Abstract
Liver receptor homologue-1 (LRH-1; NR5A2) is a nuclear receptor that regulates metabolic homeostasis in the liver. Previous studies identified phosphatidylcholines as potential endogenous agonist ligands for LRH-1. In the liver, distinct subsets of phosphatidylcholine species are generated by two different pathways: choline addition to phosphatidic acid via the Kennedy pathway, or trimethylation of phosphatidylethanolamine via Phosphatidylethanolamine N-methyl Transferase (PEMT). Here we report that a PEMT - LRH-1 pathway specifically couples methyl metabolism and mitochondrial activities in hepatocytes. We show that the loss of Lrh-1 reduces mitochondrial number, basal respiration, beta-oxidation and ATP production in hepatocytes, and decreases expression of mitochondrial biogenesis and beta-oxidation genes. In contrast, activation of LRH-1 by its phosphatidylcholine agonists exerts opposite effects. While disruption of the Kennedy pathway does not affect the LRH-1-mediated regulation of mitochondrial activities, genetic or pharmaceutical inhibition of the PEMT pathway recapitulates the effects of Lrh-1 knockdown on mitochondria. Furthermore, we show that S-adenosyl methionine, a cofactor required for PEMT, is sufficient to induce Lrh-1 transactivation and consequently mitochondrial biogenesis. Conclusion: A PEMT – LRH-1 axis regulates mitochondrial biogenesis and beta-oxidation in hepatocytes.
Keywords: phosphatidylcholine, phospholipid, methyl pool, mitochondria, S-adenosyl methionine
Introduction:
Liver receptor homolog-1 (Lrh-1) is a nuclear receptor that binds as a monomer to a specific response element (5’ TCAAGGTCA 3’) within the promoter and regulatory regions of its target genes (1). Initial studies of the x-ray crystal structure of the human LRH-1 ligand binding domain expressed in E. coli showed that it is occupied by bacterial phospholipids (2). Further studies suggested that mammalian phospholipids, including both phosphatidylcholines (PCs) (3, 4) and phosphatidyl inositols (5, 6) could function as endogenous LRH-1 agonists.
One of the primary targets of LRH-1 in the liver is the nuclear receptor SHP, which functions as a corepressor for LRH-1 and other nuclear receptors (7, 8). LRH-1 also positively regulates genes encoding bile acid production enzymes, particularly Cyp8b1. Shp gene expression is also induced by the bile acid receptor FXR, resulting in a negative feedback loop in which elevated hepatic bile acid levels suppress bile acid production via inhibition of LRH-1 transactivation (8). Particularly in the agonist bound state, LRH-1 transactivation can be positively regulated by coactivators, with recent structural evidence indicating an important role for PGC-1α (PPARGC1A) (9).
In addition to controlling bile acid homeostasis, several reports suggest that LRH-1 targets different metabolic pathways. We identified dilauroyl-phosphatidylcholine (DLPC), an unusual PC species with two saturated twelve carbon fatty acid acyl chains, as an exogenous LRH-1 agonist, and showed that DLPC treatment could attenuate fatty liver and improve insulin sensitivity by repressing lipogenesis (4). In the opposite direction, LRH-1 mRNA expression is strongly decreased in livers of human subjects with steatosis or non-alcoholic steatohepatitis (10), and a recent report confirms that acute knockout of LRH-1 in mouse liver disrupts lipid metabolism and induces fat accumulation (11). Another report identifying glucokinase (GCK) as a primary LRH-1 target links LRH-1 to glucose utilization and glycogen synthesis (12).
The nucleus contains a surprisingly large and dynamic endonuclear PC pool that is distinct from the nuclear membrane (13), suggesting that PCs could act as endogenous agonists. There are two endogenous de novo PC synthesis pathways in mammals. The ubiquitous pathway for choline transfer to phosphatidic acid depends on both choline kinase alpha (Cka) and phosphate cytidylyltransferase 1alpha (Pcyt1a) and is called the Kennedy pathway. The liver has another pathway based on S-adenosylmethionine (SAM) dependent triple-methylation of phosphatidylethanolamine (PE) via Phosphatidylethanolamine N-methyl transferase (PEMT). If one of these two pathways is dysfunctional, the other increase its activity to compensate and maintain total net amount of PC in hepatocytes (14–18). However, these two pathways produce distinct subsets of PC species that differ in fatty acid side chains, and thus the compensatory responses can alter overall composition of the PC pool (14).
The PEMT pathway is tightly linked with SAM metabolism. The ratio of SAM to its demethylated product S-adenosyl homocysteine (SAH) provides a key index of the status of the endogenous methyl pool. Choline is oxidized to betaine, which provides a methyl group to homocysteine for making methionine. Methionine is then adenylated by Methionine Adenosyltransferase-1a (Mat1a) to generate SAM, the cofactor for diverse methyltransferase enzymes. Depletion of the endogenous methyl pool via either dietary deficiency in methionine and choline or genetic defects in SAM production results in hepatic steatosis (19). For example, Mat1a KO mice are deficient in liver SAM and have impaired fatty acid beta-oxidation capacity and increased fatty acid uptake in the liver (20, 21). Interestingly, these KO mice develop hepatic steatosis with age, which is associated with decreased VLDL secretion linked to decreased PEMT activity (20). PEMT generates PC species with long acyl chains, such as arachidonic acid or docosahexanoic acid, that are important for forming VLDL particles (22). Consistently, Pemt KO rat hepatocytes also have decreased VLDL secretion (22), and Pemt polymorphisms in humans confer susceptibility to non-alcoholic fatty liver disease (NAFLD) (23).
Lrh-1 is both a sensor of the state of SAM metabolism in the liver and a critical regulator of the methyl pool. We previously showed that LRH-1 activity is decreased in the livers of mice fed on a methionine and choline deficient diet, and also that LRH-1 transactivation is decreased in cells maintained in methionine and choline deficient media (24). This is associated with decreased expression of Glycine N-methyltransferase (Gnmt) and Multi Drug Resistance protein 2 (Mdr2), two primary targets of LRH-1. GNMT is a major consumer of hepatic SAM, and the phospholipid flippase MDR2 transports phospholipids from hepatocytes to the bile duct. In Lrh-1 KO liver. The reduction in GNMT preserves the SAM/SAH ratio, and decreased MDR2 activity lowers the loss of labile methyl groups in the form of PC species. Thus, liver specific Lrh-1 KO mice are completely resistant to the inflammatory and fibrotic effects of dietary deficiency in methionine and choline (24).
In striking agreement with this methyl sensing role, we also showed that in response to environmental deficiency in methionine and its associated methyl metabolites, C. elegans accumulates fat in the intestine, the functional homolog of the mammalian liver, and this is mediated by the C. elegans LRH-1 relative, the NR5A family member NHR-25 (25). Genetic and metabolomics studies revealed that SAM metabolism and its associated PEMT pathway are required for this NHR-25-mediated regulation (25). Interestingly, nhr-25 mutant worms also showed more fragmented and less filamentous mitochondria compared to control worms (25).
