Abstract
Large doses of chemical pesticides are required to achieve effective concentrations in the rhizosphere, which results in the accumulation of harmful residues. Precision farming is needed to improve the efficacy of pesticides, but also to avoid environmental pollution, and slow-release formulations based on nanoparticles offer one solution. Here, we tested the mobility of synthetic and virus-based model nanopesticides by combining soil column experiments with computational modelling. We found that the tobacco mild green mosaic virus and cowpea mosaic virus penetrate soil to a depth of at least 30 cm, and could therefore deliver nematicides to the rhizosphere, whereas the Physalis mosaic virus remains in the first 4 cm of soil and would be more useful for the delivery of herbicides. Our experiments confirm that plant viruses are superior to synthetic mesoporous silica nanoparticles and poly(lactic-co-glycolic acid) for the delivery and controlled release of pesticides, and could be developed as the next generation of pesticide delivery systems.
Pesticides are needed to protect our crops and thus maximize crop yields1. However, the efficacy of chemical pesticides is limited by their instability and strong binding to organic matter in soil, which can render them inactive or prevent their accumulation at the root level, where many pests reside2. Large doses are applied to compensate, which results in the accumulation of pesticide residues in soil, water and agricultural products3. Long-term exposure to these chemicals is a risk to human health and threatens the biodiversity of an already fragile ecosystem4. Precision farming methods are therefore needed to deliver pesticides in a more controlled manner.
Advances in nanotechnology have led to the development of more effective drug delivery and medical imaging methods (nanomedicine), and the same innovations are now being applied to smart agrochemical delivery systems, known as nanopesticides5,6. These involve the use of nanomaterials for the adsorption, encapsulation or conjugation of pesticides to improve the biodegradability, stability, permeability and dispersion of the active pesticide ingredient. Nanopesticides have a much greater surface area than conventional pesticides, which increases their potential for interaction with target pests at lower doses. The encapsulation of pesticides within nanoparticles also prevents premature degradation and the risk of direct human exposure to the active ingredient. There is also evidence that nanopesticides and conventional pesticides differ in their environmental behaviour, so it is necessary to understand the fate of nanopesticides in detail to ensure they comply with regulatory guidelines and legislation7–9.
Most of the nanopesticides investigated thus far are based on synthetic or natural polymers, metallic compounds or liposomes, which tend to persist in the environment5. As a biodegradable alternative, nanopesticides can be developed from plant viruses10–12. One example, already approved by the US Environmental Protection Agency, is the application of tobacco mild green mosaic virus (TMGMV) as the herbicide Solvinix, which is produced by BioProdex for deployment against invasive tropical soda apple weed in the state of Florida13,14. The safety profile and possible risks of TMGMV have been reported14. TMGMV cannot self-disseminate and is not transmitted by vectors such as insects, seeds or pollen. Mechanical transmission through insects or contact between plants is thus the only route of transmission. Only plants of the Solanaceae are susceptible to TMGMV infections. Therefore, TMGMV offers a good safety profile for crops that are not part of the Solanaceae. Nonetheless, plant virus-based systems, including TMGMV, could be inactivated through ultraviolet radiation for safe use on any crop15,16.
To investigate the potential of plant virus nanoparticles (VNPs) and virus-like particles (VLPs) as nanopesticides in more detail, we compared the behaviour of three viruses and two synthetic particle formulations in soil column experiments and computational models as a way to gauge their ability to deliver pesticides to the rhizosphere and thus prevent infestation by root pests (Fig. 1). We tested two VNPs, based on the rod-like TMGMV and the icosahedral cowpea mosaic virus (CPMV), and a VLP based on the Physalis mosaic virus (PhMV). These were compared to mesoporous silica nanoparticles (MSNPs) and a poly(lactic-co-glycolic acid) (PLGA) formulation, which have already been developed as synthetic nanopesticides17,18.
Fig. 1 |. the combined experimental and computational approach to assess nanopesticide transport through soil.
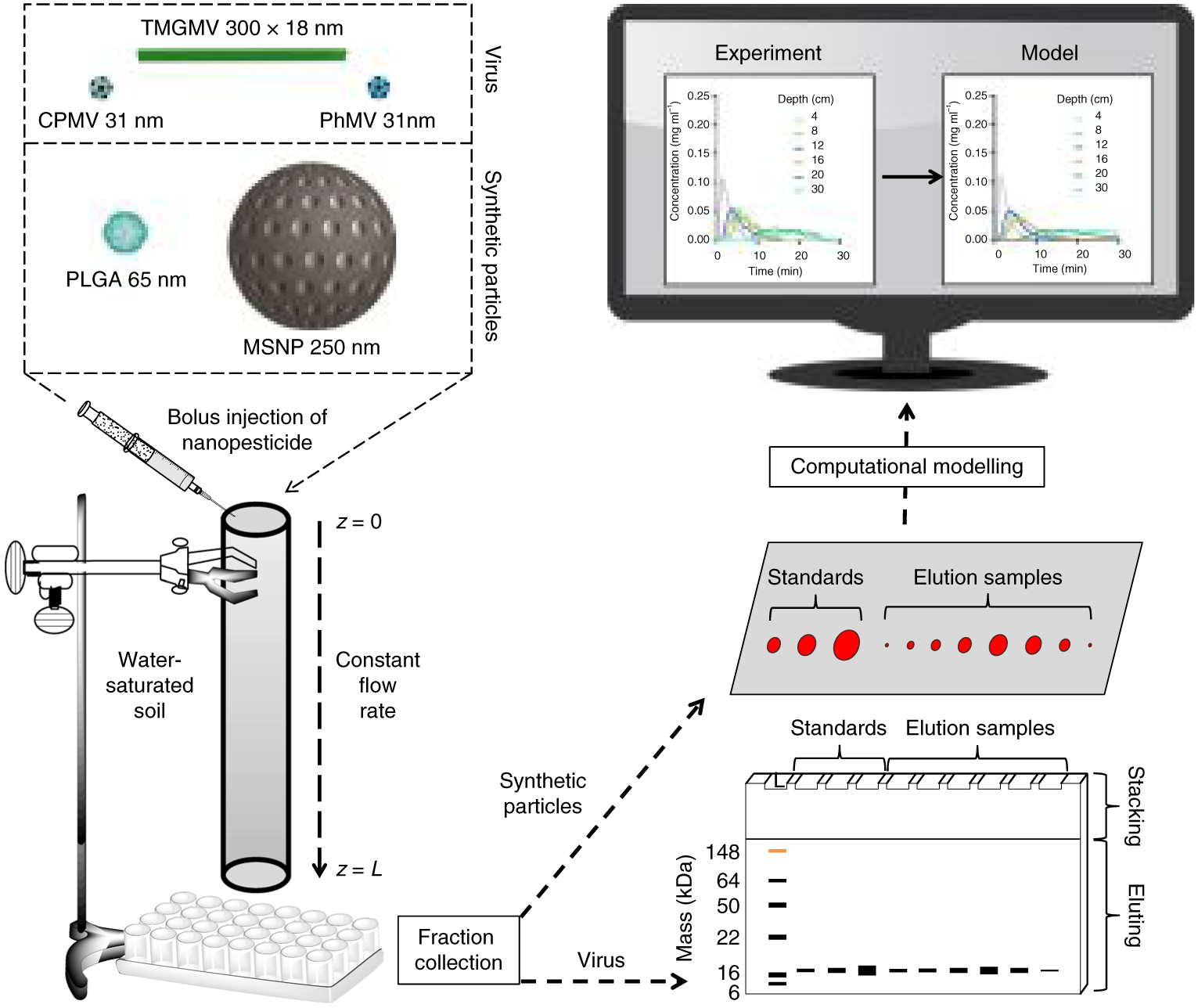
The virus-based and synthetic nanoparticles are depicted to scale in the top left. Labelled nanoparticles were injected as a bolus at the top of the soil column and moved through the column at a constant flow rate. At the bottom of the column, particles were collected as 500 μl fractions. The mass of the eluted virus-based nanoparticles was determined by SDS–PAGE and the synthetic nanoparticles were imaged as droplets on Parafilm using the FluorChem R imaging system under a MultiFluor red light. Experimental data were imported into Matlab for comparison with the output of the computational model.
