Abstract
As intracellular parasites, viruses hijack the cellular machinery to facilitate their replication and spread. This includes favouring the expression of their viral genes over host genes, appropriation of cellular molecules, and manipulation of signalling pathways, including the post-translational machinery. HIV, the causative agent of AIDS, is notorious for using post-translational modifications to generate infectious particles. Here, we discuss the mechanisms by which HIV usurps the ubiquitin and SUMO pathways to modify both viral and host factors to achieve a productive infection, and also how the host innate sensing system uses these post-translational modifications to hinder HIV replication.
Introduction to HIV and AIDS
Brief history of HIV and AIDS
Acquired Immunodeficiency Syndrome (AIDS) was first acknowledged as an infectious disease in the early 1980s, when a large number of young, previously healthy, homosexual men suffered from lymphadenopathy, contracted unusual opportunistic infections and/or experienced other malignancies, causing their imminent death (CDC, 1981a,b). The fact that this disease was initially manifested among homosexual males led the press and some authorities to mistakenly name it GRID (gay-related immunodeficiency, often referred to as gay cancer or gay plague), which further stigmatized an already marginalized gay community. Owing to collaborative efforts between American and French scientists, the causative agent for this disease was successfully identified in 1983 (Barré-Sinoussi et al., 1983; Gallo et al., 1984). The agent, a human retrovirus that substantially differed from the Human T-lymphotropic virus (HTLV) characterized by the Gallo lab, was initially named lymphadenopathy-associated virus (LAV), later known as Human Immunodeficiency Virus or HIV. Remarkably, in 1986, another human retrovirus was found to cause AIDS in West Africa, although with less severity than the virus isolated in the US and Europe. Phylogenetic and immunological analyses revealed that this African virus was related but distinct from the original LAV (Clavel et al., 1986). In consequence, LAV was termed as HIV-1 and the African retrovirus as HIV-2. In this review, we use the broad term HIV to cover general aspects of the biology of both HIV-1 and HIV-2. However, for some particular characteristics we will use their specific denomination.
Once the causative agents of the syndrome were identified, subsequent research was aimed at characterizing the biological properties of these new human viruses. These studies demonstrated that HIV could be transmitted: (a) sexually to both men and women, (b) via direct contact with infected bodily fluids (i.e. through blood transfusions, organ transplantation, syringe sharing, etc.), and (c) vertically from mother to child during birth and also while breastfeeding (Hladik and McElrath, 2008; Cohen et al., 2011b). Therefore, the population at risk of this infection was not limited to the gay community. This finding created such an alarm that the US Administration devoted ample resources for the diagnosis, prevention and treatment of this disease, although by then AIDS was already an epidemic in the USA.
Pathogenesis
AIDS is characterized by a remarkable depletion of circulating CD4+ T cells (<200 cells/μl). CD4+ T cells, or T helper lymphocytes, are essential to elicit an effective adaptive immune response. In particular, they assist in the production of antibodies against specific pathogens and support the activity of cytotoxic T lymphocytes (CTL or CD8+ T cells) as well as natural killer cells to eliminate infected and transforming cells. Therefore, the loss of CD4+ T cells caused by HIV renders affected individuals defenceless against the most trivial infections, and unable to clear abnormal cells (Swanstrom and Coffin, 2012; CDC, 2018). This is why, besides opportunistic infections, malignancies commonly diagnosed in AIDS patients are cancers, such as Kaposi’s sarcoma, cervical cancer and lymphomas (Knipe and Howley, 2013). In addition, individuals with AIDS can often experience fevers, nightly sweats, swollen lymph nodes, chills, weakness, and significant weight loss.
HIV is basically an infection of the immune system. This virus replicates in immune cells, specifically CD4+ T cells and to a lesser extent macrophages and dendritic cells, which is the main reason why the clinical manifestation of AIDS is a severe defect in eliciting immune responses (= immunodeficiency) (Knipe and Howley, 2013). The progression to AIDS after the exposure to HIV is characterized by three distinct phases (Fig. 24.1):
Figure 24.1.
Progression of the HIV infection to AIDS in the presence and absence of cART. The graph displays the HIV viral loads as RNA copies/ml of plasma, and CD4+ T cell counts as number of cells/μl. The levels of these variables in the absence of cART are represented in black and dark blue lines, while these elements are depicted in grey and light blue lines under conditions of cART adherence.
The acute HIV syndrome, in which infected individuals present flu-like symptoms for ≈2 weeks. This phase appears around 4 weeks post-infection and depicts a sharp increase in viraemia that causes an important reduction in circulating CD4+ T cells (Fig. 24.1) (Swanstrom and Coffin, 2012; Knipe and Howley, 2013; CDC, 2018).
The clinical latency. In this phase, the CD4 counts gradually decline and there is a concomitant progressive increase in viral loads (Fig. 24.1, dark blue and black curves), with no symptomatology associated. However, if the infection remains unnoticed, not only will infected individuals develop full-blown AIDS, but also they can transmit the virus to others – of note, the clinical latency can last from 2–8 years, and this largely depends on both host and viral factors (Swanstrom and Coffin, 2012; Knipe and Howley, 2013; CDC, 2018).
AIDS. This phase is characterized by viral loads close to the peak viraemia observed in the acute HIV syndrome and is accompanied by undetectable CD4+ T cells. This depletion in T helper lymphocytes renders the HIV+ patients unable to fight infections and is inevitably fatal (Swanstrom and Coffin, 2012; Knipe and Howley, 2013; CDC, 2018) (Fig. 24.1, black and dark blue curves).
However, if the infection is diagnosed early, and the individuals adhere to combination antiretroviral therapy (cART), HIV plasma loads will remain imperceptible and the CD4 counts will stay within healthy levels (Fig. 24.1, grey and light blue curves), precluding the progression to AIDS (Volberding and Deeks, 2010; Arts and Hazuda, 2012; Benítez-Gutiérrez et al., 2018; Cluck and Underwood, 2018). Importantly, the risk of HIV transmission from HIV+ individuals with viraemia suppression on cART is exceedingly low (Cohen et al., 2011a; Benítez-Gutiérrez et al., 2018).
Although HIV primarily infects T helper lymphocytes, the early loss of a large fraction of these cells in the gut-associated lymphoid tissue (GALT) does not lead to the immunodeficiency syndrome, since AIDS is caused by a massive depletion of these cells, mainly in blood, which is only observed in the last stage of the disease. In fact, the ultimate destruction of the host immune system seems to be the result of various factors. On one hand, the reduction in CD4+ T cells in the GALT makes the gastrointestinal mucosa more permeable, which, over time, facilitates the translocation of microbial products to the bloodstream, a fact that causes the characteristic chronic immune activation observed in untreated and AIDS-progressing HIV+ individuals (Marchetti et al., 2013). This,in combination with the extinguishing capacity of the host’s immune system to replenish lost CD4+ T cells, and aid in the maturation and regulation of T lymphocytes, sets the stage for the immunodeficiency disorder. Nevertheless, in the early days of the infection, the host immune system is still functional, yet unable to clear HIV from the system. In fact, antibodies against HIV can be readily detected in HIV+ individuals’ serum within weeks of the initial exposure (Weber et al., 1987), accompanied by a potent CTL response that somewhat controls viraemia (Lyles et al., 2000; Rosenberg et al., 2000). Notably, at that time the number of CD4+ T cells still exceeds plasma viral loads (Fig. 24.1). The failure to eliminate HIV and HIV-infected cells is due to several reasons:
HIV-induced MHC-I down-regulation. HIV has evolved to selectively down-regulate certain class I major histocompatibility molecules (MHC-I), preventing the recognition of HIV-infected cells by CTLs, which allows virus propagation to proceed (Schwartz et al., 1996).
Immune functional defects. Most of the HIV-specific CTL responses are functionally compromised due to the inability to either secrete multiple cytokines simultaneously, to recognize HIV-infected cells (caused by the HIV-mediated MHC-I down-regulation) or due to exhaustion caused by a chronic antigen stimulation that results in defects in generating memory CTL subsets (Knipe and Howley, 2013).
Virus evolution. As it is explained later, the HIV polymerase lacks proofreading activity, making HIV a highly mutagenic virus (Coffin, 1995). Therefore, new antigenic variants are rapidly produced, which may not be susceptible to neutralization by the circulating antibodies and T cells.
Viral latency, an event that occurs when HIV-infected and activated CD4+ T cells transition to a resting memory phenotype (Siliciano and Greene, 2011). Because HIV gene expression depends on the presence of host transcription factors that become inactivated during this differentiation process, the rate of HIV transcription significantly declines in this transition, causing HIV to become dormant. These latently infected cells are not producing virions, do not present any viral epitopes on their MHC molecules, are not responsive to the antiretroviral drugs, and are invisible to the immune system (Castro-Gonzalez et al., 2018). Whereas these latent cells appear harmless, since HIV is dormant, they have the potential to reactivate in the presence of activating stimuli (cytokines, the original pathogen that led to their initial expansion, etc.), and support a productive HIV infection, reseeding in turn viraemia. Hence, these latent reservoirs are nowadays considered the main challenge to functionally cure HIV/AIDS.
HIV/AIDS by the numbers: epidemiological data
Since the recognition of AIDS as a new disease in 1981, HIV has been estimated to infect over 77 million people around the globe, with HIV-1 being the main cause for the pandemic (UNAIDS, 2018). With the implementation of cART, the incidence of new infections has significantly decreased in developed countries, although with the current opioid crisis in the USA, a peak of new infections has been observed among injection drug users (CDC, 2018). However, in countries with limited resources, AIDS represents a major concern for public health. Africa has been particularly hit by the pandemic with ≈ 25 million infected people, which represent around 68% of all HIV+ individuals. Of note, Africa is inhabited by only 13% of the world’s population, indicating that although the mechanisms of HIV transmission are well known, the number of new HIV infections has been rising in this continent (UNAIDS, 2015, 2018). Furthermore, 3 million Africans are manifesting symptoms of advanced AIDS and are in desperate need of treatment. Besides Africa, the disease has considerably expanded to regions of Central and South America, Asia and Eastern Europe (UNAIDS, 2018). Remarkably, the most recurrent viruses in these geographical areas belong to different clades, which significantly challenge the success of vaccines and other preventative measures to efficiently protect against all HIV-1 subtypes.
According to the most recent epidemiological data from UNAIDS, there are currently 36.9 million HIV+ individuals in the world, of which 51% are women. Importantly, 15.2 million people currently do not have access to antiretroviral therapy. Most of these individuals live in low- and middle-income countries, evidencing important epidemiological inequalities for HIV/AIDS. The insufficient control measures to prevent the dissemination of the disease, together with cultural, social and economic factors have significantly limited the access of the general population to antiretroviral drugs. The combination of these factors has facilitated the spread of HIV, favouring high degree of virus diversity, which complicates even further the development of effective vaccines against all these variants (Shao and Williamson, 2012). Owing to these limitations, global efforts have been devoted to early diagnosis programs and assistance with adherence to the antiretroviral drugs in developing countries. These comprehensive measures have decreased by 53% the global death toll associated with AIDS (from its peak in 2005 of ≈ 2 million casualties/year to 0.94 million deaths in 2017). To continue diminishing these numbers, additional large-scale programs have been implemented to improve HIV prevention and provide affordable treatment, which are already reducing the transmission of HIV and in consequence the number of new infections (decreasing by 16%, currently at a rate of 1.8 million new HIV infections per year). Still, additional cooperative efforts are needed to develop a successful and inexpensive vaccine, particularly in low- and middle-income countries, so the number of new infections can be reduced to zero.
Origins of HIV
Since the discovery of HIV-1 and HIV-2 as new human viruses, the causes leading to their sudden emergence have been intensely investigated. HIV-1 and HIV-2 originated from Simian Immunodeficiency Viruses (SIVs), after their cross-species transmission from their natural hosts to humans – importantly, these SIVs are non-pathogenic, for the most part, in their reservoirs. In particular, HIV-1 is the result of four independent transmissions of SIVs infecting chimpanzees and gorillas (SIVcpz and SIVgor), whereas HIV-2 was originated after the transmission of SIVs infecting sooty man-gabeys (SIVsmm). These events are estimated to have occurred around the 1930s and were the result of human-simian encounters, most likely in the context of bushmeat hunting, in which humans were exposed to the blood or other bodily fluids of these naturally infected non-human primates (Sharp and Hahn, 2011). Although for most viruses crossspecies transmissions result in dead ends, in certain occasions the transmitted virus can adapt to replicate in the new host, becoming more pathogenic than in its natural reservoir. This first adaptation requires overcoming innate restriction factors that safeguard the host against invading pathogens. The restriction factors that HIV has adapted to for its successful spread in humans are TRIM5α, APOBEC3G, SAMHD1, and particularly BST2 (Sharp and Hahn, 2011), which are discussed extensively later in this review.
HIV replication cycle
HIV-1 and HIV-2 belong to the Retroviridae family and more specifically to the Lentivirus genus. As other retroviruses, the HIV virions enclose their genome of single-stranded RNA (ssRNA) with positive polarity (meaning that the molecule could serve as mRNA) inside of a conical capsid or core made of multiple copies of the viral capsid (CA) protein (Fig. 24.2A). In addition to this capsid, the viral genome is wrapped by a predominantly positively charged nucleocapsid protein (NC), which plays a critical role in the incorporation of viral genomes into the nascent virions during the assembly process. Inside the capsid, the viral enzymes integrase (IN) and reverse transcriptase (RT) are found associated with the genome. Surrounding the capsid, other viral proteins can be found such as Nef, Vpr, Vpx (only in HIV-2) and the viral protease (PR), essential for the maturation of the newly formed virions. Enclosing these other components is a matrix layer, made of the viral protein matrix (MA), which serves as a docking point for the viral envelope. This lipid bilayer, obtained by budding from infected cells, is embedded not only with cellular molecules but also with the viral spikes, which serve as anti-receptors. The spikes are made of two HIV protein species (gp120 and gp41) that are non-covalently linked to each other to form a trimeric spike (Freed and Martin, 2013; Knipe and Howley, 2013) (Fig. 24.2A).
Figure 24.2.
The HIV virion and its genome. (A) Schematic illustration of a HIV particle. (B) Representation of the HIV-1 and HIV-2 genomes as dsDNA. CA: capsid. IN: integrase. MA, matrix; NC, nucleocapsid; PR, protease; RT, reverse transcriptase.
The HIV genomic RNA is 9.7 Kb long and encodes for nine genes: three essential/structural genes (gag, pol and env), two regulatory genes (tat and rev) and four accessory genes (vif, vpr, vpu/vpx, and nef). The latter are dispensable for replication in vitro, but important for infectivity and pathogenesis in vivo. In addition to these coding sequences, the HIV genome harbours cis-acting elements buried in heavy secondary structures that facilitate genome incorporation into virions (packaging signal: Psi), enhance viral transcription (transactivation response element: TAR), and mRNA transport (Rev-response element: RRE) (Freed and Martin, 2013) (Fig. 24.2B, the genome is represented as proviral DNA).
Attachment and entry
As for other viruses, the HIV replication cycle starts with attachment, a process by which the viral anti-receptors bind to specific structures on the surface of their target cells. However, unlike most viruses, for attachment to be successful, HIV needs the sequential interaction of its spike with two different membrane proteins: CD4 and a chemokine receptor (either CXCR4 or CCR5, depending on the tropism of the virion) (Fig. 24.3). After binding to the CD4 molecule on the surface of CD4+ T cells, macrophages and dendritic cells, gp120 suffers a conformational change that allows the exposure of a peptide region that will recognize the chemokine co-receptor. This second interaction causes additional conformational changes in the spike, particularly in gp41, that ultimately enable the fusion between the plasma membrane and the viral envelope (Bushman et al, 2012; Wilen et al, 2012; Knipe and Howley, 2013). At this point, the contents of the virion are delivered to the cytoplasm. As soon as the capsid enters the cell, uncoating (fragmentation of the capsid) starts (Fig. 24.3). The uncoating process is extremely controlled, since a rapid uncoating will impair the subsequent steps in the HIV replication cycle. This is mainly due to the fact that uncoating is coupled with reverse transcription, the process by which the HIV RT converts the genomic RNA into a molecule of double-stranded DNA (dsDNA also referred to as proviral DNA, cDNA or provirus). Although RT is found catalytically active inside the capsid, reverse transcription can only proceed in the presence of nucleotides, which become available the moment that the capsid undergoes partial uncoating in the cytoplasm. Similar to other viruses that encode their own polymerases, RT lacks proofreading activity, a fact that makes HIV a highly mutagenic virus (1–3 mutations every 100,000 nucleotides) (Freed and Martin, 2013). Although this high mutational rate has detrimental consequences for the progeny, it is also a source for genetic diversity, and the main reason why HIV can escape the immune surveillance and develop resistance to antiretroviral drugs (Arts and Hazuda, 2012; Freed and Martin, 2013).
