Abstract
The importance of incorporating Natural Language Processing (NLP) methods in clinical informatics research has been increasingly recognized over the past years, and has led to transformative advances.
Typically, clinical NLP systems are developed and evaluated on word, sentence, or document level annotations that model specific attributes and features, such as document content (e.g., patient status, or report type), document section types (e.g., current medications, past medical history, or discharge summary), named entities and concepts (e.g., diagnoses, symptoms, or treatments) or semantic attributes (e.g., negation, severity, or temporality).
From a clinical perspective, on the other hand, research studies are typically modelled and evaluated on a patient- or population-level, such as predicting how a patient group might respond to specific treatments or patient monitoring over time. While some NLP tasks consider predictions at the individual or group user level, these tasks still constitute a minority. Owing to the discrepancy between scientific objectives of each field, and because of differences in methodological evaluation priorities, there is no clear alignment between these evaluation approaches.
Here we provide a broad summary and outline of the challenging issues involved in defining appropriate intrinsic and extrinsic evaluation methods for NLP research that is to be used for clinical outcomes research, and vice versa. A particular focus is placed on mental health research, an area still relatively understudied by the clinical NLP research community, but where NLP methods are of notable relevance. Recent advances in clinical NLP method development have been significant, but we propose more emphasis needs to be placed on rigorous evaluation for the field to advance further. To enable this, we provide actionable suggestions, including a minimal protocol that could be used when reporting clinical NLP method development and its evaluation.
Keywords: Natural Language Processing, Information extraction, Text analytics, Evaluation, Clinical informatics, Mental Health Informatics, Epidemiology, Public Health
1. Introduction
Appropriate utilization of large data sources such as Electronic Health Record (eHealth records or EHR) databases could have a dramatic impact on health care research and delivery. Owing to the large amount of free text documentation now available in EHRs, there has been a concomitant increase in research to advance Natural Language Processing (NLP) methods and applications for the clinical domain [1,2]. The field has matured considerably in recent years, addressing many of the challenges identified by Chapman et al. [3], and meeting the recommendations by Friedman et al. [4].
For example, the above include recommendations to address the key challenges of limited collaboration, lack of shared resources and evaluation-approaches of crucial tasks, such as de-identification, recognition and classification of medical concepts, semantic modifiers, and temporal information. These challenges have been addressed by the organization of several shared tasks. These include the Informatics for Integrating Biology and the Bedside (i2b2) challenges [5–9], the Conference and Labs of the Evaluation Forum (CLEF) eHealth challenges [10–13], and the Semantic Evaluation (SemEval) challenges [14–16]. These efforts have enabled a valuable platform for international NLP method development.
Furthermore, the development of open-source NLP software specifically tailored to clinical text has led to increased adoptability. Such NLP software include the clinical Text Analysis Knowledge Extraction System (cTAKES)1 and Clinical Language Annotation, Modeling, and Processing Toolkit (CLAMP),2 information extraction and retrieval infrastructure solutions such as SemEHR [17], as well as general purpose tools such as the the general architecture for text engineering (GATE)3 and Stanford CoreNLP.4 New initiatives, such as the Health Natural Language Processing (hNLP) Center,5 also aim to facilitate the sharing of resources, which would enable further progress through availability, transparency, and reproducibility of NLP methodologies.
In recent years, the field of mental health has shown a burgeoning increase in the use of NLP strategies and methods, mainly because most clinical documentation is in free-text, but also arising from the increasing availability of other types of documents providing behavioural, emotional, and cognitive indicators as well as cues on how patients are coping with different conditions and treatments. Such texts sources include social media and online fora [18–21] as well as doctor-patient interactions [22–24] and online therapy [25], to mention a few examples. However, although there have been a few shared tasks related to mental health [26–28] the field is still narrower than that of biomedical or general clinical NLP.
The maturity of NLP method development and state-of-the-art results have led to an increase in successful deployments of NLP solutions for complex clinical outcomes research. However, the methods used to evaluate and appraise NLP approaches are somewhat different from methods used in clinical research studies, although the latter often rely on the former for data preparation and extraction. There is a need to clarify these differences and to develop novel approaches and methods to bridge this gap.
This paper stems from the findings of an international one-day workshop in 2017 (see online Supplement). The objective was to explore these evaluation issues by outlining ongoing research efforts in these fields, and brought together researchers and clinicians working in the areas of NLP, informatics, mental health, and epidemiology. The workshop highlighted the need to provide an overview of requirements, opportunities, and challenges of using NLP in clinical outcomes research (particularly in the context of mental health). Our aim is to provide a broad outline of current state-of-the-art knowledge, and to make recommendations on directions going forward in this field, with a focus on considerations related to intrinsic and extrinsic evaluation issues.
2. Evaluation paradigms
All empirical research studies need to be evaluated (or validated) in order to allow for scientific assessment of a study. In clinical outcomes research, studies are usually designed as clinical trials, cohort studies or case-control studies, with the aim to assess whether a risk factor or intervention has a significant association with an outcome of interest. NLP method development, on the other hand, aims to produce computational solutions to a given problem. Studies of diagnostic tools are most similar to NLP method development - testing whether a history item, examination finding or test result is associated with a subsequent diagnosis. The most basic underlying construction for quantitative validation in both fields is a 2 × 2 contingency table (or confusion matrix), where the number of correctly and incorrectly assigned values for a given binary outcome or classification label is compared with a gold (reference) standard, i.e. the set of ’true’ or correct values. This table can then be used to calculate performance metrics such as precision (Positive Predictive Value), recall (sensitivity), accuracy, F-score, and specificity. In clinical studies, this can be used to calculate measures of association, such as risk ratio and odds ratio. There are other evaluation measures that can be used when the outcome is more complex, e.g., continuous or ranked (for NLP, see e.g., [29], for clinical prediction models, see e.g., [30]).
Validation or evaluation of clinical outcomes whether it be a trial, cohort or case-control study relies on statistical measurements of effect, and can be validated internally (measured on the original study sample) or externally (measured on a different sample) [31]. Typically, a number of predictors (variables) interact in these models, thus multivariable models are common, where it is important to account for biases to ensure model validity.
Because the goal of NLP method development is to produce computational solutions to specific problems, evaluation criteria can be intrinsic (evaluating an NLP system in terms of directly measuring its performance on attaining its immediate objective) and extrinsic (evaluating an NLP system in terms of its usefulness in fulfilling an over-arching goal where the NLP system is perhaps part of a more complex process or pipeline) [32–34,29,35]. The goal of clinical research studies, on the other hand, typically relates to assessing the effect of a treatment or intervention.
Clinical NLP method development has mainly focused on internal, intrinsic evaluation metrics. Typically, these methods have been developed and evaluated on word, sentence or document level annotations that model specific attributes and features, such as document content (e.g., patient status, or report type), document section types (e.g., current medications, past medical history, discharge or summary), named entities and concepts (e.g., diagnoses, symptoms, or treatments), or semantic attributes (e.g., negation, severity, or temporality).
Although the intrinsic evaluation metrics are important and valuable, especially when comparing different NLP methods for the same task, they are not necessarily of value or particularly informative when the task is applied on a higher-level problem (e.g., patient level) or on new data. For instance, current state-of-the-art that is achieved in medical concept classification is > 80% F-score [7], which is close to human agreement on the same task; however, if such a system was to be deployed in clinical practice, any > 0% error rate, such as the misclassification of a drug or a history of severe allergy, might be seen as unacceptable.
