Abstract
The long non-coding RNA NEAT1 locus is transcribed into two overlapping isoforms, NEAT1_1 and NEAT1_2, of which the latter is essential for the assembly of nuclear paraspeckles. NEAT1 is abnormally expressed in a wide variety of human cancers. Emerging evidence suggests that the two isoforms have distinct functions in gene expression regulation, and recently it was shown that NEAT1_2, but not NEAT1_1, expression predicts poor clinical outcome in cancer. Here, we report that NEAT1_2 expression correlates with HER2-positive breast cancers and high-grade disease. We provide evidence that NEAT1_1 and NEAT1_2 have distinct expression pattern among different intrinsic breast cancer subtypes. Finally, we show that NEAT1_2 expression and paraspeckle formation increase upon lactation in humans, confirming what has previously been demonstrated in mice.
Subject terms: Breast cancer, Breast cancer
Introduction
The long non-coding RNA (lncRNA) NEAT1 (Nuclear Paraspeckle Assembly Transcript 1) has recently gained considerable attention as it is abnormally expressed in human diseases, including cancer and neurodegenerative disorders. The NEAT1 gene is transcribed into two isoforms, NEAT1_1 of 3.7 kb and NEAT1_2 of 22.3 kb, where NEAT1_1 completely overlaps with the 5′ end of NEAT1_21. NEAT1_2 is essential for the assembly of paraspeckles, dynamic nuclear ribonucleoprotein complexes that phase-separate from the nucleoplasm to form liquid drop-like structures2–7. In contrast, NEAT1_1 expression is not sufficient to induce paraspeckle formation, and recent reports suggest that NEAT1_1 can localize to structures that are distinct from paraspeckles7,8. NEAT1 expression and paraspeckle formation are upregulated in response to a variety of cellular stressors including mitochondrial stress, proteasome inhibition, oncogene-induced replication stress, hypoxia, heat shock, and viral infections9–18. It is now generally accepted that NEAT1 and paraspeckles regulate gene expression at both transcriptional and post-transcriptional levels by acting as hubs that sequester specific gene regulatory proteins and mRNAs16–20. Several lines of evidence suggest that NEAT1 and paraspeckles play critical roles in stress response pathways in general, and at specific developmental stages. NEAT1 knockout mice display compromised mammary gland development and corpus luteum formation21,22. Moreover, it was recently shown that maternal and zygotic NEAT1-depletion frequently led to early developmental arrest at the 16- or 32-cell stage in mouse embryonic cells23.
Cancer cells are exposed to a variety of extrinsic and intrinsic stressors like hypoxia, proteotoxicity, DNA damage, and reactive metabolic intermediates24. Such malignancy-associated stress has been shown to induce NEAT1 expression and paraspeckle formation in vivo15,16. NEAT1 levels are elevated in hypoxic regions of breast cancer cell line xenografts, and skin tumors induced by genotoxic stress in mice, display increased NEAT1 expression and paraspeckle formation15,16. In consistence with these observations, NEAT1 is overexpressed in many cancers15,16,25,26. In most cases, NEAT1 expression is associated with aggressive disease and poor clinical outcomes15,16,25,26.
Breast cancer is the most common type of cancer in women and covers a broad spectrum of different malignant neoplasms with clinical and genomic heterogeneity27,28. In clinical diagnosis, breast cancer is classified according to histological grade, Ki-67 proliferative index, and to the expression of hormone and growth factor receptors estrogen receptor (ER), progesterone receptor (PgR), and human epidermal growth factor receptor 2 (HER2). The classification of breast cancer has been stratified by gene expression profiling leading to the identification of a 50-gene signature (PAM50) that groups breast cancer into luminal A, luminal B, HER2-enriched, basal-like, and normal-like intrinsic subtypes29–31. Several studies have demonstrated that NEAT1 is required for proliferation and survival of breast cancer cell lines12,16,21,32–35. Moreover, NEAT1 is frequently overexpressed in breast tumor samples compared to adjacent normal tissue and is associated with poor overall survival16,34–37. Recently, genomic analyses of 360 primary breast tumors showed that the core promoter of the NEAT1 gene is frequently mutated in cancer and most of these mutations are associated with loss of expression in in vitro assays38. In addition, focal deletions within the NEAT1 gene were found in 8% of breast cancers, and mutations are frequently found in the exonic region38,39. This suggests that NEAT1 expression might either protect or enhance cancer initiation and progression dependent on tumor stage. Moreover, a growing body of experimental evidence shows that the two NEAT1 isoforms have distinct physiological functions40,41. Therefore, it is important to address the relative contribution of NEAT1_1 and NEAT1_2 in cancer progression.
In this study, we have examined the relationship between NEAT1_2 expression and breast cancer subtypes by performing RNA-FISH analyses on core needle biopsies using probes solely recognizing the NEAT1_2 isoform. We report that NEAT1_2 expression associates with HER2-positive breast cancers, and with high tumor grade. This is verified by in silico analyses of microarray data from three independent breast cancer cohorts showing that NEAT1_2 is most highly expressed in luminal B and HER2-enriched cancers. Interestingly, we present evidence suggesting that NEAT1_1 expression shows a distinct distribution among breast cancer subtypes compared to NEAT1_2, being highest in ER-positive luminal A and luminal B cancers. This indicates that the relative expression of NEAT1_1 versus NEAT1_2 varies among the different breast cancer subclasses. Finally, we report that NEAT1_2 and paraspeckle formation are induced in vivo in human luminal epithelial cells during lactation.
