Abstract
From September to October 2019, seven patients colonised or infected with a ceftazidime-avibactam (CZA)-resistant Klebsiella pneumoniae carbapenemase (KPC)-2-producing K. pneumoniae were detected in two intensive care units of a Greek general hospital. The outbreak strain was sequence type (ST)147 and co-produced KPC-2 and the novel plasmid-borne Vietnamese extended-spectrum β-lactamase (VEB)-25 harbouring a K234R substitution associated with CZA resistance. Epidemiological investigations revealed that the resistance was probably acquired by horizontal transmission independently from previous CZA exposure.
Keywords: ceftazidime-avibactam, K.pneumoniae, KPC-2, VEB-14, VEB-25, K234R
The spread of carbapenemase-producing Klebsiella pneumoniae (CPKP) has become a significant problem worldwide. Ceftazidime-avibactam (CZA) is a novel β-lactam/β-lactamase inhibitor combination effective against strains producing serine carbapenemases, including K. pneumoniae carbapenemase (KPC)- and oxacillinase (OXA)-48-type enzymes [1]. Resistance to CZA has already been described and was mainly linked to specific mutations in bla KPC [2], alterations in Ompk35 and Ompk36 porins and/or increased expression of bla KPC [3], as well as amino acid substitutions in cefotaxime-M β-lactamase (CTX-M)-14 [4]. Moreover during editing of this report, two K. pneumoniae isolates resistant to CZA were reported from Greece due to Vietnamese extended-spectrum β-lactamase (VEB)-25 [5]. The European Centre for Disease Prevention and Control (ECDC) has identified CZA resistance as an important cross-border threat that merits careful monitoring [6]. In this article, we report an outbreak caused by a CZA-resistant K. pneumoniae strain in a hospital in Athens, Greece in 2019. The outbreak molecular investigation revealed that resistance was due to plasmid-borne VEB-25, which differs from VEB-1 by one mutation. The affected patients had not previously been treated with CZA. Another isolate detected in the same hospital in 2018 had a different bla VEB-1 variant leading to CZA-resistance but this variant did not belong to the outbreak clone.
Outbreak detection and investigation
The index hospital comprises two mixed intensive care units (ICU) and one high dependency unit (HDU) that are located in different areas of the hospital. ICU-1 includes a 14-bed open space unit and four single-patient rooms. ICU-2 consists of seven single-patient rooms, whereas HDU has 12 beds, and one isolation room. Rectum surveillance cultures are performed routinely twice weekly in all ICU patients and screened for carbapenemase producers. In November 2018, 11 months after the introduction of CZA in clinical practice (December 2017), one patient hospitalised in the HDU was found to be colonised by a CZA-resistant KPC-producing K. pneumoniae strain (KP121) after CZA treatment (Table 1 and Table 2). The patient was isolated in a single room under strict contact precautions until discharge and no more cases were detected. Since then, all surveillance strains that were KPC-producers have been routinely evaluated for CZA resistance. This practice enabled the early detection of the outbreak. From September 2019 to October 2019, four patients in ICU-1 and three patients in ICU-2 were found to be colonised by a CZA-resistant KPC-producing K. pneumoniae strain and three of them developed an infection (Table 2). The outbreak prompted an epidemiological and molecular investigation. Prevention efforts including intensification of contact precautions (provision of personal protective equipment outside the patient room and use of gloves and gown upon entering the room, minimising risk of environmental contamination by dressing patients in a gown during transport and applying all standard precautions at the receiving unit, dedicating noncritical items for single patient use and having dedicated infection control nurses overseeing strict implementation of measures), isolation of colonised patients in single rooms and strict hand hygiene practices successfully contained the outbreak as no new case was identified after October 2019.
Table 1. Characteristics of the ceftazidime-avibactam-resistant Klebsiella pneumoniae strains detected in a general hospitala as well as their transconjugants and one previously characterised transconjugant producing VEB-1, Greece, 2018 and 2019 (n = 6 strains).
