Abstract
In this Review, I present evidence supporting a multi-factorial causation of childhood acute lymphoblastic leukaemia (ALL), a major subtype of paediatric cancer. ALL evolves in two discrete steps. First, in utero initiation by fusion gene formation or hyperdiploidy generates a covert, pre-leukaemic clone. Second, in a small fraction of these cases, the post-natal acquisition of secondary genetic changes (primarily V(D)J recombination-activating protein (RAG) and activation-induced cytidine deaminase (AID)-driven copy number alterationsin the case of ETV6-RUNX1+ ALL) drives conversion to overt leukaemia. Epidemiological and modelling studies endorse a dual role for common infections. Microbial exposures earlier in life are protective but, in their absence, later infections trigger the critical secondary mutations. Risk is further modified by inherited genetics, chance and, probably, diet. Childhood ALL can be viewed as a paradoxical consequence of progress in modern societies where behavioural changes have restrained early microbial exposure. This engenders an evolutionary mismatch between historical adaptations of the immune system and contemporary lifestyles. Childhood ALL may be a preventable cancer.
Introduction
Childhood acute leukaemia is the most common paediatric cancer in developed societies, accounting for one third of all cases with a variable incidence rate of 10–45 cases per 106 children per year and a cumulative risk of ~1 in 2000 up to the age of 15 years 1. The most common paediatric leukaemia, acute lymphoblastic leukaemia (ALL), is an intrinsically lethal cancer, as evidenced by a universally adverse clinical outcome before effective therapy was developed 2. Currently, however, cure rates for ALL using combination chemotherapy are around 90% 3, making this one of the real success stories of oncology.
Whilst this is a cause for celebration, the current treatment remains toxic, traumatic for young patients and their families and with some long term health consequences 4,5. It is unfortunate that we have remained ignorant as to the cause of ALL. The open question as to whether this cancer is potentially preventable is therefore important.
Environmental exposures possibly linked to ALL are legion 6 but in many cases these associations are weak, inconsistent or lacking in biological plausibility 7. Large and multidisciplinary nationwide studies or international consortia 8,9 have provided a more enabling framework for addressing this question but, to date, the only accepted causal agent for ALL, albeit under exceptional circumstances, is ionising radiation 10–12. The causes of ALL might best be understood by using biological insights into the cancer itself as the foundation for designing, testing and validating hypotheses.
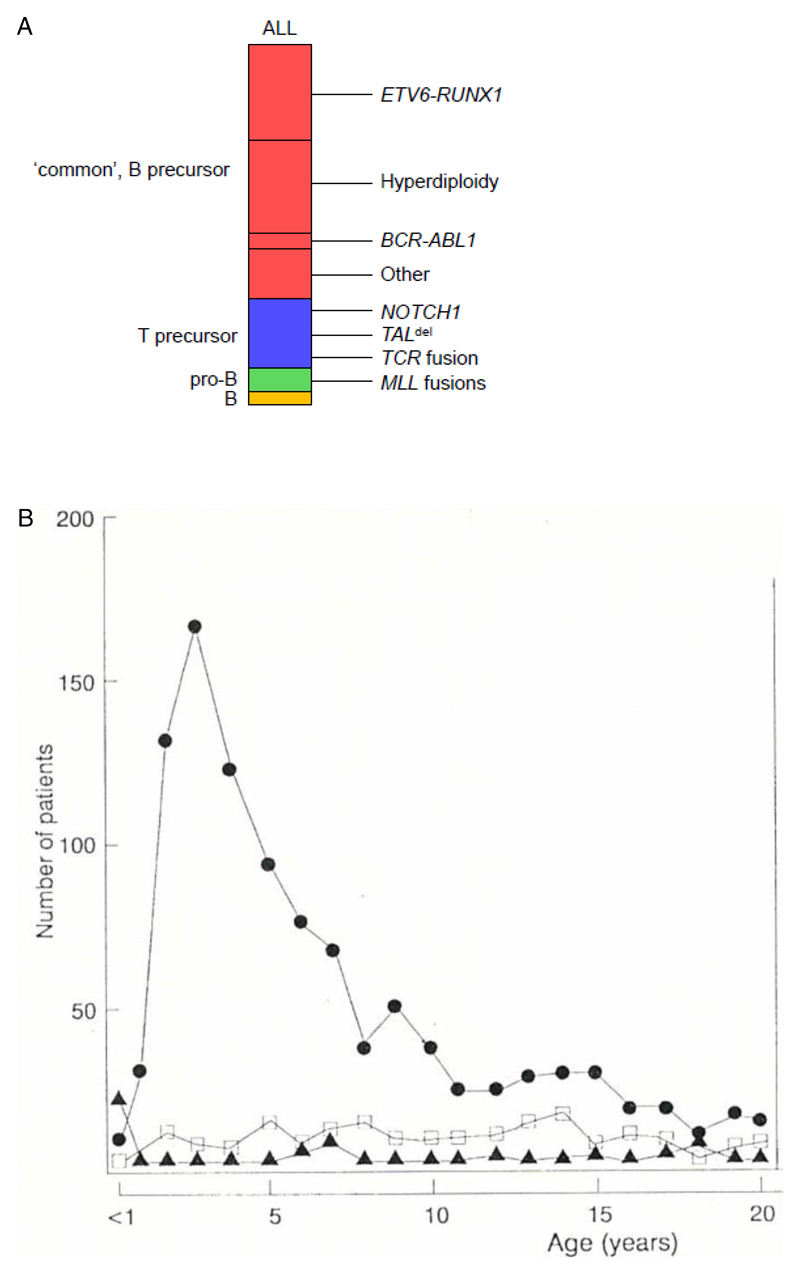
Childhood ALL includes a number of subtypes defined by cell lineage (B or T), differentiation status and genetics (Fig 1a). These differ by age distribution (Fig 1b), clinical outcome (see Fig 1 legend) and could have distinctive aetiologies. In this Review, I focus on the body of evidence – epidemiological, biological and genetic, that has accumulated, particularly over the past decade, supporting a causal mechanism that is selective for the common, or B cell precursor, subtype of childhood ALL (designated here as BCP-ALL) 7. This is suggested to be a multi-factorial mix of infectious exposure, inherited or constitutive genetics plus chance, with patterns or timing of common infection in early life identified as the critical component and a potential route for preventative intervention.
Figure 1.
a. Immunophenotype screens in the 1970’s and early 1980’s established that ALL could be divided into subsets corresponding to early developmental compartments of the B and T cell lineages, as indicated in the key Common or B cell precursor ALL (BCP-ALL) is genetically diverse (as illustrated) with the two most prevalent alterations being ETV6-RUNX1 fusion and hyperdiploidy. The rare (~2%) subtype with a mature B cell immunophenotype (and frequent IGH-MYC rearrangements) was subsequently recognised (and treated) not as ALL but as B lymphoblastic lymphoma. For more detailed descriptions of genomic diversity in ALL, see references 179–181.
b. Age distribution of ALL subtypes from a cohort of 1184 patients with ALL entered into MRC-UKALL clinical trials 182 (1975-1984). This pattern of age-associated ALL subtypes was validated in a later cohort of MRC-UKALL trials (1991-1996; 1088 patients up to 14 years of age) 8 It had been known that childhood ALL had a very marked age incidence peak at 2-5 years throughout the developed world. But this peak appeared to be diminished or absent in less developed societies and appeared in particular countries and ethnic groups at different times 182. Immunophenotypic screens, linked to clinical trials in the UK 183, and an international collaborative group study 182 documented that the peak in incidence was selectively common or BCP-ALL. Recent epidemiological data indicates that the incidence of this subtype of leukaemia in Europe has continued to increase at around 1% per year 184–186. This suggested that the increase over time could be real, rather than ascertainment bias, and that BCP-ALL might have had a distinctive aetiology. The BCP-ALL subtype was also found to have a much more favourable clinical outcome 183,187,188, emphasizing its distinct biology. Modified with permission from ref 182.
Infection hypotheses
The idea that infections might play a causal role in childhood ALL is around 100 years old 13. When it became clear that leukaemia in a number of animal species – chickens, mice, cattle and cats, were viral in origin 14, there was an expectation that a similar transforming virus might be responsible for childhood ALL, as well as for other blood cell cancers. To date, all attempts to molecularly identify or otherwise incriminate a leukaemogenic virus in ALL have failed 7.
In 1988, two hypotheses were presented that suggested a new perspective on this problem. The two models are sometimes considered as alternative or competing explanations. I believe they portray the same picture, through different lenses. Both propose that childhood leukaemia may arise as a consequence of an abnormal immune response to common infection(s). One model, advanced by epidemiologist Leo Kinlen, was based on transient and localised increases in the incidence of childhood leukaemia that could be ascribed, epidemiologically, to population mixing 15,16 (Box 1).
Box 1. The population mixing hypothesis.
The Kinlen hypothesis 15,16 was prompted by public concerns over apparent increases in childhood leukaemia in the vicinity of nuclear power plants in the UK. Kinlen’s model proposed that childhood leukaemia was caused by a rare or abnormal reaction to a common infection of low pathogenicity in a population at risk because of migration and mixing in the context of lack of herd immunity 150. By analogy with animal leukaemias, Kinlen favoured a specific virus. The model was also considered to apply across the board to childhood blood cell malignancy, irrespective of subtype and was not prescriptive with respect to timing of infection in the life of a child.
The population mixing hypothesis for childhood leukaemia has been explored by Kinlen and others in a variety of geographic and demographic settings. The data suggests that where an influx of adults and families occurs, particularly into relatively isolated or rural areas, a transient increase, in the order of twofold, occurs in incidence rates, compatible with an infectious aetiology 16,106,151–153. These studies have not been informative with respect to timing of infection in relation to the natural history of disease, the nature of the infection(s) involved or mechanistic aspects. Nevertheless, they provide an important tranche of evidence supporting a role for common infection(s) in childhood leukaemia.
The other model that I proposed was dubbed the ‘delayed infection’ hypothesis, the focus of this article, and was more biological than epidemiological in its origins and was applied specifically to BCP-ALL 7,17. Central to this were two propositions.
First, that the immune system, in both its innate and adaptive arms, had evolved to both anticipate and require microbial infectious exposure peri-natally or in infancy 18. The dynamics and composition of the microbiome and virome of infants is highly variable 19 and early, microbial exposures have lasting impacts on immune function and health 20,21. Metabolites of commensal bacteria promote regulatory T cells and impact on subsequent inflammatory signalling pathways 22. Deficits of this ‘natural’ microbial experience, especially in modern societies, result in an unmodulated or distorted immune network 23.
A consequence of an under-exposed immune network in infancy was predicted to be subsequent dysregulated responses to common infections that could promote or trigger BCP-ALL. The increased incidence in childhood ALL in developed societies was therefore considered to be a paradox of progress, and the link to infection inverted: the problem might be lack of infection. An equivalent mismatch of evolutionary adaptations and modern lifestyles may underlie the causation of several common adult cancers in the developed world 24.
The second proposition related to the natural history of the disease. The speculation was that ALL most likely developed by two critical steps: first, an initiating event in utero and second, a post-natal mutational event that promotes clinical leukaemia development. The prediction was then that an abnormal immune response to infection(s) indirectly triggered the requisite secondary mutational events. No specific infection was proposed in relation to either protection in early life or post-natal promotion and the immunological mechanism was considered to be indirect and therefore not akin to a transforming virus.
