMC5738926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, Balbin OA, Barbe S, Cai H, Casey F, Chatterjee S, Chiang DY, Chuai S, Cogan SM, Collins SD, Dammassa E, Ebel N, Embry M, Green J, Kauffmann A, Kowal C, Leary RJ, Lehar J, Liang Y, Loo A, Lorenzana E, Robert McDonald E 3rd, McLaughlin ME, Merkin J, Meyer R, Naylor TL, Patawaran M, Reddy A, Roelli C, Ruddy DA, Salangsang F, Santacroce F, Singh AP, Tang Y, Tinetto W, Tobler S, Velazquez R, Venkatesan K, Von Arx F, Wang HQ, Wang Z, Wiesmann M, Wyss D, Xu F, Bitter H, Atadja P, Lees E, Hofmann F, Li E, Keen N, Cozens R, Jensen MR, Pryer NK, Williams JA, Sellers WR. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nature medicine. 2015;21(11):1318–25. Epub 2015/10/20. doi: 10.1038/nm.3954. PubMed PMID: 26479923. [DOI] [PubMed] [Google Scholar]