| Isolate |
K. pneumoniae
ST39 strain (KP121) |
E. coli
b
RC85-pl121 |
K. pneumoniae
ST147 outbreak strain (KP67585) |
E. coli
c
RC85-pl67585 |
E. coli
d
RC85-pl52 |
E. coli
RC85 |
|---|---|---|---|---|---|---|
| Type | ||||||
| KPC-type | KPC-2 | None | KPC-2 | None | None | None |
| VEB-type | VEB-14 | VEB-14 | VEB-25 | VEB-25 | VEB-1 | None |
| Antibiotics tested | Minimum inhibitory concentration in mg/L | |||||
| Ampicillin-sulbactam | > 16 | > 16 | > 16 | > 16 | > 16 | ≤ 2 |
| Piperacillin-tazobactam | > 64 | 32 | > 64 | > 64 | > 64 | ≤ 4 |
| Cefoxitin | > 32 | ≤ 4 | > 32 | ≤ 4 | ≤ 4 | ≤ 4 |
| Ceftazidime | 2,048 | 4,096 | 1,024 | 512 | 512 | 0.25 |
| Ceftazidime-avibactam | 64 | 256 | 64 | 16 | 0.25 | 0.25 |
| Ceftriaxone | > 32 | 32 | > 32 | 8 | 32 | ≤ 1 |
| Cefepime | > 32 | > 32 | > 32 | 2 | 2 | ≤ 1 |
| Aztreonam | > 32 | > 32 | > 32 | > 32 | > 32 | ≤ 1 |
| Imipenem | 64 | 0.12 | 32 | 0.12 | 0.12 | 0.12 |
| Imipenem-relebactam | 0.5 | 0.12 | 0.5 | 0.12 | 0.12 | 0.12 |
| Meropenem | > 64 | 0.06 | 64 | 0.06 | 0.06 | 0.06 |
| Meropenem-vaborbactam | 0.5 | 0.06 | 0.25 | 0.06 | 0.06 | 0.06 |
| Amikacin | > 32 | > 32 | > 32 | > 32 | > 32 | ≤ 2 |
| Gentamicin | > 8 | > 8 | > 8 | > 8 | > 8 | ≤ 1 |
| Ciprofloxacin | > 2 | ≤ 0.25 | > 2 | ≤ 0.25 | ≤ 0.25 | ≤ 0.25 |
| Levofloxacin | > 4 | ≤ 0.12 | > 4 | ≤ 0.12 | ≤ 0.12 | ≤ 0.12 |
| Tigecycline | > 4 | 1 | 2- > 4 | 2 | ≤ 0.5 | ≤ 0.5 |
| Fosfomycin | 64 | ≤ 16 | 128 | ≤ 16 | ≤ 16 | ≤ 16 |
| Colistin | 2 | 0.5 | 64 | 0.5 | 0.5 | 0.5 |
| Trimethoprim-sulfamethoxazolee | > 8 | > 8 | > 8 | > 8 | > 8 | ≤ 1 |
| Chloramphenicol | > 128 | 128 | > 128 | 32 | 32 | 8 |
| Other β-lactamase genes | ||||||
| β-lactamase genes |
bla
SHV-11,
bla OXA-10, bla TEM-1B |
bla
OXA-10,
bla TEM-1B |
bla
SHV-11,
bla OXA-10, bla TEM-1B |
bla
OXA-10,
bla TEM-1B |
bla
OXA-10,
bla TEM-1B |
None |
| Major porin mutation | ||||||
| OmpK35 | WT | ND | PSC_aa173 | ND | ND | ND |
| OmpK36 | v3 variant | ND | v3 variant | ND | ND | ND |
| OmpK37 | PSC_aa251 | ND | WT | ND | ND | ND |
E. coli: Escherichia coli; K. pneumoniae: Klebsiella pneumoniae; KPC: K. pneumoniae carbapenemase; MIC: minimum inhibitory concentration; ND: not determined; OmpK35: OmpK35-WT (GenBank accession number: GU460162); OmpK36: OmpK36_v3 (GenBank accession number: JQ781655); OmpK37: OmpK37-WT (GenBank accession number: WP_002902433); PSC: premature stop codon; PSC_aa173: premature stop codon at amino acid 173; PSC_aa251: premature stop codon at amino acid 251; ST: sequence type; VEB: Vietnamese extended-spectrum β-lactamase; WT: wild type.
a The isolates detected in the general hospital, which are presented in the table, include one isolate (KP121) producing VEB-14 (ST39) that was discovered in 2018, as well as one isolate (KP67585) producing VEB-25 (ST147) that represents a strain responsible for the 2019 hospital outbreak, which is described in this report.
b Transconjugant E. coli RC85 isolate harbouring bla VEB-14 carrying plasmid.
c Transconjugant E. coli RC85 isolate harbouring bla VEB-25 carrying plasmid.
d Transconjugant E. coli RC85 isolate harbouring bla VEB-1 carrying plasmid. The donor isolate was an OXA-48-producing K.pneumoniae isolate carrying an IncA/C2 plasmid.
e Trimethoprim-sulfamethoxazole in the ratio 1:19. MICs are expressed as the trimethoprim concentration.
