Abstract
Relative gene expression analyses by RT-qPCR (reverse transcription-quantitative PCR) are highly dependent on the reference genes in normalizing the expression data of target genes. Therefore, inappropriate endogenous control genes will lead to inaccurate target gene expression profiles, and the selection and validation of suitable internal reference genes becomes essential. In this study, we retrieved the commonly used reference genes in transcriptome datasets of Codonopsis pilosula by RNA-Seq (unpublished data), and selected 15 candidate reference genes according to the coefficient of variation (CV) and fold change (FC) value of gene expression. The expression levels of candidate reference genes, which is at different growth stages, undergoing cold stress and drought stress, was determined by RT-qPCR. The expression stability of these genes was evaluated using software packages and algorithms including ΔCt, geNorm, NormFinder and Bestkeeper. Then appropriate reference genes were screened and validated by target gene-UDGPase (UDP glucose pyrophosphorylase). The optimal RGs combinations of C. pilosula, including PP2A59γ, CPY20-1, UBCE32, RPL5B and UBC18 for developmental stage, RPL5B, RPL13 and PP2A59γ for cold treatment, RPL13 and PP2A59γ for drought treatment, were found and proposed as reference genes for future work. This paper laid foundations for both the selection of reference genes and exploration in metabolic mechanism of C. pilosula.
Subject terms: Transcriptomics, Plant molecular biology
Introduction
Codonopsis pilosula (Franch.) Nannf) is a traditional Chinese medicinal and edible plant1,2. As a history of more than 300 years, C. pilosula is mainly distributed in four provinces of China including Gansu, Shannxi, Shanxi and Hubei2,3. It is reported that the main ingredients related to its pharmacodynamics are polysaccharide, tangshenosides, lobetyolin, atractylenolide, and so on4. By means of transcriptome sequencing, Gao J. P. et al. and Li J. et al. revealed that UDGPase (UDP glucose pyrophosphorylase) and UGDH (UDP-glucose dehydrogenase) were the key enzyme genes in C. pilosula controlling the synthesis pathway of polysaccharide5–8. Consequently, UDGPase was cloned and characterized by them adopting GAPDH as reference gene (RG)8. GAPDH and SUC4 (sucrose/H+ cotransporter 4) were also cloned and characterized by Wang X. L. et al. and Zheng Q. H. et al.5,6. In the above studies, Real-Time Quantitative PCR (RT-qPCR) was commonly used in expression analysis of those genes.
Compared with traditional quantitative analysis techniques like Northern blots or cDNA microarrays, RT-qPCR is much more popular in gene expression because of its time-saving, accuracy, low cost and high throughput characteristics9–16. RT-qPCR has played an important role in researching the molecular regulation of secondary metabolites of medicinal plants. Wu Y. N. et al. and Gao J. P. et al. analyzed several gene expression of C. pilosula, which laid foundations for the gene regulation. Quantitative method is crucial to the accuracy of gene expression in RT-qPCR, which is divided into absolute quantification and relative quantification methods10,17. The latter is well received for its easy operation characteristics. This method is highly dependent on the constant of internal reference genes (RGs). Accordingly, stable RGs without significant variance in all the tested tissues, during the different period of development, and under different cultivating conditions are required10–12,14,15,18–20. RGs, which expressed in all cells of plants as housekeeping genes, mainly include ACT (actin), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), G6PDH (glucose-6-phosphate dehydrogenase), EF (elongation factor), 18 S rRNA (18 S ribosomal RNA), UBQ (ubiquitin), TUB (tubulin), EIF (eukaryocyte initiation factor), UBC (ubiquitin conjugating enzyme), H3 (histone), PGK (phosphoglycerate kinase), TEF (translation elongation factor), CYP (Cyclophilin), NADHD (Nicotinamide adenine dinucleotide dehydrogenase), and so on9,19,21–27. Recent studies have shown that gene expression was highly tissue-specific and often varied with the physiological status of the plant or experimental conditions, and the universal reference genes have not been found at present19,21,24,28–30. Selection of RGs should be conducted amongst the same pool of samples, and carried out according to the specific tissue, developmental stages and experimental conditions, thus the selected RGs can be used for target gene expression25,31–35. In Gao J. P.’s study on the polysaccharide of C. pilosula, GAPDH was used as the internal reference gene to normalize the expression of UDGPase. This work started the exploration of synthesis mechanism for C. pilosula metabolites, and GAPDH became the first internal reference gene used in the study of molecular regulation mechanisms. In order to make clear the mechanisms, systematic selection and validation of RGs are required. Reliable internal reference genes were particularly essential for C. pilosula during the growth stages and above all under the drought and cold stress conditions, because C. pilosula is manly distributed in dry and cold areas with frequently extreme weather. The impact of extreme climates such as cold and drought on the reliable of RG was extensively reported in many plants other than C. pilosula21,24,31,32.
In this study, we retrieved the commonly used RGs of C. pilosula in transcriptome datasets (unpublished data), and selected 15 candidate RGs according to threshold method of coefficient of variation (CV) and fold change (FC) value in gene expression36,37. RT-qPCR was then adopted to determine the candidate RGs at developmental stages, under cold stress treatments and drought stress treatments. The corresponding stability was evaluated using software packages and algorithms including ΔCt38, geNorm39, NormFinder40 and Bestkeeper41. Based on the above analysis, the appropriate RGs were screened and validated by target gene-UDGPase (UDP glucose pyrophosphorylase). This research for the first time systematically explored the RGs of C. pilosula to lay foundations for the selection of RGs and analysis of gene expression.
Results
Selection of candidate reference genes based on transcriptome data in C. pilosula
Root of C. pilosula at the initial and late stages of its swelling were collected, and three tissue layers including periderm, phloem and xylem of the roots were separated as samples for transcriptome sequencing. Six replicates for each tissue. The samples were sent to Lianchuan biological co. LTD to perform the transcriptome sequencing by using paired-end sequencing technology on an Illumina Hi-Seq™ 4000 platform. After assembling and annotation, expression profile data was mapped to the transcriptome. TPM (Transcripts per kilobase of exon model per million mapped reads) was calculated by means of EdgeR package (The RSEM package), which was then used to analyze gene expression level.
According to literatures on the conventional RGs, the related genes including ACT, GAPDH, G6PDH, EF, 18 S rRNA, UBQ, TUB, EIF, UBC, H3, PGK, TEF, CYP and NADHD were retrieved from the transcriptomic data of C. pilosula. As is reported, low-expression genes would make poor RT-qPCR references due to the difficulties in detecting and quantifying their expression, thus, genes with expression levels lower than five TPM were excluded before the stability analyses. In addition, genes with log fold variance more than two in different tissues were also rejected. After the removals, a total of 207 genes were selected for further study. Calculations for mean value (MV), SD, CV and log2FC were executed in Microsoft Excel according to TPM27. Genes with low CV and log2FC were considered stable. We set 0.5 as the CV cut-off value for stable genes and ranked the selected 207 genes. The top 50 genes were displayed in Table S1. After reducing the cut-off values of CV to 0.3 and |log2FC| to 0.2 according to the literatures11,42–44, fifteen genes were selected as candidate RGs.
