Abstract
Approximately 50% of early-stage non-small cell lung cancer (NSCLC) patients that undergo surgery with curative intent will relapse within 5 years1,2. Detection of circulating tumour cells (CTCs) at the time of surgery may represent a tool to identify patients at higher risk of recurrence where more frequent monitoring is advised. Here, we asked whether CellSearch detected pulmonary venous CTCs (PV-CTCs) at surgical resection of early stage NSCLC represent subclones responsible for subsequent disease relapse. PV-CTCs were detected in 48% of 100 patients enrolled into the TRACERx study3 and were associated with lung cancer specific relapse, and remained an independent predictor of relapse in multivariate analysis adjusted for tumour stage. In a case study, genomic profiling of single PV-CTCs collected at surgery revealed a higher mutation overlap with a metastasis detected 10 months later (91%) compared to the primary tumour (79%), suggesting that early disseminating PV-CTCs were responsible for disease relapse. Together, PV-CTC enumeration and genomic profiling highlight the potential of PV-CTCs as early predictors of NSCLC recurrence after surgery. However, limited sensitivity of PV-CTCs to predict relapse suggests further studies using a larger, independent cohort are warranted to confirm and better define this potential clinical utility of PV-CTCs in early stage NSCLC.
Lung cancer is the leading cause of cancer related deaths worldwide with a 5 year relative survival rate of 4% in the metastatic setting4. NSCLC is the most common form of lung cancer. Patients presenting with early-stage NSCLC may undergo surgery with or without adjuvant chemotherapy and/or adjuvant radiotherapy in an attempt to achieve cure. However, disease recurrence following surgery is common, with 5-year relapse rates ranging from ~20% in patients with stage I disease to ~50% in those with stage III disease1,2.
Strategies to understand the biology of early dissemination and to identify patients at high risk of relapse may inform novel therapeutic approaches for adjuvant treatment to improve cure rates. CTCs are the assumed ‘foundations of metastasis’5, though this has not been formally proven in NSCLC. CTCs enriched from breast cancer, melanoma, NSCLC and small cell lung cancer (SCLC) patients’ peripheral blood can form tumours in immune compromised mice confirming their tumorigenic potential6–9. CTC number, measured using the CellSearch® platform, is a Food and Drug Administration (FDA) approved prognostic test in breast, colorectal and prostate cancers and is also prognostic in NSCLC10. Although peripheral blood CTCs (using CellSearch that captures only cells expressing EpCAM and Cytokeratin) are rare in early stage NSCLC patients, we previously demonstrated in a pilot study that CellSearch CTCs obtained from the draining pulmonary vein of the cancer-affected lung (PV-CTCs) are more frequent and we observed a trend towards worse disease-free survival (DFS) and overall survival (OS)11. To determine whether our preliminary findings that PV-CTCs at resection are associated with relapse holds in a larger patient cohort, we enumerated PV-CTCs from 100 NSCLC patients enrolled onto the TRACERx study12.
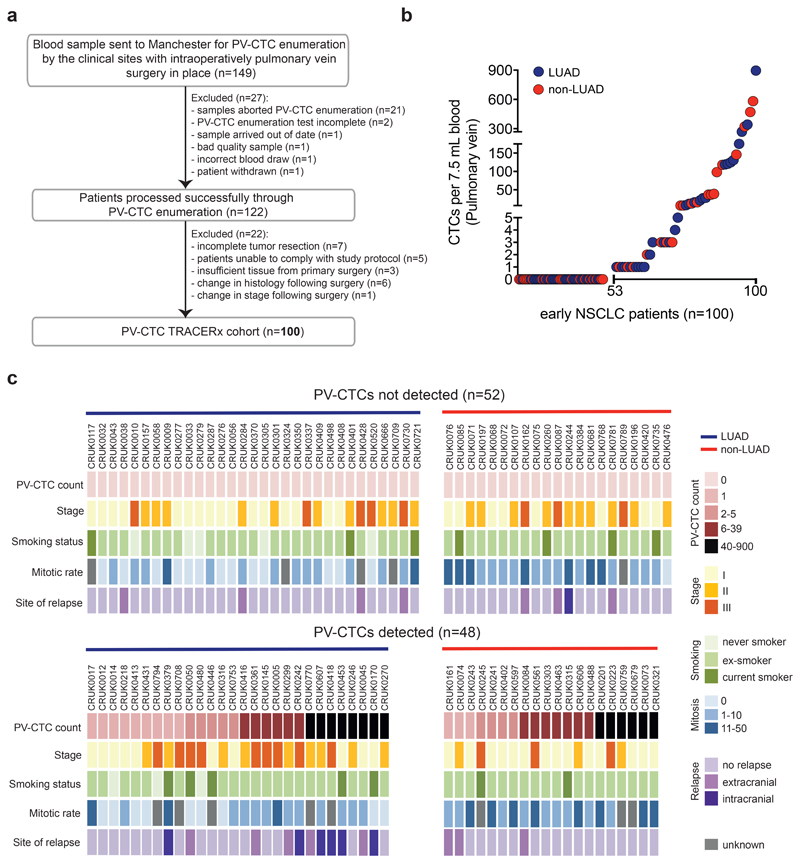
In our current cohort of 100 TRACERx patients (46% stage I, 34% stage II and 20% stage III; median follow-up 993 days), (Figure 1a, Table 1 and Supplementary Table 1), 48% (48/100) harboured at least 1 PV-CTC per/7.5mL blood (mean ± SD, 42.2 ± 127.3, median 0, range 0-896) (Figure 1b). PV-CTC count was not significantly associated with clinicopathological factors such as age, gender, pathological stage, smoking status and treatment received (Figure 1c and Supplementary Table 2). In contrast to circulating tumour DNA (ctDNA)13, PV-CTC count was not significantly different between adenocarcinoma (LUAD) and non-LUAD (p=0.554, t-test) suggesting that factors that control release of ctDNA and the dissemination of intact CTCs are distinct.
Fig. 1. PV-CTC detection in early NSCLC.
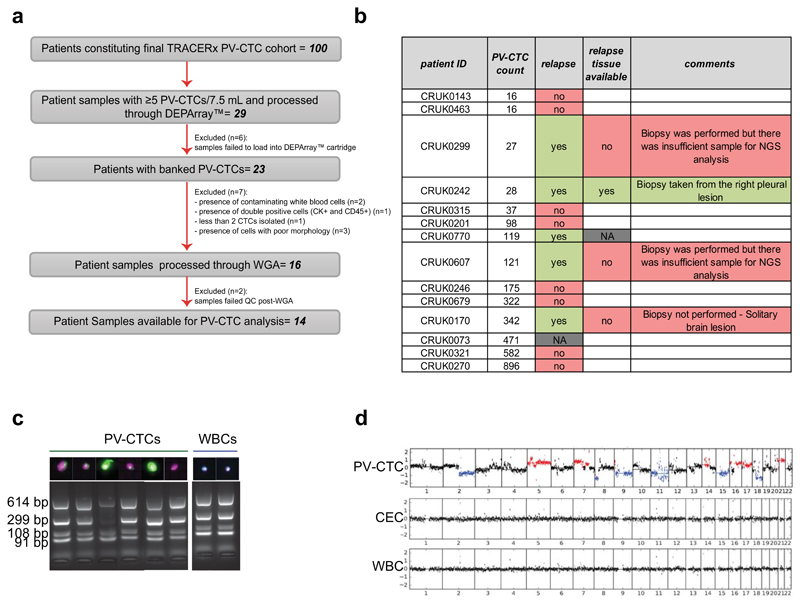
a, TRACERx consort diagram. 149 patients consented for pulmonary vein blood sampling between June 2014 and March 2017. 27 samples were excluded because of failures in CellSearch® enrichment and enumeration. 22 patients were defined ineligible post-surgery and the remaining 100 patients constituted the final cohort for PV-CTC enumeration. b, Distribution of the number of PV-CTCs enumerated by CellSearch® from 100 patients with early NSCLC. LUAD (blue circle) and non-LUAD (red circle) patients are indicated. c, Heat map showing clinicopathological and PV-CTC detection data; Patients are stratified according to PV-CTC detection. Histological disease type is indicated by coloured bar above the heatmap.
