Short abstract
Association between microRNA (miRNA) expression signatures and atrial fibrillation has been evaluated with inconsistent findings in different studies. This study aims to identify miRNAs that actually play vital role in pathophysiological process of atrial fibrillation and explore miRNA-targeted genes and the involved pathways. Relevant studies were retrieved from the electronic databases of Embase, Medline, and Cochrane Library to determine the miRNA expression profiles between atrial fibrillation subjects and non-atrial fibrillation controls. Robustness of results was assessed using sensitivity analysis. Subgroup analyses were performed based on species, miRNA detection method, sample source, and ethnicity. Quality assessment of studies was independently conducted according to QUADAS-2. Bioinformatics analysis was applied to explore the potential genes and pathways associated with atrial fibrillation, which were targeted by differentially expressed miRNAs. Form of pooled results was shown as log10 odds ratios (logORs) with 95% confidence intervals (CI), and random-effects model was used. In total, 40 articles involving 283 differentially expressed miRNAs were reported. And 51 significantly dysregulated miRNAs were identified in consistent direction, with 22 upregulated and 29 downregulated. Among above-mentioned miRNAs, miR-223-3p (logOR 6.473; P < 0.001) was the most upregulated, while miR-1-5p (logOR 7.290; P < 0.001) was the most downregulated. Subgroup analysis confirmed 53 significantly dysregulated miRNAs (21 upregulated and 32 downregulated) in cardiac tissue, with miRNA-1-5p and miRNA-223-3p being the most upregulated and downregulated miRNAs, respectively. Additionally, miR-328 and miR-1-5p were highly blood-specific, and miR-133 was animal-specific. In the detection method sub-groups, miRNA-29b and miRNA-223-3p were differentially expressed consistently. Four miRNAs, including miRNA-223-3p, miRNA-21, miRNA-328, and miRNA-1-5p, were consistently dysregulated in both Asian and non-Asian. Results of sensitivity analysis showed that 47 out of 51 (92.16%) miRNAs were dysregulated consistently. Totally, 51 consistently dysregulated miRNAs associated with atrial fibrillation were confirmed in this study. Five important miRNAs, including miR-29b, miR-328, miR-1-5p, miR-21, and miR-223-3p may act as potential biomarkers for atrial fibrillation.
Impact statement
Atrial fibrillation (AF) is considered as the most common arrhythmia, and it subsequently causes serious complications including thrombosis and heart failure that increase the social burden. The definite mechanisms underlying AF pathogenesis remain complicated and unclear. Many studies attempted to discover the transcriptomic changes using microarray technologies, and the present studies for this hot topic have assessed individual miRNAs profiles for AF. However, results of different articles are controversial and not each reported miRNA is actually associated with the pathogenesis of AF. The present systematic review and meta-analysis identified that 51 consistently dysregulated miRNAs were associated with AF. Of these miRNAs, five miRNAs (miRNA-1-5p, miRNA-328, miRNA-29b, miRNA-21, and miRNA-223-3p) may act as novel biomarkers for AF. The findings could offer a better description of the biological characteristics of miRNAs, meanwhile might serve as new target for the intervention and monitoring AF in future studies.
Keywords: microRNA, biomarker, meta-analysis, bioinformatics analysis, atrial fibrillation
Introduction
Atrial fibrillation (AF), is the most prevalent arrhythmia, which takes responsibility for the most of thromboembolism, contributing to major socioeconomic load.1 The AF prevalence was predicted to markedly increase up to 12 million in the USA and 17.9 million in Europe in the next 50 years.2,3 However, the specific molecular pathogenesis underlying AF is complex and largely remained unclear.4 The insufficient efficacy was attained for the current pharmacological interventions, and clinical treatments mainly focus on preventing complications and improving symptoms. Currently, transcriptomic changes have been studied in human and animal subjects for AF through microRNA detecting technology.5–8 The previous studies have assessed miRNA expression characteristics of AF, and several pivotal pathways associated with miRNAs have been identified, including immune and inflammatory, Ca2+-dependent signaling, as well as cell cycle and apoptotic pathways,9 and it is suggested that miRNAs may be diagnostic targets for AF.10
MiRNAs are small non-coding RNAs with 18–25 nucleotides, which negatively regulate the post-transcriptional of target gene.11 By binding to complementary site in the 3ʹ-untranslated regions (3ʹ-UTRs) of target genes, miRNAs generally result in the inhibition or degradation of translation.12 Because of the crucial role in gene regulation, miRNAs have been confirmed to be aberrantly expressed in a variety of diseases involving cancer, neurological and cardiovascular diseases.13–15 The dysregulated miRNAs may serve as new intervention target for the diseases.16,17 To date, the significant role of miRNAs has been uncovered in acute coronary syndrome, chest pain, and myocardial infarction.18–21 The level of miRNA-29 was decreased in the cardiac valve of heart failure;22 meanwhile, the administration of antagomiR-29 in mouse models can lead to an obvious improvement in fibrosis.23 In addition, cardiac-specific miRNA-208 is important in the myocardium hypertrophic response, and it has an effect on arrhythmias in mice.24,25
Growing report suggests that aberrant level of miRNA in cardiac tissue and blood is related to AF by promoting atrial remodeling.26,27 Differences in expression level of miRNAs exist among AF patients. The expression level of miRNA-486 and miRNA-638 was increased, while the expression level of miRNA-423 and miRNA-572 was decreased in the cardiac tissue of AF subjects.8,28–31 MiR-99b expression was downregulated both in blood3,32,33 and left atrial appendage (LAA).34 Additionally, miRNA expression form related to AF is various in different tissue samples. MiR-328 was detected to be downregulated in plasma,3 while upregulated in myocardial tissue.35 MiR-21 was decreased in right atrium and was increased in left atrium.36 The related article indicated that the expression of 42 and 65 miRNAs was markedly abnormal in the LAA and right atrial appendages (RAA) of AF individuals, respectively. Among these, 23 of those were detected in both atrial appendages, while 45 detected only in the RAA, and 19 detected only in the LAA.36 Considering the inconsistent findings of all the published studies, it is essential to construct this systematic review and meta-analysis to further explore and integrate the miRNA value in AF.
MiRNAs participate in regulating AF-associated remodeling and play a crucial role in the pathophysiology of AF.37 It is revealed that miRNAs are associated with structural remodeling by targeting critical genes in AF. MiR-1 could impact electrical remodeling in AF through targeting KCNE1 and KCNB2.38 miR-328 regulates myocardial electrical remodeling through targeting to CACNA1C and CACNB1,35 and miR-21 affects myocardial fibrosis through regulating the TGF-β1/Smad signaling pathway.39
Emerging findings support that a variety of dysregulated miRNAs have been confirmed to regulate the target genes related to AF.9,40 Nevertheless, inconsistency among different articles is found and not each miRNA reported is actually vital in AF. This study mainly aimed to summarize miRNA expression profiling and to predict miRNA-target genes through bioinformatics approach in the currently reported AF studies, in order to ensure the merit of miRNA-based therapy and serve as a better description for the biological features of miRNAs.
Materials and methods
Data sources and retrieval strategy
Literature review was performed using the Embase, Medline, and Cochrane Library databases to identify all potentially eligible AF-related miRNA expression profiling articles from inception to 30 April 2019 using the under-mentioned terms for the searches of the title/abstract: (microRNA or miR- or miRNA), (AF or atrial fibrillation), (expression or profiling or profile). The detailed search strategies are shown in Supplementary Table 1. Furthermore, the reference lists from retrieved studies were conducted to identify all potentially eligible articles. The databases were retrieved independently by two authors (N.S. and C.Z.), and all discrepancies were discussed to reach an agreement through dealing with the corresponding authors (Z.G. and J.W.).
Selection criteria
The included articles met the following criteria: (1) original articles that analyzed miRNA signatures between AF (case) and non-AF groups were reported (control); (2) miRNA expressed articles on human or animal subjects were associated with AF; (3) applied miRNA expression analysis including miRNA sequencing experiments or quantitative real-time PCR (qRT-PCR) technologies, etc.;41 (4) cut-off criteria and sample sizes were represented for dysregulated miRNAs. For the articles conducted in the animal models, we simultaneously collected the characteristics of models analyzed. Expert comments, reviews, conference abstracts, case, letters, and editorials with missing raw data were removed. For duplicate studies from the same data, the one most similar to the eligible standard was kept. Two investigators (C.Z. and N.S.) separately evaluated all the studies to determine their eligibility, and any discrepancies were resolved with consensus by a third reviewer (Z.G.).
