Abstract
Tooth defect and tooth loss are common clinical diseases in stomatology. Compared with the traditional oral restoration treatment, tooth regeneration has unique advantages and is currently the focus of oral biomedical research. It is known that dozens of cytokines/growth factors and other bioactive factors are expressed in a spatial-temporal pattern during tooth development. On the other hand, the technology for spatial-temporal control of drug release has been intensively studied and well developed recently, making control release of these bioactive factors mimicking spatial-temporal pattern more feasible than ever for the purpose of tooth regeneration. This article reviews the research progress on the tooth development and discusses the future of tooth regeneration in the context of spatial-temporal release of developmental factors.
Keywords: Tooth development, Spatial-temporal control of drug release, Cytokines, Biodegradable materials, Tooth regeneration
Several cytokines/growth factors are involved in the precise and directional development of specific tissues and organs. In the craniomaxillofacial region, the development of teeth depends largely on the orderly interaction between the ectodermal epithelium and the mesenchyme [1].
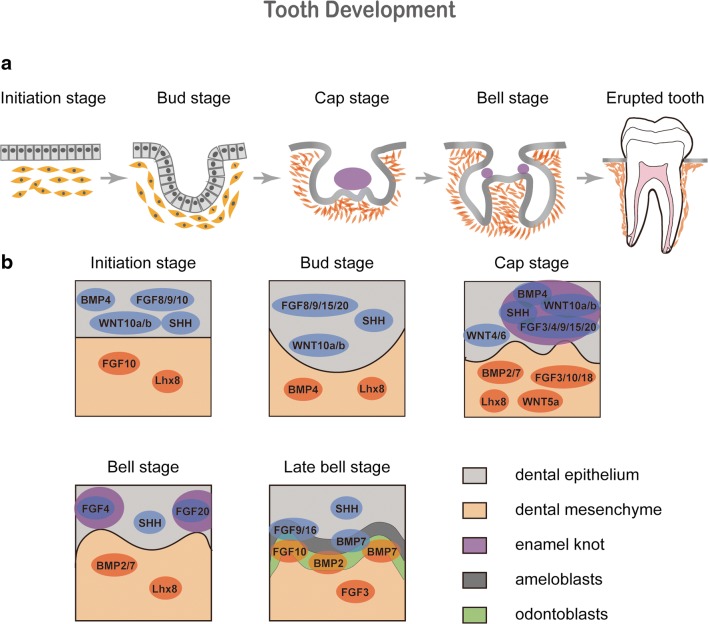
The tooth development process is generally divided into the initiation stage, the bud stage, the cap stage and the bell stage (Fig. 1). At the initiation stage, the epithelial tissue known as the dental placode, locally thickens, and continues to develop into the tooth bud [2]. Meanwhile, the mesenchymal tissue near the tooth bud, aggregates to form the tooth germ. Through the proliferation and folding of the epithelial tissue, the buds gradually evolve to the cap and bell stages. Clusters of undifferentiated epithelial cells, known as the enamel knot, can be observed at the center of the inner enamel epithelium. Each tooth germ has only one primary enamel knot. When the primary enamel knot disappears, secondary enamel knots will appear at the prospective apex of the molars. The enamel knot is considered to be the signal center that controls the shape of the cusp [3]. Subsequently, the epithelial tissue forms odontoblasts and ameloblasts, that lead to the formation of the dentin and the enamel, respectively. After the crown formation, the cervical loop of the dental epithelial cells, continues to elongate and forms a double-layered epithelial structure, found between the dental follicle and the dental papilla, and named the Hertwig’s epithelial root sheath (HERS). Conventionally, researchers believe that HERS is the signal center of the root formation [4].
Fig. 1.
Spatial-temporal expression of developmental signal molecules during tooth development. Tooth morphogenesis is divided into the initiation, bud, cap and bell stages. Expression of the fundamental signal molecules in the epithelium and mesenchyme are shown and corresponding to each stage
Many studies have shown that cytokines/growth factors such as BMPs, FGFs, SHHs, WNTs and TNFs, play an important role during this process [1]. Moreover, the expression of these cytokines is characterized by a spatial-temporal specificity [5–7] (Fig. 1). Aberrant expression may lead to tooth development abnormalities [1]. The spatio-temporal control of the developmental cues might be the future for tooth regeneration applications.
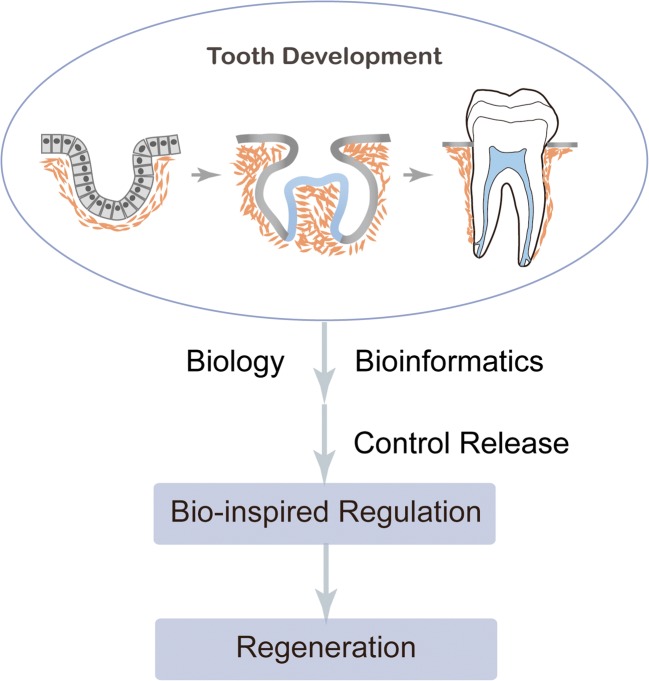
With advances in developmental biology and drug delivery, tooth regeneration would be more promising than ever before (Fig. 2). In the following sections, we summarize recent advances in developmental biology and discuss the clues for tooth regeneration in the context of the spatial-temporal control of bioactive drug release.
Fig. 2.
Schematic representation of the bio-inspired dental regeneration strategy. The gene expression pattern during tooth development is obtained by biology and bioinformatics, and the development associated with spatial-temporal specific expression could be approached by using different control release strategies for regeneration purpose, and making the goal of tooth regeneration expectable
Cytokines/ Growth Factors and Tooth Development
BMP, FGF, WNT and SHH signaling pathways are known signaling pathways in tooth development (Tables 1 and 2). Recently, other signaling pathways, such as TNF [8], YAP-Hippo [9] and mTORC1 [10], have also been found to be involved in the process.
Table 1.
Expression profile of the developmental cues involved in tooth development
| Expression stages | Signal molecules | Expression sites |
|---|---|---|
| Initiation stage | BMP4, FGF8, FGF9, WNT7b, SHH | Dental epithelium |
| FGF10 | Dental epithelium Dental mesenchyme | |
| Early bud stage | FGF8, FGF9, FGF15, FGF20 | Dental epithelium |
| BMP4 | Dental mesenchyme | |
| Late bud stage | BMP4, FGF3, FGF4, FGF9, FGF15, FGF20 | Primary enamel knot |
| FGF3, FGF10, FGF18 | Dental mesenchyme | |
| FGF16, FGF17 | Cervical loop mesenchyme | |
| Initiation and bud stages | WNT10a, WNT10b | Dental epithelium |
| Bud and cap stages | SHH | Enamel knot |
| Cap stage | BMP2, BMP7 | Dental epithelium Dental mesenchyme |
| WNT10a, WNT10b | Enamel knot | |
| WNT4, WNT6 | Dental epithelium | |
| WNT5a | Dental mesenchyme | |
| Early bell stage | BMP2, BMP7 | Dental papilla |
| Bell stage | FGF4, FGF20 | Secondary enamel knots |
| FGF9, FGF16 | Ameloblasts | |
| SHH | Inner enamel epithelium Stratum intermedium cells | |
| Late bell stage | BMP2 | Odontoblasts |
| BMP7 | Odontoblasts Ameloblasts | |
| FGF3 | Dental papilla | |
| FGF10 | Odontoblasts |
Table 2.
Functions of key cytokines during tooth development
| Signaling pathways | Key cytokines | Functions | References |
|---|---|---|---|
| BMP | BMP2 | Promotes early tooth mineralization | (Malik et al., 2018) |
| BMP4 | Coincides with the odontogenic potential; regulate the formation of the Hertwig’s epithelial root sheath |
(Jia et al., 2016) (Hosoya, Kim, Cho, & Jung, 2008) |
|
| BMP7 | Promotes early tooth mineralization | (Gao et al., 2018) | |
| BMP9 | Promotes odontoblastic differentiation and osteogenic differentiation | (Huang et al., 2019) | |
| FGF | FGF8 | Epithelial cell-originating factor | (Trumpp, Depew, Rubenstein, Bishop, & Martin, 1999) |
| FGF9 | Plays an important role in epithelial invagination and initiates ectodermal organogenesis | (Tai, Chen, Lin, Ling, & Chen, 2012) | |
| WNT | WNT7b | Positions the sites of tooth formation | (L. Sarkar et al., 2000) |
| WNT3a | Promotes cementoblast differentiation | (Nemoto et al., 2016) | |
| SHH | SHH | Stimulates epithelial cell proliferation | (Cobourne, Hardcastle, & Sharpe, 2001) |
BMPs
BMPs are a group of multifunctional homologous dimer proteins, which are members of the TGF-β superfamily. Several studies suggest that BMPs are involved at the start and during tooth development. Among BMP family members, BMP4 is a key Msx1-dependent mesenchymal odontogenic signal, that participates in the process of tooth morphogenesis through the bud-to-cap transition [11]. BMP4 expression begins in the dental lamina epithelium, and in the mesenchyme during tooth bud formation, indicating that the odontogenic potential, originally in the epithelium, is transferred to the mesenchyme [12]. Then, by the late bud stage, its expression is found in the primary knot of the dental epithelium [13]. Mechanistically, inhibitors of tooth development such as Dkk2 and Osr2 expressed in the tooth mesenchyme are suppressed by BMP4 signaling, while Msx1 synergizes with BMP4 in activating mesenchymal odontogenic potential that is essential for tooth morphogenesis [11].
