Abstract
The 26S proteasome is a multisubunit ATP-dependent peptidase complex mediating most regulated protein degradation in eukaryotes. The proteasome undergoes several coordinated conformational changes during catalysis that activate it for substrate processing and functionally couple distinct enzymatic activities during substrate degradation. Understanding the impact of substrate interactions and individual ATP binding events on these conformational changes is currently a major bottleneck in the study of proteasome function. Here, we describe a simple biochemical reporter based on engineered disulfide crosslinking for measuring the conformational distribution of the Saccharomyces cerevisiae 26S proteasome. We demonstrate its use to investigate the impact of ATP analogs and proteasome inhibitors on proteasome conformational equilibria. This reporter allows simultaneous and rapid comparison of multiple treatments or conditions on the steady-state conformational distribution of the proteasome and can be readily extended to the study of other multisubunit complexes for which multiple conformational states are known at near-atomic resolution.
Keywords: 26S proteasome, disulfide crosslinking, multisubunit complex, conformation, conformational dynamics, proteasome inhibitors, ubiquitin
1. Introduction
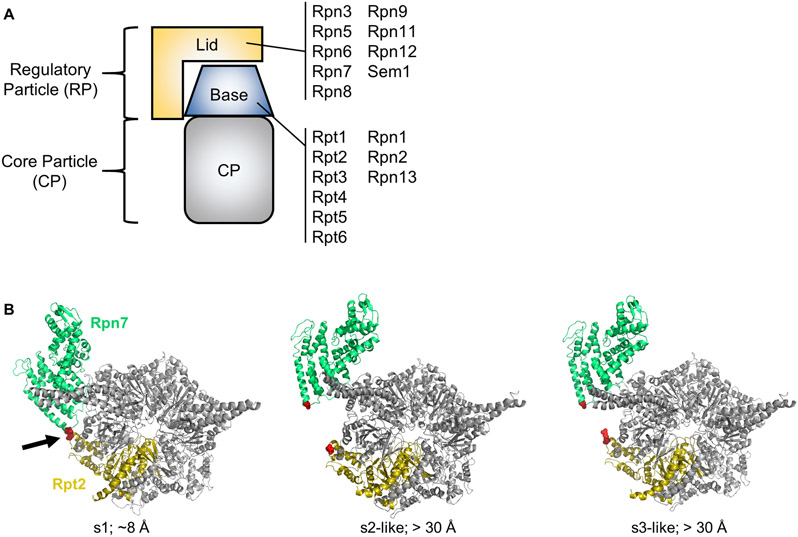
The 26S proteasome is a 2.5 MDa ATP-dependent peptidase complex responsible for most regulated intracellular degradation in eukaryotes (Bard, Goodall, Greene, Jonsson, Dong, & Martin, 2018; Finley, Chen, & Walters, 2016; Tomko & Hochstrasser, 2013). It consists of a barrel-shaped, proteolytic core particle (CP) capped on one or both ends by a 19S regulatory particle (RP) (Fig. 1A). The RP can be further subdivided into two subcomplexes, the lid and the base. The lid consists of Rpn3, 5-9, 11, 12, and Rpn15/Sem1 subunits. The base consists of non-ATPase subunits Rpn1, 2, and 13, and a hexameric ring of six ATPase subunits, Rpt1-Rpt6. Rpn1, Rpn13, and an additional subunit, Rpn10, serve as receptors for incoming substrates. During substrate processing, the Rpn11 lid subunit removes the polyubiquitin targeting signal (Verma, Aravind, Oania, McDonald, Yates, Koonin et al., 2002; Worden, Dong, & Martin, 2017; Yao & Cohen, 2002). The base unfolds the substrate and translocates it through the central pore of the ATPase ring to the peptidase active sites of the CP, where it is cleaved into short peptides.
Figure 1.
A. Cartoon depiction of the 26S proteasome. The subunit composition of the lid and base subcomplexes is shown. B. Rpn7-D123 and Rpt2-R407 are juxtaposed in the s1 conformation of the yeast 26S proteasome (red spheres indicated by arrow) (PDB 4CR2), but are distant in the s2-s6 states. In the middle panel, the s2 state (PDB 4CR3) is shown, but the s5 state is highly similar. In the right panel, the s3 state (PDB 4CR4) is shown, but the s4 and s6 states are highly similar. The approximate distances in Angstroms between the Rpn7-D123 and Rpt2-R407 alpha carbons is shown. Panel B is modified from (Eisele et al., 2018) with permission from Elsevier.
