Abstract
Background
5-Hydroxymethylcytosine (5hmC) is a novel epigenetic mark and may be involved in the mechanisms of tumorigenesis and malignant transformation. However, the role of 5hmC in ependymoma, the third most common brain tumor in children, remains unclear. The aim of this study sought to identify the characterization of 5hmC levels in pediatric posterior fossa ependymoma and to evaluate whether 5hmC levels could be a potential factor to predict clinical outcomes.
Results
Our results showed that 5hmC levels were globally decreased in posterior fossa ependymoma compared with normal cerebellum tissues (P < 0.001). Group A posterior fossa ependymomas had higher 5hmC levels than group B tumors (P = 0.007). Moreover, 5hmC levels positively correlated with Ki-67 index in posterior fossa ependymoma (r = 0.428, P = 0.003). Multivariate Cox hazards model revealed that patients with high 5hmC levels (> 0.102%) had worse PFS and OS than patients with lower 5hmC levels (< 0.102%) (PFS: HR = 3.014; 95% CI, 1.040–8.738; P = 0.042; OS: HR = 2.788; 95% CI, 0.974–7.982; P = 0.047).
Conclusions
Our findings suggest that loss of 5hmC is an epigenetic hallmark for pediatric posterior fossa ependymoma. 5hmC levels may represent a potential biomarker to predict prognosis in children with posterior fossa ependymoma.
Keywords: Pediatric ependymoma, Molecular subgroup, 5-Hydroxymethylcytosine, Epigenetics, Ki-67 index, Prognosis, 1q gain
Background
Ependymoma (EPN) is a relatively rare neuroepithelial tumor that arises throughout the whole neuraxis [1]. Intracranial EPN predominantly occurs in children and adolescents, with two third of those tumors located in posterior fossa [1, 2]. Recently, posterior fossa ependymoma (EPN_PF) has been classified into two molecular subgroups based on DNA CpG island (CpGi) methylation profiles status [3–6]. Group A ependymoma (EPN_PFA) is characterized by CpGi hypermethylation with 1q gain and occurs predominantly in infancy and young children. These subgroup tumors also exhibit global low H3K27me3 [7–9], global DNA hypomethylation [7], and high expression of EZHIP [10]. Conversely, group B ependymoma (EPN_PFB) presents with CpGi hypomethylation and primarily occurs in adolescences and young adults. Moreover, the molecular classification of EPN has provided a superior prognostic prediction and risk stratification [11]. EPN_PFA tumors are often difficult to completely resect and bear a dismal prognosis, while EPN_PFB tumors are less invasive and carry a favorable prognosis [4, 5]. It suggests that epigenetic mechanisms play an essential role in EPN_PF pathogenesis and tumor maintenance.
Abnormal DNA methylation at the 5 position of cytosine (5mC) is an epigenetic mark of cancers. Recent studies presented evidence for an active DNA demethylation pathway initiated by the ten-eleven translocation (TET) protein family, resulting in the conversion of 5mC into 5-hydroxymethylcytosine (5hmC) [12, 13]. As a new epigenetic biomarker, 5hmC is reshaping the view of the tumor epigenome. Several reports have shown that decreased 5hmC level is an indicator of poor survival in the central nervous system (CNS) tumors patients [14–17]. However, only one report studied the changes of 5hmC as well as its downstream products in two EPN cell lines, which represent a subgroup of supratentorial EPN with RELA fusion [18].
In the present study, we performed the ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS/MS) analysis and immunochemistry (IHC) staining analysis to measure global 5hmC and 5mC levels to relate this information to clinical characteristics and survival outcomes in pediatric EPN_PF.
Results
Clinical characteristics
Forty-five cases of pathologically WHO grades II/III confirmed EPN_PF (age < 18) treated in Beijing Tiantan Hospital between Jan 2010 to Dec 2017 were identified. The clinical data of the institutional cohort were summarized in Table 1. Median age at diagnosis of these children was 4 years (range 1–17). The male to female ratio was 2.8:1 (33/12). The maximum diameter of tumor ranged from 2.3 to 19.5 cm with a median size of 4.7 cm.
Table 1.