True negatives are rarely taken into consideration in NLP evaluation, often because this is intractable in text analysis [36]. Yet, specificity (the true negative ratio, i.e. the proportion of a gold standard construct that is identified by the new assessment) is often a key factor in clinical research, particularly in medical screening but also in categorisation of exposures (e.g. case status) and outcomes. Thus when using outputs from NLP approaches in clinical research studies, it is not always clear how best to incorporate and interpret NLP performance metrics.
3. Opportunities and challenges from a clinical perspective
The opportunities and potential of NLP are hugely exciting for health research generally, and for mental health research specifically. As clinical informatics resources become larger and more comprehensive as a result of text-derived meta-data, the possibility of determining outcomes, prognoses, effectiveness, and harm are all within closer reach, requiring fewer resources than would be needed to conduct primary research studies. A variety of data sources are amenable to clinical research such as social media, wearable device data, audio and video recordings of team discussions and interactions. However, EHR-derived data potentially offer the most immediate value, given the time and patient numbers over which data have already been collected, their comprehensive and now-established use across healthcare, and given the depth of clinical information potentially available in real world services. Compared to primary research cohorts, the coverage is huge and substantially more generalisable, and allows for external validation of models [37].
3.1. Clinical NLP applied on mental health records
A key issue with mental EHR data is that the most salient information for research and clinical practice tends to be entered in text fields rather than pre-structured data, with up to 70% of the record documented in free-text [38]. For instance, the Clinical Record Interactive Search (CRIS) system from the South London and Maudsley mental health trust (SLaM) contained almost 30 million event notes and correspondence letters, and more than 322, 000 patients,6 yielding an average of 90 documents per patient (even more if additional text content would be included, e.g., free-text entries in risk assessment notes). This is partly because the most important features of mental health care do not lend themselves to structured fields. Such features include the salience of the self-reported experience (i.e. mental health symptoms), determining treatment initiation and outcome evaluation, as well as the complex circumstances influencing presentations and prognoses (e.g., social support networks, recent or past stressful experiences, psychoactive substance use). Moreover, written information can be more accurate and reliable, and allows for expressiveness, which better reflects the complexity of clinical practice [39,40]. While there have been calls for increased structuring of health records, these seem to be mainly driven by convenience issues for researchers or administrators (i.e. ease of access to pre-structured data) rather than the preferences of the clinical staff actually entering data [39].
Most clinical researchers and clinicians are accustomed to research methods involving highly scrutinised de novo data collection with standardised instruments (such as the Beck Depression Inventory (BDI) or the Positive and Negative Syndrome Scale (PANNS)). These have established psychometric properties for the concepts they measure, such as symptom severity in patients with schizophrenia (e.g., positive symptoms such as delusions, hallucinations). Using NLP methods to derive and identify such concepts from EHRs holds great promise, but requires careful methodological design. If NLP algorithms are to be fit for purpose, they need to demonstrate accurate and reliable measurement, which in turn needs to be communicated in a language that is understandable across disciplines especially considering the differences in language used by the clinical and NLP academic communities. Because of the importance of information accuracy in medical practice, including the validity and reliability of tests and instruments, translating NLP system outputs to an interpretable measure is key. This way the clinical community can easily understand the basis for the underlying NLP model, allowing for the potential translation of NLP-derived observational findings into clinical interventions.
Moreover, ensuring that an NLP approach is appropriately designed for a specific clinical problem is essential. For example, timely detection of the risk of suicidal behaviour in patients using NLP approaches on EHR data is clinically important, but challenging: not only because of the various ways this can be documented in text, but also because of the complexity of the clinical construct. Suicidal behaviour is relatively rare, and current tools for assessing suicide risk are inadequate and suffer from low PPV/precision [41]. Data-driven methods hold promise as a solution to develop more accurate predictive models, but they need to be carefully designed. In one case study, < 3% of EHR documents for 200 patients had any suicide-related information documented in freetext, whilst at a patient level, 22% of the patients had at least one document with written suicide-related information [42]. Thus, for this type of use-case, it is important for method development to ensure an appropriate sample (document or patient), and to provide interpretable NLP output results.
Other key challenges in applying NLP on mental health records include moving beyond simple named-entity recognition towards ascertaining novel and more complex entities such as markers of socioeconomic status or life experiences, as well as unpicking temporality in order to reconstruct disorder and treatment pathways. In addition, there are the more computational challenges of moving beyond single-site applications to wider multi-site provision of NLP resources, as well as evaluating translation for international use. Other types of textual data such as patient-generated text (e.g., online forums, questionnaires, and feedback forms) involve additional challenges, e.g., the ability to adapt models for clinical constructs such as mood recognition from a wider population to individuals, as well as the ability to calculate mood scores over time, based on a number of linguistic features and often in the face of sparse or missing data.
3.2. Using NLP for large-sample clinical research
The capacity of NLP approaches to extract additional, non-structured information is particularly important for large-sample research studies, which are often focused on identifying as many predictors (and potential confounders) of an outcome as possible [43]. These may include factors at both the ‘macro-environment’, such as family/social circumstances, and the individual patient-level, such as tobacco use [44], and is especially important for mental health research given there may be reticence to code stigmatised conditions, such as illicit drug use, when they are not the primary reason for seeking treatment [45]. Also, structured codes cannot accommodate diagnostic uncertainty, and do not permit the recording of clinically-relevant information that supports a diagnosis, e.g., sleep or mood, but is not the specific condition for which a patient receives treatment [46]. Thus NLP approaches both enable the improvement of case identification from health records [46,47] and can provide a much richer set of data than could be achieved by the use of structured data alone.
However, this increase in the depth of data provided by NLP can come at a cost to study reproducibility and research transparency. An EHR-based study requires a clear specification of how the data recorded for each patient were collected and processed prior to analysis. In the context of EHR research this is often referred to as developing ‘phenotypes’, with the intention that the algorithms developed can be reused by others [48–50]. Incorporation of NLP output data in phenotype algorithms may make it more difficult for researchers using different EHR data to replicate results. For instance, if the underlying data that was used to develop an NLP solution to extract a phenotype such as atrial fibrillation is specific to the EHR system, geographical area and other factors, the NLP algorithm may produce different results if applied on new data for the same task.
Even if NLP methods are shared, their application may be hampered if similar source documents are not available. This issue would be compounded if multiple phenotypes are used to build the epidemiological data set. One practical solution is to adopt some of the measures suggested for clinically-focused observational research, such as the publication of study protocols and/or cohort descriptions [51].
3.3. NLP in clinical practice — towards extrinsic evaluation
Nuances of human language mean that no NLP algorithm is completely accurate, even for a seemingly straightforward task such as negation detection [52]. An error rate can be accommodated statistically in research, but to support decisions about individual patient care, results of NLP must be verified by a clinician before being used to make recommendations about patient management. Such verification might be better accepted by the users if the system provides probabilistic outputs rather than binary decisions. The difficulties in safely incorporating these uncertainties may have contributed to the gap between research applications of NLP and its use in clinical settings [2]. When algorithms are used in clinical decision support, it is important to display the information that is used to make the recommendation, and for clinicians to be aware of potential weaknesses of the algorithm. Clinical decision systems are more useful if they provide recommendations within the clinical workflow at the time and location of decision making [53].
