Abstract
Homeostatic maintenance of physiological functions is fundamental to organismal well-being. Disruption or imbalance in homeostasis results in functional disturbances at molecular, cellular, and tissue levels, leading to manifestation as physical and mental illnesses. Homeostatic imbalance is caused by a range of pathophysiological mechanisms, including disrupted reduction-oxidation (redox) reactions, inflammatory responses, metabolic disturbances, or failure in quality control of cellular proteins and organelles. However, the roles for the protein/organelle quality control in the regulation of behaviors, in particular of cognitive processes, have not been well documented until recent reports that finally support this concept. The frontline studies in neuroscience have revealed that synaptic components (e.g., synaptic proteins, organelles, neurotransmitters and their receptors) are selectively degraded by autophagy, a cellular recycling machinery implicated in surveillance and quality control of proteins and organelles responsible for the maintenance of cellular homeostasis. Apart from the canonical role of autophagy in supporting cell viability, synaptic autophagy appears to regulate synapse remodeling and plasticity. Consistently, emerging evidence suggests novel roles of autophagy in memory encoding, information processing, or cognitive functions. In this article, we will overview recent progress in understanding the roles of neuronal autophagy in homeostatic maintenance of synaptic functions, with particular focus on how disruptions in these processes may contribute to the pathophysiology of psychiatric disorders.
Keywords: Autophagy, Aggregate, Synapse, Homeostasis, Cognition, Psychiatric disorders
Introduction
A number of pathophysiological events could converge on disrupted homeostasis of physiological functions, leading to a wide range of diseases or pathological consequences (1,2). In the nervous system, redox deregulation, inflammation, metabolic disturbances, or failure in quality control of cellular proteins and organelles have been deeply implicated in neurological and neurodegenerative disorders (3–6). Neuropsychiatric disorders are no exceptions; emerging evidence suggests that the roles for the protein/organelle quality control in the regulation of behaviors, in particular of cognitive processes, and in psychiatric manifestations may become a frontline topic in biological psychiatry.
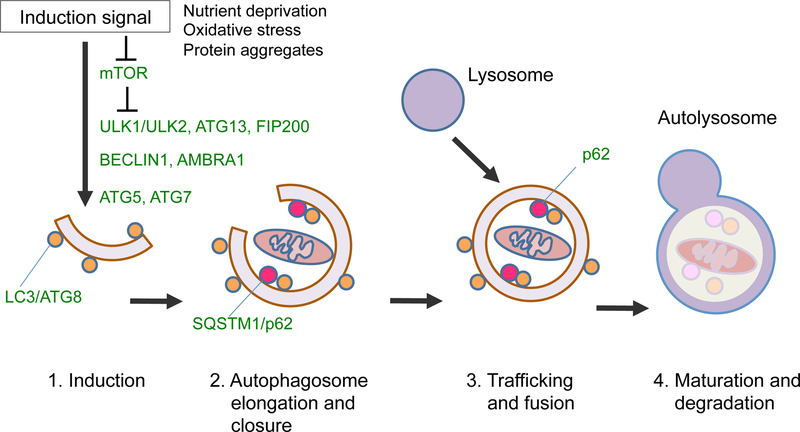
Among the mechanisms that ensure homeostasis of the protein or organelle quality, autophagy has drawn much attention in recent years. Autophagy is a Greek synonym for ‘self-eating’, originally named by Christian de Duve in 1963 based on his discovery of digestive organelles within cells, including the lysosome and the autophagic vacuole termed the autophagosome. More recently, mechanisms of autophagy started to be unveiled by Yoshinori Ohsumi two decades ago, sparked by the discovery of a series of AuTophaGy-related (ATG) genes conserved through evolution, culminating in the general concept that autophagy is fundamental to the maintenance of cellular and organismal homeostasis (7). Among three main forms of autophagy [e.g., macroautophagy, microautophagy, chaperone-mediated autophagy (CMA)], macroautophagy is the major catabolic process, in which old cytoplasmic proteins, lipids, or damaged organelles are either non-selectively or selectively engulfed into the autophagosome and subsequently delivered to the lysosome for degradation (7,8) (Fig. 1). The amino acids and lipids generated through this digestive process can then be recycled to fuel biosynthetic machinery, and contribute to continuous rejuvenation of proteins or organelles and generation of cellular energy. In addition, autophagy can be dynamically induced in response to cellular stress modalities, such as nutrient deprivation, oxidative stress, and accumulation of misfolded or aggregated proteins (9,10). Recent studies show that post-mitotic neurons are particularly vulnerable to disrupted homeostasis caused by autophagy deficiency (11,12). Reflecting these critical roles of autophagy in the maintenance of cellular homeostasis, deregulated autophagy has been implicated in a wide range of human diseases (13,14).
Fig. 1. Schematic diagram of macroautophagy.
Upon cellular stress conditions triggered by a range of stress modalities (e.g., nutrient starvation, oxidative stress, accumulation of misfolded or aggregated proteins), mammalian target of rapamycin (mTOR) signaling is suppressed, leading to a cascade of events that contribute to the induction of macroautophagy (e.g., 1. Assembly of autophagy regulatory protein complex on the isolation membrane; 2. Elongation of the isolation membrane to form autophagosomes; 3. Autophagosome trafficking to fuse with the lysosome; 4. Maturation of autolysosome and degradation). Critical autophagy regulatory proteins are depicted in green.
In this review article, we will overview: 1) current understanding of the canonical role of neuronal autophagy in the maintenance of cellular homeostasis, hence supporting neuron survival and neurodevelopment; 2) emerging evidence for neuronal autophagy in regulating synaptic functions and neuroplasticity, and then discuss 3) how neuronal autophagy regulates memory or specific dimensions of behaviors, such as cognition, as well as their dysfunctions in psychiatric disorders.
Canonical roles of autophagy in neuronal viability and neurodevelopment
Basal autophagy activity is critical to the maintenance of neuronal homeostasis and viability. Evidence supporting this view originally came from cell biology and animal studies. In post-mitotic neurons, the autophagosomes are continuously formed at the distal end of the axon and undergo unidirectional transport along microtubules toward the soma, which is enriched with lysosomes and biosynthetic machinery responsible for producing building blocks for life (e.g., amino acids, lipids) (15–18). Mice with nervous system-specific or neuronal cell type-specific ablation of core autophagy genes (i.e., Atg5, Atg7) exhibit dystrophic axonal swelling and progressive degeneration of axon termini filled with aberrant membranous structures yet no sign of autophagosome formation (19–21). Affected neurons have prominent accumulation of abnormal cytosolic proteins and ubiquitin-positive aggregates, leading to progressive neuronal death during the early postnatal development in these animals. The results indicate that continuous quality control of diffuse cytosolic proteins via basal constitutive autophagy is critical to neuronal survival. Additional evidence for the role of autophagy-related genes in neuronal viability is summarized in Table 1.
Table 1.