Clinical characteristics of pediatric posterior fossa ependymoma
| Characteristics | Value (%) |
|---|---|
| Demographics | |
| No. of patients | 45 |
| Age, years, median (range) | 4 (1–17) |
| Gender, (female/male) | 12/33 |
| Tumor size, cm, median (range) | 4.7 (2.3–19.5) |
| Extent of surgery, GTR, n (%) | 21 (46.7) |
| Radiotherapy, yes, n (%) | 29 (64.4) |
| Chemotherapy, yes, n (%) | 12 (26.7) |
| Histology, n (%) | |
| WHO II | 10 (22.2) |
| WHO III | 35 (77.8) |
| Ki-67 index, n (%) | |
| < 20% | 20 (44.4) |
| ≥ 20% | 25 (55.6) |
| Molecular subgroups, n (%) | |
| EPN_PFA | 35 (77.8) |
| EPN_PFB | 10 (22.2) |
| Chromosome 1q | |
| Gain | 16 (35.6) |
| No gain | 29 (64.4) |
| 5hmC/(C + mC) × 100%, median (range) | 0.127 (0.028–0.341) |
| 5mC/(C + mC) × 100%, mean ± SD | 3.664 ± 0.426 |
| Recurrence, n (%) | 25 (55.6) |
| Death during follow-up, n (%) | 23 (51.1) |
| Follow-up period, months, median (range) | 38 (6–60) |
GTR gross total resection, 5hmC 5- hydroxymethylcytosine, 5mC 5-methylcytosine, C cytosine, EPN_PFA Group A posterior fossa ependymoma, EPN_PFB Group B posterior fossa ependymoma
Gross total resection (GTR) was achieved in 21 (46.7%) of patients while 24 (53.3%) had a subtotal resection (STR). Histopathological diagnosis presented ten (22.2%) patients with EPN of WHO grade II and 35 (77.8%) patients with EPN of WHO grade III. We performed immunostaining of H3K27me3 to distinguish EPN_PFA from EPN_PFB (Additional file 1: Figure S1A). We found that 35 of 45 (77.8%) were negative for H3K27me3 staining and designated as EPN_PFA, while 10 of 45 (22.2%) were positive as PFB (Additional file 1: Figure S1B). EPN_PFA patients were much younger than EPN_PFB patients (P < 0.001, Additional file 1: Figure S1C). Interphase fluorescence in situ hybridization (FISH) analysis revealed that 16 tumors (35.6%) had chromosome 1q25 gain, while 29 tumors (64.4%) had a balanced chromosome 1 (Additional file 1: Figure S2A, B). A total of 29 (64.4%) patients were treated with postoperative focal radiotherapy, and 12 (26.7%) patients received chemotherapy.
5hmC levels were decreased in EPN_PF
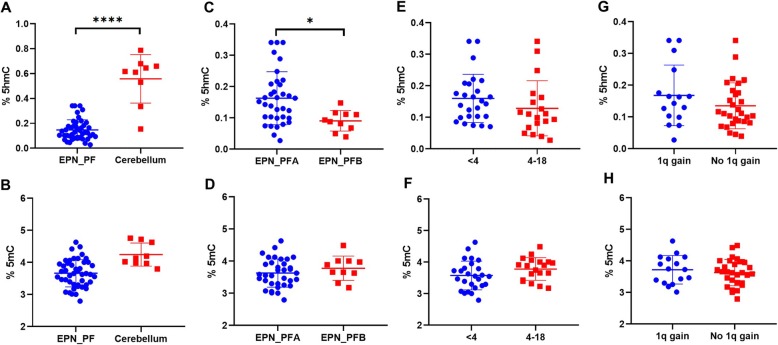
To evaluate the global changes of 5hmC and 5mC levels in pediatric EPN_PF, we first performed the UHPLC-MS/MS analysis to measure global 5hmC and 5mC levels in 45 EPN_PF and 9 normal cerebellum samples. We found that 5hmC levels significantly decreased in tumor samples compared with those in normal cerebellum tissues [EPN_PF vs. cerebellum, 0.127 (0.028–0.341) vs. 0.617 (0.154–0.788); P < 0.001] (Fig. 1a). No significant difference of global 5mC levels was observed between EPN_PF and cerebellum (EPN_PF vs. cerebellum, 3.664 ± 0.426 vs. 4.245 ± 0.361; P = 0.653) (Fig. 1b).
Fig. 1.
Comparative evaluation of 5hmC levels and 5mC levels as measured by UHPLC-MS/MS was respectively analyzed between tumors and normal cerebellum (a, b), two molecular subgroups (c, d), two age groups (e, f) and two subtypes of 1q status in (g, h). Bars, standard deviation. *P < 0.05; ****P < 0.0001. 5hmC by Mann–Whitney U test, 5mC by Student’s t test
5hmC levels were different between two molecular subgroups
We next compared the differences of 5hmC and 5mC levels between two molecular subgroups. As shown in Fig. 1c, EPN_PFA exhibited higher 5hmC levels than PFB [EPN_PFA vs. EPN_PFB, 0.152 (0.028–0.341) vs. 0.091 (0.039–0.148); P = 0.007]. No significant difference of 5mC levels between the two molecular subgroups (EPN_PFA vs. EPN_PFB, 3.630 ± 0.438 vs. 3.780 ± 0.379; P = 0.334) (Fig. 1d). Moreover, we found that the 5hmC levels and 5mC levels were similar between the age groups and subtypes of 1q status (Fig. 1e–h).