Expression profile of candidate reference genes of C. pilosula
Primers of 15 candidate RGs were used to amplify cDNA template by PCR technology, and single stripe of target amplicon could be obtained, which was consistent with the expected target stripe size. The melting curves of 15 candidate genes’ primers were plotted, all of which showed single peak, indicating that the primer had strong specificity and non-specific amplification occurred. When the length of RT-qPCR amplification product increased from 103 bp to 191 bp, the amplification efficiency of candidate RGs improved from 91.3% to 107.2% (Table 1).
Table 1.
Primer sequences and amplicon characteristics of 15 candidate RGs and validation gene for RT-qPCR analysis, *gene known to have different levels of expression in C. pilosula was used to validate the candidate RGs.
| Gene symbol | Gene description | Primer sequences (5′−3′) | Amplicon Size(bp) | TM (°C) | Efficiency (%) |
|---|---|---|---|---|---|
| CYP20-1 | Cytochrome P450 20-1 | GGACCAGACACCAATGGTTCAC | 188 | 58.0 | 91.5 |
| AAGGGTAGCTCTCCGCTGTC | |||||
| ABCC8 | ABC-transporter C family member 8 | AGCCCTAACGGGTACCCAAG | 136 | 58.0 | 96.2 |
| TGGCCTCTTGTCCTCCACAA | |||||
| ACTβ | Beta-Actin | AGAGAAAGCGCTGAAATGCCA | 103 | 58.0 | 102.1 |
| AGCCTTGGGACGAAACCCTA | |||||
| UPL-RHF2A | E3 ubiquitin-protein ligase RHF2A | GAGGGAGGGCTCAAGGAGTC | 172 | 58.0 | 92.5 |
| AGCTGCAAATGGGACAGACG | |||||
| EF1α | Elongation factor 1-alpha | GCCTGGTGACAACGTTGGAT | 191 | 58.0 | 107.2 |
| GCGAGGTGTGGCAATCAAGA | |||||
| EIF4A-14 | Eukaryotic initiation factor 4A | GTGGCAGGGAAATCGGTTGT | 187 | 58.0 | 98.1 |
| GCTGAATAGCGGAGGGCTTC | |||||
| RPL5B | 60 S ribosomal protein L5 | CAGCTGCTTACGCCCATGAA | 180 | 58.0 | 95 |
| CCGCCGGTTCAACAGAGAAA | |||||
| RPL13 | 60 S ribosomal protein L13 | AGGAACTTGCAACAGCCACA | 176 | 58.0 | 93.7 |
| CGCAGCCCTCTTAAGCCTTT | |||||
| G6PDH | glucose-6-phosphate dehydrogenase | TGGTGACGTAGTGCAGAGTC | 187 | 58.0 | 91.3 |
| AGCATCAACGTTGTCCTCAGA | |||||
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | CCAAGAAGGCAGGCATGGTT | 150 | 58.0 | 94.1 |
| CCTTTACGGCTGTGCACCTT | |||||
| TUBα3 | Alpha-tubulin | TGACTGGTGCCCTACTGGTT | 178 | 58.0 | 106.4 |
| CGCCCTCTTTGCGTACATGA | |||||
| PP2A57θ | Protein phosphatase 2A-57 | CCTGCACTGGAGAGAAACGG | 183 | 58.0 | 102.5 |
| ACGTTTCCATGCAGCTTCCC | |||||
| PP2A59γ | Protein phosphatase 2A-59 | GCGGACTCACTGGAACCAAG | 166 | 58.0 | 95.6 |
| CAGGCGCTTCCAGGTTGATT | |||||
| UBC18 | Ubiquitin-conjugating enzyme 18 | CCAGCTCCTCTACACCCTCA | 152 | 58.0 | 104.7 |
| TCTTCTGGTCGTTGCTTCGC | |||||
| UBCE32 | Ubiquitin-conjugating enzyme 32 | TGGTGACCAGGCAAACGATG | 175 | 58.0 | 91.8 |
| CCTCAAGACTGCAGGAGTGG | |||||
| UGPase* | UDP-glucose pyrophosphorylase | TGATGGCTATGTGACCCGGAA | 187 | 58.0 | 102.1 |
| ACCCTTGAGAACGACGGAAGA |
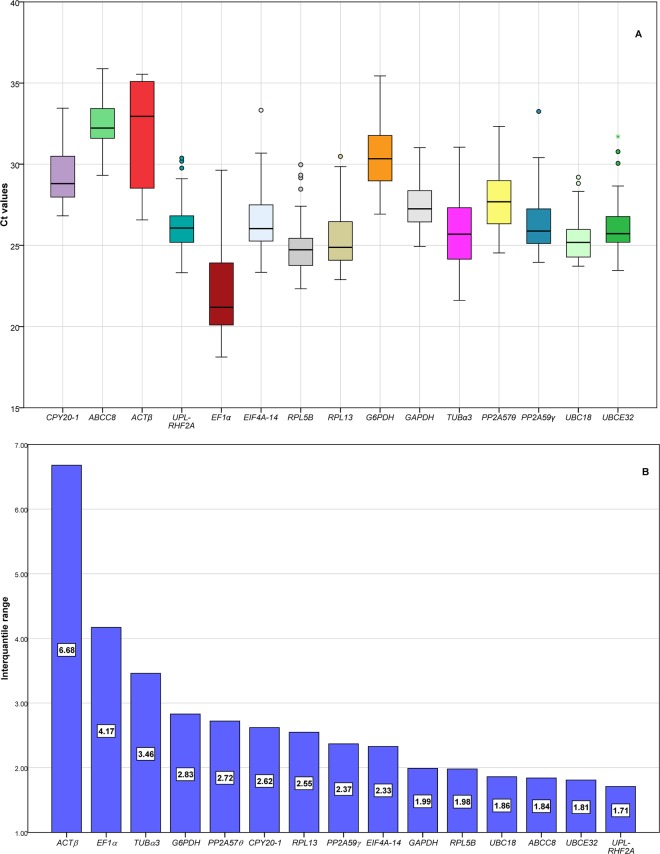
The expression level of candidate RGs was normally determined by the threshold cycle (Ct) values of RT-qPCR. Small Ct value indicated higher gene expression abundance, and vice versa. Figure 1 displayed the Ct values of RGs and their variability. The genes with lower variability were UPL-RHF2A, UBCE32, UBC18, ABCC8, and RPL5B according to the distribution of raw Ct values, thus could be selected as the RGs. The commonly used ACTβ, EF1α, and TUBα3 were the worst candidate RGs because the Ct values spanned multiple units according to the scope of the interquartile. However, analyses on the distribution of the raw Ct values and the range of the interquartile are not sufficient to assess the expression stability of candidate RGs, and other methods are required for more comprehensive and accurate evaluations.
Figure 1.