Other BMP members that closely relate to tooth development include BMP2 and BMP7. A recent study in miniature pigs showed that BMP2 and BMP7 are expressed in the epithelium and the mesenchyme during the cap stage. At early bell stage their expression decrease and are mainly present in the dental papilla [14]. During the late bell stage, the expression of BMP2 is mostly found in odontoblasts, which implies that it may participate in early tooth morphogenesis and in late odontoblast differentiation and mineral secretion [14]. Another recent study showed that BMP2 played an early temporal, non-redundant role in organic tooth mineralization [15]. BMP7 expression is similar to that of BMP2, but is also detected in ameloblasts [14]. Recently, Huang et al. found that BMP9 regulates tooth development by promoting odontoblastic differentiation and osteogenic differentiation, which was unknown. The BMP9 knockout mice displayed abraded incisor tips, smaller molar cusps and shorter molar roots [16].
FGFs
FGFs are widely expressed in invertebrates and vertebrates. They are secretory protein ligands that maintain their functions in development, tissue homeostasis and metabolism in autocrine, endocrine or paracrine manners. Several members of the FGF family are involved in odontogenesis [17]. During the initiation stage, the expression of FGF8 and FGF9 are detected in the prospective tooth region of the dental epithelium [17], suggesting that they may take part in the initiation of tooth development. Meanwhile, FGF10 is detected in the dental epithelium and the dental mesenchyme [18]. Unlike FGF8, FGF9, FGF15 and FGF20 that are expressed in the epithelium following the formation of the dental lamina, FGF10 expression is decreased [18]. The expression of FGF3,FGF4, FGF9, FGF15 and FGF20 are detected in the primary enamel knot after its formation; while, FGF3, FGF10, and FGF18 are found in the mesenchyme [19]. The expression of FGF16 and FGF17 are detected in the mesenchyme of cervical loop [19]. At the bell stage, FGF4 and FGF20 expression are restricted to the forming cusps of the secondary enamel knots; while, the expression of FGF9 and FGF16 are detected in the differentiating ameloblasts [17; 19]. The expression of FGF3 is found in the dental papilla at late bell stage; while, FGF10 is expressed in the differentiating odontoblasts [18].
A previous study has shown that FGF8 can induce the expression of Pax9 in mice, revealing the prospective odontogenesis locations, and its essential role reaching beyond the bud stage of tooth development [20]. Another recent study showed that in the first branchial arch (BA1), and using ectoderm Nestin-Cre, that conditional FGF8 knockout leads to the decrease in Pax9 expression in the expected molar region, and the inhibition of molar formation [21]. Moreover, FGF8 expression in the oral epithelium determines the rostral–caudal polarity in BA1 by inducing Lhx8 expression in neural crest–derived mesenchyme, which makes Lhx8 intensively and exclusively expressed in neural crest derived ectomesenchyme and dental mesenchyme. This expression continues to be restricted to the dental papilla and the odontoblast, and gradually decreases over time [22]. The differentiation and function of the dental mesenchyme are regulated by Lhx8 via WNT and TGFβ pathways [23]. FGF9 is also involved in epithelial invagination and initiation of ectodermal organogenesis [24].
WNTs
The WNT family consists of a group of secretory glycoproteins that are rich in a conserved cysteine sequence, and that regulate cell growth, development, migration and differentiation during embryonic development. The WNT signaling pathway can be separated into the canonical signaling pathway, namely WNT/β-catenin pathway, and the noncanonical pathway, that includes the planar cell polarity pathway and the WNT/Ca2+ pathway. It plays critical roles in the initial stage of tooth development, with most of the signaling molecules being specifically expressed in the dental epithelium [25].
WNT7b is expressed in the oral epithelium but not in the presumptive dental epithelium, when tooth forming sites and tooth patterning are defined [26]. WNT7b seems to interact with SHH signaling to delimit boundaries between the oral and the dental ectoderm, and which determine the sites of tooth formation [26]. When the dental epithelium thickens, WNT10a and WNT10b are found expressed in the dental epithelium and these expression remain during the bud stage [27]. At the cap stage, the expression of both genes can be detected in the enamel knot. Meanwhile, WNT4, WNT6 and one of the WNT receptor MFz-6 are specifically expressed in the dental epithelium; while, WNT5a, sFrp2, and sFrp3 are expressed in the dental mesenchyme [28]. A recent study in miniature pig showed that the cusp patterning and the crown calcification may depend on the spatial-temporal distribution of WNT signaling [29]. A mutation in Lef1 gene, a critical component of WNT signaling pathway, caused tooth loss [30]. Odontoblast-specific deletion of the Wls gene, a chaperone protein that regulates WNT sorting and secretion, leads to the inhibition of odontoblast maturation and root elongation via reducing the activity of the canonical WNT signaling [31].
At the initial stages of tooth development, many signaling pathways function downstream of WNT/β-catenin signaling. When the WNT/β-catenin pathway is overactivated, the epithelial markers sonic hedgehog (SHH), Epiprofin (Epfn) and FGF8 are upregulated and ectopically expressed [32]. Mutations in WNT10b, WNT10a, LRP6 and other genes involved in this pathway showed tooth agenesis with or without other ectodermal anomalies [33].
SHHs
The mammalian hedgehog (Hh) family includes the sonic hedgehog (SHH), the Indian hedgehog (IHH) and the desert hedgehog (DHH) pathways, that encode SHH, IHH and DHH proteins, respectively. Among the three members, SHH is the only Hh ligand that is expressed in teeth [34]. The expression of SHH is present in the oral epithelium prior invagination, and in the tooth epithelium during the tooth development [1]. SHH expression, that begins at the bud stage, is restricted to the enamel knot at the cap stage [35]. It is also expressed in the surrounding inner enamel epithelium and in the stratum intermedium cells during the following stages [36]. The decrease or loss of SHH expression leads to a cap stage tooth rudiment, which has a severely disrupted morphology [37]. SHH also plays vital roles in the development of periodontal tissue [38]. As described above, BMP, WNT and SHH signals are interconnected during tooth development. The differential fate of epithelial stem cells, in mouse molars and incisors, is defined by BMP/SHH signaling network [39]. When reducing SHH function in the epithelium, WNT and FGF signaling are upregulated [40].
Other Factors
The EDA (ectodysplasin A)-EDAR (ectodysplasin A receptor) system has also been found to be involved in tooth development. It regulates interactions within or between epithelial and mesenchymal cells, and tissues functions by controlling NF-κB-mediated transcription of effectors or inhibitors of the WNT, SHH, FGF and TGF-β pathways [41]. Mutation in Tabby and identified as Ectodysplasin A1 (EDAA1), displays a characterized tooth phenotype, associated with significant reduction in the size and number of molar cusps, and frequent absence of incisors and third molar in the studied mice [42]. Another recent study suggested that EDA mutations cause non-syndromic tooth agenesis [43].
Dental Regeneration Via Reactivating the Developmental Cues
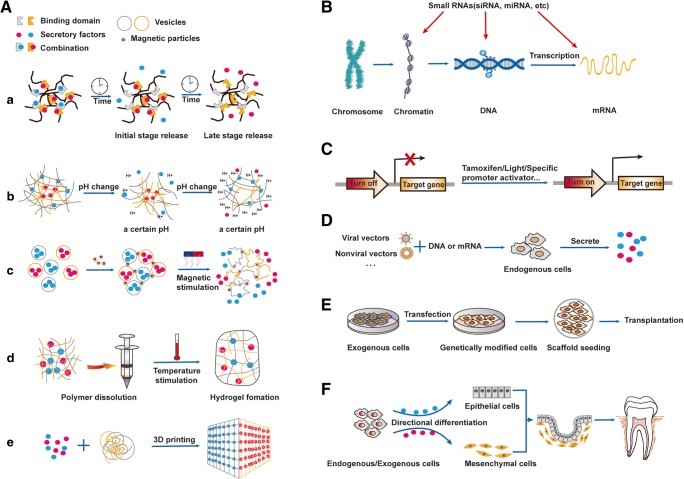
Dental regeneration medicine represents an attractive multidisciplinary approach that offsets traditional dental restoration techniques. As mentioned above, a variety of cytokines participate in different stages of tooth development and in a spatial-temporal manner [1]. The control release of the cytokines for dental regeneration is appealing and is being implemented. Its development depends on research progress in biomaterials, stem cell biology and in other scientific technologies (Fig. 3).
Fig. 3.
Strategies for tooth regeneration by reactivating developmental cues. A Different control release strategies of secretory factors based on biological materials. a) Self-degradation; b) pH-responsive release; c) Magnetic release; d) Thermal release; e) 3D printing. B Small RNAs are involved in different parts of the gene expression process. C Different turn-on/off systems for spatial-temporal control of gene expression. D In vivo delivery of gene expression system. E Transplantation of genetically modified cells. FUnder the above strategies, cells from different sources can be directed to differentiate into specific cells and eventually achieve tooth regeneration
Control Release of Secretory Factors
Biomaterial Based Control of Secretory Factors Releases
Self-degradation is based on the rate of materials degradation in a specific physiological environment, to achieve the spatial-temporal sustained release of cytokines (Fig. 3A a). Although this technique has been widely used in tissue engineering scaffolds, traditional techniques have significant drawbacks, such as high initial release and low bioactive molecular activity. In order to inhibited the burst release of cytokines and enhanced structural stability, many scholars are committed to inventing various kinds of better materials. Fahmy and his co-workers used a low dose of rBMP2 loaded on a resorbable bioactive ceramic to accelerated bone regeneration [44]. Recently, chirality-controlled enzyme-responsive protein nanocapsules were shown to alter the degradation rate by changing the constituent ratio of the material composition, resulting in enhancing wound healing and tissue repair in vivo via the delivery of multiple proteins in a spatiotemporal manner [45]. Affinity interaction is an alternative strategy to achieve sustained release of cytokines. In tissue engineering, the most common way to improve the release kinetics is through heparin-immobilized scaffolds that immobilize cytokines [46]. Wu et al. showed that heparin-based coacervate of FGF2 played a synergistic role with cell proliferation and endogenous facilitated VEGF in improving skin wound healing [47]. In addition, assembly technology like layer-by-layer self-assembly [48] and electrospinning [49] have also made it possible to sustain the release of bioactive molecules in a spatial-temporal manner.