Together, these results inspired us to investigate whether and how LRH-1 could regulate fat metabolism by tuning mitochondrial functions in response to changes in the methyl pool. We demonstrate that LRH-1 activation promotes mitochondrial biogenesis and fatty acid beta-oxidation and induces the LRH-1 coactivator PGC-1α. These responses are specifically dependent on the PEMT pathway for PC production, but not the Kennedy pathway. We also show that SAM supplementation is sufficient to transactivate LRH-1 to regulate mitochondrial activities. We conclude that LRH-1 functions as a crucial regulator of mitochondrial metabolism and a key sensor of the methyl pool, and coordinate their activities.
Materials and Methods:
For detailed and further information of materials and methods, please see Supporting Information.
Animal Studies.
See Supplementary Experimental Procedures
Cell Culture.
Primary Hepatocytes were cultured in either William’s E medium containing 10% FBS and 1% penicillin/streptomycin antibiotics or Hank’s Balanced Salt Solution (HBSS) (24020117;Invitrogen) containing 10% FBS and 1% penicillin/streptomycin antibiotics. C3A/HepG2 cells and AML12 cells were cultured in DMEM/F-12 (CM017–050;Gendepot) containing 10% FBS and 1% penicillin/streptomycin antibiotics.
Primary Hepatocyte Isolation.
Primary hepatocytes were extracted as previously reported (26). Cells were plated in 10cm plates (1.5X10^7 cells/well), 6 well plates (2.5×10^6 cells/well) or XF24 cell culture microplates (12500 cells/ well) (100777–004;Agilent). The cells were cultured in William’s E medium containing 10% FBS (12551; Invitrogen)
siRNA Transfection.
C3A/HepG2 cells on 6 well plates were transfected with siRNA targeting different genes, using RNAi max lipofectamine (13778150; Invitrogen) for 48hrs. We purchased human Lrh-1 targeting siRNA (J-003430–07; Dharmacon) and non-targeting control siRNA (D-001810-10-05; Dharmacon) from Dharmacon. siRNA targeting human Mat1a (Pooled HSS181024, HSS181023; Invitrogen), Pemt (Pooled HSS145606, HSS170611, HSS145608: InVitrogen), Chka (Pooled HSS 141030, HSS140691; Invitrogen), Pcyt1a (pooled HSS 1007689, HSS107690; Invitrogen) and their non-targeting control siRNA (12935200, 12935300, 12935400; Invitrogen) were purchased from Invitrogen.
Drug Treatment.
See Supplementary Experimental Procedures
qPCR Experiment.
Total RNA was isolated from primary hepatocytes using Quick-RNA MiniPrep kit (11–328; Zymo Research). cDNA was synthesized by qScript cDNA synthesis Kit (95047; Quanta Biosciences) with 500ng of RNA. Gene expression level was determined by real-time PCR using LightCycler 480 Real-Time PCR System (Roche) with KAPA SYBR FAST Universal qPCR Master Mix (KK 4618; Kapa Biosystems). Relative mRNA level was calculated with delta delta Ct method and normalized by 36b4, Tbp or Cyclophilin expression. Primer information is upon request.
Mitochondrial DNA Copy Number Measurement.
Luciferase Assay.
ATP and Ketone Body Measurement
Oxygen Consumption Rate Measurement.
See Supplementary Experimental Procedures
Statistical analysis.
All experiments were performed at least in biological triplicate. For comparison of multiple groups, ANOVA was used with Bonferroni’s post hoc test. For comparison of two groups, Student’s t test was used (Graph Pad PRISM program, La Jolla California, USA). A p value of less than .05 was considered significant. Error bars represent means ± standard error of means. *p-value< 0.05, **p-value<0.005.
Results:
Hepatic LRH-1 regulates mitochondrial activities.
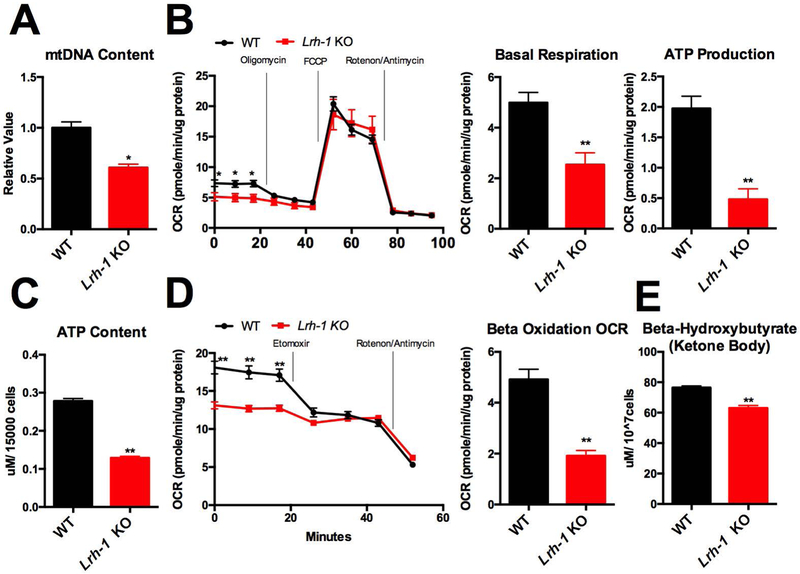
To test whether LRH-1 regulates mitochondrial functions, we first measured mitochondrial DNA copy number in primary hepatocytes extracted from wild-type (WT) and Lrh-1 liver-specific knock-out (KO) mice. We found that mitochondrial DNA copy number is reduced in Lrh-1 KO freshly isolated plated primary hepatocytes (Fig. 1A). Next, we examined the effect of LRH-1 on mitochondrial metabolic activities using SeaHorse assays. At the basal respiration level, we found that the oxygen consumption rate (OCR) is reduced by 60% in Lrh-1 KO primary hepatocytes, as assessed by the difference between initial OCR and OCR after rotenone/antimycin treatment (Fig. 1B). Upon inhibition of ATP synthase in Complex V by oligomycin treatment, OCR is reduced by 80% in Lrh-1 KO primary hepatocytes (Fig. 1B), which suggests that LRH-1 modulates ATP producing capacity in mitochondria. On the other hand we found that maximal respiration linked OCR is not affected by LRH-1, since there was no difference between WT and Lrh-1 KO upon FCCP treatment (Fig. 1B). We also directly measured ATP content in WT and Lrh-1 KO primary hepatocytes and found that it was reduced by 66% in Lrh-1 KO primary hepatocytes (Fig. 1C).
Figure 1. Hepatic Lrh-1 regulates mitochondrial functions.
