Abstract
Background
Molecular genotyping in Plasmodium serves many aims including providing tools for studying parasite population genetics and distinguishing recrudescence from reinfection. Microsatellite typing, insertion-deletion (INDEL) and single nucleotide polymorphisms is used for genotyping, but only limited information is available for Plasmodium malariae, an important human malaria species. This study aimed to provide a set of genetic markers to facilitate the study of P. malariae population genetics.
Methods
Markers for microsatellite genotyping and pmmsp1 gene polymorphisms were developed and validated in symptomatic P. malariae field isolates from Myanmar (N = 37). Fragment analysis was used to determine allele sizes at each locus to calculate multiplicity of infections (MOI), linkage disequilibrium, heterozygosity and construct dendrograms. Nucleotide diversity (π), number of haplotypes, and genetic diversity (Hd) were assessed and a phylogenetic tree was constructed. Genome-wide microsatellite maps with annotated regions of newly identified markers were constructed.
Results
Six microsatellite markers were developed and tested in 37 P. malariae isolates which showed sufficient heterozygosity (0.530–0.922), and absence of linkage disequilibrium (ISA=0.03, p value > 0.05) (N = 37). In addition, a tandem repeat (VNTR)-based pmmsp1 INDEL polymorphisms marker was developed and assessed in 27 P. malariae isolates showing a nucleotide diversity of 0.0976, haplotype gene diversity of 0.698 and identified 14 unique variants. The size of VNTR consensus repeat unit adopted as allele was 27 base pairs. The markers Pm12_426 and pmmsp1 showed greatest diversity with heterozygosity scores of 0.920 and 0.835, respectively. Using six microsatellites markers, the likelihood that any two parasite strains would have the same microsatellite genotypes was 8.46 × 10−4 and was further reduced to 1.66 × 10−4 when pmmsp1 polymorphisms were included.
Conclusions
Six novel microsatellites genotyping markers and a set of pmmsp1 VNTR-based INDEL polymorphisms markers for P. malariae were developed and validated. Each marker could be independently or in combination employed to access genotyping of the parasite. The newly developed markers may serve as a useful tool for investigating parasite diversity, population genetics, molecular epidemiology and for distinguishing recrudescence from reinfection in drug efficacy studies.
Keywords: Genotyping, INDEL, Nucleotide diversity, Tandem repeats
Background
Over the past decade, the world has experienced significant reduction in global falciparum malaria burden, but this decline was less prominent for the other human Plasmodium species [1]. Plasmodium malariae is endemic throughout parts of South America, Africa, Asia, and the Western Pacific [2–4]. It has been argued that malaria elimination programmes largely focused on Plasmodium falciparum and Plasmodium vivax may have undermined P. malariae and Plasmodium ovale endemicity [5]. Plasmodium malariae deploys a different transmission strategy, with a large parasite reservoir in asymptomatic carriers with low parasitaemia. These chronic P. malariae infections can occasionally cause anaemia and nephrotic syndrome [4, 6–8]. Use of molecular techniques such as qPCR has enabled more sensitive detection of parasite carriage compared to microscopy [3, 9]. Previous studies conducted in Cameroon [10] and Equatorial Guinea [9] using qPCR based species identification revealed that P. falciparum was the dominating species responsible for over 80.0% of malaria positive cases, followed by P. malariae (≥ 12.0%). In the Colombian Amazon region, the P. malariae infections were reported as high as 43.2% [11]. Nevertheless, P. malariae is understudied compared to the other human Plasmodium species [12], although there are recently increased efforts to describe the epidemiology of P. malariae [13–17]. However, important molecular tools for this are currently lacking, which will facilitate the description of the course of natural infections, multiplicity of infection (MOI) and anti-malarial drug resistance [18, 19]. In clinical trials on drug efficacy, genetic markers are important to distinguish between recrudescence and reinfection [20, 21].