RT-qPCR Ct values and interquartile ranges. (A) Ct values for each RG in all samples. A line across the box depicts the median. The box indicates the 25% and 75% percentiles. Whiskers represent the maximum and minimum values, circles represent outliers and asterisks indicate extremes. (B) interquartile ranges indicate variability of Ct values among 25% and 75%.
Expression stability analysis of candidate reference genes of C. pilosula
The expression stability of fifteen candidate RGs of leaf, stem, and root of C. pilosula under the conditions of different developmental stage, cold stress, drought stress, as well as the total samples were analyzed by means of Δ Ct, geNorm, NormFinder, and BestKeeper methods.
ΔCt analysis
The mean SD of ΔCt between one gene and the other 14 genes were calculated, which was used to evaluate the stability of the gene. The lower the mean SD was, the higher the stability of the gene tended to. Table 2 listed the rank of RGs and their expression stability values at different developmental stages and treatments. PP2A59γ and RPL5B were the most stable RGs at the different developmental stages. RPL5B was the most stable RG in cold stress treatment, and PP2A59γ was most stable in drought stress treatment. UBCE32 was found to be the most stable RG in the total samples, while ACTβ was the most unstable RG both at the different developmental stages and under the cold stress or drought stress treatments.
Table 2.
The rank of RGs for normalization and their expression stability values calculated by the ΔCt method.
| Rank position | Developmental stage | Cold | Drought | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | Average of STDEV | Gene | Average of STDEV | Gene | Average of STDEV | Gene | Average of STDEV | |
| 1 | PP2A59γ | 1.354 | RPL5B | 1.015 | PP2A59γ | 0.812 | UBCE32 | 1.185 |
| 2 | RPL5B | 1.354 | PP2A59γ | 1.024 | CPY20-1 | 0.857 | RPL5B | 1.223 |
| 3 | UBCE32 | 1.357 | UBCE32 | 1.055 | UBCE32 | 0.858 | PP2A59γ | 1.253 |
| 4 | UBC18 | 1.426 | RPL13 | 1.064 | RPL13 | 0.883 | RPL13 | 1.254 |
| 5 | G6PDH | 1.446 | EIF4A-14 | 1.081 | UBC18 | 0.928 | EIF4A-14 | 1.282 |
| 6 | CPY20-1 | 1.464 | PP2A57θ | 1.129 | UPL-RHF2A | 0.931 | CPY20-1 | 1.298 |
| 7 | EIF4A-14 | 1.495 | G6PDH | 1.172 | RPL5B | 0.978 | PP2A57θ | 1.308 |
| 8 | PP2A57θ | 1.499 | CPY20-1 | 1.177 | PP2A57θ | 0.990 | G6PDH | 1.353 |
| 9 | RPL13 | 1.523 | EF1α | 1.294 | EIF4A-14 | 1.020 | UBC18 | 1.386 |
| 10 | TUBα3 | 1.570 | UPL-RHF2A | 1.299 | ABCC8 | 1.060 | TUBα3 | 1.464 |
| 11 | GAPDH | 1.658 | UBC18 | 1.379 | EF1α | 1.069 | UPL-RHF2A | 1.549 |
| 12 | ABCC8 | 1.851 | TUBα3 | 1.379 | G6PDH | 1.206 | GAPDH | 1.579 |
| 13 | UPL-RHF2A | 2.077 | GAPDH | 1.519 | TUBα3 | 1.221 | ABCC8 | 1.692 |
| 14 | EF1α | 2.401 | ABCC8 | 1.815 | GAPDH | 1.364 | EF1α | 2.179 |
| 15 | ACTβ | 2.429 | ACTβ | 2.977 | ACTβ | 2.864 | ACTβ | 2.996 |
geNorm analysis
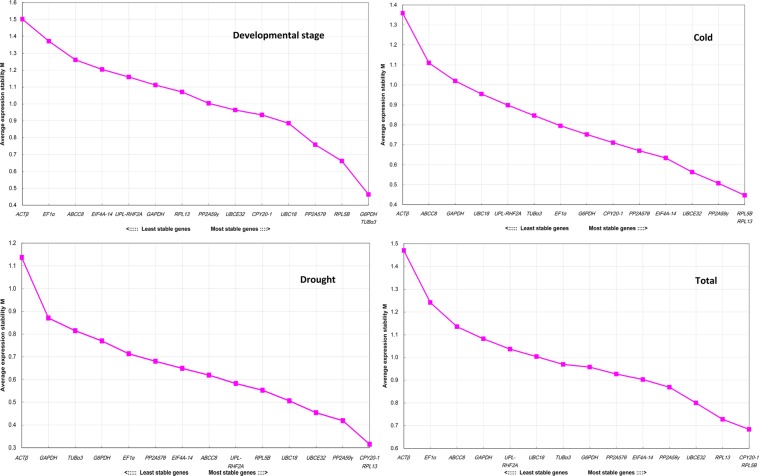
The M values of fifteen candidate RGs were calculated by using geNorm program and the stability of each candidate RG was ranked by the M value. Genes with the lowest M values are considered to have the most stable expression and denoted as the suitable RG. The samples were divided into four groups: developmental stage (ck, 15d and 30d), cold stress (ck, 4 h, 8 h and 12 h), drought stress (ck, 5d and 10d), and total (all the samples). The average expression stability values (M) and ranking of the candidate RGs was shown in Fig. 2. The optimal RGs were found to vary with the experimental conditions. CPY20-1 and UBC18 were the most credible RGs for samples collected at different developmental stages. RPL5B and RPL13 were the best RGs for samples experiencing cold stress treatment. In drought stress treatment, CPY20-1 and RPL13 were the most suitable RGs. CPY20-1 and RPL5B were the most stable RGs for all the samples.
Figure 2.
Average expression stability values (M) and ranking of the candidate RGs calculated using geNorm. A lower value of the average expression stability indicates more stable expression.
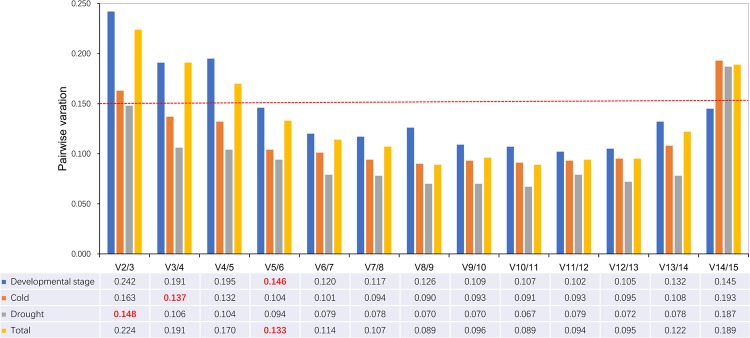
To determine the optimal number of RGs for normalization of gene expression level, the pairwise variation (Vn/n+1) of two sequential normalization factors (NFs) was computed using geNorm program. If the pairwise variation between genes is larger than or equal to 0.15, one gene should be added for reliable normalization of the pairwise variation. Once the pairwise variation is below 0.15, no additional genes are required for normalization42.