The pH-Responsive Release System
The pH release system regulates cytokines release rate by stimulating the response through pH changes (Fig. 3A b). In a recent review, the assembly structure and slow-release behavior of pH-responsive polymers were analyzed and the potential applications of this kind of materials were prospected [50]. pH-responsive release systems have special significance in tissue regeneration due to pH variations in human tissues and organs, which has begun to be applied to diagnosis [51] and treatment of some diseases [52]. Some pH sustained-release materials have been invented, but there is still a lack of in vivo experiments to prove their application in regenerative medicine [53, 54].
Magnetic and Thermal Release Technology
Nanotechnology, nanocapsules, liposomes, vesicles and other particle/nanoparticle carrier systems are widely investigated for efficient delivery of growth factors. Magneto-nano technology is one of them and its targeting ability can be divided into two different classes [55]. The first class includes magnetic fields to magnetize carriers and magnetic gradients for targeting and that are provided by external magnets. The second class relies on the combination of magnets implanted into the target area and external magnets. Controlled cytokines release can be achieved by directly and thermally heating responsive polymer particles containing magnetic nanoparticles and cytokines, or by secondary heating, where the thermally responsive particles containing cytokines are surrounded by heated magnetic particles. The local temperature increase leads to cytokines release from thermally responsive polymer particles (Fig. 3A c-d) [55]. Magnetic field and magnetic response scaffolds had been used to improve bone repair and regeneration [56]. Fan M et al. have developed nanometers of gelatin chitosan and heparin based on magnetic biopolymers for BMP-2 [57]. In another study, SAOS-2 cells cultured on the gel, combined with bFGF and human serum albumin coated Fe3O4 nanoparticles, have noticeably enhanced alkaline phosphatase activity and calcium deposition activity [58].
3D Printing Technology
The 3D printing technology is a rapid proto-typing and additive manufacturing technology, which manufactures complex architecture via a layer-by-layer building process and with high precision (Fig. 3A e) [59]. The flexibility and controllability of 3D bioprinting enable complex and customized release profiles of multiple cytokines to achieve spatial-temporal gradients that regulate cellular functions in tissue or organ regeneration [60, 61]. Moreover, many studies have promoted the application of 3D printing technology in cytokine sustained-release by improving processing [62], advancing technology [63] or allowing combinations with other forms of carriers [64]. Up to now, these materials have been successfully used in various tissue and organ regeneration experiments in vitro and in vivo, such as vascular regeneration [65], bone regeneration [63] and skin regeneration [66]. The 4D printing technology is a dynamic and time dependent manufacturing process based on advanced 3D-print features, which providing great potential for tissue and organ engineering applications [67].
Control Delivery of Small RNAs
Small RNAs including small interfering RNAs (siRNAs) and microRNAs (miRNAs), are part of the short chain RNAs in non-coding RNAs (ncRNAs) (Fig. 3B). SiRNAs are double-stranded RNAs that downregulate gene expression guided by sequence complementarity with the target mRNA. Since its first discovery in 1998 [68], its delivery strategy has developed rapidly. So far, many different siRNA delivery approaches including siRNA conjugates and lipid nanoparticles, have been applied to disease treatment and tissue regeneration [69]. For example, Zhang et al. developed a targeting system for delivering siRNAs to markedly promoted bone formation [70]. More recently, Castleberry et al. developed an ultrathin polymer coating to sustain the local delivery of siRNA so as to improve wound healing in diabetic mice [71]. Furthermore, the potential toxicities of these technology have been gradually discovered. These include but not limited to on-target effects, sequence-specific off-target effects, immune activation and toxicity associated with the delivery vehicles [72].
MiRNAs can simultaneously identify hundreds of target mRNAs with multiple miRNAs working together for the same mRNA [73]. A As post-transcriptional gene regulators, they can target and disassemble mRNAs or repress their translation [74]. Many studies have shown that miRNAs play a significant regulatory role in tissue repair and regeneration, such as wound healing [75], cardiac repair [76]. In vivo delivery of exogenous miRNAs provides an effective way to regulate gene expression during tissue repair and regeneration, which was proved and validated in mice [77] and zebrafishs [78]. To optimize miRNA delivery, Zhang et al. developed a cell-free 3D scaffold with biodegradable microspheres, that spatially regulated the release of miR-26a to repair critically-sized bone defects in osteoporotic mice [79]. Zhou et al. used miR-126-loaded electrospun membranes for miRNAs local delivery to improve blood vessel regeneration [80]. Moreover, a recent study showed that intracardiac injection of a single administration of synthetic miRNA-lipid formulations enhanced cardiac repair in mice after myocardial infarction [81].
Spatial-Temporal Delivery of Gene Expression Systems
Delivery of gene expression systems that produce locally nascent proteins in vivo, is more advantageous compared to traditional methods for products delivery. In recent years, research on genes-controlled expression has rapidly developed. Some important and potential technologies will briefly be introduced below, and their combinations will also be discussed (Figure C-D).
Spatial-Temporal Control of Gene Expression
Hormone Induction
All kinds of hormones participate in development and regeneration stages. Steroid hormones function by binding to receptor proteins in the cytoplasm of target cells to form hormone-receptor complexes, which enter the nucleus and bind to specific chromosomal sites to regulate the transcription of specific genes. For example, estrogens play pivotal roles in various physiological processes, most of which are mediated by the estrogen receptors alpha (ERα), beta (ERβ) and G protein-coupled receptor 30 (GPR30). Many studies have used estrogen-inducible promoters to modify gene expression systems to regular gene expression [82–84]. Senturk et al. optimized a CRISPR/Cas9 system by combining it with an FKBP12-derived destabilizing domain and an inducible Cre-estrogen receptor fusion domain, which enabled rapid and tunable gene editing [85].
Optogenetics Regulation
Optogenetics is a rapidly developing bioengineering technology which integrates many subjects, such as optics, software control technology, genetic engineering technology, electrophysiological technology. It was originally applied in the field of neurology and a recent review indicated that it could control nerve growth and neurotrophic factor expression in a precise spatial and temporal manner [86]. The light-based mechanisms can activate or inhibit the expression of target genes in the FGF [87], WNT/β-catenin [88] and TGF-β signaling pathways [89] by light-induced conformational change of various photoactivatable proteins or photocaging/uncaging of effectors [90]. Yang et al. created the LightON system, a light-switchable transgene system, which can initiate spatiotemporal expressions of target transgenes in mammalian cells, upon light stimulation [91]. However, potential toxicity associated with the high expression was reported by a study of zebrafish embryogenesis, which may limited its application [92]. To overcome this obstacle, the blue-light activated EL222 system, renamed TAEL was invented, and which drived the expression with minimal toxicity [93, 94]. In addition, some studies have used optical gene elements to link Cre recombinase to regulate DNA recombination [95, 96]. Recently, Nguyen et al. combined genetically encoded photo-switchable calcium actuators with dCas9 to control gene expression, overcoming some limitations of the CRISPR/Cas9 (dCas9) system [97]. Simultaneously, a CRISPR-dCas9 effector device that is activated by far-red light (FRL), engineered by Shao and his research team, efficiently promoted the differentiation of induced pluripotent stem cells (iPSCs) into functional neurons by up-regulating NEUROG2, a single neural transcription factor [98].
Dental Development-Related Specific Promoters
In the process of tooth development, some site-specific promoters like WNT1 promoter, play a vital role in regulating the orderly expression of genes. WNT1 encodes the signaling protein WNT1, involved in the canonical WNT pathway. Previous research has shown that the expression of WNT1 is restricted to the migrating neural crest cells, which contribute to tooth and mandible development [99]. Simultaneously, Chai et al. successfully constructed a transgenic model under the control of the WNT1 promoter [99]. Up to now, this conditional knockout model of transgenic mice has been widely used in the study of tooth development and regeneration [100–102].
In addition, dentin matrix protein 1 (DMP1) produced by odontoblasts and osteoblasts is mainly expressed in bone and dentin [103]. Jacob et al. showed that TCF11, which could specifically bind to the DMP1 promoter, played a significant role in regulating the transcription of DMP1 in odontoblasts and osteoblasts [103]. This provides a way to spatiotemporally regulate the expression of DMP1.
In Vivo Delivery of Gene Expression System
The in vivo gene delivery strategy can be generally divided into viral and non-viral vector delivery systems (Fig. 4). Viral vectors including oncoretroviruses, lentiviruses (LVs), adenoviruses (AVs) and adeno-associated viruses (AAVs), have relatively high efficiency. Initially, they are widely used in changing the expression of specific genes in vivo and in vitro [104]. In contrast to LVs, the nonintegrated DNA delivered by AAVs would be diluted during mitosis because of lack of integration machinery. However, it could be stably maintained in a nonintegrated form to mediate persistent gene expression in predominantly postmitotic cells [104]. With regard to damage repair and tissue regeneration, Eggers et al. used a lentiviral vector to regulate controlled expression of glial cell-line derived neurotrophic factor (GDNF), which exerts multiple effects on both Schwann cells and axons in the injured peripheral nerve [105]. Moreover, adenovirus-mediated WNT10b overexpression promoted hair follicle regeneration via the activation of the canonical WNT signaling pathway [106]. To overcome safety concerns, such as immune system activation and insertional mutagenesis, the next-generation gene therapy vectors must be developed. Hu et al. showed that a virus-biotin-avidin-biotin-material (VBABM) arrangement immobilized viral vectors on biomaterial scaffolds, that contributed to spatially control therapeutic gene delivery in bone regeneration [107]. In addition, hydrodynamic tail-vein injections of lentiviral gene delivery reduced off-target delivery and transduction in mouse liver [108].
