Abstract
Background
High-throughput transcriptomics has matured into a very well established and widely utilised research tool over the last two decades. Clinical datasets generated on a range of different platforms continue to be deposited in public repositories provide an ever-growing, valuable resource for reanalysis. Cost and tissue availability normally preclude processing samples across multiple technologies, making it challenging to directly evaluate performance and whether data from different platforms can be reliably compared or integrated.
Methods
This study describes our experiences of nine new and established mRNA profiling techniques including Lexogen QuantSeq, Qiagen QiaSeq, BioSpyder TempO-Seq, Ion AmpliSeq, Nanostring, Affymetrix Clariom S or U133A, Illumina BeadChip and RNA-seq of formalin-fixed paraffin embedded (FFPE) and fresh frozen (FF) sequential patient-matched breast tumour samples.
Results
The number of genes represented and reliability varied between the platforms, but overall all methods provided data which were largely comparable. Crucially we found that it is possible to integrate data for combined analyses across FFPE/FF and platforms using established batch correction methods as required to increase cohort sizes. However, some platforms appear to be better suited to FFPE samples, particularly archival material.
Conclusions
Overall, we illustrate that technology selection is a balance between required resolution, sample quality, availability and cost.
Keywords: FFPE, Fresh-frozen, Gene expression, Microarray, Sequencing, Transcriptomics
Background
Since their inception microarrays have been adopted as a major tool for the study of clinical samples to improve our understanding of diseases, development of molecular subtyping and prognostic signatures for clinical decision-making [1]. A crucial consideration for many clinical studies is whether new data generated can be directly compared or integrated with pre-existing datasets for robust classification and response prediction.
RNA sequencing (RNAseq) has somewhat supplanted microarrays for transcriptome analysis. However, in translational research when the focus is often restricted to identifying differentially expressed genes and pathways, rather than detecting specific isoforms and splice variants, decisions on which platform to use are often based upon cost, rather than resolution, particularly if this means more samples can be examined to maximise statistical power for a fixed budget. Indeed, RNAseq is not without its limitations, Robert and Watson recently demonstrated that RNAseq is unable to accurately measure expression of hundreds of genes in the human genome [2].
Many high-throughput profiling studies rely on sample availability and cost rather than statistical power [1]. Direct integration of datasets enables meta-analysis and has the potential to improve statistical power and the generalisability of results for robust classification and response prediction. However, non-trivial systematic bias or ‘batch effects’ can occur within and between microarray platforms [3–6]. Contrary to The MicroArray Quality Control guidelines [7], gene expression data can be directly integrated and robust results can be produced from fundamentally different technologies such as Affymetrix GeneChips and Illumina BeadChips [3]. This finding has since been supported by other studies [8, 9].
Early microarray studies involving clinical samples were dependent on relatively large amounts of high quality RNA and thus relied heavily on the availability of fresh frozen (FF) tissue. However, collection and storage of FF tissue is costly and can be logistically prohibitive. Protocols and technologies capable of generating high quality whole-genome transcriptomic data from archival formalin fixed paraffin embedded (FFPE) tissues are in demand [10]. FFPE tissues are available routinely in the clinical setting and can be stored at ambient temperature for many years, allowing easy transportation. A large number of studies have compared matched FF and FFPE samples, with some reporting reduced efficacy or numbers of detected transcripts and batch effects similar to those reported for different profiling technologies (recently reviewed [11]). Most studies conclude that the data can be compared to some extent, subject to certain considerations, accepting that RNA from FFPE samples is often degraded and continues to degrade with age [10]. Whilst earlier microarray technologies performed poorly with degraded RNA, newer kits and platforms have emerged using targeted sequencing such as Ion AmpliSeq Transcriptome and BioSpyder TempO-Seq or 3′ sequencing from Lexogen QuantSeq. Other technologies such as NanoString are promising, but are limited to panels of genes rather than whole genome transcriptome. In this study, a number of gene expression profiling platforms were compared.
Methods
Clinical samples
All patients gave informed consent and the study was approved by the local ethics committee (LREC; 2001/8/80 and 2001/8/81). RNA was extracted from primary human breast cancer samples collected over 15 years at the Edinburgh Breast Unit from post-menopausal women with estrogen receptor positive disease, treated with 3-months of neoadjuvant endocrine therapy. Sequential biopsies were taken pre-treatment, early (14-days) on-treatment and at surgery 3–6 months later (late on-treatment) from each patient. Part of the biopsy material collected was snap-frozen in liquid nitrogen and part was fixed in formalin and embedded in paraffin. RNA was extracted from fresh frozen tissue using the Qiagen miRNeasy kit and from 2 × 20 μm FFPE tissue sections using the RNeasy FFPE kit using the manufacturer’s standard protocols for each kit. Agilent RIN values for fresh frozen tissue were > 7 and for FFPE tissue were < 3.
