Abstract
This experiment was conducted to evaluate the impact of yeast and lactic acid bacteria (LAB) on mastitis and milk microbiota composition of dairy cows. Thirty lactating Holstein cows with similar parity, days in milk were randomly assigned to five treatments, including: (1) Health cows with milk SCC < 500,000 cells/mL, no clinical signs of mastitis were found, fed basal total mixed ration (TMR) without supplementation (H); (2) Mastitis cows with milk SCC > 500,000 cells/mL, fed basal TMR without supplementation (M); (3) Mastitis cows fed basal TMR supplemented with 8 g day−1 yeast (M + Y); (4) Mastitis cows fed basal TMR supplemented with 8 g day−1 LAB (M + L); (5) Mastitis cows (milk SCC > 500,000 cells/mL) fed basal TMR supplemented with 4 g day−1 yeast and 4 g day−1 LAB (M + Y + L). Blood and milk sample were collected at day 0, day 20 and day 40. The results showed efficacy of probiotic: On day 20 and day 40, milk SCC in H, M + Y, M + L, M + Y + L was significantly lower than that of M (P < 0.05). Milk concentration of TNF-α, IL-6 and IL-1β in M + Y + L were significantly reduced compared with that of M on day 40 (P < 0.05). Milk Myeloperoxidase (MPO) and N-Acetyl-β-d-Glucosaminidase (NAG) activity of M + Y, M + L, M + L + Y were lower than that of M on day 40 (P < 0.05). At genus level, Staphylococcus, Chryseobacterium and Lactococcus were dominant. Supplementation of LAB decreased abundance of Enterococcus and Streptococcus, identified as mastitis-causing pathogen. The results suggested the potential of LAB to prevent mastitis by relieving mammary gland inflammation and regulating milk microorganisms.
Keywords: Lactic acid bacteria, Yeast, Mastitis, Milk microbiota composition, Dairy cows
Introduction
Mastitis, characterized by high amounts of bacteria and mammary inflammation, is one of the most frequent disease occurred on dairy cows and has been well-recognized detrimental effects on milk quality, animal well-being and public health. A variety of bacteria pathogens were identified to be involved in the development of mastitis. Streptococcus agalactiae and Staphylococcus was considered as the mainly contagious pathogens (Ruegg 2017). Murphy (1947) demonstrated the general process of mastitis, which bacteria being established in the mammary gland as initial and followed by inflammation.
Intramammary administration of antibiotics is the widely used conventional therapy for clinical mastitis during lactating and also for prevention of new infections during dry-off. Despite the effectiveness of controlling mastitis, the development of antimicrobial resistance caused by common use of antibiotics raises society concern. In order to reduce antibiotic residues in dairy products and coincide with global requirement to limit their use in dairy cattle, the use of probiotic agents is a novel approach to cure mastitis. Probiotics are live microorganisms, which confer a health benefit to the host when administered in adequate amounts (FAO and WHO 2008). Researches have demonstrated the ability of probiotics to protect against microbial pathogens and enhance the immune functions (Cross 2002). The use of probiotics bacteria mainly focuses on gastrointestinal, vaginal tract and mammary gland (Frola et al. 2012; Deng et al. 2015; Walsh et al. 2010). The mechanism that probiotics exert beneficial effects on host health is closely associated with the capability to produce antagonistic substances, adhesion to host tissues and colonization to different sites of the host surfaces (Espeche et al. 2009). However, the dietary supplementation of probiotic on immune system and mastitis is unknown, which is more practical in dairy industrial compared with appliance to gastrointestinal tract and mammary gland. Yeast supplementation on ruminants has been shown to regulate rumen pH, microbial composition and fiber fermentation (Bach et al. 2018; Terre et al. 2015). Recent studies suggest the supplementation of yeast may also influence immune function. Zanello et al. (2011) reported that yeast inhibit the Escherichia coli—induced expression of pro-inflammation transcripts and protein, including IL-6, IL-8. Furthermore, yeast also moderate recruitment and activation of immune cells in differentiated porcine intestinal epithelial cells. Yuan et al. (2015) analyzed the effects of dietary yeast supplementation on uterine inflammatory and they found that although no treatment effect was detected for incidence of subclinical endometritis, supplementary yeast decreased uterine IL-6 mRNA abundance and improved neutrophil myeloperoxidase and neutrophil elastase expression on transition cow, as well as fecal IgA concentration, which indicating relieved uterine inflammation and strengthened immune function. To our knowledge, no study has evaluated the effects of dietary supplementation of live yeast on mastitis and milk microorganism based on the high-throughput sequencing, a new method that provide a more complete relative qualification of micro community composition. Lactic acid bacteria (LAB) is also shown as potential probiotics for therapeutic use against endometrial inflammation and mastitis and prevention for diarrhea in calf, which is capable for the ability to strengthen systemic immune function (Genis et al. 2017; Bouchard et al. 2015; Maldonado et al. 2017). Intramammary LAB incubation increased the amounts of IgG isotypes in blood and milk, as well as lymphocyte proliferation and negative the major bovine mastitis pathogens on dry-off cows, which suggests restored balance in microbiota of the mammary gland and improved systemic immune function (Pellegrino et al. 2017). However, the effects of dietary supplementation of LAB and yeast on mastitis and milk microorganisms of lactating cow is unknown. Therefore, the objective of this study is to evaluate the effect of dietary supplementation of LAB and yeast on mastitis and milk microorganism based on the high-throughput sequencing.
Materials and methods
Animals and experimental design
Thirty lactating Holstein cows with similar parity (1.5 ± 0.3) and days in milk (145 ± 2) were selected and assigned to five treatments, 6 cows each treatment, including: (1) Health cows with milk SCC < 500,000 cells/mL, no clinical signs of mastitis were found in four mammary quarters and no incidence of clinical mastitis during the last 90 days of the previous lactation, fed basal total mixed ration (TMR) without supplementation (H); (2) Mastitis cows with milk SCC > 500,000 cells/mL, fed basal TMR without supplementation (M); (3) Mastitis cows (milk SCC > 500,000 cells/mL) fed basal TMR supplemented with 8 g day−1 yeast (M + Y, company product); (4) Mastitis cows (milk SCC > 500,000 cells/mL) fed basal TMR supplemented with 8 g day−1 lactic acid bacteria (M + L, company products); (5) Mastitis cows (milk SCC > 500,000 cells/mL) fed basal TMR supplemented with 4 g day−1 yeast and 4 g day−1 lactic acid bacteria (M + Y + L). The yeast was Saccharomyces cerevisiae, with moisture contents < 6.0% and counts around 2 × 1010 CFU/g; LAB consisted of Lactococcal and maltodextrin, with moisture contents < 8.0% and counts around 2 × 109 CFU/g. Cows were housed in a free-stall barn and managed in the same manner throughout the experiment. During the morning feeding, yeast chromium and dihydropyridine were mixed with 2 kg TMR and then fed to each cow based on treatments. After the TMR mixed with supplements was consumed, the remaindering portion of TMR was delivered. Cows remained on the treatments for 40 days. Cows had ad libitum access to water and were fed at 0700, 1700 h each day. Cows were milked at 0400, 1600 h.