Within EHR systems, NLP may be used to improve the user interface, such as the ease of finding information in a patient’s record. Real-time NLP can potentially assist clinicians to enter structured observations, evaluations or instructions from free text by, for example, automatically transforming a paragraph into a diagnostic code or suggested treatment. The accuracy of such algorithms may be tested by calculating the proportion of suggested structured entries that the clinician verifies as being correct. Clinical NLP systems have not, as of yet, been developed with clinical experts in mind, and have rarely been evaluated according to extrinsic evaluation criteria. As NLP systems become more mature, usability studies will also be a necessary step in NLP method development, to ensure that clinicians’ and other non-NLP users’ input can be taken into consideration. For instance, in the 2014 i2b2 Track 3 - Software Usability Assessment, it was shown that current clinical NLP software is hard to adopt [54]. Tools such as Turf (EHR Usability Toolkit)7could be made common practice when developing NLP solutions for clinical research problems. NLP could also become more integrated into EHR systems in the future, where evaluation metrics that focus on the time a documentation task takes, documentation quality, and other aspects also need to be considered [55].The ideal evaluation would be a randomized trial in clinical practice, comparing usability and data quality between user interfaces incorporating different NLP algorithms.
4. Opportunities and challenges from a Natural Language Processing perspective
The two major approaches to NLP, as described by Friedman et al. [4], namely symbolic (based on linguistic structure and world knowledge) and statistical methods (based on frequency of occurrence and distribution patterns) methods, are still dominant in clinical NLP development. Advances in machine learning algorithms, such as neural networks, have influenced NLP applications, and there are, of course, further developments to be expected. However, many of the developments, particularly in neural network models, assume large, labeled datasets, and these are not readily available for clinical use-cases that require analysis of EHR text content. Another challenge is data availability — ethical regulations and privacy concerns need to be addressed if authentic EHR data are to be used for research, but there are also alternative methods that can be used to create novel resources (Section 4.1).
Furthermore, evaluation of NLP systems is still typically performed with standard statistical metrics based on intrinsic criteria, not necessarily optimal for the clinical research problem at hand. To address such issues, it is important to identify which level of analysis is appropriate, and model the problem accordingly (Section 4.2). Enriching informatics approaches with novel data sources, using evaluation metrics that capture novel aspects such as model interpretability or time sensitivity, and developing NLP solutions with the clinical endusers in mind (Section 4.3) could lead to considerable advances in this field.
4.1. Methods for developing shareable data
Risks for compromised privacy are particularly evident in analyzing text from health records (i.e. the inability to fully convince the relevant authorities that all explicit and implicit privacy-sensitive information has been de-identified) and in big data health research more generally (i.e. unforeseen possibilities of inferring an individual’s identity after record linkages from multiple de-identified sources). The same ethical and legal policies that protect privacy complicate the data storage, use, and exchange from one study to another, and the constraints for these data exchanges differ between jurisdictions and countries [56].
As a timely solution to these data exchange problems, synthetic clinical data has been developed. For example, a set of 301 patient cases which includes recorded spoken handover and annotated verbatim transcriptions based on synthetic patient profiles, has been released and used in shared tasks in 2015 and 2016 [12,13,57]. Similarly, synthetic clinical documents have been used in 2013 and 2014 in shared tasks on clinical NLP in Japanese [58]. Synthetic data has been successful in tasks such as dialogue generation [59] and is a promising direction at least as a complement for method development where access to data is challenging.
4.2. Intrinsic evaluation and representation levels
When considering the combination of NLP methods and clinical outcomes research, differences in granularity are a challenge. NLP methods are usually developed to identify and classify instances of some clinically relevant phenomenon at a sub-document or document level. For example, NLP methods for the extraction of a patient’s smoking status (e.g., current smoker, past smoker or non-smoker) will typically consider individual phrases that discuss smoking, of which there may be several in a single document [60]. Even in cases where an NLP method is used to classify a whole document (e.g., assigning tumor classifications to whole histopathology reports [61]), there may be several documents for an individual patient.
Typically, in evaluating clinical NLP methods, a gold standard corpus with instance annotations is developed, and used to measure whether or not an NLP approach correctly identifies and classifies these instances. If a gold standard corpus contains multiple annotations and documents for one patient, and the NLP system correctly classifies these, the evaluation score will be higher. For clinical research, on the other hand, only one of these instances may be relevant and correct. In the extreme, a small number of patients with a high number of irrelevant instances, could bias the NLP evaluation relative to the clinical research question. For instance, a gold standard corpus annotated on a mention level for positive suicide-related information (patient is suicidal) or negated (patient denies suicidal thoughts) was used to develop an NLP system [62] which had an overall accuracy of 91.9%. However, when this system was applied for a clinical research project to identify suicidal patients, implementing a document- and, more crucially, a patient-level classification based on such instance-level annotations required non-trivial assumptions, because the documents could contain several positive and negative mentions, and each patient could have several EHR documents [42].
There is thus often a gap between instance level and patient level evaluations. In order to resolve the differences in granularity between the NLP and clinical outcomes evaluations, this gap needs to be bridged somehow. Typically, some post-processing will be required, in order to filter the instances found by the NLP method, before their use in clinical outcomes research. For example, post-processing might merge instances, or might remove those that are irrelevant. For some use-cases, this post-processing procedure can be based on currently available evidence, such as case identification for certain diseases (e.g., asthma status [63]).
The gap as described does not always exist, and post-processing is not always appropriate. There are cases in which an NLP method might be used to process all of the relevant text associated with a single patient, over time, in order to directly predict a single outcome; for example, all past text (and other information) could be used in order to assign a diagnosis code [64]. Moreover, for some clinical use-cases, patient-level annotations by e.g., manual chart review of sets of notes for one patient-level clinical label might in some cases be more efficient for developing gold standards and subsequent NLP solutions. Evaluation metrics for such use-cases could be developed to measure the degree that the NLP system correctly classifies groups compared to manual review. For specific use-cases, this approach might even be more appropriate than focusing on mention- and document-level annotations. Looking further, NLP methodology that addresses clinical objectives by not only finding relevant instances, but actually summarising all relevant information over time, is desirable; however, this is a non-trivial aspiration, particularly considering methods for evaluation and conveyance of such summaries.
4.3. Beyond electronic health record data
Work on using computational language analysis on speech transcripts to study communication disturbances in patients with schizophrenia [65] or to predict onset of psychosis [66,67] has shown promising results. Further, the availability of large datasets has led to advances in the field of psycholinguistics [68].
The increasing availability online of patient related texts including social media posts and themed fora, especially around long term conditions, have also lead to an increase in NLP applications for mental health and the health domain in general. For example, recent NLP work classifies users into patient groups based on social media posts over time [21,69,70], prioritises posts for potential interventions based on topics, sentiment and the overall conversation thread [27] or identifies temporal expressions and relations within clinical texts [15,16].
Whilst this is encouraging in terms of the interaction between NLP and the health domain, these tasks are still primarily evaluated using classic NLP system performance metrics such as accuracy, recall, and F-score. Recent NLP community efforts have initiated new evaluation levels and metrics, e.g., prediction of current and future psychological health based on childhood essays from longitudinal cohort data as in the 2018 Computational Linguistics and Clinical Psychology Workshop (CLPsych) shared task,8 however the direct clinical applicability of such approaches is yet to be shown. Work by Tsakalidis et al. [71] is the first to use both language and heterogeneous mobile phone data to predict mental well-being scores of individual users over time calibrated against psychological scales. Results were promising, yet their evaluation strategy involved training and testing of data from all users, a scenario often encountered in studies involving mood score predictions using mobile phone data [72–74]. However, a more realistic evaluation scenario would involve either: (a) intra-user predictions over time, that is, for the same user calculating a mood score or other health indicator given previous data in a sequence, at consecutive time intervals or (b) predictions of some indicator, over time, for an unseen user, given a model created on the basis of other users [75].