Data extraction and quality assessment
The data were extracted and listed independently from all included studies: authors, year published, included ethnicity, baseline characteristics of patients, species, miRNA detection methods, cut-off criteria, sample types and sizes for dysregulated miRNAs, direction and number of dysregulated miRNAs were recorded. Two investigators (N.S. and C.Z.) independently rated the quality of all eligible articles according to the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2), which was conducted with eight questions. In addition, studies were checked based on Minimum Information about a Microarray Experiment (MIAME) guideline, version 2.0, or Publication of Quantitative Real-time PCR Experiments (MIQE) guideline. Finally, the studies that simultaneously meet the above guidelines were eligible for inclusion.
Statistical analysis
The random-effects model was conducted by STATA software (version 13; Statacorp, College Station, Texas, USA) in this meta-analysis. All results were shown as logORs on the basis of the numbers of dysregulation between AF and non-AF subjects along with 95% CI. In comparison with those obtained for the non-AF group, logOR values obtained for the AF group higher than 1 revealed upregulation. In comparison with the AF, a marked logOR obtained for the non-AF group higher than 1 revealed downregulation. The P-value <0.05 was considered as statistically different. Differentially expressed miRNAs of AF and non-AF comparisons were based on the importance of ranking in the order: (1) amount of consistent sub-studies; (2) total sample size; (3) P values; (4) logOR values. Subgroup analysis was conducted according to the species, detection methods, tissue types, and ethnicity. The classification of blood sample includes platelets, serum, peripheral blood mononuclear cells (PBMCs), plasma, and whole blood. Sensitivity analysis was conducted to detect the heterogeneity of the results. As a crucial determinant for sample size, this study was repeatedly performed after excluding studies with the sample size of 10 or less.
Bioinformatics analysis of miRNA target genes and pathways
To probe into the functional role of miRNAs in the pathogenesis of AF, differentially expressed miRNAs with same direction determined in AF were subjected to bioinformatics analysis to confirm the potential genes and biological pathways. The miRWalk 2.0 which was applied for miRNA target prediction was composed of miRDB, TargetScan, and miRTarBase. To reduce error, the final identification of miRNA target genes was in line with at least two of the above tools. The combination of predicted targets was applied for in-depth understanding of miRNAs. Based on the identified target genes, the network map of miRNA target genes was performed using Cytoscape software version 3.6.1.42 Cytoscape was applied to present visualizing molecular interaction networks between miRNAs and corresponding gene targets.43 Next, we conducted biological analysis of KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (Gene Ontology) analysis according to the predicted targets, and the significance of the associated biological analysis was confirmed with the evaluation of the DAVID database. Genes targeted by selected miRNAs were considered strongly enriched according to P < 0.05.
Results
Literature retrieval, features of included articles, and quality assessment
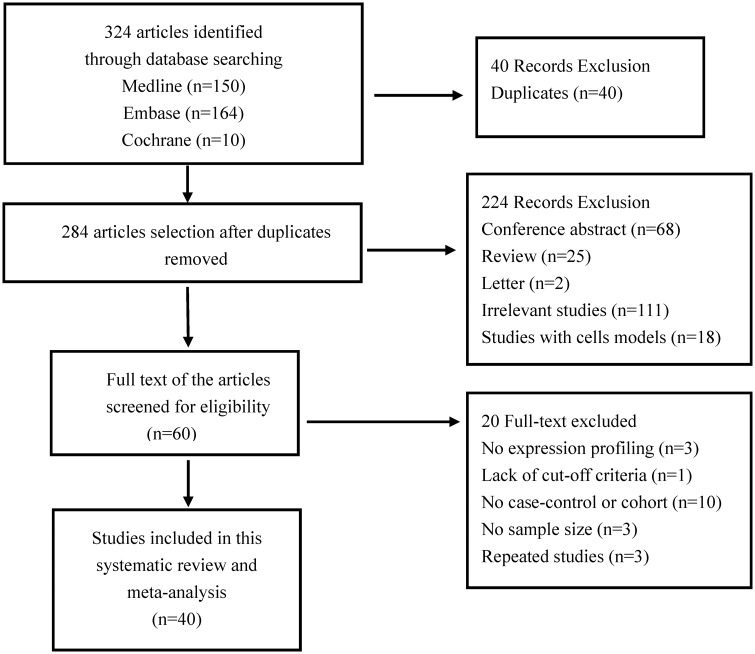
Figure 1 depicts the literature search process used for identifying the included studies. Based on the eligibility criteria, totally, 324 potential studies were initially searched from the databases (Supplementary Table 1). Ultimately, only 40 studies fulfilled the eligibility criteria for meta-analysis after excluding duplicates, conference abstracts, reviews, letters, irrelevant studies, and studies that lacked important information.3,5–10,26,28,29,31–34,36,39,40,44–66 Specific reasons for the exclusion of studies are listed in Supplementary Table 2. The list of the 40 studies characteristics is given in Tables 1 and 2. Of the 40 articles, 34 reported miRNA profiling in humans (Table 1). The patient demographics and characteristics of the 34 human studies are given in Supplementary Table 3. Only one study focused on miRNA profiles in animal samples, and five analyzed both humans and animals (Table 2). The analyzed sample sizes ranged from 8 to 224 among the studies, and the number of differentially expressed miRNAs varied from 1 to 283. The quality of each study was assessed through QUADAS-2, as mentioned in the methods section. The quality assessment results are displayed in Supplementary Table 4, and the results showed that the overall quality of articles was good.
Figure 1.
Flowchart for the search strategy in this study.
Table 1.
Main characteristics of the enrolled human miRNA expression studies.