A. Mitochondrial DNA copy number was measured in Lrh-1 KO and WT primary hepatocytes. B. Oxygen consumption rate (OCR) in WT and Lrh-1 KO primary hepatocytes was measured. Basal respiration linked OCR and ATP production linked OCR were quantified from Lrh-1 KO and WT primary hepatocytes. C. ATP level was measured from Lrh-1 KO and WT primary hepatocytes D. Oxygen consumption rate (OCR) in WT and Lrh-1 KO primary hepatocytes was measured for endogenous beta-oxidation assay. Beta-oxidation linked OCR from in Lrh-1 KO and WT primary hepatocytes was quantified. E. Beta-hydroxybutyrate level was measured from Lrh-1 KO and WT primary hepatocytes
Our previous studies showed that LRH-1 activation decreases fat accumulation in the livers of mice fed on a high fat diet (4), and also that NHR-25, the C. elegans homolog of LRH-1, regulates mitochondrial dynamics and lipid metabolism in response to different dietary inputs (25). We thus decided to use SeaHorse to analyze endogenous fatty acid beta-oxidation. We cultured primary hepatocytes in medium with 10% FBS, which contains endogenous fatty acids, and measured OCR in the presence and absence of etomoxir, which inhibits CPT1A to block fatty acid trafficking into mitochondria. We found that the etomoxir treatment reduced OCR to a much smaller degree in Lrh-1 KO primary hepatocytes (Fig. 1D), suggesting that Lrh-1 KO reduces mitochondrial beta-oxidation. This was also confirmed by directly measuring beta-hydroxybutyrate, one of the ketone bodies as an indirect beta-oxidation product. As expected, beta-hydroxybutyrate levels were decreased in freshly isolated Lrh-1 KO primary hepatocytes (Figure 1E).
Since it has been reported that mitochondria filamentation boosts beta-oxidation by increasing efficacy of fatty acids trafficking through mitochondrial network (27), we also analyzed mitochondrial morphology using mitotracker staining. We found no difference in mitochondrial morphology between WT and Lrh-1 KO primary hepatocytes, despite the reduced beta-oxidation capacity in the Lrh-1 KO (Fig.1D, Supplementary Fig. 1). In accord with previous results (27), when WT cells were cultured in starvation medium (HBSS), mitochondrial filamentation was induced, but this response was absent in Lrh-1 KO primary hepatocytes (Supplementary Fig. 1). Thus, during starvation LRH-1 is required for the induction of mitochondrial filamentation that is associated with an increased demand of mitochondrial beta-oxidation (27). However, the reduced beta-oxidation capacity of LRH-1 KO primary hepatocytes in regular medium is independent of changes in mitochondrial morphology.
LRH-1 regulates mitochondrial biogenesis and beta-oxidation genes
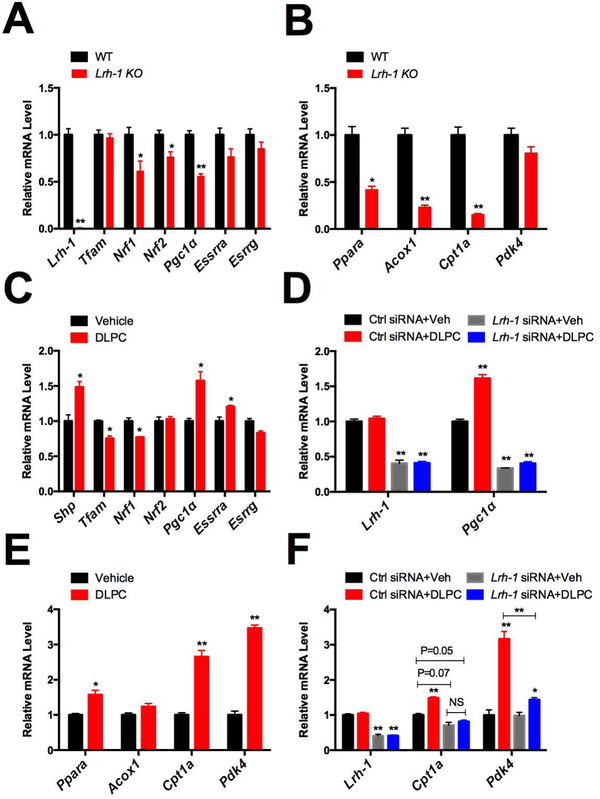
To further characterize the molecular mechanisms by which LRH-1 regulates mitochondrial activities, we extracted RNA from primary hepatocytes of WT and Lrh-1 liver specific KO mice and examined mRNA expression of genes involved in mitochondrial biogenesis and beta-oxidation. We found that mRNA expression of 3 transcriptional regulators of mitochondrial biogenesis encoded by the Nrf1, Nrf2 and Pgc-1α genes is significantly decreased more than 25% in Lrh-1 KO primary hepatocytes (p<0.05, Fig. 2A). For beta-oxidation, Acox1 and Cpt1a encode rate-limiting enzymes in the pathway, and Ppara encodes a key transcriptional activator of the pathway (28). We found that their expression levels are reduced more than 2 times in Lrh-1 KO primary hepatocytes (Fig. 2B). On the other hand, other mitochondrial biogenesis and beta-oxidation regulators, Tfam, Essrra, Esrrg and Pdk4 were unaffected by Lrh-1 KO. In accord with the SeaHorse studies, these results suggest that LRH-1 is required for the expression of specific genes involved in mitochondrial biogenesis and beta-oxidation in hepatocytes.
Figure 2. Lrh-1 regulates mitochondrial biogenesis and beta-oxidation genes.
A. The expression of mitochondrial biogenesis related genes was measured from Lrh-1 KO and WT primary hepatocytes. B. Expression of beta-oxidation related genes was measured in Lrh-1 KO and WT primary hepatocytes. C. The Lrh-1 agonist DLPC was administered to the mouse hepatocyte cell line AML12 for 16hrs and expression of genes important for mitochondria biogenesis was measured. D. DLPC was administered to control siRNA or Lrh-1 siRNA transfected C3A/HepG2 cells and Pgc1α expression was measured. E. DLPC was administered to the mouse hepatocyte cell line AML12 for 16hrs and expression of genes important for beta-oxidation was measured. F. DLPC was administered to control siRNA or Lrh-1 siRNA transfected C3A/HepG2 cells and Cpt1a and Pdk4 expression was measured.
When we asked whether WT and Lrh-1 KO livers show similar phenotypes, we found that ATP levels were significantly reduced, while mitochondrial DNA levels and mRNA expression of PGC1α and beta-oxidation genes showed trends in the expected directions that did not reach statistical significance (Supplementary Fig. 2). In contrast, analysis of WT and Lrh-1 KO primary hepatocytes prior to plating showed the same responses as freshly plated primary hepatocytes (Fig. 1C, Supplementary Fig. 2). The apparently decreased impact of Lrh-1 KO in intact livers could be due, at least in part, to the absence of the hepatocyte responses in non-parenchymal cells or the inhibition of the hepatocyte responses by non-parenchymal cells. In addition, we have previously found that LRH-1 is activated by stress responses including endoplasmic reticulum stress (26), and potentially starvation (Supplementary Fig. 1). Therefore, it is also possible that the impact of LRH-1 on mitochondrial functions is blunted in unstressed livers, but amplified by stresses associated with hepatocyte isolation.
