Abstract
We describe 2 human cases of infection with a new Neisseria species (putatively N. brasiliensis), 1 of which involved bacteremia. Genomic analyses found that both isolates were distinct strains of the same species, were closely related to N. iguanae, and contained a capsule synthesis operon similar to N. meningitidis.
Keywords: Neisseria brasiliensis, Neisseria meningititidis, meningococcal capsule, capsular switching, Neisseria species, whole genome sequencing, Brazil, surveillance, bacteria
Neisseria is a genus containing diverse organisms; most are rarely pathogenic. N. meningitidis and N. gonorrhoeae are the most clinically relevant species. The polysaccharide capsule is the most critical meningococcal virulence factor, a vaccine target, and the basis for classifying meningococci into serogroups (1). During routine laboratory-based public health surveillance in Brazil, we identified 2 cases of infection caused by a previously uncharacterized species of the Neisseria genus.
Clinicians reported 2 cases to the National Reference Laboratory, Adolfo Lutz Institute (IAL), São Paulo, Brazil. Case-patient 1 was a 64-year-old man from Rio Grande do Sul state, Brazil, who, in June 2016, had congestive heart failure with bilateral pulmonary infiltrates and pleural effusion on chest radiograph. Case-patient 2 was a 74-year-old woman with leprosy from Paraná state, Brazil, who, in February 2016, developed a polymicrobially infected ulcer of the left lower extremity. The 2 cases were separated in time and by >400 km and had no known epidemiologic link.
Overnight cultures of blood from case-patient 1 and ulcer exudate from case-patient 2 on brain–heart infusion agar containing 10% chocolate and horse blood at 37°C in the presence of 5% CO2 both revealed brownish colonies uncharacteristic of N. meningitidis. We identified both isolates (N.95-16, from case-patient 1, and N.177-16, from case-patient 2) as gram-negative glucose-fermenting diplococci with positive catalase and oxidase tests. The isolates fermented maltose, lactose, sucrose, and fructose but not mannose; they reduced nitrate and produced a starch-like polysaccharide detected with Gram’s iodine but did not produce DNase. Assessment by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy found no species match; the closest matches belonged to the Neisseria genus for both isolates.
We performed serogrouping by slide agglutination (2) with polyclonal goat or horse antisera prepared at IAL against the N. meningitidis capsule groups (ABCEWXYZ), as described previously (3), and confirmed with real-time PCR (4). Isolate N.95-16 had strong agglutination against serogroup X and nonspecific agglutination against serogroups A, B, C, W, Y, E, and Z antisera; isolate N.177-16 had nonspecific agglutination against A, B, C, W, X, Y, E, and Z antisera. Meningococcal serogroup-specific real-time PCR identified isolate 1 as N. meningitidis capsular group X and isolate 2 as capsular group B.
We extracted genomic DNA from overnight cultures and performed library preparation and whole-genome sequencing using a combination of Illumina MiSeq (https://www.illumina.com) and Oxford Nanopore MinION (https://nanoporetech.com) technologies. Sequencing reads underwent hybrid assembly using Unicycler (5), which generated a high-quality draft assembly for isolate N.95-16 and a complete genome sequence for isolate N.177-16 (GenBank accession nos. WJXO00000000 and CP046027; PubMLST [https://pubmlst.org] identification 94178–94179). We performed species investigation by querying sequencing reads and assemblies against the GenBank and PubMLST reference databases (6). We aligned gene sequences corresponding to 53 conserved ribosomal MLST (rMLST) loci (7) across the Neisseria genus and constructed a maximum likelihood phylogenetic tree using RAxML with 1,000 bootstrap replicates (Figure, panel A). We calculated average nucleotide identity (ANI) using OrthoANI (8).
Figure.