|
Differentially expressed microRNAs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| First author | Year | Country | Tissue | Method | Sample size Case/Control | Cut-off criteria | Total | Increased | Decreased |
| Adam et al.66 | 2012 | Germany | RAA | qRT-PCR | 5/5 | P < 0.05 | 1 | 1 | 0 |
| Cooley et al.34 | 2012 | Australia | RAA | microRNA array | 4/6 | P < 0.05 | 47 | 15 | 32 |
| Chiang et al.15 | 2015 | America | RAA | qRT-PCR | 4/4 | P < 0.05 | 40 | 17 | 23 |
| da Silva et al.7 | 2018 | Brazil | Plasma | qRT-PCR | 5/15 | P < 0.05 | 4 | 3 | 1 |
| Dawson et al.65 | 2013 | Canada | LA/RAA/plasma | qRT-PCR | 41/65 | P < 0.05 | 2 | 0 | 2 |
| Feldman et al.64 | 2017 | England | Venous blood | qRT-PCR | 24/24 | P < 0.05 | 3 | 0 | 3 |
| Goren et al.62 | 2014 | Israel | Platelets | qRT-PCR | 15/26 | P < 0.05 | 1 | 0 | 1 |
| Girmatsion et al.63 | 2009 | Canada | RA | qRT-PCR | 7/8 | P < 0.05 | 1 | 0 | 1 |
| Harling et al.28 | 2017 | England | RA | microRNA array | 11/11 | P < 0.05 | 15 | 10 | 5 |
| Li et al.58 | 2015 | China | RAA | qRT-PCR | 18/22 | P < 0.05 | 2 | 0 | 2 |
| Li et al.61 | 2012 | China | LA | qRT-PCR | 7/7 | P < 0.05 | 2 | 0 | 5 |
| Li et al.17 | 2014 | China | RAA | qRT-PCR | 6/5 | P < 0.05 | 5 | 0 | 5 |
| Li et al.59 | 2017 | China | RAA | qRT-PCR | 19/18 | P < 0.01 | 1 | 1 | 0 |
| Liu et al.36 | 2014 | China | RAA/LAA | qRT-PCR | 10/8 | P < 0.05 | 13 | 0 | 13 |
| Liu et al.56 | 2016 | China | Plasma | qRT-PCR | 40/40 | P < 0.05 | 2 | 0 | 2 |
| Liu et al.32 | 2012 | China | Plasma | microRNA sequencing | 10/10 | P < 0.01 | 4 | 2 | 2 |
| Lu et al.55 | 2010 | China | RAA | qRT-PCR | 12/10 | P < 0.05 | 3 | 3 | 0 |
| Lu et al.26 | 2015 | China | Plasma | Gene chips test | 112/112 | P < 0.05 | 15 | 8 | 7 |
| Ling et al.57 | 2013 | China | RAA | microRNA array | 4/4 | P < 0.05 | 21 | 14 | 7 |
| McManus et al.3 | 2014 | USA | whole blood | qRT-PCR | 107/2185 | P < 0.05 | 12 | 3 | 9 |
| McManus et al.33 | 2015 | USA | atrial tissue | qRT-PCR | 19/12 | P < 0.05 | 3 | 3 | 0 |
| Natsume et al.6 | 2018 | Japan | Serum | qRT-PCR | 50/50 | P < 0.05 | 3 | 3 | 0 |
| Nishi et al53. | 2013 | Japan | RA | qRT-PCR | 6/13 | P < 0.05 | 2 | 2 | 0 |
| Qiao et al.52 | 2017 | China | RAA | qRT-PCR | 6/4 | P < 0.01 | 1 | 0 | 1 |
| Reilly et al.10 | 2016 | UK | LA | qRT-PCR | 7/11 | P < 0.0001 | 1 | 1 | 0 |
| Slagsvold et al.29 | 2014 | Norway | RA/LA | qRT-PCR | 20/12 | P < 0.05 | 1 | 1 | 0 |
| Soeki et al.51 | 2016 | Japan | Plasma | qRT-PCR | 30/10 | P < 0.05 | 1 | 1 | 0 |
| Stillitano et al.50 | 2013 | Italy | RA | qRT-PCR | 10/10 | P < 0.05 | 1 | 0 | 1 |
| Tao et al.39 | 2018 | China | atrial tissue | qRT-PCR | 24/25 | P < 0.05 | 1 | 1 | 0 |
| Wang et al.30 | 2015 | China | RAA | qRT-PCR | 30/17 | P < 0.05 | 10 | 10 | 0 |
| Wang et al.49 | 2017 | China | Serum | qRT-PCR | 14/29 | P < 0.05 | 1 | 0 | 1 |
| Wang et al.47 | 2013 | China | Venous blood | qRT-PCR | 58/15 | P < 0.05 | 1 | 1 | 0 |
| Wei et al.46 | 2018 | China | Plasma | qRT-PCR | 18/12 | P < 0.05 | 1 | 1 | 0 |
| Wei et al.45 | 2015 | China | Venous blood | qRT-PCR | 35/32 | P < 0.01 | 1 | 0 | 1 |
| Xiao et al.31 | 2011 | China | RAA | microRNA array | 9/9 | P < 0.05 | 136 | 50 | 86 |
| Xie et al.5 | 2018 | China | RAA | qRT-PCR | 28/22 | P < 0.05 | 1 | 0 | 1 |
| Xu et al.8 | 2016 | China | PB | microRNA array | 90/90 | P < 0.05 | 6 | 3 | 3 |
| Yamac et al.40 | 2016 | Turkey | RAA | qRT-PCR | 20/43 | P < 0.05 | 2 | 0 | 2 |
| Zhao et al.44 | 2015 | China | LAA | microRNA sequencing | 3/3 | P < 0.05 | 1 | 1 | 0 |
RAA: right atrial appendage; LAA: left atrial appendage; RA: right atrium; LA: left atrium; PB: peripheral blood; qRT-PCR: quantitative real-time polymerase chain reaction.
Table 2.
Main characteristics of the enrolled animal miRNA expression studies.
|
Differentially expressed microRNAs |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| First author | Year | Country | Tissue | Method | Animal model | Sample size Case/control | Cut-off criteria | Total | Increased | Decreased |
| Lu et al.55 | 2010 | China | LA | qRT-PCR | Canine | 7/7 | P < 0.05 | 8 | 3 | 5 |
| Qiao et al.52 | 2017 | China | RAA | qRT-PCR | Canine | 6/4 | P < 0.01 | 1 | 0 | 1 |
| Luo et al.54 | 2013 | China | Atrial tissue | qRT-PCR | Canine | 9/10 | P < 0.001 | 1 | 0 | 1 |
| Dawson et al.65 | 2013 | Canada | LA | qRT-PCR | Canine | 16/16 | P < 0.001 | 1 | 0 | 1 |
| Li et al.61 | 2012 | China | LA | RT-PCR | Canine | 7/7 | P < 0.05 | 2 | 0 | 2 |
| Reilly et al.10 | 2016 | UK | LA | qRT-PCR | Goat | 7/11 | P < 0.0001 | 1 | 1 | 0 |
LA: left atrial; RAA: right atrial appendage.
Differentially expressed miRNAs in overall analysis
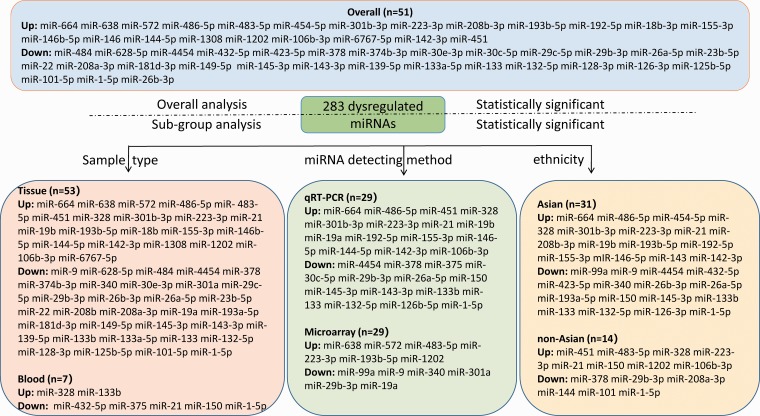
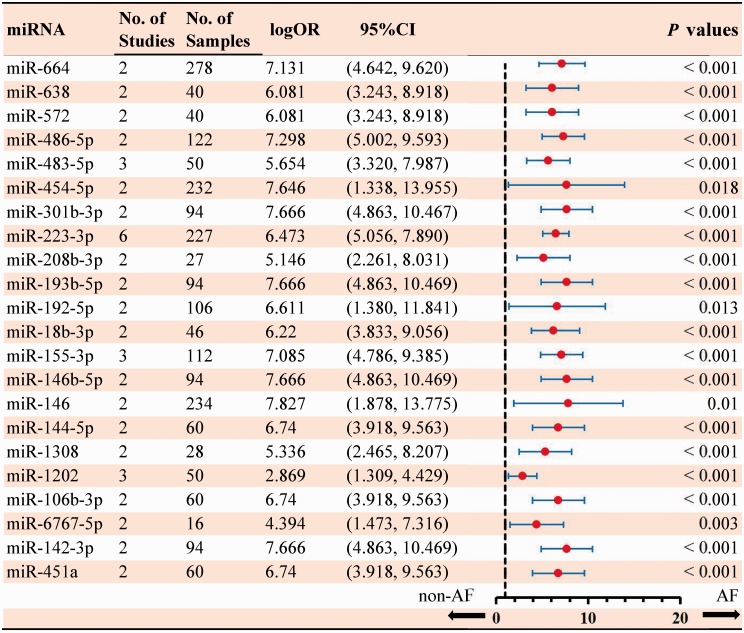
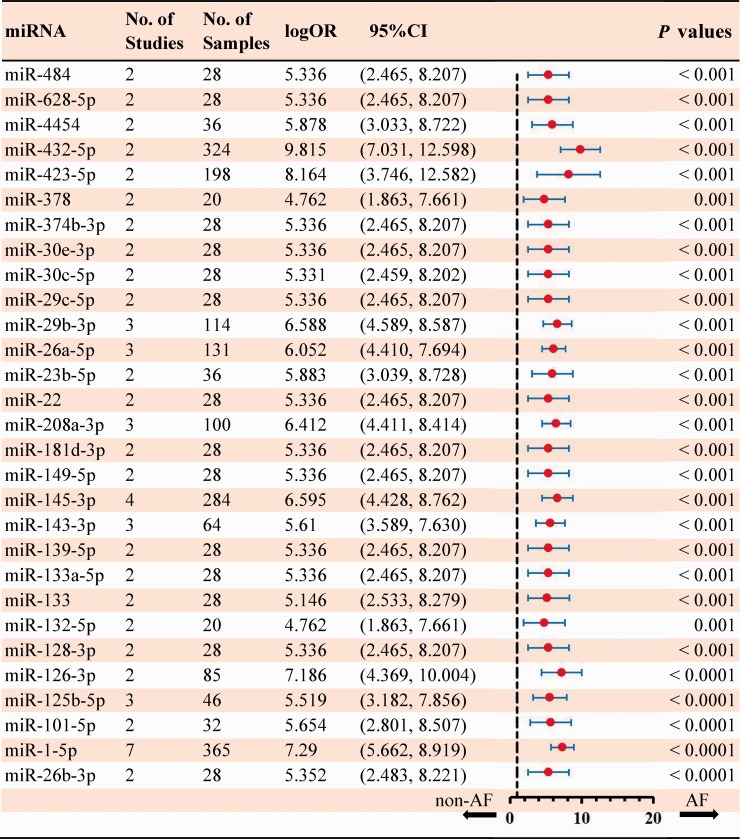
In total, 283 dysregulated miRNAs were mentioned by 40 included articles that compared AF samples with non-AF controls (Figure 2). Among these differentially expressed miRNAs, 75 aberrantly expressed miRNAs (26.5%) were identified in at least two articles. In this study, 51 miRNAs (18.7%) were identified to be consistent in terms of the direction of dysregulation (22 upregulated and 29 downregulated). The details of significant results are listed in Supplementary Tables 5 and 6. According to the results of 6 sub-studies with 227 samples and 7 sub-studies with 365 samples, miR-223-3p (logOR 6.473; P < 0.001) was determined to be the most upregulated miRNA, while miR-1-5p (logOR 7.290; P < 0.001) being the most downregulated one (Figures 3 and 4). Details of each miRNA are summarized in Supplementary Figures 1 to 51. And the inconsistently dysregulated miRNAs in the overall analysis classified by different subgroups are shown in Supplementary Table 7.