Common infections were therefore proposed to have two opposing impacts on risk of ALL, depending upon timing – antagonistic (early) or promotional (late). A parallel would be with the divergent roles of microbial infections and chronic inflammation in gastro-intestinal and other common cancers in adults 25,26.
The two-hit model of childhood ALL
With a few informative exceptions (Supplementary Information S1 (Box)), ALL has a clinically silent natural history prior to diagnosis. My colleagues and I developed three different tactics for back-tracking the origins of this covert process to before birth. These exploited the fact that common fusion genes in ALL (e.g. ETV6-RUNX1 (also known as TEL-AML1), MLL-AF4 (also known as KMT2A-AFF1, BCR-ABL1) have uniquely variable or idiosyncratic breakpoints within the intronic, breakpoint cluster regions of the two partner fusion genes involved. The genomic sequences at the gene fusion junction provide stable, sensitive and clone-specific markers 27,28.
Comparative genomics of concordant ALL in monozygotic twins
Studies on monozygotic twins have been especially informative 29. The possibility that concordance of leukaemia in identical twins might be attributable, not to co-inheritance of genetic susceptibility, but to an in utero origin in one twin was first proposed in 1962 30 and elaborated on in 1971 31. This idea was based on prior understanding that monochorionic or single placentas have vascular anastomoses permitting twin-twin blood transfusion with consequent blood cell chimaerism 32.
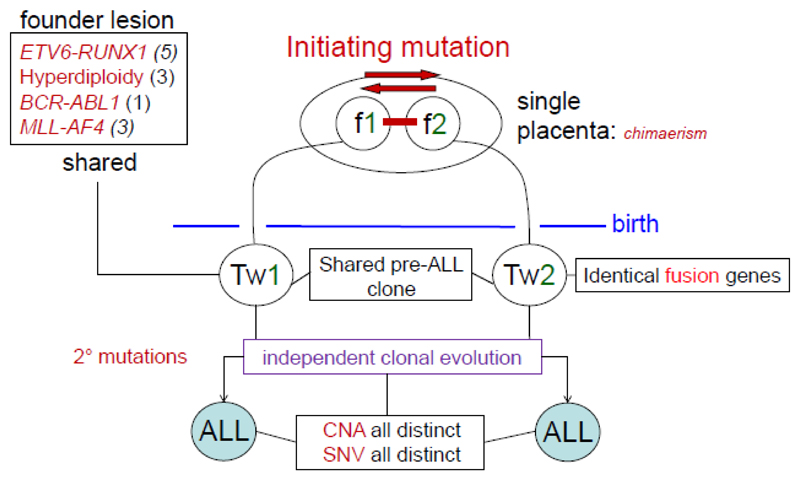
The prediction was then that ALL in both twins should arise in one twin but be monoclonal. Unambiguous evidence that this was the case derived from the finding of identical, but non-constitutive, clone-specific fusion gene breakpoints and sequences in a series of twin pairs 29,33 (Fig 2). That evidence is strengthened by the observation of shared, clone-specific immunoglobulin heavy chain (IGH) diversity-joining (DJ) or variable-diversity-joining (V(D)J) genomic sequences of concordant BCP-ALL in twins 34.
Figure 2.
Summary of comparative genomics of ALL in identical twin pairs. The figure is based on analysis of 12 monozygotic twin pairs (the number of pairs with each founder lesion is noted in parentheses) with concordant ALL 29,33,37,40,48. The sharing of a patient- and clone-specific fusion gene that is not inherited in the twins indicates that in such cases of concordant ALL, the leukaemia must have been initiated in a single cell, in one twin of the pair in utero, the clonal progeny of that cell then disseminating to the co-twin, via intra-placental anastomoses. In further support of this notion, it was noted that concordance of ALL only occurred where the twins shared a single or monochorionic placenta, providing a route for cellular transmission 29. Figure adapted from reference 189. CNAs, copy number alterations. SNVs, single nucleotide variants.
With further exploration of ALL genomes in twins, it has become clear that whilst cases with concordant ALL share the identical and singular fusion gene event, other genetic alterations present, including copy number alterations (CNAs) and single nucleotide variants (SNVs), were different in twin pairs 35,36 (Fig 2). This then suggested that such distinctive mutational events reflected independent and divergent sub-clonal evolution, post-natally. Similarly, the majority of ongoing V(D)J rearrangements in IGH are sub-clonal and distinctive in twin pairs 34. In one twin pair with concordant ETV6-RUNX1+ ALL, whole genome sequencing revealed that the fusion gene was the only shared or clonal genetic lesion 37. These data endorsed the likelihood that ETV6-RUNX1 fusion was an initiating event or founder mutation for ALL.
The concordance rate in monozygotic twins varies according to age and ALL subtype. In infants (<18 months) with pro-B ALL and MLL fusions, the rate approximates to 100% for those with a monochorionic or single placenta 29. This suggested that MLL fusion driven leukaemogenesis in such infants was essentially completed in utero and that the fusion gene, or a single mutation, was sufficient for leukaemogenesis. Subsequent genomic sequencing of such cases is compatible with this possibility even though other sub-clonal mutations, in RAS family genes for example, do occur 38,39.
Pre-leukaemic clones in healthy co-twins
The concordance rate in older children with BCP-ALL was calculated to be around 10-15%, lower than that in infants 29. A prediction for those pairs of twins with a monochorionic placenta, where only one twin develops ALL, is that the healthy co-twin should have a population of covert pre-leukaemic cells harbouring the same initiating lesion as his/her co-twin with ALL, i.e. the twins are discordant for the critical post-natal secondary, genetic event. This has been confirmed in three twin pairs with BCP-ALL with hyperdiploidy 40, BCR-ABL1 fusion 36 or ETV6-RUNX1 fusion 35,41. In this context, the healthy co-twin provides a rare ‘experiment of nature’ and unique access to the pre-leukaemic clone. Putative pre-leukaemic cells from the blood of one healthy co-twin and propagation in vitro and in vivo (in NOD/SCID mice) established that these cells have both self-renewal capacity and intact B cell differentiation capacity 41, features commensurate with a pre-leukaemic status. Equivalent pre-leukaemic stem cells for acute myeloid leukaemia (AML) have now been identified in patients with AML 42 and normal adults 43.
Backtracking early genetic events in ALL to neonatal blood spots
Less than 1% of childhood BCP-ALL cases occur in twins. However, ALL in twins is no different, in its biological and clinical features or age incidence, to that in singletons. This suggests that many or most childhood ALL cases in singletons might also be initiated in utero.
To validate this proposition, my colleagues and I exploited the fact that neonatal blood spots, or Guthrie cards, contain reasonably intact DNA. Archived blood spots from patients with ALL were screened for clone-specific fusion gene sequences identified at diagnosis. This was first carried out in three cases of infant ALL with MLL-AF4 fusion and blood spots from all three cases evaluated were positive 44. Subsequent studies with samples from children with ETV6-RUNX1+ ALL found that around 75% were positive 45. These results have been independently confirmed 46,47. Negative blood spot results are uninterpretable as this could reflect either a post-natal origin or an inadequate number of leukaemic cells in the sample. The conclusion drawn from these screens was therefore that the majority of childhood ALL cases were pre-natal in origin, though possibly not all.
The twin and blood spot studies also provided insight into persistence of pre-leukaemic stem cells and post-natal latency in ALL. The oldest twin with concordant ALL originating in utero was 14 years at diagnosis, her twin sibling having been diagnosed with ALL some nine years earlier 48. The oldest non-twin case with ALL and a positive neonatal blood spot to date was diagnosed at 9 years, 4 months old 49.
Frequency of ALL initiation in utero
The data on discordant, monozygotic twins suggested that some or possibly most individuals harbouring a pre-natally generated, covert pre-leukaemic clone never progress to overt ALL. This begs the important question, relevant to aetiology, of the frequency of initiation of ALL in utero and the frequency of its transition to overt leukaemia.
To address this issue, my colleagues and I screened a large cohort of unselected cord blood samples for ETV6-RUNX1 fusion mRNA (data summarised in Supplementary Information S2 (Fig)). The striking result was that approximately 1% of newborns (6/567) had a covert and modest sized, putative pre-leukaemic population at around 10-4 of B lineage cells 50. This result, initially challenged 51, has been independently confirmed 52–54. A ~1% incidence for ETV6-RUNX1 in relation to incidence of the leukaemia itself reflects a low transition probability of around 1% with 99% of pre-leukaemic clones initiated during foetal development never progressing to clinical ALL. This low transition probability could reflect either lack of persistence of the pre-leukaemic stem cells after birth or a severe bottleneck in acquisition of the necessary secondary genetic changes.
These data suggest that initiation of leukaemia in utero may be far more common than indicated by the incidence of disease, with implications for causation. The same may apply to some other paediatric cancers. Histological evidence and some genetic data suggest that the frequency of precursor lesions for neuroblastoma 55 and Wilms tumour 56 may also be some 100 times the incidence of clinical cancer 50.
Other subtypes of ALL
These lines of investigation were pursued using fusion genes as the predominant clonal markers of early genetic events in ALL. The most frequent subtype of BCP-ALL is, however, characterised by chromosomal hyperdiploidy, which is harder to track than fusion genes. There is evidence however that the key findings described above for the ETV6-RUNX1 subset are likely to apply to hyperdiploid ALL. Monozygotic, monochorionic twin pairs concordant for hyperdiploid ALL are described with identical karyotypes 40 and neonatal blood spots of children with hyperdiploid ALL have clone-specific IGH sequences 57–59. In one case of hyperdiploid ALL, the child’s cord blood had been frozen at birth. Retrieval of this sample led to the identification of putative pre-leukaemic cells in the cord blood with the same triploid chromosomes as in the child’s subsequent ALL 60. Hyperdiploidy, generated by a one-off abnormal mitosis resulting in trisomies 61, can therefore occur in utero as an alternative initiating event to gene fusion for BCP-ALL.
Further genomic exploration of ALL
Whole genome sequencing of a cohort of 57 cases of ETV6-RUNX1+ BCP-ALL provided an audit of all mutational changes 62. This confirmed the previous finding that the most common recurrent events were CNAs, primarily gene deletions 63. SNVs were also present but with low or undetectable recurrency62 Genomic sequencing in cancer can reveal mutational signatures of relevance to aetiology 64. Almost 50% of CNAs in ETV6-RUNX1+ BCP-ALL had partial or complete V(D)J recombination-activating protein (RAG) heptamer-nonamer recognition motifs within 20 base pairs of the breakpoints 62. This finding may explain the observation that highly recurrent CNAs in BCP-ALL are reiteratively present in subclones of individual patients 65,66. A comparison with CNAs in around 14,000 cases of breast, prostate and pancreatic cancer revealed none with RAG motifs 62. SNVs in ETV6-RUNX1+ BCP-ALL had two main mutated signatures: one was C>T transitions at CpGs and C>G and C>T mutations at TpCs, and a second was transitions and transversions in a TpC context at NpCpG trinucleotides 62. These second signatures are very common in cancer generally reflecting APOBEC cytidine deaminase activity 64,67.
These genomic studies indicate that BCP-ALL has very restricted but informative mutational signatures and a low level of background or neutral genetic alterations 62. This makes it less likely that BCP-ALL is caused by genotoxic exposures, which generally precipitate more widespread genomic instability with multiple distinctive signatures 64.