Fig. 4.
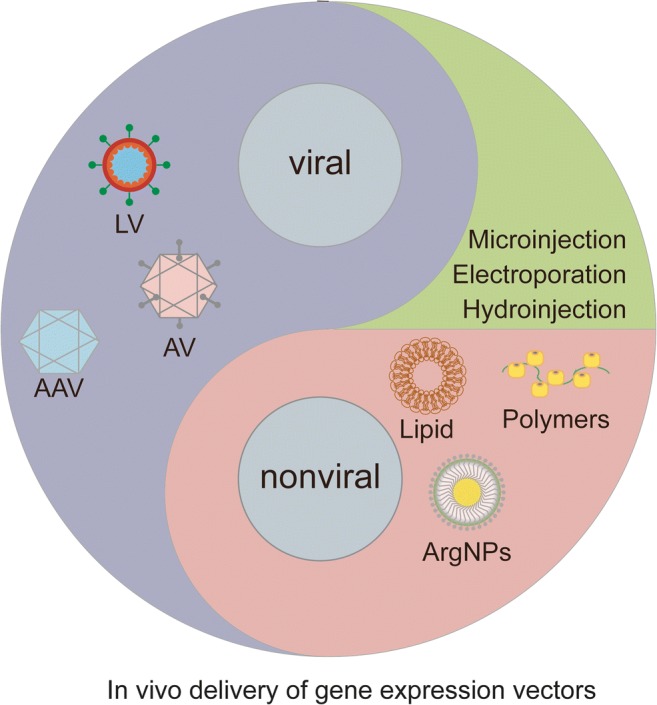
In vivo gene delivery strategies. The gene expression systems could be delivered non-virally and virally, both of which have advantages and disadvantages. LV, lentivirus; AV, adenovirus; AAV, adeno-associated virus; Lipid, liposome; ArgNPs, cationic arginine gold nanoparticles
Unlike viral vectors, non-viral vectors showed the potential to overcome many of the shortcomings of viral vectors [109]. There are a great variety of non-viral vector systems, including naked DNA or mRNA microinjection or electroporation [110], liposomes [111], cationic polymers [111], chitosan polymers [112], inorganic nanoparticles [113], transposon systems [114]. So far, the use of non-viral gene vectors has been explored in many tissue regeneration approaches, such as bone regeneration [112], cartilage regeneration [115], tendon repair [116], nerve regeneration [117]. For example, Feng et al. combined nanofibrous spongy microspheres with biodegradable nanospheres to synthesize a two-stage delivery system for plasmid DNA encoding orphan nuclear receptor 4A1 (NR4A1), and which was proved to be effective in promoting disc regeneration [118].
In recent years, exosomes have also been used as a non-viral vector of nucleic acid in regenerative medicine as an alternative to cell therapy [119]. Natural exosomes can be used to transfer small nucleic acid like siRNAs [120] and miRNAs [121], but have been shown to have issues in delivering large nucleic acids, such as plasmid DNA. Some scholars try to modify and optimize the structure of exosomes to overcome this obstacle. For example, Lin et al. developed exosome-liposome hybrid nanoparticles to deliver large nucleic acid like CRISPR-Cas9 system by increasing the binding of exosome and large nucleic acid [102].
In addition, because the messenger ribonucleic acid (mRNA) translation happens in the cytoplasm, the delivery of mRNA associated with non-viral gene delivery systems might have higher transfection efficiencies than DNA, by eliminating the need for nuclear entry. In vivo studies showed that the delivery of chemically modified ribonucleic acid (cmRNA) significantly enhances bone regeneration when compared with that of conventional plasmid DNA [122, 123].
Transplantation of Genetically Modified Cells
Due to safety concern about dissemination of the gene vectors and their adverse side-effect in non-target sites, a direct injection of the expression vector was used less than an indirect injection of cells injection. Genetically modified cell transplantation technology has been widely applied to the research of gene therapy in vitro and in vivo for many human diseases, such as junctional epidermolysis bullosa [124], metastatic cancer [125], type 1 diabetes [126]. As mentioned above, the process of tissue regeneration is regulated spatial-temporally by a variety of cytokines. Therefore, the transplantation of genetically modified cells, which express specific cytokines, could be used to promote organ and tissue regeneration (Fig. 3E). Nascent cytokines locally synthesized may have higher activity than recombinant counterparts [127].
Mesenchymal stem cells (MSCs) have many advantages in cell transplantation, such as their easy isolation and culture, secretion of a variety of cytokines, migration and homing to damaged tissues or solid tumors. Therefore, genetically modified MSCs have been used as therapeutic cytoreagents for gene therapy [128]. For example, the upregulation of BMP2 in iPSC-MSCs can promote osteogenic differentiation and bone mineralization [129]. In another study, FGF-2 transfected BMSCs effectively promoted the repair effect of avascular necrosis of femoral head in rabbits [130]. Nevertheless, MSCs are not supposed to be immune privileged, therefore the efficacy of allogeneic MSC therapies is insure in clinical treatment [131].
Both MSCs and macrophages from monocytes can produce multiple cytokines and have been used in cell therapies [132]. Ben-Mordechai et al. showed that the favorable effects of MSC therapy in myocardial infarction (MI) were mediated by macrophages [133]. Macrophage-based therapeutic strategies have been applied in regenerative medicine for a long time [134]. Cells of the monocyte-macrophage lineage play key roles in liver regeneration and function after liver cirrhosis [135]. They might act via regulating Notch and WNT signaling pathways to specify hepatic progenitor cell fate [136]. However, only few studies focused on genetically modified monocytes or macrophages. The study of Hamm et al. indicated that genetically modified macrophages expressing low levels of prolyl hydroxylase domain protein 2 (PHD2), significantly contributed to angiogenesis through the TIE2 signaling pathway [137].
Apart from MSCs and macrophages, many other cells can also be genetically modified in regenerative medicine. In a previous review, the transplantation of transfected dental pulp progenitor/stem cells with BMP genes, by electroporation or sonoporation, into the injured pulp using an appropriate scaffold could enhance reparative dentin formation [138]. Moreover, human periodontal ligament cells (hPDLCs) transfected with LV-Ctnnb lentivirus to explore the effect of its overexpression, were further confirmed to activate the canonical WNT signaling pathway and induce cementogenic differentiation in vitro and cementum regeneration in vivo [139].
Tooth Regeneration on the Way
Besides tooth tissue regeneration, studies on regenerating a whole tooth organ are also hot topics. Nakao and colleagues engineered the bioengineered incisor tooth germ by reconstituting single cells that were isolated from the epithelium and mesenchyme of the dental germ [140]. Ikeda and colleagues transplanted a bioengineered tooth germ into the alveolar socket of an adult mouse, and the results showed that the tooth successfully erupted and achieved occlusion [141]. Further studies have shown that the bioengineered tooth had masticatory properties and responded to harmful stimuli [141]. These studies on bioengineered tooth, suggest that total dental regeneration can be reached by achieving targeted differentiation and specific expression patterns of stem cells.
Notably, tooth regeneration can only be completed by specific epithelial and mesenchymal stem cells; while, the identity of these tooth-regenerating stem cells remains largely unknown, including their capacity to induce commonly available cells to the specific status. Future works defining the essential factors and the spatial-temporal pattern that induce the cells to the specific stage, and the development of an in vivo/in vitro control release system to deliver or release the factors in a spatial-temporal pattern, would certainly shed light on tooth regeneration (Fig. 3F).
Outlook
Several systems that are effective in vitro or in preclinical models may fail to translate into the clinic. At present, although many scholars devote themselves to study the spatial-temporal release system of cytokines in tooth regeneration, none of these scholars have developed a system that can simulate the release behavior of cytokines during tooth development. Due to the complexity of the spatial-temporal regulatory network of tooth development, identifying key and essential factors for tooth regeneration is still being intensively explored, and if successful, would make the regeneration practical by the release of limited factors. Moreover, the in vivo safety of these systems needs more evidences before clinical translation.
In future studies, researches should be focused on: 1) Defining the essential factors for tooth regeneration; 2) Characterizing the spatial-temporal dynamics of the above identified factors; 3) Developing in vivo/in vitro control release system to deliver or release the factors in a spatial-temporal pattern, as needed by combinatorial application of biomaterials, nanobiotechnology, 3D printing and other technologies. Hopefully, a spatial-temporal control release system of cytokines will be clinically applied to human tooth regeneration in the future, which will greatly improve the quality of life, especially for the edentulous patients.
Acknowledgements
Funding: This work was supported by grants from National Key R&D Program of China (2016YFC20160905200), National Natural Science Foundation of China (81600824), Natural Science Foundation of Guangdong Province (2016A030310220, 2018A030310278), Young Teachers Training Program of Sun Yat-sen University (16ykpy46, 17ykpy73), Science and Technology Program of Guangzhou (201707010106, 201804010459) and the Young Elite Scientist Sponsorship Program by CAST (2016QNRC001).
Authors’ Contributions
DH was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Delan Huang, Email: huangdl3@mail2.sysu.edu.cn.
Jianhan Ren, Email: renjh3@mail2.sysu.edu.cn.
Runze Li, Email: lirunze@mail2.sysu.edu.cn.