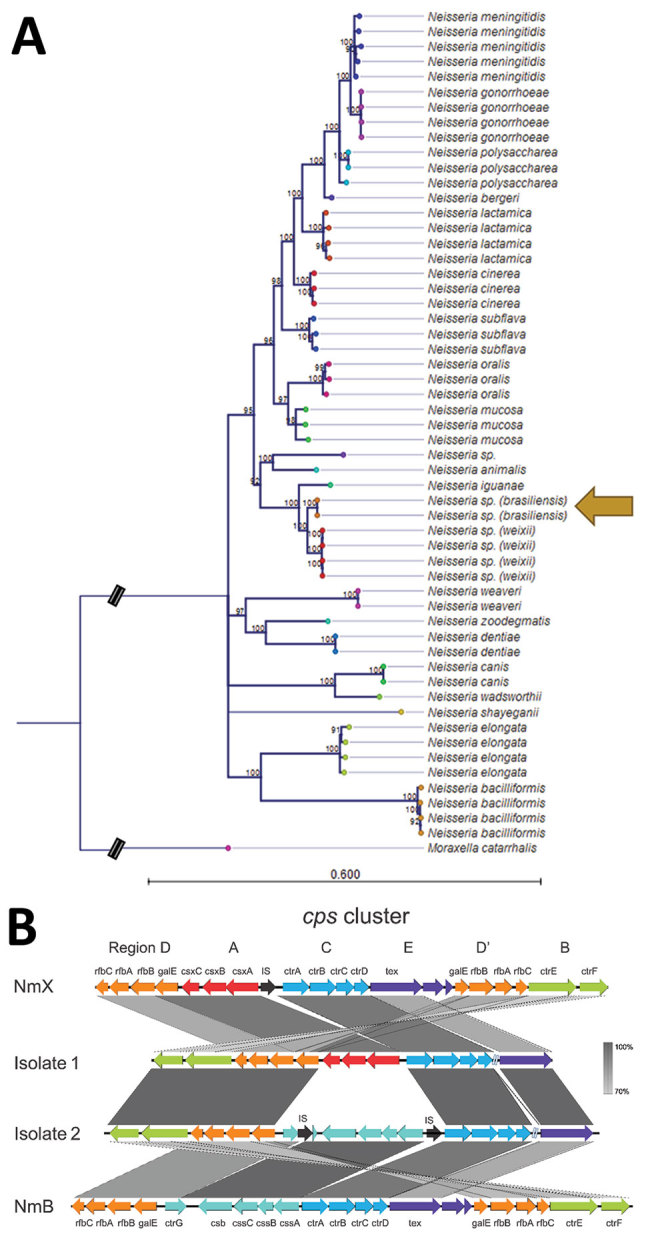
Analyses of newly characterized Neisseria species, Brazil. A) Maximum-likelihood phylogenetic tree of 53 aligned ribosomal multilocus sequence typing genes with 1,000 bootstrap replicates using Moraxella catarrhalis as an outgroup. Bootstrap support values <90% are not shown. Scale bar represents number of substitutions per site. B) cps sequences in isolates 1 (N.95-16) and 2 (N.177-16) relative to meningococcal reference genomes. Arrows represent genes, which are color coded by previously defined cps regions (1). Gray rectangles represent percentage nucleotide identity; darker shading indicates higher identity. The tex gene in both isolates is located >10 kb downstream of other cps genes. cps, capsule gene cluster; IS, insertion sequence; NmB, N. meningitidis serogroup B reference genome H44/76; NmX, N. meningitidis serogroup X reference sequence α388; IS, transposase gene.