Figure 2.
The flow diagram of miRNA categories in this meta-analysis. (A color version of this figure is available in the online journal.)
Figure 3.
Statistically significant up-regulated miRNAs in overall analysis. (A color version of this figure is available in the online journal.)
Figure 4.
Statistically significant down-regulated miRNAs in overall analysis. (A color version of this figure is available in the online journal.)
Subgroup analysis
Among the 40 included studies, 25 studies investigated miRNAs in right atrial tissue, right atrium, left atrial tissue and left atrium, 14 studies detected circulating miRNAs in plasma, venous blood, platelets, serum and whole blood, while only 1 study did research in both tissue and blood samples. Overall, 53 miRNAs (21 low expression and 32 high expression) were identified to be aberrantly expressed in tissue sample, with the most upregulated and downregulated miRNA being miR-223-3p (logOR 5.692; P < 0.001) and miR-1-5p (logOR 6.241; P < 0.001), respectively (Supplementary Table 8). A total of seven miRNAs (two upregulated, five downregulated) were statistically significant and dysregulated in blood, and the most statistically significant upregulated and downregulated was miR-328 and miR-1-5p, respectively (Supplementary Table 9).
Subgroup analysis was done by miRNA detection method, 5 and 34 sub-studies used microarray and qRT-PCR method, respectively, and only one study involved gene chip test. Therefore, a subgroup analysis with gene chip test was not performed. Totally, the aberrant expression of 12 and 29 miRNAs was discovered in the microarray and qRT-PCR assays, respectively. In microarray studies, six upregulated miRNAs and six downregulated miRNAs were significant (Supplementary Table 10). For qRT-PCR studies, 15 upregulated miRNAs and 14 downregulated miRNAs were found to be remarkable (Supplementary Table 11). Of the above-mentioned miRNAs, miRNA-223-3p and miRNA-29b were consistently upregulated and downregulated by both the detection methods.
Subgroup analyses of ethnicity with Asian and non-Asian were conducted. The subgroup analysis included 27 Asian studies and 13 non-Asian studies, respectively. In the subgroup of Asian, there were 15 upregulated miRNAs and 16 downregulated miRNAs (Supplementary Table 12). For non-Asian subgroup, eight upregulated miRNAs and six downregulated miRNAs were identified (Supplementary Table 13). Remarkably, the expression level of miR-328, miR-223-3p and miR-21 was consistently increased in both Asian and non-Asian studies, while miR-1-5p was downregulated in all studies. The above four miRNAs may have great potential value in predicting AF.
MiRNAs were differentially expressed between animals and humans, and the species subgroups of the miRNAs were only analyzed in animal studies (Supplementary Table 14). The results indicated that only miR-133 was consistently downregulated in animal models according to two studies. Several other miRNAs were only mentioned in a single study, including miRNA-26a, miRNA-449, miRNA-145, miRNA-31, miRNA-30, miRNA-664, etc.
Bioinformatic analysis
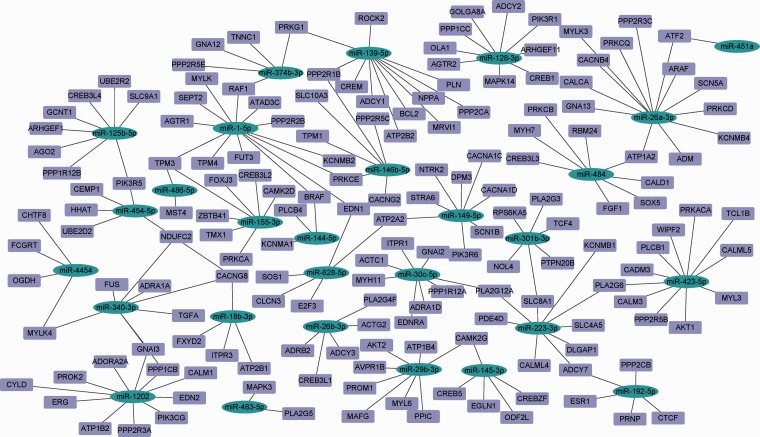
Eventually, 51 dysregulated miRNAs were used for bioinformatic analysis, and 223 potential miRNA target genes were retrieved from miRTarBase, miRDB, and TargetScan. The target genes and pathways of dysregulated miRNAs should be understood to identify their potential biological functions. The predicted human miRNA-target interaction is shown in Figure 5. Moreover, KEGG and GO analysis was carried out to analyze the target genes using the Enrichr database to further identify the involved pathways related to the selected miRNAs.
Figure 5.
Network map of miRNA-gene interactions identified in TargetScan, miRDB, and miRTarBase. (A color version of this figure is available in the online journal.)
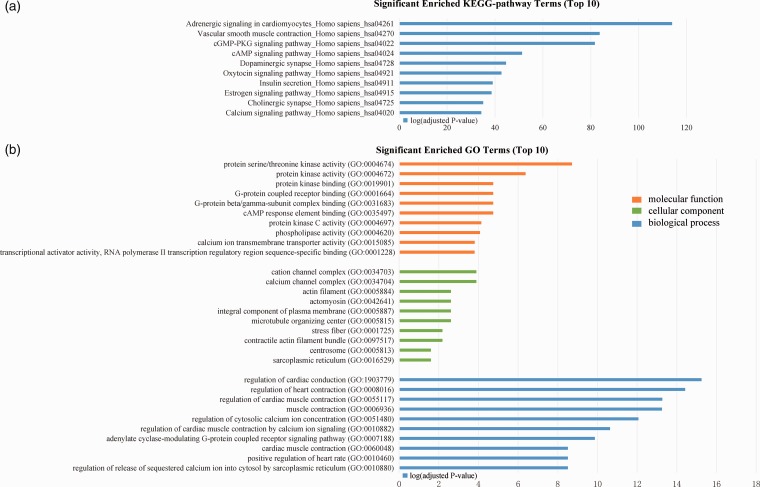
The top 10 terms identified for each enrichment analysis are presented in Figure 6. In addition, KEGG terms associated with AF pathogenesis within the functional enrichment results and significant pathways were detected, including vascular smooth muscle contraction_Homo sapiens (hsa04270) and adrenergic signaling in cardiomyocytes_Homo sapiens (hsa04261). The bioinformatic analysis for dysregulated miRNAs in subgroups according to sample type, miRNA detecting method, and ethnicity is shown in Supplementary Figures 52 to 54. The results of KEGG and GO analysis in subgroups were consistent with the overall analysis.