Results
NEAT1_2 expression is associated with high tumor grade and HER2 positive breast cancers
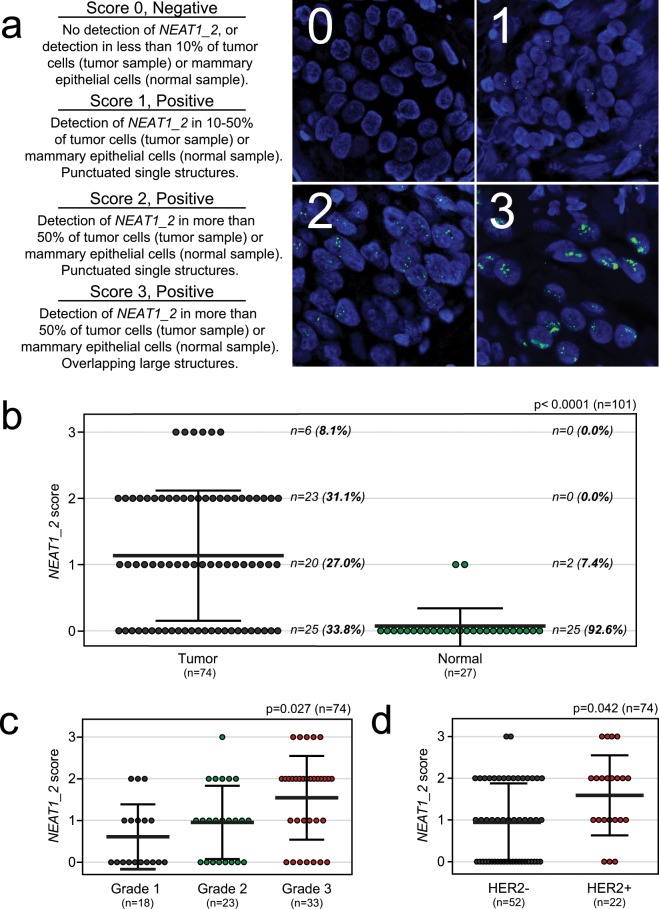
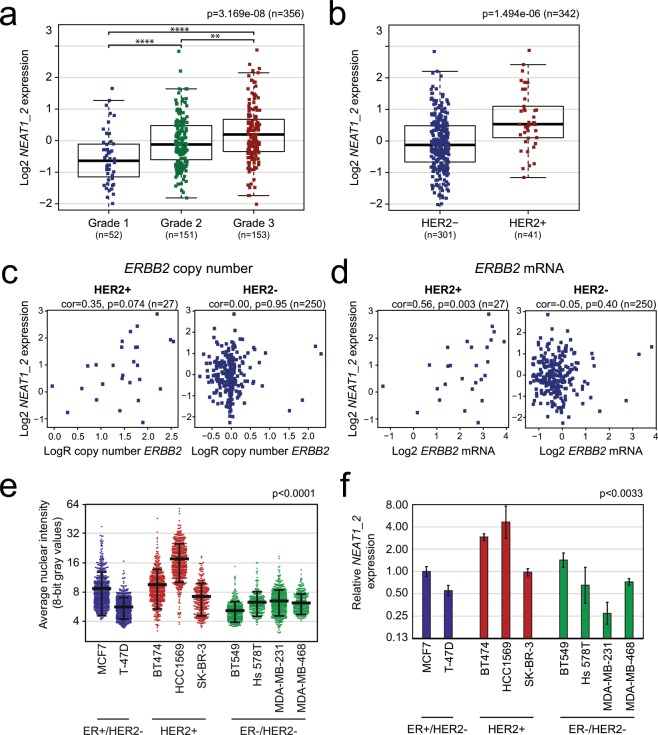
The NEAT1_2 isoform is essential for the assembly of paraspeckles that regulate the expression of specific genes at certain cellular circumstances1,16–20. Recently, it was shown that the expression of NEAT1_2, but not NEAT1_1, predicts progression-free survival of ovarian cancer treated with platinum-based chemotherapy15. This prompted us to specifically investigate the expression of NEAT1_2 in breast cancer. To determine the relationship between breast cancer subtypes and both NEAT1_2 expression and associated paraspeckle formation, we performed NEAT1_2-specifc RNA-FISH analyses on 74 formalin-fixed paraffin-embedded needle biopsies taken from females at the time of diagnosis of breast cancer. The samples were selected to represent cancers pathologically classified as luminal A (n = 23), luminal B (n = 29), triple negative/basal-like (n = 14) and HER2-positive (n = 8). We also included 27 non-cancerous breast samples in the study (23 fibroadenomas, 3 mammary reduction, and 1 BRCA1 prophylactic mastectomy). Cancer cells were identified by experienced pathologists, and NEAT1_2 expression was manually scored from “0” to “3” based on the presence and morphology of punctuated nuclear signals corresponding to paraspeckles (Fig. 1a). Samples scored as “1”, “2”, and “3” were defined as NEAT1_2-positive. Forty-nine patients (66%) were positive for NEAT1_2 expression (Fig. 1b). In all cases, the expression was strictly restricted to cancer cells, with no detectable NEAT1_2 signals in surrounding stromal tissue, infiltrating immune cells, or in unaffected breast tissue. In sharp contrast, only 2 out of 27 benign breast tissue samples were NEAT1_2-positive (7.4%), both samples being scored as “1” (Fig. 1b). Clinicopathological characteristics were acquired from each patient and correlated with NEAT1_2 expression (Table 1). NEAT1_2 levels significantly associated with tumor grade (Chi square test p = 0.027; Fig. 1c, Table 1), confirming what has previously been reported by others on total NEAT1. Interestingly, NEAT1_2 expression also associated with HER2 positive breast cancers (Chi square test p = 0.042; Fig. 1d, Table 1). To verify these results, we analyzed microarray expression data from 381 breast cancer patients (Oslo2) using data generated by a NEAT1_2-specific probe42. We confirmed that NEAT1_2 expression was associated with grade (Kruskal-Wallis test p = 3.169e-08; Fig. 2a) and HER2 status (Wilcoxon Rank-Sum test p = 1.49e-06; Fig. 2b). Intriguingly, we also found that NEAT1_2 expression was significantly lower in ER-positive tumors compared to ER-negative tumors in this cohort (Wilcoxon Rank-Sum test p = 0.005155; Supplementary Fig. 1). To further determine the relationship between NEAT1_2 and HER2 expression in the Oslo2 cohort, we investigated the correlation between NEAT1_2 levels and ERBB2 copy number and mRNA expression in HER2-positive and HER2-negative cancers. A close to significant positive correlation was found between NEAT1_2 expression and ERBB2 copy number (Spearman’s rank correlation R = 0.35, p = 0.074; Fig. 2c, left panel), and a significant positive correlation was found between NEAT1_2 and ERBB2 mRNA expression (Spearman’s rank correlation R = 0.56, p = 0.003; Fig. 2d, left panel). As expected, no correlation was found between NEAT1_2 and ERBB2 amplification or mRNA expression in HER2-negative cancers (Fig. 2c,d, right panel). Finally, we assessed the expression of NEAT1_2 by RNA-FISH and RT-qPCR in nine breast cancer cell lines classified according to the expression of hormone- and growth factor receptors into ER/PgR-positive HER2-negative cells (MCF7, T-47D), HER2-positive cells (BT474, HCC1569, SK-BR-3), and triple negative cells (BT549, Hs 578T, MDA-MB-231, MDA-MB-468)43. In consistence with previous reports, the morphology, as well as the number and size of NEAT1_2-containing paraspeckles, varied substantially between the different cell lines (Supplementary Fig. 2)44. We also observed cell-to-cell variations within each cell line. In general, both the number and size of NEAT1_2-containing punctas were hard to determine as they frequently formed clusters. We therefore measured the average intensities of NEAT_2 signals per cell in all cell lines (Fig. 2e). Interestingly, HER2-positive BT474 and HCC1569 clearly expressed the highest levels of NEAT1_2. Moreover, NEAT1_2 expression levels in HER2-positive SK-BR-3 cells were only exceeded by those in MCF7 cells. This was confirmed by RT-qPCR analyses using primers specifically amplifying the NEAT1_2 isoform (Fig. 2f). Generally, results obtained by imaging and RT-qPCR were concordant, only showing deviations for the BT549 cell line. We conclude that high NEAT1_2 expression is associated with HER2-positive breast cancer and with high-grade disease. Moreover, the presence of NEAT1_2 and paraspeckles are highly specific for cancer cells, and not present or very lowly expressed in surrounding normal cells or non-cancerous breast epithelial cells.
Figure 1.