The other genetic subtype of BCP-ALL – hyperdiploid ALL, also has recurrent CNAs that may be RAG-mediated in genes including PAX5, IKZF1 and ETV6. In contrast to ETV6-RUNX1+ ALL however, hyperdiploid ALL have recurrent mutations in receptor tyrosine kinase (RTK)-RAS pathways and histone modifiers 61,63.
Collectively, these data provide a firm cellular, genetic and mechanistic framework for the two step model for BCP-ALL and highlight both critical time windows, pre- and post-natally, and mutational mechanisms. Any proposed causative mechanism in ALL should accommodate this natural history profile. The initiating role of ETV6-RUNX1 and the postulated sequence of events in ALL is endorsed by modelling within both human and animal cells (Box 2 and further below). The timing and tissue site of B cell precursor ALL initiated by ETV6-RUNX1 or hyperdiploidy in utero is uncertain but may involve transformation of a unique, foetal liver progenitor cell (Box 3).
Box 2. Insights from modelling.
The initiating role of ETV6-RUNX1 in ALL has been modelled in zebra fish 154, mice 155–162 and with human cells 41. A consensus view is that the ETV6-RUNX1 protein functions as a weak oncogene imparting only a modest proliferative or survival advantage on pro-B or pre-B cells. This accords with the small clone size of ETV6-RUNX1 pre-leukaemic cells in cord blood and in the blood of co-twins of patients with ALL 50. ETV6-RUNX1+ ALL cells ectopically express erythropoietin (EPO) receptors and modelling with both human and mouse cells suggest that the receptors are functional and that EPO may provide a survival signal to ETV6-RUNX1-expressing pre-leukaemic cells 163,164.
The observations on identical twins and cord blood suggest that ETV6-RUNX1 fusion is insufficient by itself to induce overt or clinically evident ALL. This is endorsed by the various models (see table below). However, as anticipated, ETV6-RUNX1-expressing clones in mice do progress to BCP-ALL if additional, co-operative mutations are introduced by cross-breeding with mice heterozygous or homozygous null for the genes recurrently deleted in ETV6-RUNX1+ ALL, including Cdkn2a and Pax5 161,162. ALL also develops in some of these models if mice expressing ETV6-RUNX1 are subjected to insertional mutagenesis 160 or chemical exposure 159.
The impact of these additional genetic lesions that complement ETV6-RUNX1 is to impede or block differentiation, mirroring the early B lineage phenotypes observed in the clinical disease. This is in accord with the normal function of the transcription factors ETV6 and RUNX1, which is primarily to promote differentiation, and their loss of function or deletion in ALL 63. This interpretation is further endorsed by modelling studies in which experimental regulation of Pax5 expression and gene dosage govern both early B lineage differentiation and leukaemogenesis 165.
These murine models support the ‘two hit’ hypothesis for BCP-ALL and have also provided evidence for the role of common infections in the development of BCP-ALL (table below) 134–136.
| Initiating, transgenic lesion | Manipulation | Outcome | References |
|---|---|---|---|
| ETV6-RUNX1 | None | Covert pre-leukaemia only | 125,157 |
| ETV6-RUNX1 | Activation of AID, in vitro, with LPS, transplant to RAG1+/+ or -/- mice | BCP- ALL in RAG1+/+ background | 126 |
| ETV6-RUNX1 | Switch to non-germ-free housing | BCP- ALL | 135 |
| Pax5+/- | Switch to non-germ-free housing | BCP- ALL | 134 |
| Eμ-Ret or E2A-PBX1 | CpG ODN oligodinucleotide (at 4-8 weeks) | Delay and reduced penetrance of BCP- ALL | 136 |
Box 3. Is there a unique foetal target cell for childhood BCP-ALL?
Adults do develop BCP-ALL. But it is striking that hyperdiploid ALL and ETV6-RUNX1+ BCP-ALL decline in incidence in teenage years and are infrequent after 20 years of age. Adult BCP-ALL has a higher frequency of BCR-ABL1 fusion or a BCR-ABL1-like signalling phenotype, perhaps explaining their poorer prognosis 166. Childhood ALL in this respect appears to be a different cancer. Why should ETV6-RUNX1 or hyperdiploidy initiate BCP-ALL preferentially in early life, and especially in utero? ETV6-RUNX1 expression, driven by the endogenous Tel promoter in mice, increases quiescence and persistence of foetal liver stem cells 159. Moreover in this mouse model, foetal haemopoietic stem cells expressing ETV6-RUNX1 had a protracted but still limited lifespan as self-renewing cells and did not contribute to the early B cell progenitor pool of adults. This suggests that the low penetrance of pre-leukaemia in children and the declining incidence of ETV6-RUNX1+ (and hyperdiploid) ALL with age may be, at least in part, be due to natural attrition of the foetal pre-leukaemic clone.
One possibility is that foetal B lymphopoiesis is distinctive, in cell type and/or microenvironment, and provides a selective context in which ETV6-RUNX1 and hyperdiploidy have effective transforming functions. This appears to be the explanation for GATA1 mutations in Downs syndrome-associated acute megakaryoblastic leukaemia (AMKL) being restricted to foetal liver 167. Recently a strong candidate target cell for childhood BCP-ALL has been identified 168. This cell type is developmentally restricted to foetal lymphopoiesis, with a mixed myeloid/B lineage phenotype. In common with the putative target cell suggested by our studies in identical twins 34,41, this foetal liver myeloid/B progenitor also has DJ rearrangements of IGH 168.
Inherited susceptibility
Childhood ALL only very rarely runs in families but this observation may underplay inherited genetic risk since the disease itself is rare. Twin concordance is unhelpful in this respect since the risk has a mostly non-genetic basis – blood cell chimaerism in utero. The risk of ALL in non-identical twins is unknown but sibling risk has been estimated to be around 3.0x compared to the risk in the general population, which provides evidence of a modest but definite contribution of constitutive, genetic variation to risk 68.
Earlier, targeted gene screening approaches suggested that inherited allelic variants encoding proteins involved in DNA repair, carcinogen metabolism or the folic acid pathway might be linked to risk of childhood ALL 69,70. Unfortunately most of these studies were underpowered to detect small effects or have not been consistently replicated, so their significance remains uncertain 70.
Genome wide association studies (GWAS) have provided unambiguous evidence for multiple gene variants impacting on risk of ALL (Table 1) 70,71. The individual alleles described to date have a significant but relatively modest impact (see odds ratio in Table 1) and appear to be additive rather than synergistic, functionally. The functional logic of these associations is unclear but as most of the relevant single nucleotide polymorphisms (SNPs) lie outside coding regions, they are likely to be regulatory, impacting on levels of protein 72,73.
Table 1. Inherited alleles and risk of childhood B cell precursor ALL identified in GWASa.
| Candidate gene (chromosome)b | ALL subset bias | Odds ratioc (95% CI) | References |
|---|---|---|---|
|
IKZF1 (7p12.2) |
None detected | 1.69 (1.58 – 1.81) 1.69 (1.4 – 1.9) |
190 191 |
|
ARID5B (10q21.2) |
Hyperdiploid | 1.65 (1.54 – 1.36) 1.91 (1.6 – 2.2) |
190 191 |
|
CDKN2A and CDKN2B (9p21.3) |
None detected | 1.36 (1.18 – 1.56) 1.72 (1.50 – 1.97) 2.41 (1.99 – 2.92)b |
75 192 193 |
|
CEBPE (14q11.2) |
Hyperdiploid | 1.34 (1.22 – 1.45) 1.45 (1.30 – 1.62) |
190 194 |
|
GATA3 (10p14) |
BCR-ABL-like | 1.31 (1.21 – 1.41) 3.85 (2.7 – 5.4) |
195 196 |
|
PIP 4K2A (10p12.2) |
None detected | 1.40 (1.4 – 1.53) |
195 75 |
|
TP63 (3q28) |
ETV6-RUNX1 | 0.63 (0.52 – 0.75) | 197 |
| PTPRJ | ETV6-RUNX1 | 0.72 (0.68 – 0.89) | 197 |
|
LHBP (10q26.13) |
None detected | 1.20 (1.15 – 1.28) | 198 |
|
ELK3 (12q23.1) |
None detected | 1.19 (1.12 – 1.26) | 198 |
| 8q24.1 | None detected | 1.34 (1.21 – 1.47) | 199,200 |
| 2q22.3 | ETV6-RUNX1 | 1.32 (1.17 – 1.49) | 199 |
|
IKZF3 17q12 |
None detected | 1.18 (1.11 – 1.25) | 200 |
Cohort sizes in these studies were between ~400 and 3500 cases.
More low risk alleles are likely to be uncovered as larger cooperative group studies are pursued and other populations considered. Rare allelic variants, missed in prior studies, may also be identified which have a stronger impact on risk of ALL as described for CDKN2A (OR 2.4) 193.
p values in these studies were between 10-5 to 10-19. All cases are white, European descent in which the alleles exist at relatively high frequency (10-50%).
It is striking that in ALL, as in many other cancers, most of the candidate risk genes implicated in GWAS (Table 1) are the same genes that have acquired (non-constitutive) mutations in the same cancer type. One interpretation of this is that the inherited allelic variants interact functionally (or epistatically) with the mutated alleles to increase vulnerability of cells to transformation. A low functioning inherited allele for example would render a deletion in the other allele functionally homozygous with a potentially increased impact on cellular fitness. A prediction that follows from this is that there should be a preferential loss, by deletion, of the non-risk allele (in heterozygotes for that allele) as only that deletion would increase clonal fitness. Evidence for this has been presented with respect to risk variants of CDKN2A 74,75. However, for ARID5B there is preferential gain of the risk allele (via trisomy 10) in heterozygotes 72.
To date GWAS have provided no evidence implicating immune response gene variants, as might have been anticipated from infection-based hypotheses for the aetiology of ALL. However, prior studies examining major histocompatibility complex (HLA) genes 76–78, interferon-γ (IFNG) 79, Toll-like receptor 6 (TLR6) 80 or the presence and/or absence of specific killer-cell immunoglobulin-like receptor (KIR) family genes 81 did record significant associations with particular allelic variants. Notably, TLR6 variants and KIR genes were associated with decreased risk of all childhood ALL. It remains unclear if the large GWAS multi-cohort studies invalidate these data or whether the SNP screens in GWAS adequately detect the relevant variants. This is an important discrepancy to resolve.
Childhood ALL can also arise in a rare familial syndrome context with inherited mutations in genes also implicated as acquired mutations in leukaemia including PAX5 82 and ETV6 83. The relatively infrequent low hypodiploid subset of BCP-ALL is strongly associated with inherited TP53 mutations or Li Fraumeni syndrome 84. Further rare risk alleles, but with intermediate to high penetrance, are likely to be uncovered in ongoing, large scale studies. Children with Downs syndrome have an approximately 20-30 fold increased risk of BCP-ALL 85. Trisomy 21 in Downs syndrome is associated with over-expression of a nucleosome remodelling protein HMGN1 and enhanced self-renewal of B cell progenitors 86. In patients with ALL, this is complimented by secondary genetic changes including in CLRF2, JAK2, NRAS and KRAS 85. All told, however, familial syndromes and Downs-associated ALL are likely to account for only a small fraction of cases of childhood ALL.