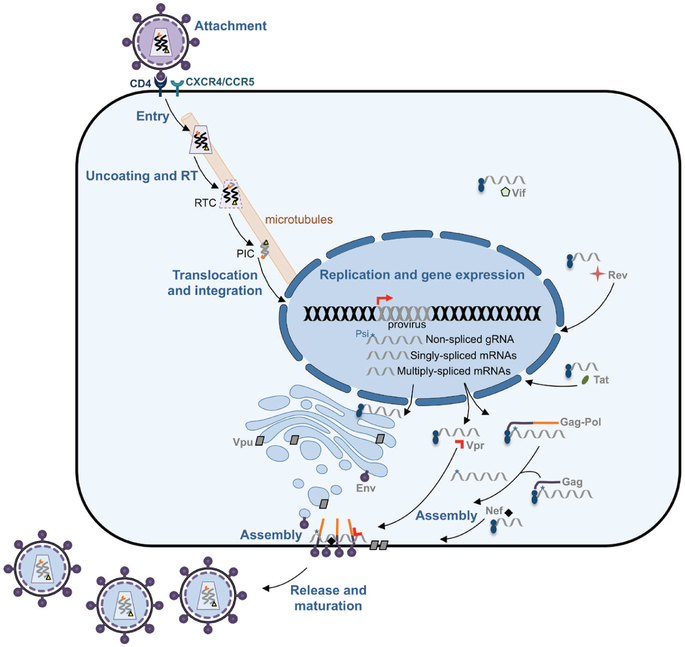
Figure 24.3.
The HIV replication cycle. The diagram illustrates how HIV infects and produces progeny in CD4+ cells. The different steps in the life cycle of HIV are indicated in bold blue and the virus proteins are labelled in grey. PIC, pre-integration complex; RT, reverse transcription; RTC, reverse transcribing complex.
Almost simultaneously with reverse transcription, the reverse-transcribing complex (RTC, which is formed by the remaining CA molecules, portions of MA, RT, the viral genome, Vpr/Vpx and IN) is transported on microtubules to the nucleus. By the time the complex reaches the nuclear pores, reverse transcription is finished and thus, the complex is referred to as the pre-integration complex (PIC) (Bushman et al., 2012; Hu and Hughes, 2012; Freed and Martin, 2013) (Fig. 24.3). The reverse transcription product, dsDNA, is larger than the original genomic RNA and is flanked by two long-terminal repeats (LTRs) that serve as promoter (the 5′LTR) and termination signal of transcription (the 3′LTR) (Fig. 24.2B). The PIC is then translocated to the nucleus, where the dsDNA is inserted into the human genome by the HIV IN, creating a provirus (Freed and Martin, 2013; Knipe and Howley, 2013) (Fig. 24.3).
Replication
For any given virus, the replication step comprises gene expression and genome replication, and HIV achieves both simply by transcribing the proviral DNA. As soon as the provirus is integrated, the upstream LTR is readily recognized by the transcriptional machinery of the cell, allowing the synthesis of genome-length transcripts. Therefore, the transcription of the provirus allows the production of new HIV genome copies that will be transported to the cytoplasm, ready for assembly. However, some of these transcripts are spliced, giving multiple splicing variants that facilitate the translation of all the HIV proteins. These proteins are synthesized in free ribosomes in the cytoplasm or in ribosomes associated with the endoplasmic reticulum (ER) (Bushman et al., 2012; Karn and Stoltzfus, 2012; Freed and Martin, 2013) (Fig. 24.3).
From HIV multiply-spliced transcripts, the regulatory proteins Tat and Rev as well as the accessory protein Nef are produced. Tat and Rev migrate to the nucleus to enhance HIV RNA synthesis through the hyper-phosphorylation of the RNA pol II (Tat), and to allow the transport of non-spliced and singly-spliced HIV transcripts to the cytoplasm (Rev) (for details read (Karn and Stoltzfus, 2012). Nef localizes in the plasma membrane where it specifically promotes immune evasion by down-regulating MHC-I molecules (Le Gall et al., 1998; Roeth et al., 2004), CD28 complexes (Swigut et al., 2001) and by overcoming the restriction factors SERINC3/5, which significantly impair the infectivity of the nascent virions (Rosa et al., 2015; Usami et al., 2015). In addition, Nef down-regulates the CD4 receptor to prevent reentry of newly formed virions and superinfection (Benson et al., 1993; Chaudhuri et al., 2007; Foster and Garcia, 2008).
The singly-spliced transcripts encoding vpu and env are translated in the ER (Fig. 24.3). The synthesis of either of these proteins is the result of ribosomal frameshifting and the use of an alternative start codon that allows the production of Env instead of Vpu (Karn and Stoltzfus, 2012). Vpu is associated with membranes of the ER, the Golgi apparatus and the plasma membrane, particularly at sites of virion assembly – although it is not incorporated into viral particles. Vpu antagonizes the antiviral effects of BST2, a membrane protein that traps nascent HIV virions on the cell surface (Neil et al., 2008; Van Damme et al., 2008). Additionally, Vpu aids in the down-regulation of CD4 (Schubert et al., 1996). Env is processed in the ER and Golgi into gp120 and gp41, which localize as spikes at sites for virion assembly in the plasma membrane. The other HIV singly-spliced transcripts are translated in the cytoplasm, producing the rest of the accessory proteins: Vpr, which causes cell cycle arrest and aids in the transport of the PIC to the nucleus in the next infection (Jowett et al., 1995; Malim and Emerman, 2008), Vpx (only in HIV-2) also contributes to the nuclear transportation of the PIC and overcomes the SAMHD1 restriction factor – which depletes the pool of cytosolic nucleotides, impacting in turn reverse transcription (Goldstone et al., 2011; Laguette et al., 2011), and Vif, which antagonizes the APOBEC3 family of restriction factors by promoting their ubiquitination and degradation (Harris et al., 2002; Marin et al., 2003; Sheehy et al., 2003; Bishop et al., 2004; Mehle et al., 2004b). APOBEC3 are a group of cytidine deaminases that can become incorporated into virions during the assembly process and promote C-to-U mutations during reverse transcription in the next round of infection, causing viral hypermutation (Harris et al., 2002). Hence, the HIV accessory proteins play a crucial role in overcoming host restriction factors and have enabled HIV to adapt for replication in humans. This is explained in more detail later in this review.
From non-spliced transcripts, the HIV structural proteins Gag (precursor of MA, CA, NC and p6) and Pol (precursor of the HIV enzymes RT, IN and PR) are produced. Of note, Pol is synthesized as a fusion protein with Gag (Gag-Pol), as a result of ribosomal frameshifting (Karn and Stoltzfus, 2012). Both, Gag and Gag-Pol localize in the plasma membrane in regions where the HIV spikes insert. Importantly, the NC region within Gag has high affinity for the genome-length transcripts, which correspond to the new genome copies of HIV, ensuring their encapsidation into the viral progeny (Sundquist and Krausslich, 2012) (Fig. 24.3).
Assembly and release
The process of assembling new virions is driven by Gag and Gag-Pol polyproteins. After their synthesis, Gag and Gag-Pol become myristoylated, a modification that renders their N-terminus highly hydrophobic and targets them to the inner leaflet of the plasma membrane. In their ‘trip’ to the cell surface, the NC portion within Gag and Gag-Pol recognizes the packaging signal (Psi) only present in genome-length non-spliced HIV RNAs, which allows the incorporation of only the genomic RNA (and not the smaller HIV transcripts) into the progeny (Fig. 24.3). At the plasma membrane, the MA portion within Gag and Gag-Pol interacts with gp41, which ensures that the spikes are successfully packaged into virions. As mentioned earlier, Nef is also associated to the plasma membrane at Gag-enriched locations, ensuring its incorporation into viral particles. Besides Nef, Vpr and Vpx are also recruited to these sites by the p6 region within Gag (Fig. 24.3). An essential feature for assembly to be fruitful involves the radial arrangement of the Gag and Gag-Pol polyproteins, exposing the late domains in p6 and thereby facilitating the recruitment of the cellular ESCRT (endosomal sorting complex required for transport) machinery to drive virion budding. As soon as the new particles exit the cell, the PR within Pol becomes catalytically active and proceeds to the cleavage of Gag and Gag-Pol to create the mature structures of the HIV virion: MA, CA, NC, RT, IN and PR (Sundquist and Krausslich, 2012; Freed and Martin, 2013). This maturation process driven by PR is essential for the infectivity of these new HIV particles.
Current treatments and challenges
After its discovery 36 years ago, the extensive studies on HIV have allowed us to gain insights into the biology of this virus: from its complete replication cycle to the pathophysiology associated with this disease. These advances in our fundamental understanding of this retrovirus have opened avenues for therapeutic intervention and vaccine development. For instance, the identification of crucial steps in the HIV’s life cycle has significantly contributed to the rational design of drugs to specifically block them. Currently, there are more than 20 FDA-approved compounds for the treatment of the HIV infection (Arts and Hazuda, 2012). These drugs are categorized in five groups according to their targets of action:
Entry inhibitors. These molecules target either the viral spike to prevent the fusion mediated by gp41, or the CCR5 chemokine receptor to thwart the association of the viral spike with this secondary receptor, which, as we mentioned above, is a crucial step to allow fusion between the viral envelope and the plasma membrane (Arts and Hazuda, 2012). In the case of CXCR4-tropic viruses, inhibition of this co-receptor also represents a promising tactic to prevent virus entry. However, unlike CCR5, CXCR4 is distributed in other tissues such as epithelium, nervous system, and the vascular system. Therefore, while preventing the entry of CXCR4-tropic HIV, blocking this chemokine receptor may have additional undesired effects on individuals (Choi et al., 2014).
Nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs). These nucleoside/nucleotide analogues can be mistakenly incorporated by the HIV RT during reverse transcription into the proviral DNA. However, owing to the absence of a 3-OH sugar, these analogues are unable to form 3′5′ phosphodiester bonds with the incoming nucleotides, leading to the premature termination of reverse transcription (Arts and Hazuda, 2012).
Non-nucleoside reverse transcriptase inhibitors (NNRTIs) on the other hand, attack a pocket close to the catalytic site of the HIV RT, causing conformational changes that reduce its polymerase activity. Although very effective, resistant HIV variants arise rapidly in response to these compounds. Therefore, NNRTIs are commonly administered in conjunction with other antiretrovirals (Arts and Hazuda, 2012).
Integrase inhibitors. IN is the most recent HIV enzyme that has been characterized and thus targeted with drugs. These drugs prevent the integration of the proviral dsDNA into the cellular genome, so HIV transcription, genome replication and virus protein production cannot proceed. In particular, these compounds target the strand transfer reaction during ligation (Arts and Hazuda, 2012).
Protease inhibitors. These molecules specifically impede the proper maturation of newly produced virions by targeting the catalytic site of HIV PR (Arts and Hazuda, 2012). Protease is in charge of processing Gag and Gag-Pol complexes into the final mature proteins, a process that is vital for the infectivity of HIV.
As noted above, the rational design of HIV ART drugs has revolutionized the treatment of this infection. Although ART is very effective, HIV variants resistant to these drugs emerge over time, since ART represents a selective pressure. These variant forms are the result of the lack of proofreading activity by the viral RT, generating mutants that, under normal conditions, would be less fit than the wild type virus, but in the presence of compounds that target these critical steps, can replicate well enough to produce offspring (Coffin, 1995). To minimize the upsurge of resistant viruses, HIV+ individuals take a combination of drugs that target different events in the HIV life cycle. This is commonly referred to as combination antiretroviral therapy (cART) or highly active antiretroviral therapy (HAART). The combination of these drugs significantly diminishes the occurrence of resistant forms of HIV and, if patients adhere to their treatment regimens, maintains viral loads to undetectable levels (Fig. 24.1, black curve). However, cART withdrawal can result in viral rebound. As explained earlier, this is due to the existence of latent HIV reservoirs that have the potential to reactivate and support a productive infection. Therefore, the advances in HIV treatment have successfully prevented patients from progressing to AIDS but cannot fully eliminate the virus from their system (Castro-Gonzalez et al, 2018). Further research is urgently needed to design strategies to specifically neutralize the latent reservoirs, so the eradication of HIV can become a reality. At the end of this article, we discuss approaches to manipulate the cellular ubiquitin and SUMO systems with the ultimate goal of reinforcing current cART as well as to effectively address the challenges posed by the latent reservoirs.
Importance of post-translational modifications for a productive HIV infection
As intracellular parasites, viruses usurp the cellular machinery to ensure their propagation. This includes favouring the expression of their viral genes, appropriation of cellular molecules, and manipulation of signalling pathways and the post-translational machinery. HIV is well known for using post-translational modifications (PTMs) to generate infectious particles. These include:
Glycosylation and processing of the HIV spikes. The spikes (gp120/gp41) need to be post-translationally modified in order to mature, and this maturation is a pre-requisite for infectivity. The precursor form of the spikes, Env (or gp160), is co-translationally glycosylated in the ER, with the main component of its glycan shield being N-linked glycosylation (Reitter et al., 1998; Wei et al., 2003). Env oligomerizes in the ER as a trimeric pre-spike that will be subsequently processed by cellular furin or furin-like proteases in Golgi to generate the mature gp120 and gp41 subunits. Gp120 and gp41 stay non-covalently associated, forming mature spikes that are transported to the plasma membrane following the secretory pathway, where they remain as integral membrane proteins, ready for their incorporation into new virions (Freed and Martin, 2013) (Fig. 24.3).
Myristoylation of Gag, Gag-Pol and Nef. Gag and Gag-Pol play key roles in the assembly of new viral particles. Almost immediately after their synthesis, they are targeted to the plasma membrane, particularly to cholesterol-enriched locations where virion assembly takes place. The selective transport of Gag and Gag-Pol to these sites is due the presence of a multipartite membrane-binding signal within the MA region in Gag and Gag-Pol polyproteins, which becomes accessible after they undergo myristoylation in their Gly2. This PTM is mediated by cellular N-myristyl transferases. Mutation of Gly2 or the inability to mediate this PTM renders HIV incapable of producing progeny, highlighting the significance of Gag and Gag-Pol myristoylation for virus infectivity (Freed and Martin, 2013). In addition to Gag and Gag-Pol, Nef is also susceptible to myristoylation in its Gly2 by cellular N-myristyl transferases, which, together with the high degree of hydrophobic residues in its N-terminal domain, makes Nef highly lipophilic and is immediately targeted to the inner leaflet of the plasma membrane. Once there, Nef remains associated with the CA region within Gag to ensure virion incorporation. Several studies have demonstrated that the mutation of Gly2 in Nef abolishes all of its functions, including the down-regulation of MHC-I, CD4 and CD28 molecules; cell activation; and infectivity enhancement through the antagonism of the restriction factors SERINC3/5 (Freed and Martin, 2013), evidencing the importance of this PTM for Nef’s functionality.
Phosphorylation of Nef. After myristoylation, Nef proteins become phosphorylated predominantly at Ser and, to a lesser extent, Thr residues by the cellular protein kinase C (PKC) (Peter, 1998). Although the relevance of this PTM in the vast roles of Nef is poorly understood, at least one study has shown that mutation of these phospho-sites abrogates Nef’s ability to enhance HIV replication (Li et al., 2005), suggesting that this PTM is necessary for HIV infectivity.
Relevance of ubiquitination and SUMOylation for a successful infection. Part I: HIV proteins
Ubiquitin is a small protein present in most cell types of all eukaryotic organisms (Goldstein et al., 1975). This molecule is attached to other proteins through a process called ubiquitination (or ubiquitination), which consists of a three-step enzymatic reaction performed by three distinct enzymes: an E1-activating enzyme, an E2-conjugating enzyme and an E3-ligating enzyme (for details on this refer to https://doi.org/10.21775/9781912530120). The attachment of ubiquitin to proteins serves as a regulatory signal that cells interpret differently depending on the type of linkage of the ubiquitin chains. The most common types of ubiquitination are mono-ubiquitination, in which only one ubiquitin molecule is ligated to one target protein residue, or poly-ubiquitination, in which several ubiquitin proteins are added to an amino acid of the protein substrate. Poly-ubiquitination takes place by connecting new ubiquitin proteins to either a Lys (K) or Met (M) residue of the previous ubiquitin molecule, creating a chain. To indicate the moiety of this reaction, the linking of new ubiquitins is designated by the amino acid and position where they are incorporated. The most common types of poly-ubiquitination reactions are K48, K29 (which normally tag proteins for proteasomal degradation), K63, K11, K6 and M1 which, together with mono-ubiquitination, are involved in trafficking, lysosomal degradation, activation/inactivation of enzymatic activities, translation, and DNA repair (Miranda and Sorkin, 2007).
SUMO (Small Ubiquitin-like Modifier) are a small family of proteins involved in SUMOylation, a PTM that closely resembles ubiquitination, since it also requires a concerted three-step enzymatic cascade. Humans encode for four different SUMO isoforms (SUMO-1 to SUMO-4), all with similar functional properties. Unlike ubiquitination, the addition of SUMO to proteins does not tag them for degradation. Instead, SUMOylation alters the pattern of protein–protein associations, protein–DNA interactions, the subcellular localization of the SUMOylated targets as well as the activation status of their enzymatic activities. For instance, SUMOylation is a common mechanism for the inactivation of transcription factors (Hay, 2005).