For these tasks to have potential clinical utility, new evaluation metrics would need to be introduced that focus on a number of other aspects, such as:
time sensitive and timely prediction: longitudinal prediction of an indicator such as a score from a psychological scale or other health indicator. These would need to be predicted over time as monitoring points rather than predictions that are independent of time, as is the case of current standard classification approaches.
personalised models: intra-user models are very useful for personalised health monitoring. However, such personalised models would require large amounts of longitudinal data for individual users which are not often available.
model interpretability: a model should provide confidence scores for its predictions and provide evidence for the prediction. Current model evaluation focusses on performance without any regard to interpretability.
5. Actionable guidance and directions for the future
NLP method development for the clinical domain has reached mature stages and has become an important part of advancing data-driven health care research. In parallel, the clinical community is increasingly seeing the value and necessity of incorporating NLP in clinical outcomes studies, particularly in domains such as mental health, where narrative data holds key information. However, for clinical NLP method development to advance further globally and, for example, become an integral part of clinical outcomes research, or have a natural place in clinical practice, there are still challenges ahead. Based on the discussions during the workshop, the main challenges include data availability, evaluation workbenches and reporting standards. We summarize these below and provide actionable suggestions to enable progress in this area.
5.1. Data availability
The lack of sufficiently large sets of shareable data is still a problem in the clinical NLP domain. We encourage the increased development of alternative data sources such as synthetic clinical notes [57,58], which alleviates the complexities involved in governance structures. However, in parallel, initiatives to make authentic data available to the research community through alternative governance models are also encouraged, like the MIMIC-III database [76]. Greater connection between NLP researchers, primary data collectors, and study participants are required. Further studies in alternative patient consent models (e.g., interactive e-consent [77]) could lead to larger availability of real-world data, which in turn could lead to substantial advances in NLP development and evaluation. Moving beyond EHR data, there is valuable information also in accessible online data sources such as social media (e.g., PatientsLikeMe), that are of particular relevance to the mental health domain, and that could also be combined with EHR data [78]. Efforts to engage users in donating their public social media and sensor data for research such as OurDataHelps9 are interesting avenues that could prove very valuable for NLP method development. Furthermore, in addition to written documentation, there is promise in the use of speech technologies, specifically for information entry at the bedside [57,79–83].
5.2. Evaluation workbenches
Current clinical NLP methods are typically developed for specific use-cases and evaluated intrinsically on limited datasets. Using such methods off-the-shelf on new use-cases and datasets leads to unknown performance. For clinical NLP method development to become more integral in clinical outcomes research, there is a need to develop evaluation workbenches that can be used by clinicians to better understand the underlying parts of an NLP system and its impact on outcomes. Work in the general NLP domain could be inspirational for such development, for instance integrating methods to analyse the effect of NLP pipeline steps in downstream tasks (extrinsic evaluation) such as the effect of dependency parsing approaches [84]. Alternatively, methods that enable analysis of areas where an existing NLP solution might need calibration when applied on a new problem, e.g., by posterior calibration [85] are an interesting avenue of progress. If clinical NLP systems are developed for non-NLP experts, to be used in subsequent clinical outcomes research, the NLP systems need to be easy to use. Facilitating the integration of domain knowledge in NLP system development can be done by providing support for formalized knowledge representations that can be used in subsequent NLP method development [86].
5.3. Reporting standards
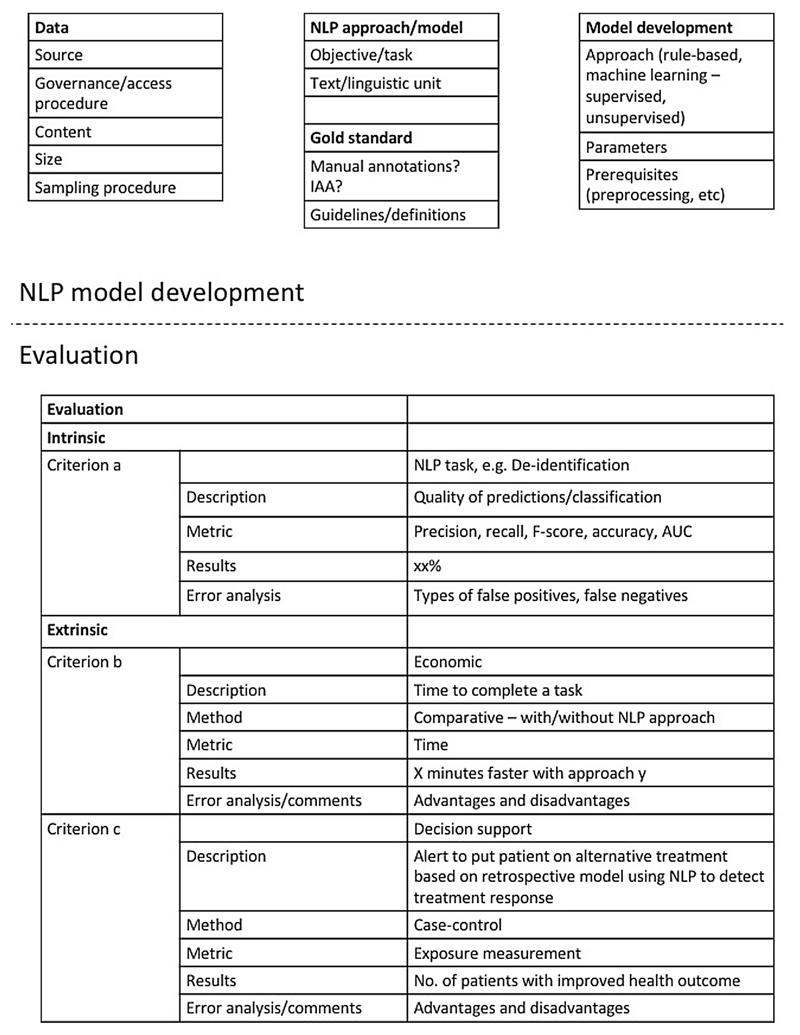
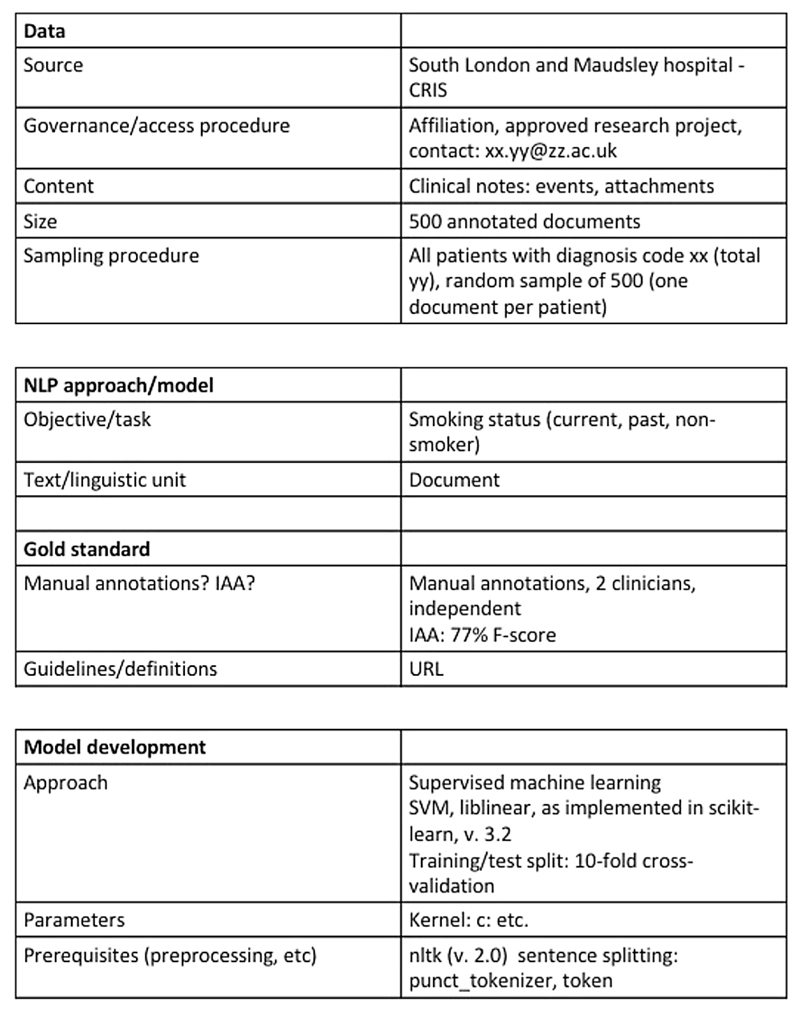
Most importantly, ensuring transparency and reproducibility of clinical NLP methods is key to advance the field. In the clinical research community, the issue of lack of scientific evidence for a majority of reported clinical studies has been raised [87]. Several aspects need to be addressed to make published research findings scientifically valid, among others replication culture and reproducibility practices [88]. This is true also for clinical NLP method development. We propose a minimal protocol inspired by [32], see Figs. 1 and 2, that outlines the key details of any clinical NLP study; by reporting on these, others can easily identify whether or not a published approach could be applicable and useful in a new study and for example, whether or not adaptations might be necessary. This could encourage further development of a comprehensive guidance framework for NLP, similar to what has been proposed for the reporting of observational studies in epidemiology in the STROBE statement [89], and other initiatives (e.g., [90,91,51]).