Nanopesticide characterization
The fluorophore Cyanine 5 (Cy5) has similar physicochemical properties to those of conventional pesticides (Supplementary Table 1), but it is easier to detect and so we used it as a model compound. Cy5 was either conjugated to the external surface of, or passively encapsulated within, TMGMV, CPMV, PhMV and MSNP particles (Supplementary Fig. 1). Degradex PLGA nanoparticles that encapsulated a red fluorophore with spectral properties similar to those of Cy5 were obtained from Phosphorex (Supplementary Fig. 2). Each formulation was characterized by a combination of transmission electron microscopy, dynamic light scattering, ultraviolet–visible (UV–vis) spectroscopy, size exclusion chromatography and denaturing gel electrophoresis (SDS–polyacrylamide gel electrophoresis (SDS–PAGE)) or agarose gel electrophoresis to confirm the particle integrity and dye loading efficiency (Supplementary Figs. 2–4). The capacity of the TMGMV was 9.9 nmol mg−1 or 390 dye molecules per TMGMV–Cy5 particle (denotes the conjugated version) but only 5.3 nmol mg−1 or 210 dye molecules per TMGMV*Cy5 particle (denotes the encapsulated version). For PhMV, the corresponding loads were 12.7 nmol mg−1 or 60 dye molecules per PhMV–Cy5 and 11.7 nmol mg−1 or 55 dye molecules per PhMV*Cy5. For CPMV, the corresponding loads were 6.2 nmol mg−1 or 35 dye molecules per CPMV-Cy5 and 2.3 nmol mg−1 or 15 dye molecules per CPMV*Cy5. The synthetic MSNP formulation was similar in capacity to CPMV (6.4 nmol mg−1 for MSNP–Cy5 and 4.3 nmol mg−1 for MSNP*Cy5), whereas the PLGA formulation had the lowest capacity (1.2 nmol mg−1 for PLGA*dye).
The release profile of passively encapsulated Cy5 (Fig. 2) was determined by dialysis. The approximate half-life, defined as the time necessary for 50% of the fluorophore to be released from its carrier, was calculated for TMGMV (t1/2 = 12 h), CPMV (t1/2 = 60 h), PhMV (t1/2 = 48 h), MSNP (t1/2 = 12 h) and PLGA (t1/2 = 72 h). Two distinct release profiles were observed that reflected the Cy5 entrapment methodology used in each formulation. For TMGMV and MSNP, Cy5 is not entrapped in a confined structure because the internal channel of TMGMV is uncapped at both ends and the mesopores of MSNP are similarly open to the surrounding medium, which potentially explains the faster release rate. For CPMV and PhMV, Cy5 is encapsulated within the protein shell and the PLGA nanoparticle encapsulates the dye in its hydrophobic core, and hence the slower release. The observed release profiles may not precisely replicate pesticide release in a real soil environment, which is rich in various minerals and organic matter that might interact with either the carriers or the pesticide molecules, as discussed below.
Fig. 2 |. Cargo release from nanoparticles during dialysis.
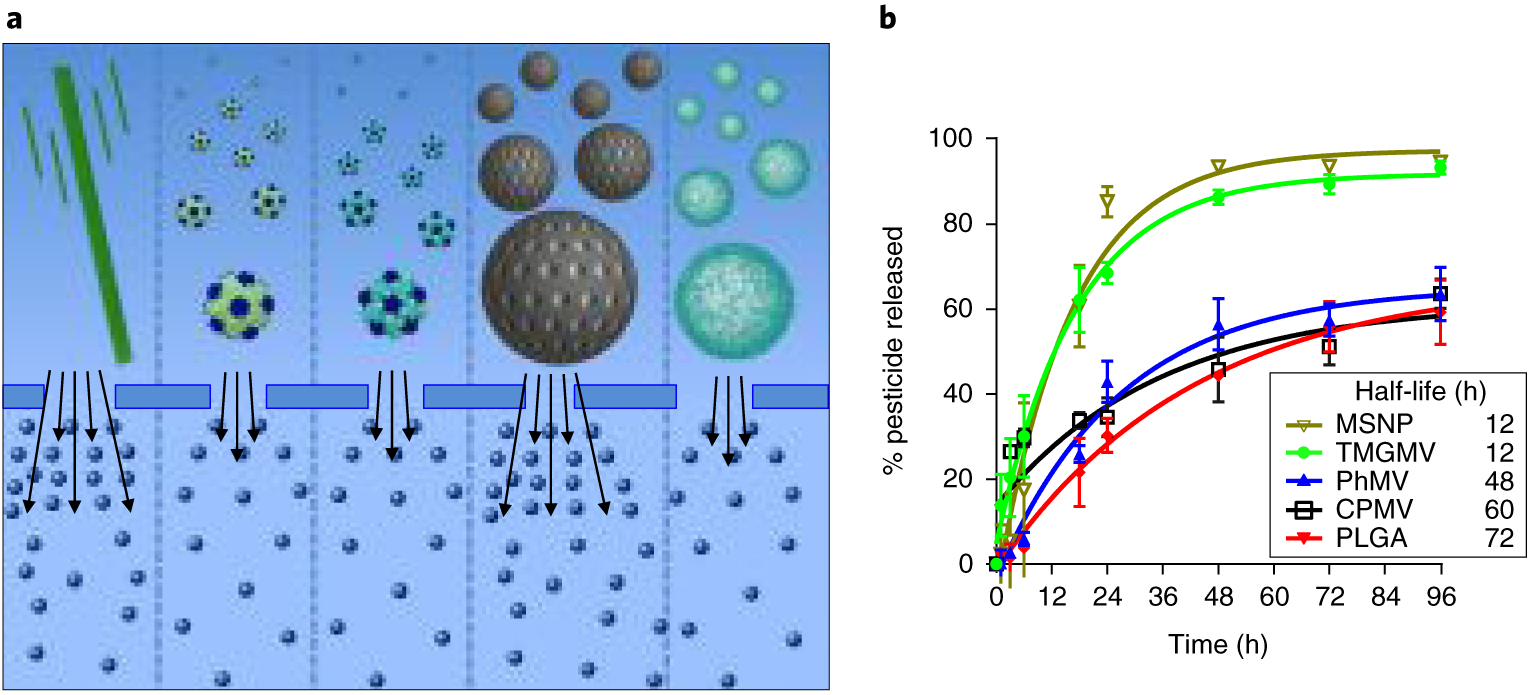
a, Schematic representation of the infused-dye release from (left to right) TMGMV, CPMV, PhMV, MSNP and PLGA. The dialysis membrane pores are large enough to allow the free movement of Cy5, but small enough to prevent nanoparticle diffusion. The number of arrows reflects the rate of dye release from each nanoparticle in a semi-quantitative manner. b, Corresponding plot of Cy5 cumulative release from each nanoparticle as a function of time. Error bars represent the s.d.