HIV takes advantage of ubiquitination and SUMOylation to modify its viral proteins and achieve a productive infection through different mechanisms. The best-characterized HIV proteins that require these PTMs for their functionality are:
p6. p6 is synthesized as part of the Gag polyprotein and comprises a 52 amino acid region at the C-terminus of Gag. This domain plays a critical role in the incorporation of Vpr and Vpx into particles, and in the budding and release events of progeny HIV virions (Freed and Martin, 2013). This is achieved through extensive PTMs in p6, including phosphorylation at Ser40 (important for its association with phospholipids at the plasma membrane) (Solbak et al., 2013), SUMOylation at Lys27 and mono-ubiquitination at Lys27 and Lys33 (Ott et al., 1998; Gottwein and Krausslich, 2005; Gurer et al., 2005; Friedrich et al., 2016). Although p6 is highly polymorphic within HIV-1, it contains several conserved regions, the so-called late domains, which allow p6 interaction with cellular molecules to facilitate virion release. The first p6 late domain, the PTAP tetrapeptide motif (Pro-Pro-Pro-Tyr), interacts with Tsg101 (tumour susceptibility gene 101) while the second late domain, the YPX3L motif, associates with ALIX (ALG-2 interacting protein X). Both Tsg101 and ALIX belong to the ESCRT machinery, whose primary role is to aid in the budding and scission of multiple vesicular bodies (MVBs) and facilitate membrane fission during cytokinesis (Carlton and Martin-Serrano, 2007). Hence, HIV has evolved to hijack the cellular ESCRT machinery to promote the release of its progeny from the plasma membrane and acquire its envelope. The association of p6 with Tsg101, ALIX and other ESCRT components is greatly enhanced if p6 is mono-ubiquitinated (Patnaik et al., 2000). However, SUMOylated p6 is significantly impaired in its budding activity (Gurer et al., 2005). As noted above, SUMOylation and mono-ubiquitination of p6 take place at the same residue (Lys27), rendering these two PTMs mutually exclusive. Hence, the SUMOylation of p6 prevents its ubiquitination, which is critical for the recruitment of Tsg101, causing severe defects in virion release (Gurer et al., 2005).
IN. The HIV IN inserts the proviral dsDNA into the host genome and also aids in the nuclear transport of the PIC, since it harbours nuclear localization signals. Besides IN, additional cellular molecules are recruited to assist in the integration process of the provirus, and their successful incorporation into this multimeric molecular complex largely depends on the presence of PTMs within IN. For instance, IN is acetylated by p300 and GCN5, which increases IN binding affinity for the proviral DNA, enhances strand transfer activity and likely regulates IN’s association with cellular dependency factors (Cereseto et al., 2005; Topper et al., 2007; Terreni et al., 2010). Additionally, IN is susceptible to become SUMOylated at three different Lys residues (Lys46, Lys136, Lys244) that are present in functionally conserved motifs within the protein. The SUMOylated status of IN does not seem to affect HIV’s ability to infect or perform reverse transcription, but SUMO-deficient IN viruses show a significant decrease in integration events compared to wild type HIV (Zamborlini et al., 2011). Remarkably, IN suffers this PTM in a step between reverse transcription and the translocation of the PIC to the nucleus, so, unlike most proteins that suffer PTMs, the SUMOylation of HIV IN does not take place on synthesis, but in the next infection (Zheng and Yao, 2013). Besides becoming SUMOylated, IN bears SUMO-interacting motifs (SIMs), which facilitate the recruitment of its SUMOylated cellular co-factors, such as lens epithelium-derived growth factor (LEDGF/p75) and p300. Similar to p6, IN is also susceptible to ubiquitination, and this PTM targets IN for proteasomal degradation. IN likely becomes poly-ubiquitinated at Lys211, Lys215, Lys219, and Lys273, although the nature of the ubiquitin linkage is currently unknown. Several E3 ligases have been proposed to mediate its ubiquitination, like the RING-finger E3 ligases UBR1, UBR2, UBR4, VBP1, VHL, and the HECT-E3 ligase Huwe1 (Zheng and Yao, 2013). Despite its rapid turnover in vitro, in the context of HIV-infected cells IN is quite stable, indicating that IN somehow evades its proteasomal degradation. This is achieved through direct association with its cellular co-factors, which mask IN ubiquitin target residues and/or the E3-binding sites. LEDGF/p75 was the first protein reported to play a protective role in IN’s stability, most likely by hiding the E3-ligase association motif (Llano et al., 2004; Zheng and Yao, 2013). Subsequent studies found that the DNA repair proteins hRad18 and Ku70 also increase IN’s half-life. Although the mechanism by which hRad18 achieves this is still unknown, Ku70 prevents IN from being directed for proteasomal degradation through its de-ubiquitinase activity: by removing ubiquitin molecules attached to IN (Zheng et al., 2011; Zheng and Yao, 2013).
Tat. The HIV transactivation of transcription protein (Tat) improves the efficiency of HIV RNA synthesis by recruiting the P-TEFb cellular factor to the virus promoter to facilitate the hyper-phosphorylation of RNA pol II, which in turn enhances its polymerase activity. The association of this molecular complex with Tat is achieved through multiple PTMs in Tat, among them K63 poly-ubiquitination at Lys71, which is mediated by the E3 ligase Mdm2. In fact, Mdm2 and Tat create a positive feedback loop in which Tat increases the stability of Mdm2 by favouring its phosphorylation, and in turn Mdm2 promotes Tat poly-ubiquitination (Raja et al., 2017). By contrast, the cellular protein ABIN1 inhibits Tat’s functionality by (1) interacting with poly-ubiquitinated Tat, preventing its association with P-TEFb, and (2) changing the subcellular distribution of Mdm2, so Tat ubiquitination is impaired (Chen et al., 2017).
Nef. Nef suffers numerous PTMs such as myristoylation and phosphorylation, essential for its functionality. In addition, Nef is susceptible to become di-ubiquitinated at Lys144, which is critical for its CD4 down-regulation activity, since Nef mutants missing this ubiquitin target site are unable to reduce the surface levels of CD4, and thus, cannot prevent virus superinfection (Jin et al., 2008).
Besides taking advantage of the cellular ubiquitination and SUMOylation machinery to modify its proteins, HIV redirects the host ubiquitin pathway to accomplish HIV-specific outcomes. Specifically, the HIV accessory protein Vpr seems to decrease the overall cellular ubiquitination to favour HIV-mediated ubiquitination of host anti-HIV factors, and target them for degradation (Arora et al., 2014). Although the underlying mechanism by which Vpr achieves this is largely unknown, these observations further highlight how viruses, and in particular HIV, appropriate the cellular machinery to secure a productive infection.
Relevance of ubiquitination and SUMOylation for a successful infection. Part II: HIV dependency factors
In addition to the PTMs of the viral proteins presented above, the replication and infectivity of HIV also relies on the ubiquitinated and SUMOylated state of several host dependency factors that are necessary for the completion of the different steps of the virus life cycle. This includes modifications to the cellular receptor, co-receptor, and the cellular transcriptional machinery.
CD4 and CXCR4. The surface levels of the CD4 receptor as well as the CXCR4 co-receptor, both required for virion attachment, can be regulated by different ubiquitin-conjugating systems. The ubiquitination of the cytoplasmic domain of the CD4 glycoprotein leads to its proteasomal degradation in a beta transducing-repeat containing protein (β-TrCP) dependent manner (Fujita et al., 1997; Margottin et al., 1998). Interestingly, this ubiquitin-mediated down-regulation of CD4 is promoted by the viral protein Vpu, which in turn increases HIV replication efficiency by reducing the re-entry of newly released virions from infected cells, and superinfection (Fujita et al., 1997; Margottin et al., 1998; Schubert et al., 1998). Specifically, Vpu and CD4 interact in the ER through their cytoplasmic domains. This association causes a conformational change in Vpu that facilitates its phosphorylation in a conserved di-serine motif (Ser52-Ser56), allowing in turn the recruitment of β-TrCP. Next, the SCFβTrCP E3 ligase complex initiates the K48-linked poly-ubiquitination at Lys and Ser/Thr residues in the cytoplasmic domain of CD4, tagging it for proteasomal degradation (Binette et al., 2007; Fujita et al., 1997; Magadán et al., 2010; Schubert et al., 1998) (Fig. 24.4). Ubiquitination is also directly related to the down-regulation of the CXCR4 HIV co-receptor (Busillo et al., 2010; Lear et al., 2017). After the interaction with its ligand, (the stromal-derived-factor-1, SDF-1, also known as CXCL12), CXCR4 undergoes phosphorylation and, to a lesser extent, ubiquitination. CXCL12 is a cytokine involved in chemotactic processes during inflammation. The CXCL12-induced phosphorylation of CXCR4 triggers responses to support migration, proliferation and homing of other immune cells (Busillo and Benovic, 2007; Döring et al., 2014). However, ligand binding-induced ubiquitination leads to the internalization and lysosomal degradation of CXCR4 through the activity of different ubiquitin-conjugating systems (Marchese and Benovic, 2001; Busillo et al., 2010) (Fig. 24.4). Among the different enzymatic complexes that play a direct role in the ubiquitination of this co-receptor, AIP4 and RNF113A are the E3 ligases predominantly used (Marchese et al., 2003) and all of them attach ubiquitin at Lys331 in CXCR4, although the moiety of the ubiquitin chains remains to be elucidated (Lear et al., 2017). Remarkably, the gp120–CXCR4 interaction, as well as CXCR4 association with agonists, also leads to the degradation of this HIV co-receptor (Marchese and Benovic, 2001; Fernandis et al., 2002). Therefore, the ubiquitin-mediated removal of CXCR4 not only precludes superinfection by other HIV virions, but also provides a mechanism of immune evasion when associated to gp120: by impeding CXCL12 from binding and signalling through CXCR4 (Tran et al., 2005).
LEDGF/p75. At a different level of the replication cycle, the SUMOylation state of the cell protein LEDGF/p75 plays a crucial role in the integration process of the proviral DNA into the host genome (Shun et al., 2007; Engelman and Cherepanov, 2008). LEDGF/p75 is a transcription co-activator that regulates gene expression in response to stress (Shinohara et al., 2002). In the context of HIV infection, LEDGF/p75 is also able to directly interact with the viral protein IN through an IN-binding domain (IBD) located in its C-terminus. This interaction allows the chromosomal association of the PIC, which ultimately promotes the integration of the HIV provirus into the host genome (Llano et al., 2006; Shun et al., 2007; Engelman and Cherepanov, 2008). Besides this primary role, the association of LEDGF/p75 with IN specifically targets the PIC to areas of the cellular genome where genes are being actively transcribed, a fact that facilitates the expression of the HIV provirus (Shun et al., 2007; Singh et al., 2015). Although the SUMOylation of LEDGF/p75 decreases its stability, it significantly increases the integration efficiency of the HIV proviral DNA (Bueno et al., 2010). This PTM is mediated by UBC9, the only known E2-conjugating SUMO enzyme – which can also function as a ligase – and occurs at Lys364 in the C-terminus of LEDGF/p75. Whereas the SUMOylation status of LEDGF/p75 has no effect on its capacity to interact with IN, the deSUMOylation of this residue reduces LEDGF’s ability to act as a co-factor in the proviral integration process, impairing the replication efficiency of HIV (Bueno et al., 2010) (Fig. 24.4).
NF-κB, NFAT and STAT5. The levels of HIV RNA synthesis, and thus, virion production, are strongly regulated by the nuclear availability of different host transcription factors. These transcription factors can bind to their responsive regulatory elements found at the 5′ LTR of the integrated provirus and drive the expression of the viral DNA (Nabel and Baltimore, 1987; DeLuca et al., 1999; Romanchikova et al., 2003; Selliah et al., 2006). In this regard, the transcription factor NF-κB plays a major role in the regulation of HIV expression (Nabel and Baltimore, 1987; DeLuca et al., 1999). Under normal conditions, NF-κB is found sequestered in the cytoplasm by the inhibitory protein IκBα. In response to the action of different stimuli, IκBα becomes phosphorylated by IKK at Ser32 and Ser36, and these PTMs make IκBα susceptible for K48-linked poly-ubiquitination (Desterro et al., 1998; Hay et al., 1999; Ferreiro and Komives, 2010). Similar to the process that causes the degradation of the CD4 receptor, the ubiquitination of IκBα is mediated by the β-TrCP E3 ligase complex, which directs this protein for proteasomal degradation, allowing in turn the nuclear translocation of NF-κB (Rothwarf and Karin, 1999; Winston et al., 1999; Hayden and Ghosh, 2012) (Fig. 24.4). However, IκBα can also be modified by SUMOylation (Desterro et al., 1998). Remarkably, both the poly-ubiquitination and SUMOylation of IκBα take place at the same site (Lys21), making these two PTMs mutually incompatible. In fact, IκBα SUMOylation renders this protein resistant to degradation and a stronger inhibitor of NF-κB (Colomer-Lluch and Serra-Moreno, 2017). Remarkably, HIV tightly controls the activation of NF-κB to ensure its propagation (Sauter et al., 2015). The HIV proteins Nef and gp41 trigger this cascade to facilitate IκBα ubiquitination and the nuclear release of NF-κB to favour HIV RNA synthesis (Postler and Desrosiers, 2012; Sauter et al., 2015) (Fig. 24.4). In addition to NF-κB, the nuclear factor of activated T cells (NFAT) is another transcription factor relevant for HIV transcription, which is also modulated by SUMOylation (Romanchikova et al., 2003). In this case, the SUMOylation of different residues within NFAT can substantially change the subcellular distribution as well as the transactivation activity of this protein. For instance, SUMOylation of Lys684 and Lys897 in NFAT causes its nuclear localization (Terui et al., 2004), while SUMOylation of Lys702 and Lys914 favours an interaction between NFAT and histone deacetylase enzymes (HDACs), which in turn leads to gene silencing through the formation of heterochromatin in NFAT-targeted areas (Nayak et al., 2009) (Fig. 24.4). Finally, STAT5, another host transcription factor that modulates the expression of HIV, is tightly regulated by phosphorylation, acetylation and SUMOylation (Van Nguyen et al., 2012; Bosque et al., 2017). On activation, STAT5 becomes phosphorylated, which drives its nuclear localization, allowing its association with the HIV 5′ LTR to drive viral transcription. However, the SUMOylation of STAT5 re-directs this protein back to the cytoplasm, where it becomes dephosphorylated and inactivated (Van Nguyen et al., 2012; Bosque et al., 2017). Therefore, STAT5 SUMOylation correlates with reduced HIV transcription and latency establishment (Bosque et al., 2017) (Fig. 24.4).
Figure 24.4.
Ubiquitination and SUMOylation of HIV dependency factors. The figure illustrates the impact of ubiquitination and SUMOylation of cellular proteins on the replication of HIV. Negative effects are indicated in red boxes. Positive effects are shown in green boxes. The characteristics of these PTMs are also illustrated: moiety (if known) is indicated in red, and the amino acid position where these chains are attached in black and italics. Virus proteins are labelled in grey. PIC: pre-integration complex.
From the examples described in these two sections (part I and part II) is evident that HIV appropriates the ubiquitin and SUMO systems to modify its proteins and ensure their functionality. However, as noted here, these PTMs can also impact the success of the replication of HIV when they are attached to its host dependency factors, having both positive as well as detrimental effects for the virus. Therefore, the knowledge gained on the roles of ubiquitination and SUMOylation in the context of HIV infection, could strategically be applied to find new targets for antiretroviral drugs, as we will discuss later.
Ubiquitination and SUMOylation as a mechanism of innate defence against HIV
Restriction factors are host cellular proteins, generally induced by interferon stimulation, that block specific steps of the replication cycle of virus pathogens, acting as potent innate barriers against their infection. In the case of HIV, the following restriction factors have been identified so far: TRIM5α (Stremlau et al., 2004; Pertel et al., 2011), APOBEC3G (Sheehy et al., 2002; Mangeat et al., 2003; Zhang et al., 2003), SAMHD1 (Laguette et al., 2011; Lahouassa et al., 2012), Mx2/MxB (Goujon et al., 2013; Kane et al., 2013; Liu et al., 2013), Tetherin/BST2 (Neil et al., 2008; Van Damme et al., 2008), SERINC3/5, IFITMs, and MARCH2/8 (Rosa et al., 2015; Tada et al., 2015; Usami et al., 2015), although new proteins may be added to this list, since this is still an emerging field. Some of these molecules attack HIV by promoting the ubiquitination or SUMOylation of viral proteins and/or HIV dependency factors, changing their functionality, and precluding HIV from completing its replication cycle. However, in other instances, the ubiquitination or SUMOylation of host restriction factors renders HIV highly vulnerable to these innate effectors.
TRIM5α and the role of ubiquitination in the premature uncoating of retroviral capsids
Tripartite motif 5-alpha (TRIM5α) nicely illustrates how the host cell employs the ubiquitin system as an innate mechanism of defence to restrict HIV. TRIM5α recognizes and binds to the incoming retroviral capsid, promoting its premature degradation by recruiting the proteasomal machinery (Anderson et al., 2006; Wu et al., 2006; Campbell et al., 2008; Rold and Aiken., 2008), a fact that will negatively impact reverse transcription, and in consequence, the subsequent steps in the HIV life cycle (Stremlau et al., 2004, 2006; Javanbakht et al., 2005; Diaz-Griffero et al., 2007; Perron et al., 2007; Li and Sodroski, 2008; Black and Aiken, 2010; Kutluay et al., 2013). Despite the potent antiretroviral function of TRIM5α, its antiviral activity is only effective in a species-specific manner. For instance, rhesus macaque TRIM5α (rhTRIM5α) potently restricts HIV-1, HIV-2, murine leukaemia virus (MLV), and equine infectious anaemia virus (EIAV), whereas it is poorly active against SIVmac . Similarly, human TRIM5α (huTRIM5α) is unable to efficiently restrict HIV) while it strongly limits the replication of MLV, SIVmac and EIAV (Hatziio-annou et al., 2004; Keckesova et al., 2004; Yap et al., 2004; Stremlau et al., 2004, 2006).