Table 2. Demographic and clinical characteristics, outcome and follow-up of the eight patients who were colonised or infected with a ceftazidime-avibactam-resistant Klebsiella pneumoniae strain, Greece, 2018 (n = 1 patient) and 2019 (n = 7 outbreak cases).
| Patient number (isolate name) | Approximate age in yearsa | Deptartment | Reason for ICU admission | Duration of stay in ICU/days before colonisation | Comorbidities; interventions | Colonisationb date | Previous antibiotics before colonisation | Previous carbapenem use (duration in days) | Type of infection from colonised strain | Antimicrobial treatment (number of days of treatment) |
Treatment outcome | Overall outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
1
(KP121) |
50 | HDU | SAH | 153/35 | Tracheostomy; MV | 1 Nov 2018 | CZA, MEM, TGC, CST | Yes (10) CZA (15) |
No | NA | NA | Discharged to rehabilitation centre |
|
2
(KP67585) |
85 | ICU-2 | Brain injury- subdural haematoma | 107/51 | CAD; tracheostomy; MV | 2 Sep 2019 | TZP, MEM, CIP | Yes (3) | CRBSI | CZA + MEM + FOS (15) | Success | Deathc |
|
3
(KP374) |
85 | ICU-1 | Thoracotomy | 51/42 | Metastatic malignancy; tracheostomy; MV | 5 Sep 2019 | MEM, CST,TGC | Yes (15) | VAP | CZA + ATM + FOS (15) |
Success | Deathc |
|
4
(KP368) |
65 | ICU-2 | SAH | 89/56 | Tracheostomy; gastrostomy; MV | 5 Sep 2019 | TZP, AMK, VAN | No (NA) | No | NA | NA | Discharged to rehabilitation centre |
|
5
(KP501) |
75 | ICU-2 | SAH | 42/21 | AH; tracheostomy; MV | 16 Sep 2019 | CRO, TZP, VAN | No (NA) | No | NA | NA | Discharged to rehabilitation centre |
|
6
(KP687) |
70 | ICU-1 | Acute coronary syndrome | 45/21 | Renal failure; CVVHDF | 26 Sep 2019 | CRO, CPT, TZP, VAN, MEM | Yes (4) | CRBSI | CZA + MEM (2) | Failure | Deathd |
|
7
(KP785) |
60 | ICU-1 | Respiratory failure | 30/11 | Metastatic malignancy COPD; AH; tracheostomy; MV | 3 Oct 2019 | TZP, MEM, VAN | Yes (10) | No | NA | NA | Deathc |
|
8
(KP842) |
55 | ICU-1 | Acute coronary syndrome | 12/12 | AH | 7 Oct 2019 | CRO | No (NA) | No | NA | NA | Discharged |
AH: arterial hypertension; AMK: amikacin; ATM: aztreonam; CAD: coronary arterial disease; CVVHDF: continuous veno-venous haemodiafiltration; CZA: ceftazidime-avibactam; CIP: ciprofloxacin; COPD: chronic obstructive pulmonary disease; CPT: ceftaroline; CRBSI: catheter related blood stream infection; CRO: ceftriaxone; CST: colistin; FOS: fosfomycin; HDU: high dependency unit; ICU: intensive care unit; MEM: meropenem; MV: mechanical ventilation; NA: not applicable; SAH: subarachnoid haemorrhage; TGC: tigecycline; TZP: piperacillin-tazobactam; VAN: vancomycin; VAP: ventilator-associated pneumonia.
a Ages are rounded to the upper limit of 5-year categories.
b Colonisation determined from analysis of a rectal sample.
c Death due to another cause than Klebsiella pneumoniae carbapenemase K. pneumoniae.
d Death due to bacteraemia and the patient was positive for Klebsiella pneumoniae carbapenemase K. pneumoniae infection.