NEAT1_2 expression and paraspeckle formation correlate with tumor grade and HER2 expression in breast cancer. (a) RNA-FISH analyses of NEAT1_2 in formalin-fixed paraffin-embedded needle biopsies from breast cancer and benign samples. NEAT1_2 expression is scored from “0” to “3” based on punctuated nuclear NEAT1_2 signals according to the indicated criteria. For tumor samples, only tumor cells were included in the scoring, while for normal samples, only epithelial cells were included. (b) NEAT1_2 is more highly expressed in tumor versus normal tissue. (c) NEAT1_2 expression associates with tumor grade. (d) NEAT1_2 expression associates with HER2 expression. Data are presented as mean (thick black line) ± standard deviation (thin black lines). Circles represent single patient scores. Statistical significance was calculated using the Mann Whitney test (b) or Chi square test (c) and (d). Data were considered statistically significant when p ≤ 0.05.
Table 1.
Clinicopathological variables and NEAT1_2 expression in breast cancer screening cohort (n = 74). The Chi square test (χ2-value) was used to calculate p-values.
| Variable, n(%) | NEAT1_2 expression | p | Total (n = 74) | ||||
|---|---|---|---|---|---|---|---|
| 0 (n = 25) | 1 (n = 20) | 2 (n = 23) | 3 (n = 6) | ||||
| Age at diagnosis | <55 | 10 (34.5) | 8 (27.6) | 8 (27.6) | 3 (10.3) | 0.920 | 29 (39.2) |
| ≥55 | 15 (33.3) | 12 (26.7) | 15 (33.3) | 3 (6.7) | 45 (60.8) | ||
| Histologic grade | 1 | 10 (55.6) | 5 (27.8) | 3 (16.7) | 0 (0.0) | 0.027* | 18 (24.3) |
| 2 | 8 (34.8) | 9 (39.1) | 5 (21.7) | 1 (4.3) | 23 (31.1) | ||
| 3 | 7 (22.2) | 6 (18.2) | 15 (45.5) | 5 (15.2) | 33 (44.6) | ||
| Tumor type | NST | 20 (29.9) | 20 (29.9) | 22 (32.8) | 5 (7.5) | 0.156 | 67 (90.5) |
| ILC | 3 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (4.1) | ||
| Other invasive carcinomaa | 2 (50.0) | 0 (0.0) | 1 (25.0) | 1 (25.0) | 4 (5.4) | ||
| Tumor diameterb | <20 mm | 14 (37.8) | 12 (32.4) | 7 (18.9) | 4 (10.8) | 0.213 | 37 (53.6) |
| ≥20 mm | 11 (34.4) | 6 (18.8) | 13 (40.6) | 2 (6.3) | 32 (46.4) | ||
| Lymph node metastasisb | Negative | 17 (35.5) | 14 (29.2) | 13 (27.1) | 4 (8.3) | 0.990 | 48 (67.6) |
| Positive | 8 (34.8) | 6 (26.1) | 7 (30.4) | 2 (8.7) | 23 (32.4) | ||
| ER | Negative (<1%) | 4 (16.7) | 7 (29.2) | 11 (45.8) | 2 (8.3) | 0.131 | 24 (32.4) |
| Positive (≥1%) | 21 (42.0) | 13 (26.0) | 12 (24.0) | 4 (8.0) | 50 (67.6) | ||
| PgR | Negative (<1%) | 6 (20.7) | 8 (27.6) | 12 (41.4) | 3 (10.3) | 0.226 | 29 (39.2) |
| Positive (≥1%) | 19 (42.2) | 12 (26.7) | 11 (24.4) | 3 (6.7) | 45 (60.8) | ||
| HER2 | Negative (0, 1+, 2+/no ISH amp) | 22 (42.3) | 13 (25.0) | 15 (28.8) | 2 (3.8) | 0.042* | 52 (70.3) |
| Positive (2+/ISH amp, 3+) | 3 (13.6) | 7 (31.8) | 8 (36.4) | 4 (18.2) | 22 (29.7) | ||
aTubulolobular carcinoma (n = 1), metaplastic squamous cell carcinoma (n = 1), mucinous carcinoma (n = 1), apocrine carcinoma (n = 1).
bPatient(s) data missing.
*P-value significant.
Invasive carcinoma of no special type (NST), invasive lobular carcinoma (ILC), in situ hybridization (ISH), amplification (amp).
Figure 2.
The association between NEAT1_2 expression and HER2 status is verified in an independent breast cancer cohort and in breast cancer cell lines. (a) NEAT1_2 expression was analyzed in microarray gene expression profiling data from patients in the Oslo2 cohort and correlated to tumor grade. The Kruskal-Wallis test was used to calculate whether any groups are significantly different from each other and Wilcoxon Rank-Sum test was used in post-testing for significant differences between pairs of groups. (***p ≤ 0.0001; **p ≤ 0.01). (b) NEAT1_2 expression levels were correlated to HER2 status. The Wilcoxon Rank-Sum test was used to test for significant differences between the groups. (c) and (d) NEAT1_2 expression positively correlates with ERBB2 copy number (c) and ERBB2 mRNA expression (d) in HER2-positive, but not in HER2-negative, patients. Correlation was calculated using Spearman’s rank correlation. (e) Breast cancer cell lines were subjected to NEAT1_2 RNA-FISH and NEAT1_2-specific signal intensity per nucleus in at least 250 cells was quantitated. Data are given as mean (thick black line) ±standard deviation (thin black lines). Circles represent single cell intensities. The Kruskal-Wallis test was used to calculate whether any groups are significantly different from each other. (f) RNA was isolated from breast cancer cell lines and the expression of NEAT1_2 was determined by RT-qPCR. The geometric means of B2M, GAPDH, and RPLP0 were used for normalization. The mean value ± standard deviation of three biological independent experiments is presented as fold change relative to MCF7 NEAT1_2 expression. Statistical significance was calculated using the Kruskal-Wallis test. Data were considered statistically significant when p ≤ 0.05.