TRIM5α was one of the first HIV restriction factors to be characterized. This protein contains: (a) an N-terminal RING zinc-binding domain with E3 ligase activity, (b) one or two B-Box domains, important for the oligomerization of TRIM5α – a factor that significantly enhances its restrictive activity (Javanbakht et al., 2006; Diaz-Griffero et al., 2009; Ganser-Pornillos et al., 2011), (c) a coiled-coil (RBCC) domain, which also facilitates TRIM5α dimerization (Langelier et al., 2008), and (d) a C-terminal SPRY domain (B30.2 or PRYSPRY domain), responsible for the species-specific interaction with retroviral capsids (Nisole et al., 2005; Sawyer et al., 2005; Sebastian and Luban, 2005; Stremlau et al., 2005, 2006). Importantly, the E3 ubiquitin ligase activity can drive TRIM5α auto-ubiquitination, which increases its antiviral efficiency. Although the underlying mechanism is not fully understood, it seems that TRIM5α promotes the premature uncoating of the retroviral cores in a proteasome-dependent manner without facilitating their ubiquitination. The most accepted model is that ubiquitinated TRIM5α bound to retroviral capsids makes this molecular complex highly susceptible to the proteasomal machinery (Diaz-Griffero et al., 2006; Towers, 2007; Lienlaf et al., 2011). Therefore, the auto-ubiquitination of TRIM5α represents an indirect mechanism of HIV restriction (Fig. 24.5).
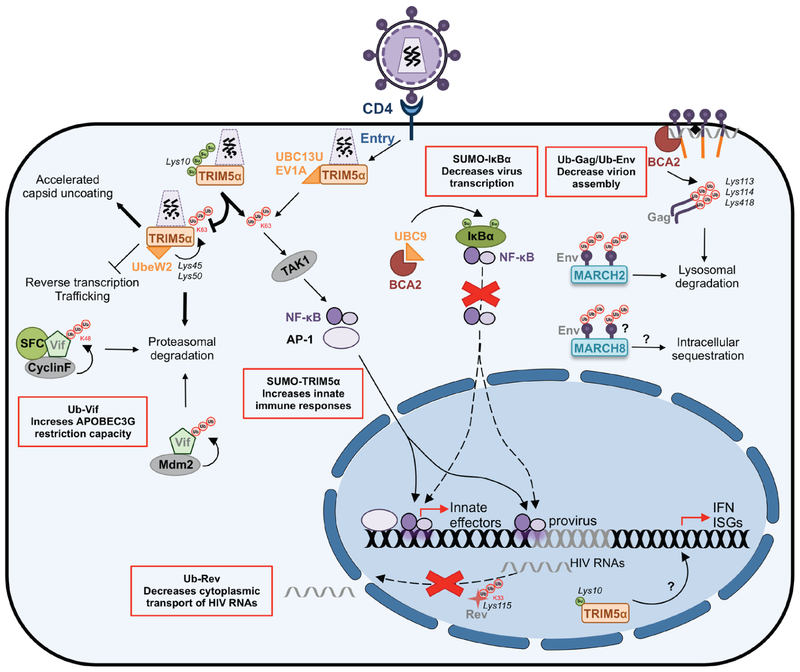
Figure 24.5.
Ubiquitination and SUMOylation as a mechanism of innate defence against HIV. The illustration shows how SUMOylation and ubiquitination are commonly used to restrict HIV replication. The red boxes indicate ubiquitination and SUMOylation events that decrease virus replication. Residues where SUMO and ubiquitin chains are attached are indicated in black and italics. The moiety of these PTMs (if known) is indicated in red. Virus proteins are labelled in grey. Question marks represent events that remain to be elucidated.
In addition to its role in accelerating capsid fragmentation, TRIM5α functions as a retroviral sensor by activating innate immune signalling pathways (Pertel et al., 2011; Tareen and Emerman, 2011; Uchil et al., 2013). Particularly, after its association with susceptible retroviral capsids, the TRIM5α RING domain recruits the UBC13-UEV1A E2-conjugating enzyme to catalyse the formation of unanchored K63-linked poly-ubiquitin chains. These free poly-ubiquitin chains activate the cellular TAK1 kinase for the downstream activation of the transcription factors AP-1 and NF-κB (Pertel et al., 2011; Tareen and Emerman, 2011; Uchil et al., 2013), which are intimately connected to the innate sensing system. The activation of AP-1 and NF-κB induces the expression of additional restriction factors and the secretion of interferon and other pro-inflammatory cytokines, which help shape the immune response (Fig. 24.5). In an analogous manner, TRIM5α uses the E2-conjugating enzyme UbeW2 for its K63-linked auto-ubiquitination at Lys45 and Lys50 (Fletcher et al., 2015), which, as mentioned above, enhances TRIM5α restrictive phenotype. In fact, the presence of Ube2W as well as TRIM5α ubiquitination are essential to block reverse transcription in a proteasome-dependent manner, although as stated above, the exact underlying mechanism remains to be confirmed (Fletcher et al, 2015; Campbell et al, 2016).
Regulation of TRIM5α activity by SUMOylation and SIMs
Accumulating evidence suggest that TRIM5α can be regulated by the SUMO machinery (Arriagada et al., 2011; Lukic et al., 2013; Nepveu-Traversy and Berthoux, 2014; Dutrieux et al., 2015). TRIM5α is susceptible to canonical mono- and poly-SUMOylation at Lys10, although its SUMOylation status does not affect its antiviral activity (Dutrieux et al., 2015). However, the fact that this SUMOylation motif (Ϋ-K-X-D/E) is highly conserved in all TRIM5α orthologues, and that it is in close proximity to the RING domain involved in the control of ubiquitination for the regulation of innate immune responses, suggests that it may play a role in TRIM5α-mediated activation of innate signalling pathways (Pertel et al., 2011; Tareen and Emerman, 2011; Uchil et al., 2013). Indeed, the SUMOylation of TRIM5α regulates its E3 ligase activity by enhancing the generation of free K63-linked poly-ubiquitin chains while decreasing its auto-ubiquitination activity, which in turn facilitates the TRIM5α-induced activation of NF-κB and AP-1, consequently triggering innate immune responses (Nepveu-Traversy and Berthoux, 2014; Dutrieux et al., 2015) (Fig. 24.5). In addition to this PTM, TRIM5α also harbours two to four SUMO-interacting motifs (SIMs) in its PRYSPRY domain, which, as mentioned above, is important for the species-specific recognition of the retroviral capsids. SIMs play an important role in the non-covalent interaction of proteins with free SUMO or with SUMO-conjugated proteins. Despite the fact that one report found that the presence of these SIMs has no effect on capsid recognition –although they increase the stability, and in consequence, the restrictive capacity of TRIM5α (Lukic et al., 2013), a more recent study found that they are essential for capsid binding, viral restriction, and innate signalling (Nepveu-Traversy et al., 2016). In particular, SIM4 is highly exposed at the surface of the PRYSPRY domain, suggesting that it is more accessible for interaction with SUMO or SUMOylated proteins for the activation of NF-κB and AP-1 cascades (Nepveu-Traversy and Berthoux, 2014; Nepveu-Traversy et al., 2016). Nevertheless, the role of SUMO and SIMs in TRIM5α restriction is still debatable, since at least one publication reported no correlation between TRIM5α SUMOylation or the presence of SIMs in the PRYSPRY domain with viral restriction (Brandariz-Nunez et al., 2013).
Besides its role in the regulation of its E3 ligase activity, the SUMOylation status of TRIM5α changes its subcellular distribution. For instance, SUMOylated TRIM5α is sequestered in the nucleus in dendritic cells, unable to interact and restrict incoming retroviruses, which may in part explain why huTRIM5α poorly restricts HIV in these cells. However, in the context of HIV infection, the nuclear accumulation of TRIM5α potently induces interferon production. This is due to the detection of reverse transcribed proviral DNA by cGAS, a molecule of the innate sensing system that recognizes cytoplasmic DNA, and signals through nuclear TRIM5α (Portilho et al., 2016) (Fig. 24.5). Therefore, although cytoplasmic TRIM5α can only partially restrict incoming HIV, its nuclear localization might benefit the host, since it aids in eliciting a strong innate response, and may represent an evolutionary adaptation of huTRIM5α to HIV.
MARCH2, MARCH8 and Env ubiquitination
MARCH2 and MARCH8 (Membrane-Associated RING-CH 2 or 8 proteins) were recently identified as anti-HIV factors. MARCH8 is a transmembrane E3-ubiquitin ligase and, similar to TRIM5α, uses the ubiquitin system to exert its restrictive function. MARCH8 interacts with HIV Env and down-modulates its expression from the plasma membrane, most likely through ubiquitination, since deletion of MARCH8 RING domain abrogates its antiviral activity. Therefore, the MARCH8-mediated down-regulation of Env reduces its incorporation into nascent virions, which significantly decreases HIV infectivity (Tada et al., 2015). Mechanistically, MARCH8 promotes the intracellular sequestration of Env rather than its proteasomal degradation (Fig. 24.5). By contrast, MARCH2, another member of the MARCH E3 ligase family, suppresses HIV infectivity by promoting Env ubiquitination and its subsequent lysosomal degradation (Zhang et al., 2018) (Fig. 24.5). Although there are still many unanswered questions on how MARCH2 and MARCH8 achieve such obstructive activities, they together with TRIM5α, perfectly exemplify how the cellular ubiquitin system can be exploited to block viral infection. This includes: (a) directing viral components for proteasomal, lysosomal degradation or intracellular sequestration, (b) enhancing the ubiquitination of cellular factors that specifically recognize viral structures to generate high molecular complexes that will be directed for degradation, and/or (c) generating free ubiquitin chains to potentiate the innate response.
Vif ubiquitination and degradation as a mechanism to enhance APOBEC3 restriction
Vif (virion infectivity factor) is one of the HIV accessory proteins and it is essential for infectivity, since it prevents the incorporation of APOBEC3 cytidine deaminases into virions. APOBEC3s drive hypermutation during reverse transcription, causing loss of the genetic integrity of the virus (Freed and Martin, 2013), thereby the importance of precluding their incorporation into HIV particles. Vif achieves this by promoting APOBEC3 ubiquitination and proteasomal degradation (explained in detail in the next section). Paradoxically, Vif is also a target for mono- and poly-ubiquitination, although the residues where these ubiquitin chains are attached remain to be elucidated. The mono-ubiquitination of Vif is driven by the HECT-E3 ligases Nedd4 and AIP4, and this PTM helps in the recruitment of Gag to sites of virion assembly and release (Dussart et al., 2004). However, the poly-ubiquitination of Vif tags the protein for proteasomal degradation (Mehle et al., 2004b; Izumi et al., 2009; Augustine et al., 2017). In this case, the cellular Cyclin F (FBXO1) protein specifically recognizes Vif’s CY motif in its C-terminus, facilitating the formation of a multimeric complex in which the SCFcyclin F E3 ligase drives Vif K48-linked poly-ubiquitination and its subsequent proteasomal degradation (Augustine et al., 2017) (Fig. 24.5). In addition to SCFcyclin F, the E3 ligase Mdm2 also causes Vif poly-ubiquitination and proteasomal degradation (Izumi et al., 2009) (Fig. 24.5). As expected, the ubiquitin-dependent removal of Vif (either through SCFcyclin F or Mdm2) correlates with more stable levels of APOBEC3s, which in turn help restore their antiviral activity. Therefore, the consequences of this innate response against Vif are two-fold: (1) to prevent the Vif-mediated degradation of APOBEC3s, and (2) to increase antigen presentation by loading Vif peptides derived from its proteasomal degradation onto MHC-I molecules. Since viruses are in a constant arms race with their hosts, it is tempting to speculate that Vif variants with substitutions in the residues susceptible to ubiquitination may arise over time. However, it is very likely that these ubiquitin target sites are also required for Vif mono-ubiquitination, and thus, the occurrence of these Vif mutants may cause infectivity defects. Studies to identify the amino acids susceptible to these PTMs will help us predict how HIV might fight back this innate response.
BCA2-mediated ubiquitination and SUMOylation: a two-hit strategy to block HIV transcription and assembly
BCA2 (Breast Cancer-Associated gene 2, Rabring7, RNF115, or ZNF364) is a RING-finger E3 ubiquitin ligase initially described as a co-factor in the restriction mediated by BST2 on HIV, although this role is independent on BCA2’s enzymatic activity (Miyakawa et al., 2009). As mentioned in the introduction section, BST2 prevents the release of nascent HIV virions by tethering them to the cell surface (Neil et al., 2008; Van Damme et al., 2008). BCA2, through its direct association with the cytoplasmic domain of BST2, targets BST2-trapped virions for lysosomal degradation (Miyakawa et al., 2009). In addition to this role, BCA2 exhibits BST2-independent anti-HIV activities in which its E3 ligase activity is critical to restrict HIV. On one hand, BCA2 promotes the lysosomal degradation of HIV Gag, essential for virion assembly, and on the other hand, BCA2 blocks HIV transcription and genome replication through the inactivation of NF-κB, a vital transcription factor for HIV (Colomer-Lluch and Serra-Moreno, 2017; Nityanandam and Serra-Moreno, 2014).
BCA2-mediated Gag ubiquitination. Studies in our lab demonstrated that the N-terminus of BCA2 interacts with the MA region in HIV Gag. This association facilitates the BCA2-mediated poly-ubiquitination of Gag at Lys113, Lys114, and Lys418, which consequently direct this protein for lysosomal degradation in a Rab7-dependent manner (Nityanandam and Serra-Moreno, 2014). Importantly, the interaction between BCA2 and Gag, and consequently, Gag ubiquitination, can only proceed if Gag is associated with cellular membranes, indicating that BCA2 specifically targets Gag proteins involved in the assembly of HIV virions (Fig. 24.5). In fact, the ubiquitination of Gag mediated by BCA2 significantly reduces the progeny size of HIV (Nityanandam and Serra-Moreno, 2014).
BCA2-dependent SUMOylation of IκBα. In a more recent study, we demonstrated that BCA2 mediates the SUMOylation of IκBα to block NF-κB, an essential cellular transcription factor to support a productive HIV infection (Colomer-Lluch and Serra-Moreno, 2017). As described above, NF-κB is a key immune modulator and enhances the expression of host restriction factors. Under normal conditions, NF-κB is sequestered in the cytoplasm by IκBα. However, in the presence of immune stimuli, IκBα is phosphorylated, which facilitates its ubiquitination and degradation, allowing the nuclear transport of NF-κB (Hay et al., 1999) (Figs. 24.4 and 24.5). Remarkably, HIV takes advantage of this innate immune pathway and uses NF-κB to aid in HIV RNA synthesis. Our studies demonstrated that BCA2 is induced by NF-κB as part of this innate response, and that in turn, BCA2 self-limits this pro-inflammatory cascade through the SUMOylation of IκBα (Fig. 24.4). As stated earlier, IκBα SUMOylation and ubiquitination occur at the same residue (Lys21), making these two PTMs mutually exclusive. Therefore, the SUMOylation of IκBα renders this molecule resistant to degradation, strongly repressing the activation of NF-κB. Remarkably, the BCA2-mediated SUMOylation of IκBα also causes a significant decrease in its phosphorylation levels, suggesting that, besides preventing its ubiquitination, SUMOylation at this position has additional consequences in other PTMs that make IκBα a stronger NF-κB inhibitor (Colomer-Lluch and Serra-Moreno, 2017). As a consequence of this, HIV transcription is significantly hindered, enforcing HIV latency (Fig. 24.5). In fact, our research shows that BCA2 is a critical host factor for the establishment and maintenance of latency in CD4+ T cells, which are considered the major reservoir for latent HIV on transitioning to a central memory phenotype.
BCA2 is an intriguing anti-HIV factor that exhibits several levels of restriction by affecting multiple steps of the virus replication cycle. Future investigations need to address whether HIV circumvents this block to establish a productive infection, and how.
Rev ubiquitination hinders its ability to transport HIV mRNAs
The HIV Rev protein is fundamental for virus replication, since it exports non-spliced and singly-spliced HIV mRNAs from the nucleus to the cytoplasm (Karn and Stoltzfus, 2012). Similar to other HIV proteins, Rev also undergoes PTMs. Whereas no signature sequences for SUMOylation have been found in Rev, this protein is susceptible to K33-poly-ubiquitination at Lys115 (Vitte et al., 2006). Although the addition of these ubiquitin chains results in a more stable form of Rev, this PTM is accompanied by a reduction in protein activity (Vitte et al., 2006), probably caused by conformational changes in Rev that affect its functionality. Therefore, in this case the host uses the ubiquitin pathway to diminish HIV gene expression by limiting the cytoplasmic transport of HIV RNAs rather than directing viral proteins for degradation (Fig. 24.5). However, additional studies are needed to provide details on the cellular molecules involved in Rev ubiquitination and how HIV circumvents this block in vivo.
Ubiquitination as a viral countermeasure to restriction factors
As part of the antiviral host defence, the cellular ubiquitin system participates in multiple processes including the proteasome-mediated degradation of virus proteins as well as the activation of the AP-1, JAK/STAT and NF-κB signalling pathways, thereby restricting viral replication and shaping the innate immune response to combat the infection (Bour et al., 2001). However, HIV, as well as other viruses, has evolved to exploit the ubiquitin machinery to induce the degradation of host antiviral factors favouring its replication, infectivity, pathogenesis, and escape from the host immune surveillance (Yu et al., 2003; Alroy et al., 2005; Luo, 2016; Seissler et al., 2017). Thus, by manipulating the ubiquitin cascade HIV either avoids ubiquitination or enhances this PTM for the inactivation of its restriction factors. HIV achieves this goal via its accessory proteins Vif, Vpr, Vpu, Vpx and Nef, which recruit cellular E3 ubiquitin ligases to target specific antiviral elements for proteasomal/lysosomal degradation, and/or sequestration into subcellular compartments, removing them from their sites of action. Besides this primary role, some of the HIV accessory proteins use ubiquitination to suppress the expression of host antiviral genes by controlling the activity of cellular transcription factors such as AP-1, STATs and NF-κB. Whereas HIV extensively appropriates the ubiquitin system, SUMOylation has not been reported as a mechanism of immune evasion for HIV, suggesting that either HIV does not exploit this machinery to block host restriction factors or that it remains to be discovered.