Sample collection and laboratory analysis
Blood sample were collected from coccygeal vessels into vacutainers without anticoagulant on day 0 and day 40 before feeding (0600 h). Blood was allowed to clot at room temperature for 20 min before centrifugation at 3000g for 30 min to collect serum and stored at − 20 °C until analysis. Concentration of serum immunoglobulin (IgA, IgG, IgM) were measured by commercially available ELISA kits (Jiancheng Biochemical Reagent Company, Nanjing, China) following the manufacturer’s instructions. Milk samples were collected from two consecutive milking and mixed equally at day 0, day 20 and day 40. Part of milk samples were analyzed for milk percentage of fat, protein, fat to protein ration, lactose, urea nitrogen, solid not fat and SCC. The other whole milk was centrifuged 8000g for 10 min and the supernatant concentration of TNF-α, IL-6, IL-1β and activity of lactic dehydrogenase (LDH), Myeloperoxidase (MPO), N-Acetyl-β-d-Glucosaminidase (NAG), alkaline phosphatase (ALP) were measured by Elisa as manufacturer’s instructions (Jiancheng Biochemical Reagent Company, Nanjing, China).
DNA extraction, PCR amplification and high-throughput sequencing
On day 40, 20 milk samples were collected (4 cows each treatment) and centrifuged. The pellet was subjected to genomic DNA extraction using GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich, Sigma Chemical Co., St. Louis, MO. USA), according to the manufacturer’s protocols. Final DNA concentration and purification were determined using a NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked using 1% agarose gel electrophoresis. The bacterial primer set of forward primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify DNA fragments of the V1–V2 region of bacterial16S rRNA genes. TransStart Fastpfu DNA Polymerase and thermocycler PCR systems (GeneAmp 9700, ABI, USA) were available for PCR. PCR was performed triplicate in a 20 μL triplicate mixture: 4 μL of FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, 10 ng of template DNA, add dH2O to 20 μL. The PCR reaction were conducted using following program: an initial denaturation step at 95 °C for 3 min, followed by 27 cycles of denaturation at 95 °C for 30 s, annulation at 55 °C for 30 s, elongation at 72 °C for 45 s with a final extension step at 72 °C for 10 min which was halted at 10 °C. The resulting PCR products were extracted from a 2% agarose gel and further purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and were quantified using a QuantiFluor-ST Real-time PCR System (Promega, USA) according to the manufacturer’s protocol, before sequencing. 16S rRNA gene sequencing was performed on an Illumina Miseq 300 Platform (Illumina, San Diego, USA) according to standard protocols by Majorbio Bio-pharm Technology Co., Ltd., Shanghai, China). The raw reads were deposited into Zenodo database (http://doi.org/10.5281/zenodo.3384327).
Data analysis
Serum concentration of IgA, IgM, IgG and milk composition, concentration of TNF-α, IL-1β, IL-6 and activity of LDH, MPO, NAG, ALP were subjected were subjected to analysis of variance (ANOVA) using SPSS statistical software (SPSS 17.0). Statistical differences among means (P < 0.05) were identified using Duncan’s multiple range test. Significance was declared at P < 0.05. Means and SEM are reported. Microbial community and group difference analyses were performed using the free online platform, Majorbio I-Sanger Cloud Platform (www.i-sanger.com).
Results
Milk composition
On day 0, cows in M, M + Y, M + L, M + Y + L had significantly higher milk SCC compared with that of H (P < 0.05, Table 1). On day 20 and day 40, milk SCC in H, M + Y, M + L, M + Y + L was significantly lower than that of M on day 20 and day 40 (P < 0.05). Milk percentage of fat in M was significantly reduced compared with that of H and M + Y + L on day 20 and day 40 (P < 0.05). Cows in M + Y + L had significantly higher percentage of milk protein than cows in other treatments on day 40 (P < 0.05). Milk fat to protein ratio and percentage of lactose in M + Y, M + L, M + Y + L was significantly increased compared with that of M on day 40 (P < 0.05). Cows in H, M + Y + L had higher milk percentage of urea nitrogen than cows in M on day 20 and day 40 (P < 0.05). There were no treatment effects on milk percentage of solid not fat (P > 0.05).
Table 1.
The effect of treatments on milk composition
| Items | Time | Treatments | SEM | P-value | ||||
|---|---|---|---|---|---|---|---|---|
| H | M | M + Y | M + L | M + Y + L | ||||
| SCC (104/mL) | Day0 | 42.00b | 229.42a | 199.33a | 217.83a | 247.16a | 21.75 | 0.05 |
| Day20 | 65.25b | 352.14a | 136.50b | 174.66b | 172.50b | 29.71 | 0.02 | |
| Day40 | 56.25B | 294.42A | 112.83B | 129.67B | 121.33B | 24.65 | 0.01 | |
| Milk fat (%) | Day0 | 3.22 | 2.68 | 2.20 | 2.47 | 2.83 | 0.12 | 0.16 |
| Day20 | 3.37A | 2.00B | 2.76AB | 2.69AB | 3.41A | 0.15 | 0.01 | |
| Day40 | 3.23B | 2.65B | 3.30B | 3.40B | 4.23A | 0.15 | 0.01 | |
| Milk protein (%) | Day0 | 3.35 | 3.52 | 3.23 | 3.29 | 3.58 | 0.06 | 0.34 |
| Day20 | 3.56 | 3.65 | 3.37 | 3.42 | 3.70 | 0.06 | 0.33 | |
| Day40 | 3.62b | 3.56b | 3.46b | 3.61b | 4.02a | 0.07 | 0.05 | |
| Fat to protein ratio | Day0 | 0.96 | 0.75 | 0.68 | 0.74 | 0.78 | 0.03 | 0.17 |
| Day20 | 0.94A | 0.54B | 0.82A | 0.78A | 0.90A | 0.04 | 0.00 | |
| Day40 | 0.89ab | 0.74b | 0.96a | 0.94a | 1.04a | 0.04 | 0.05 | |
| Lactose (%) | Day0 | 4.87 | 5.09 | 4.86 | 5.08 | 4.93 | 0.05 | 0.49 |
| Day20 | 5.21 | 4.94 | 5.01 | 5.19 | 5.03 | 0.04 | 0.18 | |
| Day40 | 5.04B | 4.80B | 5.12A | 5.28A | 5.35A | 0.05 | 0.00 | |
| Urea nitrogen (%) | Day0 | 12.12 | 12.60 | 11.43 | 11.55 | 11.51 | 0.30 | 0.68 |
| Day20 | 12.65a | 10.67b | 11.68b | 11.65b | 13.41a | 0.30 | 0.02 | |
| Day40 | 13.82a | 11.70b | 13.50a | 12.71ab | 13.71a | 0.28 | 0.07 | |
| Solids not fat (%) | Day0 | 9.06 | 9.43 | 8.87 | 9.17 | 9.18 | 0.08 | 0.23 |
| Day20 | 9.59 | 9.39 | 9.23 | 9.34 | 9.25 | 0.07 | 0.59 | |
| Day40 | 9.42 | 9.54 | 9.40 | 9.41 | 9.61 | 0.08 | 0.89 | |
In dairy cows, SCC and milk of inferior are important predictors for mastitis
abcMeans in the same row with significant difference (P < 0.05)
ABCMeans in the same row with extremely significant difference (P < 0.01)
Serum immunoglobulin
As shown in Table 2, Cows in M + Y had lower serum concentration of IgA and IgM compared with that of M on day 40 (P < 0.05). Serum concentration of IgG in M + Y, M + L and M + Y + L was significantly deceased compared with that of M on day 40 (P < 0.01).
Table 2.