An onslaught of recent cryo-electron microscopy (cryo-EM) analyses have demonstrated that 26S proteasomes can adopt at least six conformational states, herein referred to as s1-s6 (Asano, Fukuda, Beck, Aufderheide, Forster, Danev et al., 2015; Aufderheide, Beck, Stengel, Hartwig, Schweitzer, Pfeifer et al., 2015; Bashore, Dambacher, Goodall, Matyskiela, Lander, & Martin, 2015; Chen, Wu, Lu, Ma, Lee, Yu et al., 2016; de la Pena, Goodall, Gates, Lander, & Martin, 2018; Dong, Zhang, Li, Wang, Zhu, Stoilova-McPhie et al., 2018; Eisele, Reed, Rudack, Schweitzer, Beck, Nagy et al., 2018; Guo, Lehmer, Martinez-Sanchez, Rudack, Beck, Hartmann et al., 2018; Haselbach, Schrader, Lambrecht, Henneberg, Chari, & Stark, 2017; Huang, Luan, Wu, & Shi, 2016; Luan, Huang, Wu, Mei, Wang, Xue et al., 2016; Matyskiela, Lander, & Martin, 2013; Schweitzer, Aufderheide, Rudack, Beck, Pfeifer, Plitzko et al., 2016; Sledz, Unverdorben, Beck, Pfeifer, Schweitzer, Forster et al., 2013; Unverdorben, Beck, Sledz, Schweitzer, Pfeifer, Plitzko et al., 2014; Wehmer, Rudack, Beck, Aufderheide, Pfeifer, Plitzko et al., 2017; Zhu, Wang, Yu, Ouyang, Lu, & Mao, 2018) (reviewed in (Bard, Goodall, et al., 2018)). These states can be broadly grouped into three categories: s1, s2-like, and s3-like. The s1 state appears to be the ground or resting state of the proteasome, and it is believed to be the major substrate-accepting state. In the s2-like states, the lid subcomplex undergoes a rotation that positions the deubiquitinase subunit Rpn11 directly over the pore of the ATPase ring of the base for substrate deubiquitination (Matyskiela et al., 2013; Sledz et al., 2013). Although these states appear primed for deubiquitination, a mismatch between the pore of the ATPase ring and the central channel of the CP suggests that they may not be fully competent for substrate degradation (Eisele et al., 2018; Unverdorben et al., 2014). Finally, in the s3-like states, the ATPase ring undergoes movements that align its pore with the central channel of the CP, providing an unobstructed passageway for processing and delivery of the substrate into the peptidase sites (Chen et al., 2016; de la Pena et al., 2018; Dong et al., 2018; Eisele et al., 2018; Luan et al., 2016; Matyskiela et al., 2013; Sledz et al., 2013; Unverdorben et al., 2014; Wehmer et al., 2017). Thus, the s3-like states represent actively processing proteasomes (de la Pena et al., 2018; Eisele et al., 2018; Matyskiela et al., 2013). Although these cryo-EM structures have been groundbreaking in terms of visualizing the substrate processing mechanism of the proteasome, the events and signals that promote each of these rearrangements remain poorly understood (Bard, Bashore, Dong, & Martin, 2018; Bard, Goodall, et al., 2018; Eisele et al., 2018).
The large-scale repositioning of the lid relative to the base that occurs as the proteasome shifts from the resting s1 state to the s2/s3-like states results in the formation and breakage of specific molecular contacts between lid and base subunits (for example, see Fig. 1B). Such dynamic contacts can serve as reporters to discriminate the resting s1 state from the s2/s3-like states, permitting measurement of the fraction of resting vs. activated proteasomes. Engineered disulfide crosslinking has previously been used to assay static interactions between adjacent subunits within the proteasome (Tomko, Funakoshi, Schneider, Wang, & Hochstrasser, 2010; Velichutina, Connerly, Arendt, Li, & Hochstrasser, 2004). Here, we apply this approach to dynamic subunit-subunit contacts to measure the relative fraction of s1 proteasomes. We demonstrate the use of this method for assessing the impact of ATP analogs and proteasome inhibitors on the conformational distribution of 26S proteasomes in yeast whole cell extracts (WCE). This approach is simple, rapid, and can be readily implemented for studying the conformational dynamics of other large protein complexes that adopt multiple conformational states.