The general conclusion to be drawn from these genetic studies is that inherited susceptibility does contribute to risk of BCP-ALL. The attributable risk or quantitative contribution is, however, unclear. A sibling risk of three fold for ALL, seen against a background risk of 1:2000, suggest the genetic component, though real, is minor; at least compared to some other common adults cancers (prostate, breast) 87. On the other hand, there could be a complex interplay between genes and environmental exposures 73,88 in which genetic background makes a more substantial difference. This has yet to be fully explored.
Possible causes of initiating events
No epidemiological studies to date have clearly implicated exposures during pregnancy, linked to risk of ALL, that might explain how the initiating mutations for BCP-ALL arise. ETV6-RUNX1+ BCP-ALL has no mutational signatures that might implicate any particular type of aetiological exposure. There is no evidence for RAG involvement but, in common with IGH rearrangements 89, the recombination event appears to involve non-homologous end joining via micro-homologies 27.
If around 1% of unselected newborns have an in frame ETV6-RUNX1 fusion in an expanded clone derived from the appropriate cell type for BCP-ALL (B lineage progenitor), then considerably more newborns should have acquired this or other fusion genes in the wrong cell types or out of frame for a functional protein. It therefore seems likely that whatever causes this genetic alteration, it could be very common or possibly ubiquitous.
The original proposition 17 was that ALL was initiated, in utero, by a spontaneous mutation, or with no external exposure involvement. Developmental, endogenous factors such as proliferative and oxidative stress or the profound apoptotic signalling in early lymphopoiesis could be involved. Spontaneous mutations or mutations caused by endogenous processes are common during foetal development 90. Endogenously-driven double-strand breaks, required for fusion gene recombinants, occur at around 50 per cell cycle in human cells 91. It has been suggested that most paediatric cancers arise during embryonic or foetal life and can similarly be considered as developmental errors 92. In the absence of evidence to the contrary, this remains the most plausible explanation for initiation of BCP-ALL and focuses attention on the post-natal triggering of promotional events which are required for clinical disease. There is clearly scope for more research on the origins and mechanisms involved in fusion gene formation, and hyperdiploidy, in utero.
Epidemiological evidence
Epidemiological evidence suggests that patterns of infection after birth have a causal role in triggering ALL. The ‘delayed infection’ hypothesis lends itself to epidemiological evaluation in a case-control setting. A prediction of the model was that common infections in infancy should be protective against BCP-ALL. There is no prior reason to implicate any particular infectious agent (eg, bacteria, virus or parasite),and the relevant infections need not be symptomatic or pathological. A longstanding need for microbial, immune network modulation might reflect common, commensal, or ‘old friends’ organisms, such as gut microbiota, soil mycobacteria or helminth parasites 93. In this context, a surrogate of overall infectious exposure during infancy could be considered an appropriate variable. Quantifiable surrogates include social exposures of infants in the home, related to the number of siblings and birth order, or in day care centres, and breast feeding. These variables have been investigated in epidemiological case/control or cohort studies for risk associations with ALL overall and, in some instances, selectively for the major BCP-ALL subset.
Impact of day care attendance in infancy
In the 1990’s, the UK Children’s Cancer Study Group (UKCCS) was set up to test the delayed infection hypothesis, in a case-control context, in addition to an analysis of other exposures including ionising and non-ionising (EMF) radiation and chemicals 8. Day care attendance was chosen as one surrogate for infectious exposure since this was well documented as a context for enhanced social contacts facilitating spread of common infections 94. The UKCCS involved almost 1300 cases of ALL (all subtypes) and over 6000 matched controls. Although only a relatively small number of controls experienced day care in the first twelve months of life, the data showed a significant protective impact on risk of ALL overall and on BCP-ALL 95.
This association has been documented in additional studies in California 96, Scandinavia 97, France 98 and an international consortium 99, and has been endorsed by a meta-analysis 100. The meta-analysis noted significant, between study heterogeneity and one early, large scale study failed to detect any impact of day care on risk of ALL 101. No protection is afforded against childhood AML by day care attendance or, to date, any other paediatric cancer, which increases confidence that the associations seen in ALL was not confounded by social or other variables.
It was anticipated that assessing, in a case-control fashion, actual infections in infancy would be informative. This however is fraught with difficulties and has provided mixed results. Parental recall is known to be suspect or inaccurate in this respect 102. Medical records are more reliable, particularly in the UK with nationwide registration of children with general practitioners (GPs) and a free health service. One such analysis found more, rather than less, infections reported for children who subsequently developed ALL, compared with controls 102. The main difficulty here, other than possible bias in use of GP services, is that we do not know if the relevant ‘modulating’ infectious exposures in infancy are necessarily symptomatic; they might well not be. In this sense the surrogate of day care could be considered preferable. Several studies have however reported, in accord with the hypothesis, an inverse relationship between common infections in early life, including inner ear infections, and risk of ALL98,101,103–106.
Birth order and risk of ALL
A further surrogate measure of infectious exposures in infancy is the number of siblings cohabiting and, in particular, birth order. The prediction was that first-borns would be more at risk compared to later-borns who would, as infants, benefit from protective exposures via older siblings. One large UK-based study (>3000 cases of ALL and the same number of matched controls) found a striking association of birth order with risk for ALL, but not for AML 107. Other case-control studies in France 98 and California 100 also found a significantly increased risk for first (versus third) born children, as does a recent international cohort analysis (Ora Paltiel; personal communication 2018)
If natural infections early in life reduce risk of ALL, then it might be expected that some vaccinations would have an effect. The data on vaccination histories have produced null or inconsistent results. However, there is one exception: immunisation against Haemophilus influenzae B in infancy appears to confer a degree of protection against ALL 108.
If the ‘natural’ microbiota is part of a longstanding and critical interaction with the developing immune system, then antibiotic use in infancy might increase risk of ALL. This has not been systematically evaluated to date, though an earlier report from China did suggest an increased risk associated with exposure to chloramphenicol 109.
Mode of delivery, breastfeeding and risk of ALL
Mode of delivery at birth influences the early exposure of newborns to benign microbiota 110, as caesarean delivery deprives newborns of the microbial exposures associated with vaginal passage. Cohort and case/control studies have reported a significantly increased risk of ALL associated with caesarean delivery 111–114. No such increased risk was observed for brain cancer or lymphoma 111.
Breastfeeding during infancy provides nutritional support, maternal antibodies, anti-inflammatory molecules, some maternal cells, microbes (lactobacilli) and oligosaccharides that nourish the infant’s intestinal microbiome (bifidobacteria) 115. It might be anticipated that prolonged breastfeeding would have a modulating effect on the immune system of infants and reduce risk of ALL. Seventeen case/control studies of the impact of breastfeeding on ALL risk have been published 116. In the largest of these, from the USA 117 and UK 118, there was a reduced risk for ALL (10-20%) associated with breastfeeding of six months or more. Five meta-analyses have now been published with concordant conclusions and the latest of these indicated a reduced ALL risk of around 20% for breastfeeding of six months or more 116.
Clusters of ALL
Although no specific microbial agent or a unique transforming virus is suspected in ALL, there might be one circumstance where a single type of infection is involved. This is in the very rare cases of space-time clusters. A prediction of our hypothesis would be that a single cluster of cases might be associated with a single infection or microbe species but independent, space/time clusters could involve different infectious triggers.
Many putative clusters of childhood leukaemia have been reported but two stand out. The first was in of Niles, a suburb of Chicago in 1957-60 – where there were eight cases (relative risk (R.R.) 4.3) diagnosed as ALL or ‘stem cell’ leukaemia 119. All patients and/or their older siblings attended the same school. The second cluster involved 13 cases of BCP-ALL over four years (2000-2004; R.R. = 12.0), but ten cases in just ten months in 2001 in the small town of Fallon, Nevada 120.
A neglected aspect of these two clusters is that the cases, though resident in the cluster area at the time of diagnosis, were mostly born outside of that area 7. Plus the clusters involved children diagnosed with ALL at different ages (2-11 years) and a narrow time frame of diagnoses. Given what we now know of the natural history of ALL, these data then indicate any causal exposure, linked to the cluster, would have to be proximal in time to diagnosis (rather than pre-natal) and therefore promotional. The Niles cluster was linked, observationally, to an outbreak of streptococcal fever 119. The cause(s) of the Fallon cluster of ALL remain unresolved and contentious though a possible role of adenovirus was hypothesized 120.
A significant space-time cluster of BCP-ALL in Milan has recently been recorded 121. Seven cases were diagnosed in a four week period; four of these lived within one small residential area and three of these four attended a single school. The Kulldorff scan method 122 identified this as a significant space/time cluster (p = 0.017) Given the narrow time window of the diagnoses (four weeks) and the age range of the cases (2-11 years), any causal exposure, as in Niles and Fallon, would be proximal to diagnosis, promoting overt ALL evolution from a prior and covert pre-leukaemic state. The Milan cases sparked substantial public anxiety, particularly in relation to the school, and a detailed epidemiological investigation was launched. No link was found with ionising radiation, non-ionising radiation or chemicals. There was, however, an association with a particular, common infection. All seven cases had been infected with endemic AH1N1 swine flu virus during the epidemic that preceded the ALL cluster by three to six months. The infection frequency in children in Milan during the same period was relatively high, at around one third, but this still indicated the link with cases was significantly different from expected (p = 0.01) 121. Six of the seven cases were first born and none attended day care in the first year of life.
Proof of a causal role for infections in these situations is not possible and clustering of cases by chance cannot be excluded. But the observations accord with predictions of the infection hypothesis and highlight that influenza viruses are potential promoting agents for ALL. A prior study in the UK observed peaks in incidence of childhood ALL ~6 months after seasonal influenza epidemics 123. A final piece of epidemiological evidence indirectly supporting a role of common infection in childhood ALL comes from anecdotal but striking observations on rapid changes in the incidence of ALL that were preceded by major social changes in Germany and Hong Kong (Supplementary Information S3 (Box)).
There is no compelling reason for postulating an exclusive role for influenza viruses or indeed for viruses. A role for cytomegalovirus (CMV) in ALL has been postulated, but as an early, in utero, modulator of immunity, rather than as a proximal trigger 124.
In some respects it is surprising that the epidemiological data is as consistent as it is on individual factors related to infection in ALL since many variables will compound to influence patterns of microbial exposures in early life (Supplementary Information S4 (Box)).
Modelling the ‘missing link’ in ALL
There are limits to what epidemiological studies can achieve and to the robustness of the findings. Nevertheless, the associations described are compatible with the infection model proposed and their selectivity for BCP-ALL, versus AML, is striking. But associations are not necessarily causal. Functional components of the infection hypothesis are best addressed by modelling studies in mice (Box 2). These have proven very informative. One inflammatory cytokine - TGFβ, was found to induce preferential expansion of ETV6-RUNX1-driven pre-leukaemic cells of both mouse and human origin 125. Normal B cell precursor proliferation is inhibited by TGFβ via activation of the cell cycle inhibitor p27. ETV6-RUNX1 blocks this activity giving pre-leukaemic cells a fitness advantage in the presence of TGFβ 125.
The missing link in the chain of events between infection, inflammatory responses and promotion of BCP-ALL may be activation-induced cytidine deaminase (AID) 126,127. As noted above, genomic sequencing in BCP-ALL revealed that the recurrent CNAs have signatures of RAG activity 62. Physiological RAG activity in germinal centre Ig class switching or hypermutation requires AID, 128 as does illegitimate recombination between the IGH locus and oncogenes 129,130. In B cell precursors, AID is not normally co-expressed with RAGs but is inducible by infection-driven cytokine signals 131. This suggested that one route by which infection or chronic inflammation might trigger RAG-mediated CNAs and ALL is via activation of AID expression in pre-leukaemic stem cells.
