Abstract
Epigenetic transgenerational inheritance potentially impacts disease etiology, phenotypic variation, and evolution. An increasing number of environmental factors from nutrition to toxicants have been shown to promote the epigenetic transgenerational inheritance of disease. Previous observations have demonstrated that the agricultural fungicide vinclozolin and pesticide DDT (dichlorodiphenyltrichloroethane) induce transgenerational sperm epimutations involving DNA methylation, ncRNA, and histone modifications or retention. These two environmental toxicants were used to investigate the impacts of parent-of-origin outcross on the epigenetic transgenerational inheritance of disease. Male and female rats were collected from a paternal outcross (POC) or a maternal outcross (MOC) F4 generation control and exposure lineages for pathology and epigenetic analysis. This model allows the parental allelic transmission of disease and epimutations to be investigated. There was increased pathology incidence in the MOC F4 generation male prostate, kidney, obesity, and multiple diseases through a maternal allelic transmission. The POC F4 generation female offspring had increased pathology incidence for kidney, obesity and multiple types of diseases through the paternal allelic transmission. Some disease such as testis or ovarian pathology appear to be transmitted through the combined actions of both male and female alleles. Analysis of the F4 generation sperm epigenomes identified differential DNA methylated regions (DMRs) in a genome-wide analysis. Observations demonstrate that DDT and vinclozolin have the potential to promote the epigenetic transgenerational inheritance of disease and sperm epimutations to the outcross F4 generation in a sex specific and exposure specific manner. The parent-of-origin allelic transmission observed appears similar to the process involved with imprinted-like genes.
Keywords: Transgenerational, Epigenetic inheritance, Disease, DNA methylation, Allelic transmission, Vinclozolin, DDT
1. Introduction
Epigenetic transgenerational inheritance of disease has been the focus of numerous studies. Several hundred articles have been published on the topic of “environmentally induced epigenetic transgenerational inheritance and phenotype” by numerous laboratories and models to demonstrate the phenomenon. Previous observations have shown that exposing a gestating female rat to environmental toxicants such as the agricultural fungicide vinclozolin and the pesticide DDT (dichlorodiphenyltrichloroethane), during the fetal gonadal sex determination promotes a reprogramming of the male germline epigenome (Anway et al., 2005; Skinner et al., 2013). Stress, caloric restriction, high fat diets, and different toxicants have also been shown to promote the epigenetic transgenerational inheritance of disease (Soubry, 2015; Vaiserman et al., 2017; Nilsson et al., 2012). DNA methylation alterations in the sperm appear to become permanently reprogrammed, and create an abnormal epigenome in the embryo and stem cells to subsequently affect all somatic cells and tissues (Guerrero-Bosagna et al., 2010). An increased disease susceptibility develops at the adult stage associated with mammary tumors, prostate disease, kidney disease, testis abnormalities, and ovarian disease (Anway et al., 2006a; McBirney et al., 2017). The germline then transmits this altered epigenome and adult onset disease phenotype to subsequent generations (Anway et al., 2005; Nilsson et al., 2018a).
Researchers studying developmental epigenetics have been successful in linking maternal exposures (e.g. nutrition) to prenatal origins of health or behavior in the offspring (Heijmans et al., 2008; Painter et al., 2008; Tobi et al., 2009). Epigenetic modifications of the germline (sperm and egg) provides the molecular basis and origin of this non-genetic form of inheritance. The germline, by passing an altered epigenome to the early embryo stem cells, can then impact the transcriptomes and epigenetics of all subsequently derived somatic cells (Anway et al., 2005; Nilsson et al., 2018a). Epigenetics is defined as “molecular factors or processes around DNA that regulate genome activity independent of DNA sequence and which are mitotically stable” (Skinner et al., 2010). Different epigenetic molecular factors and processes include DNA methylation (Holliday and Pugh, 1975), histone modifications (Turner, 1998), non-coding RNAs (Jodar et al., 2013; Mattick, 2009), chromatin structure (Yaniv, 2014), and RNA methylation (Schaefer et al., 2010). These epigenetic processes are critical for cell type specificity and development, as well as for allowing an organism to adapt to its environment with changes in gene expression. Environmentally induced epigenetic transgenerational inheritance is found in all species investigated including plants (Quadrana and Colot, 2016), insects (Brookheart and Duncan, 2016), fish (Carvan et al., 2017), birds (Leroux et al., 2017), and mammals such as humans (Northstone et al., 2014). Numerous studies have demonstrated that these ancestral exposures can promote a variety of transgenerational pathologies and phenotypic variations (Skinner, 2014).
Exposure of a gestating female rat (F0 generation) to environmental factors also exposes the F1 generation fetus and its germline, which will generate the F2 generation. Therefore, the F3 generation becomes the first transgenerational generation with no direct exposure. Preconception adult exposures can also promote epigenetic transgenerational inheritance (Lane et al., 2015; Vassoler et al., 2014). In a variety of studies, these transgenerational epimutations in sperm have appeared to be exposure specific, indicating their possible utilization as biomarkers for ancestral toxicant exposure (Manikkam et al., 2012). Therefore, the current study utilized two different exposures, and the outcross of the transgenerational F3 generation to the F4 generation in order to investigate the parent-of-origin allelic transmission of the epigenetic alterations and pathology.
While most of the studies have focused on this first transgenerational generation (F3), this study centers on the parental origin of disease and epimutations. This is done by outcrossing the F3 generation females with F3 generation wild type males to generate an F4 generation maternal outcross (MOC) generation. Outcrossing the F3 generation males with F3 generation wild type females generates an F4 generation paternal outcross (POC) generation. This laboratory model makes it possible to determine if the paternal (sperm) or the maternal (egg) epigenome is responsible for passing down specific diseases, pathologies, and epimutations to the next generation. A previous study using methoxychlor suggested that there was a parental transgenerational transmission of disease through the female or male germline (Manikkam et al., 2012). The current study demonstrates that there are parental origins of transgenerational disease, and epimutations both through the female and the male germline. The transgenerational pathology and epimutations observed in the F3 generation intercross DDT or vinclozolin, is reduced in the F4 outcross generation in a parental allelic manner. Therefore, as observed with other epigenetic processes such as imprinted genes (Kelsey and Feil, 2013), a parent-of-origin allelic component of the epigenetic transgenerational inheritance phenomenon is shown.
2. Results
2.1. Pathology analysis
The experimental design (Fig. 1) involved a daily transient exposure of gestating female F0 generation rats during embryonic days E8–E14 to vinclozolin or DDT in dimethylsulfoxide (DMSO) as previously described (Anway et al., 2005). A separate control generation lineage was exposed during days E8–E14 of gestation to vehicle DMSO alone. Six different unrelated F0 generation gestating females from different litters for each control, vinclozolin or DDT lineage were used. The F1 generation offspring were obtained and aged to postnatal 90-day of age, and randomly selected males and females from different litters bred within the specific lineage. The F2 generation offspring were obtained and aged to 90 days, and similarly bred to unrelated males and females from different litters within the same lineage in order to generate the F3 generation. The F3 generation offspring were aged to 90 days, and males and females from different litters were selected to be outbred with either wild type males or wild type females to generate the F4 paternal outcross (POC) generation and the F4 maternal outcross (MOC) generation, Fig. 1. No sibling or cousin breeding was used to avoid any inbreeding artifacts. All the animals were sacrificed at 1 year of age for epididymal sperm collection and histopathology analysis of different tissues.
Fig. 1.
Animal breeding scheme. An intercross with the population for the F1, F2 and F3 generations and the outcross to the wild-type (green dot) for the F4 generation through paternal or maternal lines (red dot) outcross lineage. The F3 generation males from different litters were selected to be outbred with wild type females to generate the F4 paternal outcross (POC) generation, and F3 generation females from different litters were selected to be outbred with wild type males to generate the F4 maternal outcross (MOC) generation.
The testis, ovaries, prostate, kidney, and adipose tissue were collected and examined for histopathologies. To assess if there was any direct fetal exposure toxicity to vinclozolin or DDT, the litter sizes, sex ratios, and weaning body weights were measured. No significant changes were observed in the multigenerational F1 or F2 generations, or in the transgenerational F3 generation, indicating no detection of overt toxicity. However, in the F4 generation POC for DDT and vinclozolin, the male and female offspring had decreased weaning weights, Supplemental Fig. S1.