Soil transport behaviour
To establish the soil transport behaviour of each formulation, we conducted mobility studies using a cylindrical column (Methods and Supplementary Tables 2 and 3). As a reference, we ran 500 μg of free Cy5 through a soil column with a smaller diameter of 10 mm. Cy5 was unable to penetrate further than 4 cm through the soil because it bound strongly to the soil particles (Supplementary Fig. 6). About 40% of the mass of injected Cy5 was recovered from a column with a soil depth of 2 cm. These results are comparable to data reported for abamectin11, fenamiphos and oxamyl19, as well as other pesticides20. No matter which nanoparticle type was used as a carrier, the mobility of Cy5 within the column was significantly enhanced (Fig. 3). The best-performing carrier was TMGMV, which penetrated to a soil depth of 30 cm regardless of whether the cargo was conjugated or encapsulated (Fig. 3a). The spatiotemporal distribution of the nanoparticles and the Cy5 cargo was very similar, indicating that in each formulation the carrier and cargo were co-eluted (Fig. 3c–e). The quantity of encapsulated Cy5 that co-eluted with its carrier decreased with soil depth, indicating that a portion of the cargo was released over time.
Fig. 3 |. Experimental transport of nanopesticides and pesticides through soil.
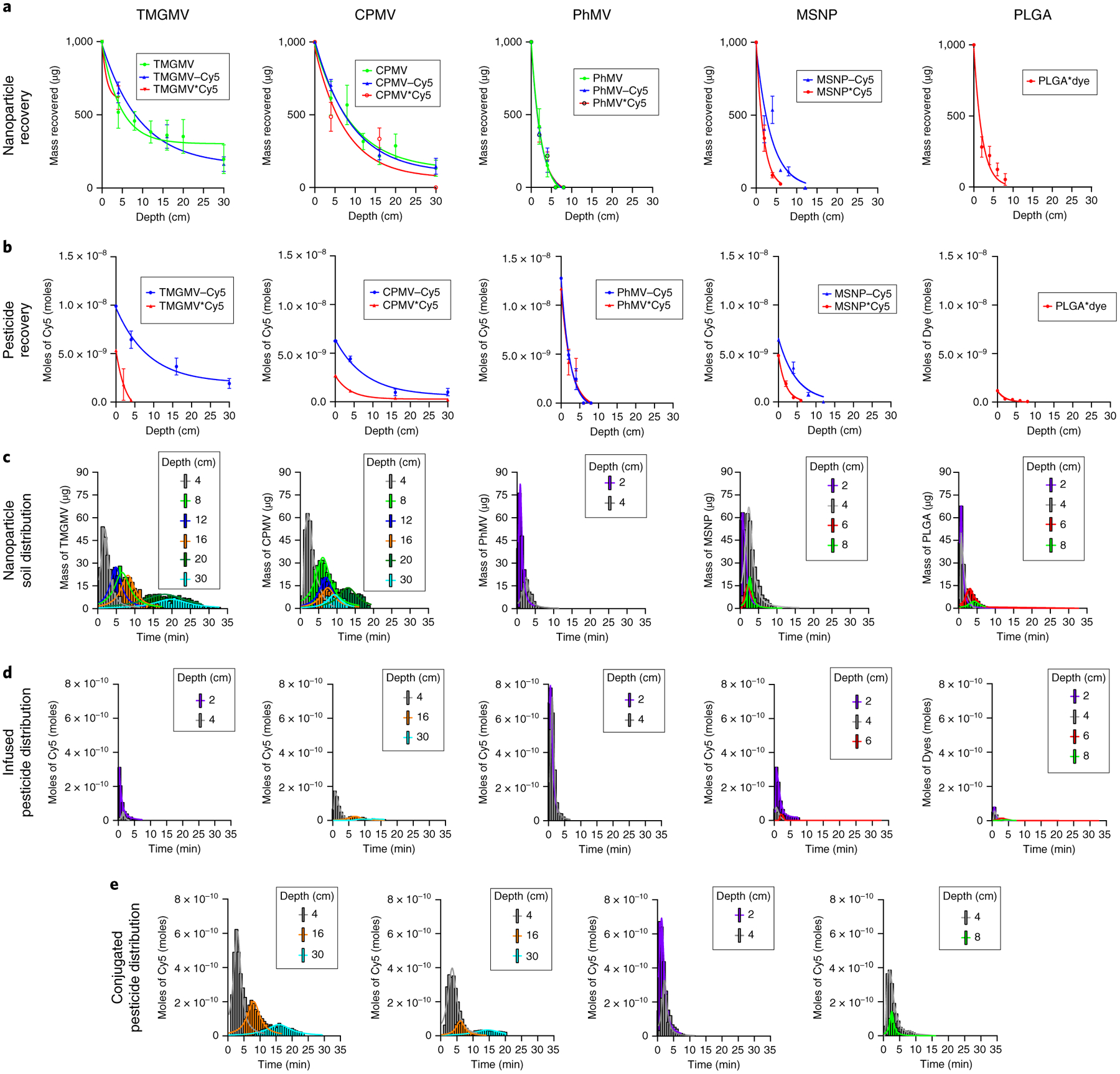
a, Cumulative mass of bare (green), Cy5-conjugated (blue) and Cy5-infused (red) nanoparticles exiting the soil column as a function of soil depth. b, The corresponding cumulative moles of conjugated Cy5 (red) and infused Cy5 (blue) exiting the soil column. Error bars represent s.d. c, Mass distribution of nanoparticles as a function of time at a given soil depth. d,e, Corresponding mole distribution of Cy5-infused (d) and Cy5-conjugated (e) particles as a function of time for a given soil depth.