The effect of treatments on serum concentration of immunoglobulin and milk concentration of inflammation cytokine and enzyme activity
| Items | Time | Treatments | SEM | P-value | ||||
|---|---|---|---|---|---|---|---|---|
| H | M | M + Y | M + L | M + Y + L | ||||
| IgA (ng/mL) | Day0 | 8.52C | 8.92BC | 8.93BC | 10.31AB | 11.23A | 0.24 | 0.00 |
| Day40 | 9.07ab | 9.77b | 8.11a | 9.20ab | 9.66b | 0.19 | 0.03 | |
| IgM (ng/mL) | Day0 | 13.20C | 16.25B | 17.48AB | 19.02A | 18.52AB | 0.46 | 0.00 |
| Day40 | 14.04a | 17.34b | 15.04a | 15.94ab | 15.60ab | 0.32 | 0.01 | |
| IgG (ng/mL) | Day0 | 15.64b | 19.48a | 19.82a | 18.39a | 18.98a | 0.47 | 0.03 |
| Day40 | 18.27AB | 20.42B | 16.67A | 15.86A | 16.66A | 0.43 | 0.00 | |
| TNF-α (pg/mL) | Day0 | 85.26 | 95.85 | 105.39 | 108.47 | 96.08 | 3.17 | 0.16 |
| Day40 | 91.30AB | 117.34A | 82.87B | 87.68B | 80.46B | 3.87 | 0.01 | |
| IL-6 (pg/mL) | Day0 | 80.80B | 97.88AB | 104.45AB | 123.08A | 105.37AB | 3.81 | 0.00 |
| Day40 | 83.87AB | 107.90A | 86.30AB | 91.69AB | 59.67B | 4.33 | 0.00 | |
| IL-1β (pg/mL) | Day0 | 157.63b | 180.19ab | 207.53a | 179.76ab | 185.76ab | 4.89 | 0.02 |
| Day40 | 161.612ab | 188.67a | 162.88ab | 160.34ab | 143.54b | 4.85 | 0.04 | |
| LDH (U/L) | Day0 | 3533.73c | 4129.25bc | 6507.89a | 6051.62ab | 5130.87abc | 342.71 | 0.02 |
| Day40 | 4529.75 | 5661.56 | 4484.27 | 3954.33 | 4029.85 | 231.52 | 0.10 | |
| ALP (U/L) | Day0 | 161.58 | 206.63 | 225.91 | 227.91 | 225.55 | 1.75 | 0.44 |
| Day40 | 212.77 | 282.17 | 187.85 | 170.22 | 150.59 | 2.59 | 0.16 | |
| MPO (U/L) | Day0 | 29.01b | 38.35ab | 38.89ab | 44.17a | 33.94ab | 1.66 | 0.04 |
| Day40 | 36.38B | 47.06A | 34.11B | 36.67B | 29.50B | 1.65 | 0.00 | |
| NAG (U/L) | Day0 | 26.21B | 111.13A | 63.48AB | 115.84A | 59.71AB | 8.33 | 0.00 |
| Day40 | 63.72BC | 130.80A | 40.00CD | 84.27B | 27.59D | 8.49 | 0.00 | |
H: healthy cows without mastitis fed basal diet; M: cows with mastitis fed basal diet; M + Y: cows with mastitis fed basal diet supplemented with 8 g day−1 yeast; M + L: cows with mastitis fed basal diet supplemented with 8 g day−1 lactic acid bacteria; M + Y + L: cows with mastitis fed basal diet supplemented with 4 g day−1 yeast and 4 g day−1 lactic acid bacteria
abcMeans in the same row with significant difference (P < 0.05)
ABCMeans in the same row with extremely significant difference (P < 0.01)
Milk inflammation cytokine and enzyme activity
Milk concentration of TNF-α in M + Y, M + L, M + Y + L significantly reduced compared with that of M on day 40 (P < 0.01, Table 2). Cows in M + Y + L had significantly lower milk concentration of IL-6 and IL-1β than cows in M on day 40 (P < 0.05). While there was no difference among other treatments (P > 0.05). Milk LDH activity in M + Y and M + L was significantly higher than that of H on day 0 (P < 0.05), while there was no significant difference between groups on day 40 (P > 0.05). Treatments didn’t affect the activity of ALP on day 0 and day 40 (P > 0.05). Milk MPO and NAG activity of M + Y, M + L, M + L + Y declined compared to M on day 40 (P < 0.05).
General DNA sequencing observation
In total, 909,264 bacterial raw 16 rRNA sequences were gained for the 20 milk samples from 5 treatments and at least 37,799 sequences were obtained per sample. Sequences with higher than 97% similarity were classified as OUT. The rarefaction curve generated reached stable level, suggesting that the number of OTUs did not increase with the expending volume of data (Additional file 1: Fig. S1). In current study, a total of 886 OUT were detected. The number of OUT shared by 5 treatments were 282, accounting for 31.82% of total OUT. The number of unique OUT for H, M, M + Y, M + L, M + Y + L were 4, 3, 17, 201 and 12 respectively (Fig. 1).
Fig. 1.
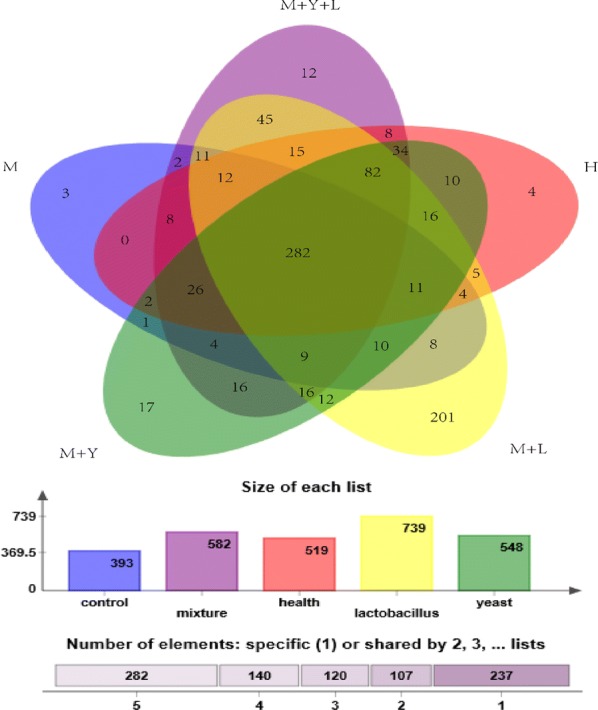
Venn diagram of OUTs from different treatments
Alpha and beta diversities of milk microbiota
Alpha diversity can be used to measure the microbiota diversity and richness. As was shown in Table 3, there was no difference on Shannon and Simpson index among groups (P > 0.05). The Ace and Chao index of M were significantly lower than that of other treatments (P < 0.05). Beta diversity can determine the microbiota diversity between different samples. PCA was conducted using genus-level taxonomic profiles to compare the composition diversity (Fig. 2). Samples from groups M, M + Y, M + L, M + Y + L were able to separate based on their clustering of microbiota, suggesting a shift in the milk microbial communities with the oral supplementation of yeast and LAB. However, there was no clear separation of milk samples from groups H and M + Y + L because of the overlap. Principal coordinate analysis (PCoA) based on weighted UniFrac distances was performed for each group and the emperor PCoA scatterplot obtained are depicted in Fig. 3. The R2 was measured to test for the percentage of variation among samples in each group using Adonis. The mean distances among the five treatments were calculated and were shown to be significantly different in the weighted analysis (R2 = 0.7114, P = 0.005), indicating a qualitatively different composition of the milk microbiota caused by the treatments.
Table 3.
