Abstract
Along with the recent spread of multidrug-resistant bacteria, outbreaks of extended-spectrum β-lactamase (ESBL) and carbapenemase-producing bacteria present a serious challenge to clinicians. β-lactam antibiotics are the most frequently used antibacterial agents and ESBLs, and carbapenemases confer resistance not only to carbapenem antibiotics but also to penicillin and cephem antibiotics. The mechanism of β-lactam resistance involves an efflux pump, reduced permeability, altered transpeptidases, and inactivation by β-lactamases. Horizontal gene transfer is the most common mechanism associated with the spread of extended-spectrum β-lactam- and carbapenem resistance among pathogenic bacterial species. Along with the increase in antimicrobial resistance, many different types of ESBLs and carbapenemases have emerged with different enzymatic characteristics. For example, carbapenemases are represented across classes A to D of the Ambler classification system. Because bacteria harboring different types of ESBLs and carbapenemases require specific therapeutic strategies, it is essential for clinicians to understand the characteristics of infecting pathogens. In this review, we summarize the current knowledge on carbapenem resistance by ESBLs and carbapenemases, such as class A carbapenemases, class C extended-spectrum AmpC (ESAC), carbapenem-hydrolyzing class D β-lactamases (CHDLs), and class B metallo-β-lactamases, with the aim of aiding critical care clinicians in their therapeutic decision making.
Keywords: β-Lactam, β-Lactamase, Carbapenemase, Classification, Multidrug resistance
Background
Among the recent spread of multidrug-resistant bacteria, outbreaks of extended-spectrum β-lactam- and carbapenem-resistant bacteria are a serious problem not only making treatment difficult but also worsening the prognosis of infected patients [1]. β-lactam antibiotics are the most frequently used antibacterial agents, and extended-spectrum β-lactams and carbapenems have been developed as specific drugs to treat bacterial species resistant to penicillins and cephems [2]. Bacterial resistance to carbapenems incorporates not only carbapenem antibiotics but also resistance to penicillin and cephem antibiotics. Therefore, resistance to carbapenems presents a significant threat to patients who are immunocompromised and are therefore susceptible to infections caused by multidrug-resistant bacteria all over the world [3]. The mechanism of resistance to extended-spectrum β-lactams and carbapenems involves an efflux pump, reduced permeability, altered transpeptidases, and inactivation by β-lactamases. Horizontal gene transfer is the most common mechanism associated with the spread of antimicrobial resistance across pathogenic bacterial species, such as carbapenemase-producing Enterobacteriaceae (CPE) [4, 5]. In various bacterial species, novel extended-spectrum β-lactamases (ESBLs) or carbapenemases with different structures or characteristic features are reported each year.
Various ESBLs and carbapenemases have been reported in the Enterobacteriaceae including Enterobacter, Klebsiella, Escherichia coli [6, 7], and other opportunistic species such as Serratia, Acinetobacter, and Pseudomonas [8]. In addition, the genetic elements by which drug-resistant genes horizontally move across bacterial species have been studied among these bacterial species. As one typical example, Pseudomonas aeruginosa, one of the major causative agents of infection in immunocompromised individuals, displays resistance to various antibacterial agents. Its resistance mechanism is, in part, derived from the organism’s natural resistance, but is also acquired through horizontal gene transfer and/or mutations within its DNA. P. aeruginosa is naturally resistant to β-lactam antibiotics, such as penicillin and cephem, and aminoglycoside antibiotics. Since the 1990s, P. aeruginosa strains have emerged that have acquired resistance to broad-spectrum penicillins, third-generation cephems, carbapenems, anti-P. aeruginosa aminoglycosides, and new quinolones [9, 10]. Among these multidrug-resistant P. aeruginosa (MDRP), one of the major clinical concerns is the spread of P. aeruginosa strains that harbor a carbapenemase because β-lactam antibiotics including carbapenems are the most frequently used antibacterial agents. An anti-methicillin-resistant Staphylococcus aureus (MRSA) drug, albekacin sulfate, a monobactam aztreonam, and polypeptide colistin appear to be effective against MDRP, but the emergence of resistant strains has also been reported including extensively drug-resistant P. aeruginosa (XDRP) and pandrug-resistant P. aeruginosa (PDRP) [10, 11].
To date, many different types of ESBLs and carbapenemases have emerged with different enzymatic characteristics [1]. Because the specific therapeutic strategy is dependent on the type of ESBL and carbapenemase, it is vital for clinicians to understand the characteristics of ESBLs and carbapenemases [12]; however, in practice, the complex biology associated with ESBLs and carbapenemases presents significant challenges in the effective control of infections. In this review, we summarize antimicrobial resistance by ESBLs and carbapenemases, with the aim of collating the current knowledge in this field to aid therapeutic decision making by critical care clinicians.
β-Lactam antibiotics, penicillin-binding protein (PBP), and β-lactamase
We will begin by reiterating the mechanism of action of β-lactam antibiotics. The main constituent of the cell wall formed in the outer membrane layer of eubacteria is peptidoglycan, which is a macromolecular structure consisting of peptide and sugar (Fig. 1). This peptidoglycan structure confers resistance to osmotic pressure and retains cell morphology and strength. It is also the target of β-lactam drugs. By inhibiting the formation of this structure, β-lactams suppress bacterial cell division (bacteriostatic action) or induce bacterial rupture against osmotic pressure (bactericidal activity). Peptidoglycan possesses a basic structural unit in which two amino sugars of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) alternate, and a longitudinal peptide chain linked to NAM forms a pillar. This peptide chain is a pentapeptide consisting of alternating d- and l-forms of l-alanine, γ-d-glutamic acid, l-lysine, d-alanine, and d-alanine residues. At the time of cross-linking, d-alanine at the carboxyl-terminal of pentapeptide is first eliminated by hydrolysis. Next, the fourth d-alanine carboxyl group and the third diaminopimelic acid, which is a ε-carboxyl derivative of lysine of the same columnar structure as the neighboring molecule, are combined with a triglycine peptide structure for horizontal reinforcement of the chain. These reactions are mediated by the bacterial enzyme known as PBP.
Fig. 1.
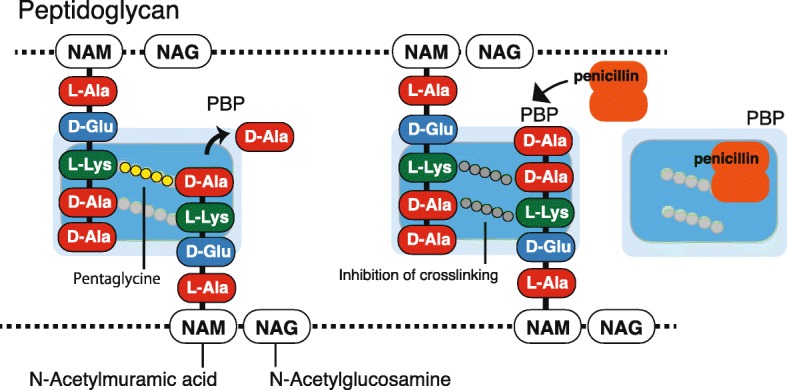
The antimicrobial action of β-lactamase against the peptidoglycan structure of the bacterial cell membrane. Peptidoglycan possesses a basic structural unit in which two amino sugars of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) alternate, and a longitudinal peptide chain linked to NAM forms a pillar. This peptide chain is a pentapeptide consisted of alternating d- and l-forms of l-alanine, γ-d-glutamic acid, l-lysine, d-alanine, and d-alanine that forms a bacterial enzyme known as penicillin-binding protein (PBP). PBP recognizes alanyl-alanine, which is an alanine dimer formed by d-alanine-d-alanine present at the end of the pentapeptide, and exerts the enzymatic action of cross-linking. Since penicillin is structurally similar to alanyl-alanine at the terminal region of the pillar structure, PBP captures penicillin, thereby inhibiting the cross-linking reaction
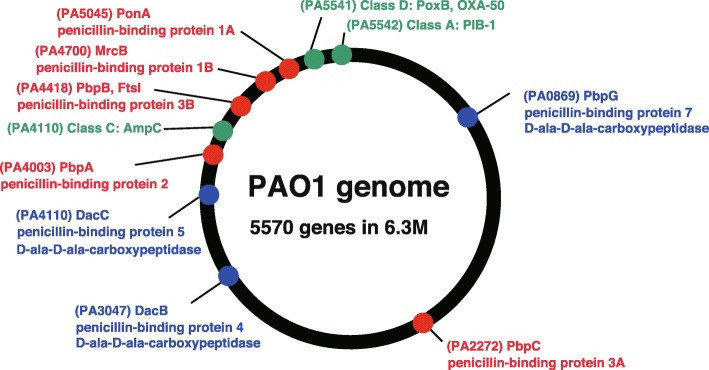
PBP recognizes alanyl-alanine, which is an alanine dimer formed by d-alanine-d-alanine present at the end of the pentapeptide, and its enzymatic activity mediates cross-linking [13, 14]. A cross-linking reaction is induced by the formation of a covalent bond between the serine residue in the active center of PBP and the carboxyl group produced by hydrolytic cleavage of d-alanine. Thereby, PBP displays adenyl-alanine endopeptidase activity. Since penicillin has a structure similar to that of alanyl-alanine at the terminal part of this pillar structure, PBP cannot distinguish between these structures and binds to both, leading to inhibition of the cross-linking reaction. Seven types of PBP genes exist in the E. coli genome, and eight types of PBP genes exist in the genome of P. aeruginosa reference strain PAO1 (Fig. 2) [15]. Among them, five types of genes (PBP1A, PBP1B, PBP2, PBP3A, and PBP3B) encode high molecular weight PBPs (HMM-PBPs, molecular weight 60,000 to 90,000) that display both transglycosylase and transpeptidase activities and play a role in cell elongation and partition formation. In cell division and morphogenesis, PBP1A and PBP1B are thought to be involved in growth and elongation, PBP2 in formation of a gonococcal form, and PBP3 in partition formation during division.