NEAT1_2 expression is associated with the HER2-enriched and luminal B breast cancer subtypes
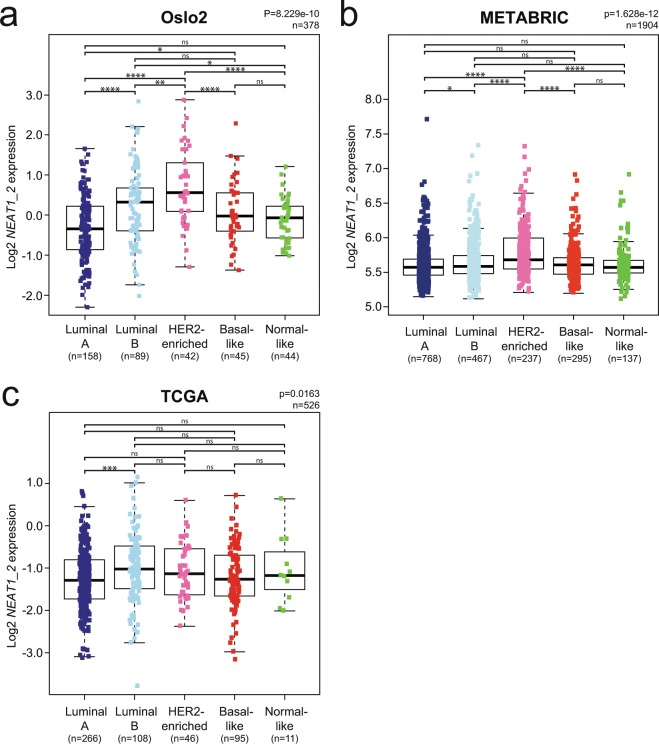
We demonstrated above that NEAT1_2 expression associates with HER2-positive breast cancer. HER2 overexpressing cancers are in most cases classified as HER2-enriched or luminal B using the PAM50 gene expression signature identifier. To assess the association between NEAT1_2 expression and intrinsic breast cancer subtypes, we analyzed microarray gene expression data derived from the Oslo2 cohort described above, and two publicly available breast cancer patient cohorts, METABRIC28 and The Cancer Genome Atlas (TCGA)45. Patients were subtyped using the PAM50 algorithm31, and only data generated from probes solely recognizing the NEAT1_2 isoform were considered. In all three cohorts, NEAT1_2 was most highly expressed in breast cancers classified as HER2-enriched and luminal B, but with different intrinsic distributions (HER2-enriched > luminal B in Oslo2 and METABRIC; luminal B > HER2-enriched in TCGA) (Fig. 3a–c). Luminal A breast cancers had the lowest expression of NEAT1_2 in all three cohorts. Taken together, these results are in accordance with the observed correlation between NEAT1_2 expression and HER2-status.
Figure 3.
NEAT1_2 is most highly expressed in HER2-enriched and luminal B intrinsic breast cancer subtypes in three independent cohorts. (a–c) NEAT1_2 expression in PAM50 intrinsic breast cancer subtypes from patients of the Oslo2 (a), METABRIC (b), and TCGA (c) breast cancer cohorts. Subtypes were determined using the PAM50 algorithm. The Kruskal-Wallis test was used to calculate whether any groups are significantly different from each other and Wilcoxon Rank-Sum test was used in post-testing for significant differences between pairs of groups. (****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, p > 0.05).
NEAT1_1 expression is highest in luminal A and luminal B breast cancers
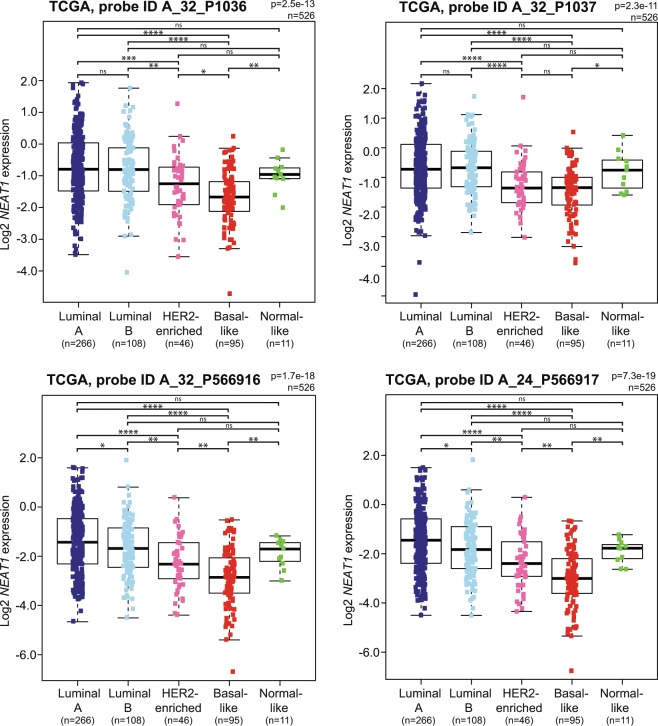
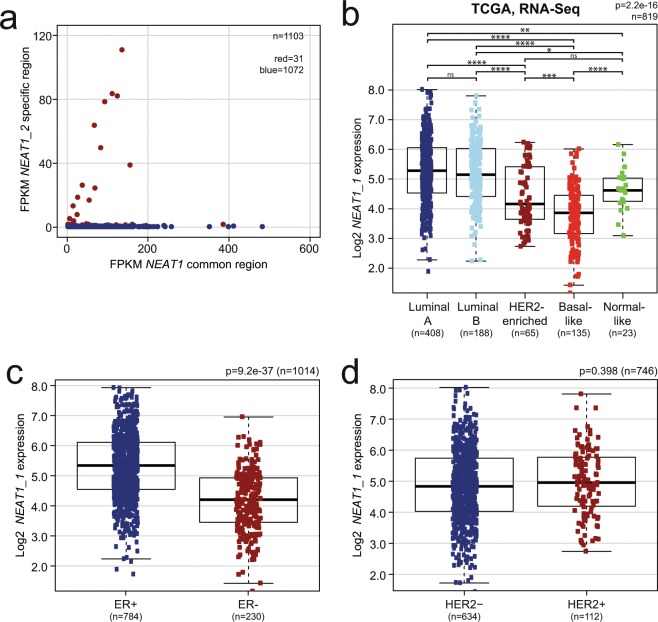
Previous reports have demonstrated that the NEAT1 gene is transcriptionally activated by ERα in both prostate and breast cancer, and the transcript participates in a gene repressor complex that induces epithelial-mesenchymal transition (EMT) in a mouse model of ER-positive breast cancer25,36. Here, we have found that the expression of the long NEAT1_2 isoform is lower in ER-positive compared to ER-negative tumors in the Oslo2 breast cancer cohort (Supplementary Fig. 1). This potential discrepancy made us analyze the expression of total NEAT1 using microarray data derived from probes binding to both NEAT1_1 and NEAT1_2 from the TCGA cohort. Interestingly, total NEAT1 expression showed a different distribution among the PAM50 subtypes compared to NEAT1_2, being most highly expressed in luminal A, luminal B, and normal-like cancers (Fig. 4). NEAT1_1 is, as opposed to NEAT1_2, a polyadenylated transcript. To more specifically investigate the expression of NEAT1_1 in breast cancer, we analyzed polyA-selected RNA-sequencing data from the TCGA breast cancer cohort. We only extracted data from samples that hardly displayed any mapping of fragments to the unique NEAT1_2 region (<1.0 FPKM (Fragments Per Kilobase Million)) (Fig. 5a). Patients were then subtyped according to the PAM50 classifier, and ER and HER2 status were extracted. NEAT1_1 showed a similar distribution among the PAM50 subtypes as total NEAT1, being highest in luminal A and luminal B breast cancers (Fig. 5b). In line with this, NEAT1_1 expression clearly associated with ER-positive tumors (Wilcoxon Rank-Sum test p = 9.2e-37; Fig. 5c). Finally, we found no association between NEAT1_1 and HER2 expression (Wilcoxon Rank-Sum test p = 0.398; Fig. 5d). To conclude, our data clearly indicate that the relative expression of NEAT1_1 versus NEAT1_2 varies among different breast cancer subclasses.
Figure 4.
Total NEAT1 is most highly expressed in the luminal subtypes in the TCGA breast cancer cohort. Expression of total NEAT1 in PAM50 intrinsic breast cancer subtypes was determined using data generated from four independent microarray probes in the TCGA cohort. The Kruskal-Wallis test was used to calculate whether any groups are significantly different from each other and Wilcoxon Rank-Sum test was used in post-testing for significant differences between pairs of groups. (****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, p > 0.05).