Transcriptomics
Building upon large scale clinical studies to investigate the effects of endocrine therapy on breast cancer using Affymetrix U133A arrays [12] and Illumina HT12-V4 BeadChips [13], this study, utilised patient-matched sets of samples across a range of transcriptomic technologies: Affymetrix Clariom S, NanoString, Ion AmpliSeq Transcriptome, BioSpyder TempO-seq [14] Lexogen QuantSeq and RNA-seq (Table 1). Microarray samples were processed as directed by the manufacturer’s instructions. Nanostring profiling was performed using nCounter technology as per the manufacturer’s instructions. Sequencing was performed as described: Ion Ampliseq samples were processed using an Ion a PI™ Chip Kit v3 and sequenced using an Ion Proton™ System. QiaSeq samples were sequenced using the NextSeq 500/550 High-Output v2 (150 cycle) Kit on the NextSeq 550 platform. For TempoSeq samples, single read (1x75bp) sequencing was performed using the NextSeq 500/550 High-Output v2 (75 cycle) Kit on the NextSeq 550 platform. For QuantSeq samples were either processed via single read (1x75bp) sequencing performed using the NextSeq 500/550 High-Output v2 (75 cycle) Kit on the NextSeq 550 platform or via Ion a PI™ Chip Kit v3 and sequenced using an Ion Proton™ System. For RNASeq samples the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold (Illumina) was used and sequencing was performed on an Illumina HiSeq 2500 using a 2x50bp configuration with an average of 136 million read pairs per sample. All data is publicly available from NCBI GEO (www.ncbi.nlm.nih.gov/geo/) under super-series accession GSE130645.
Table 1.
Comparison of traditional and new microarray platforms with sequencing approaches
| Technology | Technology/Platform | Biochemistry | Approx. Throughput | Max. no. probes/primer pairs | No. of mapped ENSG IDs | Read Depths | Input FFPE RNA (ng)* | Approx. cost per sample (£)** | Success rate of FF samples (n) | Success rate of FFPE samples (n) |
|---|---|---|---|---|---|---|---|---|---|---|
| 3′ RNA sequencing | Lexogen QuantSeq | RNA → RT, oligodT priming from 3′ end, random priming towards 3′ end → amplification and barcoding → sequencing | 96 samples per 5 days | 55,765 | 25,610 | 10 M | 500 | 90 | N/A | 98% (318) |
| QiaSeq UPX 3′ Transcriptome | RNA → RT, oligodT priming for cDNA synthesis →template switching for 2nd strand synthesis priming → fragmentation → end repair addition, adapter ligation → PCR to add indices → sequencing | 96 samples per 5 days | 42,553 | 20,000 | 15 M | 10 | 50 | N/A | 94% (48) | |
| Specific Targeted Sequencing | BioSpyder TempO-Seq | RNA → annealed 50 bp detector oligos are ligated then amplified and barcoded → sequencing | 192 samples per 4 days | 19,300 | 19,300 | 12 M | 20 μm FFPE Section | 160 | N/A | 95% (38) |
| Ion Ampliseq Transcriptome | RNA → RT, multiplex PCR → sequence barcoding → emulsion PCR → sequencing of ~ 150 bp targets | 96 samples per 5 days | 20,802 | 19,059 | 8 M | 10 | 160 | 100% (108) | 76% (76) | |
| Targeted Probes | Nanostring | RNA → hybridisation to fluorescent barcoded probes in solution → immobilised in nCounter cartridge → scan | 12 samples per day (800 genes) | 800 | 800 | N/A | 50 | 250 | N/A | 100% (12) |
| Newer Microarray | Affymetrix Clariom S | RNA → cRNA amplification → hybridisation to GeneChip → scan | 192 samples per 4 days | 211,300 | > 20,000 | N/A | 50 | 100 | 100% (3) | 100% (8) |
| Traditional Microarray | Affymetrix U133A | 192 per day | 250,833 | 11,827 | N/A | 50 | 360 | 100% (178) | 100% (286) | |
| Illumina BeadChip HT-12 v3 / v4 | RNA → RT, amplification, biotinylation (NuGEN WT Ovation kit) → hybridisation to 50 bp probes on chip → scan | 96 samples per 1.5 days | 47,323 | 22,571 | N/A | 1500 | 195 | 91% (348) | 21% (206) | |