A mouse model has tested the requirement for RAGs and AID in the transition from ETV6-RUNX1 pre-leukaemia to overt ALL 126. The data revealed that lentiviral transfection of ETV6-RUNX1 into progenitor cells leads to B cell precursor ALL when those cells are treated with a surrogate inflammatory signal (bacterial lipopolysaccharide binding to TLR4) that activates AID. Critically, mice did not develop ALL if the same experiment was conducted in a RAG1-/- genetic background. More recently, my colleagues and I have screened a series of inflammatory cytokines for their ability to trigger AID expression in human B cell precursors. The most potent was TGFβ (V Cazzaniga, AM Ford, M Greaves, unpublished data). TGFβ is known to promote other cancers, often in the context of chronic inflammation 132. In ALL, its role may include not only selective expansion of pre-leukaemic cells 125 and activation of AID but compromise of natural killer (NK) cell-based immune-surveillance 133.
If the aetiological hypothesis is correct, then it should be possible to influence risk or penetrance of ALL in murine models by timed exposure of the immune system to natural infections. Using a model of BCP-ALL it was shown that ALL developed if Pax5+/- mice were switched from a germ-free environment to one providing exposure to common microbial pathogens 134. Similarly, another study found that ~10% of mice with ETV6-RUNX1 developed BCP-ALL after exposure to common pathogens 135. The experiments provide evidence, albeit in murine models, that common infections can have, as predicted, a promotional role in ALL.
Another mouse model has provided evidence that early stimulation of the immune system can be protective. Exposure of mice transgenic for Eμ-Ret or E2A-PBX1 to oligodeoxynucleotides (which bind to TLR7, TLR8 and TLR9) at four weeks depleted both normal and pre-leukaemic precursors and both delayed and diminished risk of progression to ALL 136. This effect was dependent upon IFNγ. In contrast, binding of polyI:C, a TLR3 ligand which does not induce IFNγ, resulted in an expansion of the pre-leukaemic cell pool. These data hint that the nature of infectious exposures in infancy and responses of the innate immune system may influence not only subsequent immune responses but the fate of pre-leukaemic cells.
Conclusions: paradoxes of progress
“We incline on our evidence to the belief that the solution of the problem of leukaemia lies rather in some peculiar reaction to infection than in the existence of some specific infective agent.”
F J Poynton, H Thursfield and D Paterson
(Great Ormond Street Hospital for Sick Children), 1922 137
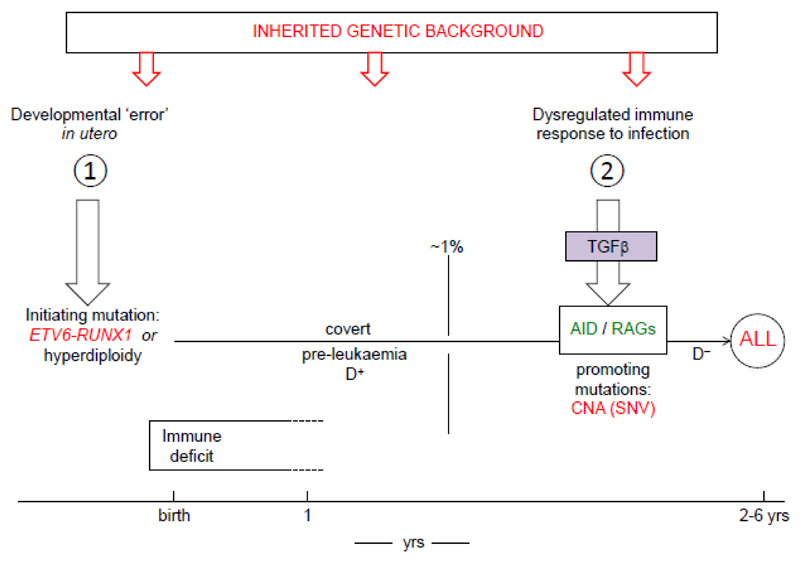
Collectively, the accumulated evidence derived from epidemiological studies, GWAS, genome sequencing, biological scrutiny of the natural history and molecular pathogenesis of BCP-ALL, plus mechanistic and modelling studies, provide us with a more substantive and credible version of the original 7,17 two hit model for childhood ALL, as summarised in Fig 3. The model applies selectively to the common, B cell precursor subset of ALL, although the evidence is, currently, more compelling for the ETV6-RUNX1+ subset of BCP-ALL than for hyperdiploid subset. The rarer pro-B ALL in infants appears likely to have a different causation and molecular pathogenesis, as does childhood AML and childhood lymphoma. There is insufficient data for T-ALL in this respect. Other causal associations in leukaemia and cancer in general might be revealed or strengthened by a focus on well-defined subtypes, as suggested for breast cancer 138.
Figure 3.
Summary of 2 hit model for role of infections in B cell precursor ALL.
Genetic, inherited risk alleles are depicted (top of figure) as impacting at any or several stages of the step-wise process of ALL.
The causal mechanism proposed here is multifactorial, involving patterns of infection, inherited genetics and other modulators of risk including chance and, probably, diet (Box 4). It has a logical coherence 139 and is grounded in the fundamental biology of leukaemia and evolutionary logic of the immune system network functions. The central thesis posits BCP-ALL as a paradox of progress in developed societies contingent upon a mismatch between the historical or evolutionary programming of the immune system and contemporary lifestyles that restrain opportunities for early life microbial exposures. Childhood ALL is probably not the only unanticipated, deleterious health consequence of diminished infectious exposure in infancy 93. Similar epidemiological associations exist for Hodgkin lymphoma in young adults 140 as well as for childhood allergies and autoimmune disease 141 (Supplementary Information S5 (Box)). In all these clinical situations, the common theme is that acquisition of common microbial infections in early life has an impact on later responses of the immune system to challenge and the subsequent presence or absence of pathology 24,93,142. Diminished exposure early in life to microbes that are pathological has been highly beneficial, reducing infant mortality, but it seems plausible that a suite of illnesses prevalent now in young people in more developed societies, including BCP-ALL, could be due to an unanticipated consequence of this advance 93.
Box 4. Other modulators of risk in childhood ALL.
In addition to patterns of infectious exposure and inherited genetics, other factors are likely to contribute to multi-factorial risk including diet and chance. For ALL as well as AML and most other paediatric cancers, risk is significantly and consistently elevated in association with higher birth weights 169,170 or, possibly, accelerated foetal growth 171. A plausible interpretation of this link is that higher weight, possibly orchestrated via IGF1 levels 172,173, may provide greater number of cells at risk. IGF1 potentiates expansion of B lineage progenitors 174. Recently, evidence has been presented, using mouse models of ALL, that a restricted diet can have a risk-reducing impact 175. Intermittent fasting was shown to block expansion of leukaemic cell populations and progression of disease. The effect operated via attenuation of leptin-receptor expression on leukaemic cells, possibly enforcing differentiation. Diet or calorie intake may, therefore, have a modulating impact on risk of ALL, reinforcing the likely multi-functional nature of causation of ALL, as in cancer in general.
Random events or chance get short shrift in cancer epidemiology but it has long been recognised that contingency and chance pervades all of biology 176. Some posit that a substantial fraction of cancers are due to chance alone 177 but this has been contentious. Chance is likely to be an ingredient in each and every cancer including childhood ALL. This is because inheritance of risk alleles is a lottery at conception, exposures including infections, at particular times, may or may not happen and mutational mechanisms alter genes independently of their function 178.
The infection hypothesis would benefit from further scrutiny including validation and extension of the animal modelling but its public health implication is clear. Most cases of childhood ALL are potentially preventable. But how? Promotion of lifestyle changes including day care attendance or protracted breastfeeding in the first year of life can be advocated but would be difficult to achieve. A more realistic prospect might be to design a prophylactic ‘vaccine’ that mimics the protective impact of natural infections in infancy, correcting the deficit in modern societies. Reconstitution or manipulation of the natural microbiome 143–146 or Helminth injections 147,148 are strategies under consideration for early life immune disorders in modern societies including autoimmune and allergic conditions. Oral administration of benign synbiotics (bacteria species such as lactobacillus and oligosaccharides) can have profound, and beneficial, modulating effects on the developing immune system 149. The results of those endeavours might inform approaches for preventing BCP-ALL. Cross collaboration of scientists working in disparate fields of early life immune dysfunction – allergy, autoimmune disease and ALL – would be beneficial.
Supplementary Material
Acknowledgements
I am very grateful for long term funding support from the Leukaemia Research Fund UK (now Bloodwise), The Kay Kendall Leukaemia Fund, the Wellcome Trust [105104/Z/14/Z] and The Institute of Cancer Research, London. My thanks to Richard Houlston for constructive comments on the manuscript.
I am indebted to many students and post-doctoral fellows in my laboratory who have researched this topic over many years, along with many scientists and clinicians who have been excellent collaborators. These include Anthony Ford, Joseph Wiemels, Ana Teresa Maia, Li Chong Chan, Caroline Bateman, Donat Alpar, Ian Titley, Sue Colman, Hiroshi Mori, Valeria Cazzaniga, Kristina Anderson, Nicola Potter, Lyndal Kearney and Richard Houlston (all at The Institute of Cancer Research, London), Andrew Lister, Tim Eden, Judith Chessells, Philip Ancliff, Ruth Jarrett, Arndt Borkhardt, Patricia Buffler, Eve Roman, Giovanni Cazzaniga, Maria Elena Cabrera, Maria Pombo de Oliveira, Andrea Biondi, John Kersey, Markus Müschen, Peter Campbell, Elli Papaemmanuil and Tariq Enver.
Footnotes
Author contributions
M. Greaves researched data for the article and wrote, reviewed and edited the manuscript.
Competing interests
The author declares no competing interests.
Reviewer information
Nature Reviews Cancer thanks K. Paulsson, S. Sallan and J. Wiemels for their contribution to the peer review of this work.