Figure 5.
NEAT1_1 is most highly expressed in the luminal breast cancer subtypes in the TCGA RNA-Seq cohort. (a) NEAT1 common region FPKM (Fragments Per Kilobase Million) is plotted against NEAT1_2 specific-region FPKM for each patient. Each dot represents one patient. Samples with more than 1 FPKM for the NEAT1_2-specific region (red dots) were excluded from further analysis. (b) NEAT1_1 is most highly expressed in luminal A and luminal B tumors. The Kruskal-Wallis test was used to calculate whether any groups are significantly different from each other and Wilcoxon Rank-Sum test was used in post-testing for significant differences between pairs of groups. (c) NEAT1_1 expression associates with ER expression status. (d) NEAT1_1 expression does not associate with HER2 status. Statistical significance was calculated in (c) and (d) using Wilcoxon Rank-Sum test. Data were considered statistically significant when p ≤ 0.05. (****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, p > 0.05).
NEAT1_2 expression is upregulated in human breast tissue during lactation
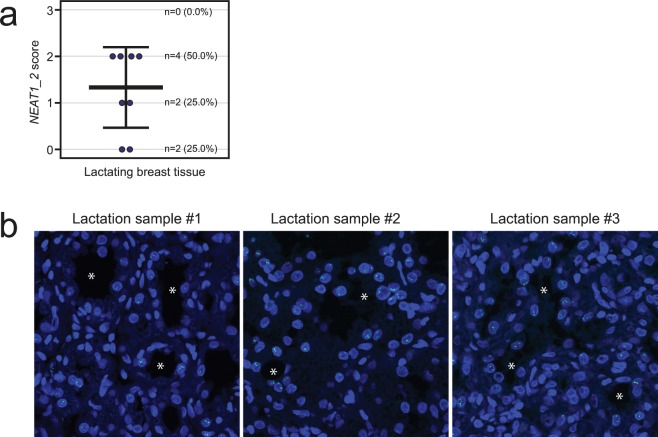
We have demonstrated that NEAT1_2 is rarely expressed in normal human breast tissue. NEAT1 female knock-out mice display compromised mammary gland development during puberty and pregnancy, and fail to lactate due to impaired proliferation of luminal alveolar cells22. This suggests that NEAT1 has an important function in mammary gland development, and during pregnancy and lactation. In order to investigate if NEAT1_2 is expressed during lactation in humans, we analyzed eight needle biopsies taken from females with lactation-related benign changes in the mammary gland. Importantly, 75% (n = 6) of the lactating breast tissue samples were positive for NEAT1_2 using the same scoring scheme as above (Figs. 1a and 6a). Of note, we also had access to one sample from a pregnant woman, which was scored as NEAT1_2 positive (score 2). In both the lactating tissue and the breast tissue from the pregnant female, the expression of NEAT1_2 was restricted to the luminal breast epithelial cells (Fig. 6b).
Figure 6.
NEAT1_2 is expressed in lactating breast tissue. (a) RNA-FISH analyses of NEAT1_2 in breast tissue from lactating females (n = 8). NEAT1_2 expression is scored from “0” to “3” based on punctuated nuclear NEAT1_2 signals according to the indicated criteria in Fig. 1a. Data are shown as mean (thick black line) ±standard deviation (thin black lines). Circles represent single patient scores. (b) RNA-FISH images from three lactating females. NEAT1_2 is visualized in green, and DAPI (blue) was used to stain the nuclei. White asterisks visualize some of the lumens within the mammary glands.
Discussion
The lncRNA NEAT1 locus is conserved in mammalian species and encodes two overlapping transcripts, NEAT1_1 and NEAT1_2, of which the latter is essential for the assembly of paraspeckles1. Early analyses in mice indicated that whereas NEAT1_1 is ubiquitously expressed, the expression pattern of NEAT1_2, and thus the presence of paraspeckles, is more restricted5. Emerging evidence now suggests that NEAT1_2 and paraspeckles play critical roles in orchestrating specific gene expression upon cellular stress and at specific developmental stages9–18. Importantly, it was recently shown that the expression of NEAT1_2, but not total NEAT1, was associated with aggressive cancers15. Here, we have specifically analyzed the expression of NEAT1_2 in breast cancer. By performing RNA-FISH on 74 breast cancer needle biopsies, we found that NEAT1_2 expression and paraspeckle formation associated with HER2-positive cancers. We verified this by inspecting microarray data generated by a NEAT1_2-specific probe from a cohort of 381 patients. Moreover, we found that NEAT1_2 is highly expressed in HER2-positive compared to HER2-negative breast cancer cell lines. Finally, in three different breast cancer cohorts, NEAT1_2 expression associated with HER2-enriched and luminal B PAM50 intrinsic subtypes.
Around 15–20% of all breast cancers overexpress the HER2 receptor, many of them due to the amplification of the ERBB2 gene on chromosome 17, and HER2-driven cancers are generally aggressive46,47. The HER2 receptor is an orphan member of the epidermal growth factor receptor family that upon overexpression forms homodimers or heterodimers with either EGFR, HER3, or HER4, which elicit signaling pathways, including the MEK-ERK and PI3-kinase-Akt pathways, that drive tumorigenesis46,47. NEAT1 expression is generally regulated at the transcriptional level, and it is reasonable to assume that HER2-signaling leads to the activation of the NEAT1 promoter. Indeed, NEAT1 transcription is activated by a series of stress-induced transcription factors including HIF2α, HSF1, and NF-κB, which have been shown to be constitutively upregulated or activated in HER2 overexpressing cells48–51. Importantly, NEAT1 is also a p53 target gene, and oncogenic stress has been shown to upregulate NEAT1 expression in a p53-dependent manner15,52,53. This might account for the relatively high expression of NEAT1_2 observed in the wild-type p53 cell line MCF7 (Fig. 2e,f). Recently, it was suggested that high NEAT1 expression is associated with good prognosis in p53 wild-type breast cancers54. However, p53 is frequently mutated in HER2-positive cancers45, and HCC1569 cells, which express the largest amount of NEAT1_2 among the cell lines included in this study, carry a p53 nonsense mutation55. The relationship between p53 mutational status, HER2, and NEAT1 expression in breast cancer should be a subject of future research.