5hmC levels positively correlate with cell proliferation
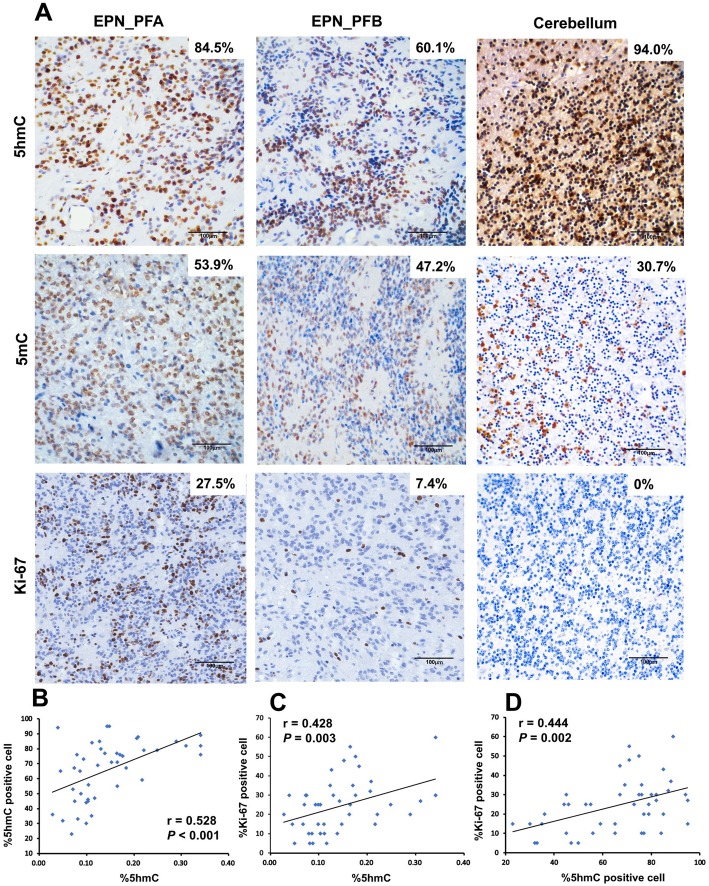
Furthermore, IHC staining yielded the similar previous results (Fig. 2a). The EPN_PF samples showed a lower nuclear positivity of 5hmC antibody (65.4% ± 19.1%) compared to the normal cerebellums (90% ± 4%, P = 0.004). The Pearson correlation analysis showed there was a significantly positive correlation between 5hmC positive cells percentage and the amount of 5hmC levels (r = 0.528, P < 0.001) (Fig. 2b). We next determine the relationship between 5hmC levels and cell proliferation using Ki-67 staining in EPN_PF. We found the tight correlation between 5hmC positive cells and Ki-67 index (r = 0.444, P = 0.002) (Fig. 2c). The results also showed that 5hmC levels were positively related with Ki-67 index (r = 0.428, P = 0.003) (Fig. 2d).
Fig. 2.
5hmC, 5mC, and Ki-67 index IHC staining in EPN_PF samples and normal cerebellum. a Representative image of 5hmC, 5mC, and Ki-67 staining in EPN_PF and cerebellum. b The Pearson correlation between nuclear positive cells of 5hmC and global 5hmC levels. c The Pearson correlation between Ki-67 index cells and global 5hmC levels. d The Pearson correlation between Ki-67 index and nuclear positive cells of 5hmC. Scale bars represent 100 μm
High 5hmC levels related to poor prognosis
In this study, the median follow-up period was 38 months (range 6–60 months). At the endpoint of the follow-up analysis, 25 (55.6%) patients presented recurrence or progressive disease. Twenty-three (51.1%) patients were dead occurred during the data collection period. The estimated 3-year PFS and OS rates were 49.9 ± 8.1% (95% CI, 34.1–65.7%) and 48.8 ± 8.4% (95% CI, 32.3–65.3%), respectively.
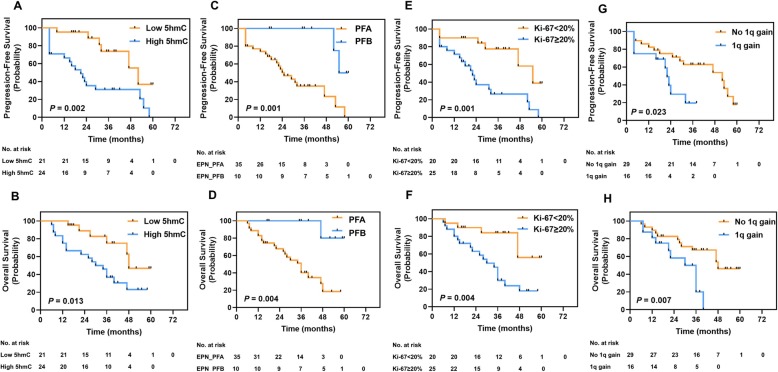
To further clarify the relationship between 5hmC level and prognostic factors, we divided patient cohorts into two subgroups according to 5hmC levels (UHPLC-MS/MS) [low 5hmC levels (< 0.102%) and high 5hmC levels (> 0.102%)] based on the Cut-off Finder [19]. We also dichotomized the Ki-67 index as high Ki-67 index (≥ 20%) and low Ki-67 (< 20%), as previous described [20]. We found that patients with low 5hmC levels (3-year PFS, 73.7 ± 11.5%; 3-year OS, 75.0 ± 11.1%) showed more favorable prognosis than those with high 5hmC levels (3-year PFS, 31.0 ± 9.7%, P = 0.002; 3-year OS, 36.7 ± 10.0%, P < 0.001; Fig. 3a, b). Patients with PFA had the worse survival rates (3-year PFS, 35.1 ± 9.0%; 3-year OS, 51.1 ± 8.9%), compared with patients with PFB (3-year PFS, 100%, P = 0.001; 3-year OS, 100%, P = 0.004; Fig. 3b, c). Patients with high Ki-67 index (3-year PFS, 26.5 ± 9.9%; 3-year OS, 45.0 ± 10.4%) had poorer prognosis compared to patients with low Ki-67 index (3-year PFS, 77.5 ± 10.0%, P = 0.001; 3-year OS, 84.0 ± 8.5%, P = 0.004; Fig. 3e, f). Moreover, 1q gain EPN_PF exhibited significantly worse PFS and OS as compared to no 1q gain EPN_PF (3-year PFS, 19.6% ± 12.2% vs. 62.8% ± 9.4%, P = 0.023; 3-year OS, 20.0 ± 12.3% vs. 67.3% ± 9.0%, P = 0.008; Fig. 3g, h).