Table 1. Baseline characteristics of 100 patients and presence of PV-CTCs.
| Characteristics | PV-CTC positive, n (%) | PV-CTC negative, n (%) |
|---|---|---|
| Age, in years | ||
| Average age | 68 | 67 |
| Range | 39-85 | 48-82 |
| Gender | ||
| Male (n=61) | 28 (46%) | 33 (54%) |
| Female (n=39) | 20 (51%) | 19 (49%) |
| Tumour Histology | ||
| Adenocarcinoma (n=59) | 28 (47%) | 31 (53%) |
| Non-adenocarcinoma (n=43) | 20 (47%) | 23 (53%) |
| Pathological Stage | ||
| I (n=47) | 22 (47%) | 25 (53%) |
| II (n=34) | 15 (44%) | 19 (56%) |
| III (n=19) | 11 (58%) | 8 (42%) |
| Smoking Status | ||
| Current smokers (n=14) | 7 (50%) | 7 (50%) |
| ex smokers (n=78) | 38 (49%) | 40 (51%) |
| never smokers (n=8) | 3 (37%) | 5 (63%) |
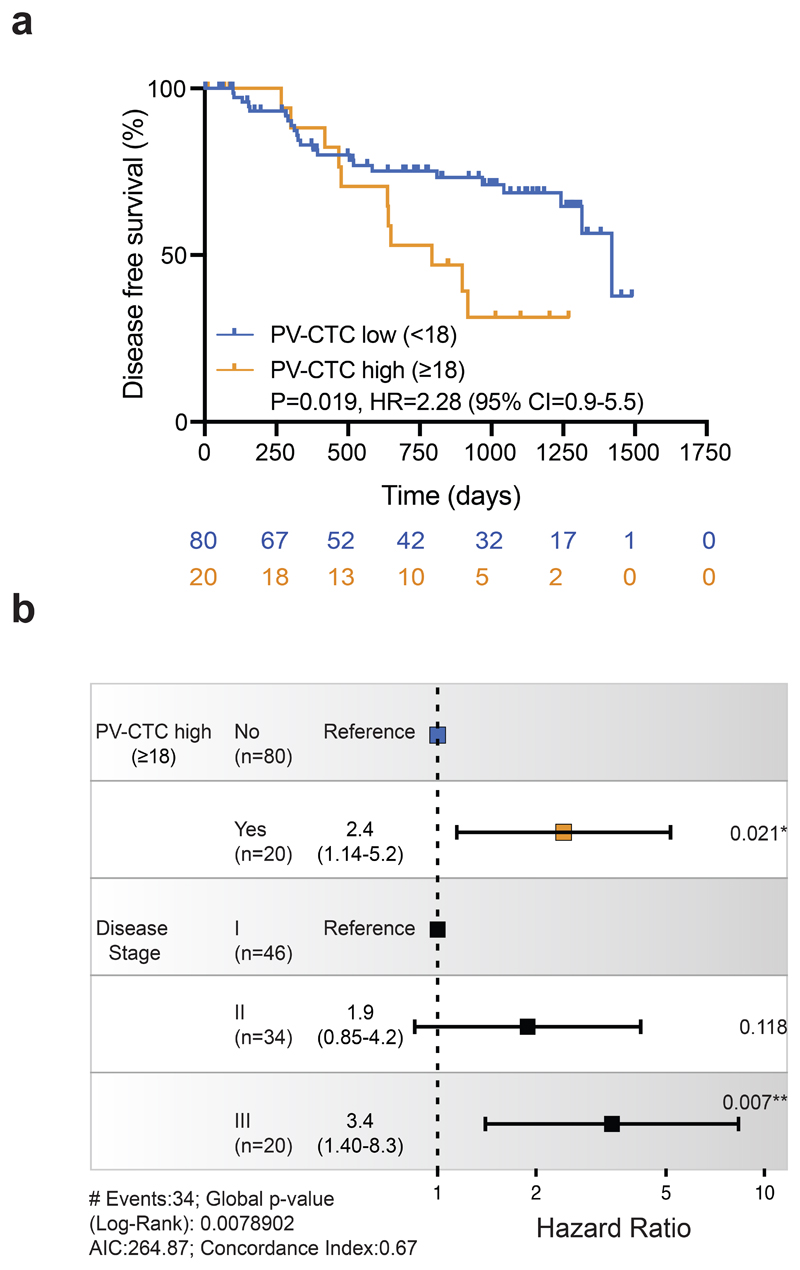
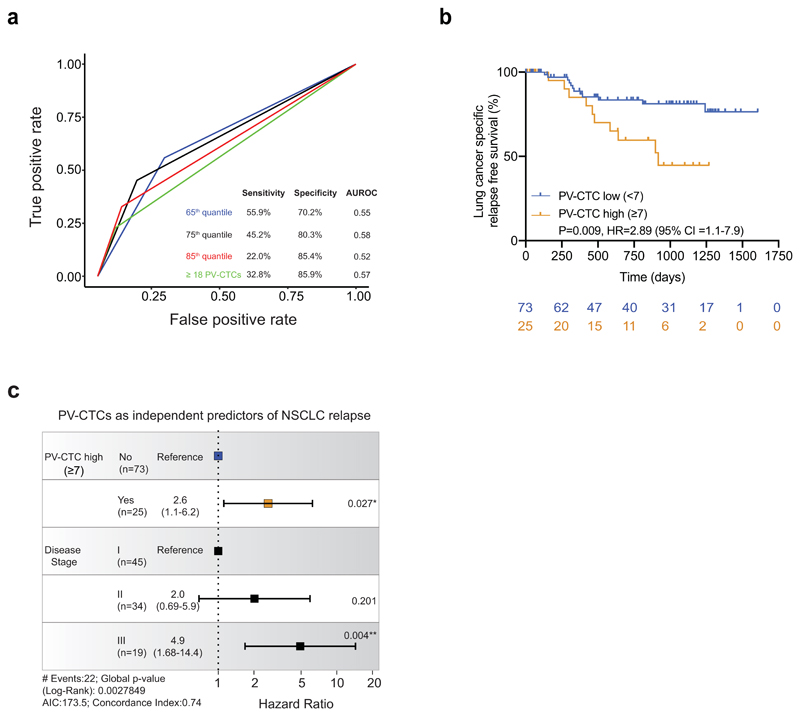
In our previous pilot study of 30 patients11, there was an association between the PV-CTC 'high' count (≥18 PV-CTCs/7.5ml blood) and DFS (p=0.055). When we applied this cutpoint in the TRACERx cohort there was a significant association with poorer DFS (p=0.019 log-rank, HR=2.28, Figure 2a) and this remained an independent predictor in multivariate analysis when adjusted for tumour stage (p=0.021, HR=2.4, 95% CI 1.14-5.2, Figure 2b). However, the performance of this cutpoint in predicting DFS, defined by time-dependent receiver operating characteristic (tdROC) curves, revealed limited sensitivity (sensitivity = 31.7%, specificity = 84.9%). We therefore conducted futher exploratory analysis to refine the ‘PV-CTC high’ cutpoint to predict lung cancer specific relapse events and investigate the biological relevance of PV-CTCs in NSCLC metastasis. Briefly, of the 37 recorded DFS events in the TRACERx cohort, 22 were due to lung cancer specific relapse. The remaining events occurred either without evidence of lung cancer relapse before death (n=9, Supplementary Table 3) due to a second non-lung primary cancer (n=4, confirmed by histology, imaging and clinical discussion, Supplementary Table 3) or lacked sufficient clinical information to determine cause (n=2, Supplementary Table 3). tdROC curves showed that the sensitivity and specificity in predicting lung cancer specific relapse at two years was optimal when a 75th quantile cutpoint was applied (≥7 PV-CTCs/7.5ml blood, Extended Data Fig.1a). A ‘PV-CTC high’ status of ≥7 PV-CTCs/7.5ml blood showed significant association with lung cancer relapse in Kaplan-Meier analysis (p=0.009 log-rank, HR=2.78, Extended Data Fig.1b) and remained an independent predictor in multivariate analysis when adjusted for tumour stage (p=0.027, HR = 2.6, 95% CI 1.1-6.2, Extended Data Fig.1c).
Fig.2. PV-CTCs as independent predictors of disease-free survival.
a, Kaplan–Meier curves showing disease-free survival (DFS) of 100 patients stratified as PV-CTC high or low based on the previously published threshold from our pilot study (≥18 PV-CTCs/7.5ml blood)11. The number of patients at risk for every time point is indicated below the time point and colour coded according to the high or low groups. P value, HR and relative 95% confidence intervals (CI) (two-sided log-rank test) are indicated. b, Forest plot showing the results of multivariable regression analysis for PV-CTC high or low patients (≥18 PV-CTCs/7.5ml blood). The x-axis represents the hazard ratio with the reference line (dashed) and significance is calculated using a Cox proportional hazards model. The estimated hazard ratios and their 95% CI are presented as error bars. The log-rank test used was two-sided.