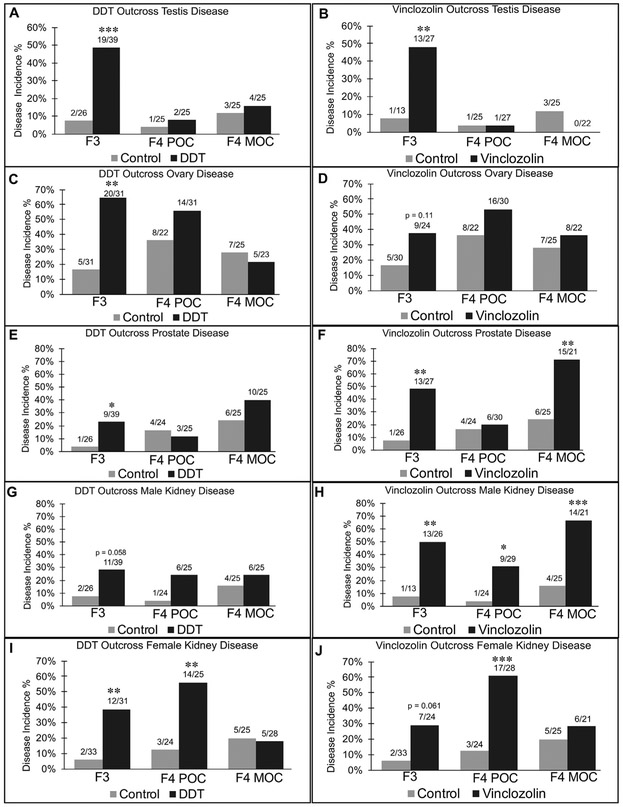
The histopathology analysis was based on examination of stained paraffin sections of isolated tissues, as described in the Methods (Skinner et al., 2013). Testis disease was characterized by the presence of histopathologies including azoospermia, atretic seminiferous tubules, presence of vacuoles in basal regions of seminiferous tubules, sloughed germ cells in the lumen of seminiferous tubules, and lack of seminiferous tubal lumen (Anway et al., 2005, 2006a, 2006b). The incidence of testis disease was higher in the vinclozolin F3 generation animals, which was not seen in the vinclozolin F4 POC and F4 MOC generations (Fig. 2B). The same phenomenon was observed for testis disease between the DDT F3 generation animals and the DDT F4 POC and F4 MOC generations (Fig. 2A). Similar results were observed with ovarian disease, Fig. 2C and D. Ovarian disease was characterized by an increase in the incidence of ovarian cysts and by a decrease in the number of oocytes present in primordial follicles (Nilsson et al., 2012). Therefore, ancestral toxicant exposure in both maternal and paternal sides appears to be required for testis and ovarian disease.
Fig. 2.
Pathology Analysis. Histological analysis of tissue sections for various histopathologies as described in the Supplemental Methods with the disease incidence (%) presented for the F3 generation, F4 generation paternal outcross (POC), and F4 generation maternal outcross (MOC) for control and DDT or vinclozolin exposure lineages. The number of individuals with disease versus total number of individuals examined is presented above each bar graph. A Fisher’s exact test was performed and indicates a p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***). (A) Testis disease DDT, (B) Testis disease vinclozolin, (C) Ovary disease DDT, (D) Ovary disease vinclozolin, (E) Prostate disease DDT, (F) Prostate disease vinclozolin, (G) Kidney disease male DDT, (H) Kidney disease male vinclozolin, (I) Kidney disease female DDT, and (J) Kidney disease female vinclozolin.
Prostate disease was characterized by atrophic or hyperplastic prostate glandular epithelium and the presence of epithelial vacuoles, as previously described (Anway and Skinner, 2008). The frequency of prostate disease was increased in the vinclozolin F3 generation compared to the control. The vinclozolin F4 generation MOC males also showed an increase in prostate disease, which suggests a maternal allelic transmission (Fig. 2F). The DDT lineage animals had an increased prostate disease frequency observed in the F3 generation animals which disappeared in the F4 POC generation and trended toward an increase in the F4 MOC generation, but it was not statistically significant (Fig. 2E).
Kidney disease was characterized by the presence of an increased number of proteinaceous fluid filled cysts, reduction in size of glomeruli, and thickening of Bowman’s capsules as previously described (Anway et al., 2006a; Manikkam et al., 2012). In the vinclozolin F4 POC and F4 MOC generation male offspring, an increase in kidney disease was observed, which suggests a paternal and maternal allelic transmission (Fig. 2H). However, no kidney disease was observed in the DDT F4 generation male POC and MOC offspring (Fig. 2G). In the DDT lineage animals, the F3 generation females displayed a significant increase of kidney disease incidence compared to the control. This kidney disease was maintained in the F4 POC generation female DDT and vinclozolin lineage animals (Fig. 2I and J). These results suggest a paternal transmission of kidney disease from father to daughter.
The adipocyte size has been found to be one of the most reliable parameters to assess metabolic disease (Chamorro-Garcia et al., 2013), and was found to increase significantly in the F3 generation DDT and vinclozolin lineage female and DDT male adipocytes (Fig. 3A-D). When the F3 generation was outcrossed, both the F4 generation MOC and POC vinclozolin and DDT lineage males were found to be obese (Fig. 3A and B), which suggests a maternal or paternal transmission of obesity to their male offspring. When the F3 generation was outcrossed the F4 generation POC vinclozolin lineage, but not the DDT lineage, females were found to be obese which suggests a paternal transmission of obesity to their female offspring (Fig. 3D). For the F3 generation DDT lineage, both males and females were found to be obese (Fig. 3A and C). Interestingly, the vinclozolin F3 generation males had negligible obesity, but both the MOC and POC F4 generation males had significant obesity (Fig. 3B). Therefore, the outcross had higher incidence of disease.
Fig. 3.
Pathology Analysis. Histological analysis of tissue sections for various histopathologies as described in the Supplemental Methods with the disease incidence (%) presented for the F3 generation, F4 generation paternal outcross (POC), and F4 generation maternal outcross (MOC) for control and DDT or vinclozolin exposure lineages. The number of individuals with disease versus total number of individuals examined is presented above each bar graph. A Fisher’s exact test was performed and indicates a p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***). (A) Obesity male DDT, (B) Obesity male vinclozolin, (C) Obesity female DDT, (D) Obesity female vinclozolin, (E) Multiple disease male DDT, (F) Multiple disease male vinclozolin, (G) Multiple disease female DDT, (H) Multiple disease female vinclozolin, (I) Mammary tumor female DDT, and (J) Mammary tumor female vinclozolin.
The incidence of multiple (≥2) types of diseases or pathologies per individual was assessed and referred to as multiple disease susceptibility (Fig. 3E-H). The DDT and vinclozolin F3 generation males and females all had significant multiple disease compared to the control lineage animals (Fig. 3E-H). The outcross F4 generation males had significant multiple disease in the MOC DDT and vinclozolin lineage (Fig. 3E and F), but no multiple disease in the POC. The outcross F4 generation females had increased multiple disease incidence in the POC and not the MOC (Fig. 3G and H). Therefore, the paternal lineage transmitted the female multiple disease and maternal lineage the male multiple disease.
The incidence of tumor development was evaluated. The primary tumors observed were mammary tumors as previously described (Anway et al., 2006a). In the current study, no significant differences in tumor development were observed in the F3 generation males or females (Fig. 3I and J and Supplemental Figs. S1E and F) for vinclozolin and DDT lineages. Negligible tumors were observed in the outcross F4 generation males (Supplemental Figs. S1E and F) for vinclozolin and DDT lineages. Interestingly, the female F4 generation MOC vinclozolin lineage outcross had a significant increase in female tumor development (Fig. 3J), but not the DDT lineage (Fig. 3I). Tumors were classified by veterinary pathologists at the Washington Animal Disease Diagnostic Laboratory (WADDL) at Washington State University. Tumors of F4 generation POC controls included two mammary fibroadenomas and one cutaneous fibrosarcoma. Tumors of F4 generation POC DDT lineage females included three mammary fibroadenomas and one pilomatricoma. Tumors of F4 generation POC vinclozolin lineage females included two mammary fibroadenomas and one mammary carcinoma. There were no tumors of F4 generation MOC control lineage animals. Tumors of F4 generation MOC DDT lineage females included three mammary fibroadenomas. Tumors of F4 generation MOC vinclozolin lineage females included three mammary fibroadenomas and one mammary adenoma.
