Abstract
Human brain networks based on neuroimaging data have already proven useful in characterizing both normal and abnormal brain structure and function. However, many brain disorders are neurodevelopmental in origin, highlighting the need to go beyond characterizing brain organization in terms of static networks. Here, we review the fast-growing literature shedding light on developmental changes in network phenotypes. We begin with an overview of recent large-scale efforts to map healthy brain development, and we describe the key role played by longitudinal data including repeated measurements over a long period of follow-up. We also discuss the subtle ways in which healthy brain network development can inform our understanding of disorders, including work bridging the gap between macroscopic neuroimaging results and the microscopic level. Finally, we turn to studies of three specific neurodevelopmental disorders that first manifest primarily in childhood and adolescence/early adulthood, namely psychotic disorders, attention-deficit/hyperactivity disorder, and autism spectrum disorder. In each case we discuss recent progress in understanding the atypical features of brain network development associated with the disorder, and we conclude the review with some suggestions for future directions.
Keywords: ADHD, Autism spectrum disorder, Development, MRI, Network neuroscience, Schizophrenia
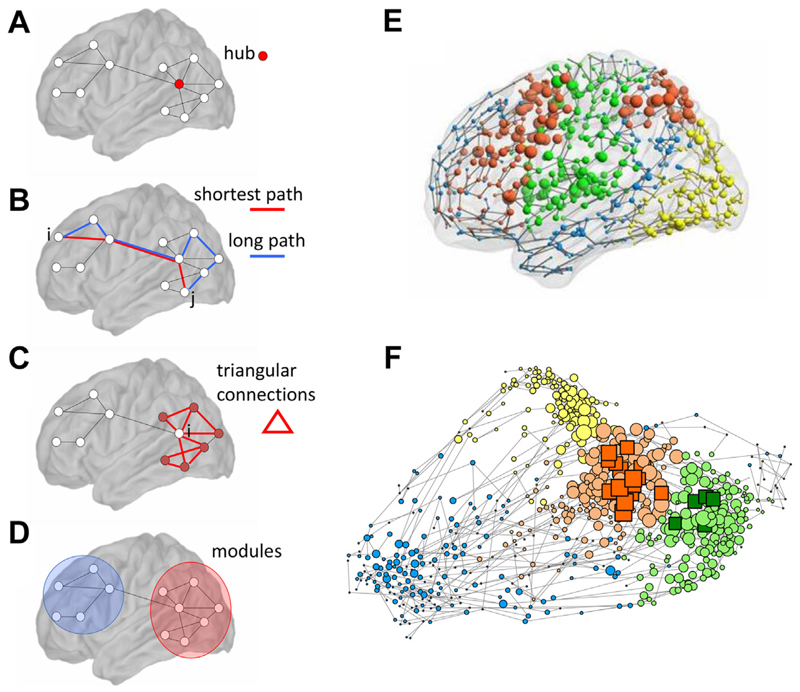
Childhood and adolescence are critical periods during which several brain disorders develop. Characterizing how atypical developmental trajectories diverge from normal brain development is essential to understand and ultimately treat these disorders. Network models of neuroimaging data have already highlighted key drivers in brain organization (1–3), for example, the role of hubs in coordinating information transfer (4) and the economic trade-off brain networks navigate between the cost of long-distance connections and their topological benefits, enabling efficient information processing by connecting distant brain regions (5). Figure 1 summarizes commonly used graph metrics. However, owing to the difficulty and cost of recruiting and scanning large cohorts, historically many network studies of neurodevelopmental disorders have focused on static patient/control differences, disregarding the changing structure of brain networks over time. What can network models tell us about healthy brain development and how developmental trajectories differ in disease? Several neuroimaging studies have begun to tackle this question, including the Human Connectome Project Development study (6), the Developing Human Connectome Project (7), the NeuroScience in Psychiatry Network (8), and the Philadelphia Neurodevelopmental Cohort (9), among others. These collaborative projects benefit from large sample sizes with several hundred participants, which is crucial to obtain enough power to detect statistically significant, potentially nonlinear changes with age. Perhaps the most exciting feature of some of these projects is the longitudinal neuroimaging data being acquired (Box 1). While cross-sectional data can provide important insights into average group changes, longitudinal data are essential to study development at an individual level because significant individual differences exist even between healthy individuals of similar ages (10,11), which may confound cross-sectional analyses (12). In atypical development, the trajectory of changes in an individual’s brain network may be a more useful biomarker than the absolute values (13).
Figure 1.
An illustration of commonly used network metrics. (A) Degree, hubs, and rich clubs: The degree of a node is the number of links or connections it makes. Hubs are nodes with a significantly higher degree than other nodes in the network. In many networks, these nodes preferentially connect to one another, forming an elite group of nodes called a rich club. (B) Path length and efficiency: the minimum path length between two nodes i and j is the minimum number of edges that need to be traversed to go from one node to another, and efficiency is inversely related to path length. (C) Clustering and local efficiency: the clustering coefficient measures the number of connections that exist between the first neighbors of a node i as a proportion of the maximum possible number of such connections. These edges form triangular connections around the node i, which increases the local efficiency by decreasing the path length locally between the node’s neighbors. (D) Modularity: Many complex networks have a modular community structure, whereby they contain subsets of highly interconnected nodes called modules. Many of these features are highlighted in a brain functional network shown in both (E) anatomical space and (F) topological space. Node size is proportional to degree and rich-club nodes are highlighted as squares in panel (F). The modular organization is highlighted by assigning different colors to nodes of different modules. Reproduced with permission from Vértes et al. (14) and Crossley et al. (134).
Box 1. From Cross-Sectional to Longitudinal Analysis.
To understand developmental change, it is crucial to distinguish individual age-related changes from subject heterogeneity. Cross-sectional studies only contain a single observation per individual in a cohort of varying age and therefore cannot make this distinction (135,136). In contrast, longitudinal studies incorporate repeat measurements of the same individuals at multiple time points. Longitudinal studies increase the complexity of design and analysis, but the scientific benefits to understanding developmental changes are substantial.
Mixed-effects models are an established approach to analyzing longitudinal data (24,137–139) and include (as special cases) other well-known techniques such as change-score regression and repeated-measures analysis of variance. However, standard mixed-effects models are not without issues, notably, assumptions of Gaussian errors and the need to specify the random-effects structure (139). Longitudinal data also bring issues of attrition, which if associated with a biomarker or outcome of interest may lead to differential missingness, also known as missing not at random (140). Another challenge is the so-called age-period-cohort problem (141)—including the difficulty of separating changes owing to aging from changes due to external factors at different calendar periods, which is especially pertinent for long-term studies.
Study design is particularly important for longitudinal studies and requires balancing robust and reliable inferences against recruitment and costs to determine not only how many participants, but also how many visits per subject are needed. A classical longitudinal design follows a single cohort of subjects for the duration of interest, observing each subject multiple times. However, this makes the duration of many developmental studies untenable.
An alternative is the accelerated longitudinal design (142), also called the cohort-sequential design (143), in which multiple cohorts are followed with overlapping follow-up. This introduces two new design considerations, the number of cohorts and the overlap, which affect the acceleration of the design. An accelerated longitudinal design with sufficient overlap can disentangle age-period-cohort effects and the number of observations per subject is the primary determinant of power to detect an age-related trajectory (144).
Here, we build on previous reviews (14–20), with particular emphasis on how healthy development can provide insights into abnormal development and the importance of longitudinal data. We begin by reviewing cross-sectional and longitudinal studies of healthy brain network development and their implications for understanding neurodevelopmental disorders. We then turn to studies of three specific neurodevelopmental disorders that first manifest primarily in childhood and adolescence/early adulthood, namely psychotic disorders, attention-deficit/hyperactivity disorder (ADHD), and autism spectrum disorder (ASD). We conclude with suggestions for future directions.
Healthy Development
The human brain undergoes extraordinary changes during development, from conception to birth and throughout childhood. Before birth, neurogenesis is largely completed by week 20 after conception; axons begin to grow and synapses start to form between neurons (19,21). After birth, a further proliferation phase in which the number of synapses continues to increase is followed by a consolidation phase characterized by cell loss, synaptic pruning, and myelination of axons, which is thought to continue into adolescence (22). Neuroimaging provides a noninvasive, albeit indirect, way to study these developmental changes. Table 1 lists cross-sectional and longitudinal studies performed in healthy children and adolescents, using structural magnetic resonance imaging (sMRI), diffusion-weighted imaging (DWI), and functional MRI (fMRI).