Analysis of PV-CTCs as a continuous variable showed that each doubling of PV-CTC count was a significant prognostic factor for DFS when modelled as a sole covariate (p=0.035, HR=1.113) and when modelled with other significant prognostic factors (p=0.040, HR=1.116, 95% CI 1.005-1.239, Wald test two-sided, Supplementary Table 4). Each doubling of PV-CTC count was also significantly associated with lung cancer specific relapse in both uni-variate (p=0.029, HR=1.148) and multi-variate analysis (p=0.024, HR=1.170, 95% CI 1.021-1.341, Wald test two-sided, Supplementary Table 5). We also noted a significant association between PV-CTCs as a continuous variable and intracranial disease present at clinical relapse (p=0.028, t-test two-sided, Supplementary Table 2).
Collectively, these data raise the possibility that patients with a ‘high’ CellSearch PV-CTC count at resection may benefit from increased minimal residual disease (MRD) monitoring post-surgery. We have shown that increasing PV-CTC count as a continuous variable is associated with poor prognosis. To use PV-CTCs in a clinical setting, a pre-defined cutpoint will be required to prospectively stratify patients. Although the previously defined cutpoint of ≥18 PV-CTCs/7.5ml blood11 was verified here and the further exploratory analysis of a ≥7 PV-CTC/7.5ml blood increased the performance of PV-CTCs in predicting lung cancer specific relapse, sensitivity remained modest (45.2% for ≥7 PV-CTCs/7.5ml blood vs 32.8% for ≥18 PV-CTCs/7.5ml blood, Extended Data Fig.2a) and further studies are clearly required before clinical utility can be evaluated.
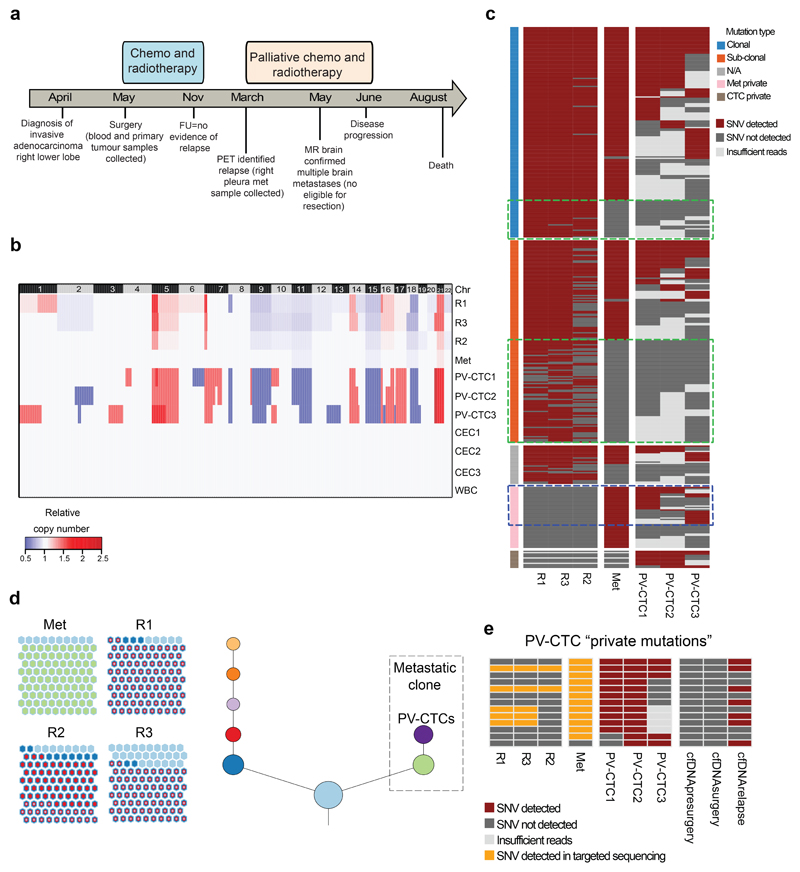
We next sought to assess the degree of genomic similarity between early-disseminated PV-CTCs and metastatic disease by comparing the primary tumour, the PV-CTCs and subsequent metastatic disease from the same patient. Single PV-CTCs and white blood cell (WBCs) controls were successfully isolated from 14 patients (Extended Data Fig. 2a and online methods). Of these 14 patients, five experienced a lung cancer specific relapse event (Extended Data Fig.2b); with an evaluable metastatic tissue biopsy available for one patient (CRUK0242). This 74-year old male was diagnosed with stage IIIA, invasive adenocarcinoma in the right lung and underwent tumour resection, at which point 28 PV-CTCs were detected. The patient received adjuvant chemotherapy and radiotherapy and at 10 months post-surgery, positron-emission tomography (PET) identified relapse involving the right pleura. At this time a biopsy from the right pleural lesion was sequenced and peripheral blood samples collected for circulating free DNA (cfDNA) analysis. After receiving palliative chemotherapy and radiotherapy, the patient progressed and died the following year (Figure 3a). In this case study, three spatially-separated primary tumour regions, PV-CTCs, cfDNA isolated from pulmonary and peripheral veins at resection and again from the periphery at disease relapse, and the pleural metastasis were genetically profiled and compared.
