Abstract
Genetic factors contribute to the risk for developing alcohol use disorder (AUD). In collaboration with the genetics company 23andMe, Inc., we performed a genome-wide association (GWAS) study of the Alcohol Use Disorder Identification Test (AUDIT), an instrument designed to screen for alcohol misuse over the past year. Our final sample consisted of 20,328 research participants of European ancestry (55.3% females; mean age = 53.8, SD = 16.1) who reported ever using alcohol. Our results showed that the ‘chip-heritability’ of AUDIT score, when treated as a continuous phenotype, was 12%. No loci reached genome-wide significance. The gene ADH1C, which has been previously implicated in AUD, was among our most significant associations (4.4 × 10−7; rs141973904). We also detected a suggestive association on chromosome 1 (2.1 × 10−7; rs182344113) near the gene KCNJ9, which has been implicated in mouse models of high ethanol drinking. Using LD score regression, we identified positive genetic correlations between AUDIT score and high alcohol consumption, and cigarette smoking. We also observed an unexpected positive genetic correlation between AUDIT and educational attainment, and additional unexpected negative correlations with BMI/obesity and attention-deficit/hyperactivity disorder (ADHD). We conclude that conducting a genetic study using responses to an online questionnaire in a population not ascertained for AUD may represent a cost-effective strategy for elucidating the etiology of AUD, in the context of alcohol consumption.
Keywords: Alcohol Use Disorder, alcohol-metabolizing enzymes, AUDIT, complex traits, genetic, GWAS
INTRODUCTION
The heritability of AUD and associated symptomatology such as high alcohol consumption has been estimated at ~50% by family and twin studies (Verhulst, Neale & Kendler 2015; Mbarek et al. Vink 2015), with a smaller proportion being attributable to additive effects of common genetic variation (e.g. 33% for AUD (Mbarek et al. 2015); 18% (Vrieze, McGue, Miller, Hicks & Iacono 2013) and 13% (Clarke et al. 2017) for alcohol consumption).
The search for specific genes that convey risk for AUD has been an active area of research for several decades. There have been numerous family-based linkage studies of AUD (Agrawal & Bierut 2012; Rietschel & Treutlein 2013; Enoch 2013; Edenberg & Foroud 2014), as well as candidate gene association studies. Robust linkage signals have been found near the cluster of aldehyde dehydrogenase (ALDH) genes on chromosome 12 (Wall, Luczak & Hiller-Sturmhöfel 2016), and alcohol dehydrogenase (ADH) genes on chromosome 4 (Long et al. 1998; Williams et al. 1999), a result that has also been identified by candidate gene association studies (Thomasson et al. 1991; Luo et al. 2006; Macgregor et al. 2009; van Beek, Willemsen, de Moor, Hottenga & Boomsma 2010; Li, Zhao & Gelernter 2011, 2012).
More recently, genome-wide association studies (GWAS) have been used to explore the genetic basis of AUD (Hart & Kranzler 2015; Tawa, Hall & Lohoff 2016). The most robust and replicated risk alleles in European, African American, and Asian ancestry populations map to alcohol-metabolizing enzyme genes on chromosome 4q22–23 and 12q24: ADH1B (Gelernter et al. 2014; Xu et al. 2015; Clarke et al. 2017), ADH1C (Treutlein et al. 2009; Edenberg et al. 2010; Frank et al. 2012; Gelernter et al. 2014; Clarke et al. 2017), ADH5 (Clarke et al. 2017), ADH7 (Park et al. 2013) and ALDH2 (Takeuchi et al. 2011; Yang et al. 2013; Park et al. 2013; Quillen et al. 2014; Jorgenson et al. 2017). More recent GWAS that target alcohol consumption rather than AUD have identified novel genes including KLB, which influence both high alcohol consumption in humans (Schumann et al. 2016; Clarke et al. 2017; Jorgenson et al. 2017) and ethanol preference in mice (Schumann et al. 2016).
In collaboration with the genetics company 23andMe, Inc., we performed a GWAS for alcohol misuse using the Alcohol Use Disorders Identification Test (AUDIT), a questionnaire developed to screen for alcohol misuse in the past year (Saunders et al. 1993). The estimated narrow-sense heritability of AUDIT dichotomous score is 60%, similar to the heritability of AUD (Mbarek et al. 2015). Self-reported AUDIT scores are predictive of future problematic drinking and higher AUD risk (Allen, Litten, Fertig & Babor 1997; Boschloo et al. 2010), with one study showing a high correlation (0.88) between AUDIT scores and AUD vulnerability (Bohn, Babor & Kranzler 1995). Indeed, high AUDIT scores (> 15) are consistent with a DSM-V diagnosis of severe AUD (Grant et al. 2015), perhaps because the AUDIT includes questions that are related to the criteria for AUD (e.g. DSM-V, criterion 2: “More than once wanted to cut down or stop drinking, or tried to, but couldn’t?” versus AUDIT, item 4: “How often during the last year have you found that you were not able to stop drinking once you had started?”). A previous GWAS of dichotomized AUDIT scores in 7,842 individuals in an unselected Dutch population did not reveal any significant associations (Mbarek et al. 2015). Here, to maximize power and to capture the dimensionality of alcohol misuse, we treated AUDIT scores from 20,328 research participants as a continuous trait rather than dichotomizing it by using a threshold score. We hypothesized that a GWAS for AUDIT scores might identify some of the same alleles that influence AUD, even though our cohort had relatively modest AUDIT scores.
MATERIALS AND METHODS
Sample
All participants included in the analyses were drawn from the customer base of 23andMe, Inc., a consumer genetics company. Participants provided informed consent and answered surveys online under a protocol approved by Ethical and Independent Review Services, an independent AAHRPP-accredited institutional review board (http://www.eandireview.com). We restricted the original sample (~25,000 individuals) to a set of unrelated participants of European ancestry (>97% as determined through an analysis of local ancestry [Durand, Do, Mountain & Macpherson 2014]; see Supplementary for additional details) for whom AUDIT data were available. Participants were not administered AUDIT if they reported that they never drank alcohol (N = 1,376; ‘Alcohol use’ questionnaire, Supplementary Table 1). The final number of participants included in the analysis was 20,328. Recruitment occurred over an approximately four-month period in 2015. Sociodemographic details are described in the Supplementary Table 1.
AUDIT scores
To evaluate alcohol misuse in the past year, participants completed the AUDIT (Saunders et al. 1993). We only included subjects who answered yes to the question “Have you ever in your life used alcohol” (i.e., “ever drinkers” vs “never drinkers”). The ten-item AUDIT questionnaire yields scores from 0 to 40. Since the scores were not normally distributed (by visual inspection, Figure 1), we used a log-10 transformation, which is frequently employed to approximate a normal distribution for AUDIT (Supplementary Table 2).
Figure 1.