To determine the quantity of particle loss from the soil transport experiment, we pooled all the elution samples that showed no evidence of nanoparticles in SDS–PAGE analysis and collected any trace amounts of the virus. We found that the residual mass of nanoparticles accounted for only ~2.5% of the overall mass of particles injected (Supplementary Table 4). Transmission electron microscopy imaging of the eluted particles revealed that they remained intact (Supplementary Fig. 7).
In terms of soil transport behaviours, TMGMV and CPMV were able to penetrate through 30 cm of soil, whereas PhMV, MSNP and PLGA only penetrated 4, 12 and 8 cm of soil, respectively. The mobility of the carriers in soil can therefore be ranked from highest to lowest: TMGMV >> CPMV >>> MSNP > PLGA > PhMV. These data suggest that the PhMV, MSNP and PLGA formulations are not suitable for pesticide delivery deep into the soil, to target the rhizosphere, but may be suitable for the delivery of pesticides that must remain close to the surface, such as herbicides. In the latter context, PhMV demonstrated the greatest pesticide delivery capability within the first 4 cm of soil (Fig. 3b). The particle size may influence the mobility of the carriers, but there was no particular trend within the size range we tested. For example, the 250 nm MSNP particles penetrated further than the 65 nm PLGA formulation, which in turn penetrated further than the 31 nm PhMV particles, but the 31 nm CPMV particles were much more mobile than all of the above. This is interesting given that CPMV and PhMV are similar in size and geometry, so the remarkable difference in mobility must reflect their surface chemistries. Both CPMV and PhMV are proteinaceous, but the distinct amino acid sequences of their coat proteins ensure that CPMV carries a negative surface charge whereas PhMV is positive (Supplementary Fig. 2). Furthermore, the rod-like (300 × 18 nm) TMGMV particles were the most mobile of all, which suggests that the elongated shape may facilitate their transport through the soil. In the field of nanomedicine, elongated nanoparticles are better at margination (migration towards blood vessels) and transport through membranes than spherical particles, which improves their tumour homing and penetration characteristics21–23. A high aspect ratio, therefore, appears to be a generally favourable property that facilitates movement between obstacles by influencing particle behaviour in flowing liquids. We therefore speculate that the field of nanopesticide delivery should further focus on the design of nanoparticles with a high aspect ratio in addition to the traditional spherical counterparts. Particles with an overall neutral to negative surface charge should also be favoured over positively charged nanoparticles to prevent early binding to soil matter.
The concentration of Cy5 as a function of soil depth was higher when the dye was conjugated to the particles rather than encapsulated (Fig. 3b). This reflects the slower release of the conjugated dye from the carrier, which allowed it to be carried further, whereas the encapsulated dye leaks more readily from the carrier and, once released, is rendered less mobile by its affinity for soil particles. Interestingly, Cy5 was released rapidly from the TMGMV*Cy5 and MSNP*Cy5 formulations (4 and 6 cm penetration, respectively), which suggests that the electrostatic forces between Cy5 and the carboxylate residues of (1) the TMGMV interior and (2) the MSNP mesopores are not strong enough to overcome the attraction between Cy5 and the soil. These results agree with the rapid loss of Cy5 observed in the dialysis assay (Fig. 2). Therefore, for field applications, the conjugated formulation appears superior to the encapsulated formulation.
Both TMGMV and CPMV were able to deliver Cy5 deep in the soil, but TMGMV–Cy5 showed by far the better performance. We previously showed that nematodes ingest nematicide-loaded TMGMV particles, which resulted in the death of 60% of the nematode population in liquid cultures within 24 h (ref.10). To increase the efficacy, future TMGMV formulations should include cleavable linkers to promote the slow and controlled release of the pesticide at the root level. However, to translate such pesticide formulations from the bench to the field, it is first necessary to establish the dose required to eradicate rhizosphere-dwelling pests. We therefore developed a mathematical model and validated it using our experimental data, as discussed below.
Computational modelling of pesticide delivery
A model column of length L (cm) and constant cross-sectional area A (cm2) was filled with a mixture of stationary soil particles and fluid (Fig. 1). The input to this model was a known mass of nanoparticles, with or without pesticide, introduced over a short period of time to the soil surface. The outputs were the concentrations of the nanoparticle ΩNP (mg cm−3), the nanoparticle–pesticide formulation CNPS (mg cm−3) and free pesticide CP (mg cm−3) at the base of the soil column as a function of time for a specific depth of soil. After the injection, fluid flow was established at the top the column at a rate Q (cm3 min−1). Nanoparticles were subsequently transported through the void volume fraction ε (dimensionless) of the saturated soil column, with an adsorption surface per soil particle volume ϕ (cm−1). The soil particle density within the column was assumed to be uniform. The rates of nanoparticle degradation and pesticide deactivation were assumed to be negligible during the experiment, as confirmed empirically (Supplementary Fig. 7). Nanoparticle binding to soil particles was modelled as a first-order irreversible reaction with rate constant kNPS (cm min−1) dependent on the nanoparticle size, aspect ratio and surface chemistry. The pesticide release rate was modelled as a first-order irreversible reaction with rate constant kPF (min−1). Although simple, using a first-order release mechanism led to comparative errors that ranged from 10−4 to 10−9 between the empirical data and the model output, which are sufficiently small to be acceptable differences (Supplementary Table 5). The resulting free pesticide may bind to soil particles through a first-order irreversible reaction with rate constant kPS (cm min−1). The interaction mechanisms are summarized in Supplementary Fig. 8 and the corresponding partial differential equations are shown in Methods. These equations were made dimensionless (Supplementary equation (1)) and solved using Matlab (Supplementary Code 1). The system contained five unknowns: the dispersion constants of the nanoparticle DNP and pesticide DP, and the rate constants of the nanoparticle absorption to soil kNPS, pesticide absorption to soil kPS and pesticide release from nanoparticles in fluid kPF. These values were obtained by comparing the model output to the empirical data and minimizing the error in Matlab. This computational model is therefore semi-empirical in nature. The resulting model outputs closely matched the empirical data (Fig. 4), although the values of DNP and kNPS differed slightly for each depth due to the experimental error caused by the need to use a new soil column in each test. Although the bulk density of the soil was kept constant across all the experiments, the soil particle distribution and the soil packing may have differed from column to column. To compensate for these variables, the average values of DNP and kNPS at different depths were computed to model the average nanoparticle soil transport profile (Fig. 4c). The nanoparticle dispersion DNP and rate of absorption to soil kNPS determine the ability of a nanoparticle to carry pesticide deep in the soil. With a greater mechanical dispersion, the nanoparticles become more widely distributed at a given soil depth over time. Therefore, mechanical dispersion greatly influences the concentration of nanoparticles at any given soil depth and time. The average DNP of each nanoparticle can be ranked from highest to lowest: TMGMV > CPMV > MSNP > PhMV > PLGA. As the absorption to the soil becomes stronger, the nanoparticles become less mobile. The average rate constant of nanoparticle absorption to soil kNPS can also be ranked from highest to lowest: MSNP >>> PLGA ≈ PhMV >> TMGMV > CPMV. The model confirms the superior mobility of TMGMV and its suitability to deliver pesticides to the rhizosphere.