Next, to test whether LRH-1 activation is sufficient to induce mitochondrial biogenesis and beta-oxidation gene expression, we turned to pharmacological gain of function studies. DLPC is an LRH-1 agonist (4), and we confirmed that DLPC treatment induces the expression of the best characterized LRH-1 target gene, Shp, in the mouse hepatocyte cell line AML12 (Fig. 2C). Importantly, DLPC treatrment induced Pgc-1α expression, and the induction level is as prominent as that of Shp (Fig. 2C). On the other hand, the expression of Nrf2 or Esrrg was not significantly affected by DLPC treatment (p>0.05), and the expression of Tfam, Nrf1, or Essrra was significantly (p<0.05) but modestly increased (20%) (Fig. 2C). Thus, we chose to focus on Pgc-1α and further investigate its response to DLPC in human C3A/HepG2 cells. We found that Pgc-1α is induced by 50% in control cells upon DLPC treatment, and this induction is fully suppressed in the cells transfected with siRNA targeting Lrh-1 (Fig. 2D). For beta-oxidation related genes, we found that Ppara, Cpt1a and Pdk4, but not Acox1 expression are induced upon DLPC treatment (Fig. 2E), and the induction of Cpt1a and Pdk4 requires Lrh-1 (Fig. 2F). Together, these studies reveal that LRH-1 regulates mitochondrial activities through controlling specific genes that are crucial for mitochondrial biogenesis and beta-oxidation, and its effects are well conserved in human hepatic cells.
The Kennedy Pathway does not contribute to LRH-1 regulation of mitochondrial activities.
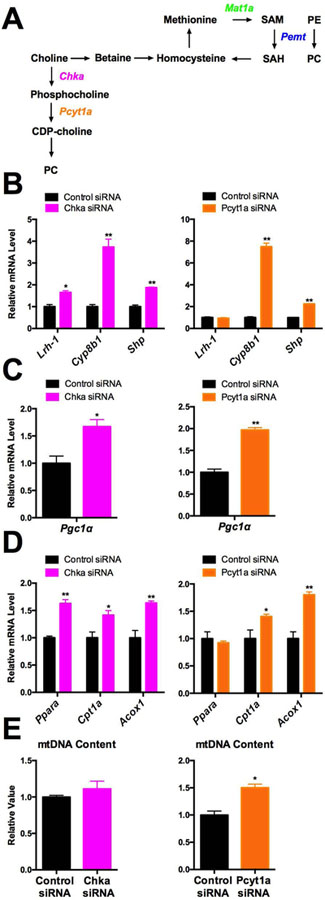
Since PC species are candidate ligands for LRH-1, managing proper levels of PC species could be crucial to control LRH-1 activity. There are two independent endogenous PC synthesis pathways in mammals (15). In the Kennedy pathway, the enzymes CKA and PCYT1A catalyze de novo synthesis of PC from choline. In the PEMT pathway, MAT1a generates SAM molecules that are used by PEMT to triply methylate PE to form PC (Fig. 3A). To investigate which pathway contributes to the production of putative PC agonists for LRH-1, we first knocked down either Pcyt1a or Chka to disrupt the Kennedy pathway. Their siRNA knock-down (KD) in C3A/HepG2 cells resulted in a compensatory increase in the expression of PEMT pathway genes (Supplementary Fig. 3). Using lipidomic profiling, we also found that Pcyt1a KD did not have a significant impact on total PC levels, but there were approximately 30 specific PC species upregulated or downregulated signficiantly upon Pcyt1a KD (Supplementary Figure 10). Knockdowns of either Pcyt1a or Chka also increased, rather than decreased the expression of Lrh-1 and its target genes, Cyp8b1 and Shp (Fig. 3B). Although Lrh-1 expression was increased only 1.5 fold upon Chka KD, the Cyp8b1 induction was up to 4 and 8 fold in Chka and Pcyt1a KD cells, respectively. Together, these results suggest increased LRH-1 transactivation in response to Kennedy pathway KD (Fig. 3B). Supporting this idea, the induction of LRH-1 target genes in Pcyt1a KD cells was reduced or abolished by the LRH-1 antagonist 505601 (29) (Supplementary Fig. 4).
Figure 3. Kennedy Pathway does not contribute to the Lrh-1 regulation of mitochondrial activities.
A. Diagram of Kennedy and PEMT pathways. B. Expression of Lrh-1 and its target genes was measured in Chka KD or Pcyt1a KD C3A/HepG2 cells. C. Expression of Pgc1α mRNA was measured in Chka KD or Pcyt1a KD C3A/HepG2 cells. D. Expression of beta-oxidation related genes was measured in Chka KD or Pcyt1a KD C3A/HepG2 cells. E. Mitochondrial DNA copy number was measured in Chka or Pcyt1a KD C3A/HepG2 cells.
Consistent with the LRH-1 transactivation, mitochondrial biogenesis and beta-oxidation genes were induced by either Pcyt1a or Chka KD. Pgc-1α and Cpt1a expression are both significantly increased in either Chka or Pcyt1a KD cells, and Ppara is significantly induced by Chka KD (p<0.05, Fig. 3C,3D). We also found that mitochondrial DNA copy number was increased in Pcyt1a KD cells (Fig. 3E). Inhibition of LRH-1 by 505601 suppressed the inducution of these mitochondrial genes in Pcyt1a KD cells (Supplementary Fig. 4). Moreover, CDP-choline is the product of PCYT1A and a cofactor required for generating Kennedy Pathway specific PC species (30). We found that CDP-choline treatment did not affect the expression of mitochondrial biogenesis or beta-oxidation genes or mitochondrial DNA content (Supplementary Fig. 5).
It is apparent that the Kennedy pathway is neither required nor sufficient for the production of PC agonists for LRH-1 to regulate mitochondrial activities. When it is disrupted, LRH-1 is activated instead to promote mitochondrial biogenesis and beta-oxidation. One possible explanation for this response is that the induction of the PEMT pathway to compensate for the loss of de novo PC synthesis (Supplemental Fig. 4) would increase the production of PC agonists for LRH-1.
PEMT pathway positively regulates the effect of Lrh-1 on mitochondria.
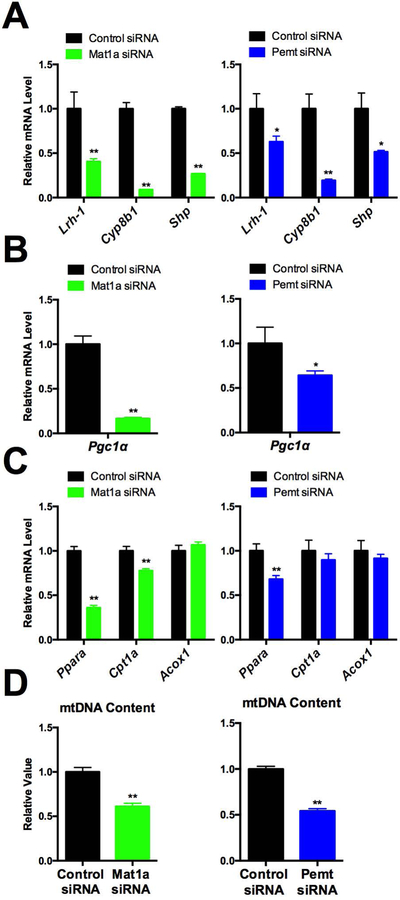
To directly assess the predicted role of the PEMT pathway, we knocked down either Mat1a or Pemt with siRNA in C3A/HepG2 cells. As expected, Mat1a or Pemt KD increased mRNA expression of two Kennedy pathway related enzymes, Chka and Pcyt1a (Supplementary Fig. 6A, 4B). Pemt KD did not significantly affect overall PC levels but did decrease levels of a large number of individual PC species, while increasing few others (Supplemental Fig. 11). Interestingly, the identities of the PC species affected by Pcyt1a KD and Pemt KD were quite different (Supplemental Fig. 13).