2. Assessing the impact of ATP analogs on 26S proteasome conformational distribution
We have found that conformation-specific crosslinking performed in whole cell extracts accurately reports on the conformation of 26S proteasomes with little to no influence from proteasome assembly intermediates. This is because the abundances of assembly intermediates are typically very low compared to the abundance of 26S proteasomes. For initial analyses, we prefer yeast strains harboring cysteine substitutions at Rpn7 D123 and Rpt2 R407 for crosslinking because these mutations have no obvious impact on proteasome structure, assembly, or function, and the spacing of these residues increases substantially between the s1 and s2-/s3-like states (Fig. 1B). However, other substitutions can also be used if the cysteine substitutions are well tolerated and are appropriately juxtaposed in the s1 state but not the s2- or s3-like states. We have also successfully utilized Rpn6(T203C) – Rpt6(G387C) and Rpn7(A236C) – Rpt6(R339C) substitution pairs ((Eisele et al., 2018) and unpublished observations). These pairs do not obviously compromise proteasome structure or function and respond to ATP analogs similarly to the Rpn7(D123C) – Rpt2(R407C) pair. Site-directed mutagenesis to introduce cysteine substitutions is not covered here.
2.1. Materials
Unless listed below, all reagents are purchased from Millipore Sigma.
Zymolyase buffer (ZB): 1.2 M d-sorbitol, 50 mM Tris-HCl, pH 8.0, and 500 μM MgCl2 dissolved in deionized water. This reagent can be stored at room temperature indefinitely.
1 M dithiothreitol (DTT): dissolve in deionized water. This reagent can be stored in small aliquots for at least a year at −20°C.
20 mg/mL Zymolyase 20T (MP Biomedicals): dissolve Zymolyase 20T in ZB. This reagent can be stored in small aliquots at −20°C for at least a year, and working stocks can be kept refrigerated for at least three months.
Lysis buffer (LB): 50 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, and 5 mM MgCl2. This reagent can be stored at room temperature indefinitely. Nucleotides are added from a concentrated stock immediately before use.
Protein concentration assay reagent (such as Bio-Rad Protein Assay Reagent or similar)
100 mM adenosine triphosphate disodium salt hydrate (ATP): dissolve in 50 mM Tris-HCl, pH 7.5. Store as single-use aliquots at −80°C. This reagent is stable for at least six months.
100 mM adenosine 5′-(3-thiotriphosphate) tetralithium salt (ATPγS): dissolve in 50 mM Tris-Cl, pH 7.5. Store as single-use aliquots at −80°C. This reagent is stable for at least six months.
100 mM adenosine 5′-(β,γ-imido)triphosphate lithium salt hydrate (AMP-PNP): dissolve in 50 mM Tris-Cl, pH 7.5. Store as single-use aliquots at −80°C. This reagent is stable for at least six months.
10 mM CuCl2 prepared in deionized water. This reagent can be stored at room temperature indefinitely.
200 mM n-ethylmaleimide (NEM) prepared in 95% ethanol. This reagent can be stored at −20°C for up to one week.
100 mM EDTA disodium salt dihydrate prepared in deionized water, pH 8.0. This reagent can be stored at room temperature indefinitely.
5X non-reducing loading buffer: 300 mM Tris-Cl, pH 6.8, 10% SDS, 50% glycerol, and 0.04% bromophenol blue. This reagent can be stored at −20°C indefinitely.
Congenic yeast strains harboring appropriate single or double cysteine substitutions, such as RTY2091 (RPN7-D123C-6xGly-V5:kanMX6), RTY2099 (RPT2-R407C:natMX4), and RTY2112 (RPN7-D123C-6xGly-V5:kanMX6 RPT2-R407C:natMX4) (Eisele et al., 2018)
Antibodies against V5 tag (Life Technologies) and glucose-6-phosphate dehydrogenase (G6PD) (Millipore Sigma)
2.2. Equipment
Absorbance spectrophotometer and appropriate cuvettes
SDS-PAGE electrophoresis equipment (such as Bio-Rad Mini-PROTEAN3 system or similar)
Electrotransfer apparatus (such as Bio-Rad Mini Trans-Blot or similar)
Chemiluminescence imaging station (such as Bio-Rad Chemidoc MP or similar)
Refrigerated microcentrifuge accepting 1.5 mL Eppendorf tubes
Heat blocks or water baths set at 25°C and 100°C
2.3. Procedure
2.3.1. Cell growth, spheroplasting, and lysis
Inoculate 3 mL of YPD medium with each reporter strain from a colony on a freshly streaked YPD plate and incubate overnight at 30°C with aeration.
The following morning, use the overnight culture to inoculate 15 mL of YPD at an optical density of OD600 ≈ 0.2. Incubate at 30°C with aeration.
Monitor cell growth via OD600. At OD600 ≈ 1.0, harvest 10 mL of cell culture in a 15 mL disposable conical tube. Centrifuge cells at 4,000 x g for two minutes at room temperature.