Table 1. Cross-Sectional and Longitudinal Neuroimaging Brain Network Studies Performed in Healthy Children and Adolescents, Using sMRI, DWI, and fMRI.
| Author | Date | No. of Subjects | Age | Longitudinal | Data Type |
|---|---|---|---|---|---|
| Alexander-Bloch et al. (42) | 2013 | 108 | 9–22 years | Yes | sMRI, rs-fMRI |
| Fan et al. (36) | 2011 | 28 | 0–2 years | Yes | sMRI |
| Khundrakpam et al. (37) | 2013 | 203 | 5–18 years | No | sMRI |
| Moura et al. (39) | 2017 | 249 | 7–14 years | No | sMRI |
| Nie et al. (38) | 2013 | 445 | 3–20 years | Yes | sMRI |
| Váša et al. (44) | 2018 | 297 | 14–24 years | No | sMRI |
| Whitaker et al. (43) | 2016 | 297 | 14–24 years | No | sMRI |
| Zielinkski et al. (40) | 2010 | 300 | 5–18 years | No | sMRI |
| Baker et al. (55) | 2015 | 31 | 15–19 years | Yes | DWI |
| Baum et al. (54) | 2017 | 882 | 8–22 years | No | DWI |
| Dennis et al. (53) | 2013 | 439 | 12–30 years | No | DWI |
| Koenis et al. (58) | 2015 | 162 | 9–15 years | Yes | DWI |
| Lim et al. (57) | 2013 | 121 | 4–40 years | No | DWI |
| Tymofiyeva et al. (51) | 2013 | 33 | 0–6 months | No | DWI |
| Wierenga et al. (52) | 2016 | 85, 38 | 7–23 years | No | DWI |
| Wierenga et al. (56) | 2017 | 146, 141 | 4–13 years, 8–13 years | Yes | DWI |
| Yap et al. (46) | 2011 | 39 | 2 weeks to 2 years | Yes | DWI |
| Hagmann et al. (49) | 2010 | 30 | 2–18 years | No | DWI/rs-fMRI |
| Uddin et al. (76) | 2011 | 23 children, 22 adults | 7–9 years, 19–22 years | No | DWI/rs-fMRI |
| Cao et al. (71) | 2014 | 126 | 7–85 years | No | rs-fMRI |
| Fair et al. (67) | 2009 | 210 | 7–31 years | No | rs-fMRI |
| Fair et al. (128) | 2007 | 139 | 7–9 years, 10–15 years, 20–31 years | No | rs-fMRI |
| Fransson et al. (68) | 2011 | 18 infants, 18 adults | 39 weeks, 22–41 years | No | rs-fMRI |
| Gao et al. (70) | 2011 | 147 | 2 weeks to 2 years | No | rs-fMRI |
| Gu et al. (78) | 2015 | 780 | 8–22 years | No | rs-fMRI |
| Hwang et al. (75) | 2013 | 99 | 10–20 years | No | rs-fMRI |
| Marek et al. (77) | 2015 | 192 | 10–26 years | No | rs-fMRI |
| Satterthwaite et al. (74) | 2013 | 780 | 8–22 years | No | rs-fMRI |
| Supekar et al. (69) | 2009 | 23 children, 22 adults | 7–9 years, 19–22 years | No | rs-fMRI |
| Wu et al. (72) | 2013 | 51 | 6–18 years | No | rs-fMRI |
| Dosenbach et al. (73) | 2010 | 115 | 7–30 years | No | fMRI |
| Kaufmann et al. (79) | 2017 | 797 | 8–22 years | No | fMRI |
Studies were identified using the PubMed search terms: brain, network, or graph theory, longitudinal or cross-sectional or development, image or imaging or MRI or DTI or scan, and healthy or normal. Studies were only included if they included subjects below 21 years of age, discussed network or graph theoretical properties, and were either cross-sectional (studying changes in age across a population) or longitudinal.
DTI, diffusion tensor imaging; DWI, diffusion-weighted imaging; fMRI, functional magnetic resonance imaging; rs-fMRI, resting-state functional magnetic resonance imaging; sMRI, structural magnetic resonance imaging.
Structural MRI
sMRI provides several metrics characterizing brain morphology, for example, gray and white matter volumes and cortical thickness (CT). The first longitudinal pediatric neuroimaging study was performed in 1999 and showed that gray matter volume increases in early childhood before decreasing in later childhood/adolescence, forming an inverted U–shaped trajectory, which peaks at different ages in different brain regions (23). These peaks occur later than expected from cross-sectional studies (13), although recent longitudinal studies have suggested that methodological issues might have confounded these results and CT may decrease from earlier in childhood (24–26). For example, Mills et al. (25) found that gray matter volume decreased from 8 to 30 years of age in four longitudinal samples. One potential confound is that in-scanner movement can lead to reduced CT and volume estimates (27,28), which can bias developmental trajectories. A recent multisample study analyzed longitudinal changes in cortical volume, surface area, and thickness from 7 to 29 years of age and found that cortical thinning is the dominant contributor to volume reductions during adolescence (29). In Giedd et al. (23), white matter volumes increased from 4 to 20 years of age, in agreement with recent results (24). Microscopically, these changes are thought to be driven by the pruning of synapses, dendrites, or cell bodies and the myelination of axons. Several studies have shown that lower-order somatosensory and visual cortices mature first, followed by higher-order association cortices (30–34), in line with the order of cognitive and behavioral changes during childhood. These studies used a variety of developmental benchmarks, e.g., age at attainment of adult brain volume or at other inflection points.
To study the relationships between brain regions, morphological metrics can be correlated between regions across subjects to create one structural covariance network per subject group (35). The network’s nodes correspond to brain regions and the edges represent cross-correlations of morphological metrics between pairs of regions taken across subjects. While this approach cannot elucidate intersubject differences, it can provide insights into average developmental changes. For example, volumetric covariance networks suggest that small-world, modular topology is already present at 1 month (36). Later work found that network measures follow nonlinear trajectories (37,38); for example, in a longitudinal study, global efficiency and CT peak at 7 years of age, and at the same time local efficiency is lowest and cortical folding becomes stable (38). Correlations between fractional anisotropy (FA) (see below for details) and CT covariance matrices suggest that white and gray matter development is synchronized (39).
Like individual morphometric measures, in structural covariance networks different brain regions follow different developmental trajectories (37,40–42). Several studies have shown early maturation of primary regions followed by protracted development of high-order regions (37,40,41). One study of 108 adolescents with three to six longitudinal scans per participant created both structural and maturational covariance networks, in which maturational networks were derived from the rate of change of CT in different regions (42). Structural and maturational covariance networks exhibited similar topological properties and spatial organization, suggesting that correlated anatomical structure between brain regions results from similarities in maturational trajectories. In other words, brain regions with high structural covariance might have developed in similar ways.
Other work has highlighted the importance of hub regions (37,43,44). Khundrakpam et al. (37) found that both the number and the distribution of hubs change over development; the number of hubs peaks in late childhood, then the distribution of hubs shifts toward frontal regions during adolescence. Whitaker et al. (43) also pointed to a key role for hubs in association cortices during adolescence, showing that they are less myelinated at 14 years of age than other regions, but have faster rates of myelination and cortical shrinkage between 14 and 24 years of age. These results are consistent with the idea that primary sensory and motor regions develop early on, while hub regions of the association cortex that are responsible for more complex integrative function mature more slowly.
Recently Seidlitz et al. (45) proposed a new approach for calculating sMRI networks at the individual level, based on interregional similarity of several morphological metrics such as CT, gray matter volumes, surface area, and curvature. Future studies based on these morphometric similarity networks could enable a deeper understanding of how individual sMRI connectivity develops.
Diffusion-Weighted Imaging
DWI exploits the diffusion of water molecules to generate contrast between different types of brain tissue. It can be used to measure mean diffusivity, axial diffusivity, or radial diffusivity, and FA, which is believed to reflect fiber density, axonal diameter, and myelination. Finally, white matter fibers can be tracked to create a tractography (diffusion tensor imaging [DTI]) network where predefined gray matter regions of interest are connected by an edge if white matter fibers are inferred between them (46). As in sMRI, DWI is susceptible to motion artifacts, particularly DTI networks, due to their long acquisition times (47,48).
During development, diffusivity decreases with age while anisotropy increases (19). These changes are believed to reflect increasing myelination. Changes occur most rapidly up to 2 years, then continue throughout childhood and adolescence, until at least 18 years of age and possibly into early adulthood (49,50).
Many global topological properties of DTI networks are already established at birth, including small-worldness and the presence of hubs (46,49). However, several studies report substantial reorganization of DTI networks during childhood and adolescence, including monotonic increases in integration and global efficiency and decreases in clustering (46,49,51–53). The studies use several approaches to calculate edge weights, including binarization, radial diffusivity, and mean diffusivity values, and span age ranges from birth to 30 years of age. The monotonic changes are consistent with increasing white matter volume. Recently, Baum et al. (54) observed that while global efficiency increases with age, surprisingly both participation coefficient and between-module connectivity decrease, suggesting that networks become both more modular and more integrated. These changes were driven by strengthening of network hub edges. Other studies agree that connectivity between hubs or association regions changes disproportionately to other regions, particularly in adolescence (52,55). Recently, Wierenga et al. (56) also showed increases in connectivity between rich-club nodes and peripheral nodes. We note that some studies observe decreasing global efficiency with age (57,58). These different results may be due to using different measures, as Lim et al. (57) used networks weighted by streamline count, while Baum et al. (54) used FA. In line with these results, in Koenis et al. (58) networks weighted by streamline count showed decreasing global efficiency with age, while FA-weighted networks showed increasing global efficiency. In Lim et al. (57), the proportion of intermodule connections increased with age, again unlike Baum et al. (54), and possibly owing to differences in network construction.
Functional MRI
In fMRI brain images, each voxel is associated with a time series. There are several approaches to analyzing fMRI networks, including 1) independent component analysis, a data-driven approach that identifies components that group voxels into regions with similar response patterns; 2) seed-based maps, which study correlations between a particular seed region and other areas of the brain; 3) regional maps, which study correlations between brain regions selected a priori; and 4) whole-brain network analysis, in which an atlas is used to parcellate the brain into regions and correlations are studied between regions. We note that there are still questions about how to preprocess fMRI data. As for other modalities, movement can bias estimates of functional connectivity (59–61). Several authors have proposed methods to address this problem (61–64); however, older studies may still be affected. The role of intersubject differences in mean connectivity and whether to perform global signal regression are also topics of debate (65,66).
In contrast to structural networks, in fMRI networks global topological metrics including efficiency and clustering generally remain stable across development (67–69), although Gao et al. (70) observed some changes in global efficiency between neonates and 1-year-olds, and Gao et al. (71) and Wu et al. (72) observed some changes in clustering at later stages of development.