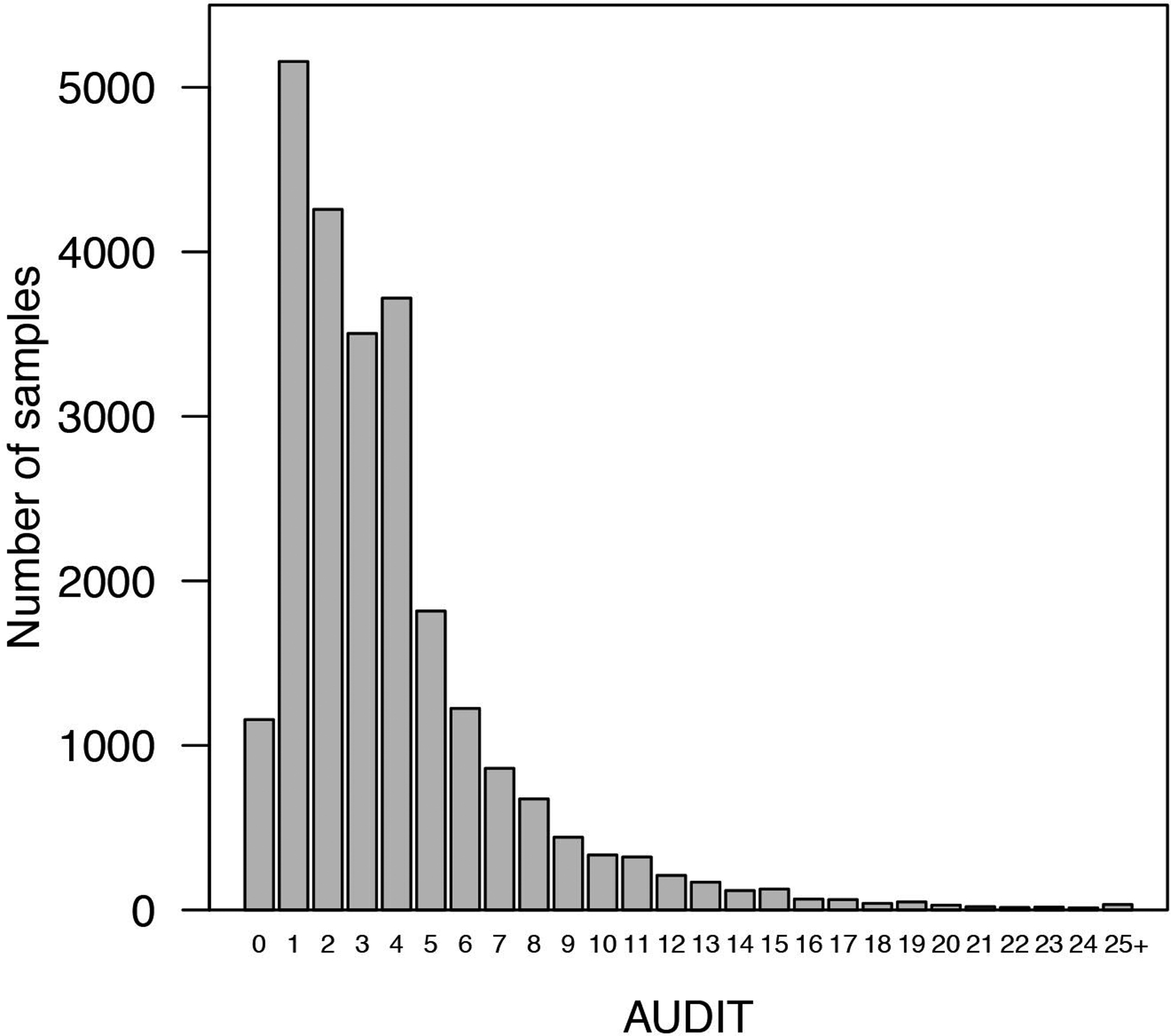
Distribution of AUDIT scores (prior to log transformation).
Genotyping, quality control and imputation
DNA extraction and genotyping were performed on saliva samples by CLIA-certified and CAP-accredited clinical laboratories of Laboratory Corporation of America. Samples were genotyped on 23andMe custom genotyping array platforms (Illumina HumanHap550+ Bead chip V1 V2, OmniExpress+ Bead chip V3, Custom array V4). Quality control of genetic variants and imputation were performed by 23andMe (see Supplementary Table 3). A full description of the methods have been reported elsewhere (Hyde et al. 2016; Lo et al. 2016).
Estimation of variance in AUDIT scores explained by the genotyped SNPs
To estimate the proportion of phenotypic variance explained (‘chip heritability’; hg2), we used a genomic restricted maximum likelihood (GREML) method implemented in Genetic Complex Trait Analysis (GCTA; Yang, Lee, Goddard & Visscher 2011). The GREML method estimates the proportion of variation in a phenotype that is due to all SNPs, and exploits the fact that genotypic similarity (i.e., “relatedness”, measured using genotyped SNPs) will be correlated with phenotypic similarity for heritable traits. Individual-level quality control was implemented, and distantly related individuals with pair-wise relationships were filtered at two thresholds (KIBS < 0.05 and KIBS < 0.03). We included age (inverse-normalized), self-reported sex (male/female), genotyping platform and top four principal components as covariates. GREML analyses were run using only directly genotyped SNPs.
Chip-heritability using LD Score Regression
We used a second method to measure chip heritability of AUDIT that is implemented by Linkage Disequilibrium Score Regression Coefficient (LDSC; Bulik-Sullivan et al. 2015b). To standardize the input file (GWAS summary statistics), we followed quality controls as implemented by the LDSC python software package. We used pre-calculated LD scores (“eur_w_ld_chr/” files (Finucane et al. 2015); MHC region excluded) for each SNP using individuals of European ancestry from the 1000 Genomes project, suitable for LD score analysis in European populations. We restricted the analysis to well-imputed SNPs: the SNPs were filtered to HapMap3 SNPs (International HapMap 3 Consortium et al. 2010), and were required to have a minor allele frequency (MAF) above 1%. InDels, structural variants, strand-ambiguous SNPs, and SNPs with extremely large effect sizes (χ2 > 80) were removed. In addition, this approach allowed us to distinguish between genomic inflation attributed to polygenic signal, from confounding biases such as population stratification or polygenicity (LD Score regression intercept > 1; Bulik-Sullivan et al. 2015b; Bulik-Sullivan et al. 2015a). As expected under polygenicity, we observed inflation of the median test statistic (Mean χ2 = 1.05), and adjusted for a genomic control inflation factor λ (the ratio of the observed median χ2 to that expected by chance) = 1.02. LD score intercept of 1.01 (SE = 0.01) suggested that deviation from the null was due to a polygenic structure rather than inflation due to population structure biases.
Genome-wide association analysis
For quality control of genotyped GWAS results, we removed SNPs with MAF of < 0.1%, a Hardy-Weinberg P <10−20 in Europeans, or a call rate of < 90%. We also removed SNPs that were only genotyped on the 23andMe V1 platform, due to limited sample size, and SNPs on chrM or chrY. Using trio data, we removed SNPs that failed a test for parent-offspring transmission; specifically, we regressed the child’s allele count against the mean parental allele count and removed SNPs with fitted β < 0.6 and P < 10−20 for a test of β < 1. We also tested genotyped SNPs for genotype date effects, and removed SNPs with P < 10−50 by ANOVA of SNP genotypes against a factor dividing genotyping date into 20 roughly equal-sized buckets. For imputed GWAS results, we removed SNPs with average r2 < 0.50 or minimum r2 < 0.30 in any imputation batch, as well as SNPs that had strong evidence of an imputation batch effect. The batch effect test is an F test from an ANOVA of the SNP dosages against a factor representing imputation batch; we removed results with P < 1 × 10−50. We also removed linear regression results for SNPs with MAF < 0.1% because tests of low frequency variants can be sensitive to violations of the regression assumption of normally distributed residuals. We performed association tests by linear regression assuming an additive model. We included age (inverse-normal transformed), sex, the top five principal components of genotype, and indicator variables for genotype platforms as covariates (Supplementary Table 4).