Aspirate the supernatant and resuspend the cell pellet in 1 mL of deionized water. Transfer the cell suspension to a 1.5 mL centrifuge tube. Centrifuge the cells at 10,000 x g for 30 seconds to pellet cells.
Repeat the wash in step 4.
Aspirate the supernatant and resuspend cells in 100 μL ZB supplemented with 3 μL of 1 M DTT. Incubate at room temperature for 15 minutes.
Pellet cells at 10,000 x g for 30 seconds at room temperature and discard the supernatant.
Resuspend the pellet in 100 μL ZB and add 4 μL of 20 mg/mL Zymolyase 20T. Mix, and incubate at 30°C for 30 minutes with gentle agitation to keep cells in suspension.
Gently pellet the spheroplasts by centrifuging at 1,200 x g for one minute at room temperature (see Note 1). Resuspend the pellet in 200 μL ZB. Break up the pellet by gently stirring with a wide-bore pipette tip and slowly pipetting up and down. Pellet the cells at 1,200 x g for one minute at room temperature. Discard the supernatant.
Add 150 μL of ice-cold LB containing 2 mM of the desired nucleotide (see Note 2). Stir the pellet with a pipette tip to partially resuspend the cells.
Lyse the spheroplasts by vortexing three times for 30 seconds at top speed, leaving on ice for one minute in between vortexings.
Pellet cell debris and unbroken cells at 21,000 x g for 10 minutes at 4°C and transfer the supernatant to a fresh 1.5 mL centrifuge tube.
Perform a protein concentration assay on a small aliquot of the cleared WCE following manufacturer's instructions to determine the protein concentration. Adjust all sample concentrations to the least concentrated sample using LB containing the appropriate nucleotide (see Note 3).
2.3.2. Crosslinking of whole cell extracts
Transfer 50 μL of the normalized WCE from subheading 2.3.1, step 13 to a fresh Eppendorf tube containing 2.5 μL of 200 mM NEM. Add 5.25 μL of 100 mM EDTA, mix well, and store sample at −80°C until analysis. This sample will serve as the non-crosslinked control.
Transfer 50 μL of the remaining WCE from subheading 2.3.1, step 13 to a fresh tube. Add 1.25 μL of 10 mM CuCl2 to the lysate and mix gently but thoroughly to initiate crosslinking (see Note 4). Immediately place tubes in a 25°C water bath to maintain temperature (see Note 5).
After 10 minutes, add 2.5 μL of 200 mM NEM and 5.25 μL of 100 mM EDTA to terminate further crosslinking (see Note 6). Mix well.
For a control to demonstrate the reversibility of disulfide formation, add 1 μL of 1 M DTT to 50 μL of crosslinked WCE. Incubate sample for 10 minutes at room temperature before adding 2.5 μL of 200 mM NEM and 5.25 μL of 100 mM EDTA as in step 3 above.
Freeze samples at −80°C until analysis or proceed directly to subheading 2.3.3.
2.3.3. Non-reducing SDS-PAGE immunoblotting and quantitation of crosslinking
Thaw frozen samples on ice if necessary.
Add 17.5 μL of 5X non-reducing SDS loading buffer to each sample. Boil for exactly three minutes (see Note 7).
Load 15 μL of each sample onto a 10% Tris-glycine SDS-polyacrylamide gel. If reduced and non-reduced samples will be loaded on the same gel, it is best to leave an empty lane between them to avoid diffusion of DTT into non-reduced sample lanes during electrophoresis.
Subsequent steps of gel electrophoresis, protein electrotransfer to polyvinylidene difluoride (PVDF) membranes (Millipore Sigma), and immunodetection follow standard procedures. Perform immunoblotting against the V5 epitope on Rpn7 and against G6PD as a loading control.
Using image quantitation software, measure the volume of the crosslinked and uncrosslinked Rpn7 signal for each lane (see Note 8).
The volume of the crosslinked band divided by the sum of the crosslinked and uncrosslinked volumes, multiplied by 100, yields the percent subunit crosslinking.