Fig. 2.
The genes encoding penicillin-binding proteins in P. aeruginosa PAO1. Eight types of PBP genes and three chromosomal β-lactamase genes for PIB-1 (class A), AmpC (class C), and PoxB (class D) exist in the genome of P. aeruginosa reference strain PAO1 [15]
The remaining three types are low molecular mass BPBs (LMM-PBPs; molecular weight of 40,000–50,000) that display d-alanine carboxypeptidase activity and are widely resistant to various β-lactams. The roles of LMM-PBPs are still under research. Both the HMM-PBP transpeptidase activity and the LMM-PBP alanine carboxypeptidase activity utilize a serine residue as an enzymatic activity center, and the β-lactam antibacterial agent exhibits enzyme inhibitory activity by binding to this serine residue, which is a common feature of class A, C, and D β-lactamases (which is discussed again later). As one mechanism of the development of resistant bacteria, the substrate specificity of PBP may be changed to reduce the ability to link to a β-lactam-based antibacterial agent. For example, MRSA produces PBP2′, which possesses a mutation in PBP, that has low affinity for β-lactams. Another example is the mechanism of resistance to the glycopeptide class of antibacterial agents, such as vancomycin. In this mechanism, Enterococci with the vanA gene cluster derived from transposon Tn1546 expresses a group of enzymes that exchange the peptidoglycan peptide longitudinal chain from d-alanine-d-alanine to d-alanine-d-serine and inhibit binding to vancomycin.
Next, we will reiterate the mechanism of action of β-lactamase. β-lactamase inhibits antibacterial activity by dissociating the –CO–NH structure of the β-lactam ring, which is part of the basic structure of β-lactams (Fig. 3). The –CO–NH structure of the β-lactam ring comprises the –CO–NH peptide in the alanyl-alanine dimer from which the peptidoglycan cross-links are formed. Thus, β-lactams mimic the alanyl-alanine of the peptidoglycan pillar structure and this is the binding region. Thus, β-lactamase is a protease (peptidase) that dissociates peptide bonds. PBP is bound by the original substrate alanyl-alanine and penicillin, which works as an inhibitor since it shares a similar structure with alanyl-alanine. These findings indicate that PBP and β-lactamase share similar structures because they exert peptidase activity, implying that β-lactamase may be evolutionarily derived from PBP.
Fig. 3.
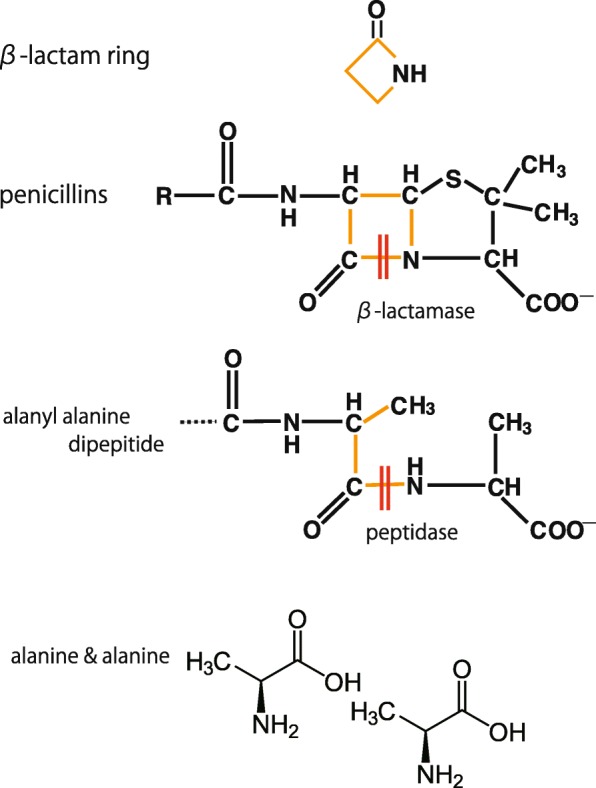
The enzymatic action of a β-lactamase to penicillins. The β-lactam ring is the basic structure of β-lactam antibiotics. β-lactamase inhibits the antibacterial action by dissociating the –CO–NH structure of the β-lactam ring. The –CO–NH structure of the β-lactam ring is similar to the –CO–NH peptide in alanyl-alanine, from which the peptidoglycan cross-links are formed. Thus, β-lactams mimic alanyl-alanine of the peptidoglycan pillar structure. Thus, β-lactamase is a protease (peptidase) that dissociates the –CO–NH structure of β-lactams
In fact, the enzyme active site motifs S-X-X-K, S-D-N, and K-S/T-G in the primary sequence of class A β-lactamases, also exist in PBP (Additional file 1: Figure S1). This indicates that the β-lactamases and the peptidoglycan cross-linking enzyme, PBP, co-evolved. At present, clinically used β-lactams can be classified into five basic structures capable of exerting different antimicrobial activities (Fig. 4). Many of them originate from natural antibacterial substances produced by actinomycetes or fungi. Thus, actinomycetes themselves are resistant to their own antimicrobial agents. This indicates that both antibacterial substances and agents that counter such substances co-evolved in actinomycetes.
Fig. 4.