Autophagy genes and neurodegeneration
| Gene | Type of gene mutation | Species | Phenotype | Ref |
|---|---|---|---|---|
| Atg5, Atg7 | Nervous system-specific knockout (KO) | Mouse | Axonal swelling and degeneration; progressive postnatal loss of neurons (pyramidal neurons in the cerebral cortex and hippocampus, Purkinje cells in the cerebellum); organismal lethality | 19,20 |
| Atg5, Atg7 | Purkinje neuron-specific KO | Mouse | Axonal swelling and degeneration; progressive postnatal loss of Purkinje cells in the cerebellum and motor coordination decline; organismal lethality | 21,120 |
| Fip200 | Nervous system-specific KO | Mouse | Purkinje cell degeneration; progressive cerebellar ataxia; organismal lethality | 121 |
| Ulk1/Ulk2 | Nervous system-specific double KO | Mouse | Progressive loss of neurons (pyramidal neurons in the hippocampus CA1 region); elevated endoplasmic reticulum stress (activation of unfolded protein response pathway); organismal lethality | 122 |
| Endophilin A | Endophilin 1/2 double KO, Endophilin 1/3 double KO | Mouse | Progressive impairment in motor coordination, ataxia, neurodegeneration (motor cortex, hippocampus) | 66 |
| Endophilin A KO | Drosophila | Loss of dopaminergic neurons | 43 | |
| Synaptojanin 1 | Point mutation (R258Q) | Drosophila | Impaired autophagosome maturation, neurodegeneration (dopaminergic neuron loss) | 44 |
| ATG5 | Homozygous missense mutation | Human | Congenital ataxia; mental retardation; developmental delay | 123 |
| WDR45 | de novo mutations | Human | Neurodegeneration with brain iron accumulation (NBIA); abnormal iron deposition in substantia nigra and globus pallidus; developmental delay; neurological deterioration (parkinsonism, dystonia, early onset dementia) | 124–126 |
Although loss of autophagy causes neuronal death (19–21), and altered autophagy functions are associated with aging-associated neurodegenerative disorders, such as Alzheimer’s, Huntington’s, and Parkinson’s diseases (AD, HD, PD) (11,22,23), autophagy deficiency may not simply represent the pathophysiological mechanisms in these disorders. Notably, the autophagosomes are reported to accumulate in vulnerable neurons in these human diseases (22,23), whereas autophagy deficiencies modeled in mice show no sign of autophagosome formation (11,19–21). Studies on human neurodegenerative disorders provide support for additional pathobiological mechanisms downstream of autophagy deregulation, including defective axonal transport (24,25) and attenuated lysosomal function (26–29), together contributing to buildup of autophagosomes and ultimate neuronal death observed in these disorders. Further discussion on this topic can be found in additional reviews (30,31).
Neuronal autophagy also plays important roles during neuronal development, for example in neural tube closure (32), axon outgrowth or pathfinding (33–35), and synapse formation (36–41), as summarized in Table 2. For example, in mice deficient for Alfy, an adaptor protein responsible for selective autophagy, some axons at the corpus callosum and anterior commissure fail to cross the midline, in part due to failure to respond to axon guidance cues, resulting in abnormal development of interhemispheric axon tracts (33). The results suggest that autophagy facilitates membrane recycling or turnover of signaling components, including the membranes carrying the axon guidance cue receptors, at the leading edge of the axon or in the migrating growth cone, together contributing to axonal development (33–36). In addition, autophagy pathways promote assembly of presynaptic compartments (38–40), in part by regulating synaptic vesicle clustering (39), and also facilitate post-synaptic maturation, by mediating elimination of excess dendritic spines (i.e., spine pruning)(41). These lines of evidence suggest the involvement of defective autophagy in neurodevelopmental disorders.
Table 2.
Autophagy genes and neurodevelopment
| Gene | Type of gene mutation | Species | Phenotype | Ref |
|---|---|---|---|---|
| Ambra1 | KO | Mouse | Neural tube closure defect; imbalance in neuronal proliferation and differentiation | 32 |
| Alfy | KO | Mouse | Abnormal development of interhemispheric axon tracts (corpus callosum, anterior commissure, hippocampal commissure); failure of axons to respond to the axon guidance cue (Netrin-1); midline crossing defects | 33 |
| Atg9 | Nervous system-specific KO | Mouse | Abnormal development of corpus callosum and anterior commissure | 34 |
| Wdr47 | KO | Mouse | Abnormal development of corpus callosum | 35 |
| Atg1 | P-element insertion and imprecise excision of the locus | Drosophila | Defective axonal transport of synaptic vesicle precursors; presynaptic assembly deficits at the neuromuscular junction (NMJ); reduced number of synaptic boutons and smaller size of NMJ | 36,37 |
| Atg1, Atg2, Atg18 | P-element insertion and imprecise excision of the locus | Drosophila | Presynaptic assembly deficits at NMJ; reduced number of synaptic boutons and smaller size of NMJ | 38 |
| Atg9 | EMS-induced point mutations | C. elegans | Defective clustering of synaptic vesicles in presynaptic specializations | 39 |
| Atg7 | Motor neuron-specific KO | Mouse | Accelerated neuromuscular denervation during early development in ALS model mice (SOD[G93A]); non-cell autonomous suppression of ALS-related phenotypes at later stages of disease progression in SOD[G93A] mice | 40 |
| Tsc2 | Heterozygous KO | Mouse | Developmental spine pruning defects; excessive dendritic spines in cortical pyramidal neurons | 41 |
Role of autophagy in synapse remodeling and synaptic plasticity
Maintaining the integrity of proteins (e.g., synaptically localized proteins, neurotransmitters, and their receptors) and organelles (e.g., synaptic vesicles, mitochondria) is crucial to sustain neuronal functionality throughout their lifetime, which could span over a century in the case of humans. Besides, highly polarized and extended morphologies of neurons pose a unique spatial challenge for coordinating the local homeostatic need for autophagic clearance. Recent cellular imaging studies revealed that the autophagosomes are not only constitutively generated in axons (17,18), but are also increased in number upon synaptic activity, either through increased local biogenesis or recruitment of pre-existing autophagosomes to the site of synaptic activation (42–45). The data raise the possibility that neuronal autophagy regulates synaptic functions and neuroplasticity by interpreting both intrinsic and extrinsic demands for maintaining homeostasis of nervous system functions.
Synaptic plasticity is defined as the activity-dependent modification or adaptation of synaptic strength to achieve lasting changes in synaptic efficacy (46). Several well-known forms of synaptic plasticity include long-term potentiation (LTP), long-term depression (LTD), and homeostatic plasticity (i.e., synaptic homeostasis) (46,47). Mechanistically, synaptic plasticity entails a broad spectrum of morphological and biochemical changes in presynaptic neurotransmitter release machinery, postsynaptic receptor composition, activity of signal transduction pathways, gene expression, as well as local proteome regulation at the synapse, including de novo protein synthesis and degradation (46–52). Protein degradation is particularly important to counterbalance protein synthesis, allowing tight control of local synaptic proteome to fine-tune synaptic activities (51,52). In addition to proteins, lipids and synaptic organelles (e.g., synaptic vesicles, mitochondria) are also important players for regulating synaptic plasticity. These synaptic components are susceptible to wear and damage due to remarkably high frequencies of neuronal firing (i.e., action potentials, being delivered up to 50 pulses per second) (53,54). Consequently, synapses are the site of high demand for cellular catabolic activities, requiring efficient degradation systems to maintain local proteome/organelle integrity and sustain synaptic functions (55,56).