As NEAT1_1 and NEAT1_2 are transcribed from the same promoter, it is logical to hypothesize that the expression pattern of NEAT1_1 mirrors that of NEAT1_2. Importantly, by analyzing microarray data derived from probes binding to both NEAT1 isoforms and polyA-enriched RNA-sequencing data, we found that NEAT1_1 expression showed a different distribution among the PAM50 subtypes compared to NEAT1_2. Whereas NEAT1_2 is most highly expressed in HER2-enriched and luminal B cancers, NEAT1_1 expression is highest in ER-positive luminal A and luminal B cancers. Thus, our analyses strongly suggest that the relative levels of NEAT1_1 versus NEAT1_2 vary in different breast cancer subtypes. Previous reports have shown that NEAT1 is transcriptionally activated by ERα in both prostate and breast cancer cell lines25,36. Recently, Li et al. found that NEAT1 participates in a transcriptional repressor complex with FOXN3 and SIN3A in ER-positive breast cancer cells36. The complex induces EMT in vitro by downregulating GATA3 expression and promotes metastasis in mouse models of ER-positive breast cancer. The FOXN3-NEAT1-SIN3A complex also binds to and represses the promoter of the ESR1 gene indicating the presence of a negative feed-back regulatory mechanism. Importantly, the authors suggest that the FOXN3-NEAT1-SIN3A complex functions independently of paraspeckles and that it is the NEAT1_1 isoform that participates in this complex. In line with this, Chakravarty et al. demonstrated that NEAT1_1, but not NEAT1_2, binds directly to histone H3 and recruits ERα to the PSMA promoter in prostate cancer cell lines25. We hypothesize that in ER-positive cancers, NEAT1_1 contributes to the tumorigenic phenotype by directly participating in transcriptional regulation at the chromatin level. This mechanism might be less important in HER2-positive cancers where increased NEAT1_2 levels and paraspeckle formation are required for their adaptation to malignancy-associated stress and survival. A recent study of pan-cancer tissue microarrays, indeed showed that 65% of human carcinomas displayed increased number of paraspeckles compared to non-malignant tissue15. More importantly, the same authors reported that the expression of NEAT1_2, but not total NEAT1, predicted progression-free survival of ovarian cancer treated with platinum-based chemotherapy. NEAT1_2 is produced when the polyadenylation signal required for the formation of NEAT1_1, is suppressed by a hnRNPK-dependent mechanism7,56. Moreover, key paraspeckle-associated proteins including NONO and SFPQ bind to and stabilize NEAT1_26. Further experiments should be undertaken to determine their expression and subcellular localization in HER2-positive breast cancers, as well as in other cancers.
We have found that NEAT1_2 is not expressed in normal tissue surrounding breast cancer cells at levels that can be detected by RNA-FISH. Furthermore, only 7.4% of benign breast tissue samples were NEAT1_2 positive. Murine Neat1 is critical for normal development of the mammary gland, and Neat1_2 and paraspeckles were detected in 30–50% of K8/K18-positive luminal cells in adult mice22. The number of Neat1_2 positive cells increased upon pregnancy and lactation. To further inspect NEAT1 expression pattern in human mammary gland development, we performed RNA-FISH on 8 benign breast tissue samples taken from lactating women. We detected NEAT1_2 and paraspeckles in 6 samples (75%). Our data strongly supports the observations done in mice and suggests that NEAT1_2 and paraspeckle formation are upregulated during lactation also in humans. However, it remains to be determined at which stage in pregnancy NEAT1_2 is upregulated. As Neat1 knockout mice display compromised proliferation of luminal alveolar epithelial cells during pregnancy22, it is reasonable to hypothesize that elevated NEAT1_2 levels are required for pregnancy-induced expansion of the epithelial compartment in humans as well. Alternatively, NEAT1_2 might be upregulated during the differentiation of luminal alveolar cells into milk-secreting cells. The epithelium of the adult mammary gland is interspersed with mammary stem cells and progenitor cells57. In the future, experiments should be undertaken to determine the expression status of NEAT1 in these cells, as this would shed light on the function of NEAT1 in both postnatal mammary gland development and breast cancer.
We provide evidence that NEAT1_2 expression associates with HER2-positive cancers and suggest that the relative expression of NEAT1_1 versus NEAT1_2 varies in breast cancer subtypes. The overlapping nature of the NEAT1_1 and NEAT1_2 hampers isoform-specific analyses and might affect the interpretation of expression data. NEAT1_2 is not polyadenylated, which needs to be taken into account when analyzing polyA-enriched RNA-sequencing data. It should also be noted that RNA stability is a technical challenge when analyzing NEAT1_2 expression in formalin fixed paraffin embedded patient samples by RNA-FISH. Nevertheless, both NEAT1_1 and NEAT1_2 are likely to contribute to breast cancer tumorigenesis, but through different mechanisms. The highly tumor-specific expression of NEAT1_2 in breast cancer, makes it a promising target for future therapeutic intervention.
Methods
Cell culture
BT474 (ATCC® HTB-20™), BT549 (ATCC® HTB-122™), HCC1569 (ATCC® CRL-2330™), Hs 578T (ATCC® HTB-126™), MDA-MB-231 (ATCC® HTB-26™), MDA-MB-468 (ATCC® HTB-132™), MCF7 (ATCC® HTB-22™), SK-BR-3 (ATCC® HTB-30™), and T-47D (ATCC® HTB-133™) cells were all purchased from the American Type Culture Collection (ATCC). BT474, BT549, HCC1569, MDA-MB-231, MDA-MB-468, SK-BR-3, and T-47D cells were cultured in RPMI 1640 (Sigma-Aldrich) supplemented with 10% Fetal bovine serum (FBS) (Biochrom) and 1% penicillin-streptomycin (Sigma-Aldrich). BT549 cells were grown in the presence of 1.0 μg/ml insulin (Sigma-Aldrich) and T-47D cells were grown in the presence of 6.0 μg/ml insulin. Hs 578 T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Sigma-Aldrich) supplemented with 10% FBS, 1% penicillin-streptomycin, and 10.0 μg/ml insulin. MCF7 cells were cultured in Minimum Essential Medium Eagle (MEM; Sigma-Aldrich) supplemented with 10% FBS, 1% penicillin-streptomycin, and 10.0 μg/ml insulin. All cell lines were incubated in a 5% CO2 humidified incubator at 37 °C.
RNA isolation, cDNA synthesis, and RT-qPCR
Cells were lysed in 300 µl Tri Reagent (Zymo Research) and heated for 10 min at 55 °C in order to prevent NEAT1 from being trapped in the protein phase during isolation44. Total RNA was isolated with Direct-zol RNA MiniPrep (Zymo Research) according to the manufacturer’s recommendation. RNA concentration was measured by NanoDrop 2000 (Thermo Fisher Scientific). cDNA synthesis of total RNA was performed with SuperScript™ IV Reverse Transcriptase (ThermoFisher Scientific). 2.5 μM of random hexamer primer (ThermoFisher Scientific) and approximately 400 ng of template were used for the reaction. Total RNA was denaturated at 65 °C for 5 min, and cDNA was synthesized at 50 °C for 10 min.
For RT-qPCR, cDNA was mixed with FastStart Essential DNA Green Master (Roche Life Science) and 0.25 μM forward and reverse primer. All primer sequences are provided in Supplementary Table 1. The LightCycler® 96 was used for quantification, and the ΔΔCq-method was used to calculate fold change using the geometric mean of GAPDH, B2M, and RPLPO as internal reference.