References
- 1.Parkin DM, et al. Vol. 87 IARC Scientific Publications; Lyon: 1988. [Google Scholar]
- 2.Pinkel D. In: White Blood. Personal journeys with childhood leukaemia. Greaves M, editor. World Scientific; 2008. pp. 13–46. [Google Scholar]
- 3.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381:1943–1955. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Essig S, et al. Risk of late effects of treatment in children newly diagnosed with standard-risk acute lymphoblastic leukaemia: a report from the Childhood Cancer Survivor Study cohort. Lancet Oncol. 2014;15:841–851. doi: 10.1016/S1470-2045(14)70265-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winther JF, Schmiegelow K. How safe is a standard-risk child with ALL? Lancet Oncol. 2014;15:782–783. doi: 10.1016/S1470-2045(14)70294-3. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia S, Robison LL. In: Hematology and Oncology of Infancy and Childhood. Orkin SH, editor. Vol. 2. Elsevier; 2015. pp. 1239–1256. [Google Scholar]
- 7.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 8.UK Childhood Cancer Study Investigators. The United Kingdom Childhood Cancer Study: objectives, materials and methods. Br J Cancer. 2000;82:1073–1102. doi: 10.1054/bjoc.1999.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metayer C, et al. The Childhood Leukemia International Consortium. Cancer Epidemiol. 2013;37:336–347. doi: 10.1016/j.canep.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preston DL, et al. Cancer incidence in atomic bomb survivors. Part III: Leukemia, lymphoma and multiple myeloma, 1950-1987. Radiat Res. 1994;137(Suppl):S68–S97. [PubMed] [Google Scholar]
- 11.Doll R, Wakeford R. Risk of childhood cancer from fetal irradiation. Br J Radiol. 1997;70:130–139. doi: 10.1259/bjr.70.830.9135438. [DOI] [PubMed] [Google Scholar]
- 12.Bartley K, Metayer C, Selvin S, Ducore J, Buffler P. Diagnostic X-rays and risk of childhood leukaemia. Int J Epidemiol. 2010;39:1628–1637. doi: 10.1093/ije/dyq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward G. The infective theory of acute leukaemia. Br J Child Dis. 1917;14:10–20. [Google Scholar]
- 14.Schulz TF, Neil JC. In: Leukemia. Henderson ES, Lister TA, Greaves MF, editors. Saunders; 2002. pp. 200–225. [Google Scholar]
- 15.Kinlen L. Evidence for an infective cause of childhood leukaemia: comparison of a Scottish New Town with nuclear reprocessing sites in Britain. Lancet. 1998;ii:1323–1327. doi: 10.1016/s0140-6736(88)90867-7. [DOI] [PubMed] [Google Scholar]
- 16.Kinlen L. Childhood leukaemia, nuclear sites, and population mixing. Br J Cancer. 2011;104:12–18. doi: 10.1038/sj.bjc.6605982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaves MF. Speculations on the cause of childhood acute lymphoblastic leukemia. Leukemia. 1988;2:120–125. [PubMed] [Google Scholar]
- 18.Torow N, Hornef MW. The neonatal window of opportunity: setting the stage for life-long host-microbial interaction and immune homeostasis. J Immunol. 2017;198:557–563. doi: 10.4049/jimmunol.1601253. [DOI] [PubMed] [Google Scholar]
- 19.Lim ES, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biesbroek G, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 22.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 24.Greaves M. Darwinian medicine: a case for cancer. Nat Rev Cancer. 2007;7:213–221. doi: 10.1038/nrc2071. [DOI] [PubMed] [Google Scholar]
- 25.Elinav E, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 26.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 27.Wiemels JL, Greaves M. Structure and possible mechanisms of TEL-AML1 gene fusions in childhood acute lymphoblastic leukemia. Cancer Res. 1999;59:4075–4082. [PubMed] [Google Scholar]
- 28.Rowley JD, Le Beau MM, Rabbitts TH. Springer International Publishing; Switzerland: 2015. [Google Scholar]
- 29.Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–2333. doi: 10.1182/blood-2002-12-3817. [DOI] [PubMed] [Google Scholar]
- 30.Wolman IJ. Parallel responses to chemotherapy in identical twin infants with concordant leukemia. J Pediatr. 1962;60:91–96. doi: 10.1016/s0022-3476(62)80014-6. [DOI] [PubMed] [Google Scholar]
- 31.Clarkson B, Boyse EA. Possible explanation of the high concordance for acute leukaemia in monozygotic twins. Lancet. 1971;i:699–701. doi: 10.1016/s0140-6736(71)92705-x. [DOI] [PubMed] [Google Scholar]
- 32.Strong SJ, Corney G. The Placenta in Twin Pregnancy. Pergamon Press; 1967. [Google Scholar]
- 33.Ford AM, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 34.Alpar D, et al. Clonal origins of ETV6-RUNX1(+) acute lymphoblastic leukemia: studies in monozygotic twins. Leukemia. 2015;29:839–846. doi: 10.1038/leu.2014.322. [DOI] [PubMed] [Google Scholar]
- 35.Bateman CM, et al. Acquisition of genome-wide copy number alterations in monozygotic twins with acute lymphoblastic leukemia. Blood. 2010;115:3553–3558. doi: 10.1182/blood-2009-10-251413. [DOI] [PubMed] [Google Scholar]
- 36.Cazzaniga G, et al. Developmental origins and impact of BCR-ABL1 fusion and IKZF1 deletions in monozygotic twins with Ph+ acute lymphoblastic leukemia. Blood. 2011;118:5559–5565. doi: 10.1182/blood-2011-07-366542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y, et al. Developmental timing of mutations revealed by whole-genome sequencing of twins with acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2013;110:7429–7433. doi: 10.1073/pnas.1221099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson AK, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–337. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobbins SE, et al. The silent mutational landscape of infant MLL-AF4 pro-B acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2013;52:954–960. doi: 10.1002/gcc.22090. [DOI] [PubMed] [Google Scholar]
- 40.Bateman CM, et al. Evolutionary trajectories of hyperdiploid ALL in monozygotic twins. Leukemia. 2015;29:58–65. doi: 10.1038/leu.2014.177. [DOI] [PubMed] [Google Scholar]
- 41.Hong D, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 42.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gale KB, et al. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci U S A. 1997;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiemels JL, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 46.Hjalgrim LL, et al. Presence of clone-specific markers at birth in children with acute lymphoblastic leukaemia. Br J Cancer. 2002;87:994–999. doi: 10.1038/sj.bjc.6600601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McHale CM, et al. Prenatal origin of TEL-AML1-positive acute lymphoblastic leukemia in children born in California. Genes Chromosomes Cancer. 2003;37:36–43. doi: 10.1002/gcc.10199. [DOI] [PubMed] [Google Scholar]
- 48.Wiemels JL, Ford AM, Van Wering ER, Postma A, Greaves M. Protracted and variable latency of acute lymphoblastic leukemia after TEL-AML1 gene fusion in utero. Blood. 1999;94:1057–1062. [PubMed] [Google Scholar]
- 49.Maia AT, et al. Protracted postnatal natural histories in childhood leukemia. Genes Chromosomes Cancer. 2004;39:335–340. doi: 10.1002/gcc.20003. [DOI] [PubMed] [Google Scholar]
- 50.Mori H, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci U S A. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lausten-Thomsen U, et al. Prevalence of t(12;21)[ETV6-RUNX1]-positive cells in healthy neonates. Blood. 2011;117:186–189. doi: 10.1182/blood-2010-05-282764. [DOI] [PubMed] [Google Scholar]
- 52.Zuna J, et al. ETV6/RUNX1 (TEL/AML1) is a frequent prenatal first hit in childhood leukemia. Blood. 2011;117:368–369. doi: 10.1182/blood-2010-09-309070. [DOI] [PubMed] [Google Scholar]
- 53.Skorvaga M, et al. Incidence of common preleukemic gene fusions in umbilical cord blood in Slovak population. PLoS One. 2014;9:e91116. doi: 10.1371/journal.pone.0091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schafer D, et al. Five percent of healthy newborns have an ETV6-RUNX1 fusion as revealed by DNA-based GIPFEL screening. Blood. 2018;131:821–826. doi: 10.1182/blood-2017-09-808402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beckwith JB, Perrin EV. In situ neuroblastomas: a contribution to the natural history of neural crest tumors. Am J Pathol. 1963;43:1089–1104. [PMC free article] [PubMed] [Google Scholar]
- 56.Charles AK, Brown KW, Berry PJ. Microdissecting the genetic events in nephrogenic rests and Wilms' tumor development. Am J Pathol. 1998;153:991–1000. doi: 10.1016/S0002-9440(10)65641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yagi T, et al. Detection of clonotypic IGH and TCR rearrangements in the neonatal blood spots of infants and children with B-cell precursor acute lymphoblastic leukemia. Blood. 2000;96:264–268. [PubMed] [Google Scholar]
- 58.Taub JW, et al. High frequency of leukemic clones in newborn screening blood samples of children with B-precursor acute lymphoblastic leukemia. Blood. 2002;99:2992–2996. doi: 10.1182/blood.v99.8.2992. [DOI] [PubMed] [Google Scholar]
- 59.Fasching K, et al. Presence of clone-specific antigen receptor gene rearrangements at birth indicates an in utero origin of diverse types of early childhood acute lymphoblastic leukemia. Blood. 2000;95:2722–2724. [PubMed] [Google Scholar]
- 60.Maia AT, et al. Identification of preleukemic precursors of hyperdiploid acute lymphoblastic leukemia in cord blood. Genes Chromosomes Cancer. 2004;40:38–43. doi: 10.1002/gcc.20010. [DOI] [PubMed] [Google Scholar]
- 61.Paulsson K, et al. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet. 2015;47:672–676. doi: 10.1038/ng.3301. [DOI] [PubMed] [Google Scholar]
- 62.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 64.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 66.Waanders E, et al. The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution. PLoS Genet. 2012;8:e1002533. doi: 10.1371/journal.pgen.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kharazmi E, et al. Familial risks for childhood acute lymphocytic leukaemia in Sweden and Finland: far exceeding the effects of known germline variants. Br J Haematol. 2012;159:585–588. doi: 10.1111/bjh.12069. [DOI] [PubMed] [Google Scholar]
- 69.Sinnett D, Krajinovic M, Labuda D. Genetic susceptibility to childhood acute lymphoblastic leukemia. Leuk Lymph. 2000;38:447–462. doi: 10.3109/10428190009059264. [DOI] [PubMed] [Google Scholar]
- 70.Vijayakrishnan J, Houlston RS. Candidate gene association studies and risk of childhood acute lymphoblastic leukemia: a systematic review and meta-analysis. Haematologica. 2010;95:1405–1414. doi: 10.3324/haematol.2010.022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moriyama T, Relling MV, Yang JJ. Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood. 2015;125:3988–3995. doi: 10.1182/blood-2014-12-580001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Studd JB, et al. Genetic and regulatory mechanism of susceptibility to high-hyperdiploid acute lymphoblastic leukaemia at 10p21.2. Nature communications. 2017;8 doi: 10.1038/ncomms14616. 14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer. 2017;17:692–704. doi: 10.1038/nrc.2017.82. [DOI] [PubMed] [Google Scholar]
- 74.Walsh KM, et al. A heritable missense polymorphism in CDKN2A confers strong risk of childhood acute lymphoblastic leukemia and is preferentially selected during clonal evolution. Cancer Res. 2015;75:4884–4894. doi: 10.1158/0008-5472.CAN-15-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, et al. Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nature communications. 2015;6 doi: 10.1038/ncomms8553. 7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor GM, et al. Strong association of the HLA-DP6 supertype with childhood leukaemia is due to a single allele, DPB1*0601. Leukemia. 2009;23:863–869. doi: 10.1038/leu.2008.374. [DOI] [PubMed] [Google Scholar]