Vif-induced ubiquitination and degradation of APOBEC3G
HIV Vif counteracts several APOBEC3 cytidine deaminases by inducing their degradation. As explained above, these proteins are incorporated into HIV particles to promote viral hypermutation, significantly restricting HIV infectivity. APOBEC3G is the best-characterized member of this family that is antagonized by Vif. In this case, the Vif-induced ubiquitination and degradation of APOBEC3G impedes its incorporation into nascent virions, preserving in turn the virus genetic integrity (Sheehy et al., 2002; Harris et al., 2003; Marin et al., 2003; Sheehy et al., 2003). By directly associating with APOBEC3G, hijacking the ubiquitin proteasome system and serving as a molecular adaptor to recruit on one hand the host transcription co-factor CBF-β, and on the other one the ElonginB-ElonginC-Cullin5 E3 ubiquitin ligase complex, Vif enhances the K48-linked poly-ubiquitination of APOBEC3G, which directs this protein for proteasomal degradation (Yu et al., 2003; Mehle et al., 2004b; Kobayashi et al., 2005; Luo et al., 2005; Shirakawa et al., 2006; Iwatani et al., 2009; Jäger et al., 2011; Zhang et al., 2011) (Table 24.1). In an analogous manner, Vif also targets APOBEC3F for degradation, suggesting that the Cullin5 E3 ubiquitin ligase complex is a shared component for Vif counteraction of APOBEC3 family members (Liu et al., 2005). Although the exact residues in APOBEC3G that are susceptible to Vif-induced poly-ubiquitination are currently unknown, mutagenesis analyses have narrowed down the potential target sites to twenty lysines (He et al., 2008; Iwatani et al., 2009; Turner et al., 2016), but additional studies are needed to reveal the relevance of each lysine residue for APOBEC3G ubiquitination and degradation.
Table 24.1.
Ubiquitination as a viral countermeasure to restriction factors. This table lists HIV proteins that overcome host restriction factors and/or molecules of the innate immune system by facilitating their ubiquitination. ISGs: Interferon-stimulated genes
| Virus protein | Substrate host factor |
Recruited complex | Type of PTM | Motif | Effect | Attributed function |
References |
|---|---|---|---|---|---|---|---|
| Vif | APOBEC3G/F | CBF-β and CUL5 E3 ubiquitin ligase complex | K48-linked poly-ubiquitination | Not known | Proteasomal degradation | APOBEC3G/F counteraction | Yu et al. (2003); Mehle et al. (2004b); Kobayashi et al. (2005); Liu et al. (2005); Luo et al. (2005); Shirakawa et al. (2006); Iwatani et al. (2009); Jäger et al. (2011); Zhang et al. (2011) |
| Vif | STAT1/3 | CUL5 E3 ubiquitin ligase complex | poly-ubiquitination | Not known | Proteasomal degradation | Inhibition of INF-α signalling and ISGs expression | Gargan et al. (2018) |
| Vpr | UNG | CUL4A- DDB1-DCAF1 E3 ubiquitin ligase complex | poly-ubiquitination | Not known | Alternative mechanism for APOBEC3G counteraction | Schröfelbauer et al. (2005); Wu et al. (2016) | |
| Vpx | SAMHD1 | CUL4A- DDB1-DCAF1 E3 ubiquitin ligase complex | poly-ubiquitination | Not known | Proteasomal degradation | SAMHD1 counteraction | Bergamaschi et al. (2009); Hrecka et al. (2011); Laguette et al. (2011); Ahn et al. (2012) |
| Vpr | SAMHD1 | CUL4A- DDB1-DCAF1 E3 ubiquitin ligase complex | poly-ubiquitination | Not known | Proteasomal degradation | SAMHD1 counteraction | Romani and Cohen (2012); Fregoso et al. (2013); Cassiday et al. (2015); DePaula-Silva et al. (2015) |
| Vpu | BST2 | β-TrCP-E3 ubiquitin ligase complex | poly-ubiquitination | Ser-Thr in N-terminus | Proteasomal degradation | BST2 down-regulation from sites of virion budding and degradation | Dubé et al. (2009); Douglas et al. (2009); Iwabu et al. (2009); Mangeat et al. (2009); Mitchell et al. (2009); Hauser et al. (2011); Schmidt et al. (2011); Tokarev and Guatelli (2011); Tokarev et al. (2011); McNatt et al. (2013) |
| K48-linked poly-ubiquitination | Lys18 and Lys21 | Proteasomal degradation | |||||
| K63-linked poly-ubiquitination | Not known | Lysosomal degradation | |||||
| mono-ubiquitination | Lys18 and Lys21 | Lysosomal degradation | |||||
| Vpu | CD4 | β-TrCP-E3 ubiquitin ligase complex | poly-ubiquitination | Not known | Proteasomal degradation | CD4 down-regulation | Margottin et al. (1998) |
| Vpu | NF-κB and AP-1 | β-TrCP-E3 ubiquitin ligase complex | poly-ubiquitination | Lys21 and not known | Proteasomal degradation | Inhibition of NF-κB and AP-1 innate signalling pathways | Bour et al. (2001); Sauter et al. (2015) |
In addition to its well established role in promoting APOBEC3G ubiquitination for the counteraction of this restriction factor, Vif might also drive the degradation of APOBEC3G in a ubiquitin-independent manner (Dang et al., 2008). Similar to TRIM5α, Vif poly-ubiquitination, rather than APOBEC3G poly-ubiquitination, would direct APOBEC3G for proteasomal degradation due to the high affinity association between these two proteins. In fact, ubiquitinated Vif has been found within the assembled Vif-Cul5 E3 ligase complex (Mehle et al., 2004a), although whether this PTM recruits the proteasome system or if it is necessary for Vif’s functionality is still unknown.
In an analogous manner to TRIM5α, the recognition of APOBEC3G by Vif is species-specific (Mariani et al., 2003). Particularly, Vif interacts with APOBEC3G through different motifs in the APOBEC3G N-terminal and C-terminal regions, which differ across species. Following this principle, HIV Vif potently overcomes the human APOBEC3G protein, but not APOBEC3G from African green monkeys. Likewise, SIV Vif efficiently counteracts APOBEC3G from African green monkeys whereas it is unable to antagonize human APOBEC3G. Therefore, the Vif-APOBEC3G couple, together with the TRIM5α–CA pair, establishes an important biological barrier to cross-species transmission. Remarkably, no Vif defective HIV or SIV primary isolates have been identified (Sheehy et al., 2003), which underpins the critical function of this accessory protein for viral infectivity.
Besides antagonizing APOBEC3s, Vif also promotes the poly-ubiquitination and proteasomal degradation of the transcription factors STAT1 and STAT3, which belong to the JAK/STAT signalling pathway. Thus, through this activity Vif inhibits the production of type I interferons and the expression of many antiviral effectors besides APOBEC3G (Gargan et al., 2018). Similar to its anti-APOBEC3G activity Vif interacts with STAT1 and STAT3 and recruits the Cullin5 E3 ligase complex to facilitate their poly-ubiquitination and degradation, supporting the notion that the Cullin5 E3 ubiquitin ligase complex is a conserved component for Vif suppression of the host innate defence (Table 24.1). However, further research is needed to elucidate which residues in STAT1 and STAT3 become ubiquitinated.
Vpr and UNG ubiquitination: a backup mechanism to prevent APOBEC3G-induced restriction
Although HIV Vif efficiently relieves the block imposed by APOBEC3G, in some cases it is unable to completely prevent APOBEC3G incorporation into virions. In this scenario, the HIV Vpr accessory protein reduces the negative effects of these few APOBEC3G molecules during reverse transcription: by promoting the poly-ubiquitination and proteasomal degradation of cellular UNGs (uracil DNA glycosylases). UNGs belong to the cellular DNA repairmachinery. They act by recognizing uracil nucleotides found in DNA and, if the error is not repaired, they promote the degradation of these DNA molecules. Similar to APOBEC3G, UNGs can be found encapsidated in HIV particles. In the context of APOBEC3G restriction, the occurrence of uridylated DNA during reverse transcription is remarkably high. These uridylated molecules are recognized by the encapsidated UNGs and ultimately directed for degradation, a fact that significantly reduces the success of proviral DNA integration (Schröfelbauer et al., 2005). Vpr prevents the degradation of these HIV DNA molecules by enhancing the poly-ubiquitination and proteasomal degradation of UNGs through its association with the CRL4A-DCAF1 E3 ubiquitin ligase complex, hampering in turn their incorporation into HIV virions (Schröfelbauer et al., 2005; Wu et al., 2016) (Table 24.1). In addition to this rather indirect mechanism of evading one of the negative effects of the deaminase activity of APOBEC3G, recent bioinformatics screenings revealed that Vpr might physically interact with APOBEC3G (Zhou et al., 2015), and facilitate its poly-ubiquitination and proteasomal degradation through Vpr’s association with DDB1-Cul4A associated factor 1 (DCAF1), but this needs further investigation.
The fact that two different HIV accessory proteins target the same host restriction factor, although through different mechanisms, evidences the importance of bypassing the host first line of defence to establish a productive infection. Intriguingly, both Vif and Vpr have converged to hijack the ubiquitin-proteasomal machinery to reach this goal, which reflects the extraordinary adaptation of HIV to its host through the manipulation of the cellular post-translational machinery.
Vpx/Vpr-mediated ubiquitination and degradation of SAMHD1
Vpx, an accessory protein uniquely expressed in the HIV-2/SIVsmm lineage, is packaged into HIV virions via its interaction with the p6 domain of the Gag polyprotein. Together with Vpr, Vpx contributes to the nuclear import of the PIC, especially in non-dividing cells (Cheng et al. 2008). Vpx was initially known to physically interact with the Cullin4 E3 ubiquitin ligase complex via association with DCAF1 and DDB1 (damage-specific DNA binding protein1) (Sharova et al., 2008; Srivastava et al., 2008), suggesting that, similar to Vif and Vpu, Vpx targets a restriction factor for degradation. However, it was not until 2011 that this enigma was resolved: Vpx counteracts the dNTP triphosphohydrolase SAM domain and HD domain-containing protein 1 (SAMHD1), an endonuclease that specifically depletes the pool of cytosolic nucleotides, a fact that inevitably affects HIV reverse transcription (Laguette et al., 2011; Lahouassa et al., 2012). SAMHD1 expression is predominantly higher in myeloid and resting CD4+ T cells than in activated CD4+ T cells, indicating that this restriction is more relevant to macrophage and dendritic-tropic HIV isolates, and that SAMHD1 may be one of the components influencing the establishment of latency in resting CD4+ T cells (Baldauf et al., 2012; Descours et al., 2012).
Vpx opposes the antiviral effects of SAMHD1 by recruiting the CUL4A-DDB1-DCAF1 E3 ubiquitin ligase complex, inducing SAMHD1 poly-ubiquitination and proteasomal degradation, thereby favouring HIV reverse transcription in both resting CD4+ T cells and myeloid cells (Bergamaschi et al., 2009; Hrecka et al., 2011; Laguette et al., 2011; Ahn et al., 2012) (Table 24.1). Even though the specific determinants in SAMHD1 necessary for Vpx-mediated degradation are largely unknown, some studies reported that C-terminal sequences on SAMHD1 are recognized by Vpx to load the restriction factor onto the CUL4A-DDB1-DCAF1 E3 ubiquitin ligase complex (Hofmann et al., 2012; Wei et al., 2012). However, others found that the linker region and HD domain are necessary for proper folding and ubiquitination (Guo et al., 2013).
Similar to Vpx, the Vpr proteins of certain primate lentiviruses can also antagonize SAMHD1, reinforcing the notion that Vpx is the result of a genetic duplication of Vpr in the HIV-2/SIVsmm lineage (Tristem et al., 1992; Lim et al., 2012; Fregoso et al., 2013; Spragg and Emerman, 2013). In these cases, Vpr also usurps the CUL4A-DDB1-DCAF1 E3 ubiquitin ligase complex to target SAMHD1 for proteasome-mediated degradation (Table 24.1). However, Vpx and Vpr recognize distinct regions of SAMHD1, implying that these accessory proteins evolved divergent mechanisms to overcome this restriction factor in different hosts (Romani and Cohen, 2012; Fregoso et al., 2013; Cassiday et al., 2015; DePaula-Silva et al., 2015). Despite the fact that Vpr can compensate for the absence of Vpx in HIV, HIV Vpr is poorly active against SAMHD1. Still, HIV efficiently replicates in humans. Therefore, either HIV uses a different, yet undiscovered, mechanism to subvert SAMHD1, or the low expression levels of SAMHD1 in activated CD4+ T have minimal effects on HIV propagation. By contrast, in the rhesus macaque/SIVmac model, Vpx-mediated antagonism of SAMHD1 is crucial in the context of CD4+ T cells replication (Shingai et al., 2015), indicating that HIV-1 and SIVmac exhibit contrasting susceptibilities to the restriction imposed by SAMHD1. Similar to the Vif-Vpr case introduced above, this Vpx-Vpr duplicated capacity to antagonize SAMHD1 underscores the importance of subverting this restriction factor for a productive infection, at least for the majority of the primate lentiviruses.
Vpu-mediated ubiquitination as a mechanism to circumvent BST2 and other immune factors
Vpu is a type I integral membrane protein that localizes primarily at the plasma membrane, the ER, the trans-Golgi network (TGN) and endosomal compartments. HIV-1 Vpu achieved notoriety for its role antagonizing BST2 to enhance virion release and pathogenesis. BST2 is a type II integral membrane protein with an unusual topology. It has a short cytoplasmic domain, followed by a transmembrane domain, a coiled-coil extracellular domain, and a glycosylphosphatidylinositol anchor (GPI), which forces its C-terminus to interact with the outer leaflet of the plasma membrane (Kupzig et al., 2003). The presence of the GPI anchor makes BST2 accumulate at cholesterol-enriched microdomains in the trans-Golgi network, but specially in the plasma membrane, which are the main sites for HIV assembly (Kupzig et al., 2003; Rollason et al., 2007). It is due to its peculiar topology that BST2 acts as a potent restriction factor against a myriad of enveloped viruses (Evans et al., 2010). In particular, BST2 becomes incorporated into nascent virions that obtain their envelope membrane by budding off from cholesterol rafts. By virtue of keeping each of its ends associated to the lipid bilayer, BST2 anchors one terminus to the plasma membrane and the other one to the virus envelope, thereby tethering nascent virions to the cell surface (Neil et al., 2008; Van Damme et al., 2008; Perez-Caballero et al., 2009). Vpu in HIV-1 opposes the antiviral effects of BST2 by different cooperative mechanisms: (1) by down-regulating BST2 from sites of virion budding and sequestering this protein in intracellular compartments. This is achieved by interfering with the trafficking and recycling pathways and requires the conserved diserine phospho-motif in the cytoplasmic tail of Vpu (Dubé et al., 2009; Hauser et al., 2011; Schmidt et al., 2011; Tokarev and Guatelli, 2011; Tokarev et al., 2011; McNatt et al., 2013). (2) By promoting the degradation of BST2. For this, Vpu directly interacts through its transmembrane domain with the transmembrane domain of BST2. This association causes conformational changes in Vpu that allow the recruitment of the β-TrCP E3 ubiquitin ligase complex, which facilitates the ubiquitination of the cytoplasmic region of BST2, targeting it in turn for proteasomal and/or lysosomal degradation (Douglas et al., 2009; Iwabu et al., 2009; Mangeat et al., 2009; Mitchell et al., 2009). Although the underlying mechanism by which Vpu achieves this is still controversial, the proposed models include: (a) Vpu-mediated poly-ubiquitination of BST2 in an unconventional Ser-Thr motif in its N-terminus (Tokarev et al., 2011), for its subsequent proteasomal degradation, (b) Vpu-mediated K48-Linked poly-ubiquitination of BST2 at Lys18 and Lys21 and proteasomal degradation, (c) Vpu-mediated K63-linked poly-ubiquitination, which would tag BST2 for lysosomal degradation, (Mangeat et al., 2009), and (d) Vpu-mediated mono-ubiquitination of BST2 at Lys18 and Lys21 to mediate its lysosomal degradation (Table 24.1). Among all the proposed models, the latter is the only scenario in which Vpu would directly interact with BST2 molecules found in the trans-Golgi network and recycling endosomes to prevent their accumulation at sites of virion assembly in the plasma membrane. Nevertheless, either by down-regulating BST2 from the plasma membrane or by affecting its intracellular trafficking, Vpu efficiently removes BST2 from HIV assembling locations, enhancing the release of HIV particles from infected cells.