Fig. 1.
Example of a suggested structured protocol with essential details for documenting NLP approaches and performed evaluations. The example includes different levels of evaluation (intrinsic and extrinsic) that could be outlined with details about the task, metrics, results, and error analysis/comments.
Fig. 2.
A minimal protocol example of details to report on the development of a clinical NLP approach for a specific problem, that would enable more transparency and ensure reproducibility.
The key details that are needed are:
Data: What type of source data was used? How was it sampled? What is the size (in terms of sentences, words) How was the data obtained? Is it available to other researchers?
NLP approach: What was the objective or task? At which type of textual unit is analysis performed (document, sentence, entities, word)? Is there a gold/reference standard? If so, how was the gold/reference standard generated? If it was manual, what was the Interrater/annotator agreement? Have the guidelines and definitions been made publicly available?
Model development: what type of approach was taken? Parameter settings? Prerequisites?
Evaluation: was the model evaluated intrinsically or extrinsically? Which metrics were used? What types of errors were common? If the evaluation was extrinsic, what assumptions were made? For example, if an NLP output provides counts of mentions for a condition, what threshold for determining whether or not someone can be considered a case was chosen, and why? How was the conversion from mention to case level done? Conversely, if an NLP system provides an output for a patient-level label based on a set of information sources (e.g., documents), what method was applied to identify and analyse disagreements?
6. Conclusions
We have sought to provide a broad outline of the current state-of-the-art, opportunities, challenges, and needs in the use of NLP for health outcomes research, with a particular focus on evaluation methods. We have outlined methodological aspects from a clinical as well as an NLP perspective and identify three main challenges: data availability, evaluation workbenches and reporting standards. Based on these, we provide actionable guidance for each identified challenge. We propose a minimal structured protocol that could be used when reporting clinical NLP method development and its evaluation, to enable transparency and reproducibility. We envision further advances particularly in methods for data access, evaluation methods that move beyond current intrinsic metrics and move closer to clinical practice and utility, and in transparent and reproducible method development.
Supplementary Material
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jbi.2018.10.005.
Acknowledgements
This work is the result of an international workshop held at the Institute of Psychiatry, Psychology and Neuroscience, King’s College London, on April 28th 2017, with financial support from the European Science Foundation (ESF) Research Networking Programme Evaluating Information Access Systems: http://eliasnetwork.eu/. SV is supported by the Swedish Research Council (2015-00359) and the Marie Skłodowska Curie Actions, Cofund, Project INCA 600398. RD is funded by a Clinician Scientist Fellowship (research project e-HOST-IT) from the Health Foundation in partnership with the Academy of Medical Sciences. JD is supported by a Medical Research Council (MRC) Clinical Research Training Fellowship (MR/L017105/1). KM is funded by the Wellcome Trust Seed Award in Science [109823/Z/15/Z]. RS and AR is part-funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. We wish to acknowledge contributions by the Data61/CSIRO Natural Language Processing Team in general, and in particular, those by Adj/Prof Leif Hanlen. David (PJ) Osborn, Joseph (F) Hayes and Anoop (D) Shah are supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. Prof Osborn is also in part supported by the NIHR Collaboration for Leadership in Applied Health Research and Care (CLAHRC) North Thames at Bart’s Health NHS Trust. ML has a fellowship by the Alan Turing Institute for data science and artificial intelligence at 40%.