Diversity indices and richness estimators of the 16rRNA gene libraries in milk microbiota of dairy cows in different treatments
| Items | Treatments | SEM | P-value | ||||
|---|---|---|---|---|---|---|---|
| H | M | M + Y | M + L | M + Y + L | |||
| Sequence | 49,642 | 47,709 | 46,315 | 40,192 | 44,501 | 2216 | 0.13 |
| Shannon | 2.65 | 2.41 | 2.18 | 2.53 | 2.73 | 0.40 | 0.70 |
| Simpson | 0.21 | 0.19 | 0.28 | 0.20 | 0.16 | 0.07 | 0.56 |
| Ace | 451.51a | 305.84b | 491.65a | 509.69a | 450.84a | 57.34 | 0.02 |
| Chao | 430.36a | 277.60b | 452.25a | 472.63a | 445.92a | 59.08 | 0.03 |
| Coverage | 0.998 | 0.997 | 0.997 | 0.998 | 0.998 | 0.00 | 0.14 |
Shannon and Simpson indices are used to assess biodiversity of milk microbiota
Ace and Chao indices are used to assess the microbiotal richness
Coverage index are used to reflect species coverage
abcMeans in the same row with significant difference (P < 0.05)
Fig. 2.
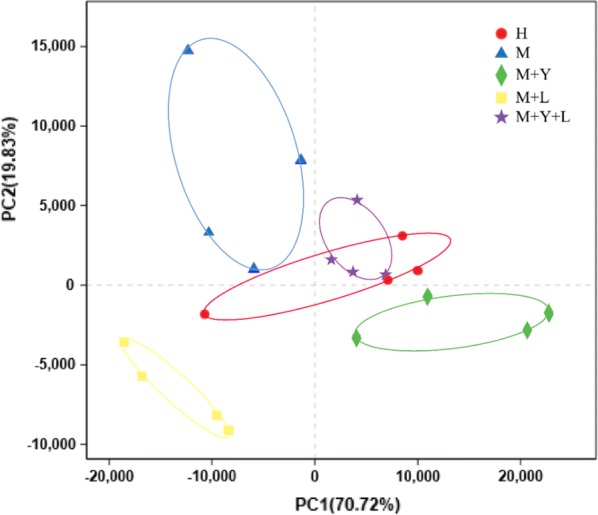
PCA analysis of milk samples microbiota of different treatments on genus level
Fig. 3.
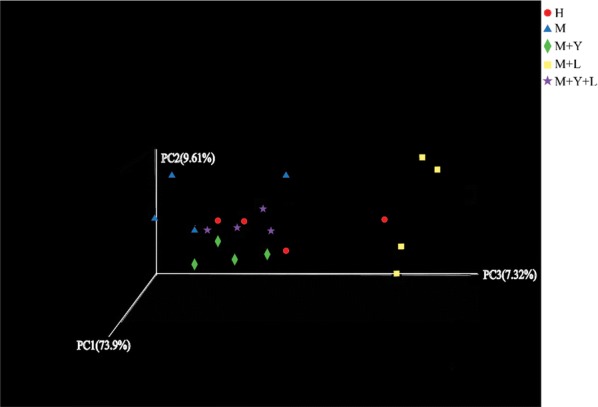
Scatterplots from principal coordinates analysis (PCoA), based on weighted UniFrac distance in bacterial communities
Milk microbiota community composition at phylum, genus, species level
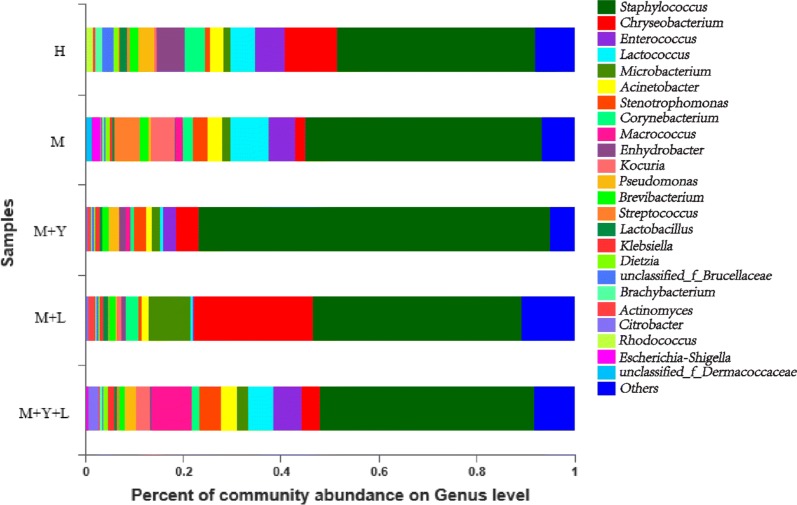
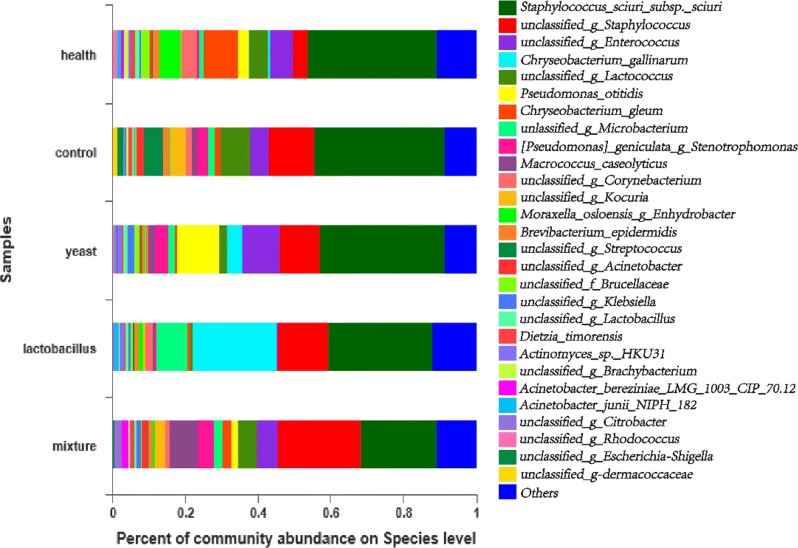
Firmicute was the major phylum with a prevalence ranging from 72.46 to 42.54% (Additional file 1: Fig. S2), followed by Proteobacteria (17.67–10.45%), Actionbacteria (17.67–10.04%). As shown in Fig. 4, Staphylococcus, Chryseobacterium and Lactococcus were dominant genera among groups, but the abundance of genera was different. At species level, the effects of supplementation of LAB and yeast on milk microorganisms community abundance were shown in Fig. 5. The most prevalent microbial species were Staphylococcus sciuri, Enterococcus and unclassified Enterococcus, but the abundance were diverse among groups.
Fig. 4.
The effect of treatments on the distribution of milk bacterial community at the genus level
Fig. 5.