Analyzation on the pairwise variation of candidate RGs was displayed in Fig. 3. For both the total samples and samples developed at different stages, V5/6 was below 0.15, accordingly, five RGs was the optimal number. For samples subjected to cold stress treatment, V3/4 was lower than 0.15, thus 3 RGs are required, while for samples subjected to drought stress treatment, 2 RGs are needed. Results of pairwise variation was consistent with that of the stability measurements. RGs of samples subjected to cold stress or drought stress treatment demonstrated lower average pairwise variation and higher stability. In contrast, RGs of both the total samples and samples at various growth stage behaved unstable and showed higher pairwise variation.
Figure 3.
Pairwise varation to determine the optimal number of control genes for accurate normalization. The pairwise variation (Vn/Vn + 1) was analyzed between the NFs (NFn and NFn+1) using geNorm software, where n is the number of genes involved in the NF. Red figures indicate the optimal number of genes for normalization.
NormFinder analysis
NormFinder software was also applied to analyze the pairwise variation of candidate RGs. Expression stability values of genes were shown in Table 3. Genes with lower SV was considered to be stable, and suitable for RGs. Stable genes in samples at different developmental stages were PP2A59γ, UBCE32 and RPL5B. RPL13 and RPL5B were the most suitable RGs in cold stress treatment. PP2A59γ was the most stable RG in drought stress treatment. PP2A59γ, RPL13, RPL5B, and UBCE32 were the most suitable RGs for total samples. However, ACTβ was the most unstable gene in all of the samples, which agreed with the results from geNorm analysis.
Table 3.
The rank of RGs for normalization and their expression stability values calculated by the NormFinder program.
| Rank position | Developmental stage | Cold | Drought | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | Stability value | Gene | Stability value | Gene | Stability value | Gene | Stability value | |
| 1 | PP2A59γ | 0.205 | RPL13 | 0.075 | PP2A59γ | 0.084 | PP2A59γ | 0.145 |
| 2 | UBCE32 | 0.238 | RPL5B | 0.095 | RPL13 | 0.252 | RPL13 | 0.157 |
| 3 | RPL5B | 0.243 | PP2A59γ | 0.157 | UBCE32 | 0.328 | UBCE32 | 0.160 |
| 4 | PP2A57θ | 0.291 | EIF4A-14 | 0.174 | CPY20-1 | 0.334 | RPL5B | 0.179 |
| 5 | RPL13 | 0.305 | UBCE32 | 0.184 | UPL-RHF2A | 0.408 | CPY20-1 | 0.208 |
| 6 | CPY20-1 | 0.313 | CPY20-1 | 0.187 | PP2A57θ | 0.443 | PP2A57θ | 0.222 |
| 7 | G6PDH | 0.313 | PP2A57θ | 0.219 | RPL5B | 0.444 | EIF4A-14 | 0.236 |
| 8 | EIF4A-14 | 0.352 | G6PDH | 0.268 | UBC18 | 0.460 | G6PDH | 0.311 |
| 9 | TUBα3 | 0.356 | EF1α | 0.281 | EIF4A-14 | 0.462 | EF1α | 0.320 |
| 10 | UBC18 | 0.367 | UPL-RHF2A | 0.333 | EF1α | 0.481 | UBC18 | 0.385 |
| 11 | EF1α | 0.494 | TUBα3 | 0.437 | ABCC8 | 0.573 | TUBα3 | 0.393 |
| 12 | GAPDH | 0.528 | UBC18 | 0.496 | TUBα3 | 0.636 | UPL-RHF2A | 0.397 |
| 13 | ABCC8 | 0.634 | GAPDH | 0.521 | G6PDH | 0.689 | GAPDH | 0.512 |
| 14 | UPL-RHF2A | 0.730 | ABCC8 | 0.647 | GAPDH | 0.819 | ABCC8 | 0.542 |
| 15 | ACTβ | 0.903 | ACTβ | 1.276 | ACTβ | 1.669 | ACTβ | 1.167 |
BestKeeper analysis
BestKeeper analysis determined stable RG according to the Ct values of each gene. Genes with high coefficient of determination (r), low SD and CV are considered stable. According to SD, CV, and r of Ct value, the expression stability of candidate RGs was ranked corresponding to the different developmental stages, cold and drought stress treatment, as well as the total samples. Then the geomean value of each RG was calculated and used to rank the fifteen RGs (Table 4). The results indicated that the optimal RG was ABCC8 for developmental stages, UBC18 for cold treatment, and RPL13 for both drought treatment and the total samples. The most unstable RG was ACTβ except for developmental stage, in which the most unreliable RG was UPL-RHF2A.
Table 4.
The rank of RGs for normalization calculated by the BestKeeper program.
| Rank position | Developmental stage | Cold | Drought | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | Geomean | Gene | Geomean | Gene | Geomean | Gene | Geomean | |
| 1 | ABCC8 | 2.466 | UBC18 | 3.302 | RPL13 | 2.000 | RPL13 | 2.154 |
| 2 | GAPDH | 3.634 | ABCC8 | 3.557 | CPY20-1 | 2.466 | CPY20-1 | 3.000 |
| 3 | RPL13 | 4.579 | GAPDH | 3.733 | UBCE32 | 3.420 | UBCE32 | 3.420 |
| 4 | CPY20-1 | 4.718 | RPL5B | 3.826 | PP2A59γ | 3.826 | UBC18 | 4.041 |
| 5 | G6PDH | 4.762 | PP2A59γ | 4.642 | UBC18 | 4.160 | RPL5B | 4.932 |
| 6 | EIF4A-14 | 4.946 | UBCE32 | 5.192 | ABCC8 | 6.782 | EIF4A-14 | 4.946 |
| 7 | UBC18 | 5.848 | EIF4A-14 | 6.415 | RPL5B | 6.910 | PP2A59γ | 5.518 |
| 8 | RPL5B | 6.316 | RPL13 | 7.114 | G6PDH | 8.173 | ABCC8 | 6.952 |
| 9 | UBCE32 | 6.542 | UPL-RHF2A | 7.343 | PP2A57θ | 8.243 | PP2A57θ | 7.319 |
| 10 | PP2A59γ | 6.604 | EF1α | 7.764 | UPL-RHF2A | 8.573 | TUBα3 | 9.524 |
| 11 | EF1α | 7.958 | PP2A57θ | 8.143 | EIF4A-14 | 10.288 | UPL-RHF2A | 10.260 |
| 12 | PP2A57θ | 9.655 | CPY20-1 | 8.243 | EF1α | 10.298 | GAPDH | 10.537 |
| 13 | ACTβ | 12.897 | G6PDH | 10.459 | TUBα3 | 10.483 | G6PDH | 11.334 |
| 14 | TUBα3 | 12.974 | TUBα3 | 12.083 | GAPDH | 10.801 | EF1α | 12.209 |
| 15 | UPL-RHF2A | 13.976 | ACTβ | 14.659 | ACTβ | 15.000 | ACTβ | 15.000 |
Comprehensive stability analysis of reference genes
Based on the ranking results of expression stability by means of the four methods (ΔCt, geNorm, NormFinder, and BestKeeper), the geomean value of each RG was calculated and applied to determine the comprehensive rank of the fifteen RGs in developmental stage, cold treatment, drought treatment, and the total samples. Results in Table 5 showed that ACTβ was the most unstable RG (single low-ranked RG). The top-ranked single RG is PP2A59γ for developmental stage, RPL5B for cold treatment, and RPL13 for both drought treatment and the total samples. According to the number of RGs suggested by geNorm and the ranking listed in Table 5, the best combination of RGs was PP2A59γ, CPY20-1, UBCE32, RPL5B, and UBC18 for developmental stage, RPL5B, RPL13, and PP2A59γ for cold treatment, RPL13 and PP2A59γ for drought treatment, and RPL13, UBCE32, RPL5B, CPY20-1 and PP2A59γ for the total samples.