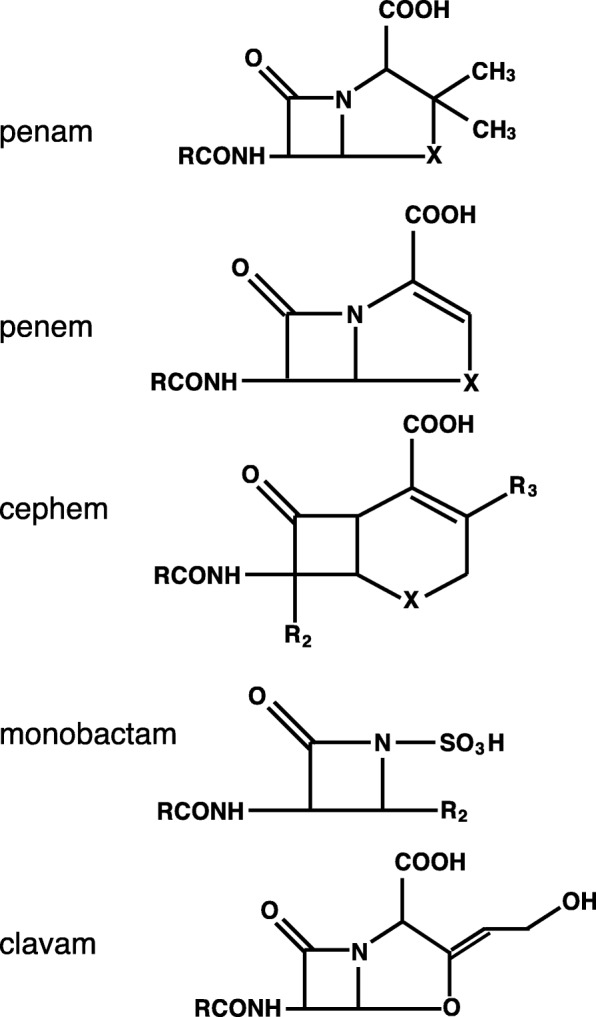
The chemical structure of β-lactam antimicrobials. Clinically used β-lactams can be classified into five basic structures capable of exerting different antimicrobial activities
Classification of β-lactamases and P. aeruginosa drug resistance
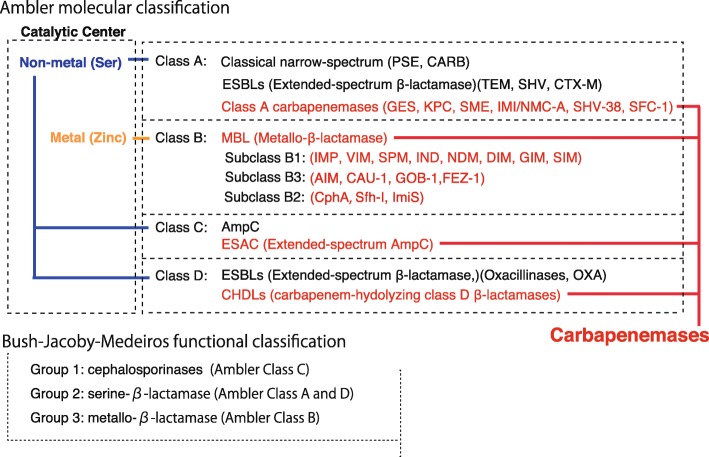
To date, β-lactamases have been classified based on the molecular structure classification of Ambler [16] and the functional classification of Bush-Jacobi-Medeiros (Fig. 5) [17, 18]. In the Ambler classification, β-lactamases are grouped into four class A, B, C, and D according to motifs composed of primary sequences constituting the protein molecules. β-lactamases of classes A, C, and D use a serine as an enzyme active center, whereas β-lactamases of class B use the metal zinc. In the functional classification of Bush-Jacobi-Medeiros, β-lactamases are classified into groups 1 to 3 depending on the degradation of β-lactam substrates and the effects of inhibitors. Group 1 comprises cephalosporinases that are classified as class C based on molecular structural classification, and the gene involved was originally chromosomal. Group 2 comprises β-lactamases other than those in group 1 having serine in the active center and includes both classes A and D according to molecular structural classification. Group 3 comprises metallo-β-lactamases (MBLs) and corresponds to class B of the molecular structural classification. Within the genome of P. aeruginosa PAO1, a standard strain of P. aeruginosa and the first strain subjected to whole-genome sequence analysis, the class A PIB-1 (PA5542), class C AmpC (PA 4410), and class D PoxB (OXA-50, PA5541) β-lactamases are encoded (Fig. 2). Thus, P. aeruginosa PAO1 is equipped with three different types of β-lactamases, only lacking a class B β-lactamase [15]. These chromosome-encoded intrinsic β-lactamase genes of P. aeruginosa are partially responsible for its natural resistance to penicillins and cephalosporins.
Fig. 5.
The classification of β-lactamases. Molecular structure classification using the Ambler method [16] and functional classification using the Bush-Jacobi-Medeiros method [17, 18] have both been employed. In the Ambler classification, β-lactamases are grouped into four classes A, B, C, and D by motifs composed of primary sequences constituting the protein molecules. β-lactamases of classes A, C, and D use a serine at the enzyme active center, whereas β-lactamases of class B use metal zinc ions. In functional classification using the Bush-Jacobi-Medeiros method, β-lactamases are classified into groups 1 to 3 depending on the degradation of β-lactam substrates and the effect of the inhibitor
ESBLs and carbapenemases
ESBLs degrade third-generation cephem systems (such as cefotaxime and ceftazidime) [17] and are characterized by whether they are inhibited by β-lactamase inhibitors, such as clavulanic acid, sulbactam, and tazobactam [17]. The appearance of ESBLs in organisms such as Klebsiella and E. coli indicates that β-lactamase-producing genes, such as TEM-type and SHV-type genes, encoded on a plasmid (R plasmid), have broadened the range of target drugs by genetic mutation [12, 19]. Because the resistance genes are derived from the plasmid, they are easily transmitted among different bacterial species belonging to the same genera, such as the Enterobacteriaceae. The repeated mutation of ESBLs has contributed to the emergence of new β-lactamases, namely carbapenemases, which hydrolyze carbapenem antibiotics. In all classes A to D, based on molecular structural classification, β-lactamases that degrade carbapenems have been identified and are discussed below.
Class A penicillinases, ESBLs, and carbapenemases
Class A penicillinases, such as PC1, which belong to the Bush-Jacoby functional subgroup 2a, hydrolyze a relatively limited spectrum of penicillins and are predominantly found in Gram-positive cocci. As class A ESBLs that degrade the early cephalosporins and belong to the Bush-Jacoby functional subgroup 2b, plasmid-mediated TEM-1, TEM-2, and SHV-1 were reported from the 1970s–1980s [20]. Soon after, mutated TEM and SHV ESBLs, such as TEM-3, SHV-2, and CTX-M, which efficiently degrade cefotaxime but remain sensitive to inhibition by clavulanic acid, were reported. These ESBLs belong to the Bush-Jacoby functional subgroup 2be, and since the 1990s, many variants such as BEL-1, BES-1, SFO-1, PER, and VEB were reported as members of this group [21]. TEM-30 and SHV-10, which hydrolyze penicillins and display relative resistance to clavulanic acid, sulbactam, and tazobactam, belong to Bush-Jacoby functional subgroup 2br. TEM-50 is a broad-spectrum lactamase that hydrolyzes extended-spectrum cephalosporins (oxyimino-β-lactams) and monobactams but shows resistance to clavulanic acid, sulbactam, and tazobactam. Enzymes in this group have acquired resistance to clavulanic acid, sulbactam, and tazobactam and belong to Bush-Jacoby functional subgroup 2ber [22, 23]. Class A carbenicillinases and cephalosporinases, which belong to Bush-Jacoby functional group 2, can be further classified into more minor subclasses, such as functional groups 2c, 2ce, and 2e [24]. β-lactamases, such as PSE-1 and CARB-3, belonging to Bush-Jacoby functional subgroup 2c, hydrolyze carbenicillin and are inhibited by clavulanic acid or tazobactam. A subclass of 2c, namely subclass 2ce, comprises one enzyme RTG-4 (CARB-10) that was recently identified as an extended-spectrum carbenicillinase, and hydrolyzes carbenicillin, cefepime, and cefpirome. Subclass 2ce, which includes CepA, hydrolyzes extended-spectrum cephalosporins and is inhibited by clavulanic acid or tazobactam but not aztreonam.
Finally, in the late 1990s, the β-lactamase classified as class A was mutated to become a carbapenemase, which degrades carbapenems [24]. As a feature of class A β-lactamases, the tetrad motif S-X-X-K, the S-D-N triad, and the K-S/T-G triad, which contain the serine of the enzyme active site in their primary sequences, are retained (Additional file 2: Figure S2) [25]. In addition, class A carbapenemases are characterized by susceptibility to inhibition by β-lactamase inhibitors such as clavulanic acid and tazobactam and are classified as Bush-Jacoby functional subgroup 2f [24]. Six types of class A carbapenemases have been reported, these include GES (Guiana extended-spectrum β-lactamase) [26], SME (Serratia marcescens enzyme) [27], SHV (sulfhydryl variable lactamase) [28], KPC (Klebsiella pneumoniae carbapenemase) [29, 30], IMI/NMC-A (imipenemase/non-metallocarbapenemase-A) [31], and SFC (Serratia fonticola carbapenemase) (Fig. 6) [35, 36]. The KPC-type is transmitted via a plasmid and is the most representative example of a class A carbapenemase, it also plays an important role in controlling the characteristics of carbapenem-resistant Enterobacteriaceae bacteria (CRE) [37]. SFC-1 and SME have been reported in Serratia and are closely related to KPC. SME and IMI/NMC-A are chromosomally encoded, and the GES gene blaGES is inserted into the class 1 integron in a plasmid of P. aeruginosa [38].