Footnotes
Counted on May 16th, 2018.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- [1].Névéol A, Zweigenbaum P. Clinical Natural Language Processing in 2014: foundational methods supporting efficient healthcare. Yearb Med Inform. 2015;10(1):194–198. doi: 10.15265/IY-2015-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Velupillai S, Mowery D, South BR, Kvist M, Dalianis H. Recent advances in clinical natural language processing in support of semantic analysis. IMIA yearb Med Inform. 2015;10:183–193. doi: 10.15265/IY-2015-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc. 2011;18(5):540–543. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Friedman C, Rindflesch TC, Corn M. Natural language processing: State of the art and prospects for significant progress, a workshop sponsored by the National Library of Medicine. J Biomed Inform. 2013;46(5):765–773. doi: 10.1016/j.jbi.2013.06.004. [DOI] [PubMed] [Google Scholar]
- [5].Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic deidentification. J Am Med Inform Assoc. 2007;14(5):550. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17(5):514. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18(5):552. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Uzuner Ö, Bodnari A, Shen S, Forbush T, Pestian J, South BR. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc. 2012;19(5):786. doi: 10.1136/amiajnl-2011-000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc. 2013;20(5):806. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Suominen H, Salanterä S, Velupillai S, Chapman W, Savova G, Elhadad N, Pradhan S, South B, Mowery D, Jones G, Leveling J, et al. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Vol. 8138. LNCS; 2013. Overview of the ShARe/CLEF eHealth evaluation lab 2013; pp. 212–231. [DOI] [Google Scholar]
- [11].Kelly L, Goeuriot L, Suominen H, Schreck T, Leroy G, Mowery D, Velupillai S, Chapman W, Martinez D, Zuccon G, Palotti J. Lecture Notes in Computer Science (including sub-series Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Vol. 8685. LNCS; 2014. Overview of the ShARe/CLEF eHealth evaluation lab 2014; pp. 172–191. [DOI] [Google Scholar]
- [12].Goeuriot L, Kelly L, Suominen H, Hanlen L, Névéol A, Grouin C, Palotti J, Zuccon G. Overview of the CLEF eHealth Evaluation Lab 2015. Springer International Publishing; 2015. pp. 429–443. [Google Scholar]
- [13].Kelly L, Goeuriot L, Suominen H, Névéol A, Palotti J, Zuccon G. Overview of the CLEF eHealth Evaluation Lab 2016. Springer International Publishing; 2016. pp. 255–266. [DOI] [Google Scholar]
- [14].Elhadad N, Pradhan S, Gorman S, Manandhar S, Chapman W, Savova G. SemEval-2015 task 14: Analysis of clinical text. Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015); Denver, Colorado. Association for Computational Linguistics; 2015. pp. 303–310. [Google Scholar]
- [15].Bethard S, Derczynski L, Savova G, Pustejovsky J, Verhagen M. SemEval-2015 task 6: Clinical TempEval. Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015); Denver, Colorado. Association for Computational Linguistics; 2015. pp. 806–814. [Google Scholar]
- [16].Bethard S, Savova G, Chen W-T, Derczynski L, Pustejovsky J, Verhagen M. Semeval-2016 task 12: Clinical tempeval. Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016); San Diego, California. Association for Computational Linguistics; 2016. pp. 1052–1062. [Google Scholar]
- [17].Wu H, Toti G, Morley KI, Ibrahim ZM, Folarin A, Jackson R, Kartoglu I, Agrawal A, Stringer C, Gale D, Gorrell G, et al. SemEHR: a general-purpose semantic search system to surface semantic data from clinical notes for tailored care, trial recruitment, and clinical research. J Am Med Inform Assoc. 2018;25(5):530–537. doi: 10.1093/jamia/ocx160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].De Choudhury M, De S. Mental health discourse on reddit: self-disclosure, social support, and anonymity. Eighth International AAAI Conference on Weblogs and Social Media; 2014. [Google Scholar]
- [19].Pavalanathan U, De Choudhury M. Identity Management and Mental Health Discourse in Social Media. Proceedings of the International World-Wide Web Conference. International WWW Conference 2015 (Companion); 2015. pp. 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mowery D, Smith H, Cheney T, Stoddard G, Coppersmith G, Bryan C, Conway M. Understanding depressive symptoms and psychosocial stressors on twitter: a corpus-based study. J Med Internet Res. 2017;19(2):e48. doi: 10.2196/jmir.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gkotsis G, Oellrich A, Velupillai S, Liakata M, Hubbard TJP, Dobson RJB, Dutta R. Characterisation of mental health conditions in social media using Informed Deep Learning. Sci Rep. 2017;7 doi: 10.1038/srep45141. 45141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Howes C, Purver M, McCabe R. Linguistic Indicators of Severity and Progress in Online Text-based Therapy for Depression. Proceedings of the Workshop on Computational Linguistics and Clinical Psychology: From Linguistic Signal to Clinical Reality; Baltimore, Maryland, USA. Association for Computational Linguistics; 2014. pp. 7–16. [Google Scholar]
- [23].Angus D, Watson B, Smith A, Gallois C, Wiles J. Visualising conversation structure across time: insights into effective doctor-patient consultations. PLOS One. 2012;7(6):1–12. doi: 10.1371/journal.pone.0038014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Althoff T, Clark K, Leskovec J. Natural Language Processing for Mental Health: Large Scale Discourse Analysis of Counseling Conversations. CoRR abs/1605.04462. URL < http://arxiv.org/abs/1605.04462>. [PMC free article] [PubMed]
- [25].Yelland E. What text mining analysis of psychotherapy records can tell us about therapy process and outcome. Ph.D. thesis; UCL (University College London): 2017. [Google Scholar]
- [26].Pestian JP, Matykiewicz P, Linn-Gust M, South B, Uzuner O, Wiebe J, Cohen KB, Hurdle J, Brew C. Sentiment analysis of suicide notes: a shared task. Biomedical Informatics Insights. 2012;5:3. doi: 10.4137/BII.S9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Milne DN, Pink G, Hachey B, Calvo RA. CLPsych 2016 shared task: triaging content in online peer-support forums. Proceedings of the Third Workshop on Computational Linguistics and Clinical Psychology; San Diego, CA, USA. Association for Computational Linguistics; 2016. pp. 118–127. [Google Scholar]
- [28].Filannino M, Stubbs A, Uzuner Ö. Symptom severity prediction from neuropsychiatric clinical records: Overview of 2016 {CEGS} N-GRID shared tasks Track 2. J Biomed Inform. 2017 doi: 10.1016/j.jbi.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Suominen H, Pyysalo S, Hiissa M, Ginter F, Liu S, Marghescu D, Pahikkala T, Back B, Karsten H, Salakoski T. Performance evaluation measures for text mining. In: Song M, Wu Y-F, editors. Handbook of Research on Text and Web Mining Technologies. II. IGI Global; Hershey, Pennsylvania, USA: 2008. pp. 724–747. (Chapter XLI) [Google Scholar]
- [30].Steyerberg E. Clinical Prediction Models. Springer-Verlag; New York: 2009. [Google Scholar]
- [31].Collins G, Reitsma J, Altman D, Moons K. Transparent reporting of a multi-variable prediction model for individual prognosis or diagnosis (tripod): The tripod statement. Ann Intern Med. 2015;162(1):55–63. doi: 10.7326/M14-0697. URL arXiv:/data/journals/aim/931895/0000605-201501060-00009.pdf. [DOI] [PubMed] [Google Scholar]
- [32].Sparck Jones K. Evaluating Natural Language Processing Systems An Analysis and Review. Lecture Notes in Computer Science, Lecture Notes in Artificial Intelligence. 1995;1083 [Google Scholar]
- [33].Paroubek P, Chaudiron S, Hirschman L. Editorial: Principles of Evaluation in Natural Language Processing. TAL. 2007;48(1):7–31. < http://www.atala.org/Editorial>. [Google Scholar]
- [34].Dybkjaer L. Evaluation of Text and Speech Systems. Text, Speech and Language Technology; 2007. p. 37. [Google Scholar]
- [35].Cohen PR, Howe AE. Toward AI research methodology: three case studies in evaluation. IEEE Trans Syst, Man Cybern. 1989;19(3):634–646. [Google Scholar]