Phenotypic and genetic correlation analyses
We examined two distinct types of correlations: phenotypic correlations, where both variables were measured in the same individuals, and genetic correlations, where we used AUDIT data from this cohort in conjunction with summary statistics for GWAS conducted in other cohorts (Supplementary Table 9). The interpretation of these is different, since phenotypic correlations can be due to a combination of genetic and non-genetic factors, whereas genetic correlations measure only genetically driven correlations.
We used bivariate correlations to examine the direct phenotypic correlations between AUDIT and several variables of interest (age, gender, race, education, annual household), and to identify significant covariates for inclusion in GWAS analysis (Supplementary Table 5).
Using LDSC, we calculated genetic correlations (rg) between AUDIT and 30 other complex traits or diseases that have been previously associated with AUDIT or alcohol-related phenotypes, for which we had access to summary statistics. We used height as a control. References for the datasets used are identified in Supplementary Table 9. Files were standardized using the steps described in the section above (“Chip-heritability using LD Score Regression”). We did not constrain the intercepts in our analysis because the degree of sample overlap was unknown. We used False Discovery Rate (FDR) to correct for multiple testing (Benjamini & Hochberg 1995).
Query for expression quantitative trait loci (eQTL)
We queried eQTL evidence for our top (P < 10−7) GWAS SNPs using public online resources. We used the Genotype-Tissue Expression Portal (GTEx) to identify eQTLs associated with the SNPs; and the RegulomeDB (Boyle et al. 2012) to identify regulatory DNA elements in non-coding and intergenic regions of the genome in normal cell lines and tissues.
RESULTS
Demographics
Demographic data are shown in Supplementary Table 1. Mean age was 53.8 years (SD = 16.1), and 55.3% were women. The annual household income ranged from less than $14,999 (13.5%) to greater than $75,000 (21.5%), and the mean years of education completed was 16.8 (SD = 2.6). About half of the participants (49.3%) were married/partnered. Participants showed low to moderate alcohol use, average frequency of alcohol use was ~9 days per month (mean = 8.78, SD = 9.82); during the period of heaviest lifetime use, subjects reported reaching an average of 13.78±10.96 days over a 30-day period. Over the prior year, 78% of the participants reported drinking 1 or 2 drinks on a single day, and only 28% reported drinking more than 6 drinks on one occasion. Also over the prior year, 92% of the participants were able to stop drinking once they started, and 85% drank alcohol without feeling guilt or remorse.
AUDIT scores
The distribution of the AUDIT scores is shown in Figure 1 and Supplementary Table 2. 472 research participants had AUDIT scores > 15, and 127 had scores > 20, with an average AUDIT response of 3.84, (SD= 3.47). The phenotypic correlations between AUDIT and demographic variables measured in the same cohort are shown in the Supplementary Table 4. Age and sex were negatively correlated with AUDIT scores; younger individuals and males showed higher AUDIT scores (r = −0.15, P < 0.0001; r = −0.17, P < 0.0001, respectively). BMI was negatively associated with AUDIT (r = −0.07, P < 0.0001), whereas household income was positively correlated with AUDIT (r = 0.07, P < 0.0001). AUDIT scores were slightly higher in unmarried individuals (r = −0.02, P = 0.013) but we did not observe significant correlations with years of education (r = 0.01, P = 0.085). Measures of alcohol use were positively correlated with AUDIT scores (r = 0.50–0.52, P < 0.0001).
Chip-heritability estimates
We estimated the chip-heritability of AUDIT at 12.05% (± 1.91%, P = 2.70 × 10−11, GCTA; 8.8% ± 2.50%, LDSC), which is lower than previous chip-heritability estimates based on dichotomized AUDIT data (30% ±12%; Mbarek et al. 2015), and considerably lower than twin based heritabilities of alcohol abuse, dependence and alcoholism (~50%, (Goldman, Oroszi & Ducci 2005; Enoch 2013), as expected (e.g. Plomin, Haworth, Meaburn, Price & Davis 2013).
GWAS of AUDIT
The Manhattan and quantile-quantile (QQ) plots for AUDIT are shown in Figure 2 and Supplementary Figure 4, respectively. The most significant association was at rs182344113, located on chromosome 1 (P = 2.10 × 10−7; β = 0.168, SE = 0.03; MAF = 0.002; Supplementary Fig. 1). The association was in an intergenic region of the gene PIGM, and near KCNJ9 (GIRK3), which has been implicated in preclinical models of ethanol sensitivity. G-protein–gated inwardly rectifying potassium (GIRK) channels, which are coupled to GABA-B receptors, can be activated by ethanol (Aryal, Dvir, Choe & Slesinger 2009; Bodhinathan & Slesinger 2013). Interestingly, Kcnj9 knock-out mice exhibit excessive alcohol drinking (Dere et al. 2015).
Figure 2.
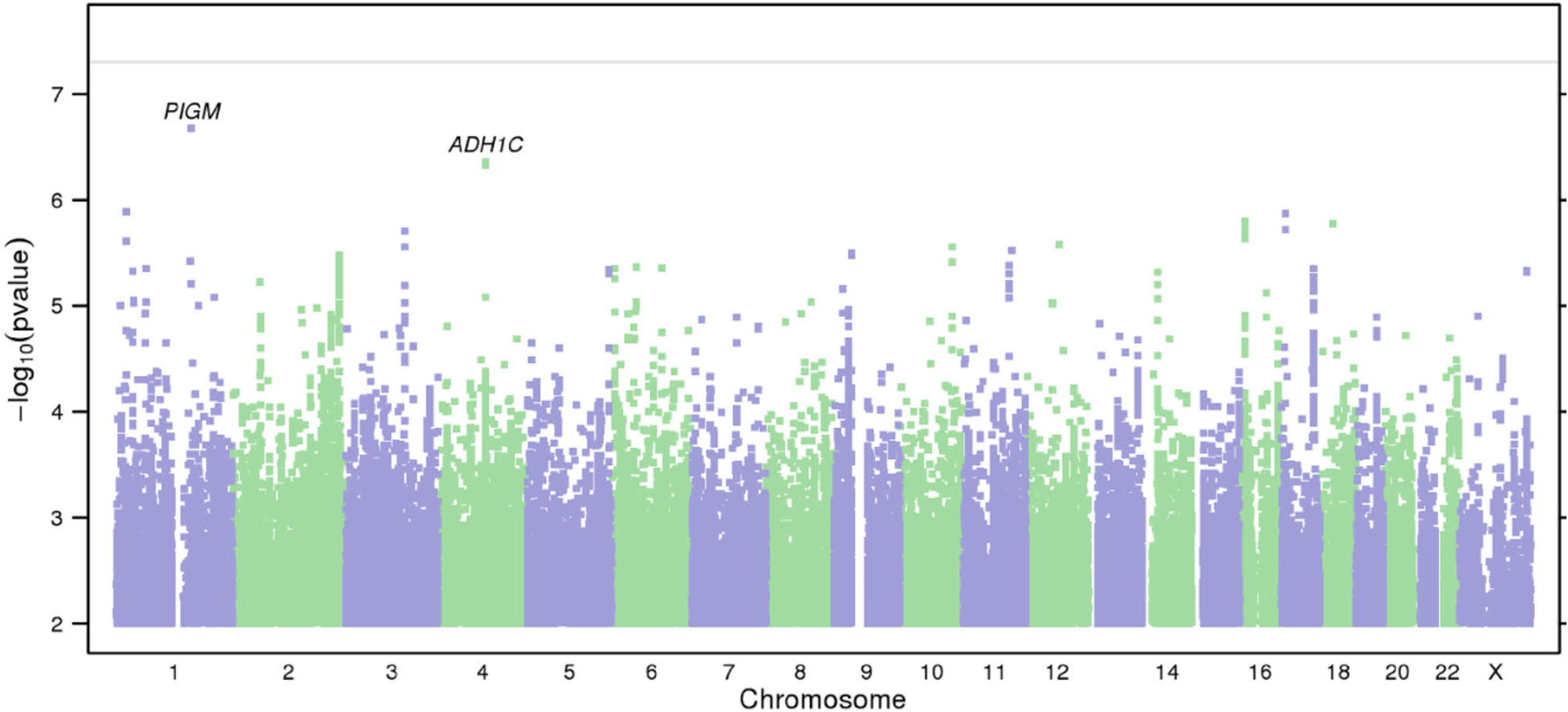
Results of GWAS on AUDIT. (a) Manhattan plot of GWAS results indicating the strongest associations between the 22 autosomes, X chromosome, and AUDIT. Line denotes genome-wide significance (P < 5 × 10−8).