Fig.3. Mutations present in the relapse tumour are detected 10 months earlier in PV-CTCs and not in the primary tumour.
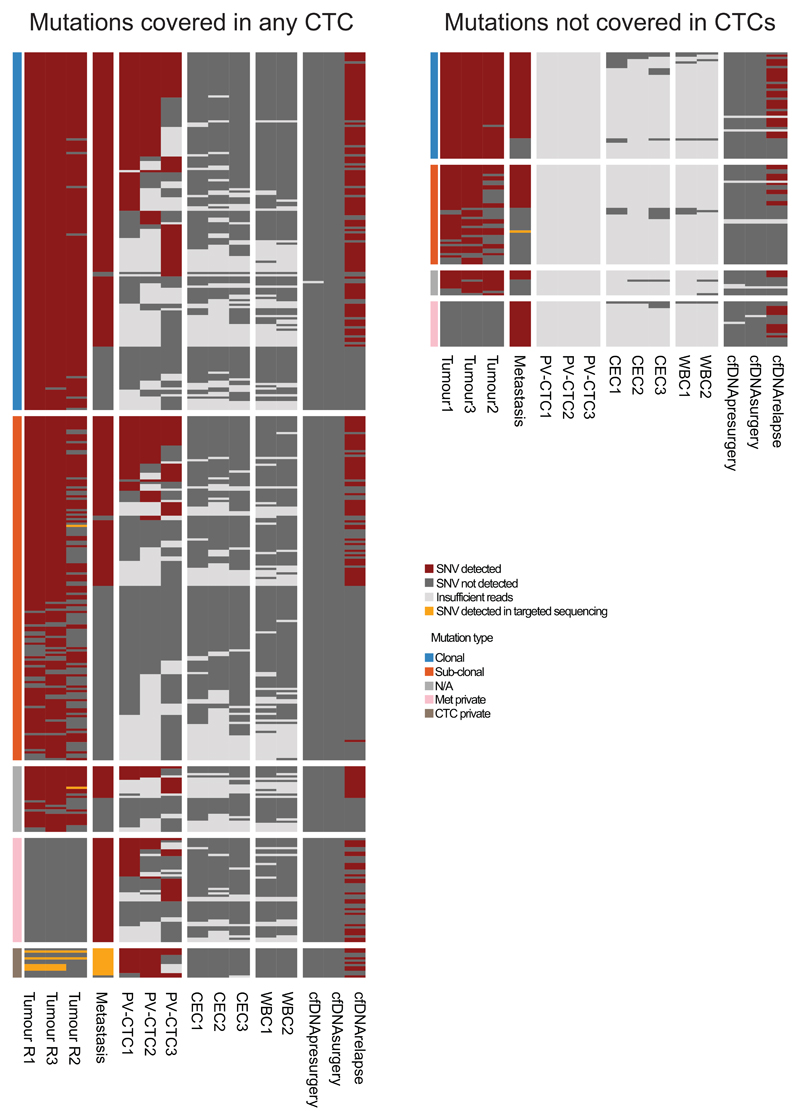
a, Patient timeline from diagnosis to death (FU=follow up; PET=positron emission tomography; MR=magnetic resonance). b, Heat map showing the comparison between CNA detected in PV-CTCs or circulating epithelial cells (CECs), in primary tumour regions (R1-3), in relapse tumour (Met) and in a WBC control. Regions of loss are coloured blue, regions of gain are coloured red. Chromosomes are indicated at the top of the figure. c, Heat map showing the comparison of SNVs detected in PV-CTCs, primary tumour regions and the metastasis. Mutations are ordered according to their clonality as established by primary tumour analysis. Green dashed boxes indicate mutations that are seen in the primary tumour, but not metastasis or PV-CTCs. Blue dashed box indicates the overlap between mutations considered metastatic private by primary tumour analysis and PV-CTCs. No mutations were found in the three CECs and two WBCs. d, Evolutionary tree encompassing tumour and PV-CTCs: the relationships between identified subclones is depicted, with size of circle reflecting the number of mutations in each subclone relative to largest. Length of lines connecting tumor subclones does not carry information. The beehive plots indicate the subclonal architecture of each tumour region, with 100 representative cells shown for each region and the nested colours corresponding to the ancestry of each cell. e, Heat map showing PV-CTC private mutations that are detected in primary tumour, metastasis and cfDNA following targeted deep sequencing.