Fig. 4 |. Theoretical transport of nanoparticles through soil.
a, The empirical output of TMGMV, CPMV, PhMV, MSNP and PLGA is used as a reference. b, Computational modelling of nanoparticle transport through soil. DNP and kNPS were optimized for each depth. c, Corresponding model of nanoparticle transport through soil using the average value of DNP and kNPS obtained in b.
To quantify the efficiency of pesticide delivery at the root level, we solved the model for the Cy5 dispersion constant DP and the rate constant of Cy5 absorption to soil kPS (Supplementary Fig. 9). We then used the average values of DNP, kNPS, DP and kPS to optimally estimate kPF (Fig. 5). Again, the model output matched the empirical data closely. The rate of Cy5 release kPF can be ranked from highest to lowest: PhMV >>> CPMV > TMGMV >> MSNP. Interestingly, these results do not match the release profile of Cy5 in the dialysis assay (Fig. 2), which suggests that the interaction between nanoparticles and soil has a major influence on the release rate.
Fig. 5 |. Theoretical transport of Cy5 through soil.
a, The empirical output of Cy5 infused into TMGMV, CPMV, PhMV and MSNP used as a reference. b, Computational modelling of Cy5 transport through soil by nanoparticle infusion. c, Corresponding model output of free Cy5 transport through soil.
To test experimentally whether a new nanoparticle formulation is a suitable candidate for pesticide delivery to the rhizosphere is time consuming and expensive; each nanoparticle described above required seven soil column experiments and we used an average of 100 SDS denaturing gels (each holding ten samples) to solve the soil transport profile of the nanocarriers through 2 to 30 cm of soil depth (note that other detection techniques would need to be developed for non-proteinaceous materials, but this is expected to be equally laborious). The model described above minimizes the time and cost required to evaluate a novel nanopesticide. In conjunction with the model, the only experimental work required is to run the new nanocarrier candidate through a 4 cm deep soil column, a mobility test well recognized and established by the Organisation for Economic Co-operation and Development24. This experimental data is necessary to establish the value of DNP and kNPS and predict the nanopesticides behaviour at any other soil depth (Supplementary Fig. 10). The VLP bacteriophage Qβ, which has been investigated as a drug carrier for medical application but not for pesticide delivery25, was tested for its ability to transport through soil (Supplementary Figs. 11 and 12). Qβ was predicted to transport through soil similarly to CPMV and TMGMV, which further confirms the superior soil mobility of VNPs and VLPs over synthetic materials.
The data obtained from the six different nanoparticles tested indicate that kNPS is linearly related to the surface area of MSNP, TMGMV, Qβ and CPMV; however, the PLGA and PhMV behaviours are different (Supplementary Fig. 13). PhMV nanoparticles have a positive zeta potential at a pH of 7.4. However, both clay and organic matter have a net negative surface charge, which may explain the enhanced soil binding of PhMV. We suspect the polymer nature of PLGA and its strong electronegativity also promote its enhanced binding to organic matter present in the soil. The linear relationship of kNPS of other nanoparticles with surface area suggests that the binding for the nanoparticles follows a mechanism that depends on the surface area, such as a mechanism based on van der Waals forces. Even though such an analysis is limited due to the limited number of particles tested, it does suggest that the model parameters have a physical basis that may be elucidated.
Testing the nanopesticide model in a real-life scenario
Nematode endoparasites infect 3,000 different plant species, which include many crops26, and are most abundant at ~24 cm beneath the soil surface27. Based on our empirical and modelling results, we selected TMGMV to deliver the nematicide abamectin28. Abamectin is insoluble in water and binds strongly to organic matter in the top layer of soil, so its effect in the rhizosphere is limited and it is an ideal candidate for nanopesticide delivery using TMGMV. We used our nanopesticide model to determine how much TMGMV formulation must be applied to maintain the half-maximum inhibitory concentration (IC50) of abamectin 24 cm beneath the surface for at least 24 h. A conjugated formulation would be better than encapsulation to avoid a premature release, and the linkage should be stable enough to allow the carrier to reach the target depth before the cargo is dispersed, such as a labile ester with a half-life release rate of four days29. The IC50 value of abamectin is 1.309 × 10−4 mg cm−3, and therefore at least this concentration must be achieved in the rhizosphere11. We modelled various flow rates to represent the typical range of crop irrigation systems, and used a common irrigation regimen of one hour three times a week, with the first irrigation taking place immediately after nanopesticide application. The values of DNP and kNPS for TMGMV were determined as above, and in place of abamectin we used the values for the chemically similar Cy5. We assumed a complete release at the root level due to the hydrolysis of the labile ester linkage over the course of a few days. The simulation output (Fig. 6) revealed that the mass of nanopesticide needed to maintain the target abamectin concentration for 24 hours was dependent on the flow rate. With no irrigation, neither free nor conjugated abamectin would achieve that concentration due to the extremely slow rate of diffusion. At a flow rate of 0.5 cm3 min−1, the lowest dose of TMGMV–abamectin required to maintain the target abamectin concentration 24 cm below the surface was 0.1056 mg cm−2. The model therefore offers a powerful tool to optimize the dose regimen that must be used to maximize the efficacy of pesticides in the rhizosphere.
Fig. 6 |. Theoretical treatment of a crop infected with nematodes using tMGMV–abamectin.
Each curve represents the temporal concentration distribution of abamectin conjugated to TMGMV at a soil depth equal to 24 cm as a function of the irrigation flow rate (Q). The corresponding minimal dose of TMGMV (m) that must be applied on the crop to maintain the IC50 of Abamectin is indicated.
Conclusions
We propose the use of VNPs and/or VLPs as carriers to deliver pesticides to the rhizosphere, where many pest species reside. Compared to icosahedral VNPs based on CPMV and VLPs based on PhMV, and synthetic counterparts with a similar geometry (PLGA and MSNP), the rod-like VNPs based on TMGMV achieved a much greater mobility in soil and also showed the highest dye loading capacity. This is the first evidence that nanoparticles with a high aspect ratio are more mobile in the soil than the spherical counterparts. It remains to be determined whether VNPs and VLPs are as cost effective as some synthetic materials. To date there is only one commercial plant VNP product, namely Solvinix (Bioprodex). In conjunction with our empirical data, we developed a computational model to predict the transport behaviour of pesticides encapsulated within or conjugated to nanoparticles. This model allowed us to calculate the optimal pesticide dose that must be applied to crops to achieve an effective dose at root level. This precision farming approach will increase the efficacy of pesticide applications, but also reduces the risk of residual chemicals to human health and the environment.
Methods
Expression and purification of nanoparticles.