In accord with the prediction that the PEMT pathway is responsible for generating endogenous LRH-1 agonists, Mat1a or Pemt KD decreased LRH-1 target gene expression, including Cyp8b1 and Shp, as well as that of Lrh-1 itself (Fig. 4A). Despite the 40% reduction in Lrh-1 expression upon Pemt KD, the decrease in Cyp8b1 was twice as much, 80% (Fig 4A). Furthermore, we found that mitochondrial biogenesis and beta-oxidation genes, Pgc-1α and Ppara are reduced by either Mat1a or Pemt KD, and Cpt1a is reduced by Mat1a KD (Fig. 4B, 4C). We also found that mitochondrial DNA content levels are reduced in both Mat1a and Pemt KD cells (Fig. 4D). Importantly, treatment with the exogenous agonist DLPC restored the expression of LRH-1 target genes, including Shp, Pgc-1α, Cpt1a and Acox1, that were downregulated by Mat1a KD, although reduced mitochondria DNA content was not rescued (Supplementary Fig. 7).
Figure 4. PEMT pathway positively regulates the effect of Lrh-1 on mitochondria.
A. Expression of Lrh-1 and its target genes including was measured in Mat1a KD and Pemt KD in C3A/HepG2 cells. B. Expression of Pgc1α was measured in Mat1a KD and Pemt KD C3A/HepG2 cells. C. Expression of beta-oxidation related genes was measured in Mat1a KD and Pemt KD C3A/HepG2 cells. D. Mitochondrial DNA copy number was measured in Mat1a KD and Pemt KD C3A/HepG2 cells.
Together, these results indicate that the PEMT pathway specifically contributes to the production of PC agonists for LRH-1 activation, and also expression of Lrh-1 itself.
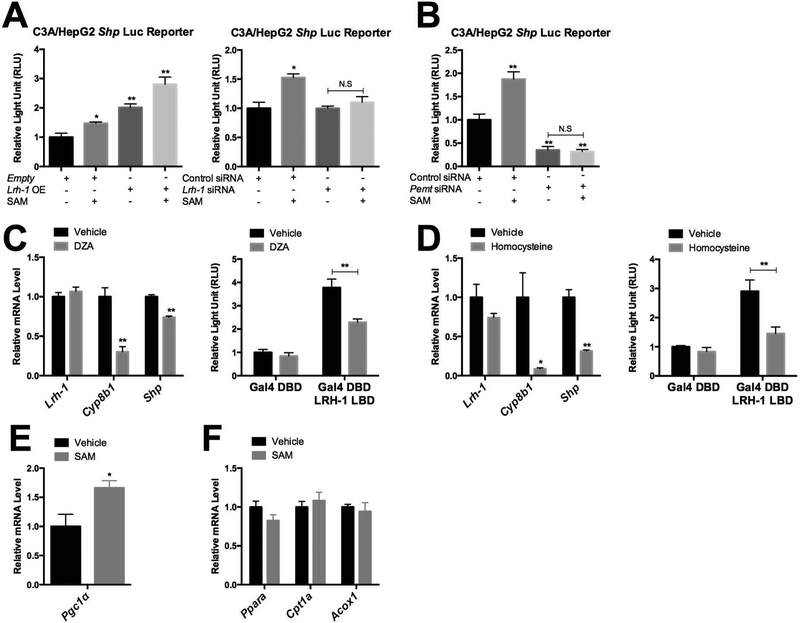
SAM supplementation increases LRH-1 transactivation in a Pemt dependent manner
The PEMT pathway requires SAM to synthesize PC from PE. Given the importance of the PEMT pathway in regulating LRH-1 activities, we tested whether and how SAM contributes to LRH-1 transactivation. We first used a luciferase reporter driven by the LRH-1 responsive Shp promoter, and showed that SAM supplementation increases luciferase activity by 1.5 fold in C3A/HepG2 cells that endogenously express Pemt and Lrh-1. When the cells were transiently transfected with an LRH-1 expression vector, basal luciferase activity increased, as expected, and SAM supplementation further increased reporter activity by 1.5 fold compared to vehicle treated LRH-1 expressing cells (Fig. 5A). In contrast, in Lrh-1 KD C3A/HepG2 cells SAM supplementation did not induce luciferase reporter activity, confirming Lrh-1 dependence of this response (Fig. 5A). Moreover, SAM supplementation also failed to induce luciferase reporter activity in Pemt KD cells (Fig. 5B), suggesting that the increased LRH-1 transactivation in response to SAM is also Pemt dependent. In addition, SAM supplementation did not alter total PC levels, but did increase and decrease levels of a large number of individual PC species (Supplementary Fig. 12). The identities of the PC species affected by SAM supplementation were quite different from those affected by Pcyt1a or Pemt KD (Supplementary Fig 13).
Figure 5. SAM mediates the transactivation of LRH-1 in a Pemt dependent manner.
A. Shp luc reporter activity was measured upon 10hrs of SAM supplementation (100μM) to Lrh-1 or empty vector transfected C3A/HepG2 cells. Shp luc reporter activity was measured upon 10hrs of SAM supplementation (100μM) to Lrh-1 KD C3A/HepG2 cells. B. Shp luc reporter activity was measured upon 10hrs of SAM supplementation (100μM) to Pemt KD C3A/HepG2 cells. C. Expression of Lrh-1 and Lrh-1 target gene expression was measured upon DZA (30μM) treatment for 6hrs. LRH-1 ligand binding domain fused Gal4 DBD reporter activity was measured upon DZA (10μM) treatment for 6hrs after C3A/HepG2 cells were transfected with luciferase reporter vectors for 12hrs. D. Expression of Lrh-1 and Lrh-1 target gene expression was measured upon homocysteine (5mM) treatment for 8hrs. LRH-1 ligand binding domain fused Gal4 DBD reporter activity was measured upon homocysteine (2mM) treatment for 6hrs after C3A/HepG2 cells were transfected with luciferase reporter vectors for 12hrs. E. Expression of Pgc-1α was measured in SAM (200μM) or vehicle treated C3A/HepG2 cells for 48hrs. F. Expression of beta-oxidation related genes was measured in SAM (200μM) or vehicle treated C3A/HepG2 cells for 48hrs.