Chenyu Guan, Email: guanchy@mail2.sysu.edu.cn.
Zhicai Feng, Email: fengzhc@mail.sysu.edu.cn.
Baicheng Bao, Email: baobch@mail.sysu.edu.cn.
Weicai Wang, Email: wangwc3@mail.sysu.edu.cn.
Chen Zhou, Email: zhouch46@mail.sysu.edu.cn.
References
- 1.Yan DZ, Zhi C, Yi QS, Chao L, Yi PC. Making a tooth: Growth factors, transcription factors, and stem cells. Cell Research. 2005;15(5):301–316. doi: 10.1038/sj.cr.7290299. [DOI] [PubMed] [Google Scholar]
- 2.Thiery JP, Duband JL, Delouvee A. Pathways and mechanisms of avian trunk neural crest cell migration and localization. Developmental Biology. 1982;93(2):324–343. doi: 10.1016/0012-1606(82)90121-x. [DOI] [PubMed] [Google Scholar]
- 3.Thesleff I, Mikkola M. The role of growth factors in tooth development. International Review of Cytology. 2002;217:93–135. doi: 10.1016/s0074-7696(02)17013-6. [DOI] [PubMed] [Google Scholar]
- 4.Ten Cate AR. The role of epithelium in the development, structure and function of the tissues of tooth support. Oral Diseases. 1996;2(1):55–62. doi: 10.1111/j.1601-0825.1996.tb00204.x. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Yu M, Tian W. An inductive signalling network regulates mammalian tooth morphogenesis with implications for tooth regeneration. Cell Proliferation. 2013;46(5):501–508. doi: 10.1111/cpr.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graf D, Malik Z, Hayano S, Mishina Y. Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine & Growth Factor Reviews. 2016;27:129–139. doi: 10.1016/j.cytogfr.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai Y, Maxson R. Recent advances in craniofacial morphogenesis. Developmental Dynamics. 2006;235(9):2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- 8.Laurikkala J, Mikkola M, Mustonen T, Aberg T, Koppinen P, Pispa J, Nieminen P, Galceran J, Grosschedl R, Thesleff I. TNF signaling via the ligand-receptor pair ectodysplasin and edar controls the function of epithelial signaling centers and is regulated by Wnt and activin during tooth organogenesis. Developmental Biology. 2001;229(2):443–455. doi: 10.1006/dbio.2000.9955. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Martin JF. Hippo pathway: An emerging regulator of craniofacial and dental development. Journal of Dental Research. 2017;96(11):1229–1237. doi: 10.1177/0022034517719886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie F, Dai Q, Liu X, Wang J. Conditional knockout of raptor/mTORC1 results in dentin malformation. Frontiers in Physiology. 2019;10:250. doi: 10.3389/fphys.2019.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia S, Zhou J, Gao Y, Baek J, Martin J, Lan Y, Jiang R. Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development. 2013;140(2):423–432. doi: 10.1242/dev.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia S, Kwon H, Lan Y, Zhou J, Liu H, Jiang R. Bmp4-Msx1 signaling and Osr2 control tooth organogenesis through antagonistic regulation of secreted Wnt antagonists. Developmental Biology. 2016;420(1):110–119. doi: 10.1016/j.ydbio.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connell D. J., Ho J. W. K., Mammoto T., Turbe-Doan A., O'Connell J. T., Haseley P. S., Koo S., Kamiya N., Ingber D. E., Park P. J., Maas R. L. A Wnt-Bmp Feedback Circuit Controls Intertissue Signaling Dynamics in Tooth Organogenesis. Science Signaling. 2012;5(206):ra4–ra4. doi: 10.1126/scisignal.2002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, Z., Wang, L., Wang, F., Zhang, C., Wang, J., He, J., & Wang, S. (2018). Expression of BMP2/4/7 during the odontogenesis of deciduous molars in miniature pig embryos. Journal of Molecular Histology. [DOI] [PubMed]
- 15.Malik Z, Alexiou M, Hallgrimsson B, Economides A, Luder H, Graf D. Bone morphogenetic protein 2 coordinates early tooth mineralization. Journal of Dental Research. 2018;97(7):835–843. doi: 10.1177/0022034518758044. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Wang F, Zhao C, Yang S, Cheng Q, Tang Y, Zhang F, Zhang Y, Luo W, Wang C, Zhou P, Kim S, Zuo G, Hu N, Li R, He TC, Zhang H. Dentinogenesis and tooth-alveolar bone complex defects in BMP9/GDF2 knockout mice. Stem Cells and Development. 2019;28(10):683–694. doi: 10.1089/scd.2018.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kettunen P, Thesleff I. Expression and function of FGFs-4, −8, and −9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Developmental Dynamics. 1998;211(3):256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Kettunen P, Laurikkala J, Itäranta P, Vainio S, Itoh N, Thesleff I. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Developmental Dynamics. 2000;219(3):322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 19.Porntaveetus T, Otsuka-Tanaka Y, Basson M, Moon A, Sharpe P, Ohazama A. Expression of fibroblast growth factors (Fgfs) in murine tooth development. Journal of Anatomy. 2011;218(5):534–543. doi: 10.1111/j.1469-7580.2011.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neubüser A, Peters H, Balling R, Martin G. Antagonistic interactions between FGF and BMP signaling pathways: A mechanism for positioning the sites of tooth formation. Cell. 1997;90(2):247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- 21.Prochazka Jan, Prochazkova Michaela, Du Wen, Spoutil Frantisek, Tureckova Jolana, Hoch Renee, Shimogori Tomomi, Sedlacek Radislav, Rubenstein John L., Wittmann Torsten, Klein Ophir D. Migration of Founder Epithelial Cells Drives Proper Molar Tooth Positioning and Morphogenesis. Developmental Cell. 2015;35(6):713–724. doi: 10.1016/j.devcel.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou C, Yang G, Chen M, He L, Xiang L, Ricupero C, Mao JJ, Ling J. Lhx6 and Lhx8: Cell fate regulators and beyond. The FASEB Journal. 2015;29(10):4083–4091. doi: 10.1096/fj.14-267500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou C, Yang G, Chen M, Wang C, He L, Xiang L, Chen D, Ling J, Mao J. Lhx8 mediated Wnt and TGFβ pathways in tooth development and regeneration. Biomaterials. 2015;63:35–46. doi: 10.1016/j.biomaterials.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai YY, Chen RS, Lin Y, Ling TY, Chen MH. FGF-9 accelerates epithelial invagination for ectodermal organogenesis in real time bioengineered organ manipulation. Cell Communication and Signaling: CCS. 2012;10(1):34. doi: 10.1186/1478-811x-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B, Li H, Liu Y, Lin X, Lin Y, Wang Y, Hu X, Zhang Y. Expression patterns of WNT/β-CATENIN signaling molecules during human tooth development. Journal of Molecular Histology. 2014;45(5):487–496. doi: 10.1007/s10735-014-9572-5. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar L, Cobourne M, Naylor S, Smalley M, Dale T, Sharpe PT. Wnt/Shh interactions regulate ectodermal boundary formation during mammalian tooth development. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(9):4520–4524. doi: 10.1073/pnas.97.9.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dassule H, McMahon A. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Developmental Biology. 1998;202(2):215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- 28.Sarkar L, Sharpe PT. Expression of Wnt signalling pathway genes during tooth development. Mechanisms of Development. 1999;85(1–2):197–200. doi: 10.1016/s0925-4773(99)00095-7. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, Li Y, Wang F, Hu L, Li Y, Wang J, Zhang C, Wang S. Spatiotemporal expression of Wnt/β-catenin signaling during morphogenesis and Odontogenesis of deciduous molar in miniature pig. International Journal of Biological Sciences. 2017;13(8):1082–1091. doi: 10.7150/ijbs.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Genderen C, Okamura R, Fariñas I, Quo R, Parslow T, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes & Development. 1994;8(22):2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 31.Bae C, Kim T, Ko S, Lee J, Yang X, Cho E. Wntless regulates dentin apposition and root elongation in the mandibular molar. Journal of Dental Research. 2015;94(3):439–445. doi: 10.1177/0022034514567198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aurrekoetxea, M., Irastorza, I., García-Gallastegui, P., Jiménez-Rojo, L., Nakamura, T., Yamada, Y., . . . Unda, F. (2016). Wnt/β-catenin regulates the activity of Epiprofin/Sp6, SHH, FGF, and BMP to coordinate the stages of Odontogenesis. Frontiers in Cell and Development Biology, 4, 25. [DOI] [PMC free article] [PubMed]
- 33.Yu P, Yang W, Han D, Wang X, Guo S, Li J, Li F, Zhang X, Wong SW, Bai B, Liu Y, du J, Sun ZS, Shi S, Feng H, Cai T. Mutations in WNT10B are identified in individuals with Oligodontia. American Journal of Human Genetics. 2016;99(1):195–201. doi: 10.1016/j.ajhg.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, R. L., & Tabin, C. (1995). The long and short hedgehog signaling. 81(3), 313-316. [DOI] [PubMed]
- 35.Vaahtokari A, Åberg T, Jernvall J, Keränen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mechanisms of Development. 1996;54(1):39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- 36.Koyama, E., Yamaai, T., Iseki, S., Ohuchi, H., Nohno, T., Yoshioka, H., . . . Noji, S. (1996). Polarizing activity, sonic hedgehog, and tooth development in embryonic and postnatal mouse. Developmental Dynamics, 206(1), 59–72. [DOI] [PubMed]