The effect of treatments on the distribution of milk bacterial community at the species level
Milk microbiota community composition difference between different groups
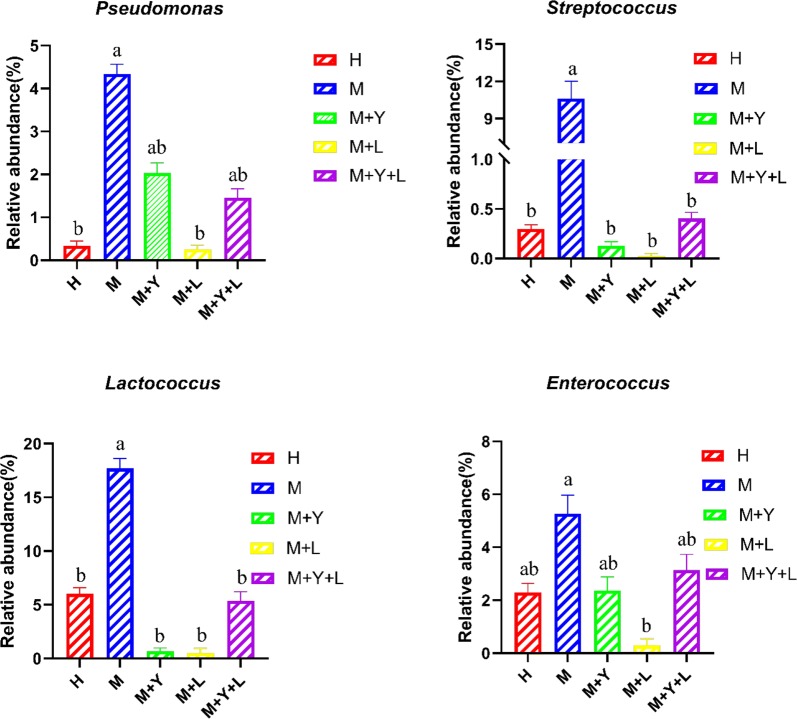
We used one-way ANOVA and T test to identify the difference in microbiota composition between groups at genus level. As is shown in Fig. 6, treatments significantly changed the relative abundance of Pseudomonas, Streptococcus, L. and Ent. (P < 0.05). The relative abundance of Ps. in group M were significantly higher than that of group H, M + L (P < 0.05). Group M + Y and M + Y + L showed the middle value in relative abundance of Ps. (P > 0.05). Supplementation of probiotic decreased the relative abundance of Strep. compared with group M (P < 0.05), but there was no difference among group M + Y, M + L and M + Y + L (P > 0.05). The relative abundance of L. in group M were significantly higher than other groups (P < 0.05). The relative abundance of Ent. were highest in group M, lowest in group M + L (P < 0.05), and intermediate for H, M + Y, M + Y + L (P > 0.05).
Fig. 6.
The relative abundance (%) of Pseudomonas, Streptococcus, Lactococcus and Enterococcus at genus level
Microbial function prediction
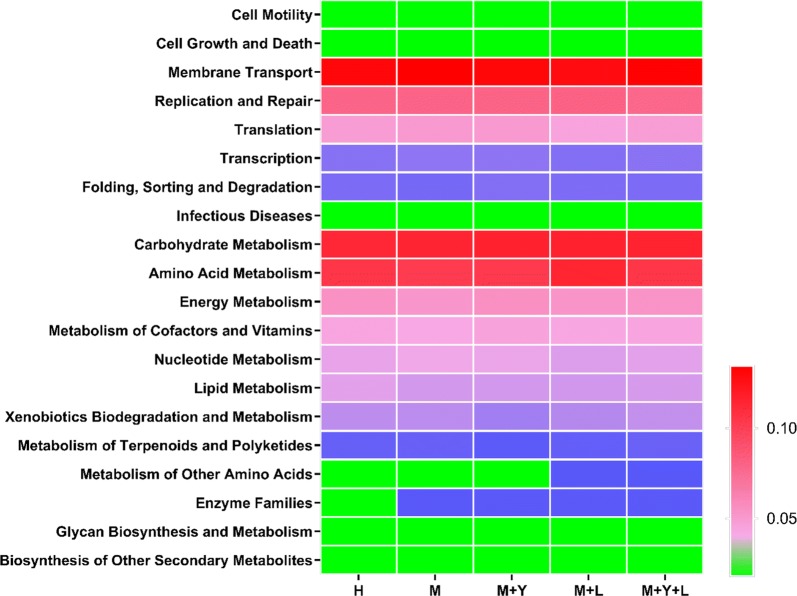
To investigate the functional profiles of milk bacterial community, PICRUSt was used to analyze the KEGG pathway compositions in milk microbiota populations. A total of 39 level 2 KEGG pathway were detected in milk samples. The top 20 abundant level 2 KEGG pathway in milk microbiota were shown in Fig. 7. The most prevalent pathway involved in milk microbiota were membrane transport, followed by carbohydrate metabolism and amino acid metabolism. As shown in Table 4, the relative abundance of 13 pathway were significantly influenced by treatments (P < 0.05). Basically, relative abundance of immune system disease and metabolic disease pathway in group H, M + Y, M + L, M + Y + L were lower than that of group M (P < 0.05). Supplementation of LAB significantly decreased the relative abundance of infectious disease pathway and increased the relative abundance of immune system pathway (P < 0.05).
Fig. 7.
Top 20 abundant of Heatmap of KEGG from different treatments on level 2
Table 4.
The relative abundance of level 2 KEGG pathway of milk microbiota from different treatments
| Items | Treatments | SEM | P-value | ||||
|---|---|---|---|---|---|---|---|
| H | M | M + Y | M + L | M + Y + L | |||
| Human disease | |||||||
| Cancers | 0.1133b | 0.0850b | 0.1012b | 0.1456a | 0.1052b | 0.0149 | 0.024 |
| Immune system diseases | 0.0519b | 0.0629a | 0.0475b | 0.0541b | 0.0529b | 0.0034 | 0.012 |
| Infectious diseases | 0.4914a | 0.5046a | 0.5477a | 0.3779b | 0.4942a | 0.0542 | 0.092 |
| Metabolic diseases | 0.0812b | 0.0956a | 0.0824b | 0.0874b | 0.0892b | 0.0039 | 0.028 |
| Neurodegenerative diseases | 0.2609a | 0.1935b | 0.2158b | 0.2304a | 0.2048b | 0.0193 | 0.049 |
| Organismal systems | |||||||
| Digestive system | 0.0567b | 0.0400c | 0.0470bc | 0.0767a | 0.0391c | 0.0069 | 0.002 |
| Environmental adaptation | 0.1018b | 0.0960b | 0.0992b | 0.1194a | 0.1028b | 0.0052 | 0.010 |
| Immune system | 0.0361b | 0.0363b | 0.0259b | 0.0628a | 0.0341b | 0.0091 | 0.022 |
| Metabolism | |||||||
| Biosynthesis of other secondary metabolites | 0.7393b | 0.7458b | 0.6535b | 1.0859a | 0.7434b | 0.1151 | 0.029 |
| Enzyme families | 1.8068b | 1.8567a | 1.8796a | 1.8767a | 1.8676a | 0.0229 | 0.055 |
| Environmental information processing | |||||||
| Signal transduction | 1.7126b | 1.7509b | 1.8880a | 1.6072c | 1.8691a | 0.0503 | 0.001 |
| Cellular process | |||||||
| Signaling molecules and Interaction | 0.1751bc | 0.1972ab | 0.1576c | 0.2010a | 0.1682c | 0.0130 | 0.031 |
| Transport and catabolism | 0.3127b | 0.2578b | 0.2635b | 0.3818a | 0.2611b | 0.0319 | 0.013 |
13 level 2 KEGG pathway of milk microbiota are influenced by treatments, which are classified into 5 classfications according to the pathway involved in
abcMeans in the same row with significant difference (P < 0.05)
Discussion
It is recently known that milk has its own microbiota and the mechanism probiotics exert benefit on milk microorganisms just starts. It is not clear that if oral intake of probiotics will pass from gut tract to mammary gland through endogenous route or pointing towards a systemic effect such as enhancing immune function, or both. Mastromarino et al. (2015) reported that oral administration of a multi-strain probiotic product (VSL#3) to women, consisting of lactobacilli, bifidobacteri and Streptococcus thermophilus, positively influenced milk microbiota through increased milk lactobacilli and bifidobacteri levels during late pregnancy and early lactation. Since high SCC and milk of inferior are the common indicator and consequence of mastitis for dairy cows, decreased SCC and improved milk quality in our study suggested the amelioration of mastitis. Higher milk percentage of fat and lactose caused by dietary supplementation of yeast has been reported in dairy goats (Ma et al. 2019) and dairy cows (Olagaray et al. 2019), while similar study with LAB are rare. Catozzi et al. (2019) reported that intramammary incubation of inactivated Lactobacillus rhamnosus caused significant increase of SCC on cows with subclinical at the beginning but decreased afterwards, as well as modulation of milk microbiota. Combined with our study, oral intake of LAB also decreased milk SCC and improved the milk quality in mastitis cows, indicating the relieving effects of LAB on mastitis.