Microsatellite markers remain an important technique in population genetics because of its high level of polymorphisms and fidelity to discriminate variants within parasite population [13, 14, 22]. Microsatellites are short tandem repeats of nucleotides usually consisting of 1 to 10 base pair (bp) unit motifs [23]. These are abundant in non-coding regions of the genome, which are generally not under selection pressure, and their molecular origin and evolution result from improper alignment, mispairing and strand-slippage events [22, 24, 25]. Heterozygosity refers to observation of two different alleles at a locus which forms fundamental bases for investigation of genetic variation in population. Since blood-stage Plasmodium is haploid, a single-genotype infection is expected to have single allele at each location while a multiple-genotype infection is expected to carry multiple alleles [14, 20]. Defining potential microsatellite markers requires assessment of their polymorphisms, and evaluation of heterozygosity and linkage disequilibrium [20, 26]. High precision fragment analyzers based on capillarity electrophoresis have a resolution of 1–2 bp. This technology in combination with analysis software like GeneScan™ provides a powerful tool for analyses of microsatellite markers. Few adequate microsatellite markers have been identified for P. malariae compared to P. falciparum the [21, 27–29] and P. vivax [20, 30–32]. The previously identified few P. malariae genotyping markers [13, 14] showed promising potential and could identify up to 10 distinct alleles for a single locus.
Recent developments in high-throughput sequencing technology has enabled Insertion-deletions (INDEL)-based gene polymorphisms to construct high-resolution genetic maps and evaluate parasite population structures [33]. The approach has been used for investigating polymorphisms in genes encoding gametocyte antigen, duffy binding protein and merozoite surface proteins (msp) [34–36]. Previous studies in P. vivax and P. falciparum have utilized gene polymorphisms in msp and circumsporozoite protein to study the parasite diversity [37–39]. The msp genes are part of a larger family that encodes the major surface antigens for invasive forms of Plasmodium during erythrocytic stages [40–42]. Functions for some of these proteins have described for P. vivax [43] and P. falciparum [44, 45]. Compared to microsatellite-based genotyping, the utility of surface protein gene-based markers are often limited due to reduced polymorphisms as a result of selective pressure by the host immune system [46, 47], the members of the msp family have been widely used as genotyping markers [36, 48]. The variable number of tandem repeats (VNTR)-related INDEL polymorphisms contributes to msp gene diversity [40, 48, 49]. The msp genes consists of exons as amino acid coding blocks interspersed between their conserved and semi-conserved blocks (Fig. 1). Sequences within these semi-conserved regions can be highly variable and thus are potential genetic markers. In this context, the performance of pfmsp1, pfmsp2, pvmsp1 and pvmsp3α genes to discriminate between recrudescence and reinfection has been studied [50, 51]. The homologue gene P. malariae merozoite surface protein 1 (pmmsp1) is equally polymorphic and a candidate for parasite genotyping [52, 53]. In particular, high allelic frequency and genetic diversity in msp1 were observed for symptomatic P. falciparum malaria patients in Africa [54], Thailand [38] and Burkina-Faso [55]. Apart from a few previous studies reported with limited samples for P. malariae infections in human [56] and monkey [57, 58], only sparse information is available on pmmsp1 INDEL gene polymorphisms. The present study developed and validated six microsatellite genotyping markers as well as markers for pmmsp1 polymorphisms, which can serve as a tool for epidemiological and other studies.
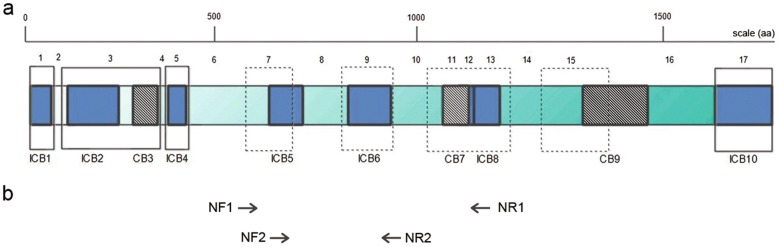
Fig. 1.