The fact that BST2 potently restricts a broad range of enveloped viruses highlights the importance for virus pathogens to antagonize BST2 to ensure their propagation. In the case of the AIDS viruses, only HIV-1 and few SIVs encode vpu in their genome, indicating that for their successful spread in their hosts, the other AIDS viruses must have evolved other countermeasures against BST2. In fact, HIV-2 and some SIVs use Env to facilitate the sequestration of BST2 in the trans-Golgi network (Gupta et al., 2009; Le Tortorec and Neil, 2009; Serra-Moreno et al., 2011). However, the majority of SIVs use Nef to counteract BST2. Specifically, Nef directly interacts with the cytoplasmic tail of BST2 and this association facilitates BST2 internalization and lysosomal degradation (Jia et al., 2009; Sauter et al., 2009; Zhang et al., 2009; Serra-Moreno et al., 2013). Remarkably, only Vpu, and not Env and Nef, seems to use the ubiquitin system to neutralize BST2.
Besides this primordial role in enhancing virion production, Vpu plays additional roles to increase viral infectivity, such as the down-regulation of the CD4 receptor (to impede superinfection) or the inhibition of NF-κB and AP-1 innate signalling pathways (Table 24.1). Although HIV uses NF-κB to increase its transcriptional capacity, the chronic activation of this cascade can also have deleterious effects on infectivity, which is the reason why HIV finely tunes the activation of this transcription factor (Sauter et al., 2015). Remarkably, Vpu achieves such multifaceted activities through its association with the β-TrCP E3 ubiquitin ligase complex, which directs all these Vpu targets for ubiquitination and proteasomal degradation (Margottin et al., 1998; Bour et al., 2001). Although Vpu is not a structural protein and is not necessary for HIV infection, it is considered an important virulence factor. In fact, HIV variants with mutations in Vpu that abolish its functions exhibit a milder phenotype in terms of infectivity and delayed progression to AIDS. The versatility of this protein to evade the BST2 block, virus superinfection and immune activation is in contrast to the fact that in order to achieve this, Vpu has converged to use the same host ubiquitin molecular complex. It is tempting to envisage that the next evolutionary adaptation in humans would involve thwarting the recruitment of the β-TrCP E3 ubiquitin ligase complex by Vpu.
Manipulation of SUMO and ubiquitin cascades as therapeutic strategies against HIV
As explained earlier, the simultaneous use of various compounds that specifically impair different steps of the HIV replication cycle is referred to as combination antiretroviral therapy or cART. These treatment regimens effectively keep the plasma viral loads below detectable levels in HIV-infected individuals, preclude the progression to AIDS, and significantly reduce the risk of transmission. Among the wide range of characterized antiretroviral agents, some are designed to target host-dependency factors, while others block HIV proteins with the common goal of preventing the replication of the virus. In line with this, our increasing understanding of the roles of ubiquitination and SUMOylation in the context of HIV infection has opened new avenues for therapeutic intervention. Below we provide examples of targets that are currently under investigation, where ubiquitin and SUMO systems are being modulated to prevent a productive HIV infection.
Blocking HIV through the modulation of the ubiquitination profile of host restriction factors and HIV proteins
The prevention of different ubiquitin-associated events related to the progression of HIV replication has been extensively explored in the design of new anti-HIV treatments. The main focus of these approaches is to enhance the action of innate immune effectors (such as TRIM5α, APOBEC3G and BST2) by reducing their ubiquitin-mediated degradation. In addition, strategies to specifically modify the ubiquitination pattern of HIV proteins, aimed at rendering HIV non-infectious, are also being investigated. Although some studies are at their earlier stages, others have already identified and characterized the mechanism of action of compounds that selectively modulate the ubiquitination profiles of these proteins.
TRIM5α. A recent study demonstrated that the expression of a more stable version of human TRIM5α conferred protection against HIV-mediated destruction of CD4+ T cells in vivo. This TRIM5α mutant not only exhibits lower auto-ubiquitination, which increases its steady-state levels, but also displays a phenotype that closely resembles that of rhesus TRIM5α (rhTRIM5α), being potently restrictive of HIV. This variant (huTRIM5αR332–323) was obtained by introducing two point mutations in the PRYSPRY domain, which improve the recognition of retroviral capsids and at the same time the stability of TRIM5α. This study indicates that efforts to stabilize host restriction factors, probably through genetic engineering, have significant therapeutic value (Richardson et al., 2014).
APOBEC3G. Approaches to enhance the activity of APOBEC3G have been widely investigated. Their common goal is to inhibit the Vif-mediated ubiquitination and degradation of APOBEC3G, or to compensate for its rapid decay. One of these investigations identified the small molecule RN-18 that specifically targets Vif for degradation when Vif is associated with APOBEC3G (Nathans et al., 2008). VEC-5 is another Vif small inhibitor that disrupts the formation of the protein complex required for Vif-mediated degradation of APOBEC3G. In an APOBEC3G-dependent manner, VEC-5 prevents the association between Vif and the ubiquitin E3 ligase enzyme ElonginC (Zuo et al., 2012). Finally, the compounds IMB-26 and IMB-35 are another example of Vif inhibitors with the ability to suppress HIV infectivity. In particular, they prevent the interaction between Vif and APOBEC3G, and thus, protect the restriction factor from Vif-mediated degradation (Cen et al., 2010). Besides the use of chemical compounds to target the Vif-APOBEC3G complex, genetic engineering represents an alternative approach to increase the restrictive capacity of APOBEC3G. For instance, the addition of transcription activation domains to the promoter of APOBEC3G induces high levels of expression of the protein, compensating for its Vif-mediated loss (Bogerd et al., 2015). All these approaches significantly augment the intracellular amounts of APOBEC3G and facilitate its incorporation into new virions, enhancing its anti-HIV activity.
BST2. Similar to Vif-APOBEC3G, the Vpu–BST2 pair represents an attractive target for antiretroviral therapy. In fact, a small molecule, IMB-LA, potently prevents the Vpu-dependent degradation of BST2 without affecting the interaction between Vpu, BST2 and the ubiquitin E3 ligase complex β-TrCP. Specifically, IMB-LA acts by blocking the sorting process of poly-ubiquitinated BST2 to lysosomes in a Vpu-specific manner, and in turn increases the surface levels of BST2. Therefore, this compound restores the antiviral activity of the restriction factor, and thus, reduces HIV infectivity (Mi et al., 2015).
In addition to these antiviral agents, whose common goal is to enhance the activity of the innate immunity, inhibitors of de-ubiquitinase enzymes (DIs) might have potential to hinder HIV replication and improve antigen presentation by preventing the removal of ubiquitin chains in HIV proteins. For instance, DIs P22077 and PR-619 impair ubiquitin-related processes involved in Gag-mediated assembly. These compounds considerably reduce HIV infectivity and, unlike other small molecules, promote MHC-I-mediated antigen presentation, which significantly enhances the clearance of HIV-infected cells by CTLs (Setz et al., 2017).
Manipulation of the SUMOylation profile of HIV and host dependency factors as a therapeutic approach against HIV
The functionality of several HIV proteins as well as host dependency factors can be modulated through their SUMOylation status. Therefore, the chemical manipulation of this PTM represents a promising approach to block HIV replication. For instance, inhibitors of HIV IN SUMOylation, as well as compounds that bind SUMOylated IN, significantly reduce the efficiency of proviral DNA integration, since this PTM facilitates the recruitment of cellular co-factors that aid in the integration process. In this regard, the agent SPI-01, an inhibitor of SUMO-specific proteases (SENP), affects the SUMOylation machinery involved in IN modification. Although the underlying mechanism is not well understood, it seems that SPI-01 would prevent the deSUMOylation of IN, most likely through direct association with SUMOylated IN. By doing this, SPI-01 would preclude IN association with LEDGF/p75 and other co-factors, and thereby, impair proviral DNA integration. In fact, treatment of HIV-infected cells with SPI-01 renders the progeny virions unable to drive proviral integration in the subsequent infections (Madu et al., 2013, 2015).
Despite the high efficiency of cART at inhibiting HIV replication, the current treatments are unable to completely eliminate HIV from the system of HIV-infected individuals, since viral rebound is frequently observed among patients who discontinue therapy. Therefore, a lifelong adherence to cART is the only possible option for HIV-infected individuals. However, long-term exposure to these drugs is often associated with toxicity, adverse interactions with other drugs used for AIDS-unrelated conditions and, since these compounds exert a strong selective pressure, the occurrence of cART-resistant HIV variants (Chun et al., 1999; Petoumenos et al., 2012; Pinoges et al., 2015). The reason why cART cannot completely eliminate HIV and HIV-infected cells is the presence of latent reservoirs, which are refractory to cART, and thus, considered the main challenge to functionally cure HIV/AIDS. Therefore, new strategies need to be developed to tackle these latently infected cells. At the moment, two main approaches are being investigated: the ‘shock and kill’ and the ‘block and lock’ methods (Castro-Gonzalez et al., 2018). Remarkably, some of the underlying strategies employed to achieve an efficient ‘shock and kill’ and/or ‘block and lock’ approach are based on the manipulation of the SUMO systems.
Preventing SUMO-modification for a ‘shock and kill’ method. The ‘shock and kill’ strategy uses latency-reversing agents (LRAs) to reactivate latently infected cells, so they support a productive HIV infection (Castro-Gonzalez et al., 2018). This, in combination with cART, renders the produced virions as well as the activated, HIV-infected cells susceptible to cART and clearance by CTLs. Different LRAs have already been tested in clinical trials, presenting different levels of efficiency in re-activating the latent reservoirs (Shan et al., 2012; Spivak and Planelles, 2016; Castro-Gonzalez et al., 2018). Among them, benzotriazoles significantly reduce the size of the HIV latent reservoir by maintaining the activated state of the host transcription factor STAT5, which as stated above, HIV uses to drive viral transcription. In particular, benzotriazoles inhibit the SUMOylation of STAT5, which is responsible for its inactivation and cytoplasmic localization. Therefore, these agents prolong the STAT5-induced transcription of HIV, preventing the establishment of latency, and aid in the reactivation of latently infected cells (Bosque et al., 2017).
Enhancing SUMOylation and ubiquitination as a ‘block and lock’ approach. The main goal of the ‘block and lock’ strategy is to achieve a drug-mediated enforcement of HIV latency, so latently infected cells remain harmless in a dormant state. Under these conditions, HIV transcription is severely impaired, and so is viral rebound, even when cART is discontinued. Different HIV latency-promoting agents (LPAs) are currently under investigation (Castro-Gonzalez et al., 2018). Among them, the host anti-HIV factor BCA2, an E3 ubiquitin and SUMO ligase, represents a promising ‘block and lock’ candidate due to its ability to (a) inhibit HIV transcription and promote HIV latency through the specific SUMOylation of the NF-κB key regulator IκBα, which represses NF-κB (Colomer-Lluch and Serra-Moreno, 2017), and (b) its ability to mediate Gag ubiquitination and degradation through its E3 ubiquitin ligase activity (Nityanandam and Serra-Moreno, 2014). By means of these two-hit functions, BCA2 potently blocks the transcription of HIV, but in case of transcriptional leaking, it also strongly represses reactivated proviruses by preventing the assembly of new progeny. This is an exciting protein that, in combination to cART, could be administered to HIV+ individuals to neutralize productively infected as well as latently infected cells. However, its potential as a ‘block and lock’ enforcer needs to be further examined, particularly in vivo.
Concluding remarks
Thirty-six years after the identification of HIV as the causative agent of AIDS, the search for effective treatments to completely eliminate the virus from affected individuals is still ongoing. To adequately design new therapeutic options, we need to increase our fundamental understanding of the HIV-host interactions, especially at the molecular level. This includes increase awareness on how the use of the cellular post-translational machinery, and in particular ubiquitination and SUMOylation, influences the fate of HIV infection, since these PTMs represent promising targets for therapeutic intervention, as evidenced throughout this review.
Acknowledgements
We would like to thank Brendan Allison (Texas Tech University) for critical revision of this review article and his helpful comments. This work has been supported in part by NIH/NIAID R21 AI106400, R21 AI138589 and Texas Tech University.
References
- Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, and Skowronski J (2012). HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem 287, 12550–12558. 10.1074/jbc.M112.340711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alroy I, Tuvia S, Greener T, Gordon D, Barr HM, Taglicht D, Mandil-Levin R, Ben-Avraham D, Konforty D, Nir A, et al. (2005). The trans-Golgi network-associated human ubiquitin-protein ligase POSH is essential for HIV type 1 production. Proc. Natl. Acad. Sci. U.S.A 102, 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, and Hope TJ (2006). Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol 80, 9754–9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Verma S, and Banerjea AC (2014). HIV-1 Vpr redirects host ubiquitination pathway. J. Virol 88, 9141–9152. 10.1128/JVI.00619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriagada G, Muntean LN, and Goff SP (2011). SUMOinteracting motifs of human TRIM5α are important for antiviral activity. PLOS Pathog. 7, e1002019 10.1371/journal.ppat.1002019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts EJ, and Hazuda DJ (2012). HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2, a007161 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine T, Chaudhary P, Gupta K, Islam S, Ghosh P, Santra MK, and Mitra D (2017). Cyclin F/FBXO1 interacts with HIV-1 viral infectivity factor (Vif) and restricts progeny virion infectivity by ubiquitination and proteasomal degradation of Vif protein through SCFcyclin F E3 ligase machinery. J. Biol. Chem. 292, 5349–5363. 10.1074/jbc.M116.765842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, et al. (2012). SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18, 1682–1687. 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, et al. (1983). Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868–871. [DOI] [PubMed] [Google Scholar]
- Benítez-Gutiérrez L, Soriano V, Requena S, Arias A, Barreiro P, and de Mendoza C (2018). Treatment and prevention of HIV infection with long-acting antiretrovirals. Expert Rev. Clin. Pharmacol. 11, 507–517. [DOI] [PubMed] [Google Scholar]
- Benson RE, Sanfridson A, Ottinger JS, Doyle C, and Cullen BR (1993). Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J. Exp. Med. 177, 1561–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi A, Ayinde D, David A, Le Rouzic E, Morel M, Collin G, Descamps D, Damond F, Brun-Vezinet F, Nisole S, et al. (2009). The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J. Virol. 83, 4854–4860. 10.1128/JVI.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binette J, Dubé M, Mercier J, Halawani D, Latterich M, and Cohen EA (2007). Requirements for the selective degradation of CD4 receptor molecules by the human immunodeficiency virus type 1 Vpu protein in the endoplasmic reticulum. Retrovirology 4, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, and Malim MH (2004). Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14, 1392–1396. 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Black LR, and Aiken C (2010). TRIM5alpha disrupts the structure of assembled HIV-1 capsid complexes in vitro. J. Virol. 84, 6564–6569. 10.1128/JVI.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Kornepati AV, Marshall JB, Kennedy EM, and Cullen BR (2015). Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc. Natl. Acad. Sci. U.S.A. 112, E7249–56. 10.1073/pnas.1516305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosque A, Nilson KA, Macedo AB, Spivak AM, Archin NM, Van Wagoner RM, Martins LJ, Novis CL, Szaniawski MA, Ireland CM, et al. (2017). Benzotriazoles Reactivate Latent HIV-1 through Inactivation of STAT5 SUMOylation. Cell Rep. 18, 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S, Perrin C, Akari H, and Strebel K (2001). The human immunodeficiency virus type 1 Vpu protein inhibits NF-kappa B activation by interfering with beta TrCP-mediated degradation of Ikappa B. J. Biol. Chem. 276, 15920–15928. 10.1074/jbc.M010533200. [DOI] [PubMed] [Google Scholar]
- Brandariz-Nuñez A, Roa A, Valle-Casuso JC, Biris N, Ivanov D, and Diaz-Griffero F (2013). Contribution of SUMO-interacting motifs and SUMOylation to the antiretroviral properties of TRIM5α. Virology 435, 463–471. 10.1016/j.virol.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MT, Garcia-Rivera JA, Kugelman JR, Morales E, Rosas-Acosta G, and Llano M (2010). SUMOylation of the lens epithelium-derived growth factor/p75 attenuates its transcriptional activity on the heat shock protein 27 promoter. J. Mol. Biol. 399, 221–239. 10.1016/j.jmb.2010.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Nabel GJ, and Swanstrom R (2012). HIV: from biology to prevention and treatment, 1st edn (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: ). [Google Scholar]
- Busillo JM, and Benovic JL (2007). Regulation of CXCR4 signaling. Biochim. Biophys. Acta 1768, 952–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busillo JM, Armando S, Sengupta R, Meucci O, Bouvier M, and Benovic JL (2010). Site-specific phosphorylation of CXCR4 is dynamically regulated by multiple kinases and results in differential modulation of CXCR4 signaling. J. Biol. Chem. 285, 7805–7817. 10.1074/jbc.M109.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Perez O, Anderson JL, and Hope TJ (2008). Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha. J. Cell Biol. 180, 549–561. 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Weingart J, Sette P, Opp S, Sastri J, O’Connor SK, Talley S, Diaz-Griffero F, Hirsch V, and Bouamr F (2016). TRIM5α-mediated ubiquitin chain conjugation is required for inhibition of HIV-1 reverse transcription and capsid destabilization. J. Virol. 90, 1849–1857. 10.1128/JVI.01948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, and Martin-Serrano J (2007). Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912. [DOI] [PubMed] [Google Scholar]
- Cassiday PA, DePaula-Silva AB, Chumley J, Ward J, Barker E, and Planelles V (2015). Understanding the molecular manipulation of DCAF1 by the lentiviral accessory proteins Vpr and Vpx. Virology 476, 19–25. 10.1016/j.virol.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Gonzalez S, Colomer-Lluch M, and Serra-Moreno R (2018). Barriers for HIV cure: the latent reservoir. AIDS Res. Hum. Retroviruses 34, 739–759. 10.1089/AID.2018.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Peng ZG, Li XY, Li ZR, Ma J, Wang YM, Fan B, You XF, Wang YP, Liu F, et al. (2010). Small molecular compounds inhibit HIV-1 replication through specifically stabilizing APOBEC3G. J. Biol. Chem. 285, 16546–16552. 10.1074/jbc.M109.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control (CDC) (1981a). Follow-up on Kaposi’s sarcoma and Pneumocystis pneumonia. MMWR Morb. Mortal Wkly Rep. 30, 409–410. [PubMed] [Google Scholar]
- Centers for Disease Control (CDC) (1981b). Kaposi’s sarcoma and Pneumocystis pneumonia among homosexual men – New York City and California. MMWR Morb. Mortal Wkly Rep. 30, 305–308. [PubMed] [Google Scholar]
- Centers for Disease Control (CDC) (2018). About HIV/AIDS. Available online: https://www.cdc.gov/hiv/default.html. Accessed June 2019.
- Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, and Giacca M (2005). Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 24, 3070–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, and Bonifacino JS (2007). Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J. Virol. 81, 3877–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yang X, Cheng W, Ma Y, Shang Y, Cao L, Chen S, Chen Y, Wang M, and Guo D (2017). Immune regulator ABIN1 suppresses HIV-1 transcription by negatively regulating the ubiquitination of Tat. Retrovirology 14, 12 10.1186/s12977-017-0338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WT, Yang Y, Xu Y, and An J (2014). Targeting chemokine receptor CXCR4 for treatment of HIV-1 infection, tumor progression, and metastasis. Curr. Top. Med. Chem. 14, 1574–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Davey RT, Engel D, Lane HC, and Fauci AS (1999). Re-emergence of HIV after stopping therapy. Nature 401, 874–875. 10.1038/44755. [DOI] [PubMed] [Google Scholar]
- Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey MA, Santos-Ferreira MO, Laurent AG, Dauguet C, Katlama C, Rouzioux C, et al. (1986). Isolation of a new human retrovirus from West African patients with AIDS. Science 233, 343–346. [DOI] [PubMed] [Google Scholar]
- Cluck DB, and Underwood RF (2018). A therapeutic perspective of living with human immunodeficiency virus/AIDS in 2017. Nurs. Clin. North Am. 53, 97–110. [DOI] [PubMed] [Google Scholar]
- Coffin JM (1995). HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267, 483–489. [DOI] [PubMed] [Google Scholar]
- Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, Hakim JG, Kumwenda J, Grinsztejn B, Pilotto JH, et al. (2011a). Prevention of HIV-1 infection with early antiretroviral therapy. N. Engl. J. Med. 365, 493–505. 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Shaw GM, McMichael AJ, and Haynes HF (2011b). Acute HIV-1 infection. N. Engl. J. Med. 364, 1943–1954. 10.1056/NEJMra1011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Lluch M, and Serra-Moreno R (2017). BCA2/Rabring7 interferes with HIV-1 proviral transcription by enhancing the SUMOylation of IºBα. J. Virol. 91, e02098–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Siew LM, and Zheng YH (2008). APOBEC3G is degraded by the proteasomal pathway in a Vifdependent manner without being polyubiquitylated. J. Biol. Chem. 253, 13124–13131. 10.1074/jbc.M708728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca C, Petropoulos L, Zmeureanu D, and Hiscott J (1999). Nuclear IkappaBbeta maintains persistent NF-kappaB activation in HIV-1-infected myeloid cells. J. Biol. Chem. 274, 13010–13016. [DOI] [PubMed] [Google Scholar]
- DePaula-Silva AB, Cassiday PA, Chumley J, Bosque A, Monteiro-Filho CMR, Mahon CS, Cone KR, Krogan N, Elde NC, and Planelles V (2015). Determinants for degradation of SAMHD1, Mus81 and induction of G2 arrest in HIV-1 Vpr and SIVagm Vpr. Virology 477, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, and Benkirane M (2012). SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9, 87 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, and Hay RT (1998). SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2, 233–239. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, Stremlau M, and Sodroski J (2006). Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 349, 300–315. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F, Kar A, Perron M, Xiang SH, Javanbakht H, Li X, and Sodroski J (2007). Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J. Virol. 81, 10362–10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Qin XR, Hayashi F, Kigawa T, Finzi A, Sarnak Z, Lienlaf M, Yokoyama S, and Sodroski J (2009). A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83, 10737–10751. 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring Y, Pawig L, Weber C, and Noels H (2014). The CXCL12/CXCR4 chemokine ligand/receptor axis in cardiovascular disease. Front. Physiol 5, 212 10.3389/fphys.2014.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Früh K, and Moses AV (2009). Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J. Virol 83, 7931–7947. 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé M, Roy BB, Guiot-Guillain P, Mercier J, Binette J, Leung G, and Cohen EA (2009). Suppression of Tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J. Virol 83, 4574–4590. 10.1128/JVI.01800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussart S, Courcoul M, Bessou G, Douaisi M, Duverger Y, Vigne R, and Decroly E (2004). The Vif protein of human immunodeficiency virus type 1 is posttranslationally modified by ubiquitin. Biochem. Biophys. Res. Commun 315, 66–72. 10.1016/j.bbrc.2004.01.023. [DOI] [PubMed] [Google Scholar]
- Dutrieux J, Portilho DM, Arhel NJ, Hazan U, and Nisole S (2015). TRIM5α is a SUMO substrate. Retrovirology 12, 28 10.1186/s12977-015-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, and Cherepanov P (2008). The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLOS Pathog. 4, e1000046 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DT, Serra-Moreno R, Singh RK, and Guatelli JC (2010). BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18, 388–396. 10.1016/j.tim.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandis AZ, Cherla RP, Chernock RD, and Ganju RK (2002). CXCR4/CCR5 down-modulation and chemotaxis are regulated by the proteasome pathway. J. Biol. Chem 277, 18111–18117. 10.1074/jbc.M200750200. [DOI] [PubMed] [Google Scholar]
- Ferreiro DU, and Komives EA (2010). Molecular mechanisms of system control of NF-kappaB signaling by IkappaBalpha. Biochemistry 49, 1560–1567. 10.1021/bi901948j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher AJ, Christensen DE, Nelson C, Tan CP, Schaller T, Lehner PJ, Sundquist WI, and Towers GJ (2015). TRIM5α requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J. 34, 2078–2095. 10.15252/embj.201490361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JL, and Garcia JV (2008). HIV-1 Nef: at the crossroads. Retrovirology 5, 84 10.1186/1742-4690-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, and Martin MA (2013). Human immunodeficiency viruses: replication In Fields Virology, Knipe DM, and Howley PM, eds. (Lippincott Williams & Wilkins, Philadelphia, PA: ), p. 59. [Google Scholar]
- Fregoso OI, Ahn J, Wang C, Mehrens J, Skowronski J, and Emerman M (2013). Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLOS Pathog. 9, e1003496 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M, Setz C, Hahn F, Matthaei A, Fraedrich K, Rauch P, Henklein P, Traxdorf M, Fossen T, and Schubert U (2016). Glutamic acid residues in HIV-1 p6 regulate virus budding and membrane association of Gag. Viruses 8, 117 10.3390/v8040117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Omura S, and Silver J (1997). Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J. Gen. Virol 78, 619–625. 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, et al. (1984). Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224, 500–503. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, and Yeager M (2011). Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U.S.A 108, 534–539. 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargan S, Ahmed S, Mahony R, Bannan C, Napoletano S, O’Farrelly C, Borrow P, Bergin C, and Stevenson NJ (2018). HIV-1 promotes the degradation of components of the type 1 IFN JAK/STAT pathway and blocks anti-viral ISG induction. EBioMedicine 30, 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, and Boyse EA (1975). Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. U.S.A 72, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, et al. (2011). HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382. 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Gottwein E, and Kräusslich HG (2005). Analysis of human immunodeficiency virus type 1 Gag ubiquitination. J. Virol 79, 9134–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, Schaller T, Hué S, Barclay WS, Schulz R, and Malim MH (2013). Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502, 559–562. 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Wei W, Wei Z, Liu X, Evans SL, Yang W, Wang H, Guo Y, Zhao K, Zhou JY, et al. (2013). Identification of critical regions in human SAMHD1 required for nuclear localization and Vpx-mediated degradation. PLOS ONE 8, e66201 10.1371/journal.pone.0066201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Mlcochova P, Pelchen-Matthews A, Petit SJ, Mattiuzzo G, Pillay D, Takeuchi Y, Marsh M, and Towers GJ (2009). Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc. Natl. Acad. Sci. U.S.A 106, 20889–20894. 10.1073/pnas.0907075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurer C, Berthoux L, and Luban J (2005). Covalent modification of human immunodeficiency virus type 1 p6 by SUMO-1. J. Virol 79, 910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, and Neuberger MS (2002). RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247–1253. [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, and Malim MH (2003). DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809. [DOI] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, and Bieniasz PD (2004). Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. U.S.A 101, 10774–10779. 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, Guatelli JC, and Cannon PM (2011). HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 8, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT (2005). SUMO: a history of modification. Mol. Cell 18, 1–12. [DOI] [PubMed] [Google Scholar]
- Hay RT, Vuillard L, Desterro JM, and Rodriguez MS (1999). Control of NF-kappa B transcriptional activation by signal induced proteolysis of I kappa B alpha. Philos. Trans. R. Soc. Lond., B, Biol. Sci 354, 1601–1609. 10.1098/rstb.1999.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, and Ghosh S (2012). NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234. 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhang W, Chen G, Xu R, and Yu XF (2008). Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol 381, 1000–1011. 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- Hladik F, and McElrath MJ (2008). Setting the stage: host invasion by HIV. Nat. Rev. Immunol 8, 447–457. 10.1038/nri2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Logue EC, Bloch N, Daddacha W, Polsky SB, Schultz ML, Kim B, and Landau NR (2012). The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol 86, 12552–12560. 10.1128/JVI.01657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, and Skowronski J (2011). Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661. 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WS, and Hughes SH (2012). HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med 2, a006882 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, Tanaka Y, Sata T, and Tokunaga K (2009). HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J. Biol. Chem 284, 35060–35072. 10.1074/jbc.M109.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Chan DS, Liu L, Yoshii H, Shibata J, Yamamoto N, Levin JG, Gronenborn AM, and Sugiura W (2009). HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc. Natl. Acad. Sci. U.S.A 106, 19539–19544. 10.1073/pnas.0906652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Takaori-Kondo A, Shirakawa K, Higashitsuji H, Itoh K, Io K, Matsui M, Iwai K, Kondoh H, Sato T, et al. (2009). MDM2 is a novel E3 ligase for HIV-1 Vif. Retrovirology 6, 1 10.1186/1742-4690-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, et al. (2011). Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375. 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H, Diaz-Griffero F, Stremlau M, Si Z, and Sodroski J (2005). The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J. Biol. Chem 280, 26933–26940. [DOI] [PubMed] [Google Scholar]
- Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, and Sodroski J (2006). Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353, 234–246. [DOI] [PubMed] [Google Scholar]
- Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, and Evans DT (2009). Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLOS Pathog. 5, e1000429 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YJ, Cai CY, Zhang X, and Burakoff SJ (2008). Lysine 144, a ubiquitin attachment site in HIV-1 Nef, is required for Nef-mediated CD4 down-regulation. J. Immunol 180, 7878–7886. [DOI] [PubMed] [Google Scholar]
- Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, and Chen IS (1995). The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol 69, 6304–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, et al. (2013). MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502, 563–566. 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J, and Stoltzfus CM (2012). Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb. Perspect. Med 2, a006916 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, and Towers GJ (2004). The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. U.S.A 101, 10780–10785. 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, and Howley P (2013). Fields Virology 6th edn. (Lippincott Williams and Wilkin, Philadelphia: ). [Google Scholar]
- Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, and Uchiyama T (2005). Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem 280, 18573–18578. [DOI] [PubMed] [Google Scholar]
- Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, and Banting G (2003). Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4, 694–709. [DOI] [PubMed] [Google Scholar]
- Kutluay SB, Perez-Caballero D, and Bieniasz PD (2013). Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLOS Pathog. 9, e1003214 10.1371/journal.ppat.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, and Benkirane M (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657. 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, et al. (2012). SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol 13, 223–228. 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, Aiken C, Olsen JC, Kar AK, Sodroski JG, et al. (2008). Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J. Virol 82, 11682–11694. 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear T, Dunn SR, McKelvey AC, Mir A, Evankovich J, Chen BB, and Liu Y (2017). RING finger protein 113A regulates C-X-C chemokine receptor type 4 stability and signaling. Am. J. Physiol., Cell Physiol 313, C584–C592. 10.1152/ajpcell.00193.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall S, Erdtmann L, Benichou S, Berlioz-Torrent C, Liu L, Benarous R, Heard JM, and Schwartz O (1998). Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8, 483–495. [DOI] [PubMed] [Google Scholar]
- Le Tortorec A, and Neil SJ (2009). Antagonism and intracellular sequestration of human tetherin by the HIV-2 envelope glycoprotein. J Virol. 83, 11966–11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PL, Wang T, Buckley KA, Chenine AL, Popov S, and Ruprecht RM (2005). Phosphorylation of HIV Nef by cAMP-dependent protein kinase. Virology 331, 367–374. [DOI] [PubMed] [Google Scholar]
- Li X, and Sodroski J (2008). The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol 82, 11495–11502. 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienlaf M, Hayashi F, Di Nunzio F, Tochio N, Kigawa T, Yokoyama S, and Diaz-Griffero F (2011). Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): structure of the RING domain of TRIM5alpha. J. Virol 85, 8725–8737. 10.1128/JVI.00497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, and Emerman M (2012). The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204. 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Sarkis PT, Luo K, Yu Y, and Yu XF (2005). Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol 79, 9579–9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, and Liang C (2013). The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14, 398–410. 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- Llano M, Delgado S, Vanegas M, and Poeschla EM (2004). Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem 279, 55570–55577. [DOI] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, and Poeschla EM (2006). An essential role for LEDGF/p75 in HIV integration. Science 314, 461–464. [DOI] [PubMed] [Google Scholar]
- Lukic Z, Goff SP, Campbell EM, and Arriagada G (2013). Role of SUMO-1 and SUMO interacting motifs in rhesus TRIM5α-mediated restriction. Retrovirology 10, 10 10.1186/1742-4690-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H (2016). Interplay between the virus and the ubiquitin-proteasome system: molecular mechanism of viral pathogenesis. Curr. Opin. Virol 17, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, and Yu XF (2005). Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U.S.A 102, 11444–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles RH, Muñoz A, Yamashita TE, Bazmi H, Detels R, Rinaldo CR, Margolick JB, Phair JP, and Mellors JW (2000). Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. J. Infect. Dis 181, 872–880. [DOI] [PubMed] [Google Scholar]
- Madu IG, Namanja AT, Su Y, Wong S, Li YJ, and Chen Y (2013). Identification and characterization of a new chemotype of noncovalent SENP inhibitors. ACS Chem. Biol 8, 1435–1441. 10.1021/cb400177q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu IG, Li S, Li B, Li H, Chang T, Li YJ, Vega R, Rossi J, Yee JK, Zaia J, et al. (2015). A novel class of HIV-1 antiviral agents targeting HIV via a SUMOylation-dependent mechanism. Sci. Rep 5, 17808 10.1038/srep17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadán JG, Pérez-Victoria FJ, Sougrat R, Ye Y, Strebel K, and Bonifacino JS (2010). Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLOS Pathog. 6, e1000869 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, and Emerman M (2008). HIV-1 accessory proteins – ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398. 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, and Trono D (2003). Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103. 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, and Piguet V (2009). HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLOS Pathog. 5, e1000574 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A, and Benovic JL (2001). Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J. Biol. Chem 276, 45509–45512. 10.1074/jbc.C100527200. [DOI] [PubMed] [Google Scholar]
- Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, and Benovic JL (2003). The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell 5, 709–722. [DOI] [PubMed] [Google Scholar]
- Marchetti G, Tincati C, and Silvestri G (2013). Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin. Microbiol. Rev 26, 2–18. 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, and Benarous R (1998). A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1, 565–574. [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, Münk C, Nymark-McMahon H, and Landau NR (2003). Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31. [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, and Kabat D (2003). HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med 9, 1398–1403. 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- McNatt MW, Zang T, and Bieniasz PD (2013). Vpu binds directly to tetherin and displaces it from nascent virions. PLOS Pathog. 9, e1003299 10.1371/journal.ppat.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Goncalves J, Santa-Marta M, McPike M, and Gabuzda D (2004a). Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18, 2861–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Strack B, Ancuta P, Zhang C, McPike M, and Gabuzda D (2004b). Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem 279, 7792–7798. 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- Mi Z, Ding J, Zhang Q, Zhao J, Ma L, Yu H, Liu Z, Shan G, Li X, Zhou J, et al. (2015). A small molecule compound IMB-LA inhibits HIV-1 infection by preventing viral Vpu from antagonizing the host restriction factor BST-2. Sci. Rep 5, 18499 10.1038/srep18499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, and Sorkin A (2007). Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol. Interv 7, 157–167. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, and Guatelli JC (2009). Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLOS Pathog. 5, e1000450 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa K, Ryo A, Murakami T, Ohba K, Yamaoka S, Fukuda M, Guatelli J, and Yamamoto N (2009). BCA2/Rabring7 promotes tetherin-dependent HIV-1 restriction. PLOS Pathog. 5, e1000700 10.1371/journal.ppat.1000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, and Baltimore D (1987). An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326, 711–713. 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Nathans R, Cao H, Sharova N, Ali A, Sharkey M, Stranska R, Stevenson M, and Rana TM (2008). Small-molecule inhibition of HIV-1 Vif. Nat. Biotechnol 26, 1187–1192. 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Glöckner-Pagel J, Vaeth M, Schumann JE, Buttmann M, Bopp T, Schmitt E, Serfling E, and Berberich-Siebelt F (2009). Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J. Biol. Chem 284, 10935–10946. 10.1074/jbc.M900465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, and Bieniasz PD (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430. 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Nepveu-Traversy MÉ, and Berthoux L (2014). The conserved sumoylation consensus site in TRIM5α modulates its immune activation functions. Virus Res. 184, 30–38. 10.1016/j.virusres.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Nepveu-Traversy MÉ, Demogines A, Fricke T, Plourde MB, Riopel K, Veillette M, Diaz-Griffero F, Sawyer SL, and Berthoux L (2016). A putative SUMO interacting motif in the B30.2/SPRY domain of rhesus macaque TRIM5α important for NF-κB/AP-1 signaling and HIV-1 restriction. Heliyon 2, e00056 10.1016/j.heliyon.2015.e00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Stoye JP, and Saïb A (2005). TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol 3, 799–808. [DOI] [PubMed] [Google Scholar]