Figure 6.
Results of bioinformatic analysis. (a) Top 10 terms of significant enriched KEGG pathways. (b) Top 10 significant enriched GO terms, including molecular functions, cellular component and biological process. (A color version of this figure is available in the online journal.)
Sensitivity analysis
The small sample size might influence the robustness of this study, which was determined by sensitivity analysis; 3 out of 40 included studies had small sample sizes that less than 10. Therefore, the remaining 37 studies were reanalyzed. As a result, 47 miRNAs were identified significant, with 18 of them upregulated and 29 downregulated (Supplementary Table 15). Our result of sensitivity analysis showed that the other four miRNAs were not significant, but significant in the overall analysis. The above results showed that a small sample size may lead to differences in miRNA expression profiling.
Discussion
For the first time, we perform a systematic review to comprehensively explore the miRNA expression signatures of AF. Due to the different technological platforms and inconsistent sample sources used, it is essential to provide specific and valuable miRNA data by a meta-analysis. Overall, 51 aberrantly expressed miRNAs with consistent direction were identified in all included studies. Based on further analysis, five miRNAs (miRNA-328, miRNA-223-3p, miRNA-21, miRNA-29b, miRNA-1-5p) were finally identified as potential biomarkers of AF.
The main pathophysiological basis of AF is generally considered to be atrial remodeling processes involving electrophysiological changes and myocardial fibrosis.67 Notably, a variety of miRNAs have been confirmed to be involved in AF-related remodeling processes.68 MiR-328, an upregulated miRNA was shown to be increased in animal models and patients with AF, with CACNA1C and CACNB1 being target genes.35 There is also a correlation between miR-21 and CACNA1C, and miR-21 participates in electrical changes in AF.69 In addition, the miR-106b-25 mediates AF-associated pathogenesis via enhancing RyR2,70 and increased level of miR-30a in myocardial fibroblasts promotes myocardial fibrosis by inducing the decreased expression of snail 1.71 Accordingly, miRNAs participate in the pathological processes of AF and may serve as new target for the intervention and monitoring of AF in the future.
The dysregulation of miR-223 has been confirmed to play vital role in the pathophysiological process of various diseases, including acute and chronic liver injury,72 diseases related to innate immunity,73 and Alzheimer's disease.74 The circulating miR-223 was associated with the formation of local thrombus and acute myocardial infarction (AMI).75 The evidence from this meta-analysis demonstrated that miRNA-223 represents the most high expression level in AF. Besides, upregulated miR-223 was also found in LAA samples of AF patients.76 However, the specific mechanism remains unclear due to scarce studies of miR-223 in AF. In addition, miRNA-1 was highly expressed in atrial tissue;77 moreover, decreased expression leads to a significantly increased level of Kir2.1 subunits63 and HCN2 expression.58 Additionally, circulating miR-1 in blood plays an important role in regulating CACNB2 protein in L-type Ca2+ channels.26 Considering that miR-223 and miR-1 changes were specifically associated with sample types, detection methods and ethnicities, miR-223, and miR-1 are stable, and could be representative in AF studies. This meta-analysis uncovered novel molecular targets for AF and indicated that miRNA-223 and miRNA-1 might become promising biomarkers to detect AF.
MiR-328 participates in regulating multiple cardiac pathophysiological processes related to AF,7 and its expression was increased in acute AF. Upregulated miR-328 in human atrial samples contributes to result in adverse electrical remodeling in AF by targeting the L-type Ca2+ channel.55 Likewise, circulating miRNA-328 was related to the AF prevalence.3 Therefore, these studies revealed important mechanisms behind the AF, and miRNA-328 was identified as a reliable biomarker for AF. Meanwhile, the high agreement of miR-328 between the blood and tissue results confirmed the reliability of miRNA-328 as biomarkers of AF. The significant dysregulation of miR-29b was confirmed in this meta-analysis. Likewise, reduced miR-29b in AF significantly increased COL1A1, COL3A1, and fibrillin expression.65 In addition, the upregulated miRNA-21 is associated with fibrosis,78 meanwhile increased miR-21 in fibroblasts also regulates the TGF-β1/Smad signaling pathway in AF-associated fibrosis via downregulating Smad7.10 Therefore, miR-29b and miR-21 are likely to regulate myocardial fibrosis reconstruction and might be the valuable biomarkers or therapeutic targets. The above miRNAs were confirmed to remain consistently aberrantly expressed among multiple studies, and they may be potential targets for future clinical applications.
The tissue-related specificity exists in the miRNA expression, and miR-375 and miR-113b expression differed between blood samples3,7,32 and tissue.36,57 A total of 54 miRNAs were consistently differentially expressed in atrial tissue, among which increased miR-328 and decreased miR-1 were identified. Noteworthily, miR-133b was upregulated in blood but downregulated in atrial tissue. A possible explanation for the difference in tissue and plasma could be the balance between tissue and blood. Intracellular miRNA was obtained from the circulating blood, resulting in a miRNA decrease in blood. Therefore, it is suggested that the uptake of circulating regulator miRNAs by affected cells to regain intracellular level might result in different miRNA levels in tissue and plasma. Nevertheless, more researches are still essential to explore the differences between sample-specific miRNAs.
Although miRNAs exist in different tissues, they predominate as biomarkers in atrial tissue of AF. Compared with tissue miRNAs, the noninvasive sampling and testing favor circulating miRNAs are preferred as biomarkers. However, few studies on blood miRNAs are related to AF. Therefore, potential new insights into the therapeutic implications of circulating miRNAs in AF are valuable. Although animal studies may be also informative regarding disease diagnosis in humans, they are still insufficient for AF research. Few animal researches were contained in this study, and only miR-133 was shown to be consistently downregulated in animal models of two studies. Consequently, more research is also required to explore differentially expressed miRNAs in animal models of AF.
MiRNAs may have potential therapeutic effects for AF. MiR-138-5p mimics was shown to suppress cell growth via regulating CYP11B2.5 Transfected cardiac fibroblasts with miR-132 directly suppressed connective tissue growth factor, which regulates fibrosis progression.52 To date, miRNA availability was only limited in cell experiments. It is discovered that the overexpression of miR-31 significantly increases AF susceptibility in vivo.10 A few of miRNA application have reached the development in clinic, including miR-34 mimic of tumor inhibitor in phase I trial, antimiRs targeting at miR-122 associated with hepatitis in phase II trial,79 which indicated that miRNA therapeutics in the clinic have great promise. This study identified stable miRNA profiles related to AF, and may offer the valuable indicator for target miRNA treatment.
Target gene prediction for significantly dysregulated miRNAs in AF was performed by using bioinformatics analysis. The potential miRNA targets were confirmed for KEGG and GO annotation, and KEGG analysis has shown that these targets were mostly enriched in adrenergic signaling in cardiomyocytes and vascular smooth muscle contraction pathways. Some reports proved that these two pathways were associated with AF.80–82 The enriched GO annotation indicated these genes were mainly involved in protein activity, cation channel complex, and regulation of cardiac conduction, which reflected the close connection between AF and related miRNAs.83 Our bioinformatics analysis suggested that dysregulated miRNAs in AF significantly regulated genes of pathways in connection with cardiovascular function and AF process, including CACNG, AKT, PRKG1, ATP2B, etc. It may contribute to verify imagination on biological function influenced by miRNAs. Furthermore, based on current findings, the experimental verification is essential in the future study.