The benefits probiotics exerted on mammary gland may be induced by relieving inflammation response. Supplementation of the mixture of LAB and yeast decreased the milk concentration of TNF-α, IL-1β and IL-6 compared with mastitis cows, while single use of yeast or LAB was unable to reduce the concentration of those inflammation cytokine concentration. Yuan et al. (2015) reported that 30 g day−1 and 60 g day−1 live yeast decreased uterine IL-6 mRNA of early lactation cows, which indicating modulated uterine inflammation signal. Similarly, study showed that adding LAB prevented E. coli infection and depressed the expression of proinflammation cytokine IL-8 and IL-1β of endometrial cells in vitro (Genis et al. 2016). MPO activity is a quantitative assessment of neutrophil infiltration into mammary gland (Kan et al. 2019), and our result showed that milk activity of MPO was significantly decreased by supplementation of yeast, LAB or the mixture. In general, combination of yeast and LAB had potential at regulating inflammation of bovine mammary gland.
During mastitis, mammary gland goes through immune response, which increases the permeability of tight junctions and compromises the milk–blood barrier (Wellnitz and Bruckmaier 2012). This causes the promoted passage of several blood components to milk. The increase appearance of LDH, ALP and NAG in milk are considered as indictors of mastitis (Chagunda et al. 2006; Guha et al. 2012). As expected, the milk activity of LDH, ALP and NAG of mastitis cows were higher than health cows, which indicates the destruction of blood–milk barrier. Compared with single supplementation of yeast or LAB, cows fed combination of yeast and LAB had the lowest milk NAG activity, suggesting the best repairing effects on milk–blood barrier. Once milk–blood barrier is damaged, gram-native pathogens enters the systemic calculation and triggers immune response, which illustrates the increased in IgA and IgM in our results (Wall et al. 2016). IgG is the major immunoglobulin contained in milk, which transfer more through blood–milk barrier because of increased permeability. According to the results, Supplementation of single yeast or the mixture of yeast and LAB decreased the serum level of IgA, IgG and IgM, but the value was still higher than health cows, suggesting improved immune condition. Our results were not consistent with Geng et al. (2018), who reported that supplementation of yeast didn’t affect serum contents of IgA, IgM and IgG.
In the present study, we observed that the common bacteria OTUs shared by five groups were 282, which accounted 31.82% of the total and suggested the dynamics of milk microbiota communities among five groups. Beta diversity analysis showed different composition of the milk microbiota among caused by treatments. The most abundant phylum in our trial is Firmicutes and Proteobacteria, which is in accordance with Patel et al. (2016). At genus level, our results are in partial agreement with observation of Derakhshani et al. (2018), who reported that Staph. was the most abundant bacterial genus throughout the first week of lactation. Staph. are predominant colonizer of bovine mammary gland commonly considered to be major pathogen responsible for subclinical mastitis (Condas et al. 2017). Chryseobacterium, another dominant Gram-negative bacteria, are known for its ability to facilitate the spoilage pasteurized milk and recontamination, which lowers the milk quality (Schmidt et al. 2012). Ent., followed by L, are reported as opportunistic pathogen causing mastitis in dairy cows (Oikonomou et al. 2014).
At genus level, supplementation of LAB lowered the mean proportions of Ps., which is the main strain responsible for the spoilage of refrigerated raw milk. Many species of Ps. are able to produce extracellular peptidases and lipases that remain active during thermal process and damage technological performances of milk and deterioration of the dairy products (Decimo et al. 2014). Similar to our results, D’Amico de Alcantara et al. (2019) also reported Lactobacillus rhamnosus inhibited the growth of Ps. in inoculated milk samples. LAB exert antimicrobial activity against the growth of deteriorating and pathogenic microorganisms through the production of various antimicrobial compounds, including bacteriocins, organic acids, and hydrogen peroxide (Prabhurajeshwar and Chandrakanth 2017; Todorov and Dicks 2005). According to our results, supplementation of mixture of yeast and LAB didn’t reduce the mean proportions of Ps., which may result from the different dose of LAB. Strept. is one of the major pathogens causing mastitis in dairy cows, mainly Streptococcus agalactiae, Streptococcus dysgalactiae, and Streptococcus uberis (Coffey et al. 2006). Our results showed the decreased mean proportion of Strep. after supplementation of probiotic, which indicates the relived mastitis. In agreement with our data, Nasiri et al. (2019) reported that supplementation of 4 g day−1 of live yeast fostered the lymphocyte proliferative response in transition dairy cows, which indicates an improved immune function. Thus, antimicrobial effects of yeast on mastitis cows may be associated with enhanced systemic immune function. L., which is considered a commensal species, has also been detected in raw milk sampled from healthy bovine mammary glands (Lafarge et al. 2004). In our trial, the mean proportions of L. in group M was higher than that of other groups, which is consistent with Patel et al. (2019) who reported that milk samples from subclinical mastitis cow had higher abundance of L. compared to healthy cows. Some strains of L. exhibit high adherence to bovine mammary epithelial cells, which means L. can also enter mammary gland of mastitis cows through teat and contaminant with pathogenic bacterial (Hagi et al. 2013). Ent., especially Enterococcus faecium and Enterococcus Faecalis, is the predominant environmental mastitis causing pathogen (Gao et al. 2019). Ent. are commonly found in animal intestines and their feces and characterized by a high level of resistance to many antibacterial substances, which limites the therapeutic result of antimicrobials (Rozanska et al. 2019). In our trial, supplementation of LAB exhibited inhibitory effect on abundance of Ent., indicating antimicrobial activity against mastitis. Based on KEGG pathway analysis of milk samples, supplementation of LAB also decreased relative abundance of pathway in immune system disease and infectious disease, while increased relative abundance of pathway in immune system, all of which are associated with mastitis and inflammation.
Supplementation of LAB and yeast show potential of relieving, while supplementation single strain LAB had best relieving the mammary inflammation and changing the milk microorganisms.
Supplementary information
Additional file 1: Fig. S1. Shannon index of OTU level of milk microbiota on different treatments. Fig. S2. The effects of treatments on the distribution of milk bacterial community at the phylum level
Acknowledgements
Authors thank the laboratory staff at the Department of Animal Nutrition and Feed science in Huazhong Agricultural University, China for technical support.
Authors’ contributions
JG, YCL and XMW designed and conducted the animal study. JG wrote the paper. JG, YW, ZLQ reviewed and edited the manuscript. The other authors helped with the lab work. All authors read and approved the final manuscript.
Funding
This work was supported by grants from National Key Research and Development Program [2018YFD0501605, 2016YFD0500507], Fundamental Research Funds for the Central Universities [2662018PY079], State Key Laboratory of Animal Nutrition Open Program [2004DA125184F1721].
Availability of data and materials
All sequences analyzed in this study can be accessed in Zenodo database (http://doi.org/10.5281/zenodo.3384327). The data supporting the conclusion of this article are included in this article.
Ethics approval and consent to participate
Experimental design and animal handling were approved by Institution Animal Care and Use Committee of Huazhong Agriculture University (Wuhan, China), and the animal trial was conducted in accordance with the National Institute of Health Guidelines for the Care and Use of Experiment Animals (Beijing, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jing Gao, Email: jinggao213@outlook.com.
Yu-Chen Liu, Email: 529125920@qq.com.
Yu Wang, Email: 467308908@qq.com.
Han Li, Email: 1426016144@qq.com.
Xiang-Ming Wang, Email: 1976972323@qq.com.
Yan Wu, Email: 1527076095@qq.com.