TMGMV was obtained from Bioprodex, DegraFluorex Fluorescent PLGA nanoparticles were purchased from Phosphorex and MSNPs functionalized with propylcarboxylic acids were obtained from Sigma-Aldrich. We resuspended 3 mg ml−1 of PLGA and 1 mg ml−1 of MSNP in distilled water and sonicated them using a Branson 2800 device (Cleanosonic) for 10 min to obtain homogeneous solutions. CPMV was propagated in Burpee black-eyed pea plants and purified as previously described30. PhMV VLPs were prepared in ClearColi BL21 (DE3) cells as previously described31. United States Department of Agriculture permits (PPQ 526) were obtained for any work with plant viruses.
Bioconjugation of Cy5 to TMGMV tyrosine residues.
TMGMV comprises 2,130 identical coat proteins arranged helically around a single-stranded RNA genome to form a hollow rigid rod that measures 300 × 18 nm with a 4 nm internal channel10. The external surface features two solvent-exposed tyrosine side chains (Tyr 2 and Tyr 139), which can be functionalized using diazonium coupling reactions. We used sulfo-Cy5-azide (Lumiprobe) to modify these Tyr residues as previously described10. Briefly, we mixed 25 μl of 0.68 M 3-ethynylaniline with 75 μl of 3 M sodium nitrite (both Sigma-Aldrich) in 400 μl of 0.3 M p-toluenesulfonic acid monohydrate (Thermo Fisher Scientific) for 1 h on ice. We then added 15 equiv. of the resulting diazonium salt to 2 mg ml−1 TMGMV in 10 mM borate buffer (pH 8.8) for 30 min on ice. The particles were centrifuged at 112,000g for 1 h on a 30% (w/v) sucrose cushion to separate the TMGMV–alkyne particles from the excess diazonium salt. The TMGMV–alkyne was resuspended in 10 mM potassium phosphate (KP) buffer (pH 7.4) overnight before adding sulfo-Cy5-azide via a Cu(i)-catalysed alkyne–azide cycloaddition reaction. We added 2 equiv. Cy5 per coat protein to 2 mg ml−1 TMGMV–alkyne in the presence of 2 mM aminoguanidine, 2 mM l-ascorbic acid sodium salt and 1 mM copper(ii) sulfate (all Sigma-Aldrich) in a 10 mM KP buffer (pH 7.4) on ice for 30 min. The particles were again centrifuged at 112,000g for 1 h on a 30% (w/v) sucrose cushion to remove excess Cy5, and resuspended in a 10 mM KP buffer (pH 7.4) overnight. Further purification to remove aggregates involved centrifugation at 16,000g for 10 min. TMGMV–Cy5 was eluted using PD Minitrap G-25 desalting columns (GE Healthcare) to remove free Cy5 dye.
Bioconjugation of Cy5 to PhMV and CPMV lysine residues.
CPMV comprises 180 coat proteins and displays a total of 300 surface-exposed lysine side chains30. PhMV also comprises 180 identical coat proteins, but each displays four surface-exposed lysine side chains to make 720 in total31. CPMV and PhMV were labelled with sulfo-Cy5-NHS (Lumiprobe) using N-hydroxysuccinimide-activated esters that target the surface lysine residues. The reactions were carried out with a 1,200-fold (CPMV) or 900-fold (PhMV) molar excess of sulfo-Cy5-NHS in a 10 mM KP buffer (pH 7.0) at room temperature overnight, with agitation.
Bioconjugation of Cy5 to MSNP carboxylate residues.
Alkynes were conjugated to carboxylate groups on the MSNP surface using 1.5 mM propargylamine (Sigma-Aldrich) per gram of MSNP and 2.5 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide in 10 mM HEPES buffer (pH 7.4). The reaction was allowed to proceed for 24 h at room temperature followed by an alkyne–azide click reaction induced by adding 250 nmol of sulfo-Cy5-azide per gram of MSNP. The components were incubated at 4 °C with gentle agitation for 30 min using 1 mg ml−1 of MSNP in 10 mM KP buffer (pH 7.4) in the presence of 1 mM CuSO4, 2 mM aminoguanidine and 2 mM ascorbate (all Thermo Fisher Scientific). MSNPs were purified by centrifugation at 7,000g for 10 min and buffer exchanged at least five times.
Encapsulation of Cy5 into TMGMV, CPMV, PhMV and MSNP particles.
Encapsulated formulations were prepared by mixing 1 mg ml−1 of TMGMV, CPMV or PhMV with a 5,000-fold molar excess of Cy5–amine, or by mixing 250 nmol of Cy5 per gram of MSNP in 10 mM KP buffer (pH 7.8) overnight at room temperature with agitation.
UV–vis spectroscopy.
The UV–vis spectra of native and modified TMGMV, CPMV, PhMV, PLGA and MSNP nanoparticles were determined using a NanoDrop Spectrophotometer (Thermo Fisher Scientific). The efficiency of Cy5 loading was determined based on the dye-to-carrier ratio and the Beer–Lambert law: TMGMV, ε260 = 3 ml mg−1 cm−1, molecular weight of 39.4 × 106 g mol−1; CPMV, ε260 = 8.1 ml mg−1 cm−1, molecular weight of 5.6 × 106 g mol−1; Cy5, ε647 = 271,000 M−1 cm−1, molecular weight of 747 g mol−1; PLGA dye, ε = 250,000 M−1 cm−1, molecular weight of 519 g mol−1.
Denaturing gel electrophoresis.
We denatured 5 μg of native TMGMV, CPMV and PhMV at 100 °C for 5 min in the presence of 4 × LDS loading dye (Thermo Fisher Scientific). The Cy5-modified particles were denatured as described above using a loading dye that lacked bromophenol blue. The samples were separated on 4–12% NuPage precast gels in 1 × MOPS buffer (Thermo Fisher Scientific) for 40 min at 200 V and 120 mA, with SeeBlue Plus2 ladder size markers (Thermo Fisher Scientific). Gels were imaged before and after staining with Coomassie Brilliant Blue (0.25% w/v) using the FluorChem R imaging system under white light and MultiFluor red light.
Agarose gel electrophoresis.
We analysed 3 μg of native CPMV, PhMV, PLGA and MSNP particles by 1.2% (w/v) agarose gel electrophoresis (1 h at 100 V) in 1 × TBE running buffer in the presence of nucleic acid gel stain (GoldBio) diluted 1:20,000. Gels were imaged before and after staining with Coomassie Brilliant Blue (0.25% w/v) as above.
Transmission electron microscopy.
Formvar copper grids coated with carbon film (Electron Microscopy Sciences) were glow discharged to render the surface more hydrophilic using the PELCO easiGlow operating system. Drops of TMGMV, CPMV, PhMV or PLGA (10 μl, 1 mg ml−1) were deposited onto the grids for 2 min at room temperature. The grids were then washed twice with deionized water for 30 s and subsequently stained twice with 2% (w/v) uranyl acetate for another 45 s. MSNP (10 μl, 1 mg ml−1) was deposited onto the grids and allowed to dry-cast overnight. A Tecnai F-30 transmission electron microscope was used to capture images of the samples at 300 kV.
Dynamic light scattering.