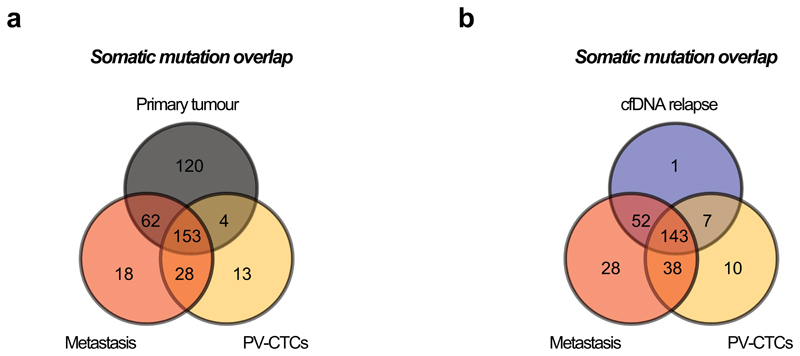
From the 28 PV-CTCs detected by CellSearch, we successfully isolated and amplified six single PV-CTCs (Extended Data Fig. 2c). Low-pass whole genome sequencing was performed which revealed that 3/6 PV-CTCs harboured copy number alterations (CNA) that matched the primary tumour. The remaining cells, although phenotypically CTC candidates by CellSearch criteria, showed flat copy number profiles as observed in WBC controls (Figure 3b, Extended Data Fig. 2d). We have termed these cells 'circulating epithelial cells' (CECs) and propose these are likely to be normal epithelial cells that enter the blood along with PV-CTCs; similar cells have recently been described in non-cancer patients14. In order to identify somatic mutations present in the PV-CTCs, we performed whole exome sequencing (WES) followed by targeted deep sequencing of the 3 PV-CTCs, 3 CECs and 2 WBC controls. This identified 198 mutations (single nucleotide variants, SNVs) in the PV-CTCs and none in the CECs (Figure 3c). After accounting for technical drop-out due to the single cell sequencing approach (loci drop-out = 102/441 in tumour, 81/342 in metastasis)15 (Supplementary Table 6 and 7, methods online), 46% (157/339) of all primary tumour mutations were also detected in PV-CTCs (Figure 3c and Extended Data Fig. 3a). Along with the CNA data this confirms the tumour origin of the PV-CTCs, but the presence of PV-CTC mutations not detected in the primary tumour suggests these cells may represent a minor subclone of the tumour. Although a resolvable tumour specific CNA pattern was not observed in the metastasis (Figure 3b), due to low tumour content, WES and targeted deep sequencing revealed 91% of the PV-CTC mutations were seen in the metastasis (181/198), which is a higher mutational overlap than between the PV-CTCs and primary tumour (157/198, 79%) (Figure 3c and Extended Data Fig.3a). In addition, 96.8% (120/124) of the primary tumour mutations that were not detected in the metastasis were also not detected in the PV-CTCs (Figure 3c). Strikingly, of the 41 PV-CTC private mutations that were not detected in the primary tumour, 28 (68.3%) were identified in the relapse biopsy WES (Figure 3c and Extended Data Fig.3a) suggesting that the PV-CTCs present in the patient’s blood at surgery share a common progenitor with the metastasis that was detected 10 months later. The evolutionary origin of the PV-CTCs and metastasis was confirmed by phylogenetic analysis that revealed both PV-CTCs and metastasis are part of the same specific branch, which is distinct from all other subclones of the primary tumour (Figure 3d). The identification of PV-CTC specific mutations that are undetectable by bulk tumour analysis, yet are present in the relapse samples, strongly suggests that the PV-CTCs belong to a minor tumour subclone which is responsible for eventual relapse.
Examination of the mutations shared between PV-CTCs and the metastatic biopsy yet absent from the primary tumour has the potential to give insight into the mechanisms of metastasis. In this patient, the 28 PV-CTC/metastatic associated mutations not detected in the primary tumour included a putative inactivating driver mutation in the tumour suppressor gene LZTS1 (p.Pro104His) (Supplementray Table 8) which has been shown to inhibit tumour migration and whose lower expression has been linked to poor overall survival in NSCLC16.
Finally, to address the question whether the 13 private PV-CTC mutations not initially detected in the primary tumour or relapse biopsy, were in fact present at low frequency, additional targeted deep-sequencing of the tumour and metastasis was performed. All 13 mutations were present in either the primary tumour (5/13), the metastasis (12/13) and/or relapse cfDNA (7/13) (Figure 3e, Extended Data Fig.3b and Extended Data Fig.4). Interestingly, even using targeted deep-sequencing none of the 520 pre-identified mutations were detected in either baseline pulmonary or peripheral blood cfDNA samples (Extended Data Fig.4), highlighting the unique aspect of molecular analysis of PV-CTCs at resection.
Previous studies have shown a genetic link between CTCs, primary tumour and metastasis with clonal and subclonal mutations detected in CTCs in both colorectal and prostate cancer17,18. However, these studies were performed in metastatic patients and to our knowledge, this case report is the first to show that CTCs at surgery are phylogenetically linked to subsequent metastatic disease. This is exemplified by the larger mutational overlap between the PV-CTCs and the metastatic tumour that arose 10 months post PV-CTC isolation, than between the PV-CTCs and primary tumour which were collected at the same time in our case study. Comprehensive molecular analysis of early disseminating PV-CTCs also raises the opportunity to identify putative mechanisms of metastatic spread from the primary tumour prior to establishment of recurrent disease.
In early-stage NSCLC disease recurrence post-surgery occurs frequently and in this scenario survival is dismal; therefore, strategies that enable the identification of patients at higher risk of recurrence are an unmet medical need. We show here PV-CTC count (using the CellSearch platform) is associated with DFS and lung cancer specific survival in the TRACERx cohort, reinforcing the biological importance of PV-CTCs as founders of NSCLC metastasis. However, the clinical strength of a PV-CTC count to predict lung cancer specific relapse is modest and requires further validation in an independent and prospective patient cohort. Reasons underpinning the modest predictive strength of PV-CTC counts for NSCLC specific relapse could include the co-existence of CECs and bonafide epithelial CTCs as seen in the blood sample of the case study. This mixed population of EpCAM positive cells could confound the true PV-CTC count and the inability of CellSearch to detect mesenchymal CTCs further reduce the sensitivity of this approach. Additional detailed investigations are warranted to differentiate between epithelial and mesenchymal CTCs and CECs and to incorporate this greater understanding into NSCLC relapse prediction models. This study highlights the benefit of combining PV-CTC, tumour and cfDNA analysis to unearth new biological insights into the process of NSCLC metastasis.
Online Methods
Patients and pathology review
The cohort of 100 patients evaluated here for PV-CTC detection within this study comprises patients analysed by the lung TRACERx study (https://clinicaltrials.gov/ct2/show/NCT01888601). Patient eligibility and exclusion criteria for TRACERx enrolment is described in Jamal-Hanjani et al12 but briefly patients had given their informed written consent to participate in the study, were at least 18 years of age, had received a diagnosis of NSCLC in stages IA through IIIA and not received previous systemic therapy. The study has received a favourable opinion from the NRES Committee London – Camden & Islington Research Ethics Committee. The clinical data used in this study was derived from the “February 2019 TRACERx data release”. The NSCLC cohort in this study consisted of lung adenocarcinoma (LUAD) (59%) and remaining 41% of non-adenocarcinoma histology (Extended Data Fig. 1b and Supplementary Tables 1 and 2). The median age of patient was 68 and the population consisted of 61 males and 39 females (Table 1).
Digital images of diagnostic tumour sections from all cases were reviewed in detail centrally by at least one pathologist, and in cases of uncertainty, by two. Histological subtype and mitotic rate (number of visible mitoses per high-power field) were evaluated on digital images from scanned diagnostic slides blinded to the PV-CTC detection status of the patient in question.
Statistical analysis
All statistical tests were 2-sided unless otherwise stated. The association of PV-CTC count with individual clinical characteristics, including gender, stage, histology, smoking status, chemotherapy received, sites of relapse were evaluated using ANOVA, while age and mitotic rate were evaluated using Pearson’s correlation. PV-CTC count was log2 transformed in all analysis.
Cox proportional hazard regression analysis
The association between PV-CTC count and patient survival (DFS or lung cancer specific relapse) was assessed by including it as a sole covariate in a Cox proportional hazards model. Assumption of proportionality was verified based on Schoenfeld residuals19. A plot of the Martingale residuals was examined for evidence of nonlinearity20. The same uni-variate analysis was carried out on each clinical characteristic. Significant covariates in the uni-variate analysis were selected for subsequent multi-variate analysis, where a backward stepwise method was applied to investigate the impact of PV-CTC count on survival with other significant clinical characteristics under control.