RNA-FISH of cells and FFPE tissue
Stellaris® NEAT1 RNA FISH probes recognizing the NEAT1_2 isoform (SMF-2037-1 conjugated with Quasar® 670) was purchased from LGC Biosearch Technologies. Preparation of cells and FFPE sections, hybridization, and mounting were performed according to the Stellaris® RNA FISH Probes manuals. In brief, cells were seeded onto circular coverslips in 12-well dishes and allowed to attach for 2–3 days. The cells were fixed with 4% formaldehyde and permeabilized with 70% EtOH. Hybridization was done at 37 °C in a humidifying chamber for at least 4 hours. FFPE tissue sections were cut fresh and placed at 60 °C for 45 min before being deparaffinized with xylene. Here, hybridization was performed overnight. Vectashield® Mounting Medium containing DAPI was used for mounting of both cells and FFPE sections. Images were generated using a Zeiss LSM780 confocal microscope. For cells, 3-dimensial Z-stack images were taken at 40x magnification (seven pictures, with 0.6 μm distance between each picture). Images of FFPE sections were taken at 20x magnification with no Z-stacking. All images were processed using ZEN 2012 (black edition) v8.0. NEAT1_2 fluorescence was quantified from maximum intensity projections of confocal z-stacks using Fiji58 running ImageJ59 version 1.52n. An automatic threshold was set in the DAPI channel in order to segment individual nuclei using the wand tool. In some cases, nuclear outlines were manually traced. The average intensity in the NEAT1_2 channel was then measured for each nucleus.
Clinical samples
Archived FFPE needle biopsies were obtained from the Department of Pathology, University Hospital of North Norway (UNN) with corresponding hematoxylin and eosin (HE) slides from all patients. Samples from 74 patients diagnosed with breast cancer (2012–2018, age range 31–84 years), 27 normal samples (2013–2015, age range 18–68 years), 8 samples from lactating females (2013–2015, age range 25–42), and 1 sample from a pregnant female (2013, age 32) were included in the study. Approval for this study, including dispensation from the requirement of patient consent, was granted by the Norwegian Regional Committee for Medical and Health Research Ethics, approval number 2014/317. We confirm that all experiments were performed in accordance with relevant guidelines and regulations. Histological tumor grade was assessed by the Nottingham Grading System60. The samples were classified by pathologists as luminal A (ER+ and/or PgR+, HER2- Ki-67 < 15%), luminal B (ER+ and/or PgR+, HER2- Ki-67 ≥ 15% or ER+ and/or PgR+, HER2+), triple negative/basal-like (ER−, PgR−, HER2−), or HER2-positive (ER−, PgR−, HER2+). The cut off values for ER and PgR were 1%. Tumors with HER2 protein overexpression (IHC 3+) or with in situ hybridization (ISH)-detected amplified HER2 gene (IHC 2+/ISH HER2 gene amplification) were considered to be HER2 positive. NEAT1_2 expression and clinicopathological characteristics were analyzed by the Chi square test (χ2-value) using SPSS version 25 (SPSS Inc., Chicago, IL, USA). P-values ≤ 0.05 (two-tailed) were considered statistically significant.
Gene expression analyses in breast cancer cohorts
NEAT1 gene expression was assessed in three independent breast cancer cohorts; Oslo242, METABRIC28, and TCGA45. Oslo2 is a multicentre study of breast cancer patients with primary operable breast cancers enrolled from hospitals in the Oslo (Norway) region (approved by the Norwegian Regional Committee for Medical and Health Research Ethics, approval number 2016/433 and 429-04148)42. Gene expression data (GEO accession number GSE80999) were obtained using SurePrint G3 Human GE 8 × 60 K microarrays (Agilent Technologies, Santa Clara, CA, USA) and data were log2-transformed, quantile-normalized, and hospital-adjusted. Probe A_33_P3263538 covered part of the unique 3′ end of NEAT1_2. ERBB2 mRNA expression values were derived from mRNA probes using the median value for the two probes matching this gene symbol (A_23_P89249 and A_33_P3292596). For HER2 copy number analysis, logR values (the log2-transformed value of the normalized intensity of the SNP) were extracted from raw CEL-files from Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA) using Affymetrix power tools. Segmented copy number values were generated and non-aberrant cell fraction and ploidy was calculated using the Allele-Specific Copy number Analysis of Tumors (ASCAT) package61. Segmented copy number data (adjusted for non-aberrant cell admixture and ploidy) were log2-transformed and made probe-centric based on the ERBB2 mRNA expression array probe location. The METABRIC cohort is composed of 1980 breast cancer patients collected at five different hospitals in the UK and Canada28. Gene expression was assessed using the Illumina HT-12 v3 microarray and downloaded from the European Genome-phenome Archive (EGA) data portal. The data were log2-transformed. Probe ILMN_1675354 covered part of the unique 3′ end of NEAT1_2. Gene expression levels for the Caucasian fraction of the TCGA cohort (n = 526) were assayed by Agilent 244 K Custom Gene Expression G4502A-07-345. The data were log2-transformed after normalization. The probe A_32_P206561 covered parts of the unique 3′ end of NEAT1_2, while probes A_32_P1036, A_32_P1037, A_24_P566917, and A_24_P566916 covered parts of the common region between NEAT1_1 and NEAT1_2. For analysis of RNA-Seq data from TCGA, bam files aligned to GRCh38 were downloaded from https://gdc.cancer.gov/. Reads mapping to the NEAT1 common region (Chr11: 65422798-65426532) and the NEAT1_2 specific region (Chr11: 65426533-65445540) were counted using the featureCounts function of the Subread package specified with the –p flag (version 1.6.1)62. Fragments spanning the end of the NEAT1 common region and the start of the NEAT1_2 specific region were excluded. FPKM was calculated as [RMg * 109]/[RMt * L], where RMg are reads mapping to each transcript, L is length of transcripts in bp, and RMt are total number of mapped reads. Samples with NEAT1_2 specific region FPKM ≥ 1.0 were filtered out, leaving 1065 tumor samples for clinical analysis.
Statistical analysis
GraphPad software version 25 (SPSS Inc., Chicago, IL, USA) was used to analyze the screening cohort (74 breast cancer needle biopsies). Analysis of significance in expression in normal versus tumor tissue was calculated using the Mann Whitney test. For analyses of clinicopathological variables and NEAT1_2 expression the Chi square test (χ2-value) were used. Data were considered statistically significant when p ≤ 0.05.
For microarray and RNA-Seq expression analyses, statistical analyses were performed in R63. NEAT1_2, NEAT1_1, and NEAT1 expression across PAM50 intrinsic subtypes and tumor grade were compared using the non-parametric Kruskal-Wallis test. For HER2 and ER expression comparison between two groups, Wilcoxon Rank-Sum test were used. Post tests between subtypes and grade were done with Wilcoxon Rank-Sum test. Spearman’s rank correlation was used for analysis of ERBB2 copy number and mRNA expression correlation with NEAT1_2 expression. Data were considered statistically significant when p ≤ 0.05.
Supplementary information
Acknowledgements
This work was supported by Northern Norway Regional Health Authority (HNF1371-17). The publication charges for this article have been funded by a grant from the publication fund of UiT - The Arctic University of Norway.
Author contributions
Conceived and designed the experiments: E.K., M.P. Performed the experiments: E.K., S.M.L., S.F., A.H. Analyzed the data: E.K., M.R.A., S.N., S.F., K.B.L., M.T.G., A.H., S.S.B., E.S.M. and M.P. Contributed reagents/materials/analysis tools: OSBREAC, A.M.B., G.M.M., T.S., E.S.M., M.P. Wrote the paper: E.K., M.P. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
A comprehensive list of consortium members appears at the end of the paper
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Silje Fismen is deceased.