A DynaPro NanoStar instrument (Wyatt Technology) was used to measure the hydrodynamic radius of the TMGMV, CPMV, PhMV, PLGA and MSNP nanoparticles. The reported hydrodynamic radii and standard derivations correspond to the average of 30 measurements, each of 100 runs.
Size exclusion chromatography.
Native and modified TMGMV, CPMV, PhMV and PLGA samples (200 μl, 1 mg ml−1) were passed through a Superose 6 Increase column on the AKTA Explorer chromatography system (GE Healthcare) at a flow rate 0.5 ml min−1 in 10 mM KP buffer (pH 7.0). The absorbances at 260 nm and 280 nm were recorded for all the particles, the absorbance at 647 nm was recorded to confirm Cy5 conjugation or encapsulation, and the absorbance at 668 nm was recorded to confirm dye encapsulation in the proprietary PLGA nanoparticles.
Fluorescent-dye release profiling.
The release of Cy5 from TMGMV, PhMV, CPMV and MSNP, and the dye release from PLGA, was evaluated using a dialysis-based assay. Slide-A-Lyzer MINI dialysis units (10,000 MWCO, Thermo Fisher Scientific) were loaded with 1 mg of particles in 10 mM KP buffer (pH 7.0) in triplicate. The particles were dialysed against 10 mM KP buffer (pH 7.0) at room temperature for 96 h. At times t = 0, 1, 3, 6, 18, 24, 48, 72 and 96 h, 10 μl was extracted from each dialysis unit and the remaining dye entrapment was measured by UV–vis spectroscopy (TMGMV, CPMV, PhMV and MSNP) or imaged under the FluorChem R imaging system (PLGA).
Soil mobility of TMGMV, CPMV, PhMV, PLGA, MSNP and free Cy5.
Garden Magic Top Soil was packed at a density of 0.32 g cm−3 into a cylindrical column (28 mm diameter, top height 30 cm) and saturated with deionized water to remove air pockets. This was the maximum density achievable under our experimental conditions, but the density of soil in real environments can be higher (0.6–1.6 g cm−3) due to compaction effects with depth and over time. We injected a bolus that contained 1 mg of each formulation with and without conjugated or infused dye molecules at the top of the soil column and saturated the column with water at a constant flow rate of 1.5 cm3 min−1 in 10 mM KP buffer (pH 7.0). The eluent was collected at the base of the column in 500 μl fractions. Up to 200 fractions were collected in each trial (two trials per depth for each formulation). The elution fractions that contained TMGMV, PhMV or CPMV were analysed by SDS–PAGE to determine the mass of nanoparticles recovered in each elution fraction. CPMV was analysed on 4–12% NuPage precast gels in 1 × MOPS buffer. TMGMV and PhMV were analysed on 4–12% NuPage polyacrylamide SDS gels cast according to the Surecast Handcast protocol (Invitrogen). We mixed 23 μl of each elution fraction with 7 μl 5× SDS loading buffer and separated the samples for 1 h at 200 V and 120 mA with SeeBlue Plus2 ladder size and three standards that contained known amounts of nanoparticles (0.5, 1 and 2 μg) for comparison. The gels were then incubated in 20% (v/v) methanol and 10% (v/v) acetic acid in water 30 min before staining with Coomassie Brilliant Blue (0.25% w/v) for an additional 30 min. The gels were imaged using the AlphaImager HP system (Protein Simple) under white light and the FluorChem R system under MultiFluor red light. The elution fractions that contained PLGA and MSNP were imaged as 20 μl droplets on Parafilm on the FluorChem R imaging system under MultiFluor red light in the presence of the nanoparticle standards described above.
All the nanoparticles were imaged in triplicate and the images were analysed using ImageJ. The area under the curve of the standards was used to create a linear standard curve that related to the area under the curve of the elution samples to the total mass of nanoparticles present in the corresponding elution fraction. Finally, fractions that appeared to contain no nanoparticles were centrifuged at 160,000g for 3 h and the pellet was resuspended in 1 ml of 10 mM KP buffer (pH 7.0) for SDS–PAGE analysis to determine the recovered mass of nanoparticles.
Computational methods.
The model equations presented here are variations of those developed for other applications32. The nanoparticle mass density distribution in fluid (that is, interstitial soil space) changes as a function of the column depth z and time t according to equation (1):
| (1) |
The irreversible rate of adsorption of nanoparticles onto the soil surface from the fluid is RNPS (mg cm−2 min−1). With and without pesticides, the nanoparticle adsorption process is assumed to be a first-order reaction as shown in equation (2):
| (2) |
The available soil surface, which changes negligibly, is incorporated into the rate constant kNPS (cm min−1). As ΩNP decreases with the reaction, the nanoparticle attachment to soil increases as shown in equation (3):
| (3) |
where ΩNPS (mg cm−3) is the mass density of nanoparticles bound to soil at any location in the column.
The pesticide mass concentration CP (mg cm−3) distribution in the fluid changes as shown in equation (4):
| (4) |
The irreversible release rate RPF (mg cm−3 min−1) of the pesticide from nanoparticles in the fluid is shown in equation (5):
| (5) |
where CNPF (mg cm−3) is the mass concentration of the pesticide bound to nanoparticles in the fluid at any location in the column. The irreversible adsorption rate RPS (mg cm−2 min–1) of free pesticide in the fluid onto the soil surface is shown in equation (6):
| (6) |
The free pesticide mass concentration CPS (mg cm−3) bound to soil changes according to equation (7):
| (7) |
where RP (min−1) is the irreversible rate of pesticide ‘transfer’ from the nanoparticles onto soil.
The concentration change of pesticide attached to nanoparticles in the fluid is therefore shown in equation (8):
| (8) |
where RPNP (mg cm−2 min−1) is the irreversible adsorption rate of pesticide attached to nanoparticles onto the soil determined according to equation (9):
| (9) |
This rate process has the same rate constant as the rate process of nanoparticles adsorption onto soil (RNPS), as defined in equation (10):
| (10) |
The pesticide concentration CNPS (mg cm−3) in nanoparticles on the soil changes according to equation (11):
| (11) |
The irreversible rate of pesticide transfer from nanoparticles to the soil with rate constant kP (min−1) is therefore shown in equation (12):
| (12) |
Initially, there is no nanoparticle or pesticide in the soil space, 0 < z < L: t=0 : ΩNP=0 ; ΩNPS=0 ; CNPF=0 ; CNPS=0 ; CP=0 ; CPS=0
At the input (top of the cylindrical column), z = 0, a solution of volume V0 with nanoparticle density and pesticide concentration is injected. The total number of nanoparticles injected is . The fluid flows through the interstitial soil space at volume rate Q. The nanoparticles are transported into the column over a time interval 0 to t1 according to equation (13):
| (13) |
Therefore, at the entrance of the cylinder the mass flow rate balances must be specified for the nanoparticles and pesticide. For the nanoparticles, the input nanoparticle mass density is derived as shown in equation (14):
| (14) |
For the pesticide, which is carried by the nanoparticle, the input mass concentration is derived as shown in equation (15):
| (15) |
As no dissolved pesticide enters, z = 0 and CP = 0. In the fluid that leaves the column, the concentrations of nanoparticle, pesticide in nanoparticle and free pesticide can be represented by X. The mass flow of X from inside the cylinder (L−) to outside the cylinder (L+) is shown in equation (16):
| (16) |
The volume flow rate, nanoparticle density and pesticide concentration are continuous across the output boundary of the cylinder, which implies that the gradients must vanish as shown in equation (17):
| (17) |
For the simultaneous numerical solution of the partial differential equations, the equations were first transformed into their dimensionless form (Supplementary equation (1)).