Despite these broadly constant global topological properties, several authors have shown that long-range connections strengthen during development and short-range connections weaken (67,69,70,73,74). Although this result may be exaggerated by motion artifacts in early studies, the effect remains significant after motion correction (74).
Consistent with structural results, several developmental changes focus on hub regions (71,75,76), and intermodular integration also increases with development (67,77). Recent evidence from 780 subjects from 8 to 22 years of age suggests that different modules follow distinct developmental trajectories: for example, the default mode system shows increasing inter- and intrasystem connectivity, while sensorimotor systems become increasingly segregated from other systems (78).
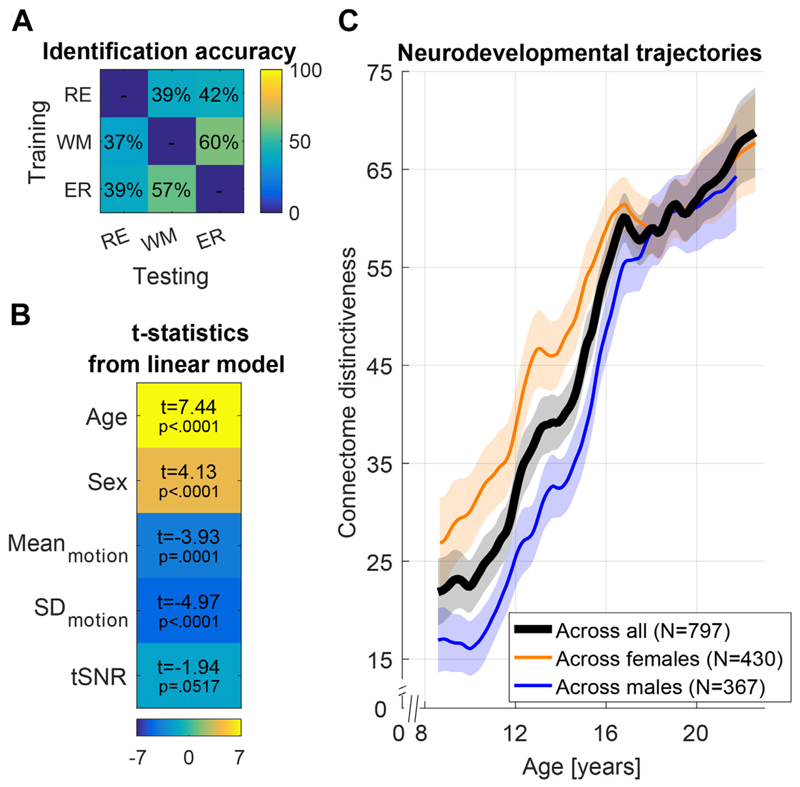
Finally, recent work found that subjects develop more stable, individual functional connectomes during adolescence (79). In particular, for 797 subjects from 8 to 22 years of age, the identification accuracy of one resting-state fMRI (rs-fMRI) or task fMRI scan from a subject based on other fMRI scans from the same subject (termed connectome distinctiveness) increases with age (see Figure 2). Machine-learning approaches like this one offer an alternative, individual-centric perspective on development.
Figure 2.
(A) Average identification accuracy using testing and training functional magnetic resonance imaging scans from a working memory task (WM), emotion recognition task (ER), and resting state (RE). (B) Statistics from a linear model of connectome distinctiveness. (C) Changes in connectome distinctiveness with age. Male subjects developed later than female subjects. Similar results were obtained using independent component analysis. Reprinted with permission from Kaufmann et al. (79). tSNR, temporal signal-to-noise ratio.
A Window into Atypical Development
Developmental studies on healthy subjects provide a crucial benchmark for neurodevelopmental disorders. They can also provide further insights, e.g., by 1) studying individuals from a general population with disorder-like traits or (subclinical) symptoms; 2) highlighting which network properties/regions change at which stages of development and linking them to the disorders separately shown to affect the same network properties/regions; and 3) associating brain development with genes that are known to be implicated in disorders.
Here, we discuss studies that take these approaches to explore what large healthy developmental cohorts can teach us about brain disorders. Above, we discussed work by Kaufmann et al. (79), which showed that connectome distinctiveness increases from 8 to 22 years of age. The authors also showed that this increase is delayed in subjects from the general population with increased clinical symptom scores for a range of disorders. This delay in connectome distinctiveness reflects both a delay in stabilization of an individual’s brain network over time and a delay in the individualization of the subject’s brain network with respect to the other subjects.
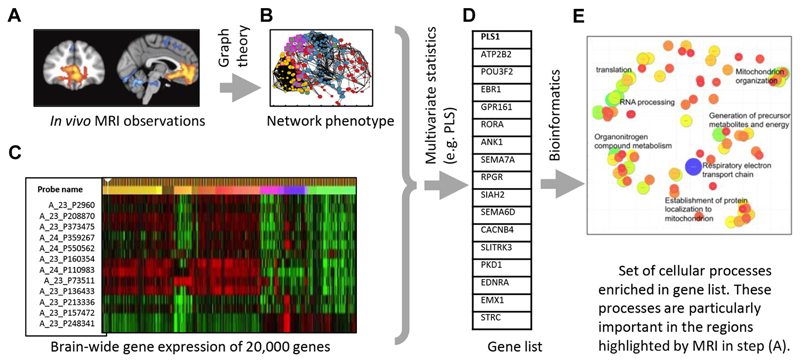
Healthy subjects can also inform our understanding of disorders by highlighting the network features that change during development and are therefore particularly vulnerable to abnormal development. As discussed above, one of the main results from several healthy developmental studies, across multiple modalities, is that developmental changes often focus on hub regions. For example, Whitaker et al. (43) showed that the greatest rates of myelination and cortical shrinking during adolescence are focused on hub regions of the association cortex. Disruptions to hub connectivity have been observed in patients: for example, a meta-analysis showed that hub regions are more likely to be anatomically abnormal than nonhub regions in nine different brain disorders, including Alzheimer’s disease and schizophrenia (80). Together, these results suggest that disorders may be linked to abnormal development of these key regions. In Whitaker et al. (43), the authors combined neuroimaging with the Allen Institute for Brain Science dataset on brainwide gene expression (81) (see Figure 3). The hub regions with greatest rates of myelination and cortical shrinkage from 14 to 24 years of age show overexpression of a set of genes enriched for both synaptic processes and schizophrenia risk-genes. The involvement of these risk genes in normal maturational processes during adolescence could explain why this period is particularly vulnerable to the onset of schizophrenia. The study provides an example of the way in which genetic and genomic datasets can link the brain development of healthy individuals and disorders for which there is a known set of risk genes (82). It also shows how macroscopic brain imaging results can be linked to a microscopic understanding of the brain (83–88). Similar methods are currently being applied to other disorders, including autism (89) and Huntington’s disease (90).
Figure 3.
Schematic of the methodology in Whitaker et al. (43). Gene expression data can be used to link magnetic resonance imaging (MRI) observations and network phenotypes to a microscopic understanding of the brain. (A) In vivo MRI observations are used to (B) identify network phenotypes. Multivariate statistical methods can then (C) combine this information with brainwide gene expression data and (D) identify genes that are associated with the network phenotype. (E) Cellular processes that are known to be enriched in those genes can provide information about the processes that are particularly important in the regions highlighted by MRI. PLS, partial least squares.
Abnormal Development
Despite the significant insights that can be gained from healthy developmental studies, ultimately building a comprehensive picture of disorders requires longitudinal studies of patients. Below we review longitudinal and cross-sectional studies on ASD, ADHD, and psychosis, which are listed in Tables 2, 3, and 4, respectively.
Table 2. Cross-Sectional and Longitudinal Neuroimaging Brain Network Studies Performed in Patients With ASD.
| Author | Date | No. of Subjects | Age | Longitudinal | Data Type |
|---|---|---|---|---|---|
| Chen et al. (103) | 2015 | 735 | 6–40 years | No | rs-fMRI |
| Nomi and Uddin (102) | 2015 | 26 children, 28 adolescents, 18 adults | 7–39 years | No | rs-fMRI |
| Ball et al. (101) | 2017 | 196, 51 | 5–86 years, 8–18 years | No | DWI |
| Ghanbari et al. (100) | 2014 | 24 ASD, 59 control subjects | 6–18 years | No | DWI |
| Lewis et al. (98) | 2017 | 260 | 6 months, 12 months | Yes | DWI |
| Watanabe and Rees (99) | 2015 | 45 ASD, 36 control subjects | 9–18 years | No | DWI |
Inclusion criteria were the same as for Table 1, except the search terms ASD and autism were used instead of healthy or normal.
ASD, autism spectrum disorder; DTI, diffusion tensor imaging; DWI, diffusion-weighted imaging; rs-fMRI, resting-state functional magnetic resonance imaging.
Table 3. Cross-Sectional and Longitudinal Neuroimaging Brain Network Studies Performed in Patients With ADHD.
| Author | Date | No. of Subjects | Age, Years | Longitudinal | Data Type |
|---|---|---|---|---|---|
| Fair et al. (112) | 2010 | 23 ADHD, 23 control subjects | 7–16 | No | rs-fMRI |
| Kessler et al. (114) | 2016 | 519, including 25 ADHD | 8–22 | No | rs-fMRI |
| Sripada et al. (113) | 2014 | 133 ADHD, 288 control subjects | 7–22 | No | rs-fMRI |
Inclusion criteria were the same as for Table 1, except the search terms ADHD and attention deficit disorder were used instead of healthy or normal.