Ding-Ran Zhang, Email: 421020384@qq.com.
Si Gao, Email: 286361154@qq.com.
Zhi-li Qi, Email: zhiliqi@mail.hzau.edu.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13568-020-0953-8.
References
- Bach A, Guasch I, Elcoso G, Chaucheyras Durand F, Castex M, Fabregas F, Garcia Fruitos E, Aris A. Changes in gene expression in the rumen and colon epithelia during the dry period through lactation of dairy cows and effects of live yeast supplementation. J Dairy Sci. 2018;101:2631–2640. doi: 10.3168/jds.2017-13212. [DOI] [PubMed] [Google Scholar]
- Bouchard DS, Seridan B, Saraoui T, Rault L, Germon P, Gonzalez Moreno C, Nader Macias FM, Baud D, Francois P, Chuat V, Chain F, Langella P, Nicoli J, Le Loir Y, Even S. Lactic acid bacteria isolated from bovine mammary microbiota: potential allies against bovine mastitis. PLoS ONE. 2015;10:e0144831. doi: 10.1371/journal.pone.0144831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catozzi C, Cuscó A, Lecchi C, De Carlo E, Vecchio D, Martucciello A, D’Angelo L, Francino O, Sanchez Bonastre A, Ceciliani F. Impact of intramammary inoculation of inactivated Lactobacillus rhamnosus and antibiotics on the milk microbiota of water buffalo with subclinical mastitis. PLoS ONE. 2019;14:e0210204. doi: 10.1371/journal.pone.0210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagunda MG, Larsen T, Bjerring M, Ingvartsen KL. l-lactate dehydrogenase and N-acetyl-β-d-glucosaminidase activities in bovine milk as indicators of non-specific mastitis. J Dairy Res. 2006;73:431–440. doi: 10.1017/s0022029906001956. [DOI] [PubMed] [Google Scholar]
- Coffey TJ, Pullinger GD, Urwin R, Jolley KA, Wilson SM, Maiden MC, Leigh JA. First insights into the evolution of Streptococcus uberis: a multilocus sequence typing scheme that enables investigation of its population biology. Appl Environ Microbiol. 2006;72:1420–1428. doi: 10.1128/aem.72.2.1420-1428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condas LAZ, De Buck J, Nobrega DB, Carson DA, Roy JP, Keefe GP, DeVries TJ, Middleton JR, Dufour S, Barkema HW. Distribution of non-aureus staphylococci species in udder quarters with low and high somatic cell count, and clinical mastitis. J Dairy Sci. 2017;100:5613–5627. doi: 10.3168/jds.2016-12479. [DOI] [PubMed] [Google Scholar]
- Cross ML. Microbes versus microbes: immune signals generated by probiotic lactobacilli and their role in protection against microbial pathogens. FEMS Immunol Med Microbiol. 2002;34:245–253. doi: 10.1111/j.1574-695x.2002.tb00632.x. [DOI] [PubMed] [Google Scholar]
- D’Amico de Alcantara AL, Bruzaroski SR, Luiz LL, Batista de Souza CH, Poli-Frederico RC, Fragnani R, Walter de Santana EH. Antimicrobial activity of Lactobacillus rhamnosus against Pseudomonas fluorescens and Pseudomonas putida from raw milk. J Food Process Preserv. 2019 doi: 10.1111/jfpp.14082. [DOI] [Google Scholar]
- Decimo M, Morandi S, Silvetti T, Brasca M. Characterization of gram-negative psychrotrophic bacteria isolated from Italian bulk tank milk. J Food Sci. 2014;79:M2081–2090. doi: 10.1111/1750-3841.12645. [DOI] [PubMed] [Google Scholar]
- Deng Q, Odhiambo JF, Farooq U, Lam T, Dunn SM, Ametaj BN. Intravaginal lactic acid bacteria modulated local and systemic immune responses and lowered the incidence of uterine infections in periparturient dairy cows. PLoS ONE. 2015;10:1–16. doi: 10.1371/journal.pone.0124167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshani H, Plaizier JC, De Buck J, Barkema HW, Khafipour E. Association of bovine major histocompatibility complex (BoLA) gene polymorphism with colostrum and milk microbiota of dairy cows during the first week of lactation. Microbiome. 2018;6:203–221. doi: 10.1186/s40168-018-0586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeche MC, Otero MC, Sesma F, Nader Macias ME. Screening of surface properties and antagonistic substances production by lactic acid bacteria isolated from the mammary gland of healthy and mastitis cows. Vet Microbiol. 2009;135:346–357. doi: 10.1016/j.vetmic.2008.09.078. [DOI] [PubMed] [Google Scholar]
- FAO, WHO (2008) Health and nutritional properties of probiotics in food, including powder milk with live lactic acid bacteria. http://www.who.int. Accessed 22 Nov 2009
- Frola ID, Pellegrino MS, Espeche MC, Giraudo JA, Nader Macias ME, Bogni CI. Effects of intramammary inoculation of Lactobacillus perolens CRL1724 in lactating cows’ udders. J Dairy Sci. 2012;79:84–92. doi: 10.1017/S0022029911000835. [DOI] [PubMed] [Google Scholar]
- Gao X, Fan C, Zhang Z, Li S, Xu C, Zhao Y, Han L, Zhang D, Liu M. Enterococcal isolates from bovine subclinical and clinical mastitis: antimicrobial resistance and integron-gene cassette distribution. Microb Pathog. 2019;129:82–87. doi: 10.1016/j.micpath.2019.01.031. [DOI] [PubMed] [Google Scholar]
- Geng CY, Ji S, Jin YH, Li CY, Xia GJ, Li YM, Zhang M. Comparison of blood immunity, antioxidant capacity and hormone indexes in finishing bulls fed active dry yeast (Saccharomyces cerevisiae) and yeast culture. Int J Agric Biol. 2018;20:2561–2568. doi: 10.17957/IJAB/15.0822. [DOI] [Google Scholar]
- Genis S, Bach A, Fabregas F, Aris A. Potential of lactic acid bacteria at regulating Escherichia coli infection and inflammation of bovine endometrium. Theriogenology. 2016;85:625–637. doi: 10.1016/j.theriogenology.2015.09.05. [DOI] [PubMed] [Google Scholar]
- Genis S, Sánchez Chardi A, Bach À, Fabregas F, Aris A. A combination of lactic acid bacteria regulates Escherichia coli infection and inflammation of the bovine endometrium. J Dairy Sci. 2017;100:479–492. doi: 10.3168/jds.2016-11671. [DOI] [PubMed] [Google Scholar]
- Guha A, Gera S, Sharma A. Evaluation of milk trace elements, lactate dehydrogenase, alkaline phosphatase and aspartate aminotransferase activity of subclinical mastitis as indicator of subclinical mastitis in riverine buffalo (Bubalus bubalis) Asian-Australas J Anim Sci. 2012;25:353–360. doi: 10.5713/ajas.2011.11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagi T, Sasaki K, Aso H, Nomura M. Adhesive properties of predominant bacteria in raw cow’s milk to bovine mammary gland epithelial cells. Folia Microbiol. 2013;58:515–522. doi: 10.1007/s12223-013-0240-z. [DOI] [PubMed] [Google Scholar]
- Kan X, Liu B, Guo W, Wei L, Lin Y, Guo Y, Gong Q, Li Y, Xu D, Cao Y, Huang B, Dong A, Ma H, Fu S, Liu J. Myricetin relieves LPS-induced mastitis by inhibiting inflammatory response and repairing the blood–milk barrier. J Cell Physiol. 2019;234:1–11. doi: 10.1002/jcp.28288. [DOI] [PubMed] [Google Scholar]