Fig. 6.
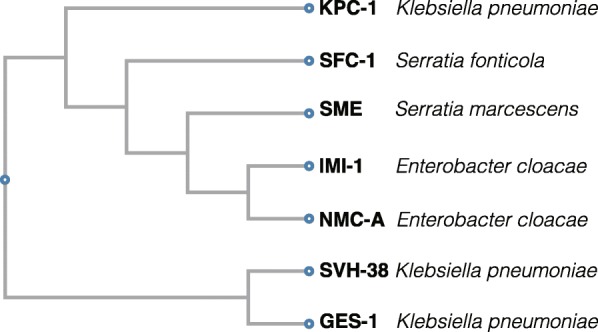
A phylogenetic tree of class A carbapenemases. A phylogenetic tree of six types of class A capbapenemases: GES (Guiana extended-spectrum β-lactamase), SME (Serratia marcescens enzyme), SHV-38 (sulfhydryl variable lactamase), KPC (Klebsiella pneumoniae carbapenemase), IMI/NMC-A (imipenemase/non-metallocarbapenemase-A), and SFC-1 (Serratia fonticola carbapenemase). Based on the primary sequences, the tree was generated using Clustal Omega [32–34] of GenomeNet at Kyoto University Bioinformatics Center (https://www.genome.jp/tools-bin/clustalw)
Class C ESAC
The β-lactamase belonging to class C is derived from the ampC gene carried on the genome of many members of the Enterobacteria genus of the Enterobacteriaceae and is functionally a cephalosporinase classified into Bush-Jacoby functional group 1 [39, 40]. It is resistant to clavulanic acid, but sensitive to cephamycins, such as cefoxitin and ceftazidime. Although the expression level of AmpC is usually low, it may be induced by administration of a penicillin system or clavulanic acid and may exhibit carbapenem resistance when it is expressed in large amounts [1]. Some β-lactamases belonging to this group, including CMY, ACT, FOX, and MIR, were encoded on a plasmid [41]. The most significant concern is that variants of AmpC contribute to the reduced sensitivity to carbapenems. These β-lactamases are known as ESAC [42]. In P. aeruginosa, AmpC mutants have been linked to reduced sensitivity to imipenem, ceftazidime, and cefepime. Such mutants, including the plasmid-encoded CMY-10, CMY-19, and CMY-37 mutants, are classified within Bush-Jacoby functional subgroup 1e [43, 44].
The class C β-lactamases share around 40–50% identity at the primary sequence level. The amino-terminal region is the most variable, whereas the carboxyl-terminal region is relatively conserved. They share three characteristic motifs of serine-reactive β-lactamases: S-X-X-K in the enzymatic active site, Y-X-N pointing into the active site, and K-T-G forming the opposite wall of the active site (Additional file 3: Figure S3) [45].
Class D ESBL oxacillinases (OXAs)
β-lactamases of Ambler class D, known as OXA enzymes (Fig. 7), which include OXA-1 and OXA-10, possess an active serine site similar to class A and C β-lactamases. These β-lactamases show cloxacillin- and oxacillin-hydrolyzing activity and are classified into Bush-Jacoby functional group 2d. The overall level of amino acid identity between class D and class A or class C β-lactamases is only about 16%. In β-lactamases such as OXA-11 and OXA-15, the degradation substrate is extended to extended-spectrum cephalosporins (oxyimino-β-lactams), but not carbapenems, and they are classified as Bush-Jacoby functional subgroup 2de [46, 47]. This type of β-lactamases is frequently encoded on plasmids and/or integrons allowing for their wide dissemination [48]. Carbapenem-hydrolyzing class D β-lactamases (CHDLs) are OXA enzymes that hydrolyze carbapenems and belong to Bush-Jacoby functional subgroup 2df. OXA enzymes with carbapenem-hydrolyzing activity have mainly been found on the chromosomes of Acinetobacter baumannii strains [49], but OXA-23 and OXA-48 have been reported on plasmids isolated from enteric bacteria [50, 51]. These OXA-type enzymes are widespread across Acinetobacter, Shewanella, Pseudomonas, and Burkholderi [48, 52]. At the time of writing, 790 OXA-variants could be found in public databases. Similar to class A β-lactamases, class D β-lactamases retain the tetrad motif S-X-X-K (including the serine of the enzyme active site), the Y-G-N/S triad, and the K-T-G triad (Additional file 4: Figure S4) [53].
Fig. 7.
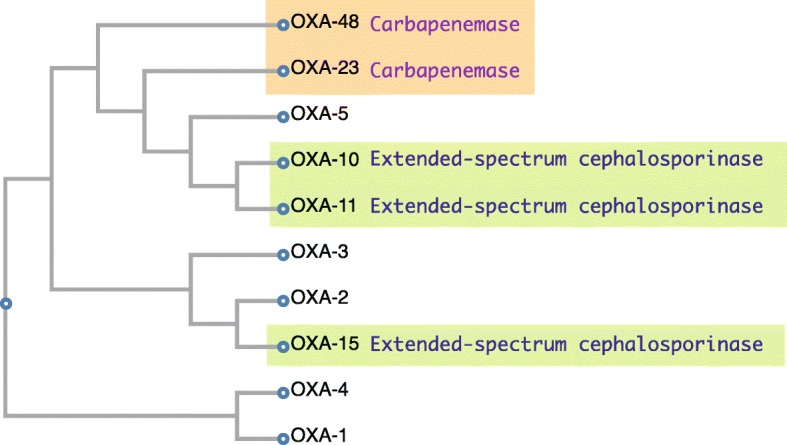
A phylogenetic tree of the class D β-lactamase OXA family (oxiacillinase). A phylogenetic tree of the OXA types of β-lactamases. OXA-10, OXA-11, and OXA-15 are recognized as extended-spectrum cephalosporinases, whereas OXA-23 and OXA-48 are recognized as carbapenem-hydrolyzing class D β-lactamases (CHDLs). Based on the primary sequences, the tree was generated using Clustal Omega [32–34] of GenomeNet at Kyoto University Bioinformatics Center (https://www.genome.jp/tools-bin/clustalw)
Class B MBL
Class B β-lactamase is a MBL that possesses the metal Zn2+ at the enzyme active center, compared with a serine residue in the same location for other classes of β-lactamase [54]. Class B β-lactamases hydrolyze carbapenems and belong to Bush-Jacoby functional group 3. P. aeruginosa carrying MBL degrades all β-lactam agents except monobactams. Since the metal is located at the enzymatic active center, its enzymatic activity is suppressed by a chelating agent, such as ethylenediaminetetraacetic acid (EDTA). The MBL gene can reside on an integron, transposon, plasmid, chromosome, or various other genetic molecules.
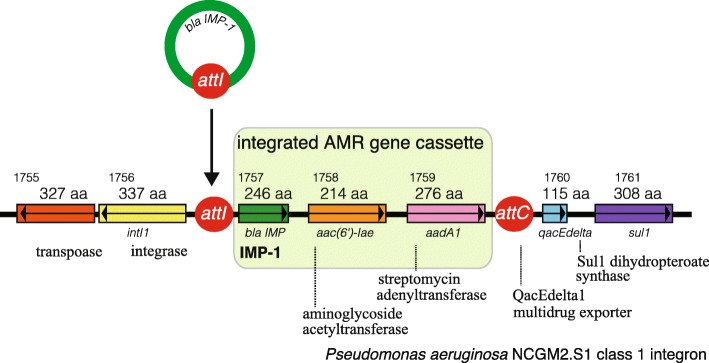
The components of an integron first reported in 1989 comprised drug-resistant genes of class B MBLs and ESBLs of classes A and D [55–57]. Gene cassettes become integrated into the genome via the interaction between two recombination sites (attI and attC), and an integrase catalyzes the genetic recombination (Fig. 8). The promoter, which is located in the integrase gene upstream of the insertion site, controls the transcription of the inserted drug-resistant genes embedded in the cassette structure.
Fig. 8.