ADHD, attention-deficit/hyperactivity disorder; rs-fMRI, resting-state functional magnetic resonance imaging.
Table 4. Cross-Sectional and Longitudinal Neuroimaging Brain Network Studies Performed in Patients With Psychotic Disorders.
| Author | Date | No. of Subjects | Age, Years | Longitudinal | Data Type |
|---|---|---|---|---|---|
| Alexander-Bloch et al. (120) | 2014 | 106 schizophrenia, 102 control subjects | 8–30 | Yes | sMRI |
| Zalesky et al. (121) | 2015 | 109 COS, 86 siblings, 102 control subjects | 12–24 | Yes | sMRI |
| Sun et al. (122) | 2016 | 31 schizophrenia, 28 control subjects | 19–54 | Yes | DWI |
| Jiang et al. (124) | 2015 | 20 adult-onset/26 early-onset schizophrenia patients, 17/25 age-matched control subjects | 26 ± 8, 15 ± 2, 30 ± 11, 14 ± 3 |
No | rs-fMRI |
| Tomasi and Volkow (123) | 2014 | 40 children/adolescents, 69 schizophrenia, 74 control subjects | 13 ± 4, 38 ± 14, 36 ± 12 |
No | rs-fMRI |
| Wang et al. (125) | 2017 | 76 ARMS nontransition, 12 ARMS transition, 48 control subjects | 21.7 ± 3.6, 19.7 ± 3.1 21.5 ± 4.2 |
Yes | rs-fMRI |
Inclusion criteria were the same as for Table 1, except the search terms psychosis and schizophrenia were used instead of healthy or normal.
ARMS, at-risk mental state; COS, childhood-onset schizophrenia; rs-fMRI, resting-state functional magnetic resonance imaging; sMRI, magnetic resonance imaging.
Autism Spectrum Disorder
ASD manifests in the first few years of childhood and strongly impacts subsequent development. Zielinski et al. (91) found that CT development in boys with ASD undergoes three phases: in childhood cortical expansion is accelerated compared with control subjects; in late childhood/adolescence cortical thinning is accelerated; and in early adulthood cortical thinning is decelerated. The wide age range studied, from 3 to 36 years of age, was made possible by an accelerated longitudinal design (see Box 1). Topologically, studies comparing adult ASD patients and control subjects have found evidence for reduced efficiency and long-range connections in patients’ fMRI and DTI networks (92,93), although increased local connectivity has also been observed (93,94). Network changes may be state dependent (e.g., sustained attention vs. at rest) (95). Disruption has also been observed in hub regions and regions related to social interaction, which is abnormal in ASD (96,97).
In DTI networks, reduced local efficiencies in low-level sensory processing regions have been observed in high-risk infants as young as 6 months of age and correlate significantly with 24-month symptom severity (98). Later in development, ASD patients fail to show the increases in rich-club organization observed in typically developing subjects (99). Nonnegative matrix factorization can be used to identify developmental subnetworks in ASD and subnetworks that discriminate between patients and control subjects. For example, Ghanbari et al. (100) identified one subnetwork with reduced interhemispheric subcortical connections that can discriminate groups but does not correlate with age, and another subnetwork involving frontal regions that is both discriminative and developmental. In contrast, other work taking a similar approach found increases in local connectivity in the cingulate cortex in ASD (101). Functional resting-state networks show a similarly complex picture. One study showed decreased default mode network (DMN) connectivity in independent component analysis networks of ASD children and adolescents, which normalizes by adulthood (102). Chen et al. (103) also observed reduced connectivity in some network edges, although other edges showed increased connectivity. A recent study suggested that rs-fMRI can predict whether 6-month-old infants develop autism at 24 months, although the sample size was relatively modest (59 infants, including 11 with ASD) (104). Further work is needed to clarify how connectivity develops in ASD.
Attention-Deficit/Hyperactivity Disorder
ADHD is one of the most common neurodevelopmental disorders and is often diagnosed between 6 and 12 years of age. Adult patients’ rs-fMRI networks show reduced integration and increased segregation compared with control subjects (105,106), and a DTI study observed local reductions in efficiency and increases in clustering (107). Several authors have highlighted the importance of development in ADHD. Patients exhibit reduced brain volumes, which normalize in some regions during development but not in others (108). There is also a delay in the age at which ADHD patients’ CT peaks, which was first established by Shaw et al. (109) using a mixed longitudinal and cross-sectional design.
Most work to date on brain network development in ADHD has focused on rs-fMRI networks, which exhibit higher clustering and lower global efficiency in children with ADHD than in age-matched control subjects (110,111), in line with results in adults. These measures correlate with behavioral symptoms including inattention and hyperactivity/impulsivity symptoms and differ between clinical subtypes (110). Early results showed that the rs-fMRI connectivity of the DMN is reduced in 23 ADHD patients compared with age-matched control subjects (112). Based on previous work on healthy development, this suggested a delay or disruption to development in ADHD patients. Later, Sripada et al. (113) showed that the connectivity within the DMN and between the DMN and task-positive networks in ADHD patients does indeed lag behind control subjects in a large cross-sectional sample from 7 to 22 years of age. Recently, Kessler et al. (114) used an independent component analysis approach to chart the development of brain connectivity networks in 519 subjects from 8 to 22 years of age, including 25 with ADHD. They found evidence for disrupted maturation in ADHD patients’ functional networks, which involves the DMN, although interestingly maturation is reduced (downshifted) in patients rather than delayed. It would be interesting to see if these results could be replicated in a larger, longitudinal sample and related to structural network changes.
Psychosis
Psychotic disorders are often associated with brain dysconnectivity. sMRI studies show reduced CT and volume in adults with schizophrenia (115), DTI network connectivity is often reduced (116), and patients’ rs-fMRI networks exhibit lower mean correlations and more randomized topologies (117). Psychotic disorders normally manifest in late adolescence/early adulthood, although earlier developmental changes could be valuable to identify individuals at risk.
Longitudinal studies show greater volume reductions in patients than in control subjects during adolescence, particularly in the frontal and temporal regions (118). Similarly, CT shows exaggerated thinning in patients (119). Alexander-Bloch et al. (120) found both age-constant deficits of CT and faster cortical thinning in childhood-onset schizophrenia patients compared with control subjects. They used structural covariance to derive five brain development modules with similar maturational trajectories in normal subjects and found that deviations in patients are primarily in the cingulofrontotemporal module. Other longitudinal work using CT structural covariance networks observed that left hemisphere occipitotemporal connectivity is significantly reduced in patients and siblings at 12 years of age compared with control subjects (121). By 17 years, siblings catch up with control subjects, but patients exhibit further delayed development, reminiscent of other disorders (79,114). Few DWI studies have focused on brain network development in psychosis, although in older subjects (19 to 54 years) longitudinal results show decreasing global efficiency over a 5-year period in patients with schizophrenia, compared with increases in the same period in control subjects (122).
There are few functional brain network studies of schizophrenia patients during childhood/adolescence. One study found that local degree, local clustering, and local path length are reduced in most cortical regions in adults compared with children/adolescents, and reduced in adults with schizophrenia compared with control subjects in the thalamus and midbrain regions (123). However not all healthy development studies show similar trends, as discussed previously. Other work has found a complex pattern of increases and decreases in local connectivity in early-onset schizophrenia patients (124). Further studies are required to resolve these differences. A recent rs-fMRI study of at-risk mental state subjects found that those who transitioned to psychosis exhibited less segregated networks and disrupted network communities (125).
An alternative approach to studying developmental disorders is using generative models (126,127), algorithms that generate synthetic networks with certain topological properties. Vértes et al. (126) proposed a generative model incorporating a term that penalizes long-distance connections and a topological term that favors connections between nodes with shared nearest neighbors. The model reproduces several topological properties of healthy rs-fMRI brain networks. Simulations of schizophrenia patients’ networks required a reduced distance penalization parameter, which increased the probability of long-distance connections, producing more randomized networks. Generative models could shed light on microscopic changes during disease. For example, increased probability of long-distance connections in disease could be related to the dysregulation of normal maturational processes such as pruning and myelination (43).
Conclusions and Future Directions
Larger sample sizes and the availability of longitudinal data are enabling more accurate assessments of the developmental changes of healthy children and adolescents across modalities. Healthy brain connectivity evolves during development, with evidence for stronger long-distance connections with age (74), more integration (52,67,128), and increasing connectome distinctiveness (79). Certain results have been observed across modalities, for example, the importance of hubs during brain network development (37,43,55), although other results vary by modality in ways that are not always clearly understood, for example, the changes in global efficiency with age observed in sMRI results are often not replicated in fMRI networks (38,67). Overall, healthy studies provide an important benchmark for neurodevelopmental disorders, for which atypical developmental trajectories may be a key biomarker.
Patient developmental studies are important to understand precisely how and when network properties change in disorders. Many results point toward delayed or disrupted development in patients (68,102,109) and to changes in network hubs and rich-club nodes, which are known to be vulnerable during development. Characteristics of specific disorders are likely to be related to their developmental timing. ASD patients exhibit a mixture of increased/decreased connectivity in different brain regions from childhood, which may be age and/or state dependent. ADHD patients show a clearer pattern of reduced local efficiency, which lags behind control subjects. In psychosis, patients exhibit increased cortical thinning during adolescence, which is likely coupled to functional changes although more work is required to elucidate this relationship.
Large, longitudinal datasets are essential to build a clearer picture, and making data openly accessible is an important step toward more reproducible research. Table 5 lists large open datasets for typical and atypical development. There are noticeable gaps, e.g., a paucity of developmental rs-fMRI data from patients with schizophrenia. Several ongoing psychosis projects will go some way to address this problem [e.g., (9,129–131)]. Similar initiatives are underway for other disorders (132,133).