- Nityanandam R, and Serra-Moreno R (2014). BCA2/Rabring7 targets HIV-1 Gag for lysosomal degradation in a tetherin-independent manner. PLOS Pathog. 10, e1004151 10.1371/journal.ppat.1004151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Copeland TD, Kane BP, Johnson DG, Sowder RC, Yoshinaka Y, Oroszlan S, Arthur LO, and Henderson LE (1998). Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72, 2962–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik A, Chau V, and Wills JW (2000). Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. U.S.A 97, 13069–13074. 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, and Bieniasz PD (2009). Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139, 499–511. 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, and Sodroski J (2007). The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol 81, 2138–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Züger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. (2011). TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365. 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter F (1998). HIV nef: the mother of all evil? Immunity 9, 433–437. [DOI] [PubMed] [Google Scholar]
- Petoumenos K, Worm SW, Fontas E, Weber R, De Wit S, Bruyand M, Reiss P, El-Sadr W, Monforte AD, Friis-Møller N, et al. (2012). Predicting the short-term risk of diabetes in HIV-positive patients: the Data Collection on Adverse Events of Anti-HIV Drugs (D:A:D) study. J. Int. AIDS Soc 15, 17426 10.7448/IAS.15.2.17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinoges L, Schramm B, Poulet E, Balkan S, Szumilin E, Ferreyra C, and Pujades-Rodríguez M (2015). Risk factors and mortality associated with resistance to first-line antiretroviral therapy: multicentric cross-sectional and longitudinal analyses. J. Acquir. Immune Defic. Syndr 68, 527–535. 10.1097/QAI.0000000000000513. [DOI] [PubMed] [Google Scholar]
- Portilho DM, Fernandez J, Ringeard M, Machado AK, Boulay A, Mayer M, Müller-Trutwin M, Beignon AS, Kirchhoff F, Nisole S, et al. (2016). Endogenous TRIM5α function is regulated by SUMOylation and nuclear sequestration for efficient innate sensing in dendritic cells. Cell Rep. 14, 355–369. 10.1016/j.celrep.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postler TS, and Desrosiers RC (2012). The cytoplasmic domain of the HIV-1 glycoprotein gp41 induces NF-κB activation through TGF-β-activated kinase 1. Cell Host Microbe 11, 181–193. 10.1016/j.chom.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja R, Ronsard L, Lata S, Trivedi S, and Banerjea AC (2017). HIV-1 Tat potently stabilises Mdm2 and enhances viral replication. Biochem. J 474, 2449–2464. 10.1042/BCJ20160825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitter JN, Means RE, and Desrosiers RC (1998). A role for carbohydrates in immune evasion in AIDS. Nat. Med 4, 679–684. [DOI] [PubMed] [Google Scholar]
- Richardson MW, Guo L, Xin F, Yang X, and Riley JL (2014). Stabilized human TRIM5α protects human T cells from HIV-1 infection. Mol. Ther 22, 1084–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeth JF, Williams M, Kasper MR, Filzen TM, and Collins KL (2004). HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J. Cell Biol 167, 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rold CJ, and Aiken C (2008). Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLOS Pathog. 4, e1000074 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollason R, Korolchuk V, Hamilton C, Schu P, and Banting G (2007). Clatharin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J. Cell Sci 120, 3850–3058. [DOI] [PubMed] [Google Scholar]
- Romanchikova N, Ivanova V, Scheller C, Jankevics E, Jassoy C, and Serfling E (2003). NFAT transcription factors control HIV-1 expression through a binding site downstream of TAR region. Immunobiology 208, 361–365. [DOI] [PubMed] [Google Scholar]
- Romani B, and Cohen EA (2012). Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase. Curr. Opin. Virol 2, 755–763. 10.1016/j.coviro.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, et al. (2015). HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526, 212–217. 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg ES, Altfeld M, Poon SH, Phillips MN, Wilkes BM, Eldridge RL, Robbins GK, D’Aquila RT, Goulder PJ, and Walker BD (2000). Immune control of HIV-1 after early treatment of acute infection. Nature 407, 523–526. 10.1038/35035103. [DOI] [PubMed] [Google Scholar]
- Rothwarf DM, and Karin M (1999). The NF-kappa B activation pathway: a paradigm in information transfer from membrane to nucleus. Sci. STKE 1999, RE1 10.1126/stke.1999.5.re1. [DOI] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Münch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, et al. (2009). Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421. 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Hotter D, Van Driessche B, Stürzel CM, Kluge SF, Wildum S, Yu H, Baumann B, Wirth T, Plantier JC, et al. (2015). Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 10, 586–599. 10.1016/j.celrep.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, and Malik HS (2005). Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A 102, 2832–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Fritz JV, Bitzegeio J, Fackler OT, and Keppler OT (2011). HIV-1 Vpu blocks recycling and biosynthetic transport of the intrinsic immunity factor CD317/tetherin to overcome the virion release restriction. MBio 2, e00036–11. 10.1128/mBio.00036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröfelbauer B, Yu Q, Zeitlin SG, and Landau NR (2005). Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol 79, 10978–10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Bour S, Ferrer-Montiel AV, Montal M, Maldarell F, and Strebel K (1996). The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol 70, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Antón LC, Bacík I, Cox JH, Bour S, Bennink JR, Orlowski M, Strebel K, and Yewdell JW (1998). CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol 72, 2280–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Maréchal V, Le Gall S, Lemonnier F, and Heard JM (1996). Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med 2, 338–342. [DOI] [PubMed] [Google Scholar]
- Sebastian S, and Luban J (2005). TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seissler T, Marquet R, and Paillart JC (2017). Hijacking of the ubiquitin/proteasome pathway by the HIV auxiliary proteins. Viruses 9, E322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selliah N, Zhang M, DeSimone D, Kim H, Brunner M, Ittenbach RF, Rui H, Cron RQ, and Finkel TH (2006). The gammac-cytokine regulated transcription factor, STAT5, increases HIV-1 production in primary CD4 T cells. Virology 344, 283–291. [DOI] [PubMed] [Google Scholar]
- Serra-Moreno R, Jia B, Breed M, Alvarez X, and Evans DT (2011). Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe 9, 46–57. 10.1016/j.chom.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Moreno R, Zimmermann K, Stern LJ, and Evans DT (2013). Tetherin/BST-2 antagonism by Nef depends on a direct physical interaction between Nef and tetherin, and on clathrin-mediated endocytosis. PLOS Pathog. 9, e1003487 10.1371/journal.ppat.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setz C, Friedrich M, Rauch P, Fraedrich K, Matthaei A, Traxdorf M, and Schubert U (2017). Inhibitors of deubiquitinating enzymes block HIV-1 replication and augment the presentation of Gag-derived MHC-I epitopes. Viruses 9, E222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, and Siliciano RF (2012). Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36, 491–501. 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, and Williamson C (2012). The HIV-1 epidemic: low- to middle-income countries. Cold Spring Harb. Perspect. Med 2, a007187 10.1101/cshperspect.a007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova N, Wu Y, Zhu X, Stranska R, Kaushik R, Sharkey M, and Stevenson M (2008). Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLOS Pathog. 4, e1000057 10.1371/journal.ppat.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, and Hahn BH (2011). Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med 1, a006841 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, and Malim MH (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650. 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, and Malim MH (2003). The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med 9, 1404–1407. 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Shingai M, Welbourn S, Brenchley JM, Acharya P, Miyagi E, Plishka RJ, Buckler-White A, Kwong PD, Nishimura Y, Strebel K, et al. (2015). The expression of functional Vpx during pathogenic SIVmac infections of Rhesus macaques suppresses SAMHD1 in CD4+ memory T cells. PLOS Pathog. 11, e1004928 10.1371/journal.ppat.1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara T, Singh DP, and Fatma N (2002). LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res 21, 341–358. [DOI] [PubMed] [Google Scholar]
- Shirakawa K, Takaori-Kondo A, Kobayashi M, Tomonaga M, Izumi T, Fukunaga K, Sasada A, Abudu A, Miyauchi Y, Akari H, et al. (2006). Ubiquitination of APOBEC3 proteins by the Vif-Cullin5-ElonginB-ElonginC complex. Virology 344, 263–266. [DOI] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, and Engelman A (2007). LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21, 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano RF, and Greene WC (2011). HIV latency. Cold Spring Harb. Perspect. Med 1, a007096 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Plumb MR, Ferris AL, Iben JR, Wu X, Fadel HJ, Luke BT, Esnault C, Poeschla EM, Hughes SH, et al. (2015). LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 29, 2287–2297. 10.1101/gad.267609.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbak SM, Reksten TR, Hahn F, Wray V, Henklein P, Henklein P, Halskau Ø, Schubert U, and Fossen T (2013). HIV-1 p6 - a structured to flexible multifunctional membrane-interacting protein. Biochim. Biophys. Acta 1828, 816–823. 10.1016/j.bbamem.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Spivak AM, and Planelles V (2016). HIV-1 Eradication: Early Trials (and Tribulations). Trends Mol. Med 22, 10–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spragg CJ, and Emerman M (2013). Antagonism of SAMHD1 is actively maintained in natural infections of simian immunodeficiency virus. Proc. Natl. Acad. Sci. U.S.A 110, 21136–21141. 10.1073/pnas.1316839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Swanson SK, Manel N, Florens L, Washburn MP, and Skowronski J (2008). Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLOS Pathog. 4, e1000059 10.1371/journal.ppat.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, and Sodroski J (2004). The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853. 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, and Sodroski J (2005). Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J. Virol 79, 3139–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, and Sodroski J (2006). Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U.S.A 103, 5514–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, and Kräusslich HG (2012). HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med 2, a006924 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R, and Coffin J (2012). HIV-1 pathogenesis: the virus. Cold Spring Harb. Perspect. Med 2, a007443 10.1101/cshperspect.a007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Shohdy N, and Skowronski J (2001). Mechanism for down-regulation of CD28 by Nef. EMBO J. 20, 1593–1604. 10.1093/emboj/20.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Zhang Y, Koyama T, Tobiume M, Tsunetsugu-Yokota Y, Yamaoka S, Fujita H, and Tokunaga K (2015). MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat. Med 21, 1502–1507. 10.1038/nm.3956. [DOI] [PubMed] [Google Scholar]
- Tareen SU, and Emerman M (2011). Human Trim5α has additional activities that are uncoupled from retroviral capsid recognition. Virology 409, 113–120. 10.1016/j.virol.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terreni M, Valentini P, Liverani V, Gutierrez MI, Di Primio C, Di Fenza A, Tozzini V, Allouch A, Albanese A, Giacca M, et al. (2010). GCN5-dependent acetylation of HIV-1 integrase enhances viral integration. Retrovirology 7, 18 10.1186/1742-4690-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terui Y, Saad N, Jia S, McKeon F, and Yuan J (2004). Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J. Biol. Chem 279, 28257–28265. 10.1074/jbc.M403153200. [DOI] [PubMed] [Google Scholar]
- Tokarev A, and Guatelli J (2011). Misdirection of membrane trafficking by HIV-1 Vpu and Nef: Keys to viral virulence and persistence. Cell. Logist 1, 90–102. 10.4161/cl.1.3.16708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarev AA, Munguia J, and Guatelli JC (2011). Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J. Virol 85, 51–63. 10.1128/JVI.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper M, Luo Y, Zhadina M, Mohammed K, Smith L, and Muesing MA (2007). Posttranslational acetylation of the human immunodeficiency virus type 1 integrase carboxyl-terminal domain is dispensable for viral replication. J. Virol 81, 3012–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GJ (2007). The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology 4, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PB, Ren D, and Miller RJ (2005). The HIV-1 coat protein gp120 regulates CXCR4-mediated signaling in neural progenitor cells. J. Neuroimmunol 160, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristem M, Marshall C, Karpas A, and Hill F (1992). Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 11, 3405–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T, Shao Q, Wang W, Wang Y, Wang C, Kinlock B, and Liu B (2016). Differential contributions of ubiquitin-modified APOBEC3G lysine residues to HIV-1 Vif-induced degradation. J. Mol. Biol 428, 3529–3539. 10.1016/j.jmb.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J, and Mothes W (2013). TRIM protein mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol 87, 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS (2015). How AIDS Changed Everything. United Nations, AIDS Division, Geneva, Switzerland. [Google Scholar]
- UNAIDS (2018). AIDSinfo, Epidemiological Data. United Nations, AIDS Division, Geneva, Switzerland. [Google Scholar]
- Usami Y, Wu Y, and Göttlinger HG (2015). SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526, 218–223. 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, and Guatelli J (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252. 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nguyen T, Angkasekwinai P, Dou H, Lin FM, Lu LS, Cheng J, Chin YE, Dong C, and Yeh ET (2012). SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol. Cell 45, 210–221. 10.1016/j.molcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte AL, Buchsbaum S, and Jalinot P (2006). Modulation of HIV-1 Rev protein abundance and activity by polyubiquitination with unconventional Lys-33 branching. FEBS Lett. 550, 6155–6160. [DOI] [PubMed] [Google Scholar]
- Volberding PA, and Deeks SG (2010). Antiretroviral therapy and management of HIV infection. Lancet 376, 49–62. 10.1016/S0140-6736(10)60676-9. [DOI] [PubMed] [Google Scholar]
- Weber JN, Clapham PR, Weiss RA, Parker D, Roberts C, Duncan J, Weller I, Carne C, Tedder RS, Pinching AJ, et al. (1987). Human immunodeficiency virus infection in two cohorts of homosexual men: neutralising sera and association of anti-gag antibody with prognosis. Lancet 1, 119–122. [DOI] [PubMed] [Google Scholar]
- Wei W, Guo H, Han X, Liu X, Zhou X, Zhang W, and Yu XF (2012). A novel D CAF1-binding motif required for Vpx-mediated degradation of nuclear SAMHD1 and Vpr-induced G2 arrest. Cell. Microbiol 14, 1745–1756. 10.1111/j.1462-5822.2012.01835.x. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. (2003). Antibody neutralization and escape by HIV-1. Nature 422, 307–312. 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wilen CB, Tilton JC, and Doms RW (2012). HIV: cell binding and entry. Cold Spring Harb. Perspect. Med 2, a006866 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, and Harper JW (1999). The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, and Hope TJ (2006). Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. U.S.A 103, 7465–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou X, Barnes CO, DeLucia M, Cohen AE, Gronenborn AM, Ahn J, and Calero G (2016). The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat. Struct. Mol. Biol 23, 933–940. 10.1038/nsmb.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Lynch C, and Stoye JP (2004). Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. U.S.A 101, 10786–10791. 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, and Yu XF (2003). Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060. 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zamborlini A, Coiffic A, Beauclair G, Delelis O, Paris J, Koh Y, Magne F, Giron ML, Tobaly-Tapiero J, Deprez E, et al. (2011). Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J. Biol. Chem 256, 21013–21022. 10.1074/jbc.M110.189274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, and Hatziioannou T (2009). Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6, 54–67. 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, and Gao L (2003). The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424, 94–98. 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Du J, Evans SL, Yu Y, and Yu XF (2011). T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379. 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu J, and Liu X (2018). MARCH2 is upregulated in HIV-1 infection and inhibits HIV-1 production through envelope protein translocation or degradation. Virology 518, 293–300. [DOI] [PubMed] [Google Scholar]
- Zheng Y, and Yao X (2013). Posttranslational modifications of HIV-1 integrase by various cellular proteins during viral replication. Viruses 5, 1787–1801. 10.3390/v5071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ao Z, Wang B, Jayappa KD, and Yao X (2011). Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J. Biol. Chem 286, 17722–17735. 10.1074/jbc.M110.184739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Wang Y, Tokunaga K, Huang F, Sun B, and Yang R (2015). The HIV-1 accessory protein Vpr induces the degradation of the anti-HIV-1 agent APOBEC3G through a VprBP-mediated proteasomal pathway. Virus Res. 195, 25–34. 10.1016/j.virusres.2014.08.021. [DOI] [PubMed] [Google Scholar]
- Zuo T, Liu D, Lv W, Wang X, Wang J, Lv M, Huang W, Wu J, Zhang H, Jin H, et al. (2012). Small-molecule inhibition of human immunodeficiency virus type 1 replication by targeting the interaction between Vif and ElonginC. J. Virol 86, 5497–5507. 10.1128/JVI.06957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]