Several other SNPs also showed suggestive associations (Supplementary Table 6), including rs141973904 (P = 4.40 × 10−7, β = −0.05, SE = 0.01; MAF = 0.02; Supplementary Fig. 2) in an intron of ADH1C, replicating previous findings for that same SNP in a GWAS of alcohol consumption in males (Clarke et al. 2017), and broadly consistent with numerous previous genetic studies of AUD (Thomasson et al. 1991; Edenberg 2007; Frank et al. 2012; Biernacka et al. 2013).
Another suggestive association was at rs8059260 (P = 1.6 × 10−6, β = 0.017, SE = 0.004; MAF = 0.160; Supplementary Fig. 3), which is near the first exon of CLEC16A. Using the Genotype-Tissue Expression Portal (GTEx) database, we identified a cis- expression quantitative trait loci (eQTLs) for CLEC16A that co-localized with rs8059260 (r2 > 0.79; see Supplementary Table 7). We also found evidence of regulatory elements associated with rs8059260 using the RegulomeDB (Boyle et al. 2012; Supplementary Table 7).
Previously studied candidate genes
Our results did not strongly support any of the previously published candidate gene studies of AUD (reviewed in Bühler et al. 2015; Supplementary Table 8); various differences including the distinction between AUD and AUDIT, demographics characteristics and especially the low prevalence of AUD in our cohort could partially account for the lack of replication.
Genetic correlations
LD score regression (Bulik-Sullivan et al. 2015b) showed a genetic overlap between AUDIT and numerous traits (Fig. 3 and Supplementary Table 9). Both alcohol consumption and AUD showed the steepest correlations with AUDIT score (rg = 0.68 for both), while the better powered alcohol consumption traits (N = 70,460; N = 112,117) yielded a significant result (P = 3.40 × 10−3; P = 9.01 × 10−10) the P value for the genetic correlation with AUD (N = 7,280) fell just short of nominal significance (P = 6.41 × 10−2). We detected a significant negative genetic correlation between AUDIT and ADHD (rg = −0.29; P = 1.43 × 10−3). We observed a positive genetic correlation between AUDIT and lifetime tobacco use (rg = 0.42; P = 1.52 × 10−3). Unexpectedly, we identified a positive correlation between AUDIT and years of education (rg = 0.27; P = 3.14 × 10−5), college attainment (rg = 0.26; P = 8.11 × 10−3) and childhood IQ (rg = 0.42; P = 6.26 × 10−3). Also surprisingly, AUDIT was negatively genetically correlated with BMI (rg = −0.25; P = 1.48 × 10−4) and adulthood obesity (rg = −0.23; P = 2.06 × 10−3). Height, which is not strongly influenced by individual behavior and thus can be viewed as a negative control, was not genetically correlated with AUDIT (rg = 0.02; P = 7.12 × 10−1).
Figure 3.
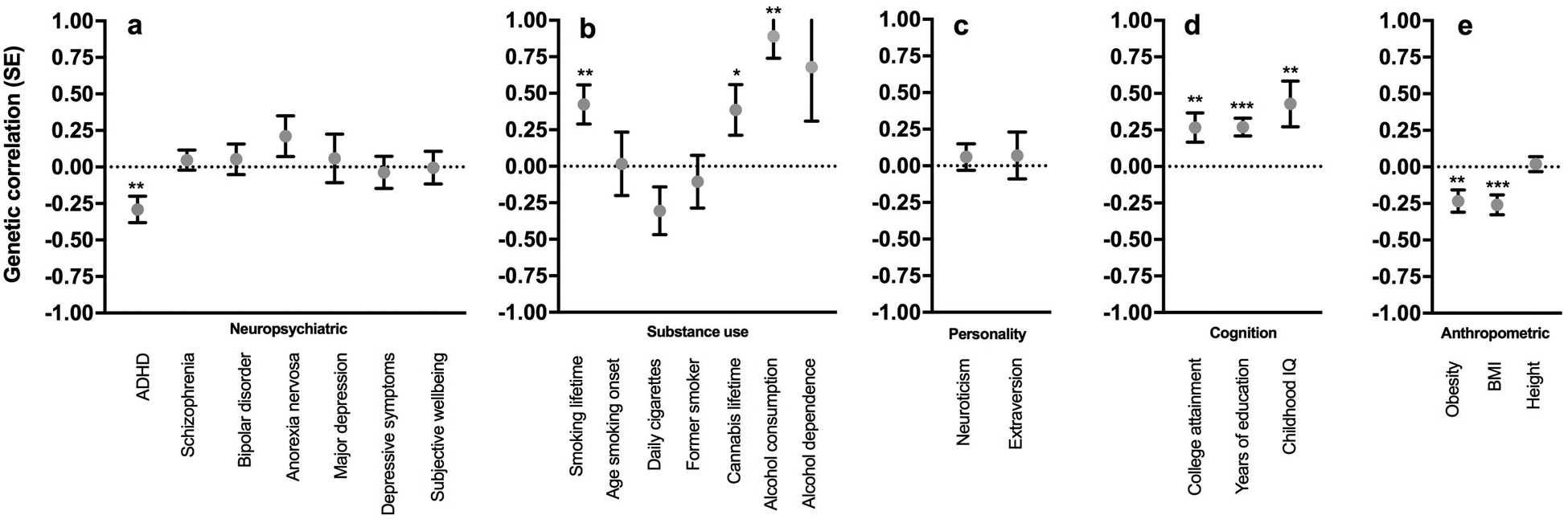
Genetic correlations between AUDIT and several traits measured in independent cohorts as described in the Supplementary Table 9: (a) neuropsychiatric, (b) smoking, (c) personality, (d) cognition, (e) anthropomorphic. * P < 0.05, ** P < 0.01, *** P < 0.0001.