WADDL performed full necropsies as required on animals that died prior to the time of scheduled sacrifice at one year. Findings for F4 generation POC control animals included a sialocoel (salivary cyst). Findings for F4 generation POC DDT lineage animals included a chronic inguinal abscess. Findings for F4 generation POC vinclozolin lineage animals included two cases with bloody vaginal discharge, both with uterine polyps or lumps. There were no early deaths among F4 generation MOC control animals. Findings for F4 generation MOC DDT lineage animals included four cases of renal disease with glomerulonephropathy and renal fibrosis, all from animals in the same litter. Findings for F4 generation MOC vinclozolin lineage animals included a case of peritonitis, and one death of unknown cause. A summary of the parental allelic transmission of disease is presented in Fig. 4A and for individual animal disease in Supplemental Tables S1-S3.
Fig. 4.
Summary paternal allelic transmission of disease and epimutations. (A) The diseases and the type of allelic transmission are presented for the DDT lineage and vinclozolin (Vinc) lineage F3 generation and F4 MOC or POC. The (+) indicates the presence and (−) the absence of a specific type of allelic transmission of disease (B) The epimutations (i.e. DMRs). overlapping with the p < 1e-05 F3 generation DMR with p < 0.05 F4 generation outcross DMR is presented for DMR number and percentage (%) of the original F3 generation DMRs. This was categorized for the POC, MOC, both alleles and not transmitted.
2.2. Sperm DNA methylation analysis
The differential DNA methylated regions (DMRs) between the control versus vinclozolin or DDT lineage male sperm were identified using a methylated DNA immunoprecipitation (MeDIP) followed by DNA next-generation sequencing (NGS) for an MeDIP-Seq analysis, as previously described (Ben Maamar et al., 2018). Initially, the sperm DNA was sheared to produce DNA fragments of 200–500 bp. A methylcytosine antibody was used to immunoprecipitate the methylated DNA fragments. Libraries were generated for sequencing 50-bp paired-end reads to assess differential levels (read depths) of DNA methylation. The DMRs for the F3 and F4 generation sperm were determined, and different threshold p values are presented in Fig. 5. A threshold p-value of p < 1e-05 was selected to compare all single 100-bp windows, as well as DMRs with multiple (≥2) adjacent windows, except for the paternal outcross (POC) vinclozolin sperm where a threshold p-value of p < 1e-07 was selected. The F4 generation MOC DDT, POC DDT and MOC vinclozolin sperm had a decreased number of DMRs compared to the F3 generation (Fig. 5A-E). The POC vinclozolin F4 generation showed an increased number of DMRs (Fig. 5F). The DMR lists are presented in the Supplemental Tables S4-S9.
Fig. 5.
DMR identification and overlap. The number of DMRs identify with different p-value cutoff thresholds. The all window column shows all DMRs and the multiple window column shows the number of adjacent 100 bp window within the DMRs containing at least two significant windows. Below is the number of DMR with each specific number of significant 100 bp windows at a p-value threshold of 1e-05 or 1e-07 indicated in bold. (A) F3 generation DDT, (B) F3 generation vinclozolin, (C) F4 generation MOC DDT, (D) F4 generation POC DDT, (E) F4 generation MOC vinclozolin, (F) F4 generation POC vinclozolin, (G) DMR overlap DDT, (H) DMR overlap vinclozolin, and (I) DMR overlap with comparison at p < 1e-05 or p < 1e-07 with other DMR sets at p < 0.05 statistical cut-off threshold with the DMR overlap number and associated percentage (%) of each comparison.
A comparison of the DMRs at p < 1e-05 for the F4 generation DDT and p < 1e-05 for the F3 generation DDT lineages demonstrated no or minimal overlap with a slightly higher level of overlap between the F3 and F4 MOC generations (Fig. 5G). Similarly, the DMRs comparison at p < 1e-05 for the F4 vinclozolin MOC and p < 1e-07 POC vinclozolin lineages also showed no or very few overlaps between the F3 and the F4 generations (Fig. 5H). In considering the high stringency threshold DMR identified, the majority of the alterations in sperm DNA methylation were unique between the generations for each exposure lineage group, and distinct between F4 generation MOC and POC.
A p-value of p < 1e-05 is usually used for the DNA methylation analysis which is very stringent. Further evaluation of the potential overlap of the DMRs between the F3 and F4 outcross generations used a reduced statistical threshold of p < 0.05 in the comparison when compared to the higher statistical threshold DMRs discussed above. Lowering the stringency to a p-value of p < 0.05 (which is statistically significant) allows the identification of more overlaps between the F4 POC generation, F4 MOC generation and the F3 generation. A comparison of the p < 1e-05, or p < 1e-07 for POC vinclozolin, with each potential comparison at p < 0.05, Fig. 5I, demonstrates a much higher overlap than shown in Fig. 5G and H. Observations indicate the outcross F4 generation DMRs appear to be derived from the F3 generation DMR in a parent-of-origin allelic manner, Fig. 5I. Analysis of the horizontal rows shows the overlapping DMR numbers and percentage (%) of the associated total DMR set at 100%, Fig. 5I. A higher overlap was observed between the F3 generation DDT and vinclozolin DMR sets of 85–90% at the low stringency comparison. A lower fraction of the F3 generation DMRs were observed in the POC or MOC suggesting a parent-of-origin allelic separation. For DDT the F4 generation POC had 11.7% overlap and MOC a 48.6% overlap. In contrast, the vinclozolin F4 generation POC had 53.2% overlap and MOC 22.2% overlap. In general, the POC were more in common with each other than the MOC, and the MOC more in common than the POC. Therefore, a parent-of-origin trend for each sex lineage. Observations indicate the inheritance of the DMRs through the parent-of-origin allele using this relaxed statistical stringency in the overlap. A follow up principle component analysis (PCA) for comparisons of all the DMR sets between the control and exposure lineage DMRs showed distinct clusters between the control and exposure lineage DMRs, Supplemental Fig. S2. Interestingly, the F3 generation control, vinclozolin, and DDT lineage DMRs clustered together and were distinct from the F4 generation outcross. The F4 generation POC from both vinclozolin and DDT lineages also clustered together and were relatively unique from the other DMR sets, Supplemental Fig. S2. The F4 generation MOC vinclozolin and DDT lineage DMRs also showed general clustering, but were more variable. The POC and MOC control lineage were similar but spread throughout PCA plot. Although some outliers are observed in the PCA (Supplemental Fig. S2), observations supported the overlap analysis presented in Fig. 5I.
The parent-of-origin transmission of the F3 generation vinclozolin lineage 632 DMRs at p < 1e-05 were found to segregate into the lower stringency (p < 0.05) F4 outcross generation with 300 DMRs through the POC, 90 DMRs through the MOC, and 78 were transmitted to both the POC and MOC and are in common, Fig. 4B. The parent-of-origin transmission of the F3 generation DDT lineage 2418 DMRs at p < 1e-05 were found to segregate into the F4 outcross generation with 156 DMRs through the POC, 1138 DMRs through the MOC, and 155 DMRs transmitted to both the POC and MOC and were in common, Fig. 4B. The F4 outcross generation DMR numbers have some replicate DMR overlaps so the total DMR numbers are higher than the total F3 generation DMRs. Therefore, as observed with the allelic transmission of disease, the DMRs also had a parent-of-origin allelic transmission of the majority of the F3 generation DMRs identified. Interestingly, the DDT lineage DMRs were predominantly transmitted through the MOC and vinclozolin lineage DMRs through the POC.
The different chromosomal locations of the DMRs for each generation and exposure lineage are shown in Fig. 6. Almost every chromosome displays DMRs as indicated by the red arrowheads, and some show clusters of DMRs indicated by black boxes. The CpG density of the DMRs for the F3 generation DDT and vinclozolin, F4 MOC and POC generations sperm is shown in Supplemental Figs. S4A-F. The CpG density of the DMRs reveals that the predominant density is one CpG per 100 bp with a range between 1 and 5 CpG. The DMRs lengths are presented in Supplemental Figs. S5A-F, and show that their predominant length is 1 kb with a range of 1–4 kb for each generation DMRs. Thus, the DMRs are associated with CpG deserts with 10–20 CpG within 1 kb (Skinner and Guerrero-Bosagna, 2014). This is typical when the DMRs are associated with CpG deserts which represents over 95% of the genome, while 5% of the genome DMRs are associated with high CpG density islands. The lists of DMRs with their chromosomal locations, size, and CpG density are presented in the Supplemental Tables S1-S6 for the F3 DDT (S1), F3 vinclozolin (S2), F4 MOC (S3) and POC (S4) DDT, and F4 MOC (S5) and POC (S6) vinclozolin generation DMRs, respectively. For the F3 generation DDT and vinclozolin DMRs, there was about 50% of the DMRs with an increase in methylation for the control and exposure lineages. The F4 generation vinclozolin MOC had 53% of the DMRs increase in DNA methylation in the exposure lineage and 47% of the DMRs increase in the control lineage. The F4 generation DDT MOC had 57% of the DMRs increase in DDT lineage and 43% of the DMRs increase in control lineage. The F4 generation vinclozolin POC had 31% of the DMRs increase in the vinclozolin and 69% of the DMRs in the control lineage. The F4 generation DDT POC had 63% of the DMRs increase in DNA methylation in the DDT lineage and 37% of the DMRs increase in the control lineage. The F4 generation DMR log-fold-change in DNA methylation is presented in Supplemental Tables S6-S9.