Until recently, little attention has been paid to the potential role of autophagy in synaptic plasticity, due in part to a classical view of autophagy as a non-selective, bulk degradation system. Accumulating evidence now shows that neuronal autophagy could achieve selective targeting of synaptic components. For example, synaptic vesicles and mitochondria are targeted by respective autophagic adaptor proteins and degraded through macroautophagy (57,58). A subset of presynaptic proteins with a specific amino acid motif (e.g., KFERQ) are selectively targeted by the molecular chaperone [i.e., Heat shock cognate 70 (Hsc70)] and degraded through microautophagy or CMA (59–61). Along with the ubiquitin/proteasomal system (UPS) that targets ubiquitinated proteins for degradation (62–64), neuronal autophagy could target a broader spectrum of synaptic components, together contributing to the regulation of neurotransmission and synaptic plasticity. Besides, given the intrinsic nature of autophagy as a mechanism for producing bioenergetic and biosynthetic materials (e.g., amino acids, lipids, and other metabolic building blocks), this machinery appears well suited to regulation of synaptic functions, allowing local autonomic control of synaptic plasticity in axons and dendrites distant from the neuronal soma (55,56).
Recent studies on local proteome in autophagy-deficient model organisms began to reveal a link between synaptic autophagy and synaptic plasticity (see schematic view in Fig. 2). In the presynaptic compartment, neuronal activities are shown to control autophagosome biogenesis and maturation, using several adaptor proteins or positive/negative regulators enriched at the presynaptic terminal [e.g., leucine-rich repeat kinase 2 (LRRK2), Endophilin A1, Synaptojanin 1, Bassoon] (42–44,65,66). For example, Endophilin A1, once phosphorylated by LRRK2, promotes local synaptic macroautophagy by recruiting core autophagy proteins necessary for autophagosome formation (43). Induced autophagosomes can selectively target synaptic vesicles (57,65,67); pharmacological acute activation of macroautophagy reduces the number of synaptic vesicles at axon terminals, and macroautophagy depletion (i.e., Atg7-knockout in dopaminergic neurons) increases evoked dopamine release in mice (67). Thus, presynaptic macroautophagy likely regulates neurotransmission by controlling the size of the synaptic vesicle pool available for neurotransmitter release. In addition, macroautophagy selectively targets mitochondria when mitochondrial outer membrane proteins are ubiquitinated (through Pink1-dependent activation of E3 ubiquitin ligase, Parkin; see additional reviews for details (68,69)). This form of selective autophagy (aka mitophagy) may regulate local energy supply or calcium buffering capacity, thereby contributing to synaptic functions and plasticity (70).
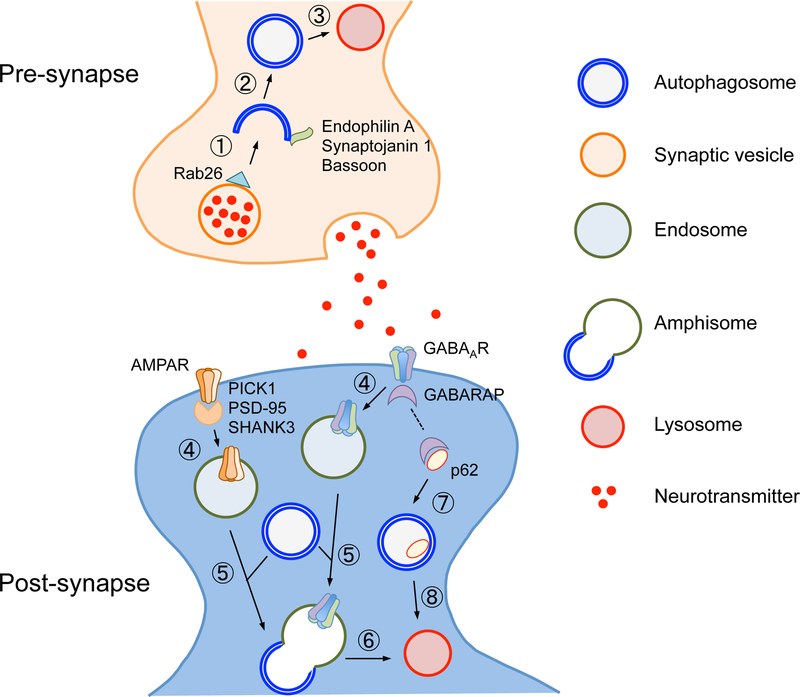
Fig. 2. Regulators of pre- and post-synaptic autophagy.
Neuronal activity upregulates autophagosome formation in the presynaptic compartment, using several presynaptically enriched adaptors (e.g., Endophilin A, Bassoon) and regulatory proteins (e.g., Synaptojanin 1). Example of the synaptic vesicle is depicted; synaptic vesicles selectively tagged by an adaptor protein (e.g., Rab26) are recognized by the pre-autophagosomal structure (①) and engulfed by the autophagosome (②), which is subsequently delivered to the lysosome for degradation (③). Endophilin A and synaptojanin 1 cooperatively recruit autophagy regulatory proteins on the autophagosomal membrane, and Bassoon negatively regulates this process by sequestering Atg5. In the postsynaptic compartment, neuronal activity also induces macroautophagy, causing endocytic removal of neurotransmitter receptors (e.g., AMPAR, GABAAR) from the plasma membrane (④), possibly via regulation of receptor adaptor proteins (e.g., PICK1, PSD-95, SHANK3). Subsequently, the endosome fuses with the autophagosome (⑤) to form the amphisome, which is delivered to the lysosome for degradation (⑥). Surface levels of GABAAR could also be regulated by the steady-state levels of p62; cytosolic p62 proteins are normally sequestered by the autophagosome (⑦) and degraded by the lysosome (⑧); however, elevated p62 expression due to reduced autophagy activity causes sequestration of GABARAP (as shown by a dotted line in the post-synaptic compartment), leading to downmodulation of surface levels of GABAAR.
Autophagy also regulates synaptic activity in the postsynaptic compartment (Fig. 2). Synaptic stimulation induces autophagosome formation or recruitment within dendrites (45). In addition, low-dose activation of N-methyl-D-aspartate (NMDA) receptors induces LTD via macroautophagy-dependent degradation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (45). Similarly, lysosomes are recruited to synapses in response to synaptic activities, leading to AMPA receptor sequestration and reduction in synaptic activity and spine density (71). Inhibitory neurotransmitter receptors (i.e., γ-aminobutyric acid (GABA)A receptors (GABAARs)) are also regulated by macroautophagy; upon denervation at the neuromuscular junction in C. elegans, GABAARs expressed on the muscle cells are selectively sorted via endocytosis to traffic from the postsynaptic membrane surface to the autophagosome, whereas acetylcholine receptors in the same cells do not (72). This suggests a role of macroautophagy in selective sorting of GABAARs, hence attenuated inhibitory neurotransmission. Further supporting a role of autophagy in synaptic plasticity, brain-derived neurotrophic factor (BDNF), an inducer of LTP, regulates autophagy in hippocampal neurons; suppression of autophagy is sufficient to rescue LTP defects and memory impairment caused by BDNF deficiency in mice (73,74). Since several post-synaptic scaffolding proteins (i.e., PICK1, PSD-95, and SHANK3) are found to be included in the autophagosomes, synaptic plasticity may be regulated by autophagic degradation of these proteins (73,74). Furthermore, recent evidence shows that TrkB, a receptor for BDNF, is localized on the autophagosomal membrane, arguing for the role of the autophagosome in transducing BDNF signals, hence neuroplasticity (75). Importantly, the above-mentioned synaptic components targeted for autophagic degradation are implicated in the regulation of synaptic plasticity (48,76).