DISCUSSION
With over 20,000 research participants, ours is by far the largest genetic study of AUDIT. By using a self-report measure of alcohol misuse, as opposed to recruiting a clinically-diagnosed population, we were able to rapidly and inexpensively ascertain a large number of participants. We identified rs141973904 (Supplementary Fig. 2) in the ADH cluster on chromosome 4q23, which has been previously associated with AUD (Edenberg et al. 2010; Frank et al. 2012; Gelernter et al. 2014; Treutlein et al. 2009). The same SNP has recently been associated (P = 1.22 × 10−15) with alcohol consumption using 53,089 males of European ancestry (Clarke et al. 2017). Furthermore, the most associated signal, rs182344113, which resides near the KCNJ9 (GIRK3) gene, was unknown, and is consistent with mouse studies of the homologous gene. The signal at rs182344113 was not significant and it will have to be replicated. We also identified a number of genetic correlations that have behavioral precedents, such as lifetime tobacco use, and several others that were unexpected, including lower BMI and obesity rates, and higher education. We found that AUDIT was genetically correlated and alcohol consumption, but fell out of significance for AUD, suggesting that a non-clinical population can be used as an alternative approach to study the genetics of AUD in the context of alcohol consumption.
In the absence of any GWAS significant results, we briefly discuss a few notable trend level results. We identified nominal associated variants in alcohol metabolizing genes. Genes influencing pharmacokinetics have previously been identified through linkage, candidate gene and genome-wide association studies for AUD and related traits (reviewed in Tawa et al. 2016). The most robust signal was located in the ADH1C gene, which contributes to ethanol oxidation. This signal was also identified in earlier GWAS studies for alcohol consumption (Clarke et al. 2017) and AUD status in both European (Clarke et al. 2017; Frank et al. 2012) and African American (Gelernter et al. 2014) populations, suggesting that pharmacokinetic factors are an important contributor to differences in both AUDIT score and AUD.
In addition to SNPs in the alcohol metabolizing genes, linkage and candidate gene studies have identified the GABRA2, OPRM1, DRD2 and ANNK1 genes, as candidate genes associated with AUD phenotypes (Bühler et al. 2015). However, we did not find robust signals for any of them (Supplementary Table 8), suggesting that previous studies may have overestimated the effects of these genes, or that these genes are associated with AUD but not AUDIT.
The strongest association we observed resides near KCNJ9; the frequency of the implicated imputed allele was very low (MAF = 0.002), suggesting the need for caution. The power to detect a significant association at this allele frequency was of XX. KCNJ9 encodes one of the G protein-activated inwardly rectifying K+ channels (GIRK3), which are expressed in the brain (Koyrakh et al. 2008), and can be directly activated by ethanol (Herman et al. 2015), even at low concentrations. In humans, two linkage studies have mapped this region for AUD (Hill et al. 2004), age of onset of drinking, harm avoidance, and novelty seeking (Dick et al. 2002). Additionally, DNA methylation levels of CpG in the promoter region of the GRIK3 gene showed altered expression in postmortem prefrontal cortex tissue of male alcoholics (Wang, Xu, Zhao, Gelernter & Zhang 2016). In mice, Kcnj9 also harbors a QTL for a variety of alcohol-related behaviors, including: ethanol preference (Tarantino, McClearn, Rodriguez & Plomin 1998), ethanol aversion (Risinger & Cunningham 1998), acute sensitivity to ethanol (Tipps, Raybuck, Kozell, Lattal & Buck 2016), and hypersensitivity to ethanol withdrawal (Kozell, Walter, Milner, Wickman & Buck 2009). Mice lacking GIRK3 in the brain have elevated alcohol drinking, without affecting the sensitivity to ethanol intoxication (Tipps, Raybuck, Kozell, Lattal & Buck 2016). Collectively, these results could provide an example of convergent results from humans and mice; however, until this non-significant observation is replicated it should be viewed with caution.
We hypothesized that the genetic risk for AUD is likely to overlap with numerous traits relevant to addiction and psychiatric phenotypes, based on previous epidemiological data (Compton, Thomas, Stinson & Grant 2007), twin studies (Kendler, Heath, Neale, Kessler & Eaves 1993; Pickens, Svikis, McGue & LaBuda 1995; Knopik, Heath, Bucholz, Madden & Waldron 2009) and recent genetic correlations between alcohol consumption and neuropsychiatric traits (Clarke et al. 2017). We showed positive genetic correlations between AUDIT and lifetime cigarette smoking, as previously observed between alcohol consumption and daily cigarettes, and tobacco initiation (Vink et al. 2014; Nivard et al. 2016). We also observed shared genetic architecture across AUDIT and other alcohol-related traits from independent cohorts: alcohol consumption (Schumann et al. 2016; Clarke et al. 2017), and AUD diagnosis (P = 0.062; Gelernter et al. 2014). With regards to other psychiatric traits, we found a negative correlation with ADHD; but there were no other significant genetic correlations between AUDIT and psychiatric traits. This result is generally consistent with a recent GWAS by Clarke et al (2017), where they did not observe strong correlations between alcohol consumption and psychiatric conditions.
Unexpectedly, we found positive genetic correlations between AUDIT and years of education, college attainment and childhood IQ. This association was suggestive for the within-sample phenotypic correlation (Supplementary Table 5) and was significant for the genetic correlation (Supplementary Table 9). Consistent with this finding, Clarke et al (2017) reported that college attainment and years of education were positively genetically correlated with alcohol consumption in females but not males.
Also unexpectedly, we observed negative genetic correlations between AUDIT and BMI and obesity, consistent with Clarke et al (2017). We also observed a phenotypic correlation between high AUDIT scores and low BMI. Previous studies have shown both positive and negative phenotypic correlations between alcohol use and BMI and obesity (Breslow & Smothers 2005; Tolstrup et al. 2005; Sobczyk-Kopciol et al. 2016), which may reflect differences in the populations used. The generalizability and biological meaning of these observations will require further research.
Our heritability estimate (12% ± 1.91) was lower the previous estimate by Mbarek et al (30% ± 12; 2015). While we cannot provide a definitive explanation for this discrepancy, we note that heritability estimates are not expected to be consistent across different populations. Differences between the two populations include geographic location (Dutch vs. US), the dichotomization of the phenotype by Mbarek et al (2015), heavier alcohol use in Mbarek’s cohort and limited AUDIT scores > 15 in our study.