Representative block diagram of merozoite surface protein 1 [40]. a The block diagram for msp1 gene is based on conserved amino acid sequences in Plasmodium vivax, P. falciparum and Plasmodium yoelii (inner block). The outer solid and dotted hollow blocks represents conserved and semi-conserved regions based on P. falciparum sequence. The solid block represents ICB with more than 48% identical sequences among three species. The hatched block represents more than 50% identical (conserved) sequences between P. falciparum and P. vivax. Open regions represents less than 45% identity among three species. ICB, interspecies conserved block; CB, Conserved block. b Schematic representation of the primary (NF1, NR1) and secondary (NF2, NR2) oligonucleotide primers used to sequence partial region of the pmmsp1 gene
Methods
Study site, DNA sampling and reference sequences
Plasmodium malariae samples from symptomatic patients were collected in 2008 in Rakhine state of Myanmar (N = 37) as part of a larger study to compare the effectiveness of five artemisinin-based combination therapy regimen [59]. Parasite DNA was extracted using QIAamp DNA mini kit (Qiagen, Germany). All samples were confirmed as P. malariae using 18 small-subunit ribosomal RNA (18S rRNA)-based PCR [60] following the Standards for the Reporting of Diagnostic Accuracy (STARD) [61–63]. Ethical approval for the study was obtained from the ethical review board of the Faculty of Tropical Medicine, Mahidol University. Reference sequences for whole genomes of P. malariae UG01, P. falciparum 3D7, P. vivax SAL-1, P. ovale curtisi GH01 and P. knowlesi STRAIN-H were downloaded from PlasmoDB [64] repository (http://plasmodb.org/common/downloads/release-36/). The reference sequence for pmmsp1 gene (Accession no. FJ824669) was obtained from NCBI nucleotide database.
Development of genotyping markers
Identification of genome-wide perfect and imperfect microsatellites was performed using Phobos version 3.3.11 [65, 66]. The detection criteria for tandem repeat was restricted to evaluation of perfect and imperfect repeats with unit motifs of 1–10 bp having a minimum threshold repeat number of 14, 7, 5, 4, 4, 4, 4, 4, 4 and 4 for mono-, di-, tri-, tetra-, penta-, hexa-, hepta-, octa-, nonan-, and deca-nucleotide microsatellites, respectively. The genome wide distribution of microsatellites in reference sequences were summarized (Additional file 1: Table S1).
Sequences of six potential microsatellite genotyping markers together with 150 bp flanking sequences were extracted and primers were designed (Table 1) using BatchPrimer3 version 1.0 (https://probes.pw.usda.gov/cgi-bin/batchprimer3/batchprimer3.cgi). The selection criteria of primers were set as: (i) PCR product should be unique to the target flanking sequence containing the microsatellite; (ii) The variation in length of PCR product should be result of deviation in microsatellite length; (iii) Microsatellites with higher copy numbers, followed by manual inspection of each primer products to avoid selection of primers that could potentially associate with experimentally verified protein coding regions of the parasite or any interspecies cross reactivity; (iv) Preference is given to primers located at physically different chromosomes with no linkage disequilibrium. Likewise, during validation steps the factors such as ease of amplification, no starter peaks and absence of non-specific bands were also considered for selection of microsatellite markers. To investigate polymorphisms in pmmsp1 gene, the partial regions of pmmsp1 genes were amplified using semi-nested PCR with specifically designed primer sets (Table 2). Allelic frequencies were determined based on length of consensus sequences for variable number of tandem repeats (VNTR) in semi-conserved regions interspersed between coding blocks (Fig. 1). Only the well-amplified VNTR in majority of samples with copy numbers greater than 2.0 were considered as alleles for INDEL polymorphisms analysis.
Table 1.
List of candidate genotyping microsatellite markers for Plasmodium malariae UG01
| S. no. | Locusa | Primer | Primer sequence | Tm (°C) | Location |
|---|---|---|---|---|---|
| 1 | Pm05_707 | Forwardb | GGTAGAAGGAGCAACGGACA | 63.5 | Chromosome 5 |
| Reverse | CGCTCGGGTCATCGTTATTA | 61.3 | |||
| 2 | Pm06_506 | Forwardb | TTGTGCGTATGCAACCTTTC | 57.6 | Chromosome 6 |
| Reverse | CAAAAGGGAAGGAGCACAAA | 57.0 | |||
| 3 | Pm07_429 | Forwardb | TTCCTTTTCATCCTCTGCAA | 59.1 | Chromosome 7 |
| Reverse | CGAATGAGAGTAGTGCGGAAA | 62.8 | |||
| 4 | Pm09_801 | Forwardb | TGACTTCGGTTAGAATATGTTTGC | 60.7 | Chromosome 9 |
| Reverse | TCACACTCCTTTCAATTTCTCA | 59.4 | |||
| 5 | Pm12_426 | Forwardb | TTCGTGTTCTCGCTTTCCTC | 62.0 | Chromosome 12 |
| Reverse | GATCACTTCGCACGGGATAG | 61.9 | |||
| 6 | Pm13_110 | Forwardb | TCAAGTGGAATAACCGCAAG | 60.0 | Chromosome 13 |
| Reverse | CAGACGAGGACTTTCATTTCG | 60.4 |
aNovel microsatellite markers for Plasmodium malariae based on fragment analysis of PCR products (N = 37)
bForward primers were labelled with 6-FAM for fragment analysis. Thermal cyclin profile: initial denaturation at 94 °C for 5 min, 40 cycles of 94 °C for 1 min, 58–63 °C for 1 min and 72 °C for 1 min, followed by a final extension at 72 °C for 5 min
Table 2.