| Full RNA Sequencing | RNA-seq | RNA → fragmentation → RT → barcoded library construction → genome-wide full RNA sequencing | 8 samples per 5 days | 20,025 | 18,57s1 | 136 M paired reads | 2000 | 250–500 | 100% (52) | 100% (87) |
*Input RNA reflects quantities used in this study – for input ranges refer to the manufacturer’s guidelines
**Estimated costs (£, UK December 2019) include library preparation and sequencing. Costs can vary by sample numbers and sequencing infrastructure
Data analysis
Illumina and Affymetrix data were pre-processed and normalised as described previously [3]. NanoString data were generated using the nSolver 3.0 software. Ion AmpliSeq Transcriptome data were generated using the AmpliSeq RNA plugin in the Torrent Suite Software and normalised using RPM (reads assigned per million mapped reads) method. QiaSeq FASTQ files were uploaded to the GeneGlobe Data Analysis Center, an online platform provided by QIAGEN. The primary analysis module for the UPX 3′ Transcriptome Kit was used to generate UMI-based gene expression estimates from the reads for all samples. QuantSeq raw data in .bcl format was transferred from the NextSeq instrument to a Linux system, where demultiplexed FASTQ files were generated using Bcl2fastq2 v2.17.1.14 software provided by Illumina. The lane-splitting feature was disabled to create a single FASTQ file for each library. FASTQ files were then uploaded to the BlueBee genomics platform (https://www.bluebee.com) and read-trimming and alignment was performed using the QuantSeq plugin. TempoSeq FASTQ files were sent to BioCalvis (the manufacturer of BioSpyder), who performed the alignment and then generated the raw (un-normalised) gene counts file using their proprietary software. For RNAseq, alignment was performed using STAR74. Transcript abundance estimates for each sample were performed using Salmon, an expectation-maximization algorithm using the UCSC gene definitions. Raw read counts for all RNAseq samples were normalized to a fixed upper quartile.
All sequence data were aligned to the human reference hg19 genome. For all data, probes or genes were then mapped to Ensembl gene annotations: Affymetrix datasets were mapped using a chip definition file (CDF) [15] and all other datasets were mapped using BioMart. All data were Log2 transformed and filtered for those expressed in 70% of samples using the cluster 3.0 software then quantile normalised using the R/Bioconductor software and packages [16]. Following data integration, correction of systematic bias was performed using ComBat as described previously [3].
Results
Performance and cost comparison of platforms for FF and FFPE tissue
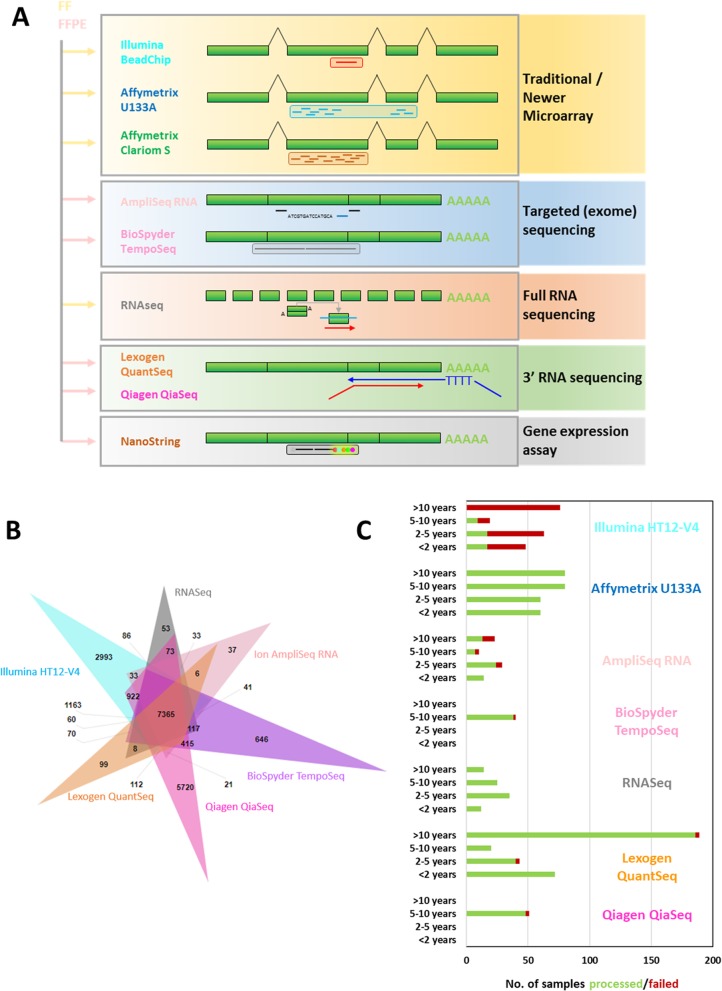
Each of the nine technologies evaluated here have different mRNA input requirements, probe designs (Fig. 1a) and protocols (summarised in Table 1). Although the total number and position of probes/primers/counts varies widely among the transcriptome-wide approaches (Table 1, Fig. 1a), a common set of 7365 Ensembl transcripts were represented across the six whole transcriptome platforms (Fig. 1b). Nanostring and Affymetrix U133 were omitted as they do not represent the whole transcriptome and the Clariom S was excluded as only three samples were processed). RNAseq may have the highest resolution, but also the highest RNA input requirement (100-4000 ng) and it is the most expensive whole transcriptome technology at two to five times the cost of other approaches (Table 1). The NanoString platform could be cost-effective for a small number of genes, but compares poorly on price for large numbers of genes (costed for maximum coverage in a single experiment: 770 genes). The newest and least expensive technologies are Affymetrix Clariom S array with WT Pico kit and Lexogen QuantSeq. Success rate is an important consideration for clinical studies, particularly with before and on-treatment