Time-dependent receiver operating characteristics (tdROC) curves were applied to evaluate the performance of predicting lung cancer specific relapse using PV-CTC counts stratified by the 65th, 75th, 85th quantiles and the previously published threshold from our pilot study11 (≥18 PV-CTCs/7.5ml blood) within 720 days post-surgery. This analysis showed the upper quartile (75th quantile) had the highest AUROC (0.58, Extended Data Fig.1a). The diagnostic odds ratio (DOR) was also calculated for each PV-CTC cutoff. In order to avoid data overfitting, these DOR values were fitted into a polynomial curve, and the optimal cutoff for PV-CTC counts was selected as the one that corresponds to the maximum point of the curve.
All analysis were performed according to REMARK guideline 21, using R version 3.5.122. R packages survival (v2.38)23, and survminer (v0.3)24 and survivalROC (v1.0.3)25 were applied.
Lung cancer specific relapse event analysis
We collected available clinical data from all 37 patients who had been reported as having experienced a DFS event (defined as the time from study enrolment until recurrence of tumour or death from any cause) in the February 2019 TRACERx data release. Clinical data was available for 35 of 37 patients, 2 patients without available data (CRUK0005 and CRUK0770) were excluded from this analysis. We defined a lung cancer specific relapse event as histological or imaging confirmed NSCLC relapse. Nine of 37 patients who experienced a DFS event died without evidence of a lung cancer specific relapse event (details in Supplementary Table 3). These patients were either censored at the point of last computed tomography (CT) scan imaging prior to death showing the absence of metastatic disease (CRUK0056, CRUK0431, CRUK0416, CRUK0260, CRUK0017, CRUK0301) or in the event of immediate post-operative death (death within 30 days of surgery), at the point of death (CRUK0196, CRUK0223, CRUK0681). Four of 37 patients experienced metastatic disease unrelated to their original lung primary (CRUK0768, CRUK0068,CRUK0759, CRUK0085) and were censored at the point of last CT imaging prior to death showing absence of metastatic NSCLC. These cases were classified as second primary malignancies based on consensus imaging, histological and clinical agreement. For 1 of 37 patients there was high clinical suspicion of a second malignancy based on CT imaging but due to lack of investigation this was not conclusively determined, therefore this patient was excluded from the analysis (CRUK0073). Initial site of clinical relapse was defined as extracranial if no brain metastasis were clinically confirmed within 60 days of clinical relapse or intracranial if a patient presented with brain metastases within 60 days of clinical relapse.
Blood collection
A blood sample (10mL) was taken intra-operatively from the cancer-draining pulmonary vein prior to vessel ligation and tumour resection for each patient. A second sample was taken from the peripheral vein of patients recruited in Manchester. Blood samples were stored at room temperature for up to 96 hours in CellSave vacutainers prior to analysis.
CTC enrichment enumeration and single cell isolation
Blood samples were processed using the CellSearch system (Menarini), according to the manufacturer's instructions. Epithelial CTCs (via EpCAM dependent capture) were classified and counted based on an intact DAPI stained nucleus and positive immunofluorescent staining for pan-cytokeratins (CK) and negative staining for the WBC marker CD45. Following CellSearch® enrichment, single cells were isolated using the DEPArray™ system (Menarini) according to the manufacturer's instructions. Images of isolated PV-CTCs were manually inspected by two independent operators to confirm that the following morphological criteria were met: (1) cells were unambiguous positive for cytokeratin, (2) had an intact nucleus and (3) were clear of contaminating WBCs. Cells that failed to meet any of the three criteria were considered “ambiguous” and excluded from all downstream analysis.
Whole genome amplification
Whole genome amplification (WGA) was performed using the Ampli1 WGA kit (Menarini) according to the manufacturer's instructions. The efficacy of WGA was then evaluated by a multiplex quality control PCR (Ampli1 QC kit, Menarini) as previously described 26 followed by visualization of PCR bands on a 1.5% (w/v) agarose gel. This quality control step allowed us to establish a Genome Integrity Index (GII) of 0–4 for each sample and single cells with GII≥2 were considered with good quality DNA and eligible for subsequent downstream analysis.
Circulating cell-free DNA and tumour samples preparation
Plasma from CellSave blood samples was separated for cfDNA extraction as previously described27. Genomic DNA from primary and relapse tumours was isolated as described in Jamal-Hanjani et al12, sheared and quantified along with cfDNA and germline samples using the TaqMan RNase P Detection Kit (Life Technologies) as per manufacturer’s instructions.
DNA library preparation, targeted enrichment and next-generation sequencing
DNA libraries for PV-CTCs and WBCs were prepared using NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs) with 50 ng of DNA added per library preparation. DNA libraries for cfDNA, tumour DNA and germline were prepared using NEBNext Ultra II End Repair/dA-Tailing Module (New England BioLabs) and KAPA Hyper Library Prep Kit (KAPA Biosystems) using an input of up to 25 ng DNA. Each library was quantified (KAPA library quantification kit, KAPA Biosystems) and equimolar amounts were pooled and shallow-depth whole genome sequencing was performed on Illumina MiSeq or NextSeq 500 desktop sequencers (paired end, 300 cycles).
PV-CTC and WBC Libraries from patient CRUK0242 were additionally subjected to targeted exome enrichment using SureSelect Human All Exon V6 (Agilent) and Whole Exome Sequencing (WES) was performed on Illumina NextSeq 500 desktop sequencer for the detection of somatic mutations (paired end, 300 cycles). WES of corresponding excised primary tumour regions was performed as previously described3. For patient CRUK0242, libraries of cfDNA, isolated at surgery and at relapse, were enriched for a panel of 520 (SureSelectXT Custom, Agilent) pre-identified mutations and sequenced as above.
Sequence alignment and data processing
After trimming of sequencing adapters, the single cell sequencing reads (fastq format) were aligned to human genome assembly 19 (hg19), using the Burrows-Wheeler Aligner (BWA) mem (v0.7.13) algorithm28 to generate SAM files. SAMtools (v0.1.19) was used to convert the SAM files to BAM files, to remove reads with low mapping quality (MQ < 10) and to merge files from the same cell. Picard tools (v1.96) was used to sort the BAM files by chromosome coordinates and to remove PCR duplicates. The BAM files were converted to BED files using Bedtools29. A combination of Picard tools, Bedtools and FastQC30 was used to generate quality control metrics.
WGA Capture-rate
To establish the capture-rate of the WGA process, we used targeted sequencing data (described above) for comparisons of the germline (GL), WGA germline (WGA-GL) and individual single cells (including WBC controls) following WGA. A list of heterozygous single nucleotide polymorphisms (SNPs) detected within the targeted regions of the germline sample was generated using Mutect (v1.1.7)31. SAMtools mpileup was then used to check which of these SNPs were detected in each WGA sample, requiring a minimum of ten reads to call the SNPs (average read depth in successfully amplified regions is ~230 reads) and a Variant Allele Frequency (VAF) of 0.2-0.8 to consider it to still be heterozygous. The WGA-GL sample shows a complete locus drop-out of 18% due to lack of amplification in the WGA process. Of the 113 heterozygous SNPs that are present in the WGA-GL, 51 and 54 are also called as heterozygous in the two WBC controls. In addition 16 loci became homozygous for the SNP in each cell, and 12 and 13 loci becoming homozygous for the reference allele due to allele drop out (Supplementary Table 6). This gives an estimate for the allele capture-rate of 58-61% of the 113 WGA-GL SNPs due to the single cell sequencing.