Fig. 3.
Kaplan–Meier survival analysis comparing PFS and OS for a, b low 5hmC group vs. high 5hmC group. c, d EPN_PFA vs. EPN_PFB. e, f Low Ki-67 index group vs. high Ki-67 index group. g, h No 1q gain group vs. 1q gain group. P value determined using the log-rank test
5hmC level was an independent prognostic factor
On univariate analysis (Table 2), EPN_PFA (PFS: HR = 8.012; 95% CI, 1.815–35.371, P = 0.006, OS: HR = 10.854; 95% CI, 1.446–81.465; P = 0.020) and high 5hmC levels (PFS: HR = 3.438; 95% CI, 1.367–8.646; P = 0.009, OS: HR = 3.030; 95% CI, 1.192–7.702; P = 0.020) as well as 1q gain (PFS: HR = 2.666; 95% CI, 1.086–6.548; P = 0.032, OS: HR = 3.148; 95% CI, 1.283–7.722; P = 0.012) were associated with worse PFS and OS. Patients with high Ki-67 index were associated with worse PFS (HR = 3.726; 95% CI, 1.475–9.427; P = 0.005) but not for OS (P = 0.086).
Table 2.
Univariate cox analysis for progression-free survival and overall survival for pediatric EPN_PF
| Progression-free survival | Overall survival | |||||
|---|---|---|---|---|---|---|
| Variable | Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value |
| Mo. subgroup (EPN_PFA vs. EPN_PFB) | 8.012 | 1.815–35.371 | 0.006 | 10.854 | 1.446–81.465 | 0.020 |
| 5hmC subgroup (high vs. low) | 3.438 | 1.367–8.646 | 0.009 | 3.030 | 1.192–7.702 | 0.020 |
| Resection (STR vs. GTR) | 1.338 | 0.602–2.972 | 0.475 | 1.445 | 0.634–3.292 | 0.381 |
| Ki-67 index (≥ 20% vs. < 20%) | 3.726 | 1.475–9.427 | 0.005 | 1.787 | 0.921–3.468 | 0.086 |
| 1q gain (yes vs. no) | 2.666 | 1.086–6.548 | 0.032 | 3.148 | 1.283–7.722 | 0.012 |
| Age, years (< 4 vs. 4–18) | 1.567 | 0.678–3.618 | 0.293 | 1.376 | 0.580–3.262 | 0.469 |
| Radiotherapy (yes vs. no) | 0.695 | 0.281–1.717 | 0.430 | 0.477 | 0.197–1.159 | 0.102 |
| Chemotherapy (yes vs. no) | 0.839 | 0.334–2.112 | 0.710 | 0.226 | 0.052–0.985 | 0.053 |
| Histology (WHO III vs. WHO II) | 2.115 | 0.760–5.885 | 0.151 | 2.938 | 0.855–10.104 | 0.087 |
| Gender (male vs. female) | 1.416 | 0.530–3.783 | 0.487 | 1.015 | 0.400–2.577 | 0.975 |
P values in italics are of statistical significance
Abbreviations: Mo. molecular, PF posterior fossa, STR subtotal resection, GTR gross total resection, EPN_PFA Group A posterior fossa ependymoma, EPN_PFB Group B posterior fossa ependymoma
A multivariate Cox proportional hazards model with backward stepwise selection for PFS and OS was fitted using the following potential prognostic factors (based on univariate analysis and published literature [21, 22]): molecular subgroup, 5hmC subgroup, Ki-67 index, 1q gain, and extent of resection. As seen in Table 3, EPN_PFA molecular subgroup (PFS: HR = 5.253; 95% CI, 1.052–26.237; P = 0.043, OS: HR = 7.496; 95% CI, 0.928–60.557; P = 0.048) and high 5hmC levels (PFS: HR = 3.014; 95% CI, 1.040–8.738; P = 0.042, OS: HR = 2.788; 95% CI, 0.974-7.982; P = 0.047) were confirmed as independent inferior prognostic indicators for PFS and OS. STR were confirmed as independent inferior prognostic indicators for OS (HR = 2.682; 95% CI, 1.168–7.664; P = 0.039). Age, WHO grade, Ki-67 index, and adjuvant treatment exhibited no association with PFS and OS.
Table 3.
Multivariate cox analysis for progression-free survival and overall survival for pediatric EPN_PF
| Progression-free survival | Overall survival | |||||
|---|---|---|---|---|---|---|
| Variable | Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value |
| Mo. subgroup (EPN_PFA vs. EPN_PFB) | 5.253 | 1.052–26.237 | 0.043 | 7.496 | 0.928–60.557 | 0.048 |
| 5hmC subgroup (high vs. low) | 3.014 | 1.040–8.738 | 0.042 | 2.788 | 0.974–7.982 | 0.047 |
| Resection (STR vs. GTR) | 2.292 | 0.919–5.719 | 0.075 | 2.682 | 1.168–7.664 | 0.039 |
P values in italics are of statistical significance
Abbreviations: Mo. molecular, STR subtotal resection, GTR gross total resection, EPN_PFAGroup A posterior fossa ependymoma, EPN_PFB Group B posterior fossa ependymoma
Furthermore, we attempted to validate whether 5hmC level was an independent prognostic factor within EPN_PFA children alone (n = 35). A multivariate analysis revealed that 5hmC levels and extent of resection were independent predictors of OS in children with EPN_PFA, but none of variables showed significant association with PFS (Additional file 1: Table S1).