Contributor Information
Maria Perander, Email: maria.perander@uit.no.
Oslo Breast Cancer Research Consortium (OSBREAC):
Jürgen Geisler, Solveig Hofvind, Tone F. Bathen, Elin Borgen, Anne-Lise Børresen-Dale, Olav Engebråten, Øystein Garred, Gry Aarum Geitvik, Anita Langerød, Bjørn Naume, Hege G. Russnes, Ellen Schlichting, Ole Christian Lingjærde, Vessela N. Kristensen, Helle Kristine Skjerven, Thomas Papathomas, Olaf-Johan Hartman-Johnsen, and Kristine Kleivi Sahlberg
Supplementary information
is available for this paper at 10.1038/s41598-020-57759-4.
References
- 1.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl Acad. Sci. USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirose, T., Yamazaki, T. & Nakagawa, S. Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip Rev RNA, e1545, 10.1002/wrna.1545 (2019). [DOI] [PubMed]
- 5.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamazaki T, et al. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell. 2018;70:1038–1053 e1037. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 7.Naganuma T, et al. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li R, Harvey AR, Hodgetts SI, Fox AH. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA. 2017;23:872–881. doi: 10.1261/rna.059477.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beeharry Y, Goodrum G, Imperiale CJ, Pelchat M. The Hepatitis Delta Virus accumulation requires paraspeckle components and affects NEAT1 level and PSP1 localization. Sci. Rep. 2018;8:6031. doi: 10.1038/s41598-018-24500-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J. Gen. Virol. 2006;87:1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4:e00596–00512. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lellahi SM, et al. The long noncoding RNA NEAT1 and nuclear paraspeckles are up-regulated by the transcription factor HSF1 in the heat shock response. J. Biol. Chem. 2018;293:18965–18976. doi: 10.1074/jbc.RA118.004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma, H. et al. The Long Noncoding RNA NEAT1 Exerts Antihantaviral Effects by Acting as Positive Feedback for RIG-I Signaling. J Virol91, 10.1128/JVI.02250-16 (2017). [DOI] [PMC free article] [PubMed]
- 14.Wang Y, et al. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 2018;20:1145–1158. doi: 10.1038/s41556-018-0204-2. [DOI] [PubMed] [Google Scholar]
- 15.Adriaens C, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 16.Choudhry H, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4546. doi: 10.1038/onc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirose T, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imamura K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa S, et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Standaert L, et al. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20:1844–1849. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hupalowska A, et al. CARM1 and Paraspeckles Regulate Pre-implantation Mouse Embryo Development. Cell. 2018;175:1902–1916 e1913. doi: 10.1016/j.cell.2018.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Chakravarty D, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol. Oncol. 2019;13:46–60. doi: 10.1002/1878-0261.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koren S, Bentires-Alj M. Breast Tumor Heterogeneity: Source of Fitness, Hurdle for Therapy. Mol. Cell. 2015;60:537–546. doi: 10.1016/j.molcel.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 28.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 30.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ke H, et al. NEAT1 is Required for Survival of Breast Cancer Cells Through FUS and miR-548. Gene Regul. Syst. Bio. 2016;10:11–17. doi: 10.4137/GRSB.S29414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X, Zhou Y, Sun AJ, Xue JL. NEAT1 contributes to breast cancer progression through modulating miR-448 and ZEB1. J. Cell Physiol. 2018;233:8558–8566. doi: 10.1002/jcp.26470. [DOI] [PubMed] [Google Scholar]
- 34.Qian K, et al. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch. Biochem. Biophys. 2017;615:1–9. doi: 10.1016/j.abb.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Zhao D, Zhang Y, Wang N, Yu N. NEAT1 negatively regulates miR-218 expression and promotes breast cancer progression. Cancer Biomark. 2017;20:247–254. doi: 10.3233/CBM-170027. [DOI] [PubMed] [Google Scholar]
- 36.Li W, et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Invest. 2017;127:3421–3440. doi: 10.1172/JCI94233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin VY, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10:270. doi: 10.1038/s41419-019-1513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rheinbay E, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adriaens C, et al. The long noncoding RNA NEAT1_1 is seemingly dispensable for normal tissue homeostasis and cancer cell growth. RNA. 2019;25:1681–1695. doi: 10.1261/rna.071456.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isobe, M. et al. Forced isoform switching of Neat1_1 to Neat1_2 leads to the loss of Neat1_1 and the hyperformation of paraspeckles but does not affect the development and growth of mice. bioRxiv, 698068, 10.1101/698068 (2019). [DOI] [PMC free article] [PubMed]
- 42.Aure MR, et al. Integrative clustering reveals a novel split in the luminal A subtype of breast cancer with impact on outcome. Breast Cancer Res. 2017;19:44. doi: 10.1186/s13058-017-0812-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chujo T, et al. Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J. 2017;36:1447–1462. doi: 10.15252/embj.201695848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutierrez C, Schiff R. HER2: biology, detection, and clinical implications. Arch. Pathol. Lab. Med. 2011;135:55–62. doi: 10.1043/2010-0454-RAR.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–6487. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarman EJ, et al. HER2 regulates HIF-2alpha and drives an increased hypoxic response in breast cancer. Breast Cancer Res. 2019;21:10. doi: 10.1186/s13058-019-1097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schulz R, et al. HER2/ErbB2 activates HSF1 and thereby controls HSP90 clients including MIF in HER2-overexpressing breast cancer. Cell Death Dis. 2014;5:e980. doi: 10.1038/cddis.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xi C, Hu Y, Buckhaults P, Moskophidis D, Mivechi NF. Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J. Biol. Chem. 2012;287:35646–35657. doi: 10.1074/jbc.M112.377481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr. Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene. 2000;19:1123–1131. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 52.Mello SS, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes. Dev. 2017;31:1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Idogawa M, Ohashi T, Sasaki Y, Nakase H, Tokino T. Long non-coding RNA NEAT1 is a transcriptional target of p53 and modulates p53-induced transactivation and tumor-suppressor function. Int. J. Cancer. 2017;140:2785–2791. doi: 10.1002/ijc.30689. [DOI] [PubMed] [Google Scholar]
- 54.Idogawa Masashi, Nakase Hiroshi, Sasaki Yasushi, Tokino Takashi. Prognostic Effect of Long Noncoding RNA NEAT1 Expression Depends on p53 Mutation Status in Cancer. Journal of Oncology. 2019;2019:1–7. doi: 10.1155/2019/4368068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr. Relat. Cancer. 2006;13:293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 56.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu Nai Yang, Nolan Emma, Lindeman Geoffrey J., Visvader Jane E. Stem Cells and the Differentiation Hierarchy in Mammary Gland Development. Physiological Reviews. 2020;100(2):489–523. doi: 10.1152/physrev.00040.2018. [DOI] [PubMed] [Google Scholar]
- 58.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 61.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc. Natl Acad. Sci. USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 63.R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.