Table 5.
The comprehensive ranking of RGs for normalization.
| Rank position | Developmental stage | Cold | Drought | Total |
|---|---|---|---|---|
| gene | gene | gene | gene | |
| 1 | PP2A59γ | RPL5B | RPL13 | RPL13 |
| 2 | CPY20-1 | RPL13 | PP2A59γ | UBCE32 |
| 3 | UBCE32 | PP2A59γ | CPY20-1 | RPL5B |
| 4 | RPL5B | UBCE32 | UBCE32 | CPY20-1 |
| 5 | UBC18 | EIF4A-14 | UBC18 | PP2A59γ |
| 6 | G6PDH | UBC18 | RPL5B | EIF4A-14 |
| 7 | RPL13 | PP2A57θ | UPL-RHF2A | PP2A57θ |
| 8 | ABCC8 | CPY20-1 | PP2A57θ | UBC18 |
| 9 | PP2A57θ | ABCC8 | ABCC8 | G6PDH |
| 10 | GAPDH | G6PDH | EIF4A-14 | TUBα3 |
| 11 | EIF4A-14 | GAPDH | EF1α | UPL-RHF2A |
| 12 | TUBα3 | EF1α | G6PDH | ABCC8 |
| 13 | EF1α | UPL-RHF2A | TUBα3 | GAPDH |
| 14 | UPL-RHF2A | TUBα3 | GAPDH | EF1α |
| 15 | ACTβ | ACTβ | ACTβ | ACTβ |
Validation on the stability of reference genes
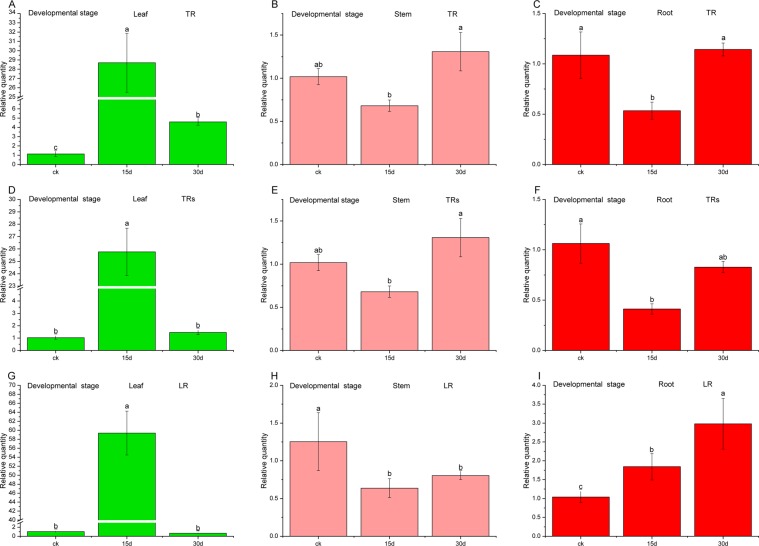
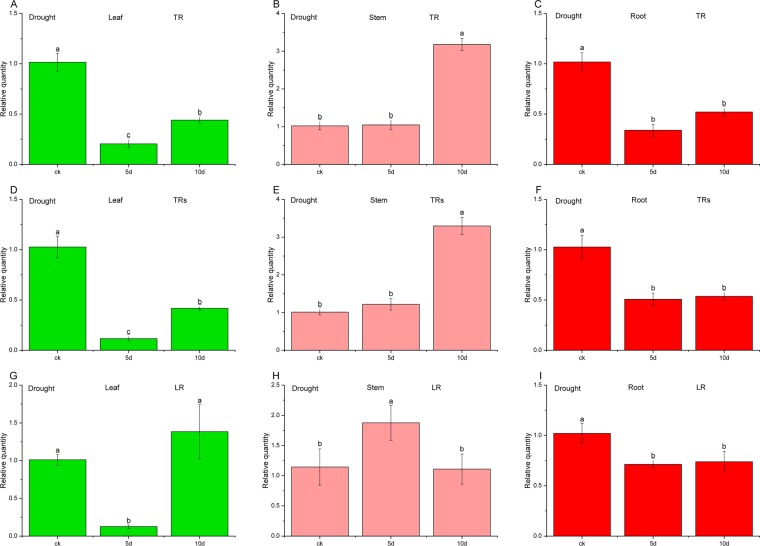
The validity of RG or RGs, denoted as relative quality in Figs. 4 to 6, was verified by normalizing the target genes UDGPase (UDP glucose pyrophosphorylase) with the selected single RG, the combination of RGs and ACTβ. For the developmental stage of C. pilosula, when PP2A59γ alone was used for target gene expression, relative expression of UDGPase in leaves increased firstly and then decreased, and the gene UDGPase was found to be overexpressed nearly 29 times on the 15th day (Fig. 4A). Relative expression of UDGPase in both the stems and roots decreased and then increased (Fig. 4B,C), reaching the highest on the 30th day for the stems. In comparison, when normalization was performed using the ACTβ gene, serious deviation produced in all tissues (leaf, stem and root), and the gene UDGPase was found to be overexpressed nearly 58 times on the 15th day (Fig. 4G–I). Using the combination of RGs including PP2A59γ, CPY20-1, UBCE32, RPL5B and UBC18, the expression characteristics of UDGPase in leaves, stems and roots was basically similar to that using PP2A59γ alone. However, the relative expression levels of UDGPase at different time points in different tissues were slightly lower than those using the most stable single RG (Fig. 4D–F).
Figure 4.
Relative quantity of UDGPase in different developmental stage with different RGs after normalization. Samples A, B, C were normalized with a single top-ranked RG (TR)- PP2A59γ. samples D, E, F were normalized with a combination of multiple top-ranked RGs (TRs)- PP2A59γ, CPY20-1, UBCE32, RPL5B and UBC18, while samples G, H, I were normalized with a single low-ranked RG (LR)- ACTβ.
Figure 6.
Relative quantity of UDGPase in drought stress with different RGs after normalization. Samples A, B, C were normalized with a single top-ranked RG (TR) - RPL13. samples D, E, F were normalized with a combination of multiple top-ranked RGs (TRs)- RPL13 and PP2A59γ, while samples G, H, I were normalized with a single low-ranked RG (LR)- ACTβ.