Although this study identified several miRNAs as possible biomarkers in AF, it does not exclude confounding factors, including the variety of biological and technical designs, inconsistent standards and technical approaches that were used. Therefore, it is difficult to obtain unified result. Several limitations of this meta-analysis should be noted. First, the discrepancies in the sample size of each subgroups, and small sample sizes may lead to errors in the results. Second, the subgroup analysis did not take the AF classification, such as paroxysmal AF, permanent AF and long-term AF, into account due to inadequate clinical details in the original studies. Finally, the findings presented are simple, and insufficient experiments were performed to verify the exact pathological role of miRNAs in AF. The further experimental verification is required to ascertain the definite mechanism of AF. This study mainly reviewed miRNA expression profiling in atrial fibrillation, and the findings presented are meaningful in this context. Based on the above described inconsistencies in miRNA expression among the included studies, this meta-analysis is essential for identifying the promising biomarkers of AF. Eventually, we identified that several miRNAs might be potential biomarkers for AF; meanwhile, bioinformatics approach demonstrated that these miRNAs were closely related to AF. Of course, the rigorous experimental verification is essential for confirming their validity in the future AF research.
Conclusion
This study identified 51 significantly dysregulated miRNAs. MiRNA-223-3p, miRNA-21, miRNA-328, miRNA-1-5p, and miRNA-29b may be potential biomarkers for AF.
Supplemental Material
Supplemental material, EBM890303 Supplemental Material for MicroRNA expression signatures of atrial fibrillation: The critical systematic review and bioinformatics analysis by Nan-Nan Shen, Chi Zhang, Zheng Li, Ling-Cong Kong, Xin-Hua Wang, Zhi-Chun Gu and Jia-Liang Wang in Experimental Biology and Medicine
Authors’ contributions
Gu and Wang were responsible for the guarantee of the whole study. Shen and Zhang contributed to conceive the research, draft and review the articles, and were the guarantors of the submission of the final version. Li, Kong, and Wang helped perform the data acquisition, analysis, and explanation of the results.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Program of Shaoxing science and technology bureau (2017B70010), Zhejiang pharmaceutical association (2016ZYY29), Shanghai municipal heath commission (20184Y0022), Cultivation fund of clinical study of Renji hospital (PY2018-III-06), Clinical Pharmacy Innovation Research Institute of Shanghai Jiao Tong University School of Medicine (CXYJY2019ZD001, CXYJY2019QN004) and Program for Key but Weak Disciplines of Shanghai Municipal Commission of Health and Family Planning (2016ZB0304).
ORCID iD
Zhi-Chun Gu https://orcid.org/0000-0002-1245-9690
SUPPLEMENTAL MATERIAL
Supplemental material for this article is available online.
References
- 1.Freedman B, Potpara TS, Lip GY. Stroke prevention in atrial fibrillation. Lancet 2016; 388:806–17 [DOI] [PubMed] [Google Scholar]
- 2.Rahman F, Kwan GF, Benjamin EJ. Global epidemiology of atrial fibrillation. Nat Rev Cardiol 2014; 11:639–54 [DOI] [PubMed] [Google Scholar]
- 3.McManus DD, Lin H, Tanriverdi K, Quercio M, Yin X, Larson MG, Ellinor PT, Levy D, Freedman JE, Benjamin EJ. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham offspring study. Heart Rhythm 2014; 11:663–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lown M, Moran P. Should we screen for atrial fibrillation? BMJ 2019; 364:l43. [DOI] [PubMed] [Google Scholar]
- 5.Xie H, Fu JL, Xie C. MiR-138-5p is downregulated in patients with atrial fibrillation and reverses cardiac fibrotic remodeling via repressing CYP11B2. Eur Rev Med Pharmacol Sci 2018; 22:4642–7 [DOI] [PubMed] [Google Scholar]
- 6.Natsume Y, Oaku K, Takahashi K, Nakamura W, Oono A, Hamada S, Yamazoe M, Ihara K, Sasaki T, Goya M, Hirao K, Furukawa T, Sasano T. Combined analysis of human and experimental murine samples identified novel circulating MicroRNAs as biomarkers for atrial fibrillation. Circ J 2018; 82:965–73 [DOI] [PubMed] [Google Scholar]
- 7.da Silva AMG, de Araujo JNG, de Oliveira KM, Novaes AEM, Lopes MB, de Sousa JCV, Filho AAA, Luchessi AD, de Rezende AA, Hirata MH, Silbiger VN. Circulating miRNAs in acute new-onset atrial fibrillation and their target mRNA network. J Cardiovasc Electrophysiol 2018; 29:1159–66 [DOI] [PubMed] [Google Scholar]
- 8.Xu G, Cui Y, Jia Z, Yue Y, Yang S. The values of coronary circulating miRNAs in patients with atrial fibrillation. PLoS One 2016; 11:e0166235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang DY, Zhang M, Voigt N, Alsina KM, Jakob H, Martin JF, Dobrev D, Wehrens XH, Li N. Identification of microRNA-mRNA dysregulations in paroxysmal atrial fibrillation. Int J Cardiol 2015; 184:190–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reilly SN, Liu X, Carnicer R, Recalde A, Muszkiewicz A, Jayaram R, Carena MC, Wijesurendra R, Stefanini M, Surdo NC, Lomas O, Ratnatunga C, Sayeed R, Krasopoulos G, Rajakumar T, Bueno-Orovio A, Verheule S, Fulga TA, Rodriguez B, Schotten U, Casadei B. Up-regulation of miR-31 in human atrial fibrillation begets the arrhythmia by depleting dystrophin and neuronal nitric oxide synthase. Sci Transl Med 2016; 8:340ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros V. The functions of animal microRNAs. Nature 2004; 431:350–5 [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015; 15:321–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861–74 [DOI] [PubMed] [Google Scholar]
- 15.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2017; 9:852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov 2016; 6:235–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 2014; 13:622–38 [DOI] [PubMed] [Google Scholar]
- 18.Ke-Gang J, Zhi-Wei L, Xin Z, Jing W, Ping S, Xue-Jing H, Hong-Xia T, Xin T, Xiao-Cheng L. Evaluating diagnostic and prognostic value of plasma miRNA133a in acute chest pain patients undergoing coronary angiography. Medicine 2016; 95:e3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan L, Liu X, Chen F, Zhang L, Chen X, Huang Q, Wu D, Yang C, Han Z. Diagnostic and prognostic value of circulating MicroRNA-133a in patients with acute myocardial infarction. Clin Lab 2016; 62:1233–41 [DOI] [PubMed] [Google Scholar]
- 20.Su T, Shao X, Zhang X, Han Z, Yang C, Li X. Circulating microRNA-1 in the diagnosis and predicting prognosis of patients with chest pain: a prospective cohort study. BMC Cardiovasc Disord 2019; 19:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parizadeh SM, Ferns GA, Ghandehari M, Hassanian SM, Ghayour-Mobarhan M, Parizadeh SMR, Avan A. The diagnostic and prognostic value of circulating microRNAs in coronary artery disease: a novel approach to disease diagnosis of stable CAD and acute coronary syndrome. J Cell Physiol 2018; 233:6418–24 [DOI] [PubMed] [Google Scholar]
- 22.Dawson K, Wakili R, Ordog B, Clauss S, Chen Y, Iwasaki Y, Voigt N, Qi XY, Sinner MF, Dobrev D, Kaab S, Nattel S. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 2013; 127:1466–75 75e128 [DOI] [PubMed] [Google Scholar]