Fig. 6.
DMR chromosomal locations. The DMR locations on the individual chromosomes. All DMRs at a p-value threshold of p < 1e-05 are shown unless specified different. (A) F3 generation DDT, (B) F3 generation vinclozolin, (C) F4 generation MOC DDT, (D) F4 generation POC DDT, with DMRs at a p-value threshold of 1e-07. (E) F4 generation MOC vinclozolin, and (F) F4 generation POC vinclozolin, with DMRs at a p-value threshold of 1e-07. The red arrow head designates a DMR and black box a cluster of DMRs.
2.3. Epimutation gene associations
The Supplemental Tables S6-S9 provide the lists of DMRs for all of the epigenetic alterations (i.e. epimutations) identified. A genomic location was used to determine the known gene associations for these epimutations which are also listed. A 10 kb flanking distance was used to allow the promoter to be included in the analysis. All of the associated genes were classified by relevant functional categories, and are presented for each generation in Fig. 7A. The top five gene pathways containing multiple genes for the F4 POC and MOC DDT and vinclozolin generations are presented. Epimutations were found predominantly in the signaling and metabolism pathways for all of the different F4 outcrosses (Fig. 7B). Similar observations were made with the F3 generation DMRs.
Fig. 7.
DMR gene associations. (A) The DMR gene associations for POC and MOC DDT or vinclozolin lineage are presented with the number of DMR associated with each gene functional category. (B) The list of the top gene associated pathways for each DMR set are listed for comparison.
3. Discussion
The current study was designed to investigate the existence of a parent-of-origin allelic transmission (paternal or maternal) of disease, and sperm epimutations following ancestral exposures to vinclozolin or DDT in an outcross lineage (F4 MOC and F4 POC). Several studies have shown that DDT and vinclozolin can induce transgenerational disease such as kidney pathology, testis disease, ovarian disease, prostate abnormalities, and anxiety behaviors in the F3 generation (Anway and Skinner, 2008; Skinner et al., 2008). Other studies have also observed that these two environmental toxicants can affect the sperm epigenome by altering concurrently the ncRNA, the DMRs, and the differential histone retention sites (DHRs) in the intercross F1, F2 and F3 generations (Ben Maamar et al., 2018; Skinner et al., 2018). An intercross within the exposure lineage population with no sibling or cousin breeding to avoid inbreeding artifacts provides the optimal phenotypes (i.e. pathology) and germline epigenetic alterations, as previously described (Anway et al., 2005; Liu et al., 2018; Dapp et al., 2015). Since both paternal and maternal allelic contributions are required for the optimal epigenetic transgenerationally inherited phenotypes and epigenetics, the transgenerational F3 generation intercross was examined (Anway et al., 2005; Nilsson et al., 2018a). Comparing the transgenerational F3 generation from our previous studies with the F4 generation outcross model (Fig. 1) permits a discrimination between the paternal and maternal allelic transmission (i.e. contribution) for disease and sperm epimutations to be assessed.
Intercrossing is based on the well-known principle that continued intercrossing of a population (while avoiding inbreeding) will increase the frequency of occurrence of the phenotype (Green et al., 1963). Therefore, intercrossing from F1 to F3 generation animals allows us to assume that the contribution of both parents to the phenotype will be present. Thus, the F3 generation animals display the optimum phenotype to study different pathologies and associated epigenetic alterations (Fig. 1). The outcrossing of the transgenerational F3 generation to generate the F4 generation permits the analysis of the paternal or maternal allelic contribution to be assessed. Outcrossing the F3 generation females with wild type males to generate the F4 MOC allows the determination of the maternal allelic contribution. Similarly, outcrossing the F3 generation males with wild type females to generate the F4 POC will then allow the determination of the paternal allelic contribution. The parent-of-origin allelic transmission of epigenetic information has previously been shown to occur with imprinted genes (Mackay and Temple, 2017; Lawson et al., 2013). This is a critical element of the molecular control of paternal or maternal imprinted genes and allows one allele to influence the other, and promote the imprinted gene expression and function (Ideraabdullah et al., 2008). The environmental induction of epigenetic transgenerational inheritance through germline epigenetic modifications was initially proposed to act in an imprinted-like gene manner due to its protection during early embryonic development from DNA methylation erasure and parent-of-origin transgenerational phenotypes (Anway et al., 2005;Jirtle and Skinner, 2007). The current study identified potential parent-of-origin epimutations in sperm that are associated with unique transgenerational pathologies.
3.1. Parent-of-origin allelic transmission
The comparison of the transgenerational F3 generation, with the outcross to the F4 generation through the paternal or maternal lineages, allows an assessment of parent-of-origin transmission of disease or pathology. Observations (Fig. 4) provided examples of the following: 1) Pathology that required combined contribution of both paternal and maternal alleles to promote disease; 2) Pathology that is derived from the opposite sex allele such as father to daughter or mother to son; 3) Pathology that is derived from either parent-of-origin alleles independently; 4) Pathology that is transmitted within the same sex, such as maternal to daughter; and 5) Pathology that is observed only following a specific parent-of-origin outcross. Examples for each of these were observed and are discussed with any previous supporting literature.
Pathology that required the combined contribution of both paternal and maternal alleles are testis disease and ovarian disease (Fig. 2A-D). Although the F3 generation had testis disease, the F4 generation POC or MOC did not have testis disease. The baseline control levels of disease increased in the F4 outcross, so this is a limitation to consider. Previous literature has suggested both maternal and paternal exposures can impact testis disease, but no predominant paternal or maternal inheritance pattern has been reported (Carouge et al., 2016). Ovarian disease such as polycystic ovarian disease (PCO) or primary ovarian insufficiency (POI) have not been shown to be derived from a specific parent, but a recent study has suggested both paternal and maternal allelic contributions are important (Kobaly et al., 2014). Therefore, some pathologies appear to require the combined contributions of both alleles to effectively develop the specific disease.
The transmission altered epigenetics to the opposite sex offspring is characterized by disease in the F4 generation males from the MOC or disease in females from the POC. Several examples of this parent-of-origin allelic transmission were observed. Prostate disease in the F3 generation was transmitted through a MOC (Fig. 2F). No previous reports of a parent-of-origin impact on prostate disease have been reported. Kidney disease in the F4 generation females was transmitted through POC for both DDT and vinclozolin lineages (Fig. 2I and J). Although specific genetic mutations have been shown to have a parent-of-origin effect in kidney disease (Hanson et al., 2001, 2013), general sex specific parent-of-origin effects have not been reported. Interestingly, the multiple disease phenotype was significant in both males and females of the F3 generation, and the POC transmitted the female multiple disease while the MOC transmitted the male multiple disease (Fig. 2E-H).
In addition, there was an increased obesity incidence in the F4 generation maternal outcross male offspring, indicating that the disease transmission was primarily passed through the female germline allele to the male offspring. In contrast, the F4 generation paternal outcross female offspring had an increased obesity incidence indicating that the disease transmission was passed through the male germline allele to the female offspring. These results are consistent with a previous study showing similar results with a methoxychlor lineage transgenerational outcross (Manikkam et al., 2014). Previous epidemiology studies in humans have indicated parental impacts on offspring obesity, but no parent-of-origin impacts have been reported (Danielzik et al., 2002, 2004).
Examples of pathology derived from either parent in the F4 generation included male kidney disease in vinclozolin lineage animals (Fig. 2H) and male obesity in the DDT and vinclozolin lineages, but to a lesser degree in the POC (Fig. 3A and B). The only same sex transmission observed was the mammary tumor development in the F4 generation MOC (Supplemental Fig. S2). Examples of pathology that was present primarily in the outcross, but not predominant in the F3 generation include the male obesity in both POC and MOC in the vinclozolin lineage (Fig. 3B) and female kidney disease in the vinclozolin lineage (Fig. 2J).