Table 5. Some Example Large Open Datasets Relevant to Healthy and Atypical Development.
| Dataset | Disorder | No. of Subjects | Age, Years | Longitudinal Neuroimaging | Data Type | Availability |
|---|---|---|---|---|---|---|
| Child Mind Institute Healthy Brain Network (145) | Population | 10,000 | 5–21 | No | fMRI, sMRI, DWI, EEG | Ongoing, 664 subjects available in first release |
| IMAGEN (146) | Population | 2000 | 14–22 | Yes | fMRI, sMRI, DWI | Open to project proposals |
| NKI/Rockland Study (147) | Population | 1000 expected | 6–85 | Yes | rs-fMRI, sMRI, DWI | Ongoing, some data available |
| PNC (9) | Population | 1000 | 8–21 | Obtained but not openly available at present | fMRI, sMRI, DWI | Available |
| ADHD-200 (148) | ADHD | 491 HC, 285 ADHD | 7–21 | No | rs-fMRI, sMRI | Available |
| ABIDE I (149) | Autism | 539 ASD, 573 HC | 7–64 | No | rs-fMRI, sMRI | Available |
| ABIDE II (132) | Autism | 521 ASD, 593 HC | 5–64 | Partially (n = 38) | rs-fMRI, sMRI, DWI | Available |
| NUSDAST (150) | Schizophrenia | 171 SCZ, 170 HC, 44 SCZ siblings, 66 HC siblings | 13–67 | Yes | sMRI | Available |
Datasets were only included if they had large numbers of subjects (N > 300), with subjects from a range of ages, including significant numbers of subjects under 21 years of age, and are (or will be) openly available.
ABIDE, Autism Brain Imaging Data Exchange; ADHD, attention-deficit/hyperactivity disorder; ASD, autism spectrum disorder; DTI, diffusion tensor imaging; DWI, diffusion-weighted imaging; EEG, electroencephalography; fMRI, functional magnetic resonance imaging; HC, healthy control; NKI, Nathan S. Kline Institute for Psychiatric Research; NUSDAST, Northwestern University Schizophrenia Data and Software Tool; PNC, Philadelphia Neurodevelopmental Cohort; rs-fMRI, resting-state functional magnetic resonance imaging; SCZ, schizophrenia; sMRI, structural magnetic resonance imaging.
Another advantage of large collaborative projects is that they often collect data from multiple modalities, enabling the relationships between modalities to be assessed. Networks provide an excellent framework to compare or combine different data types across temporal and spatial scales, for example, by using multilayer networks. Combining neuroimaging results with nonneuroimaging data also represents an exciting avenue for future research. For example, gene expression data from the Allen Institute for Brain Science has already provided a link between the developmental changes in hub regions during adolescence and genes implicated in schizophrenia (43). Ultimately, using networks to bridge the gap between macroscopic neuroimaging results and the microscopic level could open the door to a new depth of understanding and treatments for neurodevelopmental disorders.
Acknowledgments
This work was supported by Medical Research Council Grant Nos. MR/ K020706/1 (to PEV) and U105292687 (to SRW); the PSYSCAN project, which is funded by the European Commission within its 7th Framework Programme under Grant No. 603196 (to SEM); a Henslow Fellowship at Lucy Cavendish College, University of Cambridge, funded by the Cambridge Philosophical Society (to SEM); and the Behavioural and Clinical Neuroscience Institute, which is supported by the Medical Research Council (United Kingdom) and the Wellcome Trust. PEV is a Fellow of MQ: Transforming Mental Health, Grant No. MQF17_24.
Footnotes
Disclosures
ETB is employed half-time by the University of Cambridge and half-time by GlaxoSmithKline; he holds stock in GlaxoSmithKline. The other authors report no biomedical financial interests or potential conflicts of interest.
Contributor Information
Sarah E. Morgan, Behavioural and Clinical Neuroscience Institute, University of Cambridge, Cambridge
Simon R. White, Department of Psychiatry, University of Cambridge, Cambridge; MRC Biostatistics Unit, University of Cambridge, Cambridge
Edward T. Bullmore, Behavioural and Clinical Neuroscience Institute, University of Cambridge, Cambridge; Cambridgeshire and Peterborough NHS Foundation Trust, Huntingdon; ImmunoPsychiatry, Immuno-Inflammation Therapeutic Area Unit, GlaxoSmithKline R&D, Stevenage, United Kingdom
Petra E. Vértes, Behavioural and Clinical Neuroscience Institute, University of Cambridge, Cambridge
References
- 1.Bassett D, Sporns O. Network neuroscience. Nat Neurosci. 2017;20:353–364. doi: 10.1038/nn.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fornito A, Zalesky A, Bullmore E. Fundamentals of Brain Network Analysis. San Diego, CA: Academic Press; 2015. [Google Scholar]
- 3.Sporns O. Networks of the Brain. Cambridge, MA: MIT Press; 2010. [Google Scholar]
- 4.van den Heuvel M, Sporns O. Network hubs in the human brain. Trends Cogn Sci. 2013;17:683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- 6.van Essen D, Barch D, Bookheimer S, Dapretto M, Thomas K, Yacoub E, et al. HCP development. [Accessed November 14, 2017];2017 Available at: https://www.humanconnectome.org/study/hcp-lifespan-development.
- 7.Hughes E, Cordero Grande L, Murgasova M, Hutter J, Price A, Santos Gomes A, et al. The Developing Human Connectome: Announcing the first release of open access neonatal brain imaging. Presented at the Organization for Human Brain Mapp; June 25–29, 2017; Vancouver, British Columbia, Canada. 2017. [Google Scholar]
- 8.Kiddle B, Inkster B, Prabhu G, Moutoussis M, Whitaker KJ, Bullmore ET, et al. Cohort Profile: The NSPN 2400 Cohort: A developmental sample supporting the Wellcome Trust NeuroScience in Psychiatry Network. Int J Epidemiol. 2018;47:18–19g. doi: 10.1093/ije/dyx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satterthwaite T, Connolly J, Ruparel K, Calkins M, Jackson C, Elliott M, et al. The Philadelphia Neurodevelopmental Cohort: A publicly available resource for the study of normal and abnormal brain development in youth. Neuroimage. 2016;86:1115–1119. doi: 10.1016/j.neuroimage.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn E, Shen X, Scheinost D, Rosenberg M, Huang J, Chun M, et al. Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nat Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanai R, Rees G. The structural basis of inter-individual differences in human behaviour and cognition. Nat Rev Neurosci. 2011;12:231–242. doi: 10.1038/nrn3000. [DOI] [PubMed] [Google Scholar]
- 12.Kraemer H, Yesavage J, Taylor J, Kupfer D. How can we learn about developmental processes from cross-sectional studies, or can we? Am J Psychiatry. 2000;157:163–171. doi: 10.1176/appi.ajp.157.2.163. [DOI] [PubMed] [Google Scholar]
- 13.Wolff J, Piven J. Neurodevelopmental disorders: Accelerating progress in autism through developmental research. Nat Rev Neurol. 2014;10:431–432. doi: 10.1038/nrneurol.2014.126. [DOI] [PubMed] [Google Scholar]
- 14.Vértes P, Bullmore E. Growth connectomics- the organization and reorganization of brain networks during normal and abnormal development. J Child Psychol Psychiatry. 2015;56:299–320. doi: 10.1111/jcpp.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao M, Huang H, Peng Y, Dong Q, He Y. Toward developmental connectomics of the human brain. Front Neuroanat. 2016;10:25. doi: 10.3389/fnana.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menon V. Developmental pathways to functional brain networks: Emerging principles. Trends Cogn Sci. 2013;17:627–640. doi: 10.1016/j.tics.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16:159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser M. Mechanisms of connectome development. Trends Cogn Sci. 2017;21:703–717. doi: 10.1016/j.tics.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Collin G, van den Heuvel M. The ontogeny of the human connectome: Development and dynamic changes of brain connectivity across the life span. Neuroscientist. 2013;19:616–628. doi: 10.1177/1073858413503712. [DOI] [PubMed] [Google Scholar]
- 20.Tymofiyeva O, Hess C, Xu D, Barkovich A. Structural MRI connectome in development: Challenges of the changing brain. Br J Radiol. 2014;87 doi: 10.1259/bjr.20140086. 20140086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stiles J, Jernigan T. The basics of brain development. Neuropsychol Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petanjek Z, Judaš M, Šimić G, Rašin MR, Uylings H, Rakic P, Kostović I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giedd JN, Blumenthal J, Jeffries NO, Castellanos F, Liu H, Zijdenbos A, et al. Brain development during childhood and adolescence: A longitudinal MRI study. Nat Neurosci. 1999;2:861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- 24.Mills K, Tamnes C. Methods and considerations for longitudinal structural brain imaging analysis across development. Dev Cogn Neurosci. 2014;9:172–190. doi: 10.1016/j.dcn.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills K, Goddings AL, Herting M, Meuwese R, Blakemore SJ, Crone E, et al. Structural brain development between childhood and adulthood: Convergence across four longitudinal samples. Neuroimage. 2016;141:273–281. doi: 10.1016/j.neuroimage.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ducharme S, Albaugh M, Nguyen TV, Hudziak J, Mateos-Pérez J, Labbe A, et al. Trajectories of cortical thickness maturation in normal brain development: The importance of quality control procedures. Neuroimage. 2016;125:267–279. doi: 10.1016/j.neuroimage.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuter M, Dylan Tisdall M, Qureshi A, Buckner R, van der Kouwe A, Fischl B. Head motion during MRI acquisition reduces gray matter volume and thickness estimates. Neuroimage. 2015;107:107–115. doi: 10.1016/j.neuroimage.