matched samples considered in this study. Looking at the numbers of samples which have failed using different technologies based on the respective manufacturers quality control criteria we found that success rates for generating robust expression profiles from FFPE tissues were excellent (> 95%) for the latest Lexogen QuantSeq, Qiagen Qiaseq, BioSpyder TempO-Seq methods. This is despite the RNA integrity number (RIN) values for fresh frozen tissue normally being above 7, but for FFPE tissues were generally less than 3. However, success rate was moderate for the Ampliseq RNA Transcriptome (83%) and poor for the older Illumina BeadChip (22%). By comparison RNA from FF tissue had a high success rate (91–100%) with several hundred samples processed on the Illumina BeadChip, Affymetrix U133A chips and RNAseq (Table 1). As shown previously [10], older FFPE samples were found to perform very poorly with the more established technologies (Fig. 1c) whereas NanoString, Lexogen QuantSeq and RNA-seq were found to work well with old FFPE tissue-derived RNA.
Fig. 1.
Comparison of gene expression profiling approaches (a) Schematic of probe/primer designs for each technology. A table showing which samples were processed on each technology is provided in Additional file 1: Table S1. b Number of overlapping Ensembl gene identifiers detected in each dataset (Nanostring and Affymetix U133 were omitted as they do not represent the whole transcriptome and the Clariom S was excluded as only three samples were processed). c Summary of FFPE sample processing success rates by sample age using whole-transcriptome platforms
Integration of datasets across platforms while preserving biological variability
To evaluate how newer technologies with desirable features such as lower costs or RNA input requirements compared to the more established methodologies, we profiled the same RNA from a subset of samples to directly compare gene expression measurements across the platforms (Additonal file 1: Table S1). These comparisons have two purposes; firstly to determine whether the new technology provides similar quality results to the established method. Secondly, to evaluate whether it will be possible to directly integrate datasets generated on the new platform with existing local or publicly available data from another platform, as we have done previously [3, 4, 6]. Indeed, while it is altruistic to minimise measurement error by using the same platforms, with constantly evolving technologies and lower associated costs this is not often realistic. Therefore, the ability to implement approaches to increase validity across platforms is of great importance.
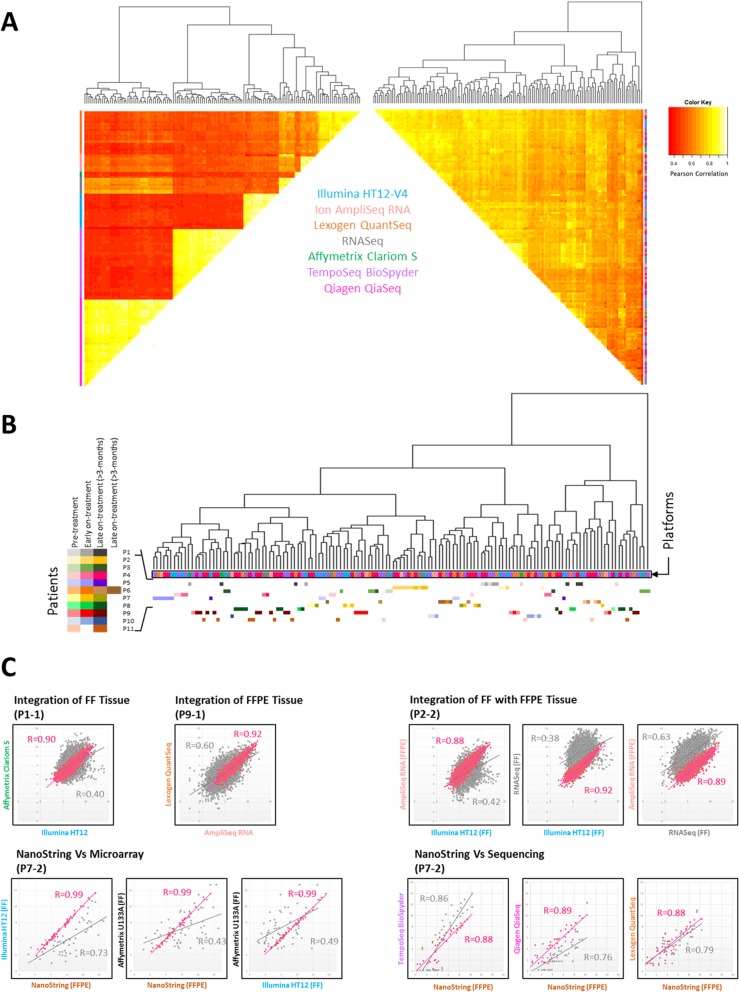
Not surprisingly, when all samples were integrated together low correlations (r = 0.4–0.6) were observed between pairs of samples processed on different technologies. Hierarchical clustering showed clearly that gene expression values group by technology and technical artefacts, rather than by genuine biology (Fig. 2a, left). Following batch correction using the well-established and highly cited ComBat method [17], correlations were much higher and the majority of ‘paired’ samples clustered together, indicating greater variation between biological samples than between gene expression measurement platforms (Fig. 2a, right). Looking more closely, instances of the same time-point processed on different platforms clustered closely (if not together) and different time points from the same patients showed variation (due to treatment), whilst also often clustering with other time points from the same patient (Fig. 2b), as has been previously shown for sequential patient-matched samples [13]. These results are consistent with our previous results showing a reduction in technical artifacts, without loss of biological variation [3].