- [36].Hripcsak G, Rothschild AS. Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc. 2005;12(3):296–298. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Goldstein BA, Navar AM, Pencina MJ, Ioannidis JPA. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc : JAMIA. 2017;24(1):198–208. doi: 10.1093/jamia/ocw042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Roberts A. Language, structure, and reuse in the electronic health record. AMA J Ethics. 2017;19(3):281–288. doi: 10.1001/journalofethics.2017.19.3.stas1-1703. [DOI] [PubMed] [Google Scholar]
- [39].Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011;18(2):181–186. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Greenhalgh T, Potts HWW, Wong G, Bark P, Swinglehurst D. Tensions and paradoxes in electronic patient record research: a systematic literature review using the meta-narrative method. Milbank Q. 2009;87(4):729–788. doi: 10.1111/j.1468-0009.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Carter G, Milner A, McGill K, Pirkis J, Kapur N, Spittal MJ. Predicting suicidal behaviours using clinical instruments: systematic review and meta-analysis of positive predictive values for risk scales. Br J Psychiatry. 2017;210(6):387–395. doi: 10.1192/bjp.bp.116.182717. < http://bjp.rcpsych.org/content/210/6/387>. [DOI] [PubMed] [Google Scholar]
- [42].Downs J, Velupillai S, Gkotsis G, Holden R, Kikoler M, Dean H, Fernandes A, Dutta R. Detection of suicidality in adolescents with autism spectrum disorders: developing a natural language processing approach for use in electronic health records. AMIA annual Symposium, AMIA; 2017. pp. 641–649. [PMC free article] [PubMed] [Google Scholar]
- [43].Gange S, Golub ET. From smallpox to big data: the next 100 years of epidemiologic methods. Am J Epidemiol. 2016;183(5):423–426. doi: 10.1093/aje/kwv150. [DOI] [PubMed] [Google Scholar]
- [44].Lynch SM, Moore JH. A call for biological data mining approaches in epidemiology. BioData Mining. 2016;9(1):1. doi: 10.1186/s13040-015-0079-8. < http://www.biodatamining.org/content/9/1/1>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bell J, Kilic C, Prabakaran R, Wang YY, Wilson R, Broadbent M, Kumar A, Curtis V. Use of electronic health records in identifying drug and alcohol misuse among psychiatric in-patients. The Psychiatrist. 2013;37(1):15–20. doi: 10.1192/pb.bp.111.038240. [DOI] [Google Scholar]
- [46].Ford E, Carroll JA, Smith HE, Scott D, Cassell JA. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc. 2016;23(5):1007–1015. doi: 10.1093/jamia/ocv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liao KP, Cai T, Savova GK, Murphy SN, Karlson EW, Ananthakrishnan AN, Gainer VS, Shaw SY, Xia Z, Szolovits P, Churchill S, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ. 2015;350:h1885. doi: 10.1136/bmj.h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20(1):117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, Basford M, Chute CG, Kullo IJ, Li R, Pacheco Ja, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Morley KI, Wallace J, Denaxas SC, Hunter RJ, Patel RS, Perel P, Shah AD, Timmis AD, Schilling RJ, Hemingway H. Defining disease phenotypes using national linked electronic health records: a case study of atrial fibrillation. PLoS One. 2014;9(11):e110900. doi: 10.1371/journal.pone.0110900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Peat G, Riley RD, Croft P, Morley KI, Kyzas Pa, Moons KGM, Perel P, Steyerberg EW, Schroter S, Altman DG, Hemingway H. Improving the transparency of prognosis research: the role of reporting, data sharing, registration, and protocols. PLoS Med. 2014;11(7):e1001671. doi: 10.1371/journal.pmed.1001671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wu S, Miller T, Masanz J, Coarr M, Halgrim S, Carrell D, Clark C. Negation’s not solved: generalizability versus optimizability in clinical natural language processing. PLOS One. 2014;9(11):1–11. doi: 10.1371/journal.pone.0112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42(5):760–772. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zheng K, Vydiswaran VGV, Liu Y, Wang Y, Stubbs A, Uzuner O, Gururaj AE, Bayer S, Aberdeen J, Rumshisky A, Pakhomov S, et al. Ease of adoption of clinical natural language processing software: an evaluation of five systems. J Biomed Inform. 2015;58(Suppl):S189–96. doi: 10.1016/j.jbi.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kaufman DR, Sheehan B, Stetson P, Bhatt AR, Field AI, Patel C, Maisel JM. Natural language processing-enabled and conventional data capture methods for input to electronic health records: a comparative usability study. JMIR Med Inform. 2016;4(4):e35. doi: 10.2196/medinform.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Suominen H, Müller H, Ohno-Machado L, Salanterä S, Schreier G, Hanlen L. Prerequisites for International Exchanges of Health Information: Comparison of Australian, Austrian, Finnish, Swiss, and US Privacy Policies. In: TBA, editor. Medinfo. Vol. 2017. 2017. [PubMed] [Google Scholar]
- [57].Suominen H, Zhou L, Hanlen L, Ferraro G. Benchmarking clinical speech recognition and information extraction: new data, methods, and evaluations. JMIR Med Inform. 2015;3(2):e19. doi: 10.2196/medinform.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Aramaki E, Morita M, Kano Y, Ohkuma T. Overview of the NTCIR-11 MedNLP task. Proceedings of the 11th NTCIR Conference; Tokyo, Japan. NII Testbeds and Community for Information access Research (NTCIR); 2014. pp. 147–154. [Google Scholar]
- [59].Serban I, Sordoni A, Lowe R, Charlin L, Pineau J, Courville A, Bengio Y. A hierarchical latent variable encoder-decoder model for generating dialogues. AAAI Conference on Artificial Intelligence; 2017. URL < https://aaai.org/ocs/index.php/AAAI/AAAI17/paper/view/14567>. [Google Scholar]
- [60].Uzuner O, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].McCowan I, Moore D, Fry M-J. Classification of cancer stage from free-text histology reports. Conf Proc IEEE Eng Med Biol Soc. 2006;1:5153–5156. doi: 10.1109/IEMBS.2006.259563. [DOI] [PubMed] [Google Scholar]
- [62].Gkotsis G, Velupillai S, Oellrich A, Dean H, Liakata M, Dutta R. Don’t let notes be misunderstood: a negation detection method for assessing risk of suicide in mental health records. Proceedings of the Third Workshop on Computational Linguistics and Clinical Psychology, Association for Computational Linguistics; San Diego, CA, USA. 2016. pp. 95–105. < http://www.aclweb.org/anthology/W16-0310>. [Google Scholar]
- [63].Kaur H, Sohn S, Wi C-I, Ryu E, Park MA, Bachman K, Kita H, Croghan I, Castro-Rodriguez JA, Voge GA, Liu H, et al. Automated chart review utilizing natural language processing algorithm for asthma predictive index. BMC Pulmonary Med. 2018;18(1):34. doi: 10.1186/s12890-018-0593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Miotto R, Li L, Kidd Brian A, Dudley Joel T. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016;6(2045–2322) doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Elvevag B, Foltz PW, Rosenstein M, Delisi LE. An automated method to analyze language use in patients with schizophrenia and their first-degree relatives. J Neurolinguist. 2010;23(3):270–284. doi: 10.1016/j.jneuroling.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Corcoran CM, Carrillo F, Fernández-Slezak D, Bedi G, Klim C, Javitt DC, Bearden CE, Cecchi GA. Prediction of psychosis across protocols and risk cohorts using automated language analysis. World Psychiatry. 2018;17(1):67–75. doi: 10.1002/wps.20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Fraser KC, Meltzer JA, Graham NL, Leonard C, Hirst G, Black SE, Rochon E. Automated classification of primary progressive aphasia subtypes from narrative speech transcripts, Language. Comput Cognit Neurosci. 2014;55:43–60. doi: 10.1016/j.cortex.2012.12.006. < http://www.sciencedirect.com/science/article/pii/S0010945212003413>. [DOI] [PubMed] [Google Scholar]
- [68].Keuleers E, Balota DA. Megastudies, crowdsourcing, and large datasets in psycholinguistics: an overview of recent developments. Quart J Exp Psychol. 2015;68(8):1457–1468. doi: 10.1080/17470218.2015.1051065. [DOI] [PubMed] [Google Scholar]