The genomic structure of the class 1 integron of P. aeruginosa NCGM2.S1. The intl gene, which encodes integrase Int1, catalyzes recombination between the attl1 site of the integron and the attC site of an antimicrobial gene cassette [56]. In multidrug-resistant P. aeruginosa NCGM2.S1 [58], three antimicrobial gene cassettes (blaIMP-1, aac(6′)-1ae, and aadA1) are integrated into the region between attI and attC. The Int1 integrase gene, the attI integration site, and qacE partially deleted in the sulI gene, which encodes resistance to sulfonamide are present
To date, the carbapenem-resistant metallo-β-lactamases, imipenemase (IMP) [59], Verona integrated-encoded MBL (VIM) [60], Sao Paulo MBL (SPM) [61], Germany imipenemase (GIM) [62], New Delhi MBL (NDM) [63], and Florence imipenemase (FIM) [64] have been reported (Fig. 9, and Additional file 5: Figure S5). Among them, the IMP- and VIM-type MBLs, first discovered in the 1990s, are the major MBLs [60, 65]. New variants of IMP- and VIM-type MBLs are constantly being identified (Additional file 6: Figure S6 and Additional file 7: Figure S7). At the time of writing, 83 IMP- and 66 VIM-variants could be found in public databases. The mutations present in variants affect the spectrum of carbapenem activities, such as activity against imipenem, meropenem, and doripenem [66]. For example, IMP-6, which has only one amino acid substitution (serine to glycine at position 214) from IMP-1 (Additional file 6: Figure S6), enhanced the resistance to meropenem, and meropenem-resistant Enterobacteriaceae, K. pneumoniae, and P. aeruginosa carrying blaIMP-6 have been spreading in Japan since 2001 [67, 68]. Similarly, VIM-4, which has only one amino acid insertion (arginine at position 44) and one amino acid substitution (serine to arginine at position 265) from VIM-1 (Additional file 7: Figure S7), enhanced the resistance to carbapemems, and P. aeruginosa carrying blaVIM-4, which was first reported in 2002, has already been detected worldwide [69, 70]. IMP and VIM are mainly included in the integron structure and are integrated into chromosomal DNA and plasmid DNA in association with the transposon [60, 65]. NDM-1 was detected in Klebsiella and E. coli isolates from patients who returned to Sweden in 2008 following a trip to India, and the blaNDM-1 gene was present on the plasmid [63]. Unlike IMP and VIM, NDM was not found in the integron structure.
Fig. 9.
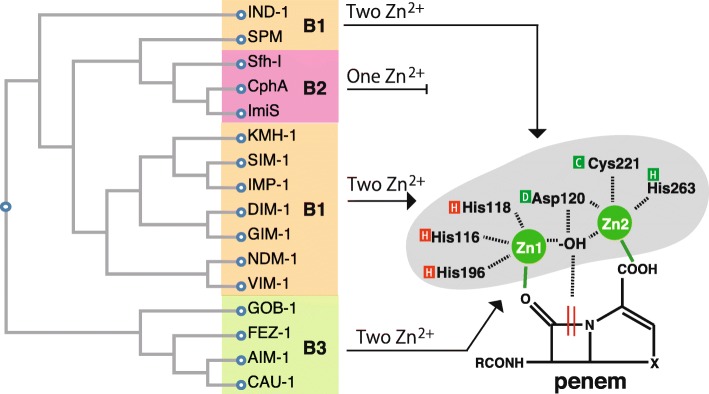
A phylogenetic tree of the class B metallo-β-lactamase family. MBLs can be classified into three subclasses (B1, B2, B3) based on their amino acid sequences. Subclasses B1 and B3 are characterized by two Zn2+ molecules in the enzyme active center (Zn1, Zn2), implying a broader range of substrates, whereas target substrates of subclass B2, which have a single Zn2+ at the active center, are narrower in range than those of classes 1 and 3 [54]. Based on the primary sequences, the tree was generated using Clustal Omega [32–34] of GenomeNet at Kyoto University Bioinformatics Center (https://www.genome.jp/tools-bin/clustalw)
MBLs can be classified into three subclasses (B1, B2, B3) based on their amino acid sequence (Fig. 9) [54, 71]. The level of amino acid identity between groups was as low as 20% or less. Subclass B1 includes IMP, VIM, NDM, and SPM, and subclass B3 includes CAU-1, GOB-1, and FEZ-1, both of which are characterized by the presence of two Zn2+ molecules in the enzyme active center (Zn1, Zn2), degradation of a broader range of substrates, and classification into Bush-Jacoby functional subgroup 3a [54]. The binding site for Zn1 of the B1 enzyme involves three histidines (His116, His118, and His196) [72]. The binding site for Zn2 of the B1 enzyme consists of aspartate, cystine, and histidine (DCH site, Asp-120, Cys-221, His-263) (Additional file 5: Figure S5) [72], whereas the target degradation substrates for CphA, Sfh-I, and ImiS, which are MBLs of subclass B2 having one Zn2+ at the active center, are narrow and these MBLs are classified into Bush-Jacoby functional subgroup 3b [73, 74].
Summary
ESBLs and carbapenemases are represented in all classes, A to D, in the Ambler classification system. Class A carbapenemases, represented by GES and KPC, are encoded on plasmids and are most frequently detected in P. aeruginosa and Klebsiella. As a characteristic feature, they exert an inhibitory effect by β-lactamase inhibitors such as clavulanic acid and tazobactam. The β-lactamases belonging to class C, including ESACs, are encoded by the ampC gene carried on the chromosome of many Enterobacteriaceae, and function as cephalosporinases. In class D, the OXA enzymes, which were originally oxacillinases, have mutated into CHDLs. These enzymes have also been found to be encoded on the chromosome of carbapenem-resistant A. baumannii and on the plasmids of intestinal bacteria such as OXA-23 and OXA-48. Class B β-lactamases are characterized as having a metal Zn2+ in their enzyme activity center. P. aeruginosa carrying an MBL degrades all β-lactam agents, except monobactams. The MBL gene is encoded on an integron, transposon, plasmid, chromosome, or various other genetic molecules. Among them, the IMP- and VIM-type enzymes, first discovered in the 1990s, are the main MBLs that fit into the integron structure. MBLs can be classified into three subclasses (B1, B2, B3) based on their amino acid sequence. Subclass B1 includes IMP, VIM, and NDM. Subclass B3 is characterized by the presence of two zinc molecules (Zn1, Zn2) in the enzyme active center, demonstrating more extensive substrate degradation, whereas subclass B2 has a single Zn2+ in the active center and shows a narrow spectrum of substrates. Figure 10 summarizes carbapenemases of the Ambler classification system [16], including the functional information based on Bush-Jacobi-Medeiros method [17, 18]. Table 1 summarizes the functional classification scheme based on the 1995 proposal by Bush-Jacobi-Medeiros [18] that was updated by Bush-Jacobi [17] in 2010. Together with the Ambler molecular structure classification, antibiotic substrates hydrolyzed by classified β-lactamases and the profiles to β-lactamase inhibitors (clavulanic acid, sulbactam, and tazobactam) are also listed in this table.
Fig. 10.
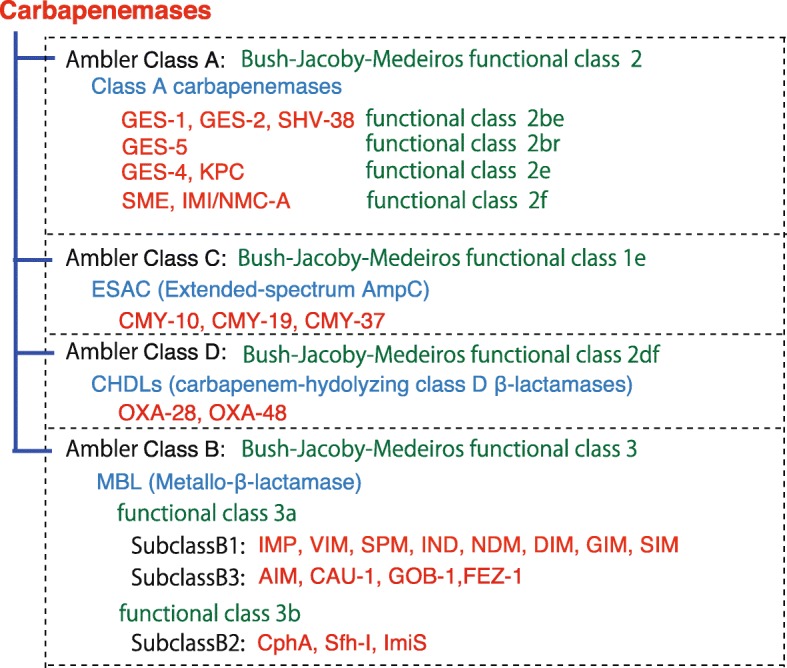
The classification of carbapenemases. Carbapenemases is represented in all classes, A to D, of the Ambler classification system [16]. Functional classification, using the Bush-Jacobi-Medeiros method [17, 18], indicated that the class A carbapenemases were represented by GES and KPC. The β-lactamases belonging to class C, which function as cephalosporinases, are encoded by the AmpC gene carried on the chromosome of many Enterobacteriaceae. ESAC enzymes are known as ESACs. In class D, OXA enzymes, which were originally oxacillinases, have mutated to become CHDLs. Class B β-lactamases are characterized by possessing a metal Zn2+ as the enzyme activity center. IMP- and VIM-type β-lactamases are the main MBLs that fit into the integron structure. MBLs can be classified into three subclasses (B1, B2, B3) based on their amino acid sequence. Subclasses B1 and B3 are characterized by two zinc molecules (Zn1, Zn2) in the enzyme active center, demonstrating more extensive substrate degradation, while subclass B2 has a single Zn2+ at the active center and displays a narrower spectrum
Table 1.