- 77.Urayama KY, et al. HLA-DP genetic variation, proxies for early life immune modulation and childhood acute lymphoblastic leukemia risk. Blood. 2012;120:3039–3047. doi: 10.1182/blood-2012-01-404723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Urayama KY, et al. SNP association mapping across the extended major histocompatibility complex and risk of B-cell precursor acute lymphoblastic leukemia in children. PLoS One. 2013;8:e72557. doi: 10.1371/journal.pone.0072557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cloppenborg T, et al. Immunosurveillance of childhood ALL: polymorphic interferon-gamma alleles are associated with age at diagnosis and clinical risk groups. Leukemia. 2005;19:44–48. doi: 10.1038/sj.leu.2403553. [DOI] [PubMed] [Google Scholar]
- 80.Miedema KG, et al. Polymorphisms in the TLR6 gene associated with the inverse association between childhood acute lymphoblastic leukemia and atopic disease. Leukemia. 2012;26:1203–1210. doi: 10.1038/leu.2011.341. [DOI] [PubMed] [Google Scholar]
- 81.Almalte Z, et al. Novel associations between activating killer-cell immunoglobulin-like receptor genes and childhood leukemia. Blood. 2011;118:1323–1328. doi: 10.1182/blood-2010-10-313791. [DOI] [PubMed] [Google Scholar]
- 82.Shah S, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noetzli L, et al. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat Genet. 2015;47:535–538. doi: 10.1038/ng.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee P, Bhansali R, Izraeli S, Hijiya N, Crispino JD. The biology, pathogenesis and clinical aspects of acute lymphoblastic leukemia in children with Down syndrome. Leukemia. 2016;30:1816–1823. doi: 10.1038/leu.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lane AA, et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat Genet. 2014;46:618–623. doi: 10.1038/ng.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 88.Rudant J, et al. ARID5B, IKZF1 and non-genetic factors in the etiology of childhood acute lymphoblastic leukemia: the ESCALE study. PLoS One. 2015;10:e0121348. doi: 10.1371/journal.pone.0121348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 90.Paashuis-Lew YR, Heddle JA. Spontaneous mutation during fetal development and post-natal growth. Mutagenesis. 1998;13:613–617. doi: 10.1093/mutage/13.6.613. [DOI] [PubMed] [Google Scholar]
- 91.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci U S A. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marshall GM, et al. The prenatal origins of cancer. Nat Rev Cancer. 2014;14:277–289. doi: 10.1038/nrc3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rook GAW. Birkhäuser; Basel: 2009. [Google Scholar]
- 94.Goodman RA, Osterholm MT, Granoff DM, Pickering LK. Infectious diseases and child day care. Pediatr. 1984;74:134–139. [PubMed] [Google Scholar]
- 95.Gilham C, et al. Day care in infancy and risk of childhood acute lymphoblastic leukaemia: findings from a UK case-control study. Brit Med J. 2005;330:1294–1297. doi: 10.1136/bmj.38428.521042.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma X, et al. Daycare attendance and risk of childhood acute lymphoblastic leukaemia. Br J Cancer. 2002;86:1419–1424. doi: 10.1038/sj.bjc.6600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kamper-Jørgensen M, et al. Childcare in the first 2 years of life reduces the risk of childhood acute lymphoblastic leukemia. Leukemia. 2007;22:189–193. doi: 10.1038/sj.leu.2404884. [DOI] [PubMed] [Google Scholar]
- 98.Ajrouche R, et al. Childhood acute lymphoblastic leukaemia and indicators of early immune stimulation: the Estelle study (SFCE) Br J Cancer. 2015;112:1017–1026. doi: 10.1038/bjc.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rudant J, et al. Childhood acute lymphoblastic leukemia and indicators of early immune stimulation: a Childhood Leukemia International Consortium study. Am J Epidemiol. 2015;181:549–562. doi: 10.1093/aje/kwu298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Urayama KY, Buffler PA, Gallagher ER, Ayoob JM, Ma X. A meta-analysis of the association between day-care attendance and childhood acute lymphoblastic leukaemia. Int J Epidemiol. 2010;39:718–732. doi: 10.1093/ije/dyp378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Neglia JP, et al. Patterns of infection and day care utilization and risk factors of childhood acute lymphoblastic leukemia. Br J Cancer. 2000;82:234–240. doi: 10.1054/bjoc.1999.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Simpson J, Smith A, Ansell P, Roman E. Childhood leukaemia and infectious exposure: a report from the United Kingdom Childhood Cancer Study (UKCCS) Eur J Cancer. 2007;43:2396–2403. doi: 10.1016/j.ejca.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 103.Rudant J, et al. Childhood acute leukemia, early common infections, and allergy: The ESCALE Study. Am J Epidemiol. 2010;172:1015–1027. doi: 10.1093/aje/kwq233. [DOI] [PubMed] [Google Scholar]
- 104.Lin JN, et al. Risk of leukaemia in children infected with enterovirus: a nationwide, retrospective, population-based, Taiwanese-registry, cohort study. Lancet Oncol. 2015;16:1335–1343. doi: 10.1016/S1470-2045(15)00060-1. [DOI] [PubMed] [Google Scholar]
- 105.Urayama KY, et al. Early life exposure to infections and risk of childhood acute lymphoblastic leukemia. Int J Cancer. 2011;128:1632–1643. doi: 10.1002/ijc.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chan LC, et al. Is the timing of exposure to infection a major determinant of acute lymphoblastic leukaemia in Hong Kong? Paediatr Perinatal Epidemiol. 2002;16:154–165. doi: 10.1046/j.1365-3016.2002.00406.x. [DOI] [PubMed] [Google Scholar]
- 107.Dockerty JD, Draper G, Vincent T, Rowan SD, Bunch KJ. Case-control study of parental age, parity and socioeconomic level in relation to childhood cancers. Int J Epidemiol. 2001;30:1428–1437. doi: 10.1093/ije/30.6.1428. [DOI] [PubMed] [Google Scholar]
- 108.Ma X, et al. Vaccination history and risk of childhood leukaemia. Int J Epidemiol. 2005;34:1100–1109. doi: 10.1093/ije/dyi113. [DOI] [PubMed] [Google Scholar]
- 109.Shu XO, et al. Chloramphenicol use and childhood leukaemia in Shanghai. Lancet. 1987;2:934–937. doi: 10.1016/s0140-6736(87)91420-6. [DOI] [PubMed] [Google Scholar]
- 110.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Greenbaum S, et al. Cesarean delivery and childhood malignancies: a single-center, population-based cohort study. J Pediatr. 2018 doi: 10.1016/j.jpeds.2017.12.049. [DOI] [PubMed] [Google Scholar]
- 112.Wang R, et al. Cesarean section and risk of childhood acute lymphoblastic leukemia in a population-based, record-linkage study in California. Am J Epidemiol. 2017;185:96–105. doi: 10.1093/aje/kww153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marcotte EL, et al. Caesarean delivery and risk of childhood leukaemia: a pooled analysis from the Childhood Leukemia International Consortium (CLIC) Lancet Haematol. 2016;3:e176–185. doi: 10.1016/S2352-3026(16)00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H. Cesarean section and chronic immune disorders. Pediatrics. 2015;135:e92–98. doi: 10.1542/peds.2014-0596. [DOI] [PubMed] [Google Scholar]
- 115.Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 116.Amitay EL, Keinan-Boker L. Breastfeeding and childhood leukemia incidence: a meta-analysis and systematic review. JAMA Pediatr. 2015;169:e151025. doi: 10.1001/jamapediatrics.2015.1025. [DOI] [PubMed] [Google Scholar]
- 117.Shu XO, et al. Breast-feeding and risk of childhood acute leukemia. J Natl Cancer Inst. 1999;91:1765–1772. doi: 10.1093/jnci/91.20.1765. [DOI] [PubMed] [Google Scholar]
- 118.UK Childhood Cancer Study Investigators. Breastfeeding and childhood cancer. Br J Cancer. 2001;85:1685–1694. doi: 10.1054/bjoc.2001.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Heath CW, Jr, Hasterlik RJ. Leukemia among children in a suburban community. Am J Med. 1963;34:796–812. doi: 10.3322/canjclin.40.1.29. [DOI] [PubMed] [Google Scholar]
- 120.Francis SS, Selvin S, Yang W, Buffler PA, Wiemels JL. Unusual space-time patterning of the fallon, Nevada leukemia cluster: evidence of an infectious etiology. Chem Biol Interact. 2012;196:102–109. doi: 10.1016/j.cbi.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cazzaniga G, et al. Possible role of pandemic AH1N1 swine flu virus in a childhood leukemia cluster. Leukemia. 2017;31:1819–1821. doi: 10.1038/leu.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kulldorff M, Nagarwalla N. Spatial disease clusters: detection and inference. Stat Med. 1995;14:799–810. doi: 10.1002/sim.4780140809. [DOI] [PubMed] [Google Scholar]
- 123.Kroll ME, Stiller CA, Murphy MF, Carpenter LM. Childhood leukaemia and socioeconomic status in England and Wales 1976-2005: evidence of higher incidence in relatively affluent communities persists over time. Br J Cancer. 2011;105:1783–1787. doi: 10.1038/bjc.2011.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Francis SS, et al. In utero cytomegalovirus infection and development of childhood acute lymphoblastic leukemia. Blood. 2017;129:1680–1684. doi: 10.1182/blood-2016-07-723148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ford AM, et al. The TEL-AML1 leukemia fusion gene dysregulates the TGFβ pathway in early B lineage progenitor cells. J Clin Invest. 2009;119:826–836. doi: 10.1172/JCI36428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Swaminathan S, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16:766–774. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Olinski R, Styczynski J, Olinska E, Gackowski D. Viral infection-oxidative stress/DNA damage-aberrant DNA methylation: separate or interrelated events responsible for genetic instability and childhood ALL development? Biochim Biophys Acta. 2014;1846:226–231. doi: 10.1016/j.bbcan.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 128.de Yébenes V, Ramiro AR. Activation-induced deaminase: light and dark sides. Trends Mol Med. 2006;12:432–439. doi: 10.1016/j.molmed.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 129.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chesi M, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rosenberg BR, Papavasiliou FN. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection. Adv Immunol. 2007;94:215–244. doi: 10.1016/S0065-2776(06)94007-3. [DOI] [PubMed] [Google Scholar]
- 132.Pickup M, Novitskiy S, Moses HL. The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rouce RH, et al. The TGF-beta/SMAD pathway is an important mechanism for NK cell immune evasion in childhood B-acute lymphoblastic leukemia. Leukemia. 2016;30:800–811. doi: 10.1038/leu.2015.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martin-Lorenzo A, et al. Infection exposure is a causal factor in B-cell precursor acute lymphoblastic leukemia as a result of Pax5-inherited susceptibility. Cancer discovery. 2015;5:1328–1343. doi: 10.1158/2159-8290.CD-15-0892. [DOI] [PubMed] [Google Scholar]
- 135.Rodriguez-Hernandez G, et al. Infection exposure promotes ETV6-RUNX1 precursor B-cell leukemia via impaired H3K4 demethylases. Cancer Res. 2017;77:4365–4377. doi: 10.1158/0008-5472.CAN-17-0701. [DOI] [PubMed] [Google Scholar]
- 136.Fidanza M, et al. Inhibition of precursor B-cell malignancy progression by toll-like receptor ligand-induced immune responses. Leukemia. 2016;30:2116–2119. doi: 10.1038/leu.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Poynton FJ, Thursfield H, Paterson D. The severe blood diseases of childhood: a series of observations from the Hospital for Sick Children, Great Ormond Street. Br J Child Dis. 1922;XIX:128–144. [Google Scholar]
- 138.Holm J, et al. Assessment of breast cancer risk factors reveals subtype heterogeneity. Cancer Res. 2017;77:3708–3717. doi: 10.1158/0008-5472.CAN-16-2574. [DOI] [PubMed] [Google Scholar]