3. Assessing the impact of proteasomal peptidase inhibitor bortezomib on 26S proteasome conformational distribution
Some proteasomal peptidase inhibitors bias the conformational distribution of 26S proteasomes toward the activated states (Haselbach et al., 2017). The methodology in subheading 2 above can be readily modified to assess the impact of such inhibitors on the conformational distribution of the 26S proteasome. We have accomplished this as described below via pre-treatment of whole cell extracts with the desired inhibitor(s) prior to crosslinking and analysis, but in principle cells could instead be treated with inhibitor prior to generation of cell extracts. By performing the crosslinking in the presence of the inhibitor and either ATP or a hydrolysis-resistant analog such as ATPγS or AMP-PNP, inhibitor bias toward or away from the s1 conformation can be assessed. Concurrent measurement of peptidase activity inhibition in an aliquot of the treated lysates using a fluorogenic substrate such as N-succinyl-leu-leu-val-tyr-7-amino-4-methylcoumarin (suc-LLVY-AMC) serves to confirm the inhibitor is bound to proteasomes under the crosslinking conditions (see Note 9). We demonstrate this approach using the FDA-approved proteasome inhibitor bortezomib as an example.
3.1. Materials
Materials described in Section 2.1
10 mM bortezomib (Sellekchem): dissolve in anhydrous dimethylsulfoxide (DMSO) and store in single-use aliquots at −80°C. This reagent is stable for at least six months.
2.5 mM suc-LLVY-AMC (R&D Systems): dissolve in anhydrous DMSO and store in single-use aliquots at −80°C. This reagent is stable for at least a year.
Black polystyrene 384-well plates (Corning Life Sciences)
3.2. Equipment
Equipment described in Section 2.2
Temperature-controlled multi-well fluorescence plate reader capable of reading 384-well plates and equipped with appropriate optics for imaging AMC fluorescence (BioTek Synergy H1 or similar)
3.3. Procedure
3.3.1. Disulfide crosslinking of vehicle- or bortezomib-treated whole cell extracts
Overnight yeast cultures are grown as in subheading 2.3.1 above, but are diluted into 30 mL of culture instead of 15 mL. One culture for DMSO vehicle and one for bortezomib treatment should be prepared.
Cell harvest is performed as in subheading 2.3.1 above, except 20 mL of culture is harvested instead of 10 mL.
Spheroplasting and lysis are performed as in subheading 2.3.1 above, except the reagent volumes listed in steps 6, 8, 9, and 10 are doubled.
Add 3 μL of DMSO or bortezomib (final concentration 100 μM) to 300 μL of the appropriate WCE and mix gently but thoroughly. Incubate on ice for 5 minutes.
Set ~160 μL of the treated WCE aside at 4°C for measurement of peptidase activity (subheading 3.3.2).
Using the remaining WCE, perform crosslinking and analysis as in subheadings 2.3.2 and 2.3.3 above.
3.3.2. Confirming proteasome inhibition via fluorogenic substrate cleavage assay
Transfer 50 μL of uncrosslinked, vehicle- or inhibitor-treated WCE from subheading 3.3.1, step 5 to each of three wells of a 384-well black polystyrene microplate.
Add 1 μL of 2.5 mM suc-LLVY-AMC to each well. Mix thoroughly by pipetting up and down without introducing bubbles.
Measure fluorescence resulting from cleavage of suc-LLVY-AMC at excitation 360 nm, emission 460 nm at 30°C every minute for sixty minutes using the multi-well fluorescence plate reader.
Average the three readings for each sample and plot the average fluorescence intensity in arbitrary units vs. time in minutes for the first twenty time points. Calculate the slope of the line to establish the peptidase rate in arbitrary fluorescence units per minute (see Note 10). Inhibition of peptidase activity by ≈ 90% is anticipated (some minimal fluorescence liberation by other cellular peptidases is typical).
4. Summary and Conclusion
Cryo-EM has been enormously powerful in documenting the conformational states of multisubunit complexes. However, it requires access to extremely expensive equipment and the preparation of highly pure samples. Further, data processing is time- and effort-intensive, resulting in limited throughput that hinders detailed comparative mechanistic studies. The crosslinking approach described here circumvents each of these limitations and thus provides a powerful means to rapidly and efficiently compare many samples in parallel. Further, because it relies on methods and equipment that are already available in most molecular biology labs, it can be broadly implemented to study any multisubunit protein complex for which high-resolution structures of multiple conformational states are known.
An additional advantage of this approach is that, due to the precise amino acid spacing necessary for disulfide bond formation (~5.9 Å between cysteine α carbons), smaller conformational changes can be measured than are possible with several other methods. For example, Förster resonance energy transfer is widely used to measure protein conformational changes, but is restricted to movements on the order of ~20-100 Å (Heyduk, 2002). In contrast, the Rpn6(T203C) – Rpt6(G387C) crosslinker pair described above discriminates a movement of ~9 Å between the s1 and s2/s3-like states (Eisele et al., 2018). Theoretically, any pair of residues whose α carbons are spaced ~5-6 Å apart in one conformation and > 10-15 Å in another can constitute a reporter pair upon cysteine substitution. In practice, these distance constraints are influenced by the local flexibilities of the protein regions in which the residues are located, and this should be considered when choosing sites for cysteine substitution.