Primary and secondary primers used for amplification of pmmsp1 gene located on chromosome seven using semi-nested PCR
| S. no. | Locus | Primer | Primer sequence | Tm (°C) |
|---|---|---|---|---|
| 1 | Primarya | PMMSP1full_F2 (N1F) | GAATTGTCGAAAGCATTGGT | 54.2 |
| PMMSP1full_OR2 (N1R) | TCAACTTCTTTCTTTTCTGCTTC | 55.0 | ||
| 2 | Secondaryb | PMMSP1VNTR_1F (NF2) | CCAAGCATACGGAACAGGAG | 58.8 |
| PMMSP1VNTR_1R (NR2) | CAAATCTAATTGGTCGCACTTC | 56.2 |
Thermal cycling profile: initial denaturation step at 95 °C for 5 min, followed by 25 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 2 min and extension at 72 °C for 2 min then last extension step at 72 °C for 5 min. 2 µL of each primary reaction was used as template for the 100 µL secondary PCR reaction. Thermal cycling profile: Initial denaturation step at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 2 min and extension at 72 °C for 2 min then final extension step at 72 °C for 5 min
aPrimary and bSecondary set of primers were used to amplify the pmmsp1 gene segment of Plasmodium malariae
Validation of markers using PCR analysis
Primers for microsatellite genotyping markers were labelled with 6-Carboxyfluorescein (6-FAM) and validated using PCR (Table 1) followed by fragment analysis. PCR were performed in Eppendorf Mastercycler® pro (Eppendorf, Germany) with a total volume of 20 μL containing 1× PCR Buffer II (Mg2+ free), 2–3 mM MgCl2, 125 μM dNTPs, 0.25 μM primers and 0.4 U/20 μL of Platinum Taq Polymerase (Invitrogen, USA). Gel electrophoresis was used to detect the amplified products on 3% agarose gel. Fragment analysis of the 6-FAM-labelled PCR products was conducted using gel capillary electrophoresis by Macrogen (Macrogen Inc., South Korea). During fragment analysis, presence of a distinct expected single peak with a minimum of 100 relative fluorescent units (RFU) were accepted as cut-off value. If multiple peaks were detected, then one-third height of dominant peak with minimum of corresponding proportionate RFUs were taken as selection threshold for scoring the multiple recessive microsatellite alleles per locus.
For pmmsp1 polymorphisms, semi-nested PCR reaction was conducted (Table 2) to increase sensitivity. Primary and secondary PCR products were generated using corresponding volumes of 20 and 100 µL reaction in the presence of 10 mM Tris–HCl, pH 8.3, 50 mM KCl, 2 mM MgCl2, 125 µM dNTPs, 125 nM of each primers and 0.4 units of Platinum Taq Polymerase (Table 2). Sequences of amplified products were obtained using high-fidelity capillary electrophoresis conducted by Macrogen. Mono infection of P. malariae DNA verified using 18S rRNA-based PCR was taken as positive control. The PCR master mix with nuclease free water instead of parasite DNA was taken as control.