- 139.Thagard P. How scientists explain disease. Princeton University Press; 1999. [Google Scholar]
- 140.Cozen W, et al. A protective role for early oral exposures in the etiology of young adult Hodgkin lymphoma. Blood. 2009;114:4014–4020. doi: 10.1182/blood-2009-03-209601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 142.Dunne DW, Cooke A. A worm's eye view of the immune system: consequences for evolution of human autoimmune disease. Nat Rev Immunol. 2005;5:420–426. doi: 10.1038/nri1601. [DOI] [PubMed] [Google Scholar]
- 143.Parker W, Ollerton J. Evolutionary biology and anthropology suggest biome reconstitution as a necessary approach toward dealing with immune disorders. Evol Med Public Health. 2013;2013:89–103. doi: 10.1093/emph/eot008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marino E, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18:552–562. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 145.Durack J, et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nature communications. 2018;9 doi: 10.1038/s41467-018-03157-4. 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 147.Elliott DE, Weinstock JV. Helminth-host immunological interactions: prevention and control of immune-mediated diseases. Ann N Y Acad Sci. 2012;1247:83–96. doi: 10.1111/j.1749-6632.2011.06292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Navarro S, et al. Hookworm recombinant protein promotes regulatory T cell responses that suppress experimental asthma. Sci Transl Med. 2016;8 doi: 10.1126/scitranslmed.aaf8807. 362ra143. [DOI] [PubMed] [Google Scholar]
- 149.Panigrahi P, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–412. doi: 10.1038/nature23480. [DOI] [PubMed] [Google Scholar]
- 150.Anderson RM, May RM. Immunisation and herd immunity. Lancet. 1990;335:641–645. doi: 10.1016/0140-6736(90)90420-a. [DOI] [PubMed] [Google Scholar]
- 151.Bellec S, et al. Childhood leukaemia and population movements in France, 1990-2003. Br J Cancer. 2008;98:225–231. doi: 10.1038/sj.bjc.6604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stiller CA, Kroll ME, Boyle PJ, Feng Z. Population mixing, socioeconomic status and incidence of childhood acute lymphoblastic leukaemia in England and Wales: analysis by census ward. Br J Cancer. 2008;98:1006–1011. doi: 10.1038/sj.bjc.6604237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dickinson HO, Hammal DM, Bithell JF, Parker L. Population mixing and childhood leukaemia and non-Hodgkin's lymphoma in census wards in England and Wales, 1966-87. Br J Cancer. 2002;86:1411–1413. doi: 10.1038/sj.bjc.6600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sabaawy HE, et al. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci, USA. 2006;103:15166–15171. doi: 10.1073/pnas.0603349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bernardin F, et al. TEL-AML1, expressed from t(12;21) in human acute lymphocytic leukemia, induces acute leukemia in mice. Cancer Res. 2002;62:3904–3908. [PubMed] [Google Scholar]
- 156.Morrow M, Horton S, Kioussis D, Brady HJM, Williams O. TEL-AML1 promotes development of specific hematopoietic lineages consistent with preleukemic activity. Blood. 2004;103:3890–3896. doi: 10.1182/blood-2003-10-3695. [DOI] [PubMed] [Google Scholar]
- 157.Tsuzuki S, Seto M, Greaves M, Enver T. Modelling first-hit functions of the t(12;21) TEL-AML1 translocation in mice. Proc Natl Acad Sci, U S A. 2004;101:8443–8448. doi: 10.1073/pnas.0402063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fischer M, et al. Defining the oncogenic function of the TEL/AML1 (ETV6/RUNX1) fusion protein in a mouse model. Oncogene. 2005;24:7579–7591. doi: 10.1038/sj.onc.1208931. [DOI] [PubMed] [Google Scholar]
- 159.Schindler JW, et al. TEL-AML1 corrupts hematopoietic stem cells to persist in the bone marrow and initiate leukemia. Cell Stem Cell. 2009;5:43–53. doi: 10.1016/j.stem.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 160.van der Weyden L, et al. Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice. Blood. 2011;118:1041–1051. doi: 10.1182/blood-2011-02-338848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.van der Weyden L, et al. Somatic drivers of B-ALL in a model of ETV6-RUNX1; Pax5(+/-) leukemia. BMC Cancer. 2015;15:585. doi: 10.1186/s12885-015-1586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li M, et al. Initially disadvantaged, TEL-AML1 cells expand and initiate leukemia in response to irradiation and cooperating mutations. Leukemia. 2013;27:1570–1573. doi: 10.1038/leu.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Inthal A, et al. Role of the erythropoietin receptor in ETV6/RUNX1-positive acute lymphoblastic leukemia. Clin Cancer Res. 2008;14:7196–7204. doi: 10.1158/1078-0432.CCR-07-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Torrano V, Procter J, Cardus P, Greaves M, Ford AM. ETV6-RUNX1 promotes survival of early B lineage progenitor cells via a dysregulated erythropoietin receptor. Blood. 2011;118:4910–4918. doi: 10.1182/blood-2011-05-354266. [DOI] [PubMed] [Google Scholar]
- 165.Liu GJ, et al. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes & development. 2014;28:1337–1350. doi: 10.1101/gad.240416.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood. 2015;125:3977–3987. doi: 10.1182/blood-2015-02-580043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li Z, et al. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37:613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- 168.Boiers C, et al. A human IPS model implicates embryonic B-myeloid fate restriction as developmental susceptibility to B acute lymphoblastic leukemia-associated ETV6-RUNX1. Dev Cell. 2018;44:362–377 e367. doi: 10.1016/j.devcel.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hjalgrim LL, et al. Birth weight as a risk factor for childhood leukemia: a meta-analysis of 18 epidemiologic studies. Am J Epidemiol. 2003;158:724–735. doi: 10.1093/aje/kwg210. [DOI] [PubMed] [Google Scholar]
- 170.Roman E, et al. Childhood acute lymphoblastic leukaemia and birthweight: insights from a pooled analysis of case-control data from Germany, the United Kingdom and the United States. Eur J Cancer. 2013;49:1437–1447. doi: 10.1016/j.ejca.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 171.Milne E, et al. Fetal growth and childhood acute lymphoblastic leukemia: findings from the childhood leukemia international consortium. Int J Cancer. 2013;133:2968–2979. doi: 10.1002/ijc.28314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chokkalingam AP, et al. Fetal growth and body size genes and risk of childhood acute lymphoblastic leukemia. Cancer Causes Control. 2012;23:1577–1585. doi: 10.1007/s10552-012-00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tower RL, Spector LG. The epidemiology of childhood leukemia with a focus on birth weight and diet. Crit Rev Clin Lab Sci. 2007;44:203–242. doi: 10.1080/10408360601147536. [DOI] [PubMed] [Google Scholar]
- 174.Gibson LF, Piktel D, Landreth KS. Insulin-like growth factor-1 potentiates expansion of Interleukin-7-dependent Pro-B cells. Blood. 1993;l82:3005–3011. [PubMed] [Google Scholar]
- 175.Lu Z, et al. Fasting selectively blocks development of acute lymphoblastic leukemia via leptin-receptor upregulation. Nat Med. 2017;23:79–90. doi: 10.1038/nm.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Monod J. Chance and Necessity. Alfred A. Knopf; 1970. [Google Scholar]
- 177.Tomasetti C, Vogelstein B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Greaves M. Evolutionary determinants of cancer. Cancer discovery. 2015;5:806–820. doi: 10.1158/2159-8290.CD-15-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35:975–983. doi: 10.1200/JCO.2016.70.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nature reviews Clinical oncology. 2015;12:344–357. doi: 10.1038/nrclinonc.2015.38. [DOI] [PubMed] [Google Scholar]
- 181.Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16:494–507. doi: 10.1038/nrc.2016.63. [DOI] [PubMed] [Google Scholar]
- 182.Greaves MF, Pegram SM, Chan LC. Collaborative group study of the epidemiology of acute lymphoblastic leukaemia subtypes: background and first report. Leuk Res. 1985;9:715–733. doi: 10.1016/0145-2126(85)90281-4. [DOI] [PubMed] [Google Scholar]
- 183.Greaves MF, Janossy G, Peto J, Kay H. Immunologically defined subclasses of acute lymphoblastic leukaemia in children: their relationship to presentation features and prognosis. Br J Haematol. 1981;48:179–197. [PubMed] [Google Scholar]
- 184.Shah A, Coleman MP. Increasing incidence of childhood leukaemia: a controversy re-examined. Br J Cancer. 2007;97:1009–1012. doi: 10.1038/sj.bjc.6603946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Spix C, Eletr D, Blettner M, Kaatsch P. Temporal trends in the incidence rate of childhood cancer in Germany 1987-2004. Int J Cancer. 2008;122:1859–1867. doi: 10.1002/ijc.23281. [DOI] [PubMed] [Google Scholar]
- 186.Steliarova-Foucher E, et al. Trends in childhood cancer incidence in Europe, 1970-99. Lancet. 2005;365:2088. doi: 10.1016/S0140-6736(05)66728-1. [DOI] [PubMed] [Google Scholar]
- 187.Chessells JM, Hardisty RM, Rapson NT, Greaves MF. Acute lymphoblastic leukaemia in children: classification and prognosis. Lancet. 1977;ii:1307–1309. doi: 10.1016/s0140-6736(77)90361-0. [DOI] [PubMed] [Google Scholar]
- 188.Sallan SE, et al. Cell surface antigens: prognostic implications in childhood acute lymphoblastic leukemia. Blood. 1980;55:395–402. [PubMed] [Google Scholar]
- 189.Greaves M. In: Nathan and Oski's Hematology and Oncology of Infancy and Childhood Vol. 2. Orkin SH, et al., editors. Ch. 39. Elsevier Saunders; 2015. pp. 1229–1238. [Google Scholar]
- 190.Papaemmanuil E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Trevino LR, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hungate EA, et al. A variant at 9p21.3 functionally implicates CDKN2B in paediatric B-cell precursor acute lymphoblastic leukaemia aetiology. Nature communications. 2016;7 doi: 10.1038/ncomms10635. 10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vijayakrishnan J, et al. The 9p21.3 risk of childhood acute lymphoblastic leukaemia is explained by a rare high-impact variant in CDKN2A. Sci Rep. 2015;5 doi: 10.1038/srep15065. 15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wiemels JL, et al. A functional polymorphism in the CEBPE gene promoter influences acute lymphoblastic leukemia risk through interaction with the hematopoietic transcription factor Ikaros. Leukemia. 2016;30:1194–1197. doi: 10.1038/leu.2015.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Migliorini G, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122:3298–3307. doi: 10.1182/blood-2013-03-491316. doi:blood-2013-03-491316 [pii] [DOI] [PubMed] [Google Scholar]
- 196.Perez-Andreu V, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet. 2013;45:1494–1498. doi: 10.1038/ng.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ellinghaus E, et al. Identification of germline susceptibility loci in ETV6-RUNX1-rearranged childhood acute lymphoblastic leukemia. Leukemia. 2012;26:902–909. doi: 10.1038/leu.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vijayakrishnan J, et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia. 2017;31:573–579. doi: 10.1038/leu.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Vijayakrishnan J, et al. Genome-wide association study identifies new susceptibility loci for B-cell childhood acute lymphoblastic leukemia. Nature Comm. 2018 doi: 10.1038/s41467-018-03178-z. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wiemels JL, et al. GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nature communications. 2018;9 doi: 10.1038/s41467-017-02596-9. 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.