To further support the link between SAM and PEMT in the induction of LRH-1 target gene expression, we used both 3-Deazaadenosine (DZA), a specific inhibitor of PEMT enzymatic activity (22), and also a high dose acute homocysteine treatment, which disrupts the flow from SAM to SAH (17). We found that both DZA and acute homocysteine treatments decrease mRNA expression of Lrh-1 target genes including Cyp8b1 and Shp (Fig. 5C, Fig. 5D). Next, we fused the Lrh-1 ligand binding domain to the Gal-4 DNA binding domain to drive an appropriate luciferase reporter and examined the effects of DZA and acute homocysteine treatments on LRH-1 transactivation. We found that both treatments reduced luciferase reporter activity (Fig. 5C, Fig. 5D). Expression of several other LRH-1 target genes, including Apoa1, Scarb1, Gls2 and Mdr3, was also reduced upon DZA or homocysteine treatment. In the opposite direction, however, only ApoA1 and Gls2 showed induction upon SAM treatment, which might be due to different transcriptional regulation or negative feedback mediated by Shp. Together, these results support that the activity of LRH-1 is regulated by SAM through the PEMT pathway.
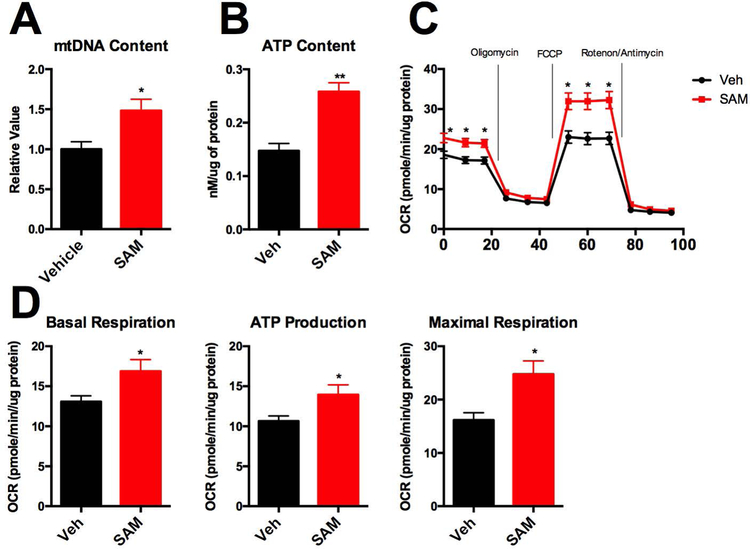
To characterize whether SAM also regulates mitochondrial activities, we measured mitochondrial biogenesis and beta-oxidation genes. Interestingly, we found that SAM administration to C3A/HepG2 cells induces Pgc-1α expression levels by 1.5 fold (Fig. 5E). On the other hand, beta-oxidation related gene expression was not significantly changed (Fig. 5F), which might be due to other effects of SAM on lipid metabolism and/or transcriptional regulation. In accord with the induction of Pgc-1α expression (Fig. 5E), SAM administration to C3A/HepG2 cells increased mitochondrial DNA content by 1.5 fold (Figure 6A). As a result of increased mitochondrial biolgenesis, SAM administration increased ATP levels by 2 fold in C3A/HepG2 cells (Figure 6B), and also increased basal respiration and maximal respiration (Figure 6C, 6D).
Figure 6. SAM supplementation promotes mitochondrial biogenesis and mitochondria functions.
A. Mitochondrial DNA copy number was measured from SAM (200 μM) or vehicle treated C3A/HepG2 cells for 48hrs. B. ATP content was measured after SAM (200 μM) treatment for 24hrs. C. Oxygen Consumption Rate (OCR) was measured in C3A/HepG2 cells after SAM (200 μM) treatment for 24hrs. Basal respiration, ATP production, and Maximal Respiration were quantified.
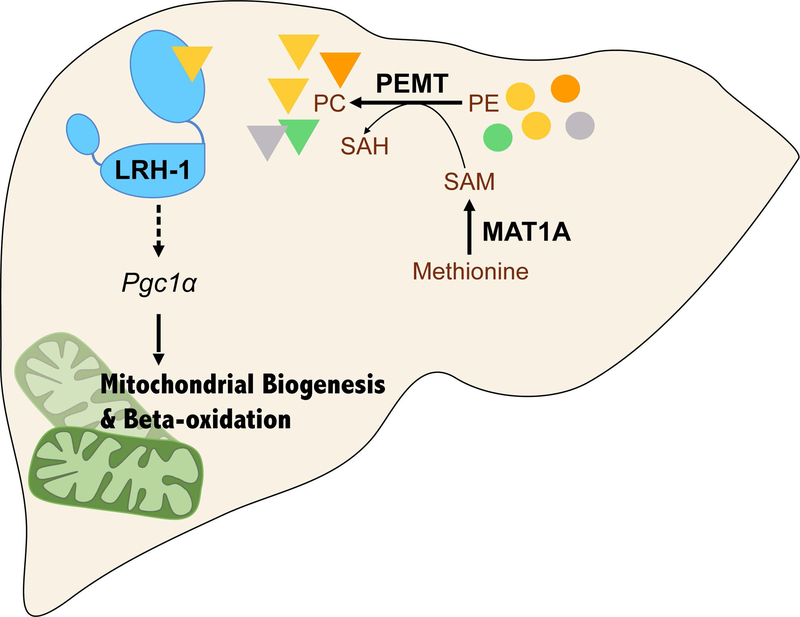
Together, our studies support a model in which the one-carbon metabolic cycle regulates SAM synthesis through Mat1a, and SAM is then fed into PC synthesis through the PEMT pathway. Specific PC species generated by PEMT act as agonists to transactivate LRH-1 and consequently regulate mitochondrial biogenesis and beta-oxidation (Fig. 7A).
Figure 7. PEMT-mediated PC synthesis trans-activates LRH-1 and promotes mitochondrial biogenesis and beta-oxidation.
A. Working Model. SAM generated by MAT1a is used by PEMT to synthesize specific PC species, which act as agonist ligands to transactivate LRH-1 to induce mitochondrial biogenesis and beta-oxidation in hepatocytes.
Discussion:
Our previous results linked LRH-1 and its C. elegans homolog NHR-25 to fat metabolism and mitochondrial dynamics (25). Here we confirm this association and extend it to mitochondrial biogenesis and beta-oxidation. We also define a new SAM – PEMT – LRH-1 pathway in which SAM metabolism couples with the PEMT pathway to tune the LRH-1-mediated regulation of mitochondrial activities. This mechanism provides a molecular basis for our previous finding that LRH-1 activity is decreased in the livers of mice fed a methionine and choline deficient diet, suggesting that LRH-1 functions as both a sensor and a regulator of methyl pool homeostasis (24). Consistently, activity of the C. elegans LRH-1 relative NHR-25 is also sensitive to the environmental methyl pool that determines endogenous SAM levels and PC synthesis through the PEMT pathway (25). Beyond the link between NHR-25 and mitochondrial morphological dynamics, our new results uncover a role for LRH-1 in regulating mitochondrial biogenesis and beta-oxidation, which are directly linked with fat metabolism.