Our study is not without limitations. Cumulative AUDIT scores reflect two distinct constructs: one measuring alcohol consumption and another measuring alcohol-related problems; thus, AUDIT scores may conflate multiple genetic signals (Bergman & Källmén 2002; Shevlin & Smith 2007). When we split AUDIT scores in three domains of consumption (items 1–3; AUDIT-C), dependence (items 4–6; AUDIT-D) and hazardous use (items 7–10; AUDIT-H), we observed higher scores for the domain of alcohol use (AUDIT-C: 2.96 ± 1.95; AUDIT-D: 0.203 ± 0.76; AUDIT-H: 0.665 ± 1.513). Our study may be tagging genetic risk for high quantity/frequency of alcohol consumption, as shown by the high genetic correlation with other alcohol-consumption traits, but may not overlap with other GWAS of AUD. In addition, our study focused on a cohort with relatively low levels of alcohol use; the unexpected positive genetic correlation between AUDIT and educational attainment, and the negative genetic correlation between AUDIT and both BMI/obesity and ADHD, may not generalize to cohorts with higher levels of alcohol use (Goldman et al. 2005) or AUD populations. Indeed, the selection of the 23andMe cohort (e.g. highly educated, with high socioeconomic background), or that of similar cohorts recruited from the general population such as UK Biobank, could have induced collider bias (Munafo, Tilling, Taylor, Evans & Smith 2017), underlying some of the paradoxical associations between AUDIT scores in our study, and high alcohol consumption in Clarke et al. (2017), and education/obesity. Another limitation of this study is the reliance on self-reported alcohol consumption, which may have induced biases and result in a more heterogeneous sample (Agrawal et al. 2012). Furthermore, AUDIT explicitly asks about alcohol use in the past year (i.e. state rather than trait), and this temporal specificity is not optimal for a genetic study. Nevertheless, AUDIT scores have a high correlation (r2=0.88) with scores on the Michigan Alcohol Screening Test (MAST), which assesses lifetime risk for AUDs, indicating that the temporal specificity of AUDIT may have a limited impact (Bohn, Babor & Kranzler 1995).
Although we used a unique and potentially powerful technique to examine the genetic basis of alcohol misuse, we recognize that alcohol consumption, misuse and dependence are influenced by numerous factors, both genetic and environmental (e.g. availability of alcohol, social norms, laws, psychosocial and personality factors, expectancies, health factors). Further, individuals vary in susceptibility at every stage of alcohol use from initiation to severe dependence, including the continued use after a first drink, the direct subjective and behavioral effects of the drug, withdrawal severity, tolerance and susceptibility to relapse, among others. Genetic factors are likely to influence variability at each of these stages, but different stages may be influenced by different sources of genetic variability. Thus, whereas we have investigated genetic variance related to an intermediate outcome measure of alcohol misuse (as measured by the AUDIT), it remains to be determined exactly how genetic sources of variation influence alcohol consumption. This ambiguity limits our ability to elucidate the underlying molecular mechanisms identified by GWAS of alcohol use and abuse.
Nonetheless, unlike studies of disease traits, which require careful diagnosis and ascertainment, we rapidly obtained a large cohort for which genotype data were available. We replicated a previously identified signal (ADH1C), and identified a novel GWAS signal (near KCNJ9) that has preclinical correlates. Our approach shows that genetic studies of AUDIT in community-based samples are an economical and effective alternative to rigorously diagnosed AUD cohorts that can nevertheless be used to gain insight into the biology of AUD, particularly aspects of alcohol drinking, and comorbid psychopathology.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the 23andMe research participants and employees for making this work possible. J.M.’s contributions were partially supported by the Peter Boris Chair in Addictions Research. S.S-R is supported by the Frontiers of Innovation Scholars Program (FISP) and the Interdisciplinary Research Fellowship in NeuroAIDS (IRFN).
Footnotes
DISCLOSURE
Pierre Fontanillas, Sarah L. Elson, and members of the 23andMe Research Team are employees of 23andMe, Inc. The opinions and assertions expressed herein are those of the author(s) and do not necessarily reflect the official policy or position of the Uniformed Services University or the Department of Defense.
Supplementary Information with nine additional tables are available on the Addiction Biology website
REFERENCES
- Agrawal A & Bierut LJ (2012) Identifying genetic variation for alcohol dependence. Alcohol Res Curr Rev 34:274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Freedman ND, Cheng Y-C, Lin P, Shaffer JR, Sun Q, Taylor K, Yaspan B, Cole JW, Cornelis MC, DeSensi RS, Fitzpatrick A, Heiss G, Kang JH, O’Connell J, Bennett S, Bookman E, Bucholz KK, Caporaso N, Crout R, Dick DM, Edenberg HJ, Goate A, Hesselbrock V, Kittner S, Kramer J, Nurnberger JI, Qi L, Rice JP, Schuckit M, van Dam RM, Boerwinkle E, Hu F, Levy S, Marazita M, Mitchell BD, Pasquale LR, Bierut LJ & GENEVA Consortium (2012) Measuring alcohol consumption for genomic meta-analyses of alcohol intake: opportunities and challenges. Am J Clin Nutr 95:539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguado C, Colón J, Ciruela F, Schlaudraff F, Cabañero MJ, Perry C, Watanabe M, Liss B, Wickman K & Luján R (2008) Cell type-specific subunit composition of G protein-gated potassium channels in the cerebellum. J Neurochem 105:497–511. [DOI] [PubMed] [Google Scholar]
- Allen J, Litten R, Fertig J & Babor T (1997) A Review of Research on the Alcohol Use Disorders Identification Test (AUDIT). Alcohol Clin Exp Res 21:613–619. [PubMed] [Google Scholar]
- Aryal P, Dvir H, Choe S & Slesinger PA (2009) A discrete alcohol pocket involved in GIRK channel activation. Nat Neurosci 12:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beek JHDA, Willemsen G, de Moor MHM, Hottenga JJ & Boomsma DI (2010) Associations between ADH gene variants and alcohol phenotypes in Dutch adults. Twin Res Hum Genet Off J Int Soc Twin Stud 13:30–42. [DOI] [PubMed] [Google Scholar]
- Benjamini Y & Hochberg MT (1995) Controlling The False Discovery Rate - A Practical And Powerful Approach To Multiple Testing. J R Stat Soc 57:289–300. [Google Scholar]
- Bergman H & Källmén H (2002) Alcohol use among Swedes and a psychometric evaluation of the alcohol use disorders identification test. Alcohol Alcohol Oxf Oxfs 37:245–251. [DOI] [PubMed] [Google Scholar]
- Biernacka JM, Geske JR, Schneekloth TD, Frye MA, Cunningham JM, Choi D-S, Tapp CL, Lewis BR, Drews MS, L Pietrzak T, Colby CL, Hall-Flavin DK, Loukianova LL, Heit JA, Mrazek DA & Karpyak VM (2013) Replication of genome wide association studies of alcohol dependence: support for association with variation in ADH1C. PloS One 8:e58798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodhinathan K & Slesinger PA (2013) Molecular mechanism underlying ethanol activation of G-protein-gated inwardly rectifying potassium channels. Proc Natl Acad Sci 110:18309–18314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn MJ, Babor TF & Kranzler HR (1995) The Alcohol Use Disorders Identification Test (AUDIT): validation of a screening instrument for use in medical settings. J Stud Alcohol 56:423–432. [DOI] [PubMed] [Google Scholar]