Multiplicity of infections
As the blood-stage malaria parasites are haploid, the presence of multiple peaks during evaluation of fragment size or VNTR analysis for one or more alleles at target locus was inferred as co-infection with two or more genetically distinct variants. This was referred to as multiplicity of infections (MOI) for the same isolate [20, 21, 67]. The mean MOI for positive samples was calculated independently for each marker by dividing the total number of P. malariae clones identified by the number of samples PCR positive for the parasite. For microsatellites, the single or predominant locus at each allele was considered for evaluating allele frequencies. The allele fragment size was interpreted using GeneScan™ version 3.1. Additional alleles were scored only if the peak height was at least one-third the height of the major peak during fragment analysis. For pmmsp1 gene, sequence data were interpreted using Bioedit version 7.0.4. Allelic frequencies for pmmsp1 gene were determined based on length of consensus sequences for variable number of tandem repeats (VNTR) in semi-conserved regions interspersed between coding blocks (Fig. 1).
Measures of diversity
The expected heterozygosity (HE) was calculated using FSTAT version 1.2 based on previously described formula where p is the frequency of the ith allele and n is the number of alleles sampled [26]. LIAN version 3.7 was used for analyzing overall multilocus linkage disequilibrium (LD) implementing a standardized index of association (). LD for candidate genotyping markers with p-values < 0.05 was considered as significant [68]. Dendogram for microsatellite fragment analysis data was constructed using ClustVis [69]. Blastx (https://blast.ncbi.nlm.nih.gov/Blast.cgi?LINK_LOC=blasthome&PAGE_TYPE=BlastSearch&PROGRAM=blastx) without low-complexity filter was used for identification of regions targeted by primers. Tandemly repeated sequences and copy numbers were identified by using TRF version 4.09 [70]. The number of haplotypes (H), haplotype diversity (Hd) and pairwise nucleotide diversity (π) were evaluated using DnaSP v5 [34]. Phylogenetic tree was constructed using Mega version 7.0 [71].
Likelihood of coinfections using genetic markers
The likelihood of two infections by same genotype were deduced by combining individual probabilities of two or more unlinked genetic markers and designated as combined probabilities (πPi), where πPi = P1 × P2 × ⋯Pi, where is individual probabilities for each markers being utilized. The assumption was based on each infections being independent and the probability of reinfection by same genotype were products of probabilities for individual markers [72].
Sensitivity and specificity
The specificity of primers was assessed on samples from symptomatic P. malariae patients (Additional file 1: Table S2) compared to the reference nested-PCR method targeting the 18S rRNA [60]. Specificity of all primer products were checked for amplification of unspecific products to access true positive results. The specificity of the markers was tested by two approaches: (i) BLAST analysis of the primers sequences against the NCBI databases and in silico PCR using default settings of UGENE version 1.30 [73] against whole genome reference database [64] of all available human infecting Plasmodium species. All markers did not belong or cross-aligned with any regions of other Plasmodium species. (ii) The samples used were verified for mono-infection of P. malariae pre-confirmed using 18S rRNA-based PCR specific. During PCR validation, all markers were specific to P. malariae only and had no cross reactivity to P. falciparum and P. vivax. (iii) Screening for sequence polymorphisms in the pmmsp1 gene by comparing to reference sequence or fragments analysis of amplified products. Non-specific amplification was not observed for any markers during assessment of qPCR products by gel electrophoresis. For microsatellite markers, inspection of potential contamination was safeguarded by routine insertion of known negative samples in each PCR run to access true negative results. For all six microsatellite markers except one sample for Pm06_506 (1/37), the target amplification was positive for all symptomatic P. malariae patients (N = 37), thus making sensitivity and specificity ~ 100% (Additional file 1: Table S3). For primers targeting the pmmsp1 gene polymorphisms, all well-sequenced samples (N = 27) exhibited VNTR polymorphisms and non-specific amplification or false positive results were not identified (Additional file 1: Table S3). Additional inspection of potential contamination involved cross-comparison of the amplified region with known sequences from P. falciparum and P. vivax. To avoid cross-contamination of samples during DNA addition and PCR processing steps, negative controls consisting of only water were added in each run.