Discussion
Epigenetic modifications are important in normal development and frequently alter during tumorigenesis [23]. Recent studies have demonstrated that the loss of 5hmC in different types of cancers might play an essential role in pathogenesis [24–28]. Kraus et al. [29] reported that loss of 5hmC was found in EPN, but only one case (1/23) in this group was EPN_PF. However, it is still unclear whether level of 5hmC is changed in pediatric EPN_PF. To the best of our knowledge, this is the first study to illustrate the alternation of 5hmC level in EPN_PF. We showed that global levels of 5hmC are strikingly decreased in pediatric EPN_PF tumor tissues compared with normal cerebellum tissues. Our results further support the previous observations of the reduced level of 5hmC that occurred in other types of CNS tumors [14, 17, 29]. Moreover, we found that 5hmC score based on IHC staining with 5hmC antibody significantly related to 5hmC levels detected by UHPLC-MS/MS, suggesting that the IHC staining approach may be a useful method to identify 5hmC level in pediatric EPN_PF.
The limited number of biomarkers reliably predicting prognosis in EPN highlights the importance of developing more robust prognostic markers [22]. Previous studies indicate that 5hmC levels are associated with clinical outcomes in multiple types of cancers [27, 30]. Analysis of multiple cohorts of intracranial ependymoma highlights a wide variance in the utility of the grade II versus grade III distinction as a prognostic marker. However, the utility of histological grading of ependymoma for risk stratification has been controversial and without consistent associations of tumor grade with patient outcome [11]. Recently, DNA methylation patterns and DNA copy number profiles can be used to predict clinical outcomes for patients with EPN [22]. In this study, our result indicated that high 5hmC level is an independent prognostic factor of poor survival in pediatric EPN_PF on PFS and OS. In most solid tumors, low 5hmC levels relate to high tumor grade and worse outcomes [17, 27, 29, 31–33]. However, our results are in line with the previous report that high levels of 5hmC independently correlate with inferior overall survival in acute myeloid leukemia [34] and suggest that 5hmC may be involved in the distinct mechanisms of tumorigenesis and malignant transformation. These findings remain to be confirmed in future studies.
Recent advances in the biological characterization of EPN_PF have demonstrated the existence of two clinically, demographically, and molecularly distinct entities [4]. EPN_PFA tumors show higher methylation of CpGi. EPN_PFA patients are younger, have laterally located tumors with an increased occurrence of chromosome 1q gain, and behave more aggressively compared with EPN_PFB patients [4]. In addition to CpGi hypermethylation, DNA hypomethylation and global H3K27me3 reduction in the absence of recurrent genetic changes in EPN_PF suggests that epigenetic mechanisms are central to EPN_PF pathogenesis [7, 8]. Moreover, several studies [7, 8, 35–37] showed that reduced H3K27me3 in EPN_PF is not genetically driven but epigenetically deregulated. Our study firstly observed distinct 5hmC levels between two molecular subgroups, suggesting that 5hmC may participate in the abnormal DNA methylation in pediatrics EPN_PF. Future researchers on EPN_PF should focus on the mechanism of epigenetic alternations.
We found a strong positive correlation between 5hmC levels and Ki-67 index in pediatric EPN_PF. Several studies demonstrate that higher Ki-67 index seems to be associated with poor prognosis in pediatrics EPN [20, 38, 39]. Our data confirm previous results and further suggest that high 5hmC levels associated with inferior outcomes. However, some studies have reported that 5hmC levels are inverse correlation of cellular proliferation in different types of cancers [17, 40–42]. These discrepancies may be explained by thedistinct tumorigenesis in this benign tumor compared with other malignant tumors. However, the mechanism of 5hmC influenced tumor cell proliferation in EPN_PF needs further research.
Our study had several limitations. The first limitation of our study was the small sample size and the relatively short follow-up period limited our ability to detect robust survival predictors. Future studies with large sample sizes and long-term follow-up are needed to confirm the results of our findings. Second, the molecular subgroup was classified by IHC. To overcome this limitation, future studies using fresh frozen tumor tissues followed by methylation arrays are needed [4].
Conclusions
In our study, we found that 5hmC is a potential prognostic predictor that may contribute to the improvement of clinical risk stratification for EPN_PF. Our results indicate that the characteristic of 5hmC level is associated with molecular subgrouping and cell proliferation. These findings suggest that the mechanisms responsible for regulating 5hmC may represent a possible future therapeutic target.
Methods
Study design and samples
The main objective of this study was to assess the clinical characteristics of DNA hydroxymethylcytosine in pediatric EPN_PF. We used UHPLC-MS/MS to investigate 5hmC abundance in EPN_PF. Further, we conducted molecular classification using IHC. A total of 45 patients (age < 18 years) who were diagnosed with EPN_PF at Beijing Tiantan Hospital between January 2010 and December 2017 were included in this study. Clinical data, including age at diagnosis, gender, tumor size, treatment, recurrence data, and survival, were collected by retrospective chart review. Two neuropathologists reviewed the histopathologic findings according to 2016 WHO classification of CNS tumors [1]. Follow-up evaluations were performed on all patients via either an outpatient consultation or a telephone interview. This study was approved by the ethics committee of Beijing Tiantan Hospital, Capital Medical University. Written informed consent was obtained.