- Lafarge V, Ogier JC, Girard V, Maladen V, Leveau JY, Gruss A, Delacroix Buchet A. Raw cow milk bacterial population shifts attributable to refrigeration. Appl Environ Microbiol. 2004;70:5644–5650. doi: 10.1128/AEM.70(9):5644-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZZ, Cheng YY, Wang SQ, Ge JZ, Shi HP, Kou JC. Positive effects of dietary supplementation of three probiotics on milk yield, milk composition and intestinal flora in Sannan dairy goats varied in kind of probiotics. J Anim Physiol Anim Nutr. 2019;00:1–12. doi: 10.1111/jpn.13226. [DOI] [PubMed] [Google Scholar]
- Maldonado NC, Chiaraviglio J, Bru E, De Chazal V, Santos V, Nader-Macias MEF. Effect of milk fermented with lactic acid bacteria on diarrheal incidence, growth performance and microbiological and blood profiles of newborn dairy calves. Probiotics Antimicrob Proteins. 2017;2017:1–9. doi: 10.1007/s12602-017-9308-4. [DOI] [PubMed] [Google Scholar]
- Mastromarino P, Capobianco D, Miccheli A, Pratico G, Campagna G, Laforgia N, Capursi T, Baldassarre ME. Administration of a multi-strain probiotic product (VSL#3) to women in the perinatal period differentially affects breast milk beneficial microbiota in relation to mode of delivery. Pharmacol Res. 2015;95–96:63–70. doi: 10.1016/j.phrs.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Murphy JM. The genesis of bovine udder infection and mastitis; the occurrence of streptococcal infection in a cow population during a seven-year period and its relationship to age. Am J Vet Res. 1947;8:29–42. [PubMed] [Google Scholar]
- Nasiri AH, Towhidi A, Shakeri M, Zhandi M, Dehghan Banadaky M, Pooyan HR, Sehati F, Rostami F, Karamzadeh A, Khani M, Ahmadi F. Effects of Saccharomyces cerevisiae supplementation on milk production, insulin sensitivity and immune response in transition dairy cows during hot season. Anim Feed Sci Technol. 2019;251:112–123. doi: 10.1016/j.anifeedsci.2019.03.007. [DOI] [Google Scholar]
- Oikonomou G, Bicalho ML, Meira E, Rossi RE, Foditsch C, Machado VS, Teixeira AG, Santisteban C, Schukken YH, Bicalho RC. Microbiota of cow’s milk; distinguishing healthy, sub-clinically and clinically diseased quarters. PLoS ONE. 2014;9:e85904. doi: 10.1371/journal.pone.0085904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagaray KE, Sivinski SE, Saylor BA, Mamedova LK, Sauls-Hiesterman JA, Yoon I, Bradford BJ. Effect of Saccharomyces cerevisiae fermentation product on feed intake parameters, lactation performance, and metabolism of transition dairy cattle. J Dairy Sci. 2019;102:8092–8107. doi: 10.3168/jds.2019-16315. [DOI] [PubMed] [Google Scholar]
- Patel RJ, Pandit RJ, Bhatt VD, Kunjadia PD, Nauriyal DS, Koringa PG, Joshi CG, Kunjadia AP. Metagenomic approach to study the bacterial community in clinical and subclinical mastitis in buffalo. Meta Gene. 2016;12:4–12. doi: 10.1016/j.mgene.2016.12.014. [DOI] [Google Scholar]
- Patel R, Kunjadia P, Koringa P, Joshi C, Kunjadiya A. Microbiological profiles in clinical and subclinical cases of mastitis in milking Jafarabadi buffalo. Res Vet Sci. 2019;125:94–99. doi: 10.1016/j.rvsc.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Pellegrino M, Berardo N, Giraudo J, Nader Macias MEF, Bogni C. Bovine mastitis prevention: humoral and cellular response of dairy cows inoculated with lactic acid bacteria at the dry-off period. Benef Microbes. 2017;8:589–596. doi: 10.3920/bm2016.0194. [DOI] [PubMed] [Google Scholar]
- Prabhurajeshwar C, Chandrakanth RK. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: an in vitro validation for the production of inhibitory substances. Biomed J. 2017;40:270–283. doi: 10.1016/j.bj.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanska H, Lewtak Pilat A, Kubajka M, Weiner M. Occurrence of enterococci in mastitic cow’s milk and their antimicrobial resistance. J Vet Res. 2019;63:93–97. doi: 10.2478/jvetres-2019-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegg PL. A 100-year review: mastitis detection, management, and prevention. J Dairy Sci. 2017;100:10381–10397. doi: 10.3168/jds.2017-13023. [DOI] [PubMed] [Google Scholar]
- Schmidt VS, Kaufmann V, Kulozik U, Scherer S, Wenning M. Microbial biodiversity, quality and shelf life of microfiltered and pasteurized extended shelf life (ESL) milk from Germany, Austria and Switzerland. Int J Food Microbiol. 2012;154:1–9. doi: 10.1016/j.ijfoodmicro.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Terre M, Maynou G, Bach A, Gauthier M. Effect of Saccharomyces cerevisiae CNCM I-1077 supplementation on performance and rumen microbiota of dairy calves. Prof Anim Sci. 2015;31:153–158. doi: 10.15232/pas.2014-01384. [DOI] [Google Scholar]
- Todorov SD, Dicks MT. Growth parameters influencing the production of Lactobacillus rhamnosus bacteriocins ST461BZ and ST462BZ. Ann Microbiol. 2005;55:283–289. [Google Scholar]
- Wall SK, Wellnitz O, Hernández Castellano LE, Ahmadpour A, Bruckmaier RM. Supraphysiological oxytocin increases the transfer of immunoglobulins and other blood components to milk during lipopolysaccharide-and lipoteichoic acid-induced mastitis in dairy cows. J Dairy Sci. 2016;99:9165–9173. doi: 10.3168/jds.2016-11548. [DOI] [PubMed] [Google Scholar]
- Walsh MC, Gardiner GE, Hart OM, Lawlor PG, Daly M, Lynch B, Richert BT, Radcliffe S, Giblin L, Hill C, Fitzgerald GF, Stanton C, Ross P. Predominance of a bacteriocin-producing Lactobacillus salivarius component of a five-strain probiotic in the porcine ileum and effects on host immune phenotype. FEMS Microbiol Ecol. 2010;64:317–327. doi: 10.1111/j.1574-6941.2008.00454.x. [DOI] [PubMed] [Google Scholar]
- Wellnitz O, Bruckmaier RM. The innate immune response of the bovine mammary gland to bacterial infection. Vet J. 2012;192:148–152. doi: 10.1016/j.tvjl.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Yuan K, Mendonca LG, Hulbert LE, Mamedova LK, Muckey MB, Shen Y, Elrod CC, Bradford BJ. Yeast product supplementation modulated humoral and mucosal immunity and uterine inflammatory signals in transition dairy cows. J Dairy Sci. 2015;98:3236–3246. doi: 10.3168/jds.2014-8469. [DOI] [PubMed] [Google Scholar]
- Zanello G, Berri M, Dupont J, Sizaret PV, Dlnca R, Salmon H, Meurens F. Saccharomyces cerevisiae modulates immune gene expressions and inhibits ETEC-mediated ERK1/2 and p38 signaling pathways in intestinal epithelial cells. PLoS ONE. 2011;6:e18573. doi: 10.1371/journal.pone.0018573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Shannon index of OTU level of milk microbiota on different treatments. Fig. S2. The effects of treatments on the distribution of milk bacterial community at the phylum level
Data Availability Statement
All sequences analyzed in this study can be accessed in Zenodo database (http://doi.org/10.5281/zenodo.3384327). The data supporting the conclusion of this article are included in this article.