| Bush-Jacoby* | Ambler | Enzymatic center | Enzymes | Distinctive substrate | Characteristic(s) | |
|---|---|---|---|---|---|---|
| 1 | 1 | C | Ser | AmpC, CMY-1, ACT-1, FOX-1, MIR-1 | Cephalosporins | Resistance to CA and TZB |
| 1e | GC1, CMY-10, CMY-19, CMY-37 | Ceftazidime, oxyimino-β-lactams ESAC | Resistance to CA and TZB reduced sensitivity to carbapenems | |||
| 2 | 2a | A | PC1 | Penicillins | Inhibited by CA, TZB | |
| 2b | TEM-1, TEM-2, SHV-1 | Penicillins, early cephalosporins | Inhibited by CA, TZB | |||
| 2be | TEM-3, SHV-2, CTX-M-15, PER-1, VEB-1 | ESCP, monobactams | Inhibited by CA, TZB | |||
| 2br | TEM-30, SHV-10 | Penicillins | Resistance to CA, SB and TZB | |||
| 2ber | TEM-50 | ESCP, monobactams | Resistance to CA, SB and TZB | |||
| 2c | PSE-1, CARB-3 | Carbenicillin | Inhibited by CA, TZB | |||
| 2ce | RTG-4 | Carbenicillin, cefepime | Inhibited by CA, TZB | |||
| 2d | D | OXA-1, OXA-10 | Cloxacillin | Variable to CA, TZB | ||
| 2de | OXA-11, OXA-15 | ESCP, oxyimino-β-lactams | ||||
| 2df | OXA-23, OXA-48 | Cloxacillin, oxacillin, carbapenems | ||||
| 2e | A | CepA | ESCP | Inhibited by CA but not aztreonam | ||
| 2f | KPC-2, IMI-1, SME-1 | Carbapenems, oxyimino-β-lactams, cephamycins | Variable to CA, TZB | |||
| 3 | 3a | B1 | Zn2+ | IMP-1, VIM-1,CcrA, IND-1 | Carbapenems |
Resistance to CA, SB and TZB Inhibited by monobactams |
| 3b | B3 | L1, CAU-1, GOB-1, FEZ-1 | ||||
| 3c | B2 | CphA, Sfh-1 | Resistance to CA, SB and TZB | |||
CA clavulanic acid, SB sulbactam, TZB tazobactam, ESCP Extended-spectrum cephalosporins
*Updated in 2010 [17]
The following is a list of important issues that clinicians should keep in mind during the daily management of infections:
Basic science: PBPs (Fig. 2), which act as adenyl-alanine endopeptidases, enzymatically target an alanyl-alanine of the peptidoglycan pillar structure and catabolize the cross-links between the peptidoglycan layers (Fig. 1). β-lactams (Fig. 4) mimic the alanyl-alanine structure (Fig. 3) and inhibit the enzymatic action of PBPs. β-lactamases share the enzyme active site motifs with the PBP family, indicating that β-lactamases and PBPs have co-evolved (Additional file 1: Figure S1).
Classification: β-lactamases have been classified in two ways: one method is based on the molecular structure classification of Ambler, and the other method is based on the functional classification of Bush-Jacobi-Medeiros (Fig. 5). Understanding both structural and functional classifications is important for clinicians in choosing the most appropriate antimicrobial agent (Table 1).
ESBLs and carbapenemases: ESBLs and carbapenemases are represented in all classes, A to D, in the Ambler classification system (Figs. 5 and 10). Carbapenemases in all classes have been found on both plasmids and chromosomal genomes. The blaGES gene of class A (Fig. 6 and Additional file 2: Figure S2), blaOXA of class D (Fig. 7 and Additional file 4: Figure S4), and the MBL genes of class B (Fig. 9 and Additional file 5: Figure S5) are frequently encoded in an integron (Fig. 8). The extended-spectrum β-lactamases belonging to class C are encoded by the ampC gene usually carried on the chromosome (Additional file 3: Figure S3).
Diversity: The accumulation of small mutations in the primary amino acid sequences of β-lactamases ultimately extends the substrate spectrum. Clinicians can gain insight into how the use of antimicrobial agents may affect the molecular mutation of lactamases by analyzing the alignment of amino acid sequences of specific MBLs, such as IMP (Additional file 6: Figure S6) and VIM (Additional file 7: Figure S7).
Diagnostic: Finally, the characterization of antimicrobial resistance is critical for the classification of β-lactam resistance. For more information, please refer to references on the detection of β-lactamase-mediated resistance [75–78], because this is beyond the scope of this review. In brief, microbiological tests such as the Etest [76], the Clinical and Laboratory Standards Institute (CLSI) method [77] and the Double Disc Synergy Test (DDST) [78] can identify the phenotypes of ESBL, MBL, and ESAC. Novel multiplex polymerase chain reaction (PCR)-based ORF typing (POT) methods that are routinely used in clinical laboratories can identify the prevalent ESBL and carbapenemase genes, such as SHV, GES, TEM, CTX-M, and KPC of class A, NDM, IMP, and VIM of class B, and OXA-48 of class D in A. baumannii [79], P. aeruginosa clones [80], and other pathogenic bacteria. Antimicrobial gene cassettes, which are sometimes integrated into the integron structure via the interaction between two recombination sites (attI and attC), can be identified by PCR amplification with a primer set specific for the consensus sequence (CS) regions (5′-CS and 3′-CS) located upstream and downstream of the insertional site and subsequent DNA sequence analysis [81–84].
Conclusion
Various ESBLs and carbapenemases belonging to one of four molecular classes have propagated and are being detected among bacteria worldwide, suggesting that careful detection and monitoring are vital when critical care clinicians are treating infections caused by ESBL- and carbapenem-resistant bacteria. For the classification of β-lactamases, the Ambler method of molecular structure classification [16] is simple and effective at organizing the various ESBLs and carbapenemases, but functional classification using the Bush-Jacobi-Medeiros method [17, 18] is also important for clinicians faced with treating patients in a critical condition due to ESBL- and carbapenem-resistant bacterial infections.
Supplementary information
Additional file 1: Figure S1. The similarities in the structures of class A β-lactamases and penicillin-binding protein (PBP).
Additional file 2: Figure S2. The alignment of class A carbapenemases based on their primary sequences.
Additional file 3: Figure S3. The alignment of class C β-lactamases.
Additional file 4: Figure S4. Alignment of the class D β-lactamase OXA family (oxacillinase).
Additional file 5: Figure S5. Alignment of the class B metallo-β-lactamase family and their zinc enzymatic active center.
Additional file 6: Figure S6. The alignment of primary sequences of the class B metallo-β-lactamase IMP family.
Additional file 7: Figure S7. The alignment of primary sequences of the class B metallo-β-lactamase VIM family.