- 23.Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, Kaminski N, van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med 2014; 6:1347–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell 2012; 149:671–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009; 119:2772–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Hou S, Huang D, Luo X, Zhang J, Chen J, Xu W. Expression profile analysis of circulating microRNAs and their effects on ion channels in Chinese atrial fibrillation patients. Int J Clin Exp Med 2015; 8:845–53 [PMC free article] [PubMed] [Google Scholar]
- 27.Nattel S, Harada M. Atrial remodeling and atrial fibrillation: recent advances and translational perspectives. J Am Coll Cardiol 2014; 63:2335–45 [DOI] [PubMed] [Google Scholar]
- 28.Harling L, Lambert J, Ashrafian H, Darzi A, Gooderham NJ, Athanasiou T. Elevated serum microRNA 483-5p levels may predict patients at risk of post-operative atrial fibrillation. Eur J Cardiothorac Surg 2017; 51:73–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slagsvold KH, Johnsen AB, Rognmo O, Hoydal MA, Wisloff U, Wahba A. Mitochondrial respiration and microRNA expression in right and left atrium of patients with atrial fibrillation. Physiol Genomics 2014; 46:505–11 [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Wang Y, Han J, Li Y, Xie C, Xie L, Shi J, Zhang J, Yang B, Chen D, Meng X. Integrated analysis of microRNA and mRNA expression profiles in the left atrium of patients with nonvalvular paroxysmal atrial fibrillation: role of miR-146b-5p in atrial fibrosis. Heart Rhythm 2015; 12:1018–26 [DOI] [PubMed] [Google Scholar]
- 31.Xiao J, Liang D, Zhang Y, Liu Y, Zhang H, Liu Y, Li L, Liang X, Sun Y, Chen YH. MicroRNA expression signature in atrial fibrillation with mitral stenosis. Physiol Genomics 2011; 43:655–64 [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Zhou C, Liu Y, Wang S, Ye P, Miao X, Xia J. The expression levels of plasma micoRNAs in atrial fibrillation patients. PLoS One 2012; 7:e44906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McManus DD, Tanriverdi K, Lin H, Esa N, Kinno M, Mandapati D, Tam S, Okike ON, Ellinor PT, Keaney JF, Jr., Donahue JK, Benjamin EJ, Freedman JE. Plasma microRNAs are associated with atrial fibrillation and change after catheter ablation (the miRhythm study). Heart Rhythm 2015; 12:3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooley N, Cowley MJ, Lin RC, Marasco S, Wong C, Kaye DM, Dart AM, Woodcock EA. Influence of atrial fibrillation on microRNA expression profiles in left and right atria from patients with valvular heart disease. Physiol Genomics 2012; 44:211–9 [DOI] [PubMed] [Google Scholar]
- 35.Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, Zhang Y, Shan H, Luo X, Bai Y, Sun L, Song W, Xu C, Wang Z, Yang B. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 2010; 122:2378–87 [DOI] [PubMed] [Google Scholar]
- 36.Liu H, Qin H, Chen GX, Liang MY, Rong J, Yao JP, Wu ZK. Comparative expression profiles of microRNA in left and right atrial appendages from patients with rheumatic mitral valve disease exhibiting sinus rhythm or atrial fibrillation. J Transl Med 2014; 12:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo X, Yang B, Nattel S. MicroRNAs and atrial fibrillation: mechanisms and translational potential. Nat Rev Cardiol 2015; 12:80–90 [DOI] [PubMed] [Google Scholar]
- 38.Jia X, Zheng S, Xie X, Zhang Y, Wang W, Wang Z, Zhang Y, Wang J, Gao M, Hou Y. MicroRNA-1 accelerates the shortening of atrial effective refractory period by regulating KCNE1 and KCNB2 expression: an atrial tachypacing rabbit model. PLoS One 2013; 8:e85639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao H, Zhang M, Yang JJ, Shi KH. MicroRNA-21 via dysregulation of WW Domain-Containing protein 1 regulate atrial fibrosis in atrial fibrillation. Heart Lung Circ 2018; 27:104–13 [DOI] [PubMed] [Google Scholar]
- 40.Yamac AH, Kucukbuzcu S, Ozansoy M, Gok O, Oz K, Erturk M, Yilmaz E, Ersoy B, Zeybek R, Goktekin O, Kilic U. Altered expression of micro-RNA 199a and increased levels of cardiac SIRT1 protein are associated with the occurrence of atrial fibrillation after coronary artery bypass graft surgery. Cardiovasc Pathol 2016; 25:232–6 [DOI] [PubMed] [Google Scholar]
- 41.Shahi S, Zununi Vahed S, Fathi N, Sharifi S. Polymerase chain reaction (PCR)-based methods: promising molecular tools in dentistry. Int J Biol Macromol 2018; 117:983–92 [DOI] [PubMed] [Google Scholar]
- 42.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011; 27:431–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucl Acids Res 2015; 43:D447–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, Huang Y, Li W, Wang Z, Zhan S, Zhou M, Yao Y, Zeng Z, Hou Y, Chen Q, Tu X, Wang QK, Huang Z. Post-transcriptional regulation of cardiac sodium channel gene SCN5A expression and function by miR-192-5p. Biochim Biophys Acta 2015; 1852:2024–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei XJ, Han M, Yang FY, Wei GC, Liang ZG, Yao H, Ji CW, Xie RS, Gong CL, Tian Y. Biological significance of miR-126 expression in atrial fibrillation and heart failure. Braz J Med Biol Res 2015; 48:983–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei J, Zhang Y, Li Z, Wang X, Chen L, Du J, Liu J, Liu J, Hou Y. GCH1 attenuates cardiac autonomic nervous remodeling in canines with atrial-tachypacing via tetrahydrobiopterin pathway regulated by microRNA-206. Pacing Clin Electrophysiol 2018; 41:459–71 [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Qiu CG, Huang ZW, Han ZY, Lu WJ, Chen XJ. [ The expression and clinical implication of plasma miR-328 in patients with atrial fibrillation]. Zhonghua Xin Xue Guan Bing Za Zhi 2013; 41:126–9 [PubMed] [Google Scholar]
- 48.Wang J, Song S, Xie C, Han J, Li Y, Shi J, Xin M, Wang J, Luo T, Meng X, Yang B. MicroRNA profiling in the left atrium in patients with non-valvular paroxysmal atrial fibrillation. BMC Cardiovasc Disord 2015; 15:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F, Zhang SJ, Yao X, Tian DM, Zhang KQ, She DM, Guo FF, Zhai QW, Ying H, Xue Y. Circulating microRNA-1a is a biomarker of graves' disease patients with atrial fibrillation. Endocrine 2017; 57:125–37 [DOI] [PubMed] [Google Scholar]
- 50.Stillitano F, Lonardo G, Giunti G, Del Lungo M, Coppini R, Spinelli V, Sartiani L, Poggesi C, Mugelli A, Cerbai E. Chronic atrial fibrillation alters the functional properties of if in the human atrium. J Cardiovasc Electrophysiol 2013; 24:1391–400 [DOI] [PubMed] [Google Scholar]
- 51.Soeki T, Matsuura T, Bando S, Tobiume T, Uematsu E, Ise T, Kusunose K, Yamaguchi K, Yagi S, Fukuda D, Yamada H, Wakatsuki T, Shimabukuro M, Sata M. Relationship between local production of microRNA-328 and atrial substrate remodeling in atrial fibrillation. J Cardiol 2016; 68:472–7 [DOI] [PubMed] [Google Scholar]
- 52.Qiao G, Xia D, Cheng Z, Zhang G. miR132 in atrial fibrillation directly targets connective tissue growth factor. Mol Med Rep 2017; 16:4143–50 [DOI] [PubMed] [Google Scholar]
- 53.Nishi H, Sakaguchi T, Miyagawa S, Yoshikawa Y, Fukushima S, Saito S, Ueno T, Kuratani T, Sawa Y. Impact of microRNA expression in human atrial tissue in patients with atrial fibrillation undergoing cardiac surgery. PLoS One 2013; 8:e73397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo X, Pan Z, Shan H, Xiao J, Sun X, Wang N, Lin H, Xiao L, Maguy A, Qi XY, Li Y, Gao X, Dong D, Zhang Y, Bai Y, Ai J, Sun L, Lu H, Luo XY, Wang Z, Lu Y, Yang B, Nattel S. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J Clin Invest 2013; 123:1939–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, Zhang Y, Shan H, Luo X, Bai Y, Sun L, Song W, Xu C, Wang Z, Yang B. Erratum: MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation (circulation (2010) 122 (2378-2387)). Circulation 2010; 124:e334. [DOI] [PubMed] [Google Scholar]