Neuronal autophagy in the regulation of memory, cognition, and psychiatric manifestations
Although we discussed the roles of autophagy in synaptic plasticity, what is the direct evidence supporting the role of autophagy in higher brain functions, such as learning, memory or cognition? Cellular autophagic activity is known to gradually decrease during normal aging (77). Spermidine, an endogenous substance with autophagy-inducing activity, has been reported to extend longevity in many species (78), and could protect from aging-associated memory impairment and metaplasticity (i.e., ultrastructural increase of presynaptic active zones) (79), by blocking the build-up of presynaptic active zone proteins (e.g., Bruchpilot, Rim-binding protein, Unc-13) and suppressing excessive neurotransmitter release (80–83). A recent study further showed that macroautophagy activity within the Drosophila learning and memory center (i.e., mushroom body) is responsible for restricting brain-wide metaplasticity by maintaining the expression of NPY, a neuropeptide expressed in interneurons (84). This suggests the non-cell autonomous role of neuronal autophagy in protecting the brain from synapse aging and highlights the critical role of the high-level brain integration center in dictating overall brain health. In addition, mice with the nervous system-specific deletion of Wdr45, an ortholog of Atg18 essential for autophagosome formation, exhibit impaired learning and memory (85). Pharmacological upregulation of autophagy using rapamycin, an inhibitor of mammalian target of rapamycin (mTOR) that normally suppresses autophagy, is shown to alleviate deficits in synaptic plasticity and improve cognition in drug- or stress-induced rodent models of cognitive impairment (86,87). Recent evidence from an auditory fear reconsolidation model in mice demonstrated that autophagy induction could enhance erasure of a reconsolidation-resistant auditory fear memory, providing a potential therapeutic target for alleviating anxiety (88). These lines of evidence suggest that autophagy has an important role in some forms of synaptic plasticity underlying memory formation.
Evidence has begun to accumulate that autophagy deficits are linked to neuropsychiatric conditions (Table 3). First evidence for a causal role of attenuated macroautophagy in biology relevant to neuropsychiatric disorders came from a study on mice heterozygous for Tsc2 loss of function (Tsc2+/−), a model for a rare variant of autism spectrum disorders (ASDs) (41). Because Tsc2 is a negative regulator of mTOR, Tsc2+/− mice have constitutively elevated mTOR activity, hence attenuated autophagy, exhibiting excessive dendritic spine formation. This parallels with increased dendritic spine density and reduced developmental spine pruning in layer V pyramidal neurons in postmortem ASD temporal lobe. Activation of autophagy by rapamycin rescues spine pruning deficits and ASD-like behaviors (i.e., impaired social interaction) in Tsc2+/− mice, but not in neuronal autophagy-deficient mice (i.e., Tsc2+/−;Atg7-conditional KO), suggesting that the deficit in mTOR-regulated macroautophagy is causal for developmental spine pruning defects and underlies the pathophysiology of ASD.
Table 3.
Autophagy genes and psychiatric conditions
| Gene | Type of gene mutation | Species | Phenotype | Ref |
|---|---|---|---|---|
| Wdr45 | Central nervous system-specific KO | Mouse | Impaired spatial working memory; impaired recall of fear memory | 85 |
| Tsc2 | Heterozygous KO | Mouse | ASD-like imapired social interaction behavior; dendritic spine pruning defect; elevated mTOR activity and reduced autophagy flux | 41 |
| Fmr1 | KO | Mouse | Aberrant dendritic spine structure; deficits in synaptic plasticity and cognition; elevated mTOR activity and reduced autophagy flux | 89 |
| Ulk1 | KO | Mouse | Ethanol exposure-dependent cognitive deficit in novel object recognition | 127 |
| Ulk2 | Heterozygous KO | Mouse | Deficits in sensorimotor gating and cognitive flexibility; elevated p62 expression in cerebral pyramidal neurons; reduced surface expression of GABAARs; imbalanced excitatory-inhibitory balance | 90 |
| Atg7 | Post-adolescent KO | Mouse | Impaired social interaction behavior; deficits in inhibitory neurotransmission; elevated p62 expression and reduced surface expression of GABAARs | 96 |
| Ulk1, Ulk2, Ulk4 | Copy number variation (CNV) | Human | CNVs of Ulk1, Ulk2, and Ulk4 genes in Icelandic schizophrenia and bipolar patients | 128 |
| Beclin1 | N/A | Human | Reduced expression of Beclin1 in hippocampus in postmortem brains of patients with schizophrenia | 113 |
| p62 | N/A | Human | Elevated expression of p62 protein in olfactory neuronal cells biopsied from patients with schizophrenia and bipolar disorder | 90 |
Furthermore, a mouse model of Fragile X syndrome (FXS), the most frequent form of heritable intellectual disability and autism, also exhibits enhanced activity of mTOR, hence reduced autophagy (89). Activation of autophagy by silencing the expression of Raptor, a component of mTOR complex 1, largely restores aberrant spine structure, synaptic plasticity, and cognition in fragile X mice, suggesting that impaired macroautophagy is causally related to the FXS phenotypes.
Recent studies show another causal link of attenuated autophagy to neurobiological and behavioral changes relevant to psychiatric manifestations. In neuronal cells sampled from living human subjects via nasal biopsy, macroautophagic activity is reported to be downregulated in patients with schizophrenia and bipolar disorder compared with healthy controls (90). In mice with reduced expression of Ulk2 (Ulk2+/−), an ortholog of Atg1 implicated in autophagy induction, expression levels of sequestosome-1/p62, an adaptor protein responsible for selective autophagy, are upregulated predominantly in pyramidal neurons of the prefrontal cortex (PFC) as a result of attenuated autophagy (90). Ulk2+/− mice exhibit behavioral deficits relevant to psychosis, including disrupted sensorimotor gating (i.e., attenuated prepulse inhibition of acoustic startle stimuli) and impaired cognitive flexibility. Mechanistically, p62 binds with GABAA receptor-associated protein (GABARAP) (91), a protein that regulates endocytic trafficking of GABAARs (92) (Fig. 2); thus, elevated p62 expression in Ulk2+/−pyramidal neurons appears to sequester a higher proportion of GABARAP and limit the amount of GABARAP available for surface presentation of GABAARs, including α5 subunit-containing GABAAR implicated in memory and cognitive functions (93,94). Consequently, Ulk2+/− pyramidal neurons have imbalanced excitatory–inhibitory neurotransmission, which may in part underlie the sensorimotor gating deficit and cognitive impairment observed. Notably, Ulk2+/− neurons selectively down-modulates surface GABAAR levels without affecting surface NMDA receptor levels (90). Because mice deficient for α5-GABAAR also show sensorimotor gating deficit and cognitive inflexibility phenotypes (94,95) similar to those seen in Ulk2+/− mice, the data suggest that Ulk2, p62 and α5-GABAAR may function together to regulate cognitive functions. Additionally, a similar mechanism by which elevated p62-dependent sequestration of GABARAP family proteins downregulates the levels of surface GABAAR is confirmed in another autophagy-deficient mouse model (i.e., post-adolescent Atg7-conditional KO), which exhibits deregulated inhibitory neurotransmission and impaired social interaction behaviors (96).