Acknowledgements
We thank Kate Fox, DPhil, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Abbreviations
- CHDL
Carbapenem-hydrolyzing class D β-lactamases
- CLSI
Clinical and Laboratory Standards Institute
- CPE
Carbapenemase-producing Enterobacteriaceae
- CS
Consensus sequence
- DDST
Double disc synergy test
- EDTA
Ethylenediaminetetraacetic acid
- ESAC
Extended-spectrum AmpC
- ESBL
Extended-spectrum β-lactamase
- FIM
Florence imipenemase
- GES
Guiana extended-spectrum β-lactamase
- GIM
Germany imipenemase
- HMW-PBP
High molecular weight penicillin-binding protein
- IMI/NMC-A
Imipenemase/non-metallocarbapenemase-A
- IMP
Imipenemase
- KPC
Klebsiella pneumoniae carbapenemase
- LMW-PBP
High molecular weight penicillin-binding protein
- MBL
Metallo-β-lactamase
- MDRP
Multidrug-resistant Pseudomonas aeruginosa
- MRSA
Methicillin-resistant Staphylococcus aureus
- NAG
N-acetylglucosamine
- NAM
N-acetylmuramic acid
- NDM
New Delhi metallo-β-lactamase
- PBP
Penicillin-binding protein
- PCR
Polymerase chain reaction
- PDRP
Pandrug-resistant Pseudomonas aeruginosa
- POT
PCR-based ORF typing
- SFC
Serratia fonticola carbapenemase
- SHV
Sulfhydryl variable lactamase
- SME
Serratia marcescens enzyme
- SPM
Sao Paulo metallo-β-lactamase
- VIM
Verona integrated-encoded MBL
- XDRP
Extensively drug-resistant Pseudomonas aeruginosa
Authors’ contributions
TS wrote the manuscript, figure legends, and tables. KK and KM contributed to preparing and revising the manuscript. All authors have read and approved the final manuscript.
Authors’ information
TS is a Professor of the Anesthesiology Department at Kyoto Prefectural University of Medicine, Japan. KK is a Chief Manager in the Division of Critical Care, Kyoto City Hospital, and a Visiting Professor of the Anesthesiology Department at Kyoto Prefectural University of Medicine, Japan. KM is an Associate Professor in the Anesthesiology Department at Kyorin University, Japan.
Funding
This work was supported by the Japan Society for the Promotion of Science and a Grant-in-Aid for Scientific Research (KAKENHI) to Teiji Sawa (No. 24390403, No. 15H05008, and No. 18H02905) from The Ministry of Education, Culture, Sports, Science and Technology, Japan.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
TS has a patent for immunization with PcrV from the Regent of the University of California (Berkeley, CA, USA). All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Teiji Sawa, Email: anesth@koto.kpu-m.ac.jp.
Kunihiko Kooguchi, Email: koochanjp@yahoo.co.jp.
Kiyoshi Moriyama, Email: mokiyo@ks.kyorin-u.ac.jp.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40560-020-0429-6.
References
- 1.Codjoe Francis, Donkor Eric. Carbapenem Resistance: A Review. Medical Sciences. 2017;6(1):1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis. 2018;66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi Y, Paterson DL. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med. 2015;36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Loon K, Voor In ’t Holt AF, Vos MC. A systematic review and meta-analyses of the clinical epidemiology of carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2017;62:e01730–e01717. doi: 10.1128/AAC.01730-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheu CC, Chang YT, Lin SY, Chen YH, Hsueh PR. Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front Microbiol. 2019;10:80. doi: 10.3389/fmicb.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CS, Doi Y. Therapy of infections due to carbapenem-resistant gram-negative pathogens. Infect Chemother. 2014;46:149–164. doi: 10.3947/ic.2014.46.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50:43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 11.Falagas ME, Koletsi PK, Bliziotis IA. The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J Med Microbiol. 2006;55:1619–1629. doi: 10.1099/jmm.0.46747-0. [DOI] [PubMed] [Google Scholar]
- 12.Bush K. Past and present perspectives on beta-lactamases. Antimicrob Agents Chemother. 2018;62:e01076–e01018. doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghuysen JM. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 14.Jamin M, Wilkin JM, Frère JM. Bacterial DD-transpeptidases and penicillin. Essays Biochem. 1995;29:1–24. [PubMed] [Google Scholar]
- 15.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 16.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond Ser B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 17.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liakopoulos A, Mevius D, Ceccarelli D. A review of SHV extended-spectrum beta-lactamases: neglected yet ubiquitous. Front Microbiol. 2016;7:1374. doi: 10.3389/fmicb.2016.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sirot D. Extended-spectrum plasmid-mediated beta-lactamases. J Antimicrob Chemother. 1995;36(Suppl A):19–34. doi: 10.1093/jac/36.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 21.Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malloy AM, Campos JM. Extended-spectrum beta-lactamases: a brief clinical update. Pediatr Infect Dis J. 2011;30:1092–1093. doi: 10.1097/INF.0b013e31823c0e9d. [DOI] [PubMed] [Google Scholar]
- 23.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walther-Rasmussen J, Hoiby N. Class A carbapenemases. J Antimicrob Chemother. 2007;60:470–482. doi: 10.1093/jac/dkm226. [DOI] [PubMed] [Google Scholar]
- 25.Philippon A, Slama P, Deny P, Labia R. A structure-based classification of class A beta-lactamases, a broadly diverse family of enzymes. Clin Microbiol Rev. 2016;29:29–57. doi: 10.1128/CMR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naas T, Vandel L, Sougakoff W, Livermore DM, Nordmann P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, SME-1, from Serratia marcescens S6. Antimicrob Agents Chemother. 1994;38:1262–1270. doi: 10.1128/aac.38.6.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barthélémy M, Peduzzi J, Verchère-Beaur C, Ben Yaghlane H, Labia R. Purification and biochemical properties of Pitton’s type 2 beta-lactamase (SHV-1) Ann Inst Pasteur Microbiol (1985) 1986;137B:19–27. doi: 10.1016/s0769-2609(86)80090-4. [DOI] [PubMed] [Google Scholar]
- 29.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J. 2011;104:40–45. doi: 10.1097/SMJ.0b013e3181fd7d5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordmann P, Mariotte S, Naas T, Labia R, Nicolas MH. Biochemical properties of a carbapenem-hydrolyzing beta-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob Agents Chemother. 1993;37:939–946. doi: 10.1128/aac.37.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sievers F, Higgins DG. Clustal Omega. Curr Protoc Bioinformatics. 2014;48:3.13.1–3.1316. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
- 34.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henriques I, Moura A, Alves A, Saavedra MJ, Correia A. Molecular characterization of a carbapenem-hydrolyzing class A beta-lactamase, SFC-1, from Serratia fonticola UTAD54. Antimicrob Agents Chemother. 2004;48:2321–2324. doi: 10.1128/AAC.48.6.2321-2324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naas T, Dortet L, Iorga BI. Structural and functional aspects of class A carbapenemases. Curr Drug Targets. 2016;17:1006–1028. doi: 10.2174/1389450117666160310144501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. GES-2, a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001;45:2598–2603. doi: 10.1128/AAC.45.9.2598-2603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22:161–182. doi: 10.1128/aac.45.9.2598-2603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomson KS. Extended-spectrum-beta-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol. 2010;48:1019–1025. doi: 10.1128/JCM.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother. 2002;46:1–11. doi: 10.1128/aac.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordmann P, Mammeri H. Extended-spectrum cephalosporinases: structure, detection and epidemiology. Future Microbiol. 2007;2:297–307. doi: 10.2217/17460913.2.3.297. [DOI] [PubMed] [Google Scholar]
- 43.Ahmed AM, Shimamoto T. Emergence of a cefepime- and cefpirome-resistant Citrobacter freundii clinical isolate harbouring a novel chromosomally encoded AmpC beta-lactamase, CMY-37. Int J Antimicrob Agents. 2008;32:256–261. doi: 10.1016/j.ijantimicag.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 44.Wachino J, Kurokawa H, Suzuki S, Yamane K, Shibata N, Kimura K, Ike Y, Arakawa Y. Horizontal transfer of blaCMY-bearing plasmids among clinical Escherichia coli and Klebsiella pneumoniae isolates and emergence of cefepime-hydrolyzing CMY-19. Antimicrob Agents Chemother. 2006;50:534–541. doi: 10.1128/AAC.50.2.534-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michaux C, Massant J, Kerff F, Frere JM, Docquier JD, Vandenberghe I, Samyn B, Pierrard A, Feller G, Charlier P, Van Beeumen J, Wouters J. Crystal structure of a cold-adapted class C beta-lactamase. FEBS J. 2008;275:1687–1697. doi: 10.1111/j.1742-4658.2008.06324.x. [DOI] [PubMed] [Google Scholar]