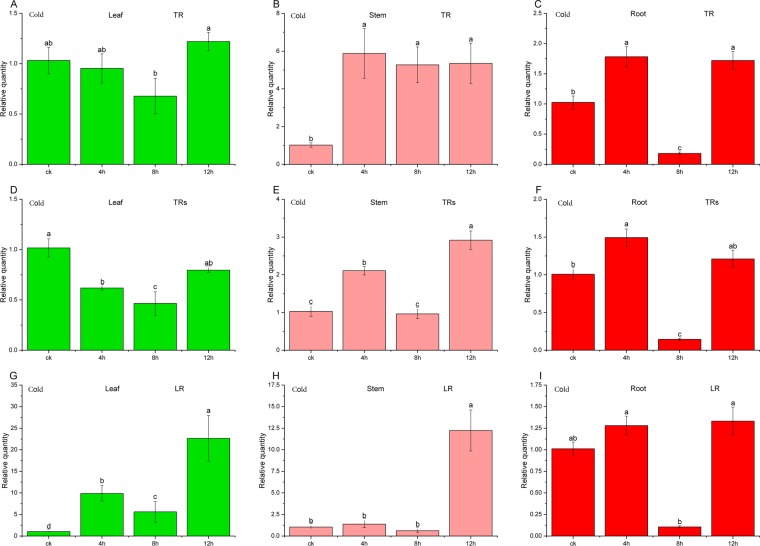
For the cold treatment, as shown in Fig. 5, the variation characteristics of relative expression of UDGPase are similar in three organs whether using the most stable RPL5B or combination of RPL5B, RPL13 and PP2A59γ. The relative expression levels of UDGPase using single stable RG were slightly higher than those using combination of RGs except for the control. Application of unstable ACTβ gene for normalizing target gene in root leaded to larger deviations. In the drought treatment, Both the most stable RPL13 and combination of RPL13 and PP2A59γ were the effective RGs for the relative expression of UDGPase of the three tissues. In contrast, using the most unstable ACTβ gene for normalization, although the expression of UDGPase in the roots was similar to that of the most stable RG or RGs, the expression bias of UDGPase appeared in the leaves and stems (Fig. 6).
Figure 5.
Relative quantity of UDGPase in cold stress with different RGs after normalization. Samples A, B, C were normalized with a single top-ranked RG (TR)- RPL5B. samples D, E, F were normalized with a combination of multiple top-ranked RGs (TRs)- RPL5B, RPL13 and PP2A59γ, while samples G, H, I were normalized with a single low-ranked RG (LR)- ACTβ.
Discussion
Although C. pilosula is a traditional Chinese medicine, insufficient researches have been conducted on the internal reference genes for gene expression normalization in biosynthesis of its main components. As a result, the synthesis mechanism is not clear by now. Fortunately, Gao, J. P. et al. initiated the study of the internal reference gene of C. pilosula8. In their work, GAPDH was selected as RG to study the expression characteristics of UDGPase gene in polysaccharide synthesis. In our study, more stable RG and RGs combinations than GAPDH were found. Based on RT-qPCR study of C. pilosula, systematic selection and validation analysis of reliable RGs for gene expression normalization were carried out. More importantly, candidate RGs, used in the selection of stable RGs, were retrieved from the transcriptome data of C. pilosula, and then screened according to the CV, log2FC values of the expression level of each gene in all samples.
RT-qPCR was used to determine the expression of the candidate RGs under the conditions of cold stress, drought stress and developmental stages, and finally the stable RGs were selected. This study confirmed that RNA-seq technology was a feasible and reliable way to select RGs for C. pilosula. To obtain suitable RG, four calculation methods including ΔCt38, geNorm39, NormFinder40 and BestKeeper41 were applied in analyzing the stability of candidate RGs. The rankings derived from the four methods were slightly different due to the changed algorithms45. Taking full consideration of the ranks obtained from the four methods, the geometric means of the four rankings were calculated and used to get an integrated ranking. Accordingly, the suitable RG or the combination of RGs were selected. Surprisingly, some RGs commonly used in other plants, such as ACTβ, EF1α and TUBα3, are considered as unstable RGs in our study9,11,12,19,31, which were consistent with the analysis results of Ct values and their range of interquartile. This indicated that genes with too high or too low expression abundance were not suitable for RGs, as well as the genes with a wide interquartile range of Ct values.
Some candidate RGs that ranked at the top of the RNA-Seq analysis were not always stable in RT-qPCR test. This study also showed that the single optimal RG is PP2A59γ for developmental stage, RPL5B for cold treatment, and RPL13 for drought treatment. Single RG can be used for gene expression analysis, as is used in most studies. However, disturbing factors such as extremely cold and drought climates during the period of growth probably leaded to deviation in gene expression by using single RG. Therefore, two or more RGs are recommended for reliable gene expression43. Suitable combinations of RGs were obtained as follows. PP2A59γ, CPY20-1, UBCE32, RPL5B, and UBC18 were the optimal combination RGs for developmental stage, RPL5B, RPL13, and PP2A59γ for cold treatment, and the combination of RPL13 and PP2A59γ for drought treatment. The combination of RGs could reduce the influencing of experimental factors on the gene expression, accordingly, it was more accurate and reliable than single RG in the normalization of gene expression9,25,45. Comprehensive analysis on the selected RGs recommended five RGs combination for gene expression of C. pilosula, which are RPL13, UBCE32, RPL5B, CPY20-1, and PP2A59.
Materials and Methods
Plant material
One-year-old seedlings of C. pilosula were collected as test plant from Tanchang, Gansu province, China on April 5th 2018. The seedlings were cultivated in sandy soil at a temperature of 25 ± 2 °C, relative humidity of 55–60%, irradiance of 300 μmol m−2 s−1 and 14 h photoperiod (6:00–20:00 h). After 30 days cultivation, test plants were selected from the uniformly grown seedlings with the length of vines between 30 and 50 cm. These plants were subjected to three treatments, which are the normal watering and fertilization for developmental stages study, cold-stress treatment and drought-stress treatment. For developmental stages study, samples of leaf, stem and root were collected after 30, 45 and 60 days. Cold-stress treatment were conducted in incubator under 4 ± 2 °C for 0, 4, 8 and 12 h, respectively. The leaf, stem and root were collected correspondingly from the treated and untreated plants. Drought-stress treatment were carried out by controlling the relative water content of soil at 50 ± 5%, with the normally irrigated soil as controls. The leaf, stem and root were collected after 5 and 10 days from the drought-stress and normally irrigated plants. Three plants were sampled for each replicate and three replicates were required for each treatment. All samples were immediately frozen in liquid nitrogen, and kept frozen at −80 °C for analyses.
RNA isolation and cDNA synthesis
Total RNA was extracted from each sample using the RNAprepPure Plant Kit DP441 (Tiangen Biotech CO., LTD, Beijing, China) according to the manufacturer’s instructions, followed by DNase I (Tiangen Biotech CO., LTD, Beijing, China) treatment to eliminate DNA contamination. The integrity of RNA was determined by 1.5% agarose gel electrophoresis. The purity and concentration of total RNA was determined using NanoDrop 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, US). RNA samples with a concentration higher than 60 ng/μL and a ratio of A260/A280 between 1.8 and 2.0 were required for cDNA preparation. Synthesis of cDNA was conducted in the PrimerScript™ RT cDNA Synthesis Kit (TaKaRa Bio Inc., Dalian, China) using 1.0 μg RNA solution. The obtained cDNA was then diluted with 10 times nuclease-free water to prepare RT-qPCR.