Microbiological and molecular analyses
Susceptibility testing performed by VITEK 2, broth microdilution and minimum inhibitory concentration (MIC) test strips revealed resistance, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints (2019, v 9.0) [7], to all antimicrobial agents tested except imipenem-relebactam, meropenem-vaborbactam, and colistin. CZA MICs (tested with a fixed avibactam concentration of 4 mg/L) ranged from 32 to 64 mg/L (Supplementary Table 1). PCR and sequencing analysis [8], showed that all isolates harboured bla KPC-2 as the sole carbapenemase gene. In addition to KPC-2, all eight isolates produced sulfhydryl reagent variable β-lactamase (SHV)-11, OXA-10, Temoniera β-lactamase (TEM)-1 and an extended spectrum β-lactamase (ESBL) of VEB-type. Isolate KP121 produced VEB-14 (T216del, per Ambler numbering scheme) [9]. All other isolates produced a novel VEB-1 variant carrying a substitution of lysine by an arginine at position 237 (K234R, per Ambler numbering scheme), due to nt A710G substitution (Supplementary Table 1). The mutant bla VEB-K237R sequence was assigned the novel allele VEB-25 (GenBank accession number: MN853159). Further acquired resistance genes justifying the resistance phenotype of studied strains are presented in Supplementary Table 1.
Whole genome sequencing (WGS) analysis, performed as described previously [10], revealed that both bla VEB-14 and bla VEB-25 were carried on conjugative plasmids of IncA/C2 incompatibility group. bla VEB14 and bla VEB-25 were present as the first gene cassette of an integron that also included an array of aadB, arr2, cmlA1, bla OXA-10, and aadA1 cassettes. An insertion sequence (IS)1999 was located upstream of both bla VEB-variants providing a strong promoter for bla VEB expression. The two whole genome sequences were deposited in the Sequence Read Archive (SRA) under the following accession numbers: PRJNA602658 (for the bla VEB-14 harbouring strain) and PRJNA602657 (for the strain with bla VEB-25).
PFGE analysis classified the studied isolates in two pulsotypes [8], with < 80% Dice similarity index. The first pulsotype included only the KP121, isolated in November 2018, while all the strains isolated in the period September–October 2019, belonged to one clone (second pulsotype). Based on WGS [10], KP121 belonged to sequence type (ST)39 and capsular type KL23 (wzc: 24; wzi: 83), while the outbreak strain belonged to clonal lineage ST147 and capsular type KL64 (wzc: 64; wzi: 64).
The ST147 outbreak strain, which was similar to one of the strains reported recently from another hospital in Athens [5], harboured a non-functional porin OmpK35 due to a premature stop codon at position 173, while ST39 isolate (KP121) had an intact OmpK35. OmpK36_v3 variant, previously associated with ST147 K. pneumoniae isolates from Greece [11], was present in both ST147 and ST39 isolates. OmpK36_v3 harbours a duplication of two amino acids, Gly134–Asp135, located at the conserved loop L3, which contributes to high-level resistance to carbapenems [11]. However, OmpK35 and OmpK36 are not the primary pathways for avibactam into the cell of K. pneumoniae [12].
bla VEB-harbouring plasmids from the ST39 (KP121), and the ST147 outbreak strain (KP67585) were transferred by conjugation to rifampicin-resistant Escherichia coli RC85 R K12. The susceptibility profile of the transconjugants (RC85/pl121 and RC85/pl67585) is shown in Table 1. The susceptibility profile of a previously studied transconjugant, RC85/pl52, harbouring bla VEB-1 is included in Table 1 for comparison. Transconjugants were resistant to β-lactams, aminoglycosides, trimethoprim/sulfamethoxazole and chloramphenicol (Table 1). PCR and sequencing confirmed the presence of bla VEB, bla OXA-10, bla TEM-1B and rmtB1 and the absence of bla KPC-2 in all transconjugants. CZA MICs determined by Liofilchem MIC Test Strips, differed between the transconjugants depending on the VEB-variant they produced. RC85/pl52 harbouring bla VEB-1 exhibited ceftazidime MIC of 512 mg/L, which was reduced to 0.25 mg/L (2,048-fold reduction) in the presence of avibactam. Transconjugant RC85/pl67585 carrying bla VEB-25, exhibited similar MIC to ceftazidime (512 mg/L), which was reduced to 16 mg/L by avibactam (32-fold reduction). Transconjugant RC85/pl121 carrying bla VEB-14, exhibited higher MIC to ceftazidime (4,096 mg/L) and to CZA (256 mg/L) (16-fold reduction by avibactam).