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savalia N, Agres P, Chan M, Feczko E, Kennedy K, Wig G. Motion-related artifacts in structural brain images revealed with independent estimates of in-scanner head motion. Human Brain Mapp. 2017;38:472–492. doi: 10.1002/hbm.23397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamnes C, Herting M, Goddings AL, Meuwese R, Blakemore SJ, Dahl R, et al. Development of the cerebral cortex across adolescence: A multisample study of inter-related longitudinal changes in cortical volume, surface area, and thickness. J Neurosci. 2017;37:3402–3412. doi: 10.1523/JNEUROSCI.3302-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenroot R, Gogtay N, Greenstein D, Wells E, Wallace G, Clasen L, et al. Sexual dimorphism of brain developmental trajectories during childhood and adolescence. Neuroimage. 2007;36:1065–1073. doi: 10.1016/j.neuroimage.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raznahan A, Shaw P, Lerch J, Clasen L, Greenstein D, Berman R, et al. Longitudinal four-dimensional mapping of subcortical anatomy in human development. Proc Natl Acad Sci U S A. 2014;111:1592–1597. doi: 10.1073/pnas.1316911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw P, Kabani N, Lerch J, Eckstrand K, Lenroot R, Gogtay N, et al. Neurodevelopmental trajectories of the human cerebral cortex. J Neurosci. 2008;28:3586–3594. doi: 10.1523/JNEUROSCI.5309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijayakumar N, Allen N, Youssef G, Dennison M, Yücel M, Simmons J, Whittle S. Brain development during adolescence: A mixed-longitudinal investigation of cortical thickness, surface area, and volume. Human Brain Mapp. 2016;37:2027–2038. doi: 10.1002/hbm.23154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander-Bloch A, Giedd J, Bullmore E. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 2013;14:322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y, Shi F, Smith J, Lin W, Gilmore J, Shen D. Brain anatomical networks in early human brain development. Neuroimage. 2011;54:1862–1871. doi: 10.1016/j.neuroimage.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khundrakpam B, Reid A, Brauer J, Carbonell F, Lewis J, Ameis S, et al. Developmental changes in organization of structural brain networks. Cereb Cortex. 2013;23:2072–2085. doi: 10.1093/cercor/bhs187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie J, Li G, Shen D. Development of cortical anatomical properties from early childhood to early adulthood. Neuroimage. 2013;76:216–224. doi: 10.1016/j.neuroimage.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moura LM, Crossley NA, Zugman A, Pan PM, Gadelha A, Del Aquilla MAG, et al. Coordinated brain development: Exploring the synchrony between changes in grey and white matter during childhood maturation. Brain Imaging Behav. 2017;11:808–817. doi: 10.1007/s11682-016-9555-0. [DOI] [PubMed] [Google Scholar]
- 40.Zielinski B, Gennatas E, Zhou J, Seeley W. Network-level structural covariance in the developing brain. Proc Natl Acad Sci U S A. 2010;107:18191–18196. doi: 10.1073/pnas.1003109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raznahan A, Lerch J, Lee N, Greenstein D, Wallace G, Stockman M, et al. Patterns of coordinated anatomical change in human cortical development: A longitudinal neuroimaging study of maturational coupling. Neuron. 2011;72:873–884. doi: 10.1016/j.neuron.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander-Bloch A, Raznahan A, Bullmore E, Giedd J. The convergence of maturational change and structural covariance in human cortical networks. J Neurosci. 2013;33:2889–2899. doi: 10.1523/JNEUROSCI.3554-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitaker K, Vértes P, Romero-Garcia R, Váša F, Moutoussis M, Prabhu G, et al. Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc Natl Acad Sci U S A. 2016;113:9105–9110. doi: 10.1073/pnas.1601745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Váša F, Seidlitz J, Romero-Garcia R, Whitaker K, Rosenthal G, Vértes P, et al. Adolescent tuning of association cortex in human structural brain networks. Cereb Cortex. 2018;28:281–294. doi: 10.1093/cercor/bhx249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seidlitz J, Váša F, Shinn M, Romero-Garcia R, Whitaker K, Vértes P, et al. Morphometric similarity networks detect microscale cortical organisation and predict inter-individual cognitive variation. Neuron. 2018;97:231–247.e7. doi: 10.1016/j.neuron.2017.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yap PT, Fan Y, Chen Y, Gilmore J, Lin W, Shen D. Development trends of white matter connectivity in the first years of life. PLoS One. 2011;6:e24678. doi: 10.1371/journal.pone.0024678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Bihan D, Poupon C, Amadon A, Lethimonnier F. Artifacts and pitfalls in diffusion MRI. J Magn Reson Imaging. 2006;24:478–488. doi: 10.1002/jmri.20683. [DOI] [PubMed] [Google Scholar]
- 48.Yendiki A, Koldewyn K, Kakunoori S, Kanwisher N, Fischl B. Spurious group differences due to head motion in a diffusion MRI study. Neuroimage. 2014;88:79–90. doi: 10.1016/j.neuroimage.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagmann P, Sporns O, Madan N, Cammoun L, Pienaar R, Wedeen V, et al. White matter maturation reshapes structural connectivity in the late developing human brain. Proc Natl Acad Sci U S A. 2010;107:19067–19072. doi: 10.1073/pnas.1009073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebel C, Beaulieu C. Longitudinal development of human brain wiring continues from childhood into adulthood. J Neurosci. 2011;31:10937–10947. doi: 10.1523/JNEUROSCI.5302-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tymofiyeva O, Hess C, Ziv E, Lee P, Glass H, Ferriero D. A DTI-based template-free cortical connectome study of brain maturation. PLoS One. 2013;8:e63310. doi: 10.1371/journal.pone.0063310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wierenga L, van den Heuvel M, van Dijk S, Rijks Y, de Reus M, Durston S. The development of brain network architecture. Human Brain Mapp. 2015;37:717–729. doi: 10.1002/hbm.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dennis E, Jahanshad N, McMahon K, de Zubicaray G, Martin N, Hickie I, et al. Development of brain structural connectivity between ages 12 and 30: A 4-Tesla diffusion imaging study in 439 adolescents and adults. Neuroimage. 2013;64:671–684. doi: 10.1016/j.neuroimage.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baum G, Ciric R, Roalf D, Betzel R, Moore T, Shinohara R, et al. Modular segregation of structural brain networks supports the development of executive function in youth. Curr Biol. 2017;27:1561–1572. doi: 10.1016/j.cub.2017.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker S, Lubman D, Yücel M, Allen N, Whittle S, Fulcher B, et al. Developmental changes in brain network hub connectivity in late adolescence. J Neurosci. 2015;35:9078–9087. doi: 10.1523/JNEUROSCI.5043-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wierenga L, van den Heuvel M, Oranje B, Giedd J, Durston S, Peper J, et al. A multisample study of longitudinal changes in brain network architecture in 4–13-year-old children. Human Brain Mapp. 2018;39:157–170. doi: 10.1002/hbm.23833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim S, Han CE, Uhlhaas PJ, Kaiser M. Preferential detachment during human brain development: Age- and sex-specific structural connectivity in diffusion tensor imaging (DTI) data. Cereb Cortex. 2013:1477–1489. doi: 10.1093/cercor/bht333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koenis M, Brouwer R, van den Heuvel M, Mandl R, van Soelen I, Kahn R, et al. Development of the brain’s structural network efficiency in early adolescence: A longitudinal DTI twin study. Human Brain Mapp. 2015;36:4938–4953. doi: 10.1002/hbm.22988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hallquist M, Hwang K, Luna B. The nuisance of nuisance regression: Spectral misspecification in a common approach to resting-state fMRI preprocessing reintroduces noise and obscures functional connectivity. Neuroimage. 2013;82:208–225. doi: 10.1016/j.neuroimage.2013.05.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel A, Kundu P, Rubinov M, Jones P, Vértes P, Ersche K, et al. A wavelet method for modeling and despiking motion artifacts from resting-state fMRI time series. Neuroimage. 2014;95:287–304. doi: 10.1016/j.neuroimage.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bright M, Murphy K. Removing motion and physiological artifacts from intrinsic BOLD fluctuations using short echo data. Neuroimage. 2013;64:526–537. doi: 10.1016/j.neuroimage.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kundu P, Brenowitz N, Voon V, Worbe Y, Vértes P, Inati S, et al. Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proc Natl Acad Sci U S A. 2013;110:16187–16192. doi: 10.1073/pnas.1301725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salimi-Khorshidi G, Douaud G, Beckmann C, Glasser M, Griffanti L, Smith S. Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van den Heuvel M, de Lange M, Zalesky A, Seguin C, Yeo T, Schmidt R. Proportional thresholding in resting-state fMRI functional connectivity networks and consequences for patient-control connectome studies: Issues and recommendations. Neuroimage. 2017;152:437–449. doi: 10.1016/j.neuroimage.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 66.Murphy K, Fox M. Towards a consensus regarding global signal regression for resting state functional connectivity MRI. Neuroimage. 2016;154:169–173. doi: 10.1016/j.neuroimage.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fair D, Cohen A, Power J, Dosenbach N, Church J, Miezin F, et al. Functional brain networks develop from a “local to distributed” organization. PLoS Comput Biol. 2009;5:e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fransson P, Aden U, Blennow M, Lagercrantz H. The functional architecture of the infant brain as revealed by resting-state fMRI. Cereb Cortex. 2011;21:145–154. doi: 10.1093/cercor/bhq071. [DOI] [PubMed] [Google Scholar]