Tumor tissues were obtained during initial surgery before radiation or any other adjuvant treatment. All samples were snap-frozen (− 80 °C) or fixed with 4% buffered formalin, paraffin-embedded. All tumor specimens were sterilely stored at Beijing Neurosurgical Institute by the Ethics Review Board of the Beijing Tiantan Hospital. As control samples, all normal cerebellums used in this study were provided by the Human Brain Bank, Chinese Academy of Medical Sciences & Peking Union Medical College, with the approval from the Institutional Review Board of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Approval Number: 009-2014).
Definition of EPN_PF molecular subgroup by immunohistochemistry
IHC analyses were performed as reported [9]. In brief, tissue sections were cut at 5 μm, followed by deparaffinization and rehydration using xylene and ethanol. Then, the slides were incubated in 3% hydrogen peroxide for 10 min in phosphate-buffered saline to block endogenous peroxidase activity. Slides were incubated overnight with rabbit monoclonal anti-H3K27me3 antibody (C36B11, Cell Signaling, Danvers, MA, USA) at a concentration of 1:150 using the standard Leica Bond protocol IHC-F. The Leica Bond Polymer Refine DAB detection kit was used according to the manufacturer’s instructions. All IHC slides were evaluated by two independent neuropathologists; the scoring methods were performed as report described [9]. H3K27me3 positive staining was defined as scored positive when more than 80% cells had nuclear positivity and scored negative when they did not.
Evaluation of global 5mC and 5hmC by UHPLC-MS/MS
The absolute amount of 5hmC and 5mC in EPN was measured as previously described [43, 44]. Briefly, DNA isolation was performed using the Wizard® Genomic DNA Purification Kit (A1620, Promega, Madison, WI, USA) according to the manufacturer’s protocol. DNA for each sample (1 μg/sample) was denatured by heating at 100 °C for 3 min and then digested by incubation at 42 °C with nuclease P1 (2U, Sigma, N8630, Darmstadt, Germany) for 6 h. Subsequently, 1 U of alkaline phosphatase (Sigma, M183A) was added and incubated at 37 °C for another 6 h. Finally, the sample was diluted to a total volume of 60 μl and filtered (0.45 μm, PALL). Nucleosides were separated by ultra-performance liquid chromatographic on T3 column (WATERS, 186003538, MA, USA) and detected using triple-4 quadrupole mass spectrometer (WATERS, ACQUITY UPLC XEVO TQ-S). The mass transitions of m/z 228.4 to 112.2 (C), m/z 242.3 to 126.1 (mC), m/z 258.2 to 124.2 (hmC) were monitored and recorded. Quantification was performed in comparison with the standard curve obtained from pure nucleoside standards running on the same batch of samples. Finally, the percentages of 5mC and 5hmC were calculated by the following formula: M (cytosine) and M (5mC) are the molar quantities of cytosine: 5mC% = M (5mC)/[M (cytosine) + M (5mC)] × 100, 5hmC% = M (5hmC)/[M(cytosine) + M(5mC)] × 100.
IHC analysis for 5hmC, 5mC, and Ki-67
The utilized primary antibodies were including 5hmC (1:800, ab214728, Abcam), 5mC (1:200, ab10805, Abcam), and Ki-67 (1:1500, ab15580, Abcam). Immunohistochemical detection of 5hmC and 5mC was performed as described above exclusive of the step of DNA denatures by 2N HCl [27]. The 5hmC and Ki-67 staining and score methods were performed according to described previously [45]. In brief, positive staining was defined as a dark brown staining pattern, confined to the nuclear region. Scant or fine granular background staining or no staining was considered as negative. The mean value of the five snapshots was calculated to represent the percentage of positive cells in each case.
1q gain by interphase FISH
Dual color interphase fluorescence in FISH analysis was performed on formalin-fixed paraffin-embedded sections as previously described using commercially available 1q25 (spectrum green) and 1p36 (spectrum orange) probe sets (ZytoVision, Germany) [46]. The evaluation criteria and scoring system adopted was based on previously described [46, 47].
Statistical analysis
All statistical analyses were performed with SPSS 23 (IBM Corp., New York, NY, USA) and two-sided P values < 0.05 were considered statistically significant. The normality of variables was assessed. Data are expressed as mean ± standard deviation (SD) or median (minimum to maximum). Differences in mean and median values were evaluated by using Student’s t test and the Mann–Whitney U test, respectively. Associations between categorical variables were assessed via Fisher’s exact test. To interpret the effect of 5hmC level in a more clinically relevant manner, 5hmC levels were dichotomized into two groups using Cutoff Finder [19]. The cutoff values (0.102%) were defined as the points with the most significant split between groups, including PFS and OS.
For the survival analysis, overall survival (OS) was calculated from the date of initial surgery that established the pathological diagnosis to the time of death. Progression-free survival (PFS) was derived from the date of initial surgery to the time of progression. Kaplan–Meier curves of OS and PFS were generated and log-rank tests were used to compare OS and PFS between demographic and clinical factors. Multivariate Cox proportional hazards regression models with backward stepwise selection were used to identify significant prognostic factors for OS and PFS. Hazard ratios with corresponding 95% confidence intervals were calculated.
Supplementary information
Additional file 1: Figure S1, S2, Table S1. Supplementary Material
Acknowledgements
Not applicable.