Epidemiological investigation
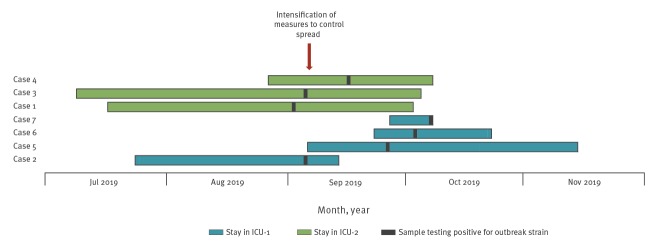
The eight patients found with a CZA-resistant KPC producing K. pneumoniae comprised five men and three women. Possible chains of transmission were investigated but no common source was identified between patients in ICU-1 and ICU-2. All colonised or infected patients had a long period of ICU stay (median: 48 days; range: 11–56 days) before colonisation (Figure). All patients were originally hospitalised in the index hospital with the exception of one patient (KP368), colonised with the outbreak strain, who had been hospitalised in another hospital for 24 hours. None had been previously received CZA, however all patients had been pre-treated or were on β-lactam therapy during hospitalisation in the ICU (Table 2). Only the patient with KP121 had received CZA before colonisation but he represented an independent case not related to the outbreak. Three patients developed an infection: two catheter-related bloodstream infections (catheter culture revealed the same pathogen in both cases) and one ventilator-associated pneumonia. The salvage therapeutic regimens were chosen based on in vitro data showing synergy of the combinations used, all including CZA, the rationale being that avibactam inactivates the class A, C, or D β-lactamases and restores susceptibility to aztreonam or meropenem [13-15]. The triple combination was successful in two of the cases at Day 14, while the combination of CZA and meropenem was reported as failure in the remaining case (Table 2); in terms of all-cause mortality, by Day 28 all infected patients had died.
Figure.
Timeline of the hospital outbreak ofsequence type (ST)147 Klebsiella pneumoniae harbouring a Vietnamese extended-spectrum β-lactamase-25, Greece, 2019 (n = 7 outbreak cases)
Cases 1, 2, 3, 4, 5, 6 and 7 correspond to patients 2, 3, 4, 5, 6, 7 and 8 respectively in Table 2.
Intensified measures included the requirement of wearing gloves and a gown upon entering the case’s room, minimal transport of this patient and, if so, prior advising the receiving unit so that necessary precautions could be taken in advance, dedicated use of noncritical items and having dedicated infection control nurses inspecting the implementation of infection control measures, isolation of colonised patients in single rooms and strict hand hygiene practices.
Discussion
CZA demonstrates high in vitro activity against non-metallo-β-lactamase-producing K. pneumoniae strains in Greece [8]. To date, there are two reports on sporadic cases of CZA resistant K. pneumoniae from Greece, due to the production of KPC-23 [10], or VEB-25 [5]. In the present study, we provide further evidence that CZA resistance may emerge through evolution of bla VEB-1 with the detection of a bla VEB-25-harbouring strain causing a hospital outbreak and another strain endowed with bla VEB-14 in a single hospitalised patient. Both VEB-14 and VEB-25 variants exhibit decreased inactivation by avibactam.
The K. pneumoniae isolates presented in this study produced KPC-2, VEB-14 or VEB-25, OXA-10, TEM-1B and the chromosomally encoded SHV-11 β-lactamases. The transconjugants carried bla OXA-10, bla TEM-1B, and bla VEB-1, or bla VEB-14 or bla VEB-25. Avibactam appeared to reduce ceftazidime MICs to a lesser extent in the presence of VEB-14 (16-fold reduction) or VEB-25 (32-fold reduction) than in the presence of VEB-1 (2,048-fold reduction).
VEB-25, which was recently described [5], harbours a substitution of lysine by an arginine at position 237 (K234R, per Ambler numbering scheme), due to nt A710G substitution, while VEB-14 [9], has a threonine deletion at position 217 (T216del, per Ambler numbering scheme).
K234 residue is highly conserved among class A β-lactamases (Supplementary Figure 1), forming strong hydrogen bonds with the sulphate group of avibactam [9]. We hypothesise that K234R substitution resulted in a structural change that attenuated avibactam's inhibitory effect by disrupting its ability to bind at the active site, thereby causing resistance. According to Papp-Wallace et al., residue K234 contributes notably to the inactivation of KPC-2 by avibactam [16], while substitution K234R in SHV enzymes has been reported to lead to resistance to inhibitors [17-20].