A previous report described altered mitochondrial function in global Pemt KO livers (31). In accord with our studies, mitochondrial area was decreased. Gluconeogenesis was significantly compromised, but in hepatocytes maintained in the absence of fatty acids glycolysis was increased. This increase was correlated with the expected decrease in the mitochondrial PC/PE ratio. Expression of mitochondrial biogenesis and fatty acid oxidation genes was not altered, perhaps due to compensatory adaptations in mitochondrial functions that are not directly linked to Pemt deficiency. In our studies, it is likely that acute disruption of PEMT by KD reveals direct effects of PEMT deficiency on mitochondrial functions. Importantly, our results from Mat1a KD and SAM supplementation are consistent with those from Pemt KD, providing strong support for the role of the SAM – PEMT – LRH-1 pathway in regulating mitochondrial functions.
Many previous studies link increased SAM levels to reduced fat accumulation in the liver, and decreased SAM levels to the opposite. Decreasing the SAM/SAH ratio or depleting the methyl pool causes fat accumulation in hepatocytes. This has been previously attributed to a reduction in VLDL mediated lipid export related to decreased PEMT activity (20). Based on our findings, we propose an additional mechanism, in which decreased methyl-pool or SAM levels decrease levels of endogenous LRH-1 agonist(s) and therefore decrease mitochondrial biogenesis and beta-oxidation, leading to fat accumulation. A high level of homocysteine, homocysteinemia, is often observed in the serum of NAFLD patients (32). High homocysteine levels inhibit SAM to SAH flow (33). Based on our result that interrupting SAM to SAH flow by Mat1a KD or acute homocysteine treatments decreases Lrh-1 signaling (Fig. 4A, Fig. 5D), it is possible that NAFLD patients with homocysteinemia might have defective Lrh-1 signaling and mitochondrial malfunction.
A recent report identified mammalian target of rapamycin complex1 (mTORC1) as another downstream target of SAM (34). SAM is directly sensed by an inhibitory mTORC1 binding partner called SAMTOR. SAM binding dissociates SAMTOR from mTORC1 and activates mTORC1 signaling. In the opposite direction, methionine starvation depletes SAM and inactivates mTORC1 signaling. Activation of mitochondrial energy metabolism in coordination with ESRRA (ERRα, NR3B1) (35) is one of the many pathways regulated by mTORC1. Our results suggest that the SAM – PEMT - LRH-1 pathway could reinforce this mTORC1 function by both increasing SAM levels and augmenting mitochondrial number and function.
Although the Kennedy pathway is thought to account for approximately 70% of PC production in hepatocytes, it is clearly dispensable for generation of the putative, but still unknown endogenous PC agonists of LRH-1. Instead, LRH-1 transactivation is increased upon the disruption of the Kennedy pathway. On the other hand, inhibition of the PEMT pathway by Mat1a or Pemt KD decreased LRH-1, strongly implicating the SAM - PEMT pathway in LRH-1 transactivation and providing a direct mechanism for LRH-1 response to the labile methyl pool. It is interesting that this pathway preferentially produces PC species with longer fatty acid side chains (36), suggesting that such species may be preferential LRH-1 agonists. However, our biochemical studies agree with previous results (37) that many different PC species are able to bind the LRH-1 ligand binding domain in vitro (data not shown). Furthermore, the lipidomics profiling failed to identify any specific candidate PC species that fit the predicted LRH-1 agonist profile: increased by Pcyt1a KD and SAM supplementation, but decreased by Pemt KD (Supplementary Fig. 13). Therefore, more functional studies are required to address the important question regarding the nature of endogenous LRH-1 agonists. Particularly in light of previous results with phosphoinositols (5, 6), it is likely that LRH-1 has multiple agonists, potentially with distinct regulatory effects.
When regulating mitochondrial biogenesis, LRH-1 activation induces an increase in mitochondrial DNA content that is associated with an increase in the expression of PGC-1α. PGC-1α is not only a well known inducer of mitochondrial biogenesis (38), but also an LRH-1 coactivator (9). This suggests the existence of a self-supporting feed forward loop in which LRH-1 activation reinforces its own transactivation via increased activity of its key coregulator. The identification of Pemt (39, 40) for PC production, and potentially several genes for generation of specific longer chain unsaturated PC species (11) as LRH-1 target gene suggests an additional loop in which LRH-1 induces expression of its own agonist(s). Upon fasting, PGC-1α levels are induced in response to PKA activation of CREB (38), and PGC-1α activity can also be increased through phosphorylation by AMPK and de-acetylation by SIRT1 (41). Thus, increased PGC-1α activity in response to nutrient deprivation could provide an additional nutritional input to regulate LRH-1 transactivation. It is intriguing that the potential PGC-1α agonist feed forward loops are strongly opposed by the previously described role of LRH-1 as an inducer of its potent negative regulator SHP (8). The existence of both positive and negative feedback loops substantially increases the complexity of LRH-1 regulatory circuits, with difficult to predict implications for the magnitudes of responses and their kinetics over time.
In summary, we conclude that Lrh-1, together with PEMT mediated PC synthesis, is required for mitochondrial biogenesis and beta-oxidation. This highlights Lrh-1 as a potential target to manipulate mitochondrial activities, with beneficial impact on type 2 diabetes and other diseases associated with elevated liver fat.
Supplementary Material
Acknowledgements:
We thank Dr. Misun Kim, Dr. Jae Man Lee, Dr. Jennifer Mamrosh and Ying Zhou for discussion and technical support. We also thank Drs. Mangelsdorf and Kliewer from UT Southwestern for Lrh-1 fl/fl mice.
Financial Support:
This work was supported by the R.P. Doherty Jr. – Welch Chair in Science Q-0022 (to DDM), NIH grants DPDK113644, R01AG045183, R01AT009050 (to MCW), National Research Foundation of Korea (NRF) grants NRF-2018R1A2A3075389, NRF-2016M3A9B6902871 (to JMS). SC and DDM were supported by USDA ARS 3092-5-001-057.