Although the above-mentioned models of monogenic mutations causally link attenuated autophagy to psychiatric conditions, a question remains as to the extent to which attenuated autophagy is prevalent in psychiatric diseases in general. Enrichment analysis (see Supplementary materials for details) indicated that Genome-wide association study (GWAS) risk genes for brain disorders [schizophrenia, bipolar disorder, psychosis, AD, PD, dementia, attention-deficit/hyperactivity disorder (ADHD), autism, cognitive decline, and depression] are over-represented in autophagy-related pathways from gene ontology (GO) biological processes: (i) negative regulation of autophagy (p-value: 0.017); (ii) positive regulation of autophagy (p-value: 9.0E-5); and (iii) umbrella pathway consisting of core autophagy genes, autophagosome maturation, autophagosome assembly, negative regulation of autophagy, and positive regulation of autophagy (p-value: 3.1E-4) (Table S1). In addition, Gene set enrichment analysis (GSEA) using RNA-Seq data from CommonMind Consortium (97) indicated that the autophagy pathway from GO biological process was significantly downregulated in patients with schizophrenia compared to healthy controls (Table S2).
Future perspective
These studies have broader implications in neurological and neuropsychiatric disorders. Elevated levels of p62 proteins or p62-positive protein aggregates in the nervous system may represent a molecular signature shared across these two disease categories. For example, a rodent model of 22q11.2 chromosomal deletions, which are at high risk of developing schizophrenia or early-onset PD (98,99), exhibits elevated expression of p62 and α-synuclein proteins in PFC or substantia nigra, presumably due to attenuated autophagy caused by elevated mTOR activity (100). Given the finding that p62 protein levels could modulate surface GABAAR levels (90,96), elevated p62 pathology may serve as a mechanism underlying deregulated GABAergic inhibitory neurotransmission and associated cognitive impairment observed across neurological and neuropsychiatric disorders, including aging (101–106). In addition, α-synuclein and p62 are often co-localized in Lewy bodies (107), a hallmark of PD pathophysiology (70). α-synuclein is a presynaptically enriched protein implicated in neurotransmitter release, and is targeted by CMA (59). Therefore, deregulated expression of α-synuclein due to deficits in the autophagic process may underlie neuropsychiatric symptoms associated with PD in general or with 22q11.2 deletion syndrome in particular.
Attenuated autophagy may have therapeutic and diagnostic implications in neurological and neuropsychiatric disorders. Reduced surface α5-GABAAR levels may provide a potential therapeutic target for remediating cognitive impairment downstream of attenuated autophagy or elevated p62 pathology, possibly via augmenting GABAergic neurotransmission using, for example, positive allosteric modulators directed at α5-GABAAR (108,109). A recent study reported that several classes of antidepressants (e.g., amitriptyline, fluoxetine) commonly activate autophagy via slow accumulation of sphingomyelin and ceramide in neurons, and that direct inhibition of sphingomyelin synthase exerts rapid accumulation of ceramide, autophagy activation, and behavioral reversal in the stress-induced depression model (110). To evaluate efficacy of such autophagy-targeting therapeutic strategies, it is critically important to monitor autophagy activity in live brains. While the current methods of monitoring neuronal autophagy largely depend on measuring LC3/Atg8 protein levels in the brain in animal models (7), this approach may not be applicable to humans. Instead, monitoring levels of these or relevant counterpart proteins in surrogate tissues may be useful. For example, we have shown clinical utility of using olfactory neuronal cells biopsied from patients (111,112), and reported attenuated autophagy (i.e., p62 overexpression) in these cells from patients with sporadic schizophrenia and bipolar disorder (90). In addition, levels of activity-dependent neuroprotective protein (ADNP), a binding partner of LC3/Atg8, are shown to be significantly elevated in the peripheral lymphocytes of patients with schizophrenia compared to healthy controls (113).
Besides autophagy, the UPS plays a major role in homeostatic control of protein quality (62,63). Although the roles of the UPS in psychiatric disorders are not clearly understood yet, a recent report showed that a subset of the postmortem brains from patients with schizophrenia display significantly elevated levels of insoluble proteins and protein ubiquitination, arguing for pathological involvement of the UPS in some cases of psychiatric disorders (114). Another recent report also showed increased aggregate formation of GABARAP family proteins in a subset of postmortem brains from patients diagnosed as ASD (96). These studies are consistent extension and generalization of a notion that elevated insolubility of specific proteins, such as DISC1, occurs in the brains with psychiatric disorders (115–117). Mechanistically, DISC1 protein has an aggregate-prone property and sequestrates its interacting proteins into insoluble fractions, leading to a loss of function of key interacting proteins such as phosphodiesterase-4 and the associated changes in emotionality (118,119). An outstanding question is how autophagy and the UPS are related with each other under an overall picture of protein quality control and, in turn, contribute to psychiatric disorders.
We have witnessed part of the strategies that neuronal autophagy uses in regulating synaptic functions and network activity. This includes activity-dependent degradation of neurotransmitters and their receptors, selective surface presentation of neurotransmitter receptors, and morphological regulation of synaptic spines. Furthermore, these recent findings provide insight into how neuronal autophagy regulates higher brain functions, such as cognition, mood, and social interaction. Future studies at molecular, cellular, circuit, and behavioral levels are warranted to provide insight into the detailed mechanisms for autophagy-mediated brain functions, and to further address how deregulated autophagy underlies specific dimensions of symptomatic outcomes in neurological and psychiatric disorders. Depending on the cell types (e.g., excitatory or inhibitory neurons), the brain regions (e.g., PFC or substantia nigra), or the circuits (e.g., corticolimbic or nigrostriatal projection) that are principally affected, we predict to see varying phenotypic consequences in specific behavioral or cognitive dimensions. Finally, in-depth understanding of the circuit-wide alterations following deregulated neuronal autophagy is critical to devise novel therapeutic approaches for these pathological conditions.
Supplementary Material
Acknowledgments
This work was supported by NIH (MH-092443, MH-094268 Silvio O. Conte center, MH-105660, and MH-107730), as well as the foundation grants of Stanley, RUSK/S-R, and NARSAD/BBRF (to A.S.); and DOD/CDMRP (W81XWH-11–1-0269 to T.T.).