- 69.Supekar K, Musen M, Menon V. Development of large-scale functional brain networks in children. PLoS Biol. 2009;7:e1000157. doi: 10.1371/journal.pbio.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao W, Gilmore J, Giovanello K, Smith J, Shen D, Zhu H. Temporal and spatial evolution of brain network topology during the first two years of life. PLoS One. 2011;6:e25278. doi: 10.1371/journal.pone.0025278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao M, Wang JH, Dai ZJ, Cao XY, Jiang LL, Fan FM, et al. Topological organization of the human brain functional connectome across the lifespan. Dev Cogn Neurosci. 2014;7:76–93. doi: 10.1016/j.dcn.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu K, Taki Y, Sato K, Hashizume H, Sassa Y, Takeuchi H, et al. Topological organization of functional brain networks in healthy children: Differences in relation to age, sex, and intelligence. PLoS One. 2013;8:e55347. doi: 10.1371/journal.pone.0055347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dosenbach N, Nardos B, Cohen A, Fair D, Power J, Church J, et al. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Satterthwaite T, Wolf D, Ruparel K, Erus G, Elliott M, Eickhoff S, et al. Heterogeneous impact of motion on fundamental patterns of developmental changes in functional connectivity during youth. Neuroimage. 2013;83:45–57. doi: 10.1016/j.neuroimage.2013.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hwang K, Hallquist M, Luna B. The development of hub architecture in the human functional brain network. Cereb Cortex. 2013;23:2380–2393. doi: 10.1093/cercor/bhs227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Uddin L, Supekar K, Ryali S, Menon V. Dynamic reconfiguration of structural and functional connectivity across core neurocognitive brain networks with development. J Neurosci. 2011;31:18578–18589. doi: 10.1523/JNEUROSCI.4465-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marek S, Hwang K, Foran W, Hallquist M, Luna B. The contribution of network organization and integration to the development of cognitive control. PLoS Biol. 2015;13:e1002328. doi: 10.1371/journal.pbio.1002328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu S, Satterthwaite T, Medaglia J, Yang M, Gur R, Gur R, Bassett D. Emergence of system roles in normative neurodevelopment. Proc Natl Acad Sci U S A. 2015;112:13681–13686. doi: 10.1073/pnas.1502829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaufmann T, Alnæs D, Doan N, Brandt C, Andreassen O, Westyle L. Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nat Neurosci. 2017;20:513–515. doi: 10.1038/nn.4511. [DOI] [PubMed] [Google Scholar]
- 80.Crossley N, Mechelli A, Scott J, Carletti F, Fox P, McGuire P, Bullmore E. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137:2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hawrylycz M, Lein E, Guillozet-Bongaarts A, Shen E, Ng L, Miller J, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lein E, Belgard TG, Hawrylycz M, Molnár Z. Transcriptomic perspectives on neocortical structure, development, evolution, and disease. Annu Rev Neurosci. 2017;40:629–652. doi: 10.1146/annurev-neuro-070815-013858. [DOI] [PubMed] [Google Scholar]
- 83.Wang G, Belgard T, Mao D, Chen L, Berto S, Preuss T, et al. Correspondence between resting-state activity and brain gene expression. Neuron. 2015;88:659–666. doi: 10.1016/j.neuron.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krienen F, Yeo B, Ge T, Buckner R, Sherwood C. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci U S A. 2016;113:469–478. doi: 10.1073/pnas.1510903113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T, et al. Correlated gene expression supports synchronous activity in brain networks. Science. 2015;348:1241–1244. doi: 10.1126/science.1255905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parkes L, Fulcher B, Yücel M, Fornito A. Transcriptional signatures of connectomic subregions of the human striatum. Genes Brain Behav. 2017;16:647–663. doi: 10.1111/gbb.12386. [DOI] [PubMed] [Google Scholar]
- 87.Romme I, de Reus M, Ophoff R, Kahn R, van den Heuvel M. Connectome disconnectivity and cortical gene expression in patients with schizophrenia. Biol Psychiatry. 2017;81:495–502. doi: 10.1016/j.biopsych.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 88.Ganglberger F, Kaczanowska J, Penninger JM, Hess A, Bühler K, Haubensak W. Predicting functional neuroanatomical maps from fusing brain networks with genetic information. Neuroimage. 2018;170:113–120. doi: 10.1016/j.neuroimage.2017.08.070. [DOI] [PubMed] [Google Scholar]
- 89.Romero-Garcia R, Warrier V, Bullmore E, Baron-Cohen S, Bethlehem RAI. Synaptic and transcriptionally downregulated genes are associated with cortical thickness differences in autism. Mol Psychiatry. 2018 doi: 10.1038/s41380-018-0023-7. [published online ahead of print Feb 26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McColgan P, Gregory S, Seunarine K, Razi A, Papoutsi M, Johnson E, et al. Brain regions showing white matter loss in Huntington’s disease are enriched for synaptic and metabolic genes. Biol Psychiatry. 2017;83:456–465. doi: 10.1016/j.biopsych.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zielinski B, Prigge M, Nielsen J, Froehlich A, Abildskov T, Anderson J, et al. Longitudinal changes in cortical thickness in autism and typical development. Brain. 2014;137:1799–1812. doi: 10.1093/brain/awu083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lewis J, Theilmann R, Townsend J, Evans A. Network efficiency in autism spectrum disorder and its relation to brain over-growth. Front Hum Neurosci. 2013;7:845. doi: 10.3389/fnhum.2013.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Müller RA, Shih P, Keehn B, Deyoe J, Leyden K, Shukla D. Underconnected, but how? A survey of functional connectivity MRI studies in autism spectrum disorders. Cereb Cortex. 2011;21:2233–2243. doi: 10.1093/cercor/bhq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kana R, Uddin L, Kenet T, Chugani D, Müller RA. Brain connectivity in autism. Front Hum Neurosci. 2014;8:349. doi: 10.3389/fnhum.2014.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.You X, Norr M, Murphy E, Kuschner E, Bal E, Gaillard W, et al. Atypical modulation of distant functional connectivity by cognitive state in children with autism spectrum disorders. Front Hum Neurosci. 2013;7:482. doi: 10.3389/fnhum.2013.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Itahashi T, Yamada T, Watanabe H, Nakamura M, Jimbo D, Shioda S, et al. Altered network topologies and hub organization in adults with autism: A resting-state fMRI study. PloS One. 2014;9:e94115. doi: 10.1371/journal.pone.0094115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bernhardt B, Valk S, Silani G, Bird G, Frith U, Singer T. Selective disruption of sociocognitive structural brain networks in autism and alexithymia. Cereb Cortex. 2014;24:3258–3267. doi: 10.1093/cercor/bht182. [DOI] [PubMed] [Google Scholar]
- 98.Lewis J, Evans A, Pruett J, Botteron K, McKinstry R, Zwaigenbaum L, et al. The emergence of network inefficiencies in infants with autism spectrum disorder. Biol Psychiatry. 2017;82:176–185. doi: 10.1016/j.biopsych.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Watanabe T, Rees G. Age-associated changes in rich-club organisation in autistic and neurotypical human brains. Sci Rep. 2015;5 doi: 10.1038/srep16152. 16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ghanbari Y, Smith A, Schultz R, Verma R. Identifying group discriminative and age regressive sub-networks from DTI-based connectivity via a unified framework of nonnegative matrix factorization and graph embedding. Med Image Anal. 2014;18:1337–1348. doi: 10.1016/j.media.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ball G, Beare R, Seal M. Network component analysis reveals developmental trajectories of structural connectivity and specific alterations in autism spectrum disorder. Human Brain Mapp. 2017;38:4169–4184. doi: 10.1002/hbm.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nomi J, Uddin L. Developmental changes in large-scale network connectivity in autism. Neuroimage Clin. 2015;7:732–741. doi: 10.1016/j.nicl.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen H, Kelly C, Castellanos FX, He Y, Zuo X, Reiss P. Quantile rank maps: A new tool for understanding individual brain development. Neuroimage. 2015;11:454–463. doi: 10.1016/j.neuroimage.2014.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Emerson R, Adams C, Nishino T, Heather H, Wolff J, Zwaigenbaum L, et al. Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aag2882. eeag2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castellanos F, Aoki Y. Intrinsic functional connectivity in attention deficit/hyperactivity disorder: A science in development. Biol Psychiatry Cogn Neurosci Neuroimaging. 2016;1:253–261. doi: 10.1016/j.bpsc.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin P, Sun J, Yu G, Wu Y, Yang Y, Liang M, Liu X. Global and local brain network reorganization in attention-deficit/hyperactivity disorder. Brain Imaging Behav. 2014;8:558–569. doi: 10.1007/s11682-013-9279-3. [DOI] [PubMed] [Google Scholar]
- 107.Sidlauskaite J, Caeyenberghs K, Sonuga-Barke E, Roeyers H, Wiersema J. Whole-brain structural topology in adult attention-deficit/hyperactivity disorder: Preserved global – disturbed local network organization. Neuroimage Clin. 2015;9:506–512. doi: 10.1016/j.nicl.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Castellanos F, Lee P, Sharp W, Jeffries N, Greenstein D, Clasen L, et al. Developmental trajectories of brain volume abnormalities in children and adolescents with attention-deficit/hyperactivity disorder. JAMA. 2002;288:1740–1748. doi: 10.1001/jama.288.14.1740. [DOI] [PubMed] [Google Scholar]