Fig. 2.
Batch correction allows robust direct integration of transcriptomic data across platforms. a Dissimilarity heatmaps based upon Pearson correlations ranging from 0.4 (red) through shades of orange and yellow to 1.0 (white). Left triangle shows the combined dataset of 6844 genes across 7 gene expression platforms. Right triangle shows the same data following batch correction with Combat. Coloured bars below dendrograms denote the platform. b Enlargement of the dendrogram to demonstrate that the majority of the same time-point patient samples processed on different platforms cluster together following batch correction. c Scatter plots before (grey) and after batch correction (pink) of the same sample, either FF or FFPE processed across different platforms. In each case the Pearson correlations increase substantially following batch correction. Patient samples are denoted − 1 for pre-treatment, − 2 for early on-treatment
Clear batch effects were evident when comparing mRNA extracted from FF samples across Illumina HT12, Ion Ampliseq Transcriptome and Affymetrix Clariom S, with low Pearson correlations (r = 0.4–0.58). However standard batch correction approaches such as ComBat [17] minimised technical bias effect and increased correlation to r > 0.9 for paired samples. Similar low correlations and improved correlations following batch correction were observed for different technologies with FFPE samples and for comparisons of matched FF and FFPE or for the same sample across different platforms (Fig. 2c). Comparison of measurements of the 56 overlapping genes assayed using NanoString, whole-genome (Illumina HT12) and part-genome (Affymetrix U133A) expression microarrays were also significantly improved following batch correction.
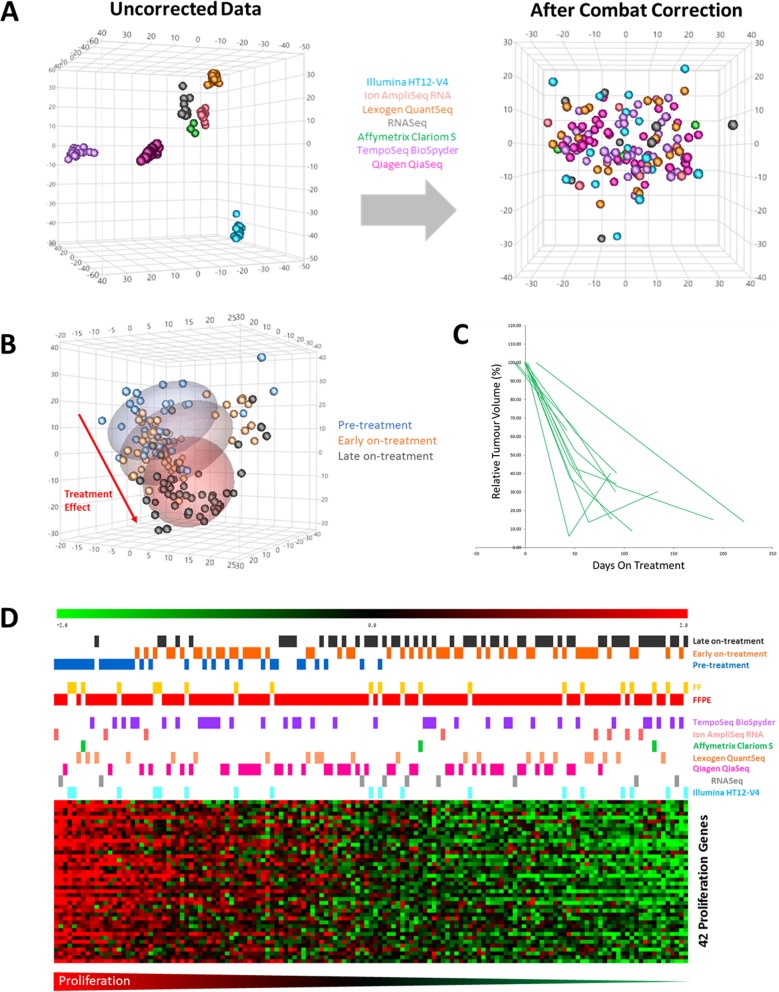
Looking at the samples more closely by multi-dimensional scaling it is clear that whilst they cluster by platform before batch correction (Fig. 3a), afterwards they do not (Fig. 3b) and more importantly, instead they cluster by time point (Fig. 3c). Pre-treatment samples are most clearly separated from late on-treatment samples, with early on treatment samples in-between, as would be expected.