Both isolate genomes were 2.5 Mb and had 49.2% guanine-cytosine (GC) content. ANI was 99.3% between the 2 genomes and <86% relative to all other Neisseria species. The closest genome matches were N. iguanae and the proposed N. weixii, isolated from the intestinal contents of a Tibetan Plateau pika (Figure, panel A) (PubMLST identification 56407–56409; GenBank accession no. CP023429). Both genomes shared identical rMLST profiles (rST 61343); the 4 proposed N. weixii genomes shared only 1–2 alleles of 53 rMLST loci with these isolate genomes, and N. iguanae shared no rMLST alleles. Both genomes contained an intact capsule gene cluster (cps) that was similar in gene organization and sequence identity to N. meningitidis (Figure, panel B). The ctrA-cssA/csxA promoter region was conserved in both isolates. However, both genomes contained only 1 copy of galE-rfbCAB (Region D), compared with 2 copies found in meningococcal reference genomes; the tex gene was located >10 kb outside cps, upstream from ctrD (Figure, panel B). The 2 isolates differed in their sequence of sialic acid biosynthesis genes within region A; isolate N.95-16 contained csxABC genes that shared 98% amino acid identity with the meningococcal serogroup X reference strain α388 (1), and isolate N.177-16 contained cssABC-csb genes that shared 99% amino acid identity with serogroup B reference strain H44/76 (Figure, panel B). The cps differences observed between the isolates were similar to the mosaic recombination pattern associated with meningococcal capsular switching (9). Taken together, the presence of cps genes sharing substantial similarity to meningococcal homologs suggests that both isolates have the potential to synthesize meningococcal-like capsules.
In summary, we describe 2 sporadic cases of a new Neisseria species (which we propose to name Neisseria brasiliensis), 1 of which also involved bacteremia. Both genomes contain an intact repertoire of genes for capsule synthesis, a key meningococcal virulence factor. The significance of capsule genes and potential capsule synthesis in nonmeningococcal Neisseria is unknown (10). Continued surveillance is required to establish the pathogenic potential and host range for this apparent new species.
Acknowledgments
We thank Maria Vaneide PB. dos Santos, Gladys Maria Zubaran, and Maria Cristina P. Cecconi for assistance in microbiological tests, and Fatima Ali for microbiological and clinical data.
Biography
Dr. Mustapha is a research assistant professor in the department of medicine and a member of the Microbial Genomic Epidemiology Laboratory at the University of Pittsburgh School of Medicine, Pittsburgh, PA, USA. His research focuses on the genomic epidemiology of bacterial diseases.
Footnotes
Suggested citation for this article: Mustapha MM, Lemos APS, Griffith MP, Evans DR, Marx R, Coltro ESF, et al. Two cases of newly characterized Neisseria species. Emerg Infect Dis. 2020 Feb [date cited]. https://doi.org/10.3201/eid2602.190191
References
- 1.Harrison OB, Claus H, Jiang Y, Bennett JS, Bratcher HB, Jolley KA, et al. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis. 2013;19:566–73. 10.3201/eid1904.111799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castillo D, Harcourt B, Hatcher C, Jackson M, Katz L, Mair R, et al. Laboratory methods for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae, second edition [cited 2019 Dec10]. https://www.cdc.gov/meningitis/lab-manual/index.html
- 3.Alkmin MG, Shimizu SH, Landgraf IM, Gaspari EN, Melles CE. Production and immunochemical characterization of Neisseria meningitidis group B antiserum for the diagnosis of purulent meningitis. Braz J Med Biol Res. 1994;27:1627–34. [PubMed] [Google Scholar]
- 4.Mothershed EA, Sacchi CT, Whitney AM, Barnett GA, Ajello GW, Schmink S, et al. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J Clin Microbiol. 2004;42:320–8. 10.1128/JCM.42.1.320-328.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett JS, Jolley KA, Earle SG, Corton C, Bentley SD, Parkhill J, et al. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology. 2012;158:1570–80. 10.1099/mic.0.056077-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM, et al. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology. 2012;158:1005–15. 10.1099/mic.0.055459-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–3. 10.1099/ijsem.0.000760 [DOI] [PubMed] [Google Scholar]
- 9.Mustapha MM, Marsh JW, Krauland MG, Fernandez JO, de Lemos APS, Dunning Hotopp JC, et al. Genomic investigation reveals highly conserved, mosaic, recombination events associated with capsular switching among invasive Neisseria meningitidis serogroup W sequence type (ST)-11 strains. Genome Biol Evol. 2016;8:2065–75. 10.1093/gbe/evw122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemence MEA, Maiden MCJ, Harrison OB. Characterization of capsule genes in non-pathogenic Neisseria species. Microb Genom. 2018;4. 10.1099/mgen.0.000208 [DOI] [PMC free article] [PubMed] [Google Scholar]