- [69].Coppersmith G, Dredze M, Harman C, Hollingshead K, Mitchell M. CLPsych 2015 Shared Task: Depression and PTSD on Twitter. Proceedings of the 2nd Workshop on Computational Linguistics and Clinical Psychology: From Linguistic Signal to Clinical Reality, Association for Computational Linguistics; Denver, Colorado. 2015. pp. 31–39. [Google Scholar]
- [70].Benton A, Mitchell M, Hovy D. Multitask learning for mental health conditions with limited social media data. Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics, Long Papers; Valencia, Spain. Association for Computational Linguistics; 2017. pp. 152–162. [Google Scholar]
- [71].Tsakalidis A, Liakata M, Damoulas T, Jellinek B, Guo W, Cristea A. Combining Heterogeneous User Generated Data to Sense Well-being. Proceedings of COLING 2016, the 26th International Conference on Computational Linguistics: Technical Papers; Osaka, Japan. The COLING 2016 Organizing Committee; 2016. pp. 3007–3018. [Google Scholar]
- [72].Canzian L, Musolesi M. Trajectories of depression: unobtrusive monitoring of depressive states by means of smartphone mobility traces analysis; Proceedings of the 2015 ACM International Joint Conference on Pervasive and Ubiquitous Computing; New York, NY, USA. UbiComp ’15, ACM; 2015. pp. 1293–1304. [DOI] [Google Scholar]
- [73].Jaques N, Taylor S, Sano A, Picard R. Multi-task, multi-kernel learning for estimating individual wellbeing. Proceedings of NIPS Workshop on Multimodal Machine Learning; 2015. [Google Scholar]
- [74].Jaques N, Rudovic O, Taylor S, Sano A, Picard R. Predicting tomorrow’s mood, health, and stress level using personalized multitask learning and domain adaptation. Proc IJCAI; 2017. [Google Scholar]
- [75].Tsakalidis A, Liakata M, Damoulas T, Cristea A. Can we assess mental health through social media and smart devices? Addressing bias in methodology and evaluation. Proceedings of ECML-PKDD 2018, the European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases; Dublin, Ireland. The ECML-PKDD Organizing Committee; 2018. [Google Scholar]
- [76].Johnson AE, Pollard TJ, Shen L, Lehman L-wH, Feng M, Ghassemi M, Moody B, Szolovits P, Anthony Celi L, Mark RG. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3 doi: 10.1038/sdata.2016.35. 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Harle CA, Golembiewski EH, Rahmanian KP, Krieger JL, Hagmajer D, Mainous AG, 3rd, Moseley RE. Patient preferences toward an interactive e-consent application for research using electronic health records. J Am Med Inform Assoc. 2018;25(3):360–368. doi: 10.1093/jamia/ocx145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Suominen H, Hanlen L, Paris C. Twitter for health — seeking to understand and curate laypersons’ personal experiences: building a social media search engine to improve search, summarization, and visualization. In: Neustein A, editor. Speech Technology and Natural Language Processing in Medicine and Healthcare. De Gruyter; Berlin, Germany: 2014. pp. 134–174. (Chapter 6) [Google Scholar]
- [79].Johnson M, Lapkin S, Long V, Sanchez P, Suominen H, Basilakis J, Dawson L. A systematic review of speech recognition technology in health care. BMC Med Inform Decis Mak. 2014;14:94. doi: 10.1186/1472-6947-14-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Goeuriot L, Kelly L, Suominen H, Hanlen L, Névéol A, Grouin C, Palotti JRM, Zuccon G. Overview of the CLEF eHealth Evaluation Lab 2015. In: Mothe J, Savoy J, Kamps J, Pinel-Sauvagnat K, Jones GJF, SanJuan E, Cappellato L, Ferro N, editors. Experimental IR Meets Multilinguality, Multimodality, and Interaction, Proceedings of the Sixth International Conference of the CLEF Association (CLEF 2015), Lecture Notes in Computer Science (LNCS) 9283; Heidelberg, Germany. Springer; 2015. pp. 429–443. [Google Scholar]
- [81].Hodgson T, Coiera E. Risks and benefits of speech recognition for clinical documentation: a systematic review. J Am Med Inform Assoc. 2016;23(e1):e169–e179. doi: 10.1093/jamia/ocv152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Suominen H, Johnson M, Zhou L, Sanchez P, Sirel R, Basilakis J, Hanlen L, Estival D, Dawson L, Kelly B. Capturing patient information at nursing shift changes: methodological evaluation of speech recognition and information extraction. J Am Med Inform Assoc. 2015;22(e1):e48–66. doi: 10.1136/amiajnl-2014-002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hodgson T, Magrabi F, Coiera E. Evaluating the usability of speech recognition to create clinical documentation using a commercial electronic health record. Int J Med Inform. 2018;113:38–42. doi: 10.1016/j.ijmedinf.2018.02.011. < http://www.sciencedirect.com/science/article/pii/S1386505618300820>. [DOI] [PubMed] [Google Scholar]
- [84].Mollá D, Hutchinson B. Intrinsic versus extrinsic evaluations of parsing systems. Proceedings of the EACL 2003 Workshop on Evaluation Initiatives in Natural Language Processing: Are Evaluation Methods, Metrics and Resources Reusable?, Evalinitiatives ’03; Stroudsburg, PA, USA. Association for Computational Linguistics; 2003. pp. 43–50. < http://dl.acm.org/citation.cfm?id=1641396.1641403>. [Google Scholar]
- [85].Nguyen K, O’Connor B. Posterior calibration and exploratory analysis for natural language processing models. Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing; Lisbon, Portugal. Association for Computational Linguistics; 2015. pp. 1587–1598. < http://aclweb.org/anthology/D15-1182>. [Google Scholar]
- [86].Scuba W, Tharp M, Mowery D, Tseytlin E, Liu Y, Drews FA, Chapman WW. Knowledge Author: facilitating user-driven, domain content development to support clinical information extraction. J Biomed Semant. 2016;7(1):42. doi: 10.1186/s13326-016-0086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ioannidis JPA. Why most clinical research is not useful. PLOS Med. 2016;13(6):1–10. doi: 10.1371/journal.pmed.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ioannidis JPA. How to make more published research true. PLoS Med. 2014;11(10):e1001747. doi: 10.1371/journal.pmed.1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet (London, England) 2007;370(9596):1453–1457. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- [90].Benchimol EI, Smeeth L, Guttmann A, Harron K, Moher D, Petersen I, Sørensen HT, von Elm E, Langan SM, R.W. Committee The reporting of studies conducted using observational routinely-collected health data (record) statement. PLOS Med. 2015;12(10):1–22. doi: 10.1371/journal.pmed.1001885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Gilbert R, Lafferty R, Hagger-Johnson G, Harron K, Zhang L-C, Smith P, Dibben C, Goldstein H. Guild: Guidance for information about linking data sets. J Public Health. 2018;40(1):191–198. doi: 10.1093/pubmed/fdx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The lack of sufficiently large sets of shareable data is still a problem in the clinical NLP domain. We encourage the increased development of alternative data sources such as synthetic clinical notes [57,58], which alleviates the complexities involved in governance structures. However, in parallel, initiatives to make authentic data available to the research community through alternative governance models are also encouraged, like the MIMIC-III database [76]. Greater connection between NLP researchers, primary data collectors, and study participants are required. Further studies in alternative patient consent models (e.g., interactive e-consent [77]) could lead to larger availability of real-world data, which in turn could lead to substantial advances in NLP development and evaluation. Moving beyond EHR data, there is valuable information also in accessible online data sources such as social media (e.g., PatientsLikeMe), that are of particular relevance to the mental health domain, and that could also be combined with EHR data [78]. Efforts to engage users in donating their public social media and sensor data for research such as OurDataHelps9 are interesting avenues that could prove very valuable for NLP method development. Furthermore, in addition to written documentation, there is promise in the use of speech technologies, specifically for information entry at the bedside [57,79–83].