Fig. 3.
Robust gene expression measurement across platforms following batch correction. Correction of systematic platform bias and integration of data from fresh frozen and FFPE tissues. a 3D multi-dimensional scaling (MDS) before (left) and after (right) batch correction of 6844 common genes. Samples coloured by platform and shapes indicates time point. b MDS plot of the batch corrected data with samples coloured by time-point clearly demonstrates a consistent treatment effect seen across sequential patient-matched samples. c Ultrasound measurements of the eleven breast tumours which relate to the sequential patient-matched samples indicating consistent reductions in tumour volume over time across the patients. d Ranking patient samples by the expression of 42 common proliferation genes (listed in Additional file 2: Table S2) illustrates consistent changes resulting from endocrine therapy, which appears to be independent from profiling platform. Pre-treatment samples tend to have relatively high proliferation, whilst as expected early, and particularly late on-treatment samples have lower proliferation. Heatmap colours are Red = High, Green = low
For further confirmation of the validity of the batch-corrected data, we ranked samples by expression of 42 proliferation genes, previously reported by us [12] that change with endocrine therapy (list of gene provided in Additional file 2: Table S2). Molecular changes in the tumours reflect the ultrasound measurements across the eleven breast tumours, concordant with consistent reductions in tumour volume over time across the patients (Fig. 3c). Ranked by proliferation genes the samples are ordered by time point, consistent with our previous results [12], and not by platform or preservation method (Fig. 3b). These results suggest that comparable gene expression profiles can be generated across the platforms using FFPE material and FFPE is a reliable alternative to FF (Fig. 3d).
Discussion
Overall we find that gene expression data from the newer technologies is largely concordant with that from the more established methods. The newer 3′ sequencing approaches from Lexogen and Qiagen appear highly reliable and cost effective for old FFPE samples, this potentially allows valuable data to be generated from clinical samples that would not have been previously possible. The TempO-Seq method [14] from BioSpyder is an interesting approach as you can analyse expression without pre-amplification directly from a micro-dissected area of interest taken from a single FFPE section, maximizing utilization of precious or limited samples. Full RNAseq analysis is often considered the gold standard, however when tissue samples are particularly small or there is a desire to perform a range of assays or multi-omic approaches, the newer targeted sequencing approaches with many fold smaller input requirements may be a much more attractive proposition. A number of previous studies have conducted comparisons of the same samples generated from fresh and archived tissues [18, 19]. The numbers of detected genes from FFPE samples has previously been shown to be lower than from fresh tissue [19], however protocols have continued to improve [10]. It is important to remember that in all pairwise tissue comparisons where RNA is extracted separately that they cannot represent exactly the same material and are only ever adjacent, leading to inevitable potential minor variations in tissue composition. Despite this, the well-established Combat method for batch correction [17] was again found to perform well to integrate data from different sample types or technologies, this approach has been found to be superior in a many of previous studies [20].
A general finding of most platform comparison approaches is that although the correlation values between different microarray or sequencing approaches may be poor to moderate, which may relate to differences in dynamic range of the technologies, there is generally very high concordance when considering differentially expressed genes [3, 6, 21]. A comprehensive study of TCGA data found only 1.2% of genes were inconsistent by fold change [21]. A wider issue with transcriptomic studies that there is no optimal analysis pipeline for every single analysis [22].
This single study perhaps considers the widest range of gene expression technologies using FF and FFPE tissues published to date, but we acknowledge that this study documents a translational research group’s experiences, rather than being a definitive, comparison study. Not every sample was tested on every platform and some leading technologies remain to be tested, including Agilent, TaqMan and Fluidigm - due to local availability and opportunities.
Conclusion
This study highlights the relative merits and limitations of a range of new and established gene expression profiling platforms and demonstrates that transcriptomic data from FFPE archival samples can be reliably integrated with data from FF samples, even if different measurement platforms are used. Ultimately, the choice of technology will depend upon the required resolution and coverage, throughput, sample quality, availability and budget.