- 37.Dassule H, Lewis P, Bei M, Maas R, McMahon A. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127(22):4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 38.Bae WJ, Auh QS, Lim HC, Kim GT, Kim HS, Kim EC. Sonic hedgehog promotes Cementoblastic differentiation via activating the BMP pathways. Calcified Tissue International. 2016;99(4):396–407. doi: 10.1007/s00223-016-0155-1. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Feng J, Liu Y, Ho T, Grimes W, Ho H, Park S, Wang S, Chai Y. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Developmental Cell. 2015;33(2):125–135. doi: 10.1016/j.devcel.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho S.-W., Kwak S., Woolley T. E., Lee M.-J., Kim E.-J., Baker R. E., Kim H.-J., Shin J.-S., Tickle C., Maini P. K., Jung H.-S. Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop for spatial patterning of the teeth. Development. 2011;138(9):1807–1816. doi: 10.1242/dev.056051. [DOI] [PubMed] [Google Scholar]
- 41.Kowalczyk-Quintas C, Schneider P. Ectodysplasin a (EDA) - EDA receptor signalling and its pharmacological modulation. Cytokine & Growth Factor Reviews. 2014;25(2):195–203. doi: 10.1016/j.cytogfr.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Pispa J, Jung H, Jernvall J, Kettunen P, Mustonen T, Tabata M, Kere J, Thesleff I. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Developmental Biology. 1999;216(2):521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- 43.Shen W, Wang Y, Liu Y, Liu H, Zhao H, Zhang G, Snead ML, Han D, Feng H. Functional study of Ectodysplasin-a mutations causing non-Syndromic tooth agenesis. PLoS One. 2016;11(5):e0154884. doi: 10.1371/journal.pone.0154884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fahmy R, Mahmoud N, Soliman S, Nouh S, Cunningham L, El-Ghannam A. Acceleration of alveolar ridge augmentation using a low dose of recombinant human bone morphogenetic Protein-2 loaded on a Resorbable bioactive ceramic. Journal of Oral and Maxillofacial Surgery. 2015;73(12):2257–2272. doi: 10.1016/j.joms.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Zhu S, Nih L, Carmichael S, Lu Y, Segura T. Enzyme-responsive delivery of multiple proteins with spatiotemporal control. Adv. Mater. Weinheim. 2015;27(24):3620–3625. doi: 10.1002/adma.201500417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knaack S, Lode A, Hoyer B, Rösen-Wolff A, Gabrielyan A, Roeder I, Gelinsky M. Heparin modification of a biomimetic bone matrix for controlled release of VEGF. Journal of Biomedical Materials Research. Part A. 2014;102(10):3500–3511. doi: 10.1002/jbm.a.35020. [DOI] [PubMed] [Google Scholar]
- 47.Wu J, Ye J, Zhu J, Xiao Z, He C, Shi H, Wang Y, Lin C, Zhang H, Zhao Y, Fu X, Chen H, Li X, Li L, Zheng J, Xiao J. Heparin-based Coacervate of FGF2 improves dermal regeneration by asserting a synergistic role with cell proliferation and endogenous facilitated VEGF for cutaneous wound healing. Biomacromolecules. 2016;17(6):2168–2177. doi: 10.1021/acs.biomac.6b00398. [DOI] [PubMed] [Google Scholar]
- 48.Shah N, Hyder M, Quadir M, Dorval Courchesne N, Seeherman H, Nevins M, Spector M, Hammond P. Adaptive growth factor delivery from a polyelectrolyte coating promotes synergistic bone tissue repair and reconstruction. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(35):12847–12852. doi: 10.1073/pnas.1408035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mei L, Wang Y, Tong A, Guo G. Facile electrospinning of an efficient drug delivery system. Expert Opinion on Drug Delivery. 2016;13(5):741–753. doi: 10.1517/17425247.2016.1142525. [DOI] [PubMed] [Google Scholar]
- 50.Kocak G, Tuncer C, Bütün V. pH-responsive polymers. Polymer Chemistry. 2016;8(1):144–176. [Google Scholar]
- 51.Baliga S, Muglikar S, Kale R. Salivary pH: A diagnostic biomarker. Journal of Indian Society of Periodontology. 2013;17(4):461–465. doi: 10.4103/0972-124X.118317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalhapure RS, Jadhav M, Rambharose S, Mocktar C, Singh S, Renukuntla J, Govender T. pH-responsive chitosan nanoparticles from a novel twin-chain anionic amphiphile for controlled and targeted delivery of vancomycin. Colloids & Surfaces B Biointerfaces. 2017;158(4):650. doi: 10.1016/j.colsurfb.2017.07.049. [DOI] [PubMed] [Google Scholar]
- 53.Halacheva SS, Adlam DJ, Hendow EK, Freemont TJ, Hoyland J, Saunders BR. Injectable biocompatible and biodegradable pH-responsive hollow particle gels containing poly(acrylic acid): The effect of copolymer composition on gel properties. Biomacromolecules. 2014;15(5):1814–1827. doi: 10.1021/bm5002069. [DOI] [PubMed] [Google Scholar]
- 54.Alvarez Echazú María Inés, Olivetti Christian Ezequiel, Peralta Ignacio, Alonso Maria Rosario, Anesini Claudia, Perez Claudio Javier, Alvarez Gisela Solange, Desimone Martin Federico. Development of pH-responsive biopolymer-silica composites loaded with Larrea divaricata Cav. extract with antioxidant activity. Colloids and Surfaces B: Biointerfaces. 2018;169:82–91. doi: 10.1016/j.colsurfb.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 55.Sensenig R, Sapir Y, Macdonald C, Cohen S, Polyak B. Magnetic nanoparticle-based approaches to locally target therapy and enhance tissue regeneration in vivo. Nanomedicine. 2012;7(9):1425–1442. doi: 10.2217/nnm.12.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li, Y., Ye, D., Li, M., Ma, M., & Gu, N. (2018). Adaptive materials based on Iron oxide nanoparticles for bone regeneration. Chemphyschem A European Journal of Chemical Physics & Physical Chemistry. [DOI] [PubMed]
- 57.Fan M, Yan J, Tan H, Miao Y, Hu X. Magnetic biopolymer nanogels via biological assembly for vectoring delivery of biopharmaceuticals. Journal of Materials Chemistry B. 2014;2(47):8399–8405. doi: 10.1039/c4tb01106f. [DOI] [PubMed] [Google Scholar]
- 58.Karahaliloğlu, Z., Yalçın, E., Demirbilek, M., & Denkbaş, E. B. (2017). Magnetic silk fibroin e-gel scaffolds for bone tissue engineering applications. Journal of Bioactive\s&\scompatible Polymers, 32(6), 088391151769363.
- 59.O'Brien CM, Holmes B, Faucett S, Zhang LG. Three-dimensional printing of nanomaterial scaffolds for complex tissue regeneration. Tissue Engineering. Part B, Reviews. 2015;21(1):103–114. doi: 10.1089/ten.teb.2014.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Legemate K, Tarafder S, Jun Y, Lee C. Engineering human TMJ discs with protein-releasing 3D-printed scaffolds. Journal of Dental Research. 2016;95(7):800–807. doi: 10.1177/0022034516642404. [DOI] [PubMed] [Google Scholar]
- 61.Hamlet SM, Vaquette C, Shah A, Hutmacher DW, Ivanovski S. 3-dimensional functionalized polycaprolactone-hyaluronic acid hydrogel constructs for bone tissue engineering. Journal of Clinical Periodontology. 2016;44(4):428. doi: 10.1111/jcpe.12686. [DOI] [PubMed] [Google Scholar]
- 62.Huang K, Lin Y, Shie M, Lin C. Effects of bone morphogenic protein-2 loaded on the 3D-printed MesoCS scaffolds. Journal of the Formosan Medical Association. 2018;117(10):879–887. doi: 10.1016/j.jfma.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 63.Wang C, Zhao Q, Wang M. Cryogenic 3D printing for producing hierarchical porous and rhBMP-2-loaded Ca-P/PLLA nanocomposite scaffolds for bone tissue engineering. Biofabrication. 2017;9(2):025031. doi: 10.1088/1758-5090/aa71c9. [DOI] [PubMed] [Google Scholar]
- 64.Li S, Xu Y, Yu J, Becker ML. Enhanced osteogenic activity of poly(ester urea) scaffolds using facile post-3D printing peptide functionalization strategies. Biomaterials. 2017;141:176–187. doi: 10.1016/j.biomaterials.2017.06.038. [DOI] [PubMed] [Google Scholar]
- 65.Wagner, E. R., Parry, J., Dadsetan, M., Bravo, D., Riester, S. M., Wijnen, A. J. V., . . . Kakar, S. (2018). VEGF-mediated angiogenesis and vascularization of a fumarate-crosslinked polycaprolactone (PCLF) scaffold. Connective Tissue Research, 1-8. [DOI] [PubMed]
- 66.Xiong S, Zhang X, Lu P, Wu Y, Wang Q, Sun H, Heng BC, Bunpetch V, Zhang S, Ouyang H. A gelatin-sulfonated silk composite scaffold based on 3D printing technology enhances skin regeneration by stimulating epidermal growth and dermal neovascularization. Scientific Reports. 2017;7(1):4288. doi: 10.1038/s41598-017-04149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miao Shida, Castro Nathan, Nowicki Margaret, Xia Lang, Cui Haitao, Zhou Xuan, Zhu Wei, Lee Se-jun, Sarkar Kausik, Vozzi Giovanni, Tabata Yasuhiko, Fisher John, Zhang Lijie Grace. 4D printing of polymeric materials for tissue and organ regeneration. Materials Today. 2017;20(10):577–591. doi: 10.1016/j.mattod.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 69.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nature Materials. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 70.Zhang G, Guo B, Wu H, Tang T, Zhang BT, Zheng L, He Y, Yang Z, Pan X, Chow H, To K, Li Y, Li D, Wang X, Wang Y, Lee K, Hou Z, Dong N, Li G, Leung K, Hung L, He F, Zhang L, Qin L. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nature Medicine. 2012;18(2):307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 71.Castleberry SA, Almquist BD, Li W, Reis T, Chow J, Mayner S, Hammond PT. Self-assembled wound dressings silence MMP-9 and improve diabetic wound healing in vivo. Advanced Materials. 2016;28(9):1809–1817. doi: 10.1002/adma.201503565. [DOI] [PubMed] [Google Scholar]