Selection of candidate reference genes
Transcriptome sequencing was performed on C. pilosula samples collected from different growth stages adopting paired-end sequencing technology on an Illumina Hi-Seq™ 4000 platform. After assembling and annotating the transcriptome data, the gene expression level (GEL) was analyzed by calculating TPM (Transcripts per kilobase of exon model per million mapped reads) with the help of EdgeR package46. Multiples of gene expression difference was calculated from the log fold change (FC) value. The related data has not been officially published. According to the literature36,37, the conventional RGs include ACT, GAPDH, G6PDH, EF, 18 S rRNA, UBQ, TUB, EIF, UBC, H3, PGK, TEF, CYP, and NADHD. The related genes were retrieved from the transcriptomic data of C. pilosula, and the gene expression levels including values of CV(coefficient of variation) and log FC were calculated. In general, the lower the CV value is, the more stable the expression of relevant genes becomes. Under the conditions of CV < 0.5 and |log2FC| < 0.2, the candidate reference RGs were screened from the genes which ranked high in stability.
Primer design and RT-qPCR
Primers were designed using Primer5 software based on the sequences retrieved from the RNA-Seq dataset of C. pilosula. The criteria for primer design were as follows: primer lengths in 20–24 bp, GC content of 45–55%, melting temperature at 58 °C, and amplicon lengths in 100–250 bp. Primer information for the candidate RGs was listed in Table 2. Reactions were conducted using TB Green Premix Ex Taq II kit (Takara) and amplified on QuantStudioTM 6 Flex Real-Time PCR system (ABI Technologies) in a final volume of 10 μL according to the manufacturer’s instructions. Melting curve analyses were done following the amplification cycles in order to examine the specificity of the reactions. Amplification efficiencies were calculated using the QuantStudioTM Real-Time PCR software (ver 1.1). The PCR efficiency of each primer pair was determined by the slope of the standard curve according to the equation E = [10(−1/slope) − 1] × 100%47. Efficiency values should be between 90% and 110%.
Data analysis
The four most commonly used methods including ΔCt38, geNorm39, NormFinder40 and BestKeeper41, were adopt to calculate and identify the expression stability of candidate reference genes. The ΔCt method displays the pairwise comparisons by calculating the standard deviation (SD) of each pair candidate RGs and the average SD of each gene. The lower the average SD is, the more stable the RG performs. The geNorm algorithm calculates the expression stability value (M value) and pairwise variation (Vn/n + 1) for all candidate genes. Lower M values suggests higher stability of gene expression. The Vn/n + 1 value determines the optimal number of RGs for accurate normalization. A cut-off value of Vn/n + 1 < 0.15 indicates that an additional RG makes no significant contribution to the normalization42. NormFinder program is based on the ANOVA model, which reveals expression variation by calculating the stability value (SV) when using RGs for normalization. Lower SV indicates higher stability. The BestKeeper method is an Excel-based tool using pairwise correlations, which discloses the expression level of all candidate genes by calculating the SD, coefficient of correlation (r) and coefficient of variance (CV). After rejecting the SD values of RGs higher than one, the candidate RGs was ranked according to the three indexes. The most stable gene expression exhibits the lowest CV ± SD value. Based on the results derived from the four methods, the geometric mean of each candidate gene was calculated to obtain its overall final ranking. A lower geometric mean of ranking indicates more stable expression.
Validation of reference genes
In order to validate the selected RGs under different treatment conditions, relative expression analyses of UDGPase with published data were performed using the 2−ΔΔCt method17. The effect of the single RG and the combination of RGs in normalizing the relative expression level of the target gene were compared to determine whether the combination of internal RGs can improve the reliability and accuracy of RT-qPCR results. Gene expression was also normalized using the most unstable ACTβ gene for comparison.
Conclusions
This research provided the optimal single RG and combination of RGs for C. pilosula. The combination of PP2A59γ, CPY20-1, UBCE32, RPL5B, and UBC18 are suitable for developmental stage, RPL5B, RPL13, and PP2A59γ for cold treatment, and in drought treatment, RPL13 and PP2A59γ were the optimal combination of RGs. These combinations of RGs are recommended as reference genes in RT-qPCR analysis of C. pilosula. This study also demonstrated the validity of using RNA-Seq expression data for the determination of candidate reference genes for RT-qPCR analysis.
Supplementary information
Acknowledgements
This research was funded by the 13th five-year plan of Ministry of science and technology of the People’s Republic of China, grant number 2015BAC-01B-03.
Author contributions
Z.L. supervised the project and gained financial support; L.L., H.Y. and Z.H. conceived the experiments; L.L., E.W., X.Z., B.Z., C.Z. and Z.L. performed the experiments; L.L. and H.Y. analysed the data; L.L. and Z.H. wrote and edited the manuscript; Z.L. and C.Z. modified the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-58328-5.
References
- 1.Commission, C. P. Pharmacopoeia of the People’s Republic of China. Vol. I (China Medical Science Press, 2015).