Results
Identification of novel markers
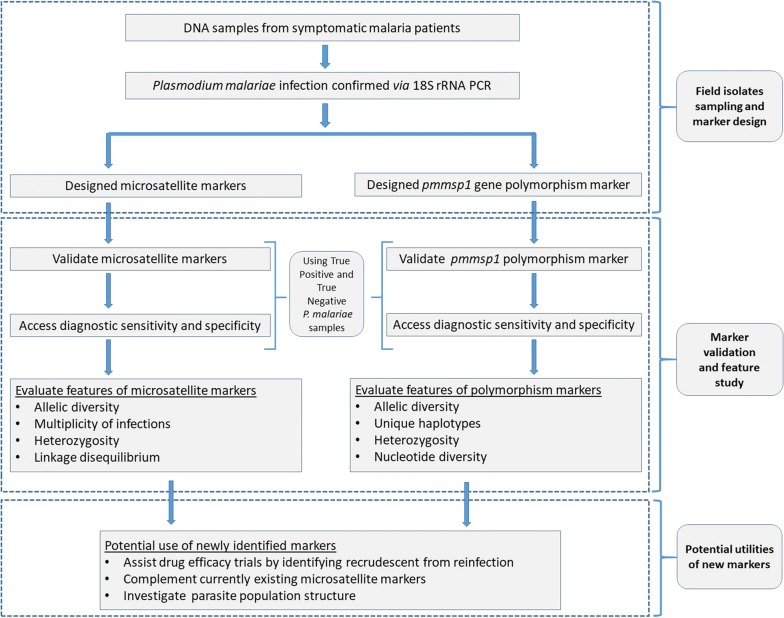
Figure 2 summarizes the approach we used to develop and validate genetic markers in P. malariae. To determine suitable P. malariae genotyping markers, the prevalence of polymorphisms for potential markers in 37 symptomatic P. malariae-positive blood samples from Myanmar was evaluated (Additional file 1: Table S2). Six promising genotyping markers, namely, Pm05_707, Pm06_506, Pm07_429, Pm09_801, Pm12_426 and Pm13_110 (Table 1) were identified. For all markers polymorphic fragments could be generated in most P. malariae-positive test samples (Additional file 1: Table S3). Blastx analysis showed that the regions covered by these markers were not linked to known CDS or experimentally verified protein expressing regions of the parasite genome. For the pmmsp1 gene, a gene sequence could be generated in 27 out of the 37 parasite samples, in which VNTR-related INDEL polymorphisms could be evaluated. The majority of the VNTRs belonged to a semi-conserved region in block 8 of pmmsp1 gene (Fig. 1) and contained imperfect repeats of the consensus sequence with a length of 27 bp and with copy numbers in range 2.4–23.9 resulting in allele length in range 64–645 bp (Additional file 2: Supplementary information). In the remaining samples, lack of PCR efficiency was the most likely cause of failure to amplify the pmmsp1 gene.
Fig. 2.
Schematic overview of the Plasmodium malariae molecular marker development and validation used in the current study and their potential implementation
Allelic frequencies and measures of marker diversity
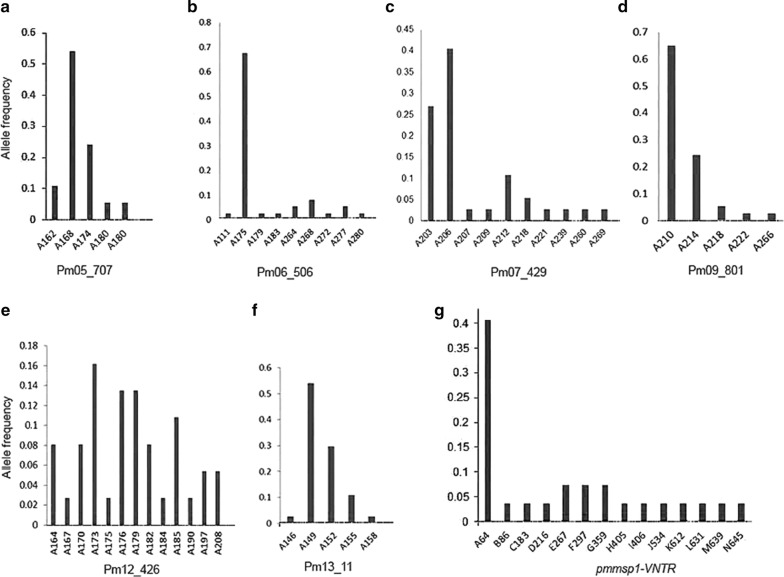
The six microsatellite markers, namely, Pm05_707, Pm06_506, Pm07_429, Pm09_801, Pm12_426 and Pm13_110 displayed corresponding HE (distinct alleles) of 0.649(5), 0.542(9), 0.764(10), 0.530(5), 0.922(13) and 0.623(5), respectively. Observed allele size ranged from 146 to 269 bp resulting in high diversity of allele frequencies (Fig. 3a–f). Candidate genotyping markers were situated on different chromosomes (Table 1, Fig. 4). Linkage analysis showed a ISA = 0.03 (p-value > 0.05), suggesting absence of a linkage disequilibrium. Sequence analysis of VNTR-based allelic frequencies in semi-conserved regions of pmmsp1 gene interspersed between coding blocks identified 14 distinct alleles (Fig. 3g). A copy number of 2.4 with consensus sequence “GAACAAGCAGAAACAACGGGAACAACA” located at pmmsp1 nucleotide position 2312–2374 bp was the most frequent observed VNTR (40%). Considerable nucleotide diversity (π = 0.0976) and haplotype gene diversity (Hd = 0.698) were observed. Linkage disequilibrium was significant (p-value < 0.01) for segregating sites. The combined likelihood that any two samples by chance would have the same genotypes was as low as 8.46 × 10−4 for six microsatellites markers and further reduced to 1.68 × 10−4 when pmmsp1 polymorphisms were included (Table 3).
Fig. 3.
Genetic richness and allele frequency. Allele frequencies observed for each microsatellite markers (a) Pm05_707, (b) Pm06_506, (c) Pm07_429, (d) Pm09_801, (e) Pm12_426 and (f) Pm13_11, respectively. g Allele frequency based on VNTR observed for P. malariae msp1 gene. VNTR, variable number of tandem repeats; msp, merozoite surface protein
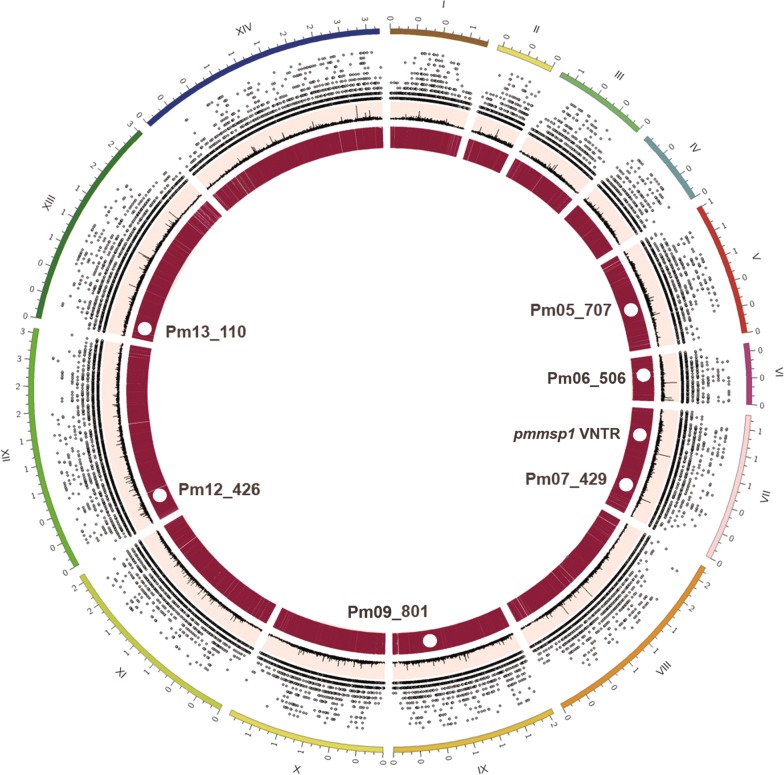
Fig. 4.
Location of the genotyping markers in Plasmodium malariae UG01. Different features indicated by the map for microsatellite distribution from outermost to innermost ring can be interpreted as: chromosome 1-14 (I-XIV) in Mbp, scatter plota for genomic microsatellite distribution based on unit motif length which corresponds to the height of spot from base of its ring, line plot with peaks indicating regions with long repeat length and heatmapb corresponding to the aggregate genomic microsatellite for the region. aSpots and regions in scatter plot and bheatmap may appear overlapped due to high density but are physically apart in sequence. The approximate location of newly identified genotyping markers are indicated with white circles and labeled accordingly; Mbs, mega base pairs
Table 3.