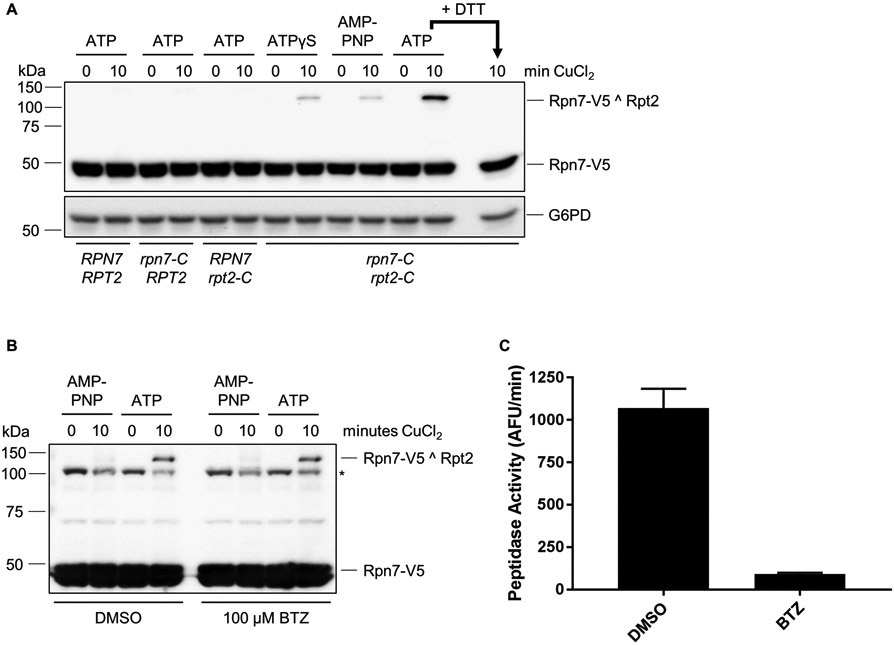
Typical results for ATP analogs (Fig. 2A) and for bortezomib treatment (Fig. 2B, C) are shown. Crosslinking efficiency of the Rpn7(D123C) – Rpt2(R407C) reporter typically ranges from < 3% in the presence of high concentrations of ATPγS or AMP-PNP to approximately 30% in the presence of ATP and is highly reproducible. Incomplete crosslinking in the presence of ATP analogs is likely due to a combination of incomplete conformational switching, imperfect alignment or spacing of cysteines for disulfide formation (see below), or spontaneous oxidation of cysteines prior to crosslinking. Generally, our efforts to further enhance the efficiency via extended crosslinking time courses or use of other oxidants have not been effective.
Figure 2.
A. Conformation-dependent crosslinking of Rpn7 and Rpt2. Crosslinking requires both engineered cysteines and is regulated by nucleotide. Crosslinking was induced in the presence of 2 mM of the indicated nucleotide. For the last lane, the WCE was pre-incubated with DTT to reduce disulfide crosslinks prior to loading. B. Bortezomib (BTZ) treatment does not influence Rpn7-Rpt2 crosslinking. Extracts from DMSO or BTZ (100 μM) treated cells were crosslinked as in (A) in the presence of 2 mM ATP or AMP-PNP as shown. Asterisk, nonspecific cross-reacting band occasionally seen with V5 antibody. C. Measurement of proteasomal peptidase activity in extracts from (B). Cell extract (20 μg) from DMSO- or BTZ-treated cells was incubated with 50 μM suc-LLVY-AMC for 60 minutes at 30°C and fluorescence from liberated AMC was measured. The rate of fluorescence liberation is shown. AFU, arbitrary fluorescence units. Error bars indicate standard deviations (N = 5). Panel A is modified from (Eisele et al., 2018) with permission from Elsevier.
Although the utility of this crosslinking-based approach has been established (Eisele et al., 2018), there are two potential limitations that should be carefully considered before use. First, the assay is not time-resolved. Specifically, it reports on all proteasomes cycling through the s1 state over the course of crosslinking. Thus, fast rearrangements may be difficult to capture and quantitate accurately via this approach. For this reason, we have had the greatest success using this approach in combination with chemicals or mutations that arrest particular enzymatic activities of the proteasome, as they often trap particular conformational states or distributions.