Footnotes
Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Cannon WB (1932): The Wisdom of the Body. New York: W.W.Norton pp; 177–201. [Google Scholar]
- 2.Kotas ME, Medzhitov R (2015): Homeostasis, inflammation, and disease susceptibility. Cell 160:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnham KJ, Masters CL, Bush AI (2004): Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov 3:205–214. [DOI] [PubMed] [Google Scholar]
- 4.Yin F, Yao J, Brinton RD, Cadenas E (2017): Editorial: The metabolic–inflammatory axis in brain aging and neurodegeneration. Front Aging Neurosci 9:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankowski R, Mader S, Valdés-Ferrer SI (2015): Systemic inflammation and the brain: novel roles of genetic, molecular, and environmental cues as drivers of neurodegeneration. Front Cell Neurosci 9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas PM, Dillin A (2010): Protein homeostasis and aging in neurodegeneration. J Cell Biol 190:719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Komatsu M (2011): Autophagy: renovation of cells and tissues. Cell 147:728–741. [DOI] [PubMed] [Google Scholar]
- 8.Ariosa AR, Klionsky DJ (2016): Autophagy core machinery: overcoming spatial barriers in neurons. J Mol Med (Berl) 94:1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Klionsky DJ (2010): Eaten alive: a history of macroautophagy. Nat Cell Biol 12:814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider JL, Cuervo AM (2014): Autophagy and human disease: emerging themes. Curr Opin Genet Dev 26:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto A, Yue Z (2014): Autophagy and its normal and pathogenic states in the brain. Annu Rev Neurosci 37:55–78. [DOI] [PubMed] [Google Scholar]
- 12.Kulkarni A, Chen J, Maday S (2018): Neuronal autophagy and intercellular regulation of homeostasis in the brain. Curr Opin Neurobiol 51:29–36. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N (2018): A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol 20:521–527. [DOI] [PubMed] [Google Scholar]
- 14.Levine B, Kroemer G (2019): Biological Functions of Autophagy Genes: A disease perspective. Cell 176:11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenbeck PJ (1993): Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol 121:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue Z (2007): Regulation of neuronal autophagy in axon: implication of autophagy in axonal function and dysfunction/degeneration. Autophagy 3:139–141. [DOI] [PubMed] [Google Scholar]
- 17.Maday S, Wallace KE, Holzbaur EL (2012): Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 196:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maday S, Holzbaur EL (2014): Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell 30:71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. (2006): Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. (2006): Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884. [DOI] [PubMed] [Google Scholar]
- 21.Komatsu M, Wang QJ, Holstein GR, Friedrich VL Jr, Iwata J, Kominami E, et al. (2007): Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A 104:14489–14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. (2005): Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 64:113–122. [DOI] [PubMed] [Google Scholar]
- 23.Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, et al. (2017): Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93:1015–1034. [DOI] [PubMed] [Google Scholar]
- 24.Millecamps S, Julien JP (2013): Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci 14:161–176. [DOI] [PubMed] [Google Scholar]
- 25.Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL (2014): Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84:292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. (2010): Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141:1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Sato Y, Nixon RA (2011): Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J Neurosci 31:7817–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Y, Zhou B, Lin MY, Wang S, Foust KD, Sheng ZH (2015): Endolysosomal deficits augment mitochondria pathology in spinal motor neurons of asymptomatic fALS mice. Neuron 87:355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, et al. (2015): Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci U S A 112:E3699–E3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frake RA, Ricketts T, Menzies FM, Rubinsztein DC (2015): Autophagy and neurodegeneration. J Clin Invest 125:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lie PPY, Nixon RA (2019): Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol Dis 122:94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. (2007): Ambra1 regulates autophagy and development of the nervous system. Nature 447:1121–1125. [DOI] [PubMed] [Google Scholar]
- 33.Dragich JM, Kuwajima T, Hirose-Ikeda M, Yoon Ms, Eenjes E, Bosco JR, et al. (2016): Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife 5:pii:e14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi J, Suzuki C, Nanao T, Kakuta S, Ozawa K, Tanida I, et al. (2018): Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy 14:764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kannan M, Bayam E, Wagner C, Rinaldi B, Kretz PF, Tilly P, et al. (2017): WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy. Proc Natl Acad Sci U S A 114:E9308–E9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toda H, Mochizuki H, Flores R 3rd, Josowitz R, Krasieva TB, Lamorte VJ, et al. (2008): UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev 22:3292–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wairkar YP, Toda H, Mochizuki H, Furukubo-Tokunaga K, Tomoda T, Diantonio A (2009): Unc-51 controls active zone density and protein composition by downregulating ERK signaling. J Neurosci 29:517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen W, Ganetzky B (2009): Autophagy promotes synapse development in Drosophila. J Cell Biol 187:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stavoe AK, Hill SE, Hall DH, Colon-Ramos DA (2016): KIF1A/UNC-104 transports ATG-9 to regulate neurodevelopment and autophagy at synapses. Dev Cell 38:171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudnick ND, Griffey CJ, Guarnieri P, Gerbino V, Wang X, Piersaint JA, et al. (2017): Distinct roles for motor neuron autophagy early and late in the SOD1G93A mouse model of ALS. Proc Natl Acad Sci U S A 114: E8294–E8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, et al. (2014): Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, Martin S, Papadopulos A, Harper CB, Mavlyutov TA, Niranjan D, et al. (2015): Control of autophagosome axonal retrograde flux by presynaptic activity unveiled using botulinum neurotoxin type a. J Neurosci 35:6179–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soukup SF, Kuenen S, Vanhauwaert R, Manetsberger J, Hernandez-Diaz S, Swerts J, et al. (2016): A LRRK2-dependent Endophilin A phosphoswitch is critical for macroautophagy at presynaptic terminals. Neuron 92:829–844. [DOI] [PubMed] [Google Scholar]
- 44.Vanhauwaert R, Kuenen S, Masius R, Bademosi A, Manetsberger J, Schoovaerts N, et al. (2017): The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J 36:1392–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, InokuchI K (2012): Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci 32:10413–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bliss TVP, Collingridge GL, Morris RGM (2014): Synaptic plasticity in health and disease: Introduction and overview. Philos Trans R Soc Lond B Biol Sci 369: 20130129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Rourke NA, Weiler NC, Micheva KD, Smith SJ (2012): Deep molecular diversity of mammalian synapses: Why it matters and how to measure it. Nat Rev Neurosci 13: 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi T, Duszkiewicz AJ, Morris RGM (2014): The synaptic plasticity and memory hypothesis: encoding, storage and persistence. Philos Trans R Soc Lond B Biol Sci 369:20130288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nägerl UV (2006): A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52:239–245. [DOI] [PubMed] [Google Scholar]