Abbreviations
- 5hmC
5-Hydroxymethylcytosine
- 5mC
5-Methylcytosine
- CNS
Central nervous system
- CpGi
CpG island
- EPN
Ependymoma
- EPN_PF
Posterior fossa ependymoma
- EPN_PFA
Group A posterior fossa ependymoma
- EPN_PFB
Group B posterior fossa ependymoma
- FISH
Fluorescence in situ hybridization
- GTR
Gross total resection
- IHC
Immunohistochemistry
- OS
Overall survival
- PFS
Progression-free survival
- STR
Subtotal resection
- UHPLC-MS/MS
Ultra-high-performance liquid chromatography-mass spectrometry
Authors’ contributions
The study was conceived and designed by FZ, PL, TW, and YN. TW and ZZ conducted the experiments. Sample collection was performed by BW, ZY, and CL. Bioinformatics and statistical analyses were performed by TW, ZJ, SL, and ZZ. The manuscript was written and reviewed by TW, FZ, and YN. All authors read and approved the final manuscript.
Funding
This study was partly supported by the Capital Health Development and Research Special Projects of Beijing (Grant number: 2018-2-1073 to CL) and Medical Epigenetics Research Center, Chinese Academy of Medical Sciences (Grant number: 2019PT310017, to YN).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study has been approved by the ethics committee of Beijing Tiantan Hospital, Capital Medical University. All samples and records were de-identified and anonymized prior to the study. Consent was obtained from all individuals for publication. No individual-level data are reported.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yamei Niu, Email: niuym@ibms.pumc.edu.cn.
Pinan Liu, Email: pinanliu@ccmu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13148-020-0809-8.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Kilday JP, Rahman R, Dyer S, Ridley L, Lowe J, Coyle B, Grundy R. Pediatric ependymoma: biological perspectives. Mol Cancer Res. 2009;7(6):765–786. doi: 10.1158/1541-7786.MCR-08-0584. [DOI] [PubMed] [Google Scholar]
- 3.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, Benner A, Hielscher T, Milde T, Remke M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20(2):143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramaswamy V, Hielscher T, Mack SC, Lassaletta A, Lin T, Pajtler KW, Jones DT, Luu B, Cavalli FM, Aldape K, et al. Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: a retrospective multicohort analysis. J Clin Oncol. 2016;34(21):2468–2477. doi: 10.1200/JCO.2015.65.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wani K, Armstrong TS, Vera-Bolanos E, Raghunathan A, Ellison D, Gilbertson R, Vaillant B, Goldman S, Packer RJ, Fouladi M, et al. A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012;123(5):727–738. doi: 10.1007/s00401-012-0941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, Sabari B, Margol AS, Panwalkar P, Parolia A, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med. 2016;8(366):366ra161. doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, Wang X, Gallo M, Garzia L, Zayne K, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panwalkar P, Clark J, Ramaswamy V, Hawes D, Yang F, Dunham C, Yip S, Hukin J, Sun Y, Schipper MJ, et al. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 2017;134(5):705–714. doi: 10.1007/s00401-017-1752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain SU, Do TJ, Lund PJ, Rashoff AQ, Diehl KL, Cieslik M, Bajic A, Juretic N, Deshmukh S, Venneti S, et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27 M-like mechanism. Nat Commun. 2019;10(1):2146. doi: 10.1038/s41467-019-09981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison DW, Kocak M, Figarella-Branger D, Felice G, Catherine G, Pietsch T, Frappaz D, Massimino M, Grill J, Boyett JM, et al. Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed. 2011;10:7. doi: 10.1186/1477-5751-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroeze LI, van der Reijden BA, Jansen JH. 5-Hydroxymethylcytosine: an epigenetic mark frequently deregulated in cancer. Biochim Biophys Acta. 2015;1855(2):144–154. doi: 10.1016/j.bbcan.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Vasanthakumar A, Godley LA. 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet. 2015;208(5):167–177. doi: 10.1016/j.cancergen.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Bezerra Salomao K, Cruzeiro GAV, Bonfim-Silva R, Geron L, Ramalho F, Pinto Saggioro F, Serafini LN, Antunes Moreno D, de Paula Queiroz RG, Dos Santos AS, et al. Reduced hydroxymethylation characterizes medulloblastoma while TET and IDH genes are differentially expressed within molecular subgroups. J Neuro-Oncol. 2018;139(1):33–42. doi: 10.1007/s11060-018-2845-1. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez AF, Bayon GF, Sierra MI, Urdinguio RG, Torano EG, Garcia MG, Carella A, Lopez V, Santamarina P, Perez RF, et al. Loss of 5hmC identifies a new type of aberrant DNA hypermethylation in glioma. Hum Mol Genet. 2018;27(17):3046–3059. doi: 10.1093/hmg/ddy214. [DOI] [PubMed] [Google Scholar]