- Boschloo L, Vogelzangs N, Smit J, van den Brink W, Veltman D, Beekman A & Penninx B (2010) The performance of the Alcohol Use Disorder Identification Test (AUDIT) in detecting alcohol abuse and dependence in a population of depressed or anxious persons. J Affect Disord 126:441–446. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM & Snyder M (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow RA & Smothers BA (2005) Drinking patterns and body mass index in never smokers: National Health Interview Survey, 1997–2001. Am J Epidemiol 161:368–376. [DOI] [PubMed] [Google Scholar]
- Bühler K-M, Giné E, Echeverry-Alzate V, Calleja-Conde J, de Fonseca FR & López-Moreno JA (2015) Common single nucleotide variants underlying drug addiction: more than a decade of research. Addict Biol 20:845–871. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, Duncan L, Perry JRB, Patterson N, Robinson EB, Daly MJ, Price AL & Neale BM (2015a) An atlas of genetic correlations across human diseases and traits. Nat Genet 47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL & Neale BM (2015b) LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T-K, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ & McIntosh AM (2017) Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112,117). Molecular Psychiatry: doi: 10.1038/mp.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton WM, Thomas YF, Stinson FS & Grant BF (2007) Prevalence, correlates, disability, and comorbidity of DSM-IV drug abuse and dependence in the United States: results from the national epidemiologic survey on alcohol and related conditions. Arch Gen Psychiatry 64:566–576. [DOI] [PubMed] [Google Scholar]
- Dere E, Winkler D, Ritter C, Ronnenberg A, Poggi G, Patzig J, Gernert M, Müller C, Nave K-A, Ehrenreich H & Werner HB (2015) Gpm6b deficiency impairs sensorimotor gating and modulates the behavioral response to a 5-HT2A/C receptor agonist. Behav Brain Res 277:254–263. [DOI] [PubMed] [Google Scholar]
- Dick DM, Nurnberger J, Edenberg HJ, Goate A, Crowe R, Rice J, Bucholz KK, Kramer J, Schuckit MA, Smith TL, Porjesz B, Begleiter H, Hesselbrock V & Foroud T (2002) Suggestive linkage on chromosome 1 for a quantitative alcohol-related phenotype. Alcohol Clin Exp Res 26:1453–1460. [DOI] [PubMed] [Google Scholar]
- Durand EY, Do CB, Mountain JL & Macpherson JM (2014) Ancestry Composition: A Novel, Efficient Pipeline for Ancestry Deconvolution. bioRxiv:010512. [Google Scholar]
- Edenberg HJ (2007) The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health J Natl Inst Alcohol Abuse Alcohol 30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ & Foroud T (2014) Chapter 32 - Genetics of alcoholism In: Handbook of Clinical Neurology. Alcohol and the Nervous System, E.V.S. and Pfefferbaum A (ed). Elsevier; pp. 561–571. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B & Foroud T (2010) Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res 34:840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M-A (2013) Genetic influences on the development of alcoholism. Curr Psychiatry Rep 15:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, Ripke S, Day FR, Purcell S, Stahl E, Lindstrom S, Perry JRB, Okada Y, Raychaudhuri S, Daly MJ, Patterson N, Neale BM & Price AL (2015) Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Mössner R, Gaebel W, Dahmen N, Scherbaum N, Schmäl C, Steffens M, Lucae S, Ising M, Müller-Myhsok B, Nöthen MM, Mann K, Kiefer F & Rietschel M (2012) Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster: ADH gene cluster and alcohol dependence. Addict Biol 17:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H & Farrer LA (2014) Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 19:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G & Ducci F (2005) The genetics of addictions: uncovering the genes. Nat Rev Genet 6:521–532. [DOI] [PubMed] [Google Scholar]
- Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, Pickering RP, Ruan WJ, Smith SM, Huang B & Hasin DS (2015) Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA Psychiatry 72:757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MA, Strong M, Razak F, Subramanian SV, Relton C & Bissell P (2016) Who are the obese? A cluster analysis exploring subgroups of the obese. J Public Health 38:258–264. [DOI] [PubMed] [Google Scholar]
- Hart AB & Kranzler HR (2015) Alcohol Dependence Genetics: Lessons Learned From Genome-Wide Association Studies (GWAS) and Post-GWAS Analyses. Alcohol Clin Exp Res 39:1312–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Sidhu H, Stouffer DG, Kreifeldt M, Le D, Cates-Gatto C, Munoz MB, Roberts AJ, Parsons LH, Roberto M, Wickman K, Slesinger PA & Contet C (2015) GIRK3 gates activation of the mesolimbic dopaminergic pathway by ethanol. Proc Natl Acad Sci 112:7091–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M & Allan W (2004) A genome wide search for alcoholism susceptibility genes. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet 128B:102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde CL, Nagle MW, Tian C, Chen X, Paciga S, Wendland JR, Tung JY, Hinds DA, Perlis RH & Winslow AR (2016) Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 48:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap 3 Consortium, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PIW, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJR, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD & McEwen JE (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C & Choquet H (2017) Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Heath AC, Neale MC, Kessler RC & Eaves LJ (1993) Alcoholism and major depression in women. A twin study of the causes of comorbidity. Arch Gen Psychiatry 50:690–698. [DOI] [PubMed] [Google Scholar]
- Knopik VS, Heath AC, Bucholz KK, Madden PAF & Waldron M (2009) Genetic and environmental influences on externalizing behavior and alcohol problems in adolescence: a female twin study. Pharmacol Biochem Behav 93:313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyrakh L, Luján R, Colón J, Karschin C, Kurachi Y, Karschin A & Wickman K (2005) Molecular and cellular diversity of neuronal G-protein-gated potassium channels. J Neurosci Off J Soc Neurosci 25:11468–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozell LB, Walter NAR, Milner LC, Wickman K & Buck KJ (2009) Mapping a barbiturate withdrawal locus to a 0.44 Mb interval and analysis of a novel null mutant identify a role for Kcnj9 (GIRK3) in withdrawal from pentobarbital, zolpidem, and ethanol. J Neurosci Off J Soc Neurosci 29:11662–11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H & Gelernter J (2011) Strong Association of the Alcohol Dehydrogenase 1B Gene (ADH1B) with Alcohol Dependence and Alcohol-Induced Medical Diseases. Biol Psychiatry 70:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H & Gelernter J (2012) Strong Protective Effect of The Aldehyde Dehydrogenase Gene (ALDH2) 504lys (*2) Allele Against Alcoholism And Alcohol-Induced Medical Diseases in Asians. Hum Genet 131:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M-T, Hinds DA, Tung JY, Franz C, Fan C-C, Wang Y, Smeland OB, Schork A, Holland D, Kauppi K, Sanyal N, Escott-Price V, Smith DJ, O’Donovan M, Stefansson H, Bjornsdottir G, Thorgeirsson TE, Stefansson K, McEvoy LK, Dale AM, Andreassen OA & Chen C-H (2016) Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat Genet 49:152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH & Goldman D (1998) Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet 81:216–221. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang B & Gelernter J (2006) ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 31:1085–1095. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PAF, Richter MM, Montgomery GW, Martin NG, Heath AC & Whitfield JB (2009) Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet 18:580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbarek H, Milaneschi Y, Fedko IO, Hottenga J-J, de Moor MHM, Jansen R, Gelernter J, Sherva R, Willemsen G, Boomsma DI, Penninx BW & Vink JM (2015) The genetics of alcohol dependence: Twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet 168:739–748. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Tilling K, Taylor AE, Evans DM & Smith GD (2017) Collider Scope: When selection bias can substantially influence observed associations. bioRxiv:079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivard MG, Verweij KJH, Minică CC, Treur JL, International Cannabis Consortium, Vink JM & Boomsma DI (2016) Connecting the dots, genome-wide association studies in substance use. Mol Psychiatry 21:733–735. [DOI] [PubMed] [Google Scholar]
- Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, Kang T-C, Nam Y-W, Kim G-B, Shin HD & Choi I-G (2013) Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum Genet 132:657–668. [DOI] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M & LaBuda MC (1995) Common genetic mechanisms in alcohol, drug, and mental disorder comorbidity. Drug Alcohol Depend 39:129–138. [DOI] [PubMed] [Google Scholar]
- Plomin R, Haworth CMA, Meaburn EL, Price TS & Davis OSP (2013) Common DNA Markers Can Account for More Than Half of the Genetic Influence on Cognitive Abilities. Psychol Sci 24:562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen EE, Chen X-D, Almasy L, Yang F, He H, Li X, Wang X-Y, Liu T-Q, Hao W, Deng H-W, Kranzler HR & Gelernter J (2014) ALDH2 is associated to alcohol dependence and is the major genetic determinant of ‘daily maximum drinks’ in a GWAS study of an isolated rural Chinese sample. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet 165B:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M & Treutlein J (2013) The genetics of alcohol dependence. Ann N Y Acad Sci 1282:39–70. [DOI] [PubMed] [Google Scholar]
- Risinger FO & Cunningham CL (1998) Ethanol-induced conditioned taste aversion in BXD recombinant inbred mice. Alcohol Clin Exp Res 22:1234–1244. [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, de la Fuente JR & Grant M (1993) Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption--II. Addict Abingdon Engl 88:791–804. [DOI] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Segura Lepe M, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke T-K, Enroth S, Fischer K, Hällfors J, Harris SE, Hieber S, Hofer E, Hottenga J-J, Johansson Å, Joshi PK, Kaartinen N, Laitinen J, Lemaitre R, Loukola A, Luan J ‘an, Lyytikäinen L-P, Mangino M, Manichaikul A, Mbarek H, Milaneschi Y, Moayyeri A, Mukamal K, Nelson C, Nettleton J, Partinen E, Rawal R, Robino A, Rose L, Sala C, Satoh T, Schmidt R, Schraut K, Scott R, Smith AV, Starr JM, Teumer A, Trompet S, Uitterlinden AG, Venturini C, Vergnaud A-C, Verweij N, Vitart V, Vuckovic D, Wedenoja J, Yengo L, Yu B, Zhang W, Zhao JH, Boomsma DI, Chambers J, Chasman DI, Daniela T, de Geus E, Deary I, Eriksson JG, Esko T, Eulenburg V, Franco OH, Froguel P, Gieger C, Grabe HJ, Gudnason V, Gyllensten U, Harris TB, Hartikainen A-L, Heath AC, Hocking L, Hofman A, Huth C, Jarvelin M-R, Jukema JW, Kaprio J, Kooner JS, Kutalik Z, Lahti J, Langenberg C, Lehtimäki T, Liu Y, Madden PAF, Martin N, Morrison A, Penninx B, Pirastu N, Psaty B, Raitakari O, Ridker P, Rose R, Rotter JI, Samani NJ, Schmidt H, Spector TD, Stott D, Strachan D, Tzoulaki I, van der Harst P, van Duijn CM, Marques-Vidal P, Vollenweider P, Wareham NJ, Whitfield JB, Wilson J, Wolffenbuttel B, Bakalkin G, Evangelou E, Liu Y, Rice KM, Desrivières S, Kliewer SA, Mangelsdorf DJ, Müller CP, Levy D & Elliott P (2016) KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci U S A 113:14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevlin M & Smith GW (2007) The factor structure and concurrent validity of the alcohol use disorder identification test based on a nationally representative UK sample. Alcohol Alcohol 42:582–587. [DOI] [PubMed] [Google Scholar]
- Sobczyk-Kopciol A, Broda G, Wojnar M, Kurjata P, Jakubczyk A, Klimkiewicz A & Ploski R (2011) Inverse association of the obesity predisposing FTO rs9939609 genotype with alcohol consumption and risk for alcohol dependence. Addict Abingdon Engl 106:739–748. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, Kobayashi S, Ogihara T, Yamori Y, Fujioka A & Kato N (2011) Confirmation of ALDH2 as a Major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J Off J Jpn Circ Soc 75:911–918. [DOI] [PubMed] [Google Scholar]
- Tarantino LM, McClearn GE, Rodriguez LA & Plomin R (1998) Confirmation of quantitative trait loci for alcohol preference in mice. Alcohol Clin Exp Res 22:1099–1105. [PubMed] [Google Scholar]
- Tawa EA, Hall SD & Lohoff FW (2016) Overview of the Genetics of Alcohol Use Disorder. Alcohol Alcohol Oxf Oxfs 51:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB & Yin SJ (1991) Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet 48:677–681. [PMC free article] [PubMed] [Google Scholar]
- Tipps ME, Raybuck JD, Kozell LB, Lattal KM & Buck KJ (2016) G Protein-Gated Inwardly Rectifying Potassium Channel Subunit 3 Knock-Out Mice Show Enhanced Ethanol Reward. Alcohol Clin Exp Res 40:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstrup JS, Heitmann BL, Tjønneland AM, Overvad OK, Sørensen TIA & Grønbaek MN (2005) The relation between drinking pattern and body mass index and waist and hip circumference. Int J Obes 2005 29:490–497. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann H-E, Schreiber S, Dragano N, Sommer W, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nöthen MM, Kiefer F, Spanagel R, Mann K & Rietschel M (2009) Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 66:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst B, Neale MC & Kendler KS (2015) The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med 45:1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Hottenga JJ, de Geus EJC, Willemsen G, Neale MC, Furberg H & Boomsma DI (2014) Polygenic risk scores for smoking: predictors for alcohol and cannabis use? Addict Abingdon Engl 109:1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, McGue M, Miller MB, Hicks BM & Iacono WG (2013) Three Mutually Informative Ways to Understand the Genetic Relationships Among Behavioral Disinhibition, Alcohol Use, Drug Use, Nicotine Use/Dependence, and Their Co-occurrence: Twin Biometry, GCTA, and Genome-Wide Scoring. Behav Genet 43:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall TL, Luczak SE & Hiller-Sturmhöfel S (2016) Biology, Genetics, and Environment: Underlying Factors Influencing Alcohol Metabolism. Alcohol Res Curr Rev 38:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xu H, Zhao H, Gelernter J & Zhang H (2016) DNA co-methylation modules in postmortem prefrontal cortex tissues of European Australians with alcohol use disorders. Sci Rep 6:19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L & Blangero J (1999) Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet 65:1148–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, Zhao H, Farrer LA & Gelernter J (2015) Genomewide Association Study for Maximum Number of Alcoholic Drinks in European Americans and African Americans. Alcohol Clin Exp Res 39:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee H, Goddard ME & Visscher PM (2011) GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Hum Genet 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lu X, Wang L, Chen S, Li J, Cao J, Chen J, Hao Y, Li Y, Zhao L, Li H, Liu D, Wang L, Lu F, Shen C, Yu L, Wu X, Zhao Q, Ji X, Guo D, Peng X, Huang J & Gu D (2013) Common variants at 12q24 are associated with drinking behavior in Han Chinese. Am J Clin Nutr 97:545–551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