- 50.Ho VM, Lee J-A, Martin KC (2011): The cell biology of synaptic plasticity. Science 334:623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alvarez-Castelao B, Schuman EM (2015): The regulation of synaptic protein turnover. J Biol Chem 290:28623–28630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen LD, Ziv NE (2017): Recent insights on principles of synaptic protein degradation. F1000Research 6:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bean BP (2007): The action potential in mammalian central neurons. Nat Rev Neurosci 8:451–465. [DOI] [PubMed] [Google Scholar]
- 54.Häusser M, Raman IM, Otis T, Smith SL, Nelson A, du Lac S (2004): The beat goes on: spontaneous firing in mammalian neuronal microcircuits. J Neurosci 24:9215–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vijayan V, Verstreken P (2017): Autophagy in the presynaptic compartment in health and disease. J Cell Biol 216:1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang YC, Lauwers E, Verstreken P (2017): Presynaptic protein homeostasis and neuronal function. Curr Opin Genet Dev 44:38–46. [DOI] [PubMed] [Google Scholar]
- 57.Binotti B, Pavlos NJ, Riedel D, Wenzel D, Vorbrüggen G, Schalk AM, et al. (2015): The GTPase Rab26 links synaptic vesicles to the autophagy pathway. Elife 4:e05597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong YC, Holzbaur ELF (2014): Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A 111:E4439–E4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D (2004): Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305:1292–1295. [DOI] [PubMed] [Google Scholar]
- 60.Orenstein SJ, Kuo S-H, Tasset I, Arias E, Koga H, Fernandez-Carasa I, et al. (2013): Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci 16:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uytterhoeven V, Lauwers E, Maes I, Miskiewicz K, Melo MN, Swerts J, et al. (2015): Hsc70–4 deforms membranes to promote synaptic protein turnover by endosomal microautophagy. Neuron 88:735–748. [DOI] [PubMed] [Google Scholar]
- 62.Bingol B, Sheng M (2011): Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron 69:22–32. [DOI] [PubMed] [Google Scholar]
- 63.Tsai N-P (2014): Ubiquitin proteasome system-mediated degradation of synaptic proteins: An update from the postsynaptic side. Biochim Biophys Acta 1843:2838–2842. [DOI] [PubMed] [Google Scholar]
- 64.Lip PZY, Demasi M, Bonatto D (2017): The role of the ubiquitin proteasome system in the memory process. Neurochem Int 102:57–65. [DOI] [PubMed] [Google Scholar]
- 65.Okerlund ND, Schneider K, Leal-Ortiz S, Montenegro-Venegas C, Kim Sa, Garner LC, et al. (2017): Bassoon controls presynaptic autophagy through Atg5. Neuron 93:897–913.e7. [DOI] [PubMed] [Google Scholar]
- 66.Murdoch JD, Rostosky CM, Gowrisankaran S, Arora AS, Soukup SF, Vidal R, et al. (2016): Endophilin-A deficiency induces the Foxo3a-Fbxo32 network in the brain and causes dysregulation of autophagy and the ubiquitin-proteasome system. Cell Rep 17:1071–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hernandez D, Torres CA, Setlik W, Cebrián C, Mosharov EV, Tang G, et al. (2012): Regulation of presynaptic neurotransmission by macroautophagy. Neuron 74:277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pickrell AM, Youle RJ (2015): The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85:257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamano K, Matsuda N, Tanaka K (2016): The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep 17:300–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soukup SF, Vanhauwaert R, Verstreken P (2018): Parkinson’s disease: convergence on synaptic homeostasis. EMBO J 37:e98960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goo MS, Sancho L, Slepak N, Boassa D, Deerinck TJ, Ellisman MH, et al. (2017): Activity-dependent trafficking of lysosomes in dendrites and dendritic spines. J Cell Biol 216:2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA (2006): Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci 26:1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nikoletopoulou V, Sidiropoulou K, Kallergi E, Dalezios Y, Tavernarakis N (2017): Modulation of autophagy by BDNF underlies synaptic plasticity. Cell Metab 26:230–242.e5. [DOI] [PubMed] [Google Scholar]
- 74.Nikoletopoulou V, Tavernarakis N (2018): Regulation and roles of autophagy at synapses. Trends Cell Biol 28:646–661. [DOI] [PubMed] [Google Scholar]
- 75.Kononenko NL, Claßen GA, Kuijpers M, Puchkov D, Maritzen T, Tempes A, et al. (2017): Retrograde transport of TrkB-containing autophagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat Commun 8:14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luscher C, Malenka RC (2012): NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD). Cold Spring Harb Perspect Biol 4:pii:a005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vilchez D, Saez I, Dillin A (2014): The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun 5:5659. [DOI] [PubMed] [Google Scholar]
- 78.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. (2009): Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11:1305–1314. [DOI] [PubMed] [Google Scholar]
- 79.Gupta VK, Scheunemann L, Eisenberg T, Mertel S, Bhukel A, Koemans TS, et al. (2013): Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat Neurosci 16:1453–1460. [DOI] [PubMed] [Google Scholar]
- 80.Gupta VK, Pech U, Bhukel A, Fulterer A, Ender A, Mauermann SF, et al. (2016): Spermidine suppresses age-associated memory impairment by preventing adverse increase of presynaptic active zone size and release. PLoS Biol 14:e1002563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhukel A, Madeo F, Sigrist SJ (2017): Spermidine boosts autophagy to protect from synapse aging. Autophagy 13:444–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Petzoldt AG, Lützkendorf J, Sigrist SJ (2016): Mechanisms controlling assembly and plasticity of presynaptic active zone scaffolds. Curr Opin Neurobiol 39:69–76. [DOI] [PubMed] [Google Scholar]
- 83.Liang Y, Sigrist S (2018): Autophagy and proteostasis in the control of synapse aging and disease. Curr Opin Neurobiol 48:113–121. [DOI] [PubMed] [Google Scholar]
- 84.Bhukel A, Beuschel CB, Maglione M, Lehmann M, Juhász G, Madeo F, et al. (2019): Autophagy within the mushroom body protects from synapse aging in a non-cell autonomous manner. Nat Commun 10:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao YG, Sun L, Miao G, Ji C, Zhao H, Sun H, et al. (2015): The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy 11:881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fu J, Wang H, Gao J, Yu M, Wang R, Yang Z, et al. (2017): Rapamycin effectively impedes melamine-induced impairments of cognition and synaptic plasticity in Wistar rats. Mol Neurobiol 54:819–832. [DOI] [PubMed] [Google Scholar]
- 87.Zhai B, Shang X, Fu J, Li F, Zhang T (2018): Rapamycin relieves anxious emotion and synaptic plasticity deficits induced by hindlimb unloading in mice. Neurosci Lett 677:44–48. [DOI] [PubMed] [Google Scholar]
- 88.Shehata M, Abdou K, Choko K, Matsuo M, Nishizono H, Inokuchi K (2018): Autophagy enhances memory erasure through synaptic destabilization. J Neurosci 38:3809–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan J, Porch MW, Court-Vazquez B, Bennett MVL, Zukin RS (2018): Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc Natl Acad Sci U S A 115:E9707–E9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sumitomo A, Yukitake H, Hirai K, Horike K, Ueta K, Chung Y, et al. (2018): Ulk2 controls cortical excitatory-inhibitory balance via autophagic regulation of p62 and GABAA receptor trafficking in pyramidal neurons. Hum Mol Genet 27:3165–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. (2007): p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145. [DOI] [PubMed] [Google Scholar]
- 92.Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW (1999): GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature 397:69–72. [DOI] [PubMed] [Google Scholar]