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Science Foundation CAREER DMR 1841848 (to N.F.S.) and NIH EB021911 (to H.B.). We thank H. Hu for providing the PhMV particles used in this study.
Footnotes
online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41565-019-0453-7.
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41565-019-0453-7.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The following raw data can be found in the Supplementary Information: the composition of the soil used to produce all experimental data (Supplementary Tables 2 and 3); the SDS PAGE required to reproduce the data presented in Fig. 3 (Supplementary Fig. 5); the Matlab code required to reproduce the data presented in Figs. 4–6 (Supplementary Code 1).
Code availability
These dimensionless equations were solved using partial differential equation solver function ‘pdepe’ (Matlab). All code was made available in Supplementary Code 1.
References
- 1.Aktar MW, Sengupta D & Chowdhury A Impact of pesticides use in agriculture: their benefits and hazards. Interdisc. Toxicol 2, 1–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamberth C, Jeanmart S, Luksch T & Plant A Current challenges and trends in the discovery of agrochemicals. Science 341, 742–746 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Atwood D & Paisley-Jones C Pesticide Industry Sales and Usage: 2008–2012 Market Estimates (US Environmental Protection Agency, 2017). [Google Scholar]
- 4.Damalas CA & Eleftherohorinos IG Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Pub. He 8, 1402–1419 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nuruzzaman M, Rahman MM, Liu Y & Naidu R Nanoencapsulation, nano-guard for pesticides: a new window for safe application. J. Agr. Food Chem. 64, 1447–1483 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Vurro M, Miguel-Rojas C & Pérez-de-Luque A Safe nanotechnologies for increasing the effectiveness of environmentally friendly natural agrochemicals. Pest. Manag. Sci 10.1002/ps.5348 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Kah M, Kookana RS, Gogos A & Bucheli TD A critical evaluation of nanopesticides and nanofertilizers against their conventional analogues. Nat. Nanotechnol 13, 677–684 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Kookana RS et al. Nanopesticides: guiding principles for regulatory evaluation of environmental risks. J. Agr. Food Chem 62, 4227–4240 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Walker GW et al. Ecological risk assessment of nano-enabled pesticides: perspective on problem formulation. J. Agr. Food Chem 66, 6480–6486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chariou PL & Steinmetz NF Delivery of pesticides to plant parasitic nematodes using tobacco mild green mosaic virus as a nanocarrier. ACS Nano 11, 4719–4730 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Cao J et al. Development of abamectin loaded plant virus nanoparticles for efficacious plant parasitic nematode control. ACS Appl. Mater. Interfaces 7, 9546–9553 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Guenther RH, Lommel SA, Opperman CH & Sit TL in Virus-Derived Nanoparticles for Advanced Technologies: Methods and Protocols (eds Wege C & Lomonossoff GP) 203–214 (Springer, 2018). [Google Scholar]
- 13.Charudattan R, Pettersen M & Hiebert E Use of tobacco mild green mosaic virus (TMGMV) mediated lethal hypersensitive response (HR) as a novel method of weed control. US patent 6689718 (2009).
- 14.Charudattan R & Hiebert E A plant virus as a bioherbicide for tropical soda apple, Solanum viarum. Outlook Pest Manag. 18, 167–171 (2007). [Google Scholar]
- 15.Umekawa M & Oshima N Sensitivity of tobacco mosaic virus to ultraviolet irradiation. Jpn J. Microbiol 16, 441–443 (1972). [DOI] [PubMed] [Google Scholar]
- 16.Rae C et al. Chemical addressability of ultraviolet-inactivated viral nanoparticles (VNPs). PLoS ONE 3, e3315 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mir M, Ahmed N & ur Rehman A Recent applications of PLGA based nanostructures in drug delivery. Colloids Surf. B 159, 217–231 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Torney F, Trewyn BG, Lin VS-Y & Wang K Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat. Nanotechnol 2, 295–300 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Hassan MEM, Zawam HS, El-Nahas SEM & Desoukey AF Comparison study between silver nanoparticles and two nematicides against Meloidogyne incognita on tomato seedlings. Plant Pathol. J 15, 144–151 (2016). [Google Scholar]
- 20.Pestovsky YS & Martínez-Antonio A The use of nanoparticles and nanoformulations in agriculture. J. Nanosci. Nanotechnol 17, 8699–8730 (2017). [Google Scholar]
- 21.Truong NP, Whittaker MR, Mak CW & Davis TP The importance of nanoparticle shape in cancer drug delivery. Expert Opin. Drug Deliv. 12, 129–142 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Barua S & Mitragotri S Challenges associated with penetration of nanoparticles across cell and tissue barriers: a review of current status and future prospects. Nano Today 9, 223–243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toy R The Effect of Particle Size and Shape on the in vivo Journey of Nanoparticles. PhD thesis, Case Western Reserve University; (2014). [Google Scholar]
- 24.OECD Guidelines Test for the Testing of Chemicals No. 312: Leaching in Soil Columns (OECD, 2004). [Google Scholar]
- 25.Chen Z, Li N, Chen L, Lee J & Gassensmith JJ Dual functionalized bacteriophage Qβ as a photocaged drug carrier. Small 12, 4563–4571 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Quentin M, Abad P & Favery B Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Front. Plant Sci. 4, 53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godfrey GH The depth distribution of the root-knot nematode, Heterodera radicicola, in Florida soils. J. Agri. Res 24, 93–98 (1924). [Google Scholar]
- 28.Putter I et al. Avermectins: novel insecticides, acaricides and nematicides from a soil microorganism. Experientia 37, 963–964 (1981). [Google Scholar]
- 29.Schoenmakers RG, van de Wetering P, Elbert DL & Hubbell JA The effect of the linker on the hydrolysis rate of drug-linked ester bonds. J. Control Release 95, 291–300 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Steinmetz NF & Manchester M Viral Nanoparticles: Tools for Materials Science and Medicine (Pan Stanford, 2015). [Google Scholar]
- 31.Masarapu H et al. Physalis mottle virus-like particles as nanocarriers for imaging reagents and drugs. Biomacromolecules 18, 4141–4153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ultman JS, Baskaran H & Saidel GM Biomedical Mass Transport and Chemical Reaction: Physicochemical Principles and Mathematical Modeling (Wiley, 2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