- 72.Wittrup A, Lieberman J. Knocking down disease: A progress report on siRNA therapeutics. Nature Reviews. Genetics. 2015;16(9):543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 74.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 75.Li Dongqing, Wang Aoxue, Liu Xi, Meisgen Florian, Grünler Jacob, Botusan Ileana R., Narayanan Sampath, Erikci Erdem, Li Xi, Blomqvist Lennart, Du Lei, Pivarcsi Andor, Sonkoly Enikö, Chowdhury Kamal, Catrina Sergiu-Bogdan, Ståhle Mona, Landén Ning Xu. MicroRNA-132 enhances transition from inflammation to proliferation during wound healing. Journal of Clinical Investigation. 2015;125(8):3008–3026. doi: 10.1172/JCI79052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, Fisch S, Unno K, Sereti KI, Liao R. MicroRNA-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circulation Research. 2015;117(5):450–459. doi: 10.1161/circresaha.117.305962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 78.Aguirre Aitor, Montserrat Nuria, Zacchigna Serena, Nivet Emmanuel, Hishida Tomoaki, Krause Marie N., Kurian Leo, Ocampo Alejandro, Vazquez-Ferrer Eric, Rodriguez-Esteban Concepcion, Kumar Sachin, Moresco James J., Yates John R., Campistol Josep M., Sancho-Martinez Ignacio, Giacca Mauro, Izpisua Belmonte Juan Carlos. In Vivo Activation of a Conserved MicroRNA Program Induces Mammalian Heart Regeneration. Cell Stem Cell. 2014;15(5):589–604. doi: 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X, Li Y, Chen YE, Chen J, Ma PX. Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects. Nature Communications. 2016;7:10376. doi: 10.1038/ncomms10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou F, Jia X, Yang Y, Yang Q, Gao C, Hu S, Zhao Y, Fan Y, Yuan X. Nanofiber-mediated microRNA-126 delivery to vascular endothelial cells for blood vessel regeneration. Acta Biomaterialia. 2016;43:303–313. doi: 10.1016/j.actbio.2016.07.048. [DOI] [PubMed] [Google Scholar]
- 81.Lesizza P, Prosdocimo G, Martinelli V, Sinagra G, Zacchigna S, Giacca M. Single-dose Intracardiac injection of pro-regenerative MicroRNAs improves cardiac function after myocardial infarction. Circulation Research. 2017;120(8):1298–1304. doi: 10.1161/circresaha.116.309589. [DOI] [PubMed] [Google Scholar]
- 82.Pyo KH, Lim SM, Kim HR, Sung YH, Yun MR, Kim SM, Kim H, Kang HN, Lee JM, Kim SG, Park CW, Chang H, Shim HS, Lee HW, Cho BC. Establishment of a conditional transgenic mouse model recapitulating EML4-ALK-positive human non-small cell lung Cancer. Journal of Thoracic Oncology. 2017;12(3):491–500. doi: 10.1016/j.jtho.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 83.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, J Lin SC, Aronow BJ, Tallquist MD, Molkentin JD. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nature Communications. 2016;7:12260. doi: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Molkentin JD, Bugg D, Ghearing N, Dorn LE, Kim P, Sargent MA, Gunaje J, Otsu K, Davis J. Fibroblast-specific genetic manipulation of p38 mitogen-activated protein kinase in vivo reveals its central regulatory role in fibrosis. Circulation. 2017;136(6):549–561. doi: 10.1161/circulationaha.116.026238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Senturk S, Shirole NH, Nowak DG, Corbo V, Pal D, Vaughan A, Tuveson DA, Trotman LC, Kinney JB, Sordella R. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nature Communications. 2017;8:14370. doi: 10.1038/ncomms14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park S, Koppes RA, Froriep UP, Jia X, Achyuta AKH, Mclaughlin BL, Anikeeva P. Optogenetic control of nerve growth. Scientific Reports. 2015;5:9669. doi: 10.1038/srep09669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim N, Kim JM, Lee M, Kim CY, Chang KY, Heo WD. Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chemistry & Biology. 2014;21(7):903–912. doi: 10.1016/j.chembiol.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 88.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nature Methods. 2013;10(3):249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 89.Li Y, Lee M, Kim N, Wu G, Deng D, Kim J, Liu X, Heo WD, Zi Z. Spatiotemporal control of TGF-β signaling with light. ACS Synthetic Biology. 2018;7(2):443–451. doi: 10.1021/acssynbio.7b00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beyer HM, Naumann S, Weber W, Radziwill G. Optogenetic control of signaling in mammalian cells. Biotechnology Journal. 2015;10(2):273–283. doi: 10.1002/biot.201400077. [DOI] [PubMed] [Google Scholar]
- 91.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nature Methods. 2012;9(3):266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 92.Distel M, Wullimann MF, Köster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mottamena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, Gardner KH. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nature Chemical Biology. 2014;10(3):196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reade A, Motta-Mena LB, Gardner KH, Stainier DY, Weiner OD, Woo S. TAEL: A zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control. Development. 2017;144(2):345–355. doi: 10.1242/dev.139238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schindler, S. E., Mccall, J. G., Ping, Y., Hyrc, K. L., Li, M., Tucker, C. L., . . . Diamond, M. I. (2015). Photo-activatable Cre recombinase regulates gene expression in vivo. Scientific Reports, 5, 13627. [DOI] [PMC free article] [PubMed]
- 96.Kawano F, Okazaki R, Yazawa M, Sato M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nature Chemical Biology. 2016;12(12):1059–1064. doi: 10.1038/nchembio.2205. [DOI] [PubMed] [Google Scholar]
- 97.Nguyen N, He L, Martinez-Moczygemba M, Huang Y, Zhou Y. Rewiring calcium signaling for precise transcriptional reprogramming. ACS Synthetic Biology. 2018;7(3):814–821. doi: 10.1021/acssynbio.7b00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shao J, Wang M, Yu G, Zhu S, Yu Y, Heng B, Wu J, Ye H. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(29):E6722–E6730. doi: 10.1073/pnas.1802448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chai, Y., Jiang, X., Ito, Y., Bringas, P., Jr., Han, J., Rowitch, D. H., . . . Sucov, H. M. (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development, 127(8), 1671–1679. [DOI] [PubMed]
- 100.Liu B, Chen S, Cheng D, Jing W, Helms JA. Primary cilia integrate hedgehog and Wnt signaling during tooth development. Journal of Dental Research. 2014;93(5):475–482. doi: 10.1177/0022034514528211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo S, Zhang Y, Zhou T, Wang D, Weng Y, Wang L, Ma J. Role of GATA binding protein 4 (GATA4) in the regulation of tooth development via GNAI3. Scientific Reports. 2017;7(1):1534. doi: 10.1038/s41598-017-01689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen H, Guo S, Xia Y, Yuan L, Lu M, Zhou M, Fang M, Meng L, Xiao Z, Ma J. The role of rho-GEF trio in regulating tooth root development through the p38 MAPK pathway. Experimental Cell Research. 2018;372(2):158–167. doi: 10.1016/j.yexcr.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 103.Jacob A, Zhang Y, George A. Transcriptional regulation of dentin matrix protein 1 (DMP1) in odontoblasts and osteoblasts. Connective Tissue Research. 2014;55(Suppl 1):107–112. doi: 10.3109/03008207.2014.923850. [DOI] [PubMed] [Google Scholar]
- 104.Kotterman MA, Chalberg TW, Schaffer DV. Viral vectors for gene therapy: Translational and clinical outlook. Annual Review of Biomedical Engineering. 2015;17:63–89. doi: 10.1146/annurev-bioeng-071813-104938. [DOI] [PubMed] [Google Scholar]
- 105.Eggers R, de Winter F, Hoyng SA, Roet KC, Ehlert EM, Malessy MJ, Verhaagen J, Tannemaat MR. Lentiviral vector-mediated gradients of GDNF in the injured peripheral nerve: Effects on nerve coil formation, Schwann cell maturation and myelination. PLoS One. 2013;8(8):e71076. doi: 10.1371/journal.pone.0071076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li YH, Zhang K, Yang K, Ye JX, Xing YZ, Guo HY, et al. Adenovirus-mediated Wnt10b overexpression induces hair follicle regeneration. The Journal of Investigative Dermatology. 2013;133(1):42–48. doi: 10.1038/jid.2012.235. [DOI] [PubMed] [Google Scholar]
- 107.Hu WW, Wang Z, Krebsbach PH. Virus immobilization on biomaterial scaffolds through biotin-avidin interaction for improving bone regeneration. Journal of Tissue Engineering and Regenerative Medicine. 2016;10(2):E63–E72. doi: 10.1002/term.1774. [DOI] [PubMed] [Google Scholar]
- 108.Dalsgaard T, Cecchi CR, Askou AL, Bak RO, Andersen PO, Hougaard D, Jensen TG, Dagnæs-Hansen F, Mikkelsen JG, Corydon TJ, Aagaard L. Improved Lentiviral gene delivery to mouse liver by hydrodynamic vector injection through tail vein. Mol Ther Nucleic Acids. 2018;12:672–683. doi: 10.1016/j.omtn.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nature Reviews. Genetics. 2014;15(8):541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 110.Mun JY, Shin KK, Kwon O, Yong TL, Oh DB. Minicircle microporation-based non-viral gene delivery improved the targeting of mesenchymal stem cells to an injury site. Biomaterials. 2016;101:310–320. doi: 10.1016/j.biomaterials.2016.05.057. [DOI] [PubMed] [Google Scholar]