Supplementary information
Additional file 1 : Table S1. Table demonstrating the directly overlapping samples across the nine gene expression platforms coloured by sample type, Pink = FFPE, yellow = fresh frozen.
Additional file 2 : Table S2. List of the 42 proliferation-related genes showing reduction on endocrine treatment [12].
Acknowledgements
We are very grateful to all the patients for providing tissue samples. We would like to acknowledge the contribution of The Genetics Core at the Edinburgh Clinical Research Facility at the University of Edinburgh who performed the Illumina microarray processing and library preparation and sequencing for the Ion AmpliSeq Transcriptome, BioSpyder TempO-seq and Lexogen QuantSeq cohorts. We would also like to acknowledge the HTPU service at the University of Edinburgh who performed the NanoString processing and the Perou lab at the University of North Carolina, who performed the TruSeq RNA-sequencing.
Abbreviations
- FF
fresh frozen
- FFPE
formalin-fixed paraffin embedded
- RNA
Ribonucleic acid
Authors’ contributions
Conception and design: AKT, CS, CM-P, AHS, JMD, CMP, LAC, JDF. Collection of clinical samples: LR, JK, JMD. Generation of data: AKT, CS, CM-P, AF1, XH, AFM, LM, AF2, RC, AC. Data analysis and interpretation: AKT, CS, MT, JDF, AHS. Manuscript writing: All authors. Final approval of manuscript: All authors.
Authors’ information
Not applicable
Funding
We are grateful to Breast Cancer Now, the Breast Cancer Research Trust and Breast Cancer Institute for funding to AKT, AHS and JMD. This work was partly funded by European Commission H2020 Marie Sklodowska Curie Action Individual Fellowship [H2020-MSCA-IF, 658170] and Welcome Trust Institutional Fund (ISSF3) to CS and AHS. The funders did not have any role or influence in the study design, execution, analyses, interpretation of the data or writing of the manuscript.
Availability of data and materials
All data is publicly available from NCBI GEO (www.ncbi.nlm.nih.gov/geo/) under super-series accession GSE130645.
Ethics approval and consent to participate
All patients provided written informed consent and sample collection was approved by the local research ethics committee (Lothian NHS Local Research Ethics Committee 03, REC Reference number 07/S1103/26, approval date 13/08/2007).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Arran K. Turnbull, Email: A.Turnbull@ed.ac.uk
Cigdem Selli, Email: Cigdem.Selli@ed.ac.uk.
Carlos Martinez-Perez, Email: Carlos.Martinez-Perez@igmm.ed.ac.uk.
Anu Fernando, Email: Anu.Fernando@ed.ac.uk.
Lorna Renshaw, Email: Lorna.Renshaw@nhslothian.scot.nhs.uk.
Jane Keys, Email: Jane.Keys@nhslothian.scot.nhs.uk.
Jonine D. Figueroa, Email: Jonine.figueroa@ed.ac.uk
Xiaping He, Email: xiaping_he@med.unc.edu.
Maki Tanioka, Email: maki@unc.edu.
Alison F. Munro, Email: alison.munro@ed.ac.uk
Lee Murphy, Email: Lee.Murphy@ed.ac.uk.
Angie Fawkes, Email: A.Fawkes@ed.ac.uk.
Richard Clark, Email: rclark2@exseed.ed.ac.uk.
Audrey Coutts, Email: aduncan5@exseed.ed.ac.uk.
Charles M. Perou, Email: chuck_perou@med.unc.edu
Lisa A. Carey, Email: Lisa_Carey@med.unc.edu
J. Michael Dixon, Email: Mike.Dixon@ed.ac.uk.
Andrew H. Sims, Email: andrew.sims@ed.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12859-020-3365-5.