Copy Number Analysis
Illumina whole-genome data for PV-CTCs, WBCs and tumour samples were aligned to the human genome using BWA. For CNA analysis we only analysed samples with a minimum of 2 million reads (after duplicate removal). Copy number alterations were identified using the R Bioconductor package HMMcopy (v1.18)32 with the genome divided into 1 Mb windows. Reads in each window were normalized by GC-content and mappability, and a Hidden Markov Model-based approach was used to segment the data into regions of similar copy-number profile and to predict a CNA event for each segment.
Somatic Mutation analysis from whole exome and targeted sequencing data
For the tumour WES, high-confidence variant calls from tumour were obtained as previously described3, using a combination of Varscan2 and MuTect.
MuTect (v1.1.7)31 was used to detect SNVs utilising annotation files contained in GATK bundle. All variants called by MuTect were filtered according to the filter parameter ‘PASS’ in the judgement column. All variants were annotated using ANNOVAR33. Only variants with at least 20 reads were considered for further filtering.
To generate a high-confidence set of variant calls from PV-CTCs, the following filters were applied:
Using the annotations as provided by ANNOVAR, all variants that were present in either 1000g or the Exac03 databases are removed.
A blacklist filter, relating to the genomic location of the variant, was applied. The blacklisted genomic regions were obtained from UCSC Genome Table Browser and include regions excluded from the Encode project (both DAC and Duke list), simple repeats, segmental duplications and microsatellite regions.
Variants with VAF < 0.2 are removed.
Variants had to be either present in the Tumour tissue (Primary or Relapse) or in at least one other single PV-CTC.
Lastly, if any variant is called in any of the WBC controls, then those were filtered out.
Supplementary Tables 9-10 and Supplementary Table 11 contain the information relative to coverage and VAF for each mutation detected in the primary tumour and single cells by WES and targeted deep sequencing respectively.
For the two WBC controls from patient CRUK0242, the first three filtering steps give 134 and 253 variants, none of which are shared with the tumour or any other single cells while the non-matching three CECs have 254, 260 and 307 private SNVs. The rate of false positives due to sequencing artefacts, with a range of 134-1960 variants seen in white blood samples from two other patients (see Supplementary Table 12) has very little overlap between them. The requirement that a mutation must be present in two or more samples (whether tumour or single cell) therefore eliminates the vast majority of false positives as a very conservative proceedure.
Regions containing mutations detected in the primary tumour or metastasis which were not covered in at least 1 of the three PV-CTC samples were removed for the calculation of overlaps, although they are shown in the Extended Data Fig.5.
All somatic variants detected in PV-CTCs were analysed by using cancer genome interpreter platform34 to interpret whether the variants detected had potential as drivers in NSCLC as well as in other solid cancers.
Phylogenetic Analysis
Phylogenetic analysis was performed as previously described3. In brief, using the pigeon-hole principle (if the average cancer cell fraction of two subclones sums to more than 1, the smaller subclone must be nested within the larger) as well as the crossing rule (if the cancer cell fraction of subclone A and subclone B sums to less than 1 and the cancer cell fraction of subclone A exceeds that of subclone B in one region but the inverse is true in another region, subclone A and B must exist on separate branches of the evolutionary tree), the evolutionary relationships between subclones was determined and a phylogenetic tree inferred.
Extended Data
Extended Data Fig.1.
a, Time-dependent receiver operating characteristic (ROC) curves showing true positive and false positive rates for the 65th, 75th, 85th PV-CTC quantiles (≥3, ≥7 and ≥39 PV-CTCs/7.5ml blood respectively) alongside the previously published threshold from our pilot study (≥18 PV-CTCs/7.5ml blood)11. All predictions were made at 720 days. Sensitivity and specificity of each category is shown along with area under ROC curve (AUROC) value. b, Kaplan–Meier curve showing lung cancer specific relapse free survival for 98 patients stratified as PV-CTC high or low according to the 75th quantile (≥7 PV-CTCs/7.5ml blood). The number of patients at risk for every time point is indicated below the time point and colour coded according to the high or low groups. P value, HR and relative 95% confidence intervals (CI) (two-sided log-rank test) are indicated. c, Forest plot showing the results of multivariable regression analysis for PV-CTC high or low patients (≥7 PV-CTCs/7.5ml blood). The x-axis represents the hazard ratio with the reference line (dashed) and significance is calculated using a Cox proportional hazards model.
Extended Data Fig.2.
a, Consort diagram describing samples used for downstream analysis. Only patients with ≥5 PV-CTCs (29) were processed through single cell isolation (DEPArray™). Single cells were not isolated from 6 out of the 29 samples due to failures during sample loading into the DEPArray™ machine. From the remaining 23 samples, 7 patients whose single CTCs isolated did not meet morphology criteria (see methods) were excluded. 16 samples were processed for whole genome amplification (WGA) and 2 patients whose CTCs did not show good quality genomic integrity index in QC post-WGA were removed (see methods). b, Table showing cases of relapse among the patients with single PV-CTCs isolated. c, Agarose gel showing results of a QC–PCR assay used to determine the genome integrity of each sample. 0–4 bands determine the overall DNA integrity of each sample. DEPArray images of corresponding PV-CTC (cytokeratin (CK)+ stained green, CD45+ stained blue, DAPI+ stained purple) are shown above. d, Examples of copy number profiles detected in single PV-CTCs, CECs and WBC control. Blue and red indicate regions of copy number loss and gain respectively.
Extended Data Fig.3.
a, Venn diagram showing the overlap of somatic mutations detected between single PV-CTCs, primary and metastatic tumour. b, Venn diagram showing the overlap of somatic mutations detected between single PV-CTCs, metastatic tumour and cfDNA isolated at the time of relapse.
Extended Data Fig.4.
Heat map showing the comparison of SNVs detected in primary tumour regions, metastasis, PV-CTCs, CECs, WBCs, and cfDNA samples (cfDNA pre-surgery was isolated from peripheral blood, cfDNA surgery was isolated from the pulmonary vein and cfDNA relapse was isolated at the time of relapse). Mutations are ordered according to their clonality established by primary tumour analysis.
Supplementary Material
Acknowledgements
We sincerely thank the patients and their families for donating of blood samples for research. We thank Ekram Aidaros-Talbot for administrative assistance with the manuscript. We also thank Professor Jacqui Shaw for kindly providing plasma (relapse time point) of patient CRUK0242. TRACERx is funded by Cancer Research UK (grant number C11496/A17786). This research was supported by Cancer Research UK - Core funding to CRUK Manchester Institute (A27412) Centre funding to Manchester (A25254), and funding of the CRUK Lung Cancer Centre of Excellence. Support was also received from the Manchester Experimental Cancer Medicine Centres and Manchester NIHR Biomedical Research Centre, FC is funded by the CANCER-ID Consortium (115749- Cancer-ID). BM is funded by Menarini Biomarkers Singapore PTE Ltd. CSK is funded by The Manchester MRC Single Cell Research Centre (MR/M008908/1). CS is Royal Society Napier Research Professor. This work was supported by the Francis Crick Institute that receives its core funding from Cancer Research UK (FC001169,FC001202), the UK Medical Research Council (FC001169, FC001202), and the Wellcome Trust (FC001169, FC001202). CS is funded by Cancer Research UK (TRACERx, PEACE and CRUK Cancer Immunotherapy Catalyst Network), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rosetrees Trust, NovoNordisk Foundation (ID16584) and the Breast Cancer Research Foundation (BCRF).