- 109.Shaw P, Eckstrand K, Sharp W, Blumenthal J, Lerch J, Greenstein D, et al. Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc Natl Acad Sci U S A. 2007;104:19649–19654. doi: 10.1073/pnas.0707741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cao M, Shu N, Cao Q, Wang Y, He Y. Imaging functional and structural brain connectomics in attention-deficit/hyperactivity disorder. Mol Neurobiol. 2014;50:1111–1123. doi: 10.1007/s12035-014-8685-x. [DOI] [PubMed] [Google Scholar]
- 111.Konrad K, Eickhoff S. Is the ADHD brain wired differently? A review on structural and functional connectivity in attention deficit hyperactivity disorder. Human Brain Mapp. 2010;31:904–916. doi: 10.1002/hbm.21058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fair D, Posner J, Nagel B, Bathula D, Costa Dias T, Mills K, et al. Atypical default network connectivity in youth with ADHD. Biol Psychiatry. 2010;68:1084–1091. doi: 10.1016/j.biopsych.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sripada C, Kessler D, Angstadt M. Lag in maturation of the brain’s intrinsic functional architecture in attention-deficit/hyperactivity disorder. Proc Natl Acad Sci U S A. 2014;111:14259–14264. doi: 10.1073/pnas.1407787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kessler D, Angstadt M, Sripada C. Growth charting of brain connectivity networks and the identification of attention impairment in youth. JAMA Psychiatry. 2016;73:481–489. doi: 10.1001/jamapsychiatry.2016.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gupta C, Calhoun V, Rachakonda S, Chen J, Patel V, Liu J, et al. Patterns of gray matter abnormalities in schizophrenia based on an international mega-analysis. Schizophr Bull. 2014;41:1133–1142. doi: 10.1093/schbul/sbu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zalesky A, Fornito A, Seal M, Cocchi L, Westin C, Bullmore E, et al. Disrupted axonal fiber connectivity in schizophrenia. Biol Psychiatry. 2011;69:80–89. doi: 10.1016/j.biopsych.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu Q, Allen E, Sui J, Arbabshirani M, Pearlson G, Calhoun V. Brain connectivity networks in schizophrenia underlying resting state functional magnetic resonance imaging. Curr Top Med Chem. 2012;12:2415–2425. doi: 10.2174/156802612805289890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rapoport J, Giedd J, Blumenthal J, Hamburger S, Jeffries N, Fernandez T, et al. Progressive cortical change during adolescence in childhood-onset schizophrenia. Arch Gen Psychiatry. 1999;56:649–654. doi: 10.1001/archpsyc.56.7.649. [DOI] [PubMed] [Google Scholar]
- 119.van Haren N, Schnack H, Cahn W, van den Heuvel M, Lepage C, Collins L, et al. Changes in cortical thickness during the course of illness in schizophrenia. Arch Gen Psychiatry. 2011;68:871–880. doi: 10.1001/archgenpsychiatry.2011.88. [DOI] [PubMed] [Google Scholar]
- 120.Alexander-Bloch A, Reiss PT, Rapoport J, McAdams H, Giedd J, Bullmore E, Gogtay N. Abnormal cortical growth in schizophrenia targets normative modules of synchronized development. Biol Psychiatry. 2014;76:438–446. doi: 10.1016/j.biopsych.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zalesky A, Pantelis C, Cropley V. Delayed development of brain connectivity in adolescents with schizophrenia and their unaffected siblings. JAMA Psychiatry. 2015;72:900–908. doi: 10.1001/jamapsychiatry.2015.0226. [DOI] [PubMed] [Google Scholar]
- 122.Sun Y, Chen Y, Lee R, Bezerianos A, Collinson S, Sim K. Disruption of brain anatomical networks in schizophrenia: A longitudinal, diffusion tensor imaging based study. Schizophr Res. 2016;171:149–157. doi: 10.1016/j.schres.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 123.Tomasi D, Volkow N. Mapping small-world properties through development in the human brain: Disruption in schizophrenia. PLoS One. 2014;9:e96176. doi: 10.1371/journal.pone.0096176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jiang L, Xu Y, Zhu X, Yang Z, Li H, Zuo X. Local-to-remote cortical connectivity in early- and adulthood-onset schizophrenia. Transl Psychiatry. 2015;5:e566. doi: 10.1038/tp.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang C, Lee J, Ho NF, Lim JKW, Poh JS, Rekhi G, et al. Large-scale network topology reveals heterogeneity in individuals with at risk mental state for psychosis: Findings from the longitudinal Youth-at-Risk Study. Cereb Cortex. 2017 doi: 10.1093/cercor/bhx278. [published online ahead of print Oct 27] [DOI] [PubMed] [Google Scholar]
- 126.Vértes P, Alexander-Bloch A, Gogtay N, Giedd J, Rapoport J, Bullmore E. Simple models of human brain functional networks. Proc Natl Acad Sci U S A. 2012;109:5868–5873. doi: 10.1073/pnas.1111738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Betzel R, Bassett D. Generative models for network neuroscience: Prospects and promise. J R Soc Interface. 2017;14 doi: 10.1098/rsif.2017.0623. 20170623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fair D, Dosenbach N, Church J, Cohen A, Brahmbhatt S, Miezin F, et al. Development of distinct control networks through segregation and integration. Proc Natl Acad Sci U S A. 2007;104:13507–13512. [Google Scholar]
- 129.PRONIA. [Accessed November 14, 2017]; Available at: https://www.pronia.eu/.
- 130.PSYSCAN. [Accessed November 14, 2017]; Available at: http://psyscan.eu/.
- 131.University of California, San Francisco. North American Prodrome Longitudinal Study. [Accessed November 14, 2017]; Available at: http://napls.ucsf.edu/.
- 132.Di Martino A, O’Connor D, Chen B, Alaerts K, Anderson J, Assaf M, et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci Data. 2017;4 doi: 10.1038/sdata.2017.10. 170010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Loth E, Charman T, Mason L, Tillmann J, Jones E, Wooldridge C, et al. The EU-AIMS Longitudinal European Autism Project (LEAP): Design and methodologies to identify and validate stratification biomarkers for autism spectrum disorders. Mol Autism. 2017;8:24. doi: 10.1186/s13229-017-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Crossley N, Mechelli A, Vértes P, Winton-Brown T, Patel A, Ginestet C, et al. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc Natl Acad Sci U S A. 2013;110:11583–11588. doi: 10.1073/pnas.1220826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu Z, Shen X, Pan W. Alzheimer’s Disease Neuroimaging Initiative Longitudinal analysis is more powerful than cross-sectional analysis in detecting genetic association with neuroimaging phenotypes. PLoS One. 2014;9:e102312. doi: 10.1371/journal.pone.0102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pfefferbaum A, Sullivan E. Cross-sectional versus longitudinal estimates of age-related changes in the adult brain: Overlaps and discrepancies. Neurobiol Aging. 2015;36:2563–2567. doi: 10.1016/j.neurobiolaging.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Diggle P, Heagerty P, Liang KY, Zeger S. Analysis of Longitudinal Data. London, UK: Clarendon Press; 1994. [Google Scholar]
- 138.Fitzmaurice G, Laird N, Ware J. Applied Longitudinal Analysis. New York: Wiley; 2004. [Google Scholar]
- 139.Ombao H, Lindquist M, Thompson W, Aston J. Handbook of Neuroimaging Data Analysis. London: Chapman & Hall/CRC Handbooks fo Modern Statistical Methods; 2016. [Google Scholar]
- 140.Little RJA, Rubin DB. Statistical Analysis with Missing Data. New York: Wiley; 2002. [Google Scholar]
- 141.Bell A, Jones K. Age, period and cohort processes in longitudinal and life course analysis: A multilevel perspective. In: Burton-Jeangros C, Cullati S, Sacker A, Blane D, editors. A Life Course Perspective on Health Trajectories and Transitions. Berlin: Springer; 2015. pp. 197–213. [PubMed] [Google Scholar]
- 142.Verhulst FC, Koot KM. The Epidemiology of Child and Adolescent Psychopathology. New York: Oxford University Press; 1995. Accelerated longitudinal designs; pp. 385–405. [Google Scholar]
- 143.Prinzie P, Onghena P. Encyclopedia of Statistics in Behavioral Science. Vol. 1. New York: Wiley; 2005. Cohort sequential design; pp. 319–322. [Google Scholar]
- 144.Galbraith S, Bowden J, Mander A. Accelerated longitudinal designs: An overview of modellings, power, costs and handling missing data. Stat Methods Med Res. 2017;26:374–398. doi: 10.1177/0962280214547150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Alexander L, Escalera J, Ai L, Andreotti C, Febre K, Mangone A, et al. The Healthy Brain Network Biobank: An open resource for transdiagnostic research in pediatric mental health and learning disorders. bioRxiv. 2017 doi: 10.1038/sdata.2017.181. [published online ahead of print Jun 13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Imagen Europe: IMAGEN study. [Accessed November 14, 2017];2016 Available at: https://imagen-europe.com.
- 147.Nooner K, Colcombe S, Tobe R, Mennes M, Benedict M, Moreno A, et al. The NKI-Rockland sample: A model for accelerating the pace of discovery science in psychiatry. Front Neurosci. 2012;6:152. doi: 10.3389/fnins.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Milham M, Fair D, Mennes M, Mostofsky S. The ADHD-200 Consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Front Syst Neurosci. 2012;6:62. doi: 10.3389/fnsys.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 2014;19:659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang L, Kogan A, Cobia D, Alpert K, Kolasny A, Miller M, Marcus D. Northwestern University Schizophrenia Data and Software Tool (NUSDAST) Front Neuroinform. 2013;7:25. doi: 10.3389/fninf.2013.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]