- 16.Orr BA, Haffner MC, Nelson WG, Yegnasubramanian S, Eberhart CG. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One. 2012;7(7):e41036. doi: 10.1371/journal.pone.0041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Liu Y, Zhang Z, Li J, Wan Y, Zhang L, Wang Y, Li X, Xu Y, Fu X, et al. 5-hydroxymethylcytosine loss is associated with poor prognosis for patients with WHO grade II diffuse astrocytomas. Sci Rep. 2016;6:20882. doi: 10.1038/srep20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsawhook A, Lewis L, Coyle B, Ruzov A. Medulloblastoma and ependymoma cells display increased levels of 5-carboxylcytosine and elevated TET1 expression. Clin Epigenetics. 2017;9:18. doi: 10.1186/s13148-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7(12):e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfsberger S, Fischer I, Hoftberger R, Birner P, Slavc I, Dieckmann K, Czech T, Budka H, Hainfellner J. Ki-67 immunolabeling index is an accurate predictor of outcome in patients with intracranial ependymoma. Am J Surg Pathol. 2004;28(7):914–920. doi: 10.1097/00000478-200407000-00011. [DOI] [PubMed] [Google Scholar]
- 21.Marinoff AE, Ma C, Guo D, Snuderl M, Wright KD, Manley PE, Al-Sayegh H, Sinai CE, Ullrich NJ, Marcus K, et al. Rethinking childhood ependymoma: a retrospective, multi-center analysis reveals poor long-term overall survival. J Neuro-Oncol. 2017;135(1):201–211. doi: 10.1007/s11060-017-2568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pajtler KW, Mack SC, Ramaswamy V, Smith CA, Witt H, Smith A, Hansford JR, von Hoff K, Wright KD, Hwang E, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017;133(1):5–12. doi: 10.1007/s00401-016-1643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2(8):627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32(5):663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150(6):1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraus TF, Globisch D, Wagner M, Eigenbrod S, Widmann D, Munzel M, Muller M, Pfaffeneder T, Hackner B, Feiden W, et al. Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int J Cancer. 2012;131(7):1577–1590. doi: 10.1002/ijc.27429. [DOI] [PubMed] [Google Scholar]
- 30.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71(24):7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frycz BA, Murawa D, Borejsza-Wysocki M, Marciniak R, Murawa P, Drews M, Kolodziejczak A, Tomela K, Jagodzinski PP. Decreased expression of ten-eleven translocation 1 protein is associated with some clinicopathological features in gastric cancer. Biomed Pharmacother. 2014;68(2):209–212. doi: 10.1016/j.biopha.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Navarro A, Yin P, Ono M, Monsivais D, Moravek MB, Coon JS, Dyson MT, Wei JJ, Bulun SE. 5-Hydroxymethylcytosine promotes proliferation of human uterine leiomyoma: a biological link to a new epigenetic modification in benign tumors. J Clin Endocrinol Metab. 2014;99(11):E2437–E2445. doi: 10.1210/jc.2014-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y, Xu Z, Li Z, Liu Y, Ye X, et al. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016;26(1):103–118. doi: 10.1038/cr.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroeze LI, Aslanyan MG, van Rooij A, Koorenhof-Scheele TN, Massop M, Carell T, Boezeman JB, Marie JP, Halkes CJ, de Witte T, et al. Characterization of acute myeloid leukemia based on levels of global hydroxymethylation. Blood. 2014;124(7):1110–1118. doi: 10.1182/blood-2013-08-518514. [DOI] [PubMed] [Google Scholar]
- 35.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506(7489):451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6(12):e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14(3):R25. doi: 10.1186/gb-2013-14-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamecnik J, Snuderl M, Eckschlager T, Chanova M, Hladikova M, Tichy M, Kodet R. Pediatric intracranial ependymomas: prognostic relevance of histological, immunohistochemical, and flow cytometric factors. Mod Pathol. 2003;16(10):980–991. doi: 10.1097/01.MP.0000087420.34166.B6. [DOI] [PubMed] [Google Scholar]
- 39.Bennetto L, Foreman N, Harding B, Hayward R, Ironside J, Love S, Ellison D. Ki-67 immunolabelling index is a prognostic indicator in childhood posterior fossa ependymomas. Neuropathol Appl Neurobiol. 1998;24(6):434–440. doi: 10.1046/j.1365-2990.1998.00143.x. [DOI] [PubMed] [Google Scholar]
- 40.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraus TF, Kolck G, Greiner A, Schierl K, Guibourt V, Kretzschmar HA. Loss of 5-hydroxymethylcytosine and intratumoral heterogeneity as an epigenomic hallmark of glioblastoma. Tumour Biol. 2015;36(11):8439–8446. doi: 10.1007/s13277-015-3606-9. [DOI] [PubMed] [Google Scholar]
- 43.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao F, Zhang J, Li P, Zhou Q, Zhang S, Zhao C, Wang B, Yang Z, Li C, Liu P. Prognostic value of Ki-67 index in adult medulloblastoma after accounting for molecular subgroup: a retrospective clinical and molecular analysis. J Neuro-Oncol. 2018;139(2):333–340. doi: 10.1007/s11060-018-2865-x. [DOI] [PubMed] [Google Scholar]
- 46.Rajeshwari M, Sharma MC, Kakkar A, Nambirajan A, Suri V, Sarkar C, Singh M, Saran RK, Gupta RK. Evaluation of chromosome 1q gain in intracranial ependymomas. J Neuro-Oncol. 2016;127(2):271–278. doi: 10.1007/s11060-015-2047-z. [DOI] [PubMed] [Google Scholar]
- 47.Modena P, Buttarelli FR, Miceli R, Piccinin E, Baldi C, Antonelli M, Morra I, Lauriola L, Di Rocco C, Garre ML, et al. Predictors of outcome in an AIEOP series of childhood ependymomas: a multifactorial analysis. Neuro-Oncology. 2012;14(11):1346–1356. doi: 10.1093/neuonc/nos245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1, S2, Table S1. Supplementary Material
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.