- 56.Liu T, Zhong S, Rao F, Xue Y, Qi Z, Wu S. Catheter ablation restores decreased plasma miR-409-3p and miR-432 in atrial fibrillation patients. Europace 2016; 18:92–9 [DOI] [PubMed] [Google Scholar]
- 57.Ling TY, Wang XL, Chai Q, Lau TW, Koestler CM, Park SJ, Daly RC, Greason KL, Jen J, Wu LQ, Shen WF, Shen WK, Cha YM, Lee HC. Regulation of the SK3 channel by microRNA-499–potential role in atrial fibrillation. Heart Rhythm 2013; 10:1001–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li YD, Hong YF, Yusufuaji Y, Tang BP, Zhou XH, Xu GJ, Li JX, Sun L, Zhang JH, Xin Q, Xiong J, Ji YT, Zhang Y. Altered expression of hyperpolarization-activated cyclic nucleotide-gated channels and microRNA-1 and -133 in patients with age-associated atrial fibrillation. Mol Med Rep 2015; 12:3243–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li S, Jiang Z, Wen L, Feng G, Zhong G. MicroRNA-208a-3p contributes to connexin40 remolding in human chronic atrial fibrillation. Exp Ther Med 2017; 14:5355–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li N, Chiang D, Beavers D, Sun Q, Dobrev D, Martin JF. Loss of microRNA-106b-25 cluster promotes atrial fibrillation by enhancing ryanodine receptor type-2 mediated calcium release. Heart Rhythm 2014; 11:S163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Li S, Yu B, Liu S. Expression of miR-133 and miR-30 in chronic atrial fibrillation in canines. Mol Med Rep 2012; 5:1457–60 [DOI] [PubMed] [Google Scholar]
- 62.Goren Y, Meiri E, Hogan C, Mitchell H, Lebanony D, Salman N, Schliamser JE, Amir O. Relation of reduced expression of MiR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am J Cardiol 2014; 113:976–81 [DOI] [PubMed] [Google Scholar]
- 63.Girmatsion Z, Biliczki P, Bonauer A, Wimmer-Greinecker G, Scherer M, Moritz A, Bukowska A, Goette A, Nattel S, Hohnloser SH, Ehrlich JR. Changes in microRNA-1 expression and IK1 up-regulation in human atrial fibrillation. Heart Rhythm 2009; 6:1802–9 [DOI] [PubMed] [Google Scholar]
- 64.Feldman A, Moreira DAR, Gun C, Wang HL, Hirata MH, de Freitas Germano J, Leite GGS, Farsky P. Analysis of circulating miR-1, miR-23a, and miR-26a in atrial fibrillation patients undergoing coronary bypass artery grafting surgery. Ann Hum Genet 2017; 81:99–105 [DOI] [PubMed] [Google Scholar]
- 65.Dawson K, Wakili R, Ordog B, Clauss S, Qi XY, Cardin S, Sinner M, Kääb S, Nattel S. MicroRNA29 a mechanistic contributor and potential biomarker in atrial fibrillation. Heart Rhythm 2013; 8:S255–S56 [DOI] [PubMed] [Google Scholar]
- 66.Adam O, Lohfelm B, Thum T, Gupta SK, Puhl SL, Schafers HJ, Bohm M, Laufs U. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol 2012; 107:278. [DOI] [PubMed] [Google Scholar]
- 67.Tousoulis D. Biomarkers in atrial fibrillation; from pathophysiology to diagnosis and treatment. Curr Med Chem 2019; 26:762–4 [DOI] [PubMed] [Google Scholar]
- 68.Mun D, Kim H, Kang JY, Park H, Park H, Lee SH, Yun N, Joung B. Expression of miRNAs in circulating exosomes derived from patients with persistent atrial fibrillation. FASEB J 2019; 33:5979–89 [DOI] [PubMed] [Google Scholar]
- 69.Barana A, Matamoros M, Dolz-Gaiton P, Perez-Hernandez M, Amoros I, Nunez M, Sacristan S, Pedraz A, Pinto A, Fernandez-Aviles F, Tamargo J, Delpon E, Caballero R. Chronic atrial fibrillation increases microRNA-21 in human atrial myocytes decreasing L-type calcium current. Circ Arrhythm Electrophysiol 2014; 7:861–8 [DOI] [PubMed] [Google Scholar]
- 70.Chiang DY, Kongchan N, Beavers DL, Alsina KM, Voigt N, Neilson JR, Jakob H, Martin JF, Dobrev D, Wehrens XH, Li N. Loss of microRNA-106b-25 cluster promotes atrial fibrillation by enhancing ryanodine receptor type-2 expression and calcium release. Circ Arrhythm Electrophysiol 2014; 7:1214–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan CT, Li XX, Cheng QJ, Wang YH, Wang JH, Liu CL. MiR-30a regulates the atrial fibrillation-induced myocardial fibrosis by targeting snail 1. Int J Clin Exp Pathol 2015; 8:15527–36 [PMC free article] [PubMed] [Google Scholar]
- 72.Schueller F, Roy S, Loosen SH, Alder J, Koppe C, Schneider AT, Wandrer F, Bantel H, Vucur M, Mi QS, Trautwein C, Luedde T, Roderburg C. miR-223 represents a biomarker in acute and chronic liver injury. Clin Sci 2017; 131:1971–87 [DOI] [PubMed] [Google Scholar]
- 73.Yuan X, Berg N, Lee JW, Le TT, Neudecker V, Jing N, Eltzschig H. MicroRNA miR-223 as regulator of innate immunity. J Leukoc Biol 2018; 104:515–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia LH, Liu YN. Downregulated serum miR-223 servers as biomarker in alzheimer's disease. Cell Biochem Funct 2016; 34:233–7 [DOI] [PubMed] [Google Scholar]
- 75.Li L, Li S, Wu M, Chi C, Hu D, Cui Y, Song J, Lee C, Chen H. Early diagnostic value of circulating microRNAs in patients with suspected acute myocardial infarction. J Cell Physiol 2019; 234:13649–13658 [DOI] [PubMed] [Google Scholar]
- 76.Wang JG, Meng X, Han J, Li Y, Luo TG, Wang J, Xin M, Xi JZ. [Differential expressions of miRNAs in patients with nonvalvular atrial fibrillation]. Zhonghua Yi Xue Za Zhi 2012; 92:1816–9 [PubMed] [Google Scholar]
- 77.Kakimoto Y, Tanaka M, Kamiguchi H, Hayashi H, Ochiai E, Osawa M. MicroRNA deep sequencing reveals chamber-specific miR-208 family expression patterns in the human heart. Int J Cardiol 2016; 211:43–8 [DOI] [PubMed] [Google Scholar]
- 78.Huang Z, Chen XJ, Qian C, Dong Q, Ding D, Wu QF, Li J, Wang HF, Li WH, Xie Q, Cheng X, Zhao N, Du YM, Liao YH. Signal transducer and activator of transcription 3/MicroRNA-21 feedback loop contributes to atrial fibrillation by promoting atrial fibrosis in a rat sterile pericarditis model. Circ Arrhythm Electrophysiol 2016; 9:e003396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017; 16:203–22 [DOI] [PubMed] [Google Scholar]
- 80.Hsieh CS, Huang PS, Chang SN, Wu CK, Hwang JJ, Chuang EY, Tsai CT. Genome-wide copy number variation association study of atrial fibrillation related thromboembolic stroke. J Clin Med 2019; 8: pii: E332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suita K, Fujita T, Hasegawa N, Cai W, Jin H, Hidaka Y, Prajapati R, Umemura M, Yokoyama U, Sato M, Okumura S, Ishikawa Y. Norepinephrine-Induced adrenergic activation strikingly increased the atrial fibrillation duration through beta1- and alpha1-Adrenergic Receptor-Mediated signaling in mice. PLoS One 2015; 10:e0133664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marsh JD, Telemaque S, Rhee SW, Stimers JR, Rusch NJ. Delivery Of ion channel genes to treat cardiovascular diseases. Transac Am Clin Climatol Assoc 2008; 119:171–82; discussion 82–3 [PMC free article] [PubMed] [Google Scholar]
- 83.Andrade J, Khairy P, Dobrev D, Nattel S. The clinical profile and pathophysiology of atrial fibrillation: relationships among clinical features, epidemiology, and mechanisms. Circ Res 2014; 114:1453–68 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, EBM890303 Supplemental Material for MicroRNA expression signatures of atrial fibrillation: The critical systematic review and bioinformatics analysis by Nan-Nan Shen, Chi Zhang, Zheng Li, Ling-Cong Kong, Xin-Hua Wang, Zhi-Chun Gu and Jia-Liang Wang in Experimental Biology and Medicine