T216 residue, although in close proximity, has no direct interaction with avibactam [9]. In an E. coli DH5a strain carrying isogenically expressed VEB-14, avibactam reduced ceftazidime MIC from 64 to 1 mg/L, and VEB-14 was inhibited by avibactam in a concentration-dependent manner [9]. We hypothesise that in KP121, CZA resistance was due to increased expression of VEB-14 due to an IS1999 located upstream of the gene.
Previous epidemiological analyses (data not shown) indicated that a conjugative plasmid co-harbouring bla VEB-1, bla OXA-10, bla TEM-1B and rmtB is present in 8% of all bla KPC-positive isolates in Greece [21]. This already established vehicle can potentially enhance dissemination of VEB-mediated CZA resistance in Greek hospitals. The emergence of CZA resistance dramatically limits treatment options against carbapenemase-producing Enterobacterales. In this report, infected patients received salvage combination treatment, which was successful in two of the three cases.
Limitations of this study include the lack of environmental sampling during the outbreak investigation, which could have revealed potential environmental sources, and the lack of biochemical studies of VEB-14 and VEB-25 enzymes, which could have further elucidated the basis of the resistance phenotype.
In conclusion, we have shown that alterations in the ESBL VEB-1 enzyme can significantly reduce CZA susceptibility in K. pneumoniae co-producing KPC-2 and emergence of this resistance mechanism was independent from previous CZA exposure. Further biochemical studies are needed to reveal the basis of the resistance phenotype, conferred by the two VEB-variants. The rigorous implementation of hospital infection control precautions resulted in successful containment of the outbreak, highlighting the importance of early awareness in the fight against antimicrobial resistance.
Acknowledgements
The Authors would like to acknowledge Irene Karantani from Infectious Diseases Laboratory of Hygeia Hospital, Panagiota Adamou and Anastasia Molla from the Infectious Diseases Laboratory of the 4th Department of Internal Medicine, for outstanding technical work and Konstantina Nafplioti for providing strain RC85/pl52.
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: IG: whole genome sequencing and data analysis, manuscript writing. IK: outbreak investigation, manuscript writing. MS: manuscript writing. VP: detection and susceptibility testing of bacterial isolates. LG, AG and HG: outbreak investigation team. AA: manuscript correction. All authors contributed to the revision of the draft manuscript and approved the final version.
References
- 1. Karaiskos I, Galani I, Souli M, Giamarellou H. Novel β-lactam-β-lactamase inhibitor combinations: expectations for the treatment of carbapenem-resistant Gram-negative pathogens. Expert Opin Drug Metab Toxicol. 2019;15(2):133-49. 10.1080/17425255.2019.1563071 [DOI] [PubMed] [Google Scholar]
- 2. Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, et al. Emergence of Ceftazidime-Avibactam Resistance Due to Plasmid-Borne bla KPC-3 Mutations during Treatment of Carbapenem-Resistant Klebsiella pneumoniae Infections. Antimicrob Agents Chemother. 2017;61(3):e02097-16. 10.1128/AAC.02097-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, et al. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in bla KPC-2-harboring ceftazidime-avibactam resistance through distinct genomic adaptations in bla KPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother. 2018;62(3):e02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Both A, Büttner H, Huang J, Perbandt M, Belmar Campos C, Christner M, et al. Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. J Antimicrob Chemother. 2017;72(9):2483-8. 10.1093/jac/dkx179 [DOI] [PubMed] [Google Scholar]
- 5. Voulgari E, Kotsakis SD, Giannopoulou P, Perivolioti E, Tzouvelekis LS, Miriagou V. Detection in two hospitals of transferable ceftazidime-avibactam resistance in Klebsiella pneumoniae due to a novel VEB β-lactamase variant with a Lys234Arg substitution, Greece, 2019. Euro Surveill. 2020;25(2):1900766 10.2807/1560-7917.ES.2020.25.2.1900766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.European Centre for Disease Prevention and Control (ECDC). Emergence of resistance to ceftazidime-avibactam in carbapenem-resistant Enterobacteriaceae – 12 June 2018. Stockholm; ECDC; 2018. Available from: https://ecdc.europa.eu/en/publications-data/rapid-risk-assessment-emergenceresistance-ceftazidime-avibactam-carbapenem
- 7.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0. Växjö: EUCAST; 2019. Available from; http://www.eucast.org.