The second consideration is that disulfide formation is dependent on precise juxtaposition of cysteine residues. Although this means that crosslinking is highly selective for a particular conformation or subunit juxtaposition, it also means that small perturbations to the positioning of an engineered cysteine-containing subunit may alter the crosslinking efficiency due to relatively small changes in the positions of the engineered cysteines. The resultant loss or gain of crosslinking could thus be misidentified as global changes to the conformational state. For this reason, we recommend using multiple pairs of engineered cysteines, ideally present in different subunits and regions of the multisubunit complex, to validate the initial results whenever possible (Eisele et al., 2018).
Table 1:
Concentrations of key reagents used during extract preparation and crosslinking
| Reagent | Stock concentration | Final concentration |
|---|---|---|
| For spheroplasting | ||
| DTT | 1 M | 30 mM |
| Zymolyase 20T | 20 mg/mL | 0.8 mg/mL |
| For cell lysis | ||
| ATP | 100 mM | 2 mM |
| ATPγS | 100 mM | 2 mM |
| AMP-PNP | 100 mM | 2 mM |
| For crosslinking | ||
| CuCl2 | 10 mM | 250 μM |
| NEM | 200 mM | 10 mM |
| EDTA | 100 mM | 10 mM |
| DTT (reduced control only) | 1 M | 20 mM |
Acknowledgements
We thank members of the Tomko laboratory for critical reading of the manuscript. Work from our laboratory that led to the development of this approach was supported by NIH grant 1RO1GM118600 to R. J. T.
Footnotes
The cells are no longer protected by the cell wall and are now very fragile. Handle gently to prevent premature cell lysis.
Samples should be kept on ice at all times from this step forward to limit proteolysis and spontaneous oxidation of cysteines.
To ensure reproducible crosslinking efficiency both between samples and from experiment to experiment, we routinely dilute our extracts to 4 mg/mL prior to crosslinking.
Although we prefer CuCl2 as the mild oxidant to induce disulfide formation, many other commercially available oxidants can be used. Examples include tetrathionite and aqueous iodine, both of which have been used successfully to induce disulfide crosslinks in proteasome complexes (Snoberger, Brettrager, & Smith, 2018; Velichutina et al., 2004).
Some engineered cysteines are prone to spontaneous disulfide formation even in the absence of exogenous oxidant, which can confound interpretation of crosslinking results. This spontaneous oxidation can be minimized by incubating the samples on ice instead of 25°C during crosslinking. We have generally found that there is no need to extend the crosslinking time to accommodate for the decrease in temperature.
We have generally found that near-maximal crosslinking occurs in less than 5 minutes, and that nonspecific crosslinking increases over time. For this particular crosslink pair, 10 minutes consistently provides near-maximal site-specific crosslinking efficiency with minimal nonspecific crosslinking. If desired, a crosslinking time course can be used to optimize the results for this and other cysteine pairs.
Despite the presence of NEM and EDTA that should prevent further oxidation of free protein thiols, we have found that extensive boiling, as well as repeated boiling, of non-reducing SDS-PAGE samples leads to additional nonspecific disulfide formation over time, possibly due to incomplete reaction of NEM with free thiols. For this reason, we carefully control the boiling time and make aliquots of crosslinked samples prior to freezing if multiple analyses are planned.
It is critical to use images in which the signals for both the uncrosslinked and crosslinked subunits are unsaturated to ensure accurate calculation of crosslinking efficiency. Most modern biomedical imaging systems have very large dynamic ranges and automatically detect and alert the user to signal saturation. For this reason, such imagers are preferred over autoradiography film.
Bortezomib preferentially inhibits the chymotryptic-like activity of the proteasome. The fluorogenic substrate suc-LLVY-AMC reports on this chymotryptic-like activity. If inhibitors of the trypsin-like or caspase-like activities of the proteasome will be assayed instead, fluorogenic substrates suitable for those activities should be substituted for suc-LLVY-AMC.
It is important that peptidase assays are performed under conditions where substrate is in great molar excess of proteasomes to ensure a linear relationship between fluorescence liberation and time. If substantial deviation from linearity is observed over the first 20 minutes of the assay, then the concentration of the extracts used in the assay should be reduced.