In summary, the current study suggests that parent-of-origin allelic transmission of pathology (Fig. 4) should be more thoroughly investigated, and suggests a role for epigenetic transgenerational inheritance in the disease etiology. All potential types of parent-of-origin allelic transmission were observed, but the predominant observation was a transmission between sexes with either a POC and subsequent female disease, or an MOC and male disease. This interesting observation suggests the opposite parent-of-origin allele may have a critical role in altering the epigenetic programing to promote disease.
3.2. Parent-of-origin allelic sperm epimutation transmission
The induction of transgenerational phenotypes has been extensively studied with numerous ancestral exposures such as chemicals, diet or behavioral stress (Skinner, 2014). Sperm DNA methylation alterations was the first epigenetic process shown in a transgenerational study (1). Most DMRs identified in the F4 generation lineages whether they are from the paternal outcross or the maternal outcross are unique, unless the statistical stringency of the comparison is reduced, Fig. 5I. As previously described (Ben Maamar et al., 2018; Skinner et al., 2018), this shows once again that the transgenerational sperm DMRs are specific to each generation. Our results demonstrate that more DMRs were found in the F3 generations compared to the F4 generations in the maternal outcross for DDT and vinclozolin lineages, and also the F4 generation paternal outcross for the DDT lineage. However, we observed an increase in DMRs in the paternal outcross vinclozolin.
Exposures with vinclozolin (Anway et al., 2005) or high fat diet (Dunn and Bale, 2011) have previously shown transgenerational transmission of increased incidence of disease through the male germline whereas with methoxychlor was shown to transmit diseases (kidney and obesity) through the female germline (Manikkam et al., 2014). In contrast, another study with DDT determined that male obesity was transmitted through the female germline and female obesity through the male germline (Skinner et al., 2013). Our current results are consistent with these studies outlining the fact that the female germline (allelic) transmission of environmentally induced epigenetic transgenerational phenotypes appear to be as stable as male germline transmission, and that sometimes a combination of both paternal and maternal alleles is needed to transmit certain diseases such as testis disease to the male offspring (Fig. 2A and B).
Previous studies have reported a link between imprinted genes and a parent-of-origin effect on cell or tissue biology. Morison et al. (2001) created a database of imprinted gene parent-of-origin effects in animals (http://igc.otago.ac.nz/home.html). Imprinted genes are a special class of genes since their DNA methylation patterns are unchanged over the generation and are not affected by the methylation erasure occurring early in development (Constancia et al., 1998). Imprinted genes carry a molecular memory of their parent-of-origin allele (Surani, 2001). This molecular memory is associated with differential methylation patterns between the two alleles, which affect parent-of-origin monoallelic gene expression (Costello and Plass, 2001). During the establishment of the germline development in the embryo, these allelic differences in methylation are established (Constancia et al., 1998). The current study provides insight into the phenomenon of parent-of-origin allelic transmission of disease with an epigenetic perspective. Similar to imprinted genes, the epigenetic transgenerational germline epimutations appear to have a methylation erasure in the primordial germ cells involving an epigenetic molecular memory, which has a role in the parent-of-origin allelic transmission for epigenetic transgenerational phenomenon.
Recently, Onuchi et al. (Onuchic et al., 2018) analyzed allelic epigenome maps which link mechanistically sequence-dependent allelic imbalances of the epigenome, stochastic switching at gene regulatory loci, and disease-associated genetic variation. The allelic epigenome map reveals CpG methylation imbalances which exceeds 30% differences at 5% of the loci. These results are consistent with ours, and suggest that all promoters demonstrate epigenetic buffering (Onuchic et al., 2018). In addition, in mammals many genes show monoallelic expression often in a lineage or tissue-specific manner. The antigen receptor (AgR) loci, developing B or T lymphocyte, or the olfactory receptor genes are examples. In the olfactory receptor genes case, they are regulated such that only one gene is randomly “chosen” to become activated, and on only one allele (Chess et al., 1994). This monoallelic expression of the olfactory genes is entirely regulated by epigenetic processes (Monahan and Lomvardas, 2015). In addition, allelic transcription-associated acquisition of DNA methylation occurs in the oocytes at several somatic DMRs of imprinted domains (Koerner et al., 2012; Mehta et al., 2015; Greenberg et al., 2017). These different studies show that not only genetic modes of monoallelic expression exist, but also epigenetic mechanisms. The current study clearly shows the role of parent-of-origin transgenerational epigenetic inheritance of diseases and epimutations in the offspring.
Studies have established that the epigenetic programming of the primordial germ cells (PGCs) initiates during the gonadal sex determination developmental period (Tang et al., 2016). Following fertilization, a DNA methylation erasure occurs to help generate the stem cells of the early embryo, which then undergo a remethylation to generate the different somatic cell types (Tang et al., 2016; Surani and Hajkova, 2010). This erasure of DNA methylation allows deleterious epigenetic alterations in the germline to be reset (Iqbal et al., 2015). The transgenerational epigenetic alterations in the germline appear to be permanently reprogrammed like imprinted genes, and appear protected from this DNA methylation erasure and reprogramming at fertilization in the subsequent generations (Plasschaert and Bartolomei, 2014). Future studies are required to confirm the protection of the transgenerational epimutations during the early development DNA methylation erasure. Recent analysis of the developmental origins of the sperm epimutations demonstrate appearance at each stage of gametogenesis with the majority in the prospermatogonia, spermatogonia and spermatogenic stages (Ben Maamar et al., 2019; Skinner et al., 2019). The current study extends these observations to demonstrate a parent-of-origin allelic transmission of the epimutations, similar to imprinted genes. No classic known imprinted genes correlate with the environmentally induced DMRs identified. Comparably to imprinted genes, the DMRs identified appeared to be permanently reprogrammed and protected from the methylation erasure, but also demonstrate a parent-of-origin allelic transmission. Similar mechanisms appear to occur in the egg, but no study has investigated the epigenetics of the oocyte differential DNA methylation due to the limitation of obtaining sufficient numbers of eggs in mammals.
The current study clearly demonstrates a parent-of-origin allelic transmission of the pathology and sperm epigenetics observed. Further investigations are now needed to determine if a cross of the F4 generation MOC females with the F4 generation POC males to generate the F5 generation will develop pathologies similar to that in the F3 generation. Although the pathology disappeared in the outcrossed F4 generation, it may reappear in the combined cross in the F5 generation. In addition, comparing the F5 outcross animals with the F4 outcross would be interesting to assess the stability of the transgenerational sperm epigenome. The analysis of the stability of the transgenerational germline epigenetic alterations is important to assess in understanding the molecular processes involved in this non-genetic form of inheritance.
4. Materials and methods
4.1. Animals studies and breeding
Female and male rats of an outbred strain Hsd:Sprague Dawley®™SD®™ obtained from Harlan/Envigo (Indianapolis, IN) at about 70–100 days of age were maintained in ventilated (up to 50 air exchanges/hour) isolator cages (cages with dimensions of 10 ¾″ W×19 ¼″ D×10 ¾″ H, 143 square inch floor space, fitted in Micro-vent 36-cage rat racks; Allentown Inc., Allentown, NJ) containing Aspen Sani chips (pinewood shavings from Harlan) as bedding, and a 14 h light: 10 h dark regimen, at a temperature of 70 °F and humidity of 25%–35%. The mean light intensity in the animal rooms ranged from 22 to 26 ft-candles. Rats were fed ad lib with standard rat diet (8640 Teklad 22/5 Rodent Diet; Harlan), and ad lib tap water for drinking. To obtain time-pregnant females, the female rats in proestrus were pair-mated with male rats. The sperm-positive (i.e. sperm plug present) (day 0) rats were monitored for diestrus and body weight. On days 8 through 14 of gestation (Kobaly et al., 2014), the females received daily intraperitoneal injections of DDT (25 mg/kg body weight/day) or vinclozolin (100 mg/kg body weight/day Chem Services, Westchester PA, USA) or dimethyl sulfoxide (DMSO). The DDT (dichlorodiphenyltrichloroethane) was obtained from Chem Service Inc. (West Chester, PA) and reported to have a purity of 98.2%. DDT were dissolved and injected in DMSO vehicle as previously described (Hanson et al., 2001). Treatment lineages were designated ‘control’ or ‘DDT’ lineages. The treated gestating female rats were designated as the F0 generation. The offspring of the F0 generation rats were the F1 generation. Non-littermate females and males aged 70–90 days from the F1 generation of control or DDT or vinclozolin lineages were bred to obtain F2 generation offspring. The F2 generation rats were bred to obtain F3 generation offspring.