- 93.Rudolph U, Möhler H (2014): GABAA receptor subtypes: Therapeutic potential in Down syndrome, affective disorders, schizophrenia, and autism. Annu Rev Pharmacol Toxicol 54:483–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Engin E, Zarnowska ED, Sigal M, Keist R, Zeller A, Pearce RA, et al. (2013): Alpha5-containing GABAA receptors in dentate gyrus enable cognitive flexibility. FASEB J 27:661.7–661.7. [Google Scholar]
- 95.Hauser J, Rudolph U, Keist R, Möhler H, Feldon J, Yee BK (2005): Hippocampal alpha5 subunit-containing GABAA receptors modulate the expression of prepulse inhibition. Mol Psychiatry 10:201–207. [DOI] [PubMed] [Google Scholar]
- 96.Hui KK, Takashima N, Watanabe A, Chater TE, Matsukawa H, Nekooki-Machida Y, et al. (2019): GABARAPs dysfunction by autophagy deficiency in adolescent brain impairs GABAA receptor trafficking and social behavior. Sci Adv 5:eaau8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. (2016): Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19:1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Butcher NJ, Kiehl TR, Hazrati LN, Chow EW, Rogaeva E, Lang AE, et al. (2013): Association between early-onset Parkinson disease and 22q11.2 deletion syndrome: identification of a novel genetic form of Parkinson disease and its clinical implications. JAMA Neurol 70:1359–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schneider M, Debbané M, Bassett AS, Chow EW, Fung WL, van den Bree M, et al. (2014): Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry 171:627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sumitomo A, Horike K, Hirai K, Butcher N, Boot E, Sakurai T, et al. (2018): A mouse model of 22q11.2 deletions: Molecular and behavioral signatures of Parkinson’s disease and schizophrenia. Sci Adv 4:eaar6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guzman BCF, Vinnakota C, Govindpani K, Waldvogel HJ, Faull RLM, Kwakowsky A (2018): The GABAergic system as a therapeutic target for Alzheimer’s disease. J Neurochem 146:649–669. [DOI] [PubMed] [Google Scholar]
- 102.Garret M, Du Z, Chazalon M, Cho YH, Baufreton J (2018): Alteration of GABAergic neurotransmission in Huntington’s disease. CNS Neurosci Ther 24:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Błaszczyk JW (2016): Parkinson’s disease and neurod egeneration: GABA-collapse hypothesis. Front Neurosci 10:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hoftman GD, Datta D, Lewis DA (2017): Layer 3 Excitatory and inhibitory circuitry in the prefrontal cortex: developmental trajectories and alterations in schizophrenia. Biol Psychiatry 81:862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fee C, Banasr M, Sibille E (2017): Somatostatin-positive gamma-aminobutyric acid interneuron deficits in depression: cortical microcircuit and therapeutic perspectives. Biol Psychiatry 82:549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Porges EC, Woods AJ, Edden RA, Puts NA, Harris AD, Chen H, et al. (2017): Frontal gamma-aminobutyric acid concentrations are associated with cognitive performance in older adults. Biol Psychiatry Cogn Neurosci Neuroimaging 2:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ferrer I, Martinez A, Blanco R, Dalfó E, Carmona M (2011): Neuropathology of sporadic Parkinson disease before the appearance of parkinsonism: preclinical Parkinson disease, J Neural Transm 118:821–839. [DOI] [PubMed] [Google Scholar]
- 108.Koh MT, Rosenzweig-Lipson S, Gallagher M (2013): Selective GABA(A) α5 positive allosteric modulators improve cognitive function in aged rats with memory impairment. Neuropharmacology 64:145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prevot TD, Li G, Vidojevic A, Misquitta KA, Fee C, Santrac A, et al. (2019): Novel benzodiazepine-like ligands with various anxiolytic, antidepressant, or pro-cognitive profiles. Mol Neuropsychiatry 5:84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gulbins A, Schumacher F, Becker KA, Wilker B, Soddemann M, Boldrin F, et al. (2018): Antidepressants act by inducing autophagy controlled by sphingomyelin–ceramide. Mol Psychiatry 23:2324–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lavoie J, Sawa A, Ishizuka K (2017): Application of olfactory tissue and its neural progenitors to schizophrenia and psychiatric research. Curr Opin Psychiatry 30:176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lavoie J, Gassó Astorga P, Segal-Gavish H, Wu YC, Chung Y, Cascella NG, et al. (2017): The olfactory neural epithelium as a tool in neuroscience. Trends Mol Med 23:100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Merenlender-Wagner A, Malishkevich A, Shemer Z, Udawela M, Gibbons A, Scarr E, et al. (2015): Autophagy has a key role in the pathophysiology of schizophrenia. Mol Psychiatry 20: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nucifora LG, MacDonald ML, Lee BJ, Peters ME, Norris AL, Orsburn B, et al. (2019): Increased protein insolubility in brains from a subset of patients with schizophrenia. Am J Psychiatry appiajp201918070864. [DOI] [PubMed]
- 115.Korth C (2012): Aggregated proteins in schizophrenia and other chronic mental diseases: DISC1opathies. Prion 6:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trossbach SV, Bader V, Hecher L, Pum ME, Prikulis I, Schäble S, et al. (2016): Misassembly of non-mutant Disrupted-in-schizophrenia 1 (DISC1) protein is linked to altered dopamine homeostasis and behavioral deficits. Mol Psychiarty 21:1561–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Müller-Schiffmann A, Herring A, Abdel-Hafiz L, Schäble S, Wedel D, Tchepkova AN, et al. (2016): Aβ dimers in the absence of plaque pathology are sufficient to impair learning and synaptic plasticity. Brain 139(Pt 2):509–525. [DOI] [PubMed] [Google Scholar]
- 118.Tanaka M, Ishizuka K, Nekooki-Machida Y, Endo R, Takashima N, Sasaki H, et al. (2017): Aggregation of scaffolding protein DISC1 dysregulates phosphodiesterase 4 in Huntington’s disease. J Clin Invest 127:1438–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Endo R, Takashima N, Nekooki-Machida Y, Komi Y, Hui KK, Takao M, et al. (2018): TAR DNA-binding protein 43 and disrupted in schizophrenia 1 coaggregation disrupts dendritic local translation and mental function in frontotemporal lobar degeneration. Biol Psychiatry 84:509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nishiyama J, Miura E, Mizushima N, Watanabe M, Yuzaki M (2007): Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy 3:591–596. [DOI] [PubMed] [Google Scholar]
- 121.Liang CC, Wang C, Peng X, Gan B, Guan JL (2010): Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem 285:3499–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Joo JH, Wang B, Frankel E, Ge L, Xu L, Iyengar R, et al. (2016): The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol Cell 62:491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, et al. (2016): Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife 5:pii:e12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Haack TB, Hogarth P, Kruer MC, Gregory A, Wieland T, Schwarzmayr T, et al. (2012): Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am J Hum Genet 91:1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hayflick SJ, Kruer MC, Gregory A, Haack TB, Kurian MA, Houlden HH, et al. (2013): β-Propeller protein-associated neurodegeneration: A new X-linked dominant disorder with brain iron accumulation. Brain 136:1708–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, et al. (2013): De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45:445–449. [DOI] [PubMed] [Google Scholar]
- 127.Sumitomo A, Ueta K, Mauchi S, Hirai K, Horike K, Hikida T, et al. (2017): Ulk1 protects against ethanol-induced neuronal stress and cognition-related behavioral deficits. Neurosci Res 117:54–61. [DOI] [PubMed] [Google Scholar]
- 128.Lang B, Pu J, Hunter I, Liu M, Martin-Granados C, Reilly TJ, et al. (2014): Recurrent deletions of ULK4 in schizophrenia: a gene crucial for neuritogenesis and neuronal motility. J Cell Sci 127(Pt 3):630–640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.