- 111.Molla MR, Levkin PA. Combinatorial approach to Nanoarchitectonics for nonviral delivery of nucleic acids. Advanced Materials. 2016;28(6):1159–1175. doi: 10.1002/adma.201502888. [DOI] [PubMed] [Google Scholar]
- 112.Curtin CM, Tierney EG, Mcsorley K, Cryan SA, Duffy GP, O'Brien FJ. Combinatorial gene therapy accelerates bone regeneration: Non-viral dual delivery of VEGF and BMP2 in a collagen-nanohydroxyapatite scaffold. Advanced Healthcare Materials. 2015;4(2):223. doi: 10.1002/adhm.201400397. [DOI] [PubMed] [Google Scholar]
- 113.Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, Rotello VM. Direct cytosolic delivery of CRISPR/Cas9-Ribonucleoprotein for efficient gene editing. ACS Nano. 2017;11(3):2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xiang D, Liu CC, Wang MJ, Li JX, Chen F, Yao H, Yu B, Lu L, Borjigin U, Chen YX, Zhong L, Wangensteen KJ, He ZY, Wang X, Hu YP. Non-viral FoxM1 gene delivery to hepatocytes enhances liver repopulation. Cell Death & Disease. 2014;5:e1252. doi: 10.1038/cddis.2014.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee YH, Wu HC, Yeh CW, Kuan CH, Liao HT, Hsu HC, Tsai JC, Sun JS, Wang TW. Enzyme-crosslinked gene-activated matrix for the induction of mesenchymal stem cells in osteochondral tissue regeneration. Acta Biomaterialia. 2017;63:210–226. doi: 10.1016/j.actbio.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 116.Yang QQ, Shao YX, Zhang LZ, Zhou YL. Therapeutic strategies for flexor tendon healing by nanoparticle-mediated co-delivery of bFGF and VEGFA genes. Colloids and Surfaces. B, Biointerfaces. 2018;164:165–176. doi: 10.1016/j.colsurfb.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 117.Zhang N, Chin JS, Chew SY. Localised non-viral delivery of nucleic acids for nerve regeneration in injured nervous systems. Experimental Neurology. 2019;319:112820. doi: 10.1016/j.expneurol.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 118.Feng G, Zhang Z, Dang M, Zhang X, Doleyres Y, Song Y, Chen D, Ma PX. Injectable nanofibrous spongy microspheres for NR4A1 plasmid DNA transfection to reverse fibrotic degeneration and support disc regeneration. Biomaterials. 2017;131:86–97. doi: 10.1016/j.biomaterials.2017.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Azoidis I, Cox SC, Davies OG. The role of extracellular vesicles in biomineralisation: Current perspective and application in regenerative medicine. J Tissue Eng. 2018;9:2041731418810130. doi: 10.1177/2041731418810130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boorn JG, Den V, Martin S, Christoph C, Gunther H. SiRNA delivery with exosome nanoparticles. Nature Biotechnology. 2011;29(4):325–326. doi: 10.1038/nbt.1830. [DOI] [PubMed] [Google Scholar]
- 121.Rani S, Ritter T. The exosome - a naturally secreted nanoparticle and its application to wound healing. Advanced Materials. 2016;28(27):5542–5552. doi: 10.1002/adma.201504009. [DOI] [PubMed] [Google Scholar]
- 122.Elangovan S, Khorsand B, Do AV, Hong L, Dewerth A, Kormann M, Ross RD, Sumner DR, Allamargot C, Salem AK. Chemically modified RNA activated matrices enhance bone regeneration. Journal of Controlled Release. 2015;218:22–28. doi: 10.1016/j.jconrel.2015.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khorsand B, Elangovan S, Hong L, Dewerth A, Kormann MS, Salem AK. A comparative study of the bone regenerative effect of chemically modified RNA encoding BMP-2 or BMP-9. The AAPS Journal. 2017;19(2):438–446. doi: 10.1208/s12248-016-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mavilio Fulvio, Pellegrini Graziella, Ferrari Stefano, Di Nunzio Francesca, Di Iorio Enzo, Recchia Alessandra, Maruggi Giulietta, Ferrari Giuliana, Provasi Elena, Bonini Chiara, Capurro Sergio, Conti Andrea, Magnoni Cristina, Giannetti Alberto, De Luca Michele. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nature Medicine. 2006;12(12):1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- 125.Lu Yong-Chen, Parker Linda L., Lu Tangying, Zheng Zhili, Toomey Mary Ann, White Donald E., Yao Xin, Li Yong F., Robbins Paul F., Feldman Steven A., van der Bruggen Pierre, Klebanoff Christopher A., Goff Stephanie L., Sherry Richard M., Kammula Udai S., Yang James C., Rosenberg Steven A. Treatment of Patients With Metastatic Cancer Using a Major Histocompatibility Complex Class II–Restricted T-Cell Receptor Targeting the Cancer Germline Antigen MAGE-A3. Journal of Clinical Oncology. 2017;35(29):3322–3329. doi: 10.1200/JCO.2017.74.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gerace D, Martiniellowilks R, Nassif NT, Lal S, Steptoe R, Simpson AM. CRISPR-targeted genome editing of mesenchymal stem cell-derived therapies for type 1 diabetes: A path to clinical success? Stem Cell Research & Therapy. 2017;8(1):62. doi: 10.1186/s13287-017-0511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Evans CH, Huard J. Gene therapy approaches to regenerating the musculoskeletal system. Nature Reviews Rheumatology. 2015;11(4):234–242. doi: 10.1038/nrrheum.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hamada H, Kobune M, Nakamura K, Kawano Y, Kato K, Honmou O, Houkin K, Matsunaga T, Niitsu Y. Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Science. 2005;96(3):149–156. doi: 10.1111/j.1349-7006.2005.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu J, Chen W, Zhao Z, Xu HH. Reprogramming of mesenchymal stem cells derived from iPSCs seeded on biofunctionalized calcium phosphate scaffold for bone engineering. Biomaterials. 2013;34(32):7862–7872. doi: 10.1016/j.biomaterials.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang F, Peng WX, Wang L, Zhang J, Dong WT, Wu JH, Zhang H, Wang JB, Zhao Y. Role of FGF-2 transfected bone marrow Mesenchymal stem cells in engineered bone tissue for repair of avascular necrosis of femoral head in rabbits. Cellular Physiology and Biochemistry. 2018;48(2):773–784. doi: 10.1159/000491906. [DOI] [PubMed] [Google Scholar]
- 131.Ankrum JA, Ong JF, Karp JM. Mesenchymal stem cells: immune evasive, not immune privileged. Nature Biotechnology. 2014;32(3):252–260. doi: 10.1038/nbt.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Terai S, Tsuchiya A. Status of and candidates for cell therapy in liver cirrhosis: Overcoming the "point of no return" in advanced liver cirrhosis. Journal of Gastroenterology. 2016;52(2):1–12. doi: 10.1007/s00535-016-1258-1. [DOI] [PubMed] [Google Scholar]
- 133.Ben-Mordechai T, Holbova R, Landa-Rouben N, Harel-Adar T, Feinberg MS, Abd Elrahman I, Blum G, Epstein FH, Silman Z, Cohen S, Leor J. Macrophage subpopulations are essential for infarct repair with and without stem cell therapy. Journal of the American College of Cardiology. 2013;62(20):1890–1901. doi: 10.1016/j.jacc.2013.07.057. [DOI] [PubMed] [Google Scholar]
- 134.Spiller, K. L., & Koh, T. J. (2017). Macrophage-based therapeutic strategies in regenerative medicine. Advanced Drug Delivery Reviews. [DOI] [PMC free article] [PubMed]
- 135.Thomas, J. A., Pope, C., Wojtacha, D., Robson, A. J., Gordon-Walker, T. T., Hartland, S., . . . Iredale, J. P. (2011). Macrophage therapy for murine liver fibrosis recruits host effector cells improving fibrosis, regeneration, and function &dagger. Hepatology, 53(6), 2003–2015. [DOI] [PubMed]
- 136.Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, Ridgway RA, Seo SS, Spee B, van Rooijen N, Sansom OJ, Iredale JP, Lowell S, Roskams T, Forbes SJ. Macrophage-derived Wnt opposes notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nature Medicine. 2012;18(4):572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hamm A, Veschini L, Takeda Y, Costa S, Delamarre E, Squadrito ML, Henze AT, Wenes M, Serneels J, Pucci F, Roncal C, Anisimov A, Alitalo K, de Palma M, Mazzone M. PHD2 regulates arteriogenic macrophages through TIE2 signalling. EMBO Molecular Medicine. 2013;5(6):843–857. doi: 10.1002/emmm.201302695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakashima M, Reddi AH. The application of bone morphogenetic proteins to dental tissue engineering. Nature Biotechnology. 2003;21(9):1025–1032. doi: 10.1038/nbt864. [DOI] [PubMed] [Google Scholar]
- 139.Han P, Ivanovski S, Crawford R, Xiao Y. Activation of the canonical Wnt signaling pathway induces Cementum regeneration. Journal of Bone and Mineral Research. 2015;30(7):1160–1174. doi: 10.1002/jbmr.2445. [DOI] [PubMed] [Google Scholar]
- 140.Nakao K, Morita R, Saji Y, Ishida K, Tomita Y, Ogawa M, Saitoh M, Tomooka Y, Tsuji T. The development of a bioengineered organ germ method. Nature Methods. 2007;4(3):227–230. doi: 10.1038/nmeth1012. [DOI] [PubMed] [Google Scholar]
- 141.Ikeda E, Morita R, Nakao K, Ishida K, Nakamura T, Takano-Yamamoto T, Ogawa M, Mizuno M, Kasugai S, Tsuji T. Fully functional bioengineered tooth replacement as an organ replacement therapy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13475–13480. doi: 10.1073/pnas.0902944106. [DOI] [PMC free article] [PubMed] [Google Scholar]