- 8. Galani I, Karaiskos I, Karantani I, Papoutsaki V, Maraki S, Papaioannou V, et al. On Behalf Of The Study Collaborators Epidemiology and resistance phenotypes of carbapenemase-producing Klebsiella pneumoniae in Greece, 2014 to 2016. Euro Surveill. 2018;23(31):1700775. 10.2807/1560-7917.ES.2018.23.30.1700775 [DOI] [PubMed] [Google Scholar]
- 9. Lahiri SD, Alm RA. Identification of Novel VEB β-Lactamase Enzymes and Their Impact on Avibactam Inhibition. Antimicrob Agents Chemother. 2016;60(5):3183-6. 10.1128/AAC.00047-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galani I, Antoniadou A, Karaiskos I, Kontopoulou K, Giamarellou H, Souli M. Genomic characterization of a KPC-23-producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime-avibactam. Clin Microbiol Infect. 2019;25(6):763.e5-8. 10.1016/j.cmi.2019.03.011 [DOI] [PubMed] [Google Scholar]
- 11. Papagiannitsis CC, Giakkoupi P, Kotsakis SD, Tzelepi E, Tzouvelekis LS, Vatopoulos AC, et al. OmpK35 and OmpK36 porin variants associated with specific sequence types of Klebsiella pneumoniae. J Chemother. 2013;25(4):250-4. 10.1179/1973947813Y.0000000075 [DOI] [PubMed] [Google Scholar]
- 12. Pagès JM, Peslier S, Keating TA, Lavigne JP, Nichols WW. Role of the Outer Membrane and Porins in Susceptibility of β-Lactamase-Producing Enterobacteriaceae to Ceftazidime-Avibactam. Antimicrob Agents Chemother. 2016;60(3):1349-59. 10.1128/AAC.01585-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mikhail S, Singh NB, Kebriaei R, Rice SA, Stamper KC, Castanheira M, et al. Evaluation of the Synergy of Ceftazidime-Avibactam in Combination with Meropenem, Amikacin, Aztreonam, Colistin, or Fosfomycin against Well-Characterized Multidrug-Resistant Klebsiella pneumoniae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;63(8):e00779-19. 10.1128/AAC.00779-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaibani P, Lewis RE, Volpe SL, Giannella M, Campoli C, Landini MP, et al. In vitro interaction of ceftazidime-avibactam in combination with different antimicrobials against KPC-producing Klebsiella pneumoniae clinical isolates. Int J Infect Dis. 2017;65:1-3. 10.1016/j.ijid.2017.09.017 [DOI] [PubMed] [Google Scholar]
- 15. Marshall S, Hujer AM, Rojas LJ, Papp-Wallace KM, Humphries RM, Spellberg B, et al. Can Ceftazidime-Avibactam and Aztreonam Overcome β-Lactam Resistance Conferred by Metallo-β-Lactamases in Enterobacteriaceae? Antimicrob Agents Chemother. 2017;61(4):e02243-16. 10.1128/AAC.02243-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. Variants of β-lactamase KPC-2 that are resistant to inhibition by avibactam. Antimicrob Agents Chemother. 2015;59(7):3710-7. 10.1128/AAC.04406-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dubois V, Poirel L, Demarthe F, Arpin C, Coulange L, Minarini LA, et al. Molecular and biochemical characterization of SHV-56, a novel inhibitor-resistant beta-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2008;52(10):3792-4. 10.1128/AAC.00387-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mendonça N, Manageiro V, Robin F, Salgado MJ, Ferreira E, Caniça M, et al. The Lys234Arg substitution in the enzyme SHV-72 is a determinant for resistance to clavulanic acid inhibition. Antimicrob Agents Chemother. 2008;52(5):1806-11. 10.1128/AAC.01381-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mendonça N, Ferreira E, Louro D, Caniça M, Caniça M, ARSIP Participants Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int J Antimicrob Agents. 2009;34(1):29-37. 10.1016/j.ijantimicag.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 20. Manageiro V, Ferreira E, Albuquerque L, Bonnet R, Caniça M. Biochemical study of a new inhibitor-resistant beta-lactamase, SHV-84, produced by a clinical Escherichia coli strain. Antimicrob Agents Chemother. 2010;54(5):2271-2. 10.1128/AAC.01442-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galani I, Nafplioti K, Adamou P, Karaiskos I, Giamarellou H, Souli M, Study Collaborators Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect Dis. 2019;19(1):167. 10.1186/s12879-019-3801-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.