References
- 1.Sims AH. Bioinformatics and breast cancer: what can high-throughput genomic approaches actually tell us? J Clin Pathol. 2009;62:879–885. doi: 10.1136/jcp.2008.060376. [DOI] [PubMed] [Google Scholar]
- 2.Robert C, Watson M. Errors in RNA-Seq quantification affect genes of relevance to human disease. Genome Biol. 2015;16:177. doi: 10.1186/s13059-015-0734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbull AK, Kitchen RR, Larionov A, Renshaw L, Dixon JM, Sims AH. Direct integration of intensity-level data from Affymetrix and Illumina microarrays improves statistical power for robust reanalysis. BMC Med Genet. 2012;5:35. doi: 10.1186/1755-8794-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims AH, Smethurst GJ, Hey Y, Okoniewski MJ, Pepper SD, Howell A, Miller CJ, Clarke RB. The removal of multiplicative, systematic bias allows integration of breast cancer gene expression datasets - improving meta-analysis and prediction of prognosis. BMC Med Genet. 2008;1:42. doi: 10.1186/1755-8794-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitchen RR, Sabine VS, Simen AA, Dixon JM, Bartlett JM, Sims AH. Relative impact of key sources of systematic noise in Affymetrix and Illumina gene-expression microarray experiments. BMC Genomics. 2011;12:589. doi: 10.1186/1471-2164-12-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. The MicroArray quality control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Shu-Hong, Beane Lauren, Chasse Dawn, Zhu Kevin W., Mathey-Prevot Bernard, Chang Jeffrey T. Cross-Platform Prediction of Gene Expression Signatures. PLoS ONE. 2013;8(11):e79228. doi: 10.1371/journal.pone.0079228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen MJ, Thomassen M, Tan Q, Sørensen KP, Kruse TA. Microarray-based RNA profiling of breast Cancer: batch effect removal improves cross-platform consistency. Biomed Res Int. 2014;2014:1–11. doi: 10.1155/2014/651751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: a valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank. 2013;11:101–106. doi: 10.1089/bio.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart JP, Richman S, Maughan T, Lawler M, Dunne PD, Salto-Tellez M. Standardising RNA profiling based biomarker application in cancer—the need for robust control of technical variables. Biochim Biophys Acta - Rev Cancer. 2017;1868:258–272. doi: 10.1016/j.bbcan.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Miller WR, Larionov A, Renshaw L, Anderson TJ, Walker JR, Krause A, Sing T, Evans DB, Dixon JM. Gene expression profiles differentiating between breast cancers clinically responsive or resistant to letrozole. J Clin Oncol. 2009;27:1382–1387. doi: 10.1200/JCO.2008.16.8849. [DOI] [PubMed] [Google Scholar]
- 13.Turnbull AK, Arthur LM, Renshaw L, Larionov AA, Kay C, Dunbier AK, Thomas JS, Dowsett M, Sims AH, Dixon JM. Accurate prediction and validation of response to endocrine therapy in breast Cancer. J Clin Oncol. 2015;33:2270–2278. doi: 10.1200/JCO.2014.57.8963. [DOI] [PubMed] [Google Scholar]
- 14.Yeakley Joanne M., Shepard Peter J., Goyena Diana E., VanSteenhouse Harper C., McComb Joel D., Seligmann Bruce E. A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLOS ONE. 2017;12(5):e0178302. doi: 10.1371/journal.pone.0178302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai M. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Research. 2005;33(20):e175–e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 18.Linton KM, Hey Y, Saunders E, Jeziorska M, Denton J, Wilson CL, Swindell R, Dibben S, Miller CJ, Pepper SD, et al. Acquisition of biologically relevant gene expression data by Affymetrix microarray analysis of archival formalin-fixed paraffin-embedded tumours. Br J Cancer. 2008;98:1403–1414. doi: 10.1038/sj.bjc.6604316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linton K, Hey Y, Dibben S, Miller C, Freemont A, Radford J, Pepper S. Methods comparison for high-resolution transcriptional analysis of archival material on Affymetrix plus 2.0 and exon 1.0 microarrays. Biotechniques. 2009;47:587–596. doi: 10.2144/000113169. [DOI] [PubMed] [Google Scholar]
- 20.Lazar C., Meganck S., Taminau J., Steenhoff D., Coletta A., Molter C., Weiss-Solis D. Y., Duque R., Bersini H., Nowe A. Batch effect removal methods for microarray gene expression data integration: a survey. Briefings in Bioinformatics. 2012;14(4):469–490. doi: 10.1093/bib/bbs037. [DOI] [PubMed] [Google Scholar]
- 21.Guo Yan, Sheng Quanhu, Li Jiang, Ye Fei, Samuels David C., Shyr Yu. Large Scale Comparison of Gene Expression Levels by Microarrays and RNAseq Using TCGA Data. PLoS ONE. 2013;8(8):e71462. doi: 10.1371/journal.pone.0071462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco, J.L., Gestal, M., Dorado, J. and Fernandez-Lozano, C. (2019) Differential gene expression analysis of RNA-seq data using machine learning for Cancer research. In Tsihrintzis, G.A., Virvou, M., Sakkopoulos, E., Jain, L.C. (eds), Machine Learning Paradigms. Cham: Springer, pp. 27–65.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 : Table S1. Table demonstrating the directly overlapping samples across the nine gene expression platforms coloured by sample type, Pink = FFPE, yellow = fresh frozen.
Additional file 2 : Table S2. List of the 42 proliferation-related genes showing reduction on endocrine treatment [12].
Data Availability Statement
All data is publicly available from NCBI GEO (www.ncbi.nlm.nih.gov/geo/) under super-series accession GSE130645.