Abbreviations:
- LRH-1
liver receptor homolog-1
- Pgc-1α
peroxisomal proliferator gamma coactivator-1-alpha
- Cpt1a
carnitine palmitoyltransferase 1-alpha
- Pdk4
pyruvate dehydrogenase kinase 4
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- Chka
Choline Kinase alpha
- Pcyt1a
Phosphate Cytidylyltransferase 1alpha
- Mat1a
methionine adenosyltransferase 1a
- NAFLD
non-alcoholic fatty liver disease
- Pemt
phosphatidylethanolamine-n-methyltransferase
References:
- 1.Ueda H, Sun GC, Murata T, Hirose S. A novel DNA-binding motif abuts the zinc finger domain of insect nuclear hormone receptor FTZ-F1 and mouse embryonal long terminal repeat-binding protein. Mol Cell Biol 1992;12:5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortlund EA, Lee Y, Solomon IH, Hager JM, Safi R, Choi Y, Guan Z, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol 2005;12:357–363. [DOI] [PubMed] [Google Scholar]
- 3.Musille PM, Pathak M, Lauer JL, Hudson WH, Griffin PR, Ortlund EA. Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation. Nat Struct Mol Biol 2012;19:532–S532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JM, Lee YK, Mamrosh JL, Busby SA, Griffin PR, Pathak MC, Ortlund EA, et al. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature 2011;474:506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 2005;120:343–355. [DOI] [PubMed] [Google Scholar]
- 6.Sablin EP, Blind RD, Uthayaruban R, Chiu HJ, Deacon AM, Das D, Ingraham HA, et al. Structure of Liver Receptor Homolog-1 (NR5A2) with PIP3 hormone bound in the ligand binding pocket. J Struct Biol 2015;192:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science 1996;272:1336–1339. [DOI] [PubMed] [Google Scholar]
- 8.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 2000;6:517–526. [DOI] [PubMed] [Google Scholar]
- 9.Mays SG, Okafor CD, Tuntland ML, Whitby RJ, Dharmarajan V, Stec J, Griffin PR, et al. Structure and Dynamics of the Liver Receptor Homolog 1-PGC1alpha Complex. Mol Pharmacol 2017;92:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahini N, Borlak J. Genomics of human fatty liver disease reveal mechanistically linked lipid droplet-associated gene regulations in bland steatosis and nonalcoholic steatohepatitis. Transl Res 2016;177:41–69. [DOI] [PubMed] [Google Scholar]
- 11.Miranda DA, Krause WC, Cazenave-Gassiot A, Suzawa M, Escusa H, Foo JC, Shihadih DS, et al. LRH-1 regulates hepatic lipid homeostasis and maintains arachidonoyl phospholipid pools critical for phospholipid diversity. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oosterveer MH, Mataki C, Yamamoto H, Harach T, Moullan N, van Dijk TH, Ayuso E, et al. LRH-1-dependent glucose sensing determines intermediary metabolism in liver. J Clin Invest 2012;122:2817–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt AN. Dynamic lipidomics of the nucleus. J Cell Biochem 2006;97:244–251. [DOI] [PubMed] [Google Scholar]
- 14.Watkins SM, Zhu X, Zeisel SH. Phosphatidylethanolamine-N-methyltransferase activity and dietary choline regulate liver-plasma lipid flux and essential fatty acid metabolism in mice. J Nutr 2003;133:3386–3391. [DOI] [PubMed] [Google Scholar]
- 15.Vance DE. Phospholipid methylation in mammals: from biochemistry to physiological function. Biochim Biophys Acta 2014;1838:1477–1487. [DOI] [PubMed] [Google Scholar]
- 16.Noga AA, Vance DE. A gender-specific role for phosphatidylethanolamine N-methyltransferase-derived phosphatidylcholine in the regulation of plasma high density and very low density lipoproteins in mice. J Biol Chem 2003;278:21851–21859. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Vance DE. Phosphatidylcholine and choline homeostasis. J Lipid Res 2008;49:1187–1194. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs RL, Devlin C, Tabas I, Vance DE. Targeted deletion of hepatic CTP:phosphocholine cytidylyltransferase alpha in mice decreases plasma high density and very low density lipoproteins. J Biol Chem 2004;279:47402–47410. [DOI] [PubMed] [Google Scholar]
- 19.Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, Avila MA, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A 2001;98:5560–5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cano A, Buque X, Martinez-Una M, Aurrekoetxea I, Menor A, Garcia-Rodriguez JL, Lu SC, et al. Methionine adenosyltransferase 1A gene deletion disrupts hepatic very low-density lipoprotein assembly in mice. Hepatology 2011;54:1975–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alonso C, Fernandez-Ramos D, Varela-Rey M, Martinez-Arranz I, Navasa N, Van Liempd SM, Lavin Trueba JL, et al. Metabolomic Identification of Subtypes of Nonalcoholic Steatohepatitis. Gastroenterology 2017;152:1449–1461 e1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noga AA, Zhao Y, Vance DE. An unexpected requirement for phosphatidylethanolamine N-methyltransferase in the secretion of very low density lipoproteins. J Biol Chem 2002;277:42358–42365. [DOI] [PubMed] [Google Scholar]
- 23.Song J, da Costa KA, Fischer LM, Kohlmeier M, Kwock L, Wang S, Zeisel SH. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD). FASEB J 2005;19:1266–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner M, Choi S, Panzitt K, Mamrosh JL, Lee JM, Zaufel A, Xiao R, et al. Liver receptor homolog-1 is a critical determinant of methyl-pool metabolism. Hepatology 2016;63:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin CJ, Wang MC. Microbial metabolites regulate host lipid metabolism through NR5A-Hedgehog signalling. Nat Cell Biol 2017;19:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mamrosh JL, Lee JM, Wagner M, Stambrook PJ, Whitby RJ, Sifers RN, Wu SP, et al. Nuclear receptor LRH-1/NR5A2 is required and targetable for liver endoplasmic reticulum stress resolution. Elife 2014;3:e01694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 2015;32:678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res 2010;2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benod C, Carlsson J, Uthayaruban R, Hwang P, Irwin JJ, Doak AK, Shoichet BK, et al. Structure-based discovery of antagonists of nuclear receptor LRH-1. J Biol Chem 2013;288:19830–19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui Z, Houweling M. Phosphatidylcholine and cell death. Biochim Biophys Acta 2002;1585:87–96. [DOI] [PubMed] [Google Scholar]
- 31.van der Veen JN, Lingrell S, da Silva RP, Jacobs RL, Vance DE. The concentration of phosphatidylethanolamine in mitochondria can modulate ATP production and glucose metabolism in mice. Diabetes 2014;63:2620–2630. [DOI] [PubMed] [Google Scholar]
- 32.de Carvalho SC, Muniz MT, Siqueira MD, Siqueira ER, Gomes AV, Silva KA, Bezerra LC, et al. Plasmatic higher levels of homocysteine in non-alcoholic fatty liver disease (NAFLD). Nutr J 2013;12:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem 2000;275:29318–29323. [DOI] [PubMed] [Google Scholar]
- 34.Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, et al. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017;358:813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaveroux C, Eichner LJ, Dufour CR, Shatnawi A, Khoutorsky A, Bourque G, Sonenberg N, et al. Molecular and genetic crosstalks between mTOR and ERRalpha are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell Metab 2013;17:586–598. [DOI] [PubMed] [Google Scholar]
- 36.Pynn CJ, Henderson NG, Clark H, Koster G, Bernhard W, Postle AD. Specificity and rate of human and mouse liver and plasma phosphatidylcholine synthesis analyzed in vivo. J Lipid Res 2011;52:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musille PM, Kossmann BR, Kohn JA, Ivanov I, Ortlund EA. Unexpected Allosteric Network Contributes to LRH-1 Co-regulator Selectivity. J Biol Chem 2016;291:1411–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puigserver P Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha. Int J Obes (Lond) 2005;29 Suppl 1:S5–9. [DOI] [PubMed] [Google Scholar]
- 39.Chong HK, Biesinger J, Seo YK, Xie X, Osborne TF. Genome-wide analysis of hepatic LRH-1 reveals a promoter binding preference and suggests a role in regulating genes of lipid metabolism in concert with FXR. BMC Genomics 2012;13:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong HK, Infante AM, Seo YK, Jeon TI, Zhang Y, Edwards PA, Xie X, et al. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res 2010;38:6007–6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta 2011;1813:1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.