4.2. Tissue harvest and histology processing
Twelve-month-old rats were euthanized via CO2 inhalation and cervical dislocation prior to tissue harvest. Body weight and length were measured at dissection time. Testis, prostate, ovary, kidney, and gonadal fat were collected and prepared for histopathological examination as previously described (Nilsson et al., 2018b).
4.3. Histopathology examination and disease classification
The Washington Animal Disease Diagnostic Laboratory (WADDL) at the Washington State University College of Veterinary Medicine has board certified veterinary pathologists and assisted in initially establishing the criteria for the pathology analyses and identifying parameters to assess (Anway et al., 2006a). WADDL performed full necropsies as required on animals that died prior to the time of scheduled sacrifice at one year, and performed tumor classifications in the current study. Histopathology readers were trained to recognize the specific abnormalities evaluated for this study in rat testis, ventral prostate, ovary and kidney (see below). Three different readers were used for each tissue and blinded to the treatment groups. A set of quality control (QC) slides was generated for each tissue and was read by each reader prior to evaluating any set of experimental slides. WADDL was consulted when any questions developed.
As previously described (Nilsson et al., 2018a), testis histopathology criteria included the presence of vacuoles in the seminiferous tubules, azoospermic atretic seminiferous tubules and ‘other’ abnormalities including sloughed spermatogenic cells in center of the tubule and a lack of a tubule lumen. As previously described (Anway and Skinner, 2008), prostate histopathology criteria included the presence of vacuoles in the glandular epithelium, atrophic glandular epithelium encompassing more than one third of a gland, and hyperplasia of prostatic gland epithelium. Kidney histopathology criteria included markedly reduced size of glomerulus, thickened Bowman’s capsule and the presence of proteinaceous fluid-filled cysts >50 μm in diameter. Ovary sections were assessed for the two pathologies of primordial follicle loss and ovarian cysts as previously described (Nilsson et al., 2012). Ovarian cysts have little or no granulosa cell layer, a smooth border, and are 50–250 μm (small cysts) or > 250 μm (large cysts) in diameter. A cut-off was established to declare a tissue ‘diseased’ based on the mean number of histopathological abnormalities plus two standard deviations from the mean of control group tissues as assessed by each of the three individual observers blinded to the treatment groups. This number was used to classify rats into those with and without testis, ovary, prostate or kidney disease in each lineage. A rat tissue section was finally declared ‘diseased’ only when at least two of the three readers marked the same tissue section ‘diseased’.
4.4. Obesity phenotype analysis
Obesity was assessed with an increase in adipocyte size (area), body mass index (BMI), and abdominal adiposity as previously described (Nilsson et al., 2018b). Previous studies have used these parameters to assess toxicant impacts on transgenerational obesity (Skinner et al., 2013; McBirney et al., 2017; Manikkam et al., 2013; Tracey et al., 2013). The parameters for the adipocyte area in females is less than 2618 μm2 for lean, between 2618 μm2–4643 μm2 for non-obese, and greater than 4643 μm2 for obese. The parameters for the adipocyte area in males is less than 2526 μm2 for lean, non-obese area between 2526 μm2–3979 μm2, and obese is greater than 3979 μm2. The parameters for BMI in females is less than 0.6081 g/cm2 for lean, between 0.6081–0.7971 g/cm2 for non-obese, and greater than 0.7971 g/cm2 for obese. The parameters for BMI in males is less than 0.8196 g/cm2 for lean, between 0.8196 g/cm2 and 1.0354 g/cm2 for non-obese and greater than 1.0354 g/cm2 for obese.
4.5. Statistical analyses for histopathological analysis
Pubertal age and behavioral parameters were analyzed using a Student’s t-test. Disease and pathology parameters were analyzed using a Fisher’s exact test. The incidence of disease in rats from each lineage was assessed and the proportion of individual disease and multiple disease incidences was computed. For the individual diseases, only those rats that had histopathology assessed were included in the computation. For the multiple diseases, the total number of diseases for each rat was assessed, and the number was added up for each of the rats. The single or multiple disease proportions are listed in Supplemental Tables S1, S2, and S3.
4.6. Epididymal sperm collection and DNA isolation
The epididymis was dissected free of connective tissue, a small cut made to the cauda, and the tissue placed in 5 ml of 1X PBS solution for up to 2 h at 4 °C. The epididymal tissue was minced and the released sperm was centrifuged at 6,000×g. Then the supernatant was removed, and the pellet was resuspended in NIM buffer, to be stored at −80 °C until further use. One hundred μl of sperm suspension was sonicated to destroy somatic cells and tissue, spun down at 6,000×g, the sperm pellet was washed with 1X PBS once, and then combined with 820 μl DNA extraction buffer and 80 μl 0.1M DTT. The sample was incubated at 65 °C for 15 min. Following this incubation, 80 μl proteinase K (20 mg/ml) was added and the sample incubated at 55 °C for at least 2 h under constant rotation. Then 300 μl of protein precipitation solution (Promega, A7953) was added, the sample was mixed thoroughly, and then incubated for 15 min on ice. The sample was centrifuged at 12,500×g for 30 min at 4 °C. One ml of the supernatant was transferred to a 2 ml tube and 2 μl of glycoblue, and 1 ml of cold 100% isopropanol was added. The sample was mixed well by inverting the tube several times, then left in −20 °C freezer for at least 1 h. After precipitation, the sample was centrifuged at 12,500×g for 20 min at 4 °C. The supernatant was taken off and discarded without disturbing the (blue) pellet. The pellet was washed with 70% cold ethanol by adding 500 μl of 70% ethanol to the pellet, and returning the tube to the freezer for 20 min. After the incubation the tube was centrifuged for 10 min at 4 °C at 12,500×g and the supernatant was discarded. The tube was briefly spun again to collect residual ethanol to bottom of tube, and then as much liquid as possible was removed with gel loading tip. The pellet was air-dried at RT until it looked dry (about 5 min). The pellet was then resuspended in 100 μl of nuclease free water.
4.7. Methylated DNA immunoprecipitation MeDIP
Methylated DNA Immunoprecipitation (MeDIP) with genomic DNA was performed as follows: rat sperm DNA pools were generated using the appropriate amount of genomic DNA from each individual for 6 pools each of paternal and maternal outcross control, paternal and maternal outcross DDT, and paternal and maternal outcross vinclozolin lineage animals. Genomic DNA pools were sonicated using the Covaris M220 the following way: the pooled genomic DNA was diluted to 130 μl with TE buffer into the appropriate Covaris tube. Covaris was set to 300 bp program, and the program was run for each tube in the experiment. 10 μl of each sonicated DNA was run on 1.5% agarose gel to verify fragment size. The sonicated DNA was transferred from the Covaris tube to a 1.7 ml microfuge tube, and the volume was measured. The sonicated DNA was then diluted with TE buffer (10 mM Tris HCl, pH7.5; 1 mM EDTA) to 400 μl, heat-denatured for 10 min at 95 ° C, then immediately cooled on ice for 10 min. Then 100 μl of 5X IP buffer and 5 μg of antibody (monoclonal mouse anti 5-methyl cytidine; Diagenode #C15200006) were added to the denatured sonicated DNA. The DNA-antibody mixture was incubated overnight on a rotator at 4 °C.
The following day magnetic beads (Dynabeads M-280 Sheep anti-Mouse IgG; 11201D) were pre-washed as follows: The beads were resuspended in the vial, then the appropriate volume (50 μl per sample) was transferred to a microfuge tube. The same volume of Washing Buffer (at least 1 mL PBS with 0.1% BSA and 2 mM EDTA) was added, and the bead sample was resuspended. The tube was then placed into a magnetic rack for 1–2 min, and the supernatant was discarded. The tube was removed from the magnetic rack, and the beads were washed once. The washed beads were resuspended in the same volume of IP buffer (50 mM sodium phosphate ph7.0, 700 mM NaCl, 0.25% TritonX-100) as the initial volume of beads. 50 μl of beads were added to the 500 μl of DNA-antibody mixture from the overnight incubation, then incubated for 2 h on a rotator at 4 °C.
After the incubation, the bead-antibody-DNA complex was washed three times with IP buffer as follows: The tube was placed into magnetic rack for 1–2 min, and the supernatant was discarded, and then washed with IP buffer 3 times. The washed bead-DNA solution was then resuspended in 250 μl digestion buffer with 3.5 μl Proteinase K (20 mg/ml). The sample was then incubated for 2–3 h on a rotator at 55 °C. was 250 μl of buffered Phenol-Chloroform-Isoamylalcohol solution was added to the supe, and the tube vortexed for 30 s then centrifuged at 12,500×g for 5 min at room temperature. The aqueous supernatant was carefully removed and transferred to a fresh microfuge tube. Then 250 μl chloroform were added to the supernatant from the previous step, vortexed for 30 s, and then centrifuged at 12,500×g for 5 min at room temperature. The aqueous supernatant was removed and transferred to a fresh microfuge tube. 20 μl of 5M NaCl and 500 μl ethanol were added to the supernatant 2 μl of glycoblue (20 mg/ml), and mixed well, then precipitated in −20 °C freezer for 1 h to overnight.