References
- Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, & Baumeister W (2015). Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science, 347(6220), 439–442. doi: 10.1126/science.1261197 [DOI] [PubMed] [Google Scholar]
- Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, … Forster F (2015). Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A, 112(28), 8626–8631. doi: 10.1073/pnas.1510449112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JAM, Bashore C, Dong KC, & Martin A (2018). Deconvolution of substrate processing by the 26S proteasome reveals a selective kinetic gateway to degradation. BioRxiv. doi: 10.1101/359695 [DOI] [Google Scholar]
- Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, & Martin A (2018). Structure and Function of the 26S Proteasome. Annu Rev Biochem, 87, 697–724. doi: 10.1146/annurev-biochem-062917-011931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, & Martin A (2015). Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol, 22(9), 712–719. doi: 10.1038/nsmb.3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wu J, Lu Y, Ma YB, Lee BH, Yu Z, … Mao, Y. (2016). Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci U S A, 113(46), 12991–12996. doi: 10.1073/pnas.1614614113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pena AH, Goodall EA, Gates SN, Lander GC, & Martin A (2018). Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science. doi: 10.1126/science.aav0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Zhang S, W Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, … Mao Y (2018). Visualizing ATP-dependent substrate-processing dynamics of the human 26S proteasome at near-atomic resolution. BioRxiv. doi: 10.1101/419051 [DOI] [Google Scholar]
- Eisele MR, Reed RG, Rudack T, Schweitzer A, Beck F, Nagy I, … Sakata E (2018). Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell Rep, 24(5), 1301–1315 e1305. doi: 10.1016/j.celrep.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Chen X, & Walters KJ (2016). Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem Sci, 41(1), 77–93. doi: 10.1016/j.tibs.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, … Fernandez-Busnadiego R (2018). In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell, 172(4), 696–705 e612. doi: 10.1016/j.cell.2017.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbach D, Schrader J, Lambrecht F, Henneberg F, Chari A, & Stark H (2017). Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs. Nat Commun, 8, 15578. doi: 10.1038/ncomms15578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk T (2002). Measuring protein conformational changes by FRET/LRET. Curr Opin Biotechnol, 13(4), 292–296. [DOI] [PubMed] [Google Scholar]
- Huang X, Luan B, Wu J, & Shi Y (2016). An atomic structure of the human 26S proteasome. Nat Struct Mol Biol, 23(9), 778–785. doi: 10.1038/nsmb.3273 [DOI] [PubMed] [Google Scholar]
- Luan B, Huang X, Wu J, Mei Z, Wang Y, Xue X, … Wang F (2016). Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc Natl Acad Sci U S A, 113(10), 2642–2647. doi: 10.1073/pnas.1601561113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Lander GC, & Martin A (2013). Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol, 20(7), 781–788. doi: 10.1038/nsmb.2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer A, Aufderheide A, Rudack T, Beck F, Pfeifer G, Plitzko JM, … Baumeister W (2016). Structure of the human 26S proteasome at a resolution of 3.9 A. Proc Natl Acad Sci U S A, 113(28), 7816–7821. doi: 10.1073/pnas.1608050113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Forster F, & Baumeister W (2013). Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A, 110(18), 7264–7269. doi: 10.1073/pnas.1305782110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoberger A, Brettrager EJ, & Smith DM (2018). Conformational switching in the coiled-coil domains of a proteasomal ATPase regulates substrate processing. Nat Commun, 9(1), 2374. doi: 10.1038/s41467-018-04731-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ Jr., Funakoshi M, Schneider K, Wang J, & Hochstrasser M (2010). Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell, 38(3), 393–403. doi: 10.1016/j.molcel.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ Jr., & Hochstrasser M (2013). Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem, 82, 415–445. doi: 10.1146/annurev-biochem-060410-150257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unverdorben P, Beck F, Sledz P, Schweitzer A, Pfeifer G, Plitzko JM, … Forster F (2014). Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci U S A, 111(15), 5544–5549. doi: 10.1073/pnas.1403409111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina I, Connerly PL, Arendt CS, Li X, & Hochstrasser M (2004). Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO J, 23(3), 500–510. doi: 10.1038/sj.emboj.7600059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR 3rd, Koonin EV, & Deshaies RJ (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science, 298(5593), 611–615. doi: 10.1126/science.1075898 [DOI] [PubMed] [Google Scholar]
- Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, … Sakata E (2017). Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci U S A, 114(6), 1305–1310. doi: 10.1073/pnas.1621129114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden EJ, Dong KC, & Martin A (2017). An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Mol Cell, 67(5), 799–811 e798. doi: 10.1016/j.molcel.2017.07.023 [DOI] [PubMed] [Google Scholar]
- Yao T, & Cohen RE (2002). A cryptic protease couples deubiquitination and degradation by the proteasome. Nature, 419(6905), 403–407. doi: 10.1038/nature01071 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang WL, Yu D, Ouyang Q, Lu Y, & Mao Y (2018). Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome. Nat Commun, 9(1), 1360. doi: 10.1038/s41467-018-03785-w [DOI] [PMC free article] [PubMed] [Google Scholar]