- 2.Wang J, Deng C-Q, Shi L, Deng M-L. Progress of modern research on Codonopsis taxa. Guide of China Medicine. 2011;9:279–281. doi: 10.15912/j.cnki.gocm.2011.31.068. [DOI] [Google Scholar]
- 3.Liang L-J, et al. Study on Hybrid Characteristics of Medicinally Used Cultivated Codonopsis Species Using Ribosomal Internal Transcribed Spacer (ITS) Sequencing. Molecules. 2018;23:1565. doi: 10.3390/molecules23071565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He JY, et al. The genus Codonopsis (Campanulaceae): a review of phytochemistry, bioactivity and quality control. Journal of Natural Medicines. 2015;69:1–21. doi: 10.1007/s11418-014-0861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qing-hong Z, Jing L, Jiao-jiao J, Jian-kuan L, Jian-ping G. Cloning and Expression Analysis of the CpSUC4 Gene in Codonopsis pilosula. Journal of Shanxi University(Nat. Sci.Ed.) 2018;41:172–179. doi: 10.13451/j.cnki.shanxi.univ(nat.sci.).2018.03.27.002. [DOI] [Google Scholar]
- 6.Xiao-lin W, Jiao-jiao J, Jian-ping G. Clone and expression of CpGAPDH gene in Codonopsis pilosula. Journal of Chinese Materia Medica. 2018;43:712–720. doi: 10.19540/j.cnki.cjcmm.20180105.009. [DOI] [PubMed] [Google Scholar]
- 7.Jing L, Qiong-qiong G, Hai-feng S, Jian-ping G. Cloning, sequence analysis, and prokaryotic expression of CpUGPase gene in Codonopsis Radix. Chinese Traditional and Herbal Drugs. 2016;47:3876–3883. doi: 10.7501/j.issn.0253-2670.2016.21.021. [DOI] [Google Scholar]
- 8.Gao JP, Wang D, Cao LY, Sun HF. Transcriptome Sequencing of Codonopsis pilosula and Identification of Candidate Genes Involved in Polysaccharide Biosynthesis. PLoS One. 2015;10:e0117342. doi: 10.1371/journal.pone.0117342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T, et al. Selection and Validation of Appropriate Reference Genes for qRT-PCR Analysis in Isatis indigotica Fort. Frontiers in Plant Science. 2017;8:139. doi: 10.3389/fpls.2017.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z-J, Tian C, Jiang Q, Li X-H, Zhuang J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis) Scientific Reports. 2016;6:19748. doi: 10.1038/srep19748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma R, Xu S, Zhao Y, Xia B, Wang R. Selection and Validation of Appropriate Reference Genes for Quantitative Real-Time PCR Analysis of Gene Expression in Lycoris aurea. Frontiers in Plant Science. 2016;7:536. doi: 10.3389/fpls.2016.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian C, et al. Selection of Suitable Reference Genes for qPCR Normalization under Abiotic Stresses and Hormone Stimuli in Carrot Leaves. PLoS One. 2015;10:e0117569. doi: 10.1371/journal.pone.0117569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao X, et al. Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plant (Camellia sinensis (L.) O. Kuntze) International Journal of Molecular Sciences. 2014;15:22155–22172. doi: 10.3390/ijms151222155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, et al. Reference Gene Selection for Quantitative Real-time PCR Normalization in Caragana intermedia under Different Abiotic Stress Conditions. PLoS One. 2013;8:e53196. doi: 10.1371/journal.pone.0053196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekkers BJW, et al. Identification of Reference Genes for RT-qPCR Expression Analysis in Arabidopsis and Tomato Seeds. Plant and Cell Physiology. 2012;53:28–37. doi: 10.1093/pcp/pcr113. [DOI] [PubMed] [Google Scholar]
- 16.Maroufi A, Bockstaele EV, Loose MD. Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. Bmc Molecular Biology. 2010;11:15. doi: 10.1186/1471-2199-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Liang W, et al. Selection and evaluation of reference genes for qRT-PCR analysis in Euscaphis konishii Hayata based on transcriptome data. Plant Methods. 2018;14:42. doi: 10.1186/s13007-018-0311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozera B, Rapacz M. Reference genes in real-time PCR. Journal of Applied Genetics. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang E, et al. Selection of Reference Genes for Quantitative Gene Expression Studies in Platycladus orientalis (Cupressaceae) Using Real-Time PCR. PLoS One. 2012;7:e33278. doi: 10.1371/journal.pone.0033278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dan-dan L, Bo H, Qing W, Wei W. The Research on Reference Genes in Medicinal Plant. Molecular Plant Breeding. 2017;15:903–910. doi: 10.13271/j.mpb015.000903. [DOI] [Google Scholar]
- 22.Xiao-fu Z, Jin W, Hong-wei S, Hong-wei X. The Exploration of 18 S rRNA for Quantitative RT-PCR as Reference Gene in Plant. Journal of Jilin Normal University (Natural Science Edition) 2016;37:115–119. doi: 10.16862/j.cnki.issn1674-3873.2016.02.025. [DOI] [Google Scholar]
- 23.Yu-Fang Z, Li-Juan Z, You-Ling Z. Selection and Application of Reference Genes for Gene Expression Studies. Plant Physiology Journal. 2014;50:1119–1125. doi: 10.13592/j.cnki.pp-14.0201. [DOI] [Google Scholar]
- 24.Joseph JT, Poolakkalody NJ, Shah JM. Plant reference genes for development and stress response studies. Journal of Biosciences. 2018;43:173–187. doi: 10.1007/s12038-017-9728-z. [DOI] [PubMed] [Google Scholar]
- 25.Chapman JR, Waldenstrom J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS One. 2015;10:e0141853. doi: 10.1371/journal.pone.0141853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long XY, et al. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Molecular Biology. 2010;74:307–311. doi: 10.1007/s11103-010-9666-8. [DOI] [PubMed] [Google Scholar]
- 27.Jonge HJMD, et al. Evidence Based Selection of Housekeeping Genes. PLoS One. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong S-Y, Seo PJ, Yang M-S, Xiang F, Park C-M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. Bmc Plant Biology. 2008;8:112. doi: 10.1186/1471-2229-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez L, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription‐polymerase chain reaction (RT‐PCR) analysis in plants. Plant Biotechnology Journal. 2010;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- 30.Gimeno J, Eattock N, Deynze AV, Blumwald E. Selection and validation of reference genes for gene expression analysis in switchgrass (Panicum virgatum) using quantitative real-time RT-PCR. PLoS One. 2014;9:e91474. doi: 10.1371/journal.pone.0091474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shivhare R, Lata C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Scientific Reports. 2016;6:23036. doi: 10.1038/srep23036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maroufi A. Selection of reference genes for real-time quantitative PCR analysis of gene expression in Glycyrrhiza glabra under drought stress. Biologia Plantarum. 2016;60:1–10. doi: 10.1007/s10535-016-0601-y. [DOI] [Google Scholar]
- 33.Lekshmy S, Jha SK. Selection of reference genes suitable for qRT-PCR expression profiling of biotic stress, nutrient deficiency and plant hormone responsive genes in bread wheat. Indian Journal of Plant Physiology. 2017;22:101–106. doi: 10.1007/s40502-017-0282-3. [DOI] [Google Scholar]
- 34.Luo M, et al. Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Scientific Reports. 2018;8:4444. doi: 10.1038/s41598-018-22743-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martins PK, et al. Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Scientific reports. 2016;6:28348. doi: 10.1038/srep28348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, et al. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene. 2014;538:113–122. doi: 10.1016/j.gene.2013.12.057. [DOI] [PubMed] [Google Scholar]
- 37.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:research0034–0031. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen CL, Jensen JL, Ørntoft TF. Normalization of realtime quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data set. Cancer Research. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnology Letters. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 42.Stanton KA, et al. A Whole-Transcriptome Approach to Evaluating Reference Genes for Quantitative Gene Expression Studies: A Case Study in Mimulus. G3 (Bethesda). 2017;7:1085–1095. doi: 10.1534/g3.116.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J, et al. Genome-Wide Constitutively Expressed Gene Analysis and New Reference Gene Selection Based on Transcriptome Data: A Case Study from Poplar/Canker Disease Interaction. Frontiers in Plant Science. 2017;8:1876. doi: 10.3389/fpls.2017.01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. Bmc Plant Biology. 2010;10:4. doi: 10.1186/1471-2229-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabeh M, Duceppe M-O, St-Arnaud M, Mimee B. Transcriptome-wide selection of a reliable set of reference genes for gene expression studies in potato cyst nematodes (Globodera spp.) PLoS One. 2018;13:e0193840. doi: 10.1371/journal.pone.0193840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radonić A, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.