The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/ ERC grant agreement n°FP7 – 617844 (PROTEUS) and Marie Curie Network PloidyNet. Support was also provided to CS by the National Institute for Health Research, the University College London Hospitals Biomedical Research Centre, and the Cancer Research UK University College London Experimental Cancer Medicine Centre. All funders and sponsors had no role in this study.
Footnotes
Data Availability Statement
The majority of data generated or analysed during this study are included in this published article. The sequencing data are available through the Cancer Research UK & University College London Cancer Trials Centre for non-commercial research purposes and access will be granted upon review of a project proposal that will be evaluated by a TRACERx data access committee and entering into an appropriate data access agreement subject to any applicable ethical approvals.
Authors Contributions
CS, CD, PC, DGR and GB developed the clinical study, directed research, and co-wrote the manuscript. FC designed and conducted experiments, analysed data and drafted the manuscript with assistance of DGR and NMG. SG, SPP, GW, NB, NMG, CSK, SF, CM and MD provided bioinformatic support for the study. CA provided support for the clinical interpretation of the data. CZ performed statistical analysis. CA and DM performed centrally pathology review. DB, DST and BM provided support for single cell isolation. MJ-H, JP, FG, RS, MAB, CH, SV, YS, PC, SW, DB, JT, FB and AH provided support for patients’ recruitment, samples ‘management and clinical support for the study.
Competing Interests statement
CD receives research grants/support from Menarini and research grants are also received from AstraZeneca, Astex Pharmaceuticals, Bioven, Amgen, Carrick Therapeutics, Merck AG, Taiho Oncology, GSK, Bayer, Boehringer Ingelheim, Roche, BMS, Novartis, Celgene, Epigene Therapeutics Inc., all outside the scope of this paper. CD acts in a consultant or advisory role for Biocartis and AstraZeneca, again outside the scope of this work. CS has received honoraria, consultancy, or SAB Member fees for Pfizer, Novartis, GlaxoSmithKline, MSD, BMS, Celgene, AstraZeneca, Illumina, Sarah Canon Research Institute, Genentech, Roche-Ventana and GRAIL.Advisor for Dynamo Therapeutics. CS has also received research grants/support from Pfizer, AstraZeneca, BMS, Ventana, Roche and is a stock shareholder of Apogen Biotechnologies, Epic Bioscience, Achilles Therapeutics and GRAIL.
References
- 1.Uramoto H, Tanaka F. Recurrence after surgery in patients with NSCLC. Translational Lung Cancer Research. 2014;3:242–249. doi: 10.3978/j.issn.2218-6751.2013.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor MD, et al. Tumor recurrence after complete resection for non-small cell lung cancer. Ann Thorac Surg. 2012;93:1813–1820. doi: 10.1016/j.athoracsur.2012.03.031. discussion 1820-1811. [DOI] [PubMed] [Google Scholar]
- 3.Jamal-Hanjani M, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. 2017 doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.Aceto N, Toner M, Maheswaran S, Haber DA. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends in Cancer. 2015;1:44–52. doi: 10.1016/j.trecan.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Hodgkinson C, et al. Circulating tumour cells from small cell lung cancer patients are tumourigenic. Lung Cancer. 2015;87:S1–S1. [Google Scholar]
- 7.Morrow CJ, et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Annals of Oncology. 2016;27:1155–1160. doi: 10.1093/annonc/mdw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baccelli I, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 9.Girotti MR, et al. Application of Sequencing, Liquid Biopsies, and Patient-Derived Xenografts for Personalized Medicine in Melanoma. Cancer Discovery. 2016;6:286–299. doi: 10.1158/2159-8290.CD-15-1336. [DOI] [PubMed] [Google Scholar]
- 10.Mohan S, Chemi F, Brady G. Challenges and unanswered questions for the next decade of circulating tumour cell research in lung cancer. Transl Lung Cancer Res. 2017;6:454–472. doi: 10.21037/tlcr.2017.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosbie PA, et al. Circulating Tumor Cells Detected in the Tumor-Draining Pulmonary Vein Are Associated with Disease Recurrence after Surgical Resection of NSCLC. Journal of Thoracic Oncology. 2016;11:1793–1797. doi: 10.1016/j.jtho.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamal-Hanjani M, et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbosh C, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero-Palacios PJ, et al. Liquid biopsy beyond of cancer: Circulating pulmonary cells as biomarkers of COPD aggressivity. Crit Rev Oncol Hematol. 2019;136:31–36. doi: 10.1016/j.critrevonc.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Deleye L, Vander Plaetsen AS, Weymaere J, Deforce D, Van Nieuwerburgh F. Short Tandem Repeat analysis after Whole Genome Amplification of single B-lymphoblastoid cells. Scientific reports. 2018;8:1255. doi: 10.1038/s41598-018-19509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin CW, et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nature communications. 2013;4:1877. doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 17.Heitzer E, et al. Complex Tumor Genomes Inferred from Single Circulating Tumor Cells by Array-CGH and Next-Generation Sequencing. Cancer Research. 2013;73:2965. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 18.Lohr JG, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotech. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill R, Schumacher M. A Simple Test of the Proportional Hazards Assumption. Biometrika. 1987;74:289–300. doi: 10.2307/2336143. [DOI] [Google Scholar]
- 20.Therneau TM, Grambsch PM, Fleming TR. Martingale-Based Residuals for Survival Models. Biometrika. 1990;77:147–160. doi: 10.2307/2336057. [DOI] [Google Scholar]
- 21.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Team, R. C. R: A language and environment for statistical computing. 2013 [Google Scholar]
- 23.Therneau T. A Package for Survival Analysis in S. 2015. version 2.38. [Google Scholar]
- 24.Kassambara A, Kosinski M, Biecek P. survminer: Drawing Survival Curves using'ggplot2'. R package version 0.3. 2017;1 [Google Scholar]
- 25.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 26.Mesquita B, et al. Molecular analysis of single circulating tumour cells following long-term storage of clinical samples. Mol Oncol. 2017;11:1687–1697. doi: 10.1002/1878-0261.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothwell DG, et al. Genetic profiling of tumours using both circulating free DNA and circulating tumour cells isolated from the same preserved whole blood sample. Molecular Oncology. 2016;10:566–574. doi: 10.1016/j.molonc.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 [Google Scholar]
- 31.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31 doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha G, et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome research. 2012;22:1995–2007. doi: 10.1101/gr.137570.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamborero D, et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018;10:25. doi: 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.