The precipitate was centrifuged at 12,500×g for 20 min at 4 °C and the supernatant removed, while not disturbing the pellet. The pellet was washed with 500 μl cold 70% ethanol in −20 °C freezer for 15 min, then centrifuged again at 12,500×g for 5 min at 4 °C, and then the supernatant was discarded. The tube was spun again briefly to collect residual ethanol to bottom of tube, and as much liquid as possible was removed with gel loading tip. The pellet was air-dried at RT until it looked dry (about 5 min), then resuspended in 20 μl H2O or TE. DNA concentration was measured in Qubit (Life Technologies) with ssDNA kit (Molecular Probes Q10212).
4.8. MeDIP-seq analysis
The MeDIP pools were used to create libraries for next generation sequencing (NGS) using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, San Diego, CA), starting at step 1.4 of the manufacturer’s protocol to generate double stranded DNA. After this step, the manufacturer’s protocol was followed. Each pool received a separate index primer. NGS was performed at WSU Spokane Genomics Core using the Illumina HiSeq 2500 with a PE50 application, with a read size of approximately 50 bp and approximately 30 million reads per pool. Five to six libraries were run in one lane. The DMRs were previously validated and confirmed through Bisulfite-MS and pyrosequencing (Guerrero-Bosagna et al., 2010).
4.9. Statistics and bioinformatics
For the DMR analyses, the basic read quality was verified using summaries produced by the FastQC program http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. The raw reads were trimmed and filtered using Trimmomatic (10.1093/bioinformatics/btu170). The reads for each MeDIP sample were mapped to the Rnor 6.0 rat genome using Bowtie2 (Bolger et al., 2014) with default parameter options. The mapped read files were then converted to sorted BAM files using SAMtools (Langmead and Salzberg, 2012). The F3 samples were taken from two previous studies (Nilsson et al., 2018b; King et al., 2019) focusing on individual rats. These samples were subsampled and pooled in silico using SAMtools to create six pooled samples with similar read depths as the F4 outcross samples. To identify DMRs, the reference genome was broken into 100 bp windows. The MEDIPS (Lienhard et al., 2014) and edgeR (Robinson et al., 2010). R packages were used to calculate differential coverage between control and exposure sample groups. The edgeR p-value was used to determine the relative difference between the two groups for each genomic window. Windows with an edgeR p-value less than an arbitrarily selected threshold were considered DMRs. To calculate the differential DNA methylation, we report a log fold change, Supplemental Tables S6-S9. This is shown as the maximum log-fold change (maxLFC) column. It is also calculated with the mean raw read number of genomic windows with an edgeR p < 1e-05 where the differential methylation is about 20–100%. The DMR edges were extended until no genomic window with a p-value < 0.1 remained within 1000 bp of the DMR. CpG density and other information was then calculated for the DMR based on the reference genome. DMRs from a set of analyses were considered overlapping if they shared any genomic coordinate in common. An additional expanded overlap analysis was also performed. For this analysis, each DMR in the first analysis was considered to be present in the second analysis if any genomic window at the DMR location in the second analysis had an edgeR p-value ≤ 0.05. DMRs were annotated using the biomaRt R package (Durinck et al., 2009) to access the Ensembl database (Cunningham et al., 2015). The genes that fell within 10kbp of the DMR edges were then inputed into the KEGG pathway search (Kanehisa and Goto, 2000; Kanehisa et al., 2014) to identify associated pathways. The associated genes were then sorted into functional groups by consulting information provided by the DAVID (Huang da et al., 2009), and Panther (Mi et al., 2013), databases then incorporated into an internal curated database (www.skinner.wsu.edu under genomic data). All molecular data has been deposited into the public database at NCBI (GEO # GSE130258).
Supplementary Material
Acknowledgments
We acknowledge Ms. Margaux McBirney, Ms. Deepika Kubsad, and Mr. Ryan Thompson for technical assistance. We acknowledge Ms. Amanda Quilty for editing and Ms. Heather Johnson for assistance in preparation of the manuscript. We thank the Genomics Core laboratory at WSU Spokane for sequencing data. This study was supported by John Templeton Foundation (50183 and 61174) (https://templeton.org/) grants to MKS and NIH (ES012974) (https://www.nih.gov/) grant to MKS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Declaration of competing interest
The authors declare no competing interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2019.10.030.
Data archiving
All molecular data has been deposited into the public database at NCBI (GEO # GSE130258), and R code computational tools are available at GitHub (https://github.com/skinnerlab/MeDIP-seq) and www.skinner.wsu.edu.
References
- Anway MD, Skinner MK, 2008. Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. The Prostate 68 (5), 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK, 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308 (5727), 1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Leathers C, Skinner MK, 2006. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 147 (12), 5515–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Memon MA, Uzumcu M, Skinner MK, 2006. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J. Androl 27 (6), 868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Maamar M, Sadler-Riggleman I, Beck D, McBirney M, Nilsson E, Klukovich R, Xie Y, Tang C, Yan W, Skinner MK, 2018. Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate vinclozolin-induced epigenetic transgenerational inheritance of disease. Environ. Epigene 4 (2), 1–19 dvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Maamar M, Nilsson E, Sadler-Riggleman I, Beck D, McCarrey JR, Skinner MK, 2019. Developmental origins of transgenerational sperm DNA methylation epimutations following ancestral DDT exposure. Dev. Biol 445 (2), 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookheart RT, Duncan JG, 2016. Drosophila melanogaster: an emerging model of transgenerational effects of maternal obesity. Mol. Cell. Endocrinol 435, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carouge D, Blanc V, Knoblaugh SE, Hunter RJ, Davidson NO, Nadeau JH, 2016. Parent-of-origin effects of A1CF and AGO2 on testicular germ-cell tumors, testicular abnormalities, and fertilization bias. Proc. Natl. Acad. Sci. U. S. A 113 (37), E5425–E5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvan M.J.r., Kalluvila TA, Klingler RH, Larson JK, Pickens M, Mora-Zamorano FX, Connaughton VP, Sadler-Riggleman I, Beck D, Skinner MK, 2017. Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS One 12 (5), 1–26 e0176155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-Garcia R, Sahu M, Abbey RJ, Laude J, Pham N, Blumberg B, 2013. Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ. Health Perspect 121 (3), 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R, 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell 78 (5), 823–834. [DOI] [PubMed] [Google Scholar]
- Constancia M, Pickard B, Kelsey G, Reik W, 1998. Imprinting mechanisms. Genome Res. 8 (9), 881–900. [DOI] [PubMed] [Google Scholar]
- Costello JF, Plass C, 2001. Methylation matters. J. Med. Genet 38 (5), 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kahari AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P, Ensembl, 2015, 2015. Nucleic Acids Res. 43, D662–D669. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielzik S, Langnase K, Mast M, Spethmann C, Muller MJ, 2002. Impact of parental BMI on the manifestation of overweight 5–7 year old children. Eur. J. Nutr 41 (3), 132–138. [DOI] [PubMed] [Google Scholar]
- Danielzik S, Czerwinski-Mast M, Langnase K, Dilba B, Muller MJ, 2004. Parental overweight, socioeconomic status and high birth weight are the major determinants of overweight and obesity in 5–7 y-old children: baseline data of the Kiel Obesity Prevention Study (KOPS). Int. J. Obes. Relat. Metab. Disord 28 (11), 1494–1502. [DOI] [PubMed] [Google Scholar]
- Dapp M, Reinders J, Bediee A, Balsera C, Bucher E, Theiler G, Granier C, Paszkowski J, 2015. Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids. Native Plants 1, 15092. [DOI] [PubMed] [Google Scholar]
- Dunn GA, Bale TL, 2011. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology 152 (6), 2228–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W, 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc 4 (8), 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EL, Doolittle DP, 1963. Theoretical consequences of systems of mating used in mammalian genetics In: Burdette WJ (Ed.), Methodology in Mammalian Genetics. Holden-Day, San Francisco, pp. 3–41. [Google Scholar]
- Greenberg MV, Glaser J, Borsos M, Marjou FE, Walter M, Teissandier A, Bourc’his D, 2017. Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat. Genet 49 (1), 110–118. [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker B, Skinner M, 2010. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 5 (9), 1–17 e0184306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RL, Kobes S, Lindsay RS, Knowler WC, 2001. Assessment of parent-of-origin effects in linkage analysis of quantitative traits. Am. J. Hum. Genet 68 (4), 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RL, Guo T, Muller YL, Fleming J, Knowler WC, Kobes S, Bogardus C, Baier LJ, 2013. Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes 62 (8), 2984–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH, 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. U. S. A 105 (44), 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE, 1975. DNA modification mechanisms and gene activity during development. Science 187 (4173), 226–232. [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4 (1), 44–57. [DOI] [PubMed] [Google Scholar]