- 46.Hall LM, Livermore DM, Gur D, Akova M, Akalin HE. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danel F, Hall LM, Gur D, Livermore DM. OXA-15, an extended-spectrum variant of OXA-2 beta-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob Agents Chemother. 1997;41:785–790. doi: 10.1128/AAC.41.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antunes NT, Fisher JF. Acquired class D beta-lactamases. Antibiotics (Basel) 2014;3:398–434. doi: 10.3390/antibiotics3030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zander E, Fernandez-Gonzalez A, Schleicher X, Dammhayn C, Kamolvit W, Seifert H, Higgins PG. Worldwide dissemination of acquired carbapenem-hydrolysing class D beta-lactamases in Acinetobacter spp. other than Acinetobacter baumannii. Int J Antimicrob Agents. 2014;43:375–377. doi: 10.1016/j.ijantimicag.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Donald HM, Scaife W, Amyes SG, Young HK. Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44:196–199. doi: 10.1128/AAC.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/aac.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walther-Rasmussen J, Hoiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57:373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 53.Paetzel M, Danel F, de Castro L, Mosimann SC, Page MG, Strynadka NC. Crystal structure of the class D beta-lactamase OXA-10. Nat Struct Biol. 2000;7:918–925. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- 54.Palzkill T. Metallo-beta-lactamase structure and function. Ann N Y Acad Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 56.Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev. 2014;78:257–277. doi: 10.1128/MMBR.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng Y, Bao X, Ji L, Chen L, Liu J, Miao J, Chen D, Bian H, Li Y, Yu G. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14:45. doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyoshi-Akiyama T, Kuwahara T, Tada T, Kitao T, Kirikae T. Complete genome sequence of highly multidrug-resistant Pseudomonas aeruginosa NCGM2.S1, a representative strain of a cluster endemic to Japan. J Bacteriol. 2011;193:7010. doi: 10.1128/JB.06312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–1590. doi: 10.1128/AAC.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toleman MA, Simm AM, Murphy TA, Gales AC, Biedenbach DJ, Jones RN, Walsh TR. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J Antimicrob Chemother. 2002;50:673–679. doi: 10.1093/jac/dkf210. [DOI] [PubMed] [Google Scholar]
- 62.Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR. Molecular characterization of a beta-lactamase gene, blaGIM-1, encoding a new subclass of metallo-beta-lactamase. Antimicrob Agents Chemother. 2004;48:4654–4661. doi: 10.1128/AAC.48.12.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier JD, Rossolini GM. FIM-1, a new acquired metallo-beta-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob Agents Chemother. 2013;57:410–416. doi: 10.1128/AAC.01953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laraki N, Galleni M, Thamm I, Riccio ML, Amicosante G, Frere JM, Rossolini GM. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:890–901. doi: 10.1128/AAC.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. Epidemiology and characteristics of metallo-beta-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;47:81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. Plasmid-encoded metallo-beta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother. 2001;45:1343–1348. doi: 10.1128/AAC.45.5.1343-1348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohno Y, Nakamura A, Hashimoto E, Matsutani H, Abe N, Fukuda S, Hisashi K, Komatsu M, Nakamura F. Molecular epidemiology of carbapenemase-producing Enterobacteriaceae in a primary care hospital in Japan, 2010-2013. J Infect Chemother. 2017;23:224–229. doi: 10.1016/j.jiac.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 69.Pournaras S, Tsakris A, Maniati M, Tzouvelekis LS, Maniatis AN. Novel variant (bla(VIM-4)) of the metallo-beta-lactamase gene bla(VIM-1) in a clinical strain of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2002;46:4026–4028. doi: 10.1128/aac.46.12.4026-4028.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karampatakis T, Antachopoulos C, Tsakris A, Roilides E. Molecular epidemiology of carbapenem-resistant Pseudomonas aeruginosa in an endemic area: comparison with global data. Eur J Clin Microbiol Infect Dis. 2018;37:1211–1220. doi: 10.1007/s10096-018-3244-4. [DOI] [PubMed] [Google Scholar]
- 71.Rasmussen BA, Bush K. Carbapenem-hydrolyzing beta-lactamases. Antimicrob Agents Chemother. 1997;41:223–232. doi: 10.1128/AAC.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cadag E, Vitalis E, Lennox KP, Zhou CL, Zemla AT. Computational analysis of pathogen-borne metallo beta-lactamases reveals discriminating structural features between B1 types. BMC Res Notes. 2012;5:96. doi: 10.1186/1756-0500-5-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fonseca F, Bromley EH, Saavedra MJ, Correia A, Spencer J. Crystal structure of Serratia fonticola Sfh-I: activation of the nucleophile in mono-zinc metallo-beta-lactamases. J Mol Biol. 2011;411:951–959. doi: 10.1016/j.jmb.2011.06.043. [DOI] [PubMed] [Google Scholar]
- 74.Garau G, Bebrone C, Anne C, Galleni M, Frere JM, Dideberg O. A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J Mol Biol. 2005;345:785–795. doi: 10.1016/j.jmb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 75.Livermore DM, Brown DF. Detection of beta-lactamase-mediated resistance. J Antimicrob Chemother. 2001;48(Suppl 1):59–64. doi: 10.1093/jac/48.suppl_1.59. [DOI] [PubMed] [Google Scholar]
- 76.Cormican MG, Marshall SA, Jones RN. Detection of extended-spectrum beta-lactamase (ESBL)-producing strains by the Etest ESBL screen. J Clin Microbiol. 1996;34:1880–1884. doi: 10.1128/JCM.34.8.1880-1884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.CLSI: Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne: Clinical and Laboratory Standards Institute; 2017. p. 102–43.
- 78.Drieux L., Brossier F., Sougakoff W., Jarlier V. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clinical Microbiology and Infection. 2008;14:90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki M, Hosoba E, Matsui M, Arakawa Y. New PCR-based open reading frame typing method for easy, rapid, and reliable identification of Acinetobacter baumannii international epidemic clones without performing multilocus sequence typing. J Clin Microbiol. 2014;52:2925–2932. doi: 10.1128/JCM.01064-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suzuki M, Yamada K, Aoki M, Hosoba E, Matsumoto M, Baba H, Iinuma Y. Applying a PCR-based open-reading frame typing method for easy genotyping and molecular epidemiological analysis of Pseudomonas aeruginosa. J Appl Microbiol. 2016;120:487–497. doi: 10.1111/jam.13016. [DOI] [PubMed] [Google Scholar]
- 81.Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, Kato H, Kai K, Arakawa Y. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003;41:5407–5413. doi: 10.1128/JCM.41.12.5407-5413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/jcm.41.12.5407-5413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kainuma A, Momiyama K, Kimura T, Akiyama K, Inoue K, Naito Y, Kinoshita M, Shimizu M, Kato H, Shime N, Fujita N, Sawa T. An outbreak of fluoroquinolone-resistant Pseudomonas aeruginosa ST357 harboring the exoU gene. J Infect Chemother. 2018;24:615–622. doi: 10.1016/j.jiac.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 84.Mihara T, Kimura T, Momiyama K, Kainuma A, Akiyama K, Ohara J, Inoue K, Kinoshita M, Shimizu M, Moriyama K, Fujita N, Sawa T. Secondary in-hospital epidemiological investigation after an outbreak of Pseudomonas aeruginosa ST357. J Infect Chemother. 2019. 10.1016/j.jiac.2019.09.014. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The similarities in the structures of class A β-lactamases and penicillin-binding protein (PBP).
Additional file 2: Figure S2. The alignment of class A carbapenemases based on their primary sequences.
Additional file 3: Figure S3. The alignment of class C β-lactamases.
Additional file 4: Figure S4. Alignment of the class D β-lactamase OXA family (oxacillinase).
Additional file 5: Figure S5. Alignment of the class B metallo-β-lactamase family and their zinc enzymatic active center.
Additional file 6: Figure S6. The alignment of primary sequences of the class B metallo-β-lactamase IMP family.
Additional file 7: Figure S7. The alignment of primary sequences of the class B metallo-β-lactamase VIM family.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.