- Ideraabdullah FY, Vigneau S, Bartolomei MS, 2008. Genomic imprinting mechanisms in mammals. Mutat. Res 647 (1–2), 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Tran DA, Li AX, Warden C, Bai AY, Singh P, Wu X, Pfeifer GP, Szabo PE, 2015. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol. 16, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK, 2007. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet 8 (4), 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA, 2013. The presence, role and clinical use of spermatozoal RNAs. Hum. Reprod. Update 19, 604–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M, 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G, Feil R, 2013. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci 368 (1609), 20110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SE, McBirney M, Beck D, Sadler-Riggleman I, Nilsson E, Skinner MK, 2019. Sperm epimutation biomarkers of obesity and pathologies following DDT induced epigenetic transgenerational inheritance of disease. Environ. Epigenet 5 (2), 1–15 dvz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobaly K, Vellanki P, Sisk RK, Armstrong L, Lee JY, Lee J, Hayes MG, Urbanek M, Legro RS, Dunaif A, 2014. Parent-of-origin effects on glucose homeostasis in polycystic ovary syndrome. J. Clin. Endocrinol. Metab 99 (8), 2961–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner MV, Pauler FM, Hudson QJ, Santoro F, Sawicka A, Guenzl PM, Stricker SH, Schichl YM, Latos PA, Klement RM, Warczok KE, Wojciechowski J, Seiser C, Kralovics R, Barlow DP, 2012. A downstream CpG island controls transcript initiation and elongation and the methylation state of the imprinted Airn macro ncRNA promoter. PLoS Genet. 8 (3), e1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M, Zander-Fox DL, Robker RL, McPherson NO, 2015. Peri-conception parental obesity, reproductive health, and transgenerational impacts. Trends Endocrinol. Metab 26 (2), 84–90. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson HA, Cheverud JM, Wolf JB, 2013. Genomic imprinting and parent-of-origin effects on complex traits. Nat. Rev. Genet 14 (9), 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux S, Gourichon D, Leterrier C, Labrune Y, Coustham V, Riviere S, Zerjal T, Coville JL, Morisson M, Minvielle F, Pitel F, 2017. Embryonic environment and transgenerational effects in quail. Genet. Sel. Evol 49 (1), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L, 2014. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30 (2), 284–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu C, Tang X, Pei S, Jin D, Guo M, Yang M, Zhang Y, 2018. Genomic methylation and transcriptomic profiling provides insights into heading depression in inbred Brassica rapa L. ssp. pekinensis. Gene 665, 119–126. [DOI] [PubMed] [Google Scholar]
- Mackay DJG, Temple IK, 2017. Human imprinting disorders: principles, practice, problems and progress. Eur. J. Med. Genet 60 (11), 618–626. [DOI] [PubMed] [Google Scholar]
- Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK, 2012. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One 7 (2), 1–12 e31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner M, 2013. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 8 (1), 1–18 e55387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Haque MM, Guerrero-Bosagna C, Nilsson E, Skinner MK, 2014. Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult onset disease through the female germline. PLoS One 9 (7), 1–19 e0184306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, 2009. The genetic signatures of noncoding RNAs. PLoS Genet. 5 (4), e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBirney M, King SE, Pappalardo M, Houser E, Unkefer M, Nilsson E, Sadler-Riggleman I, Beck D, Winchester P, Skinner MK, 2017. Atrazine induced epigenetic transgenerational inheritance of disease, lean phenotype and sperm epimutation pathology biomarkers. PLoS One 12 (9), 1–37 e0184306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Williamson CM, Ball S, Tibbit C, Beechey C, Fray M, Peters J, 2015. Transcription driven somatic DNA methylation within the imprinted Gnas cluster. PLoS One 10 (2), e0117378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD, 2013. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc 8 (8), 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K, Lomvardas S, 2015. Monoallelic expression of olfactory receptors. Annu. Rev. Cell Dev. Biol 31, 721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison IM, Paton CJ, Cleverley SD, 2001. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 29 (1), 275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Larsen G, Manikkam M, Guerrero-Bosagna C, Savenkova M, Skinner M, 2012. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PLoS One 7 (5), 1–18 e36129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Sadler-Riggleman I, Skinner MK, 2018. Environmentally induced epigenetic transgenerational inheritance of disease. Environ. Epigene. 4 (2), 1–13 dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, King SE, McBirney M, Kubsad D, Pappalardo M, Beck D, Sadler-Riggleman I, Skinner MK, 2018. Vinclozolin induced epigenetic transgenerational inheritance of pathologies and sperm epimutation biomarkers for specific diseases. PLoS One 13 (8), 1–29 e0202662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northstone K, Golding J, Davey Smith G, Miller LL, Pembrey M, 2014. Prepubertal start of father’s smoking and increased body fat in his sons: further characterisation of paternal transgenerational responses. Eur. J. Hum. Genet 22 (12), 1382–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuchic V, Lurie E, Carrero I, Pawliczek P, Patel RY, Rozowsky J, Galeev T, Huang Z, Altshuler RC, Zhang Z, Harris RA, Coarfa C, Ashmore L, Bertol JW, Fakhouri WD, Yu F, Kellis M, Gerstein M, Milosavljevic A, 2018. Allele-specific epigenome maps reveal sequence-dependent stochastic switching at regulatory loci. Science (6409), 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter RC, Westendorp RG, de Rooij SR, Osmond C, Barker DJ, Roseboom TJ, 2008. Increased reproductive success of women after prenatal undernutrition. Hum. Reprod 23 (11), 2591–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert RN, Bartolomei MS, 2014. Genomic imprinting in development, growth, behavior and stem cells. Development 141 (9), 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Colot V, 2016. Plant transgenerational epigenetics. Annu. Rev. Genet 50, 467–491. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK, 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1), 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F, 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24 (15), 1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, 2014. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol. Cell. Endocrinol 398 (1–2), 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Guerrero-Bosagna C, 2014. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics 15 (1), 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Anway, Savenkova MI, Gore AC, Crews D, 2008. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One 3 (11), 1–11 e3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Guerrero-Bosagna C, 2010. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol. Metab 21 (4), 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Tracey R, Guerrero-Bosagna C, Haque MM, Nilsson E, 2013. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med. 11 (228), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W, 2018. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenet. Chromatin 11 (1), 1–24, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Nilsson E, Sadler-Riggleman I, Beck D, McCarrey JR, 2019. Transgenerational sperm DNA methylation epimutation developmental origins following ancestral vinclozolin exposure. Epigenetics 14 (7), 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A, 2015. Epigenetic inheritance and evolution: a paternal perspective on dietary influences. Prog. Biophys. Mol. Biol 118 (1–2), 79–85. [DOI] [PubMed] [Google Scholar]
- Surani MA, 2001. Reprogramming of genome function through epigenetic inheritance. Nature 414 (6859), 122–128. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hajkova P, 2010. Epigenetic reprogramming of mouse germ cells toward totipotency. Cold Spring Harbor Symp. Quant. Biol 75, 211–218. [DOI] [PubMed] [Google Scholar]
- Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA, 2016. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet 17 (10), 585–600. [DOI] [PubMed] [Google Scholar]
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT, 2009. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum. Mol. Genet 18 (21), 4046–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey R, Manikkam M, Guerrero-Bosagna C, Skinner M, 2013. Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod. Toxicol 36, 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, 1998. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell. Mol. Life Sci 54 (1), 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiserman AM, Koliada AK, Jirtle RL, 2017. Non-genomic transmission of longevity between generations: potential mechanisms and evidence across species. Epigenet. Chromatin 10 (1), 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, Byrnes EM, Pierce RC, 2014. The impact of exposure to addictive drugs on future generations: physiological and behavioral effects. Neuropharmacology 269–275, 76 Pt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv M, 2014. Chromatin remodeling: from transcription to cancer. Cancer Genet. 207 (9), 352–357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.