Probabilities of the coincidence of the same Plasmodium malariae genotype
| S. no. | Molecular markera | Probability () for each marker | Combined probabilities | Combined probability values (πPi)b |
|---|---|---|---|---|
| 1 | Pm12_426 | 0.102 | P1 = PPm12_426 | 1.02000 ×10−1 |
| 2 | Pm07_429 | 0.255 | P2 = P2 × PPm07_429 | 2.60100 ×10−2 |
| 3 | Pm05_707 | 0.369 | P3 = P3 × PPm05_707 | 9.59769 ×10−3 |
| 4 | Pm13_110 | 0.394 | P4 = P4 × PPm13_110 | 3.78149 ×10−3 |
| 5 | Pm06_506 | 0.462 | P5 = P5 × PPm06_506 | 1.74705 ×10−3 |
| 6 | Pm09_801 | 0.484 | P6 = P6 × PPm09_801 | 8.45571 ×10−4 |
| 7 | pmmsp1 | 0.196 | P7 = P7 × Ppmmsp1 | 1.65732 ×10−4 |
aOrdered according to increasing power of P for microsatellite genotyping markers except for pmmsp1 due to difference in marker type
bCombined cumulative probabilities πPi calculated as πPi = P1 × P2 × …Pi
Identification of MOI by genetic markers
Genotyping markers identified up to 13 distinct alleles for microsatellite markers and 14 for pmmsp1 markers (Fig. 3). Pm12_426 displayed the highest mean MOI (1.216) followed by Pm05_707 (Table 4). Because of their high allelic diversity and heterozygosity, the Pm12_426 and pmmsp1 markers were most potent for detecting MOI (Fig. 3, Table 3).
Table 4.
Characteristics of polymorphic microsatellite loci detected in Plasmodium malariae (N = 37)
| S. no. | Locus | Repeat unita | Allele size range (bp) | Total no. of alleles detectedb | No. of distinct alleles | Expected heterozygosity (HE)c | Mean MOI |
|---|---|---|---|---|---|---|---|
| 1 | Pm05_707 | (AAT)9 | 162–180 | 42 | 5 | 0.649 | 1.135 |
| 2 | Pm06_506 | (ACAT)40 | 175–280 | 41 | 9 | 0.542 | 1.081 |
| 3 | Pm07_429 | (AAT)14 | 203–269 | 40 | 10 | 0.764 | 1.081 |
| 5 | Pm09_801 | (ACAT)9 | 210–266 | 40 | 5 | 0.530 | 1.081 |
| 4 | Pm12_426 | (ATC)21 | 164–208 | 45 | 13 | 0.922 | 1.216 |
| 6 | Pm13_110 | (AAT)10 | 146–158 | 39 | 5 | 0.623 | 1.054 |
| 7 | pmmsp1 | VNTRd | 64–645 | 27 | 14 | 0.835 | 1.000 |
aThe repeat number of each microsatellite unit motif
bTotal numbers of alleles including both dominant and minor alleles detected
cExpected Heterozygosity (HE) was calculated from a restricted data set containing only the dominant allele in each sample
dThe allele based on consensus sequence of variable number of tandem repeats
Discussion
The genetic epidemiology of P. malaria is largely unknown. Availability of a comprehensive set of appropriate genetic markers is a prerequisite for advancing this field. The present study identifies six genotyping microsatellites and a set of VNTR-based pmmsp1 INDEL polymorphism markers in P. malariae. In contrast, allelic frequencies of microsatellites are generally higher and more evenly distributed due to absence of selective pressure. The newly identified six genotyping markers for P. malariae, supplement the widely used microsatellite markers developed for P. falciparum [21, 28] and P. vivax [20, 74]. Unlike some of the previous markers [13], all presently identified markers were located in different chromosomes and did not show any significant linkage disequilibrium, suggesting better discriminative potentials of these markers. Microsatellite markers are valuable tools for multilocus genotyping, and newly identified markers were able to clearly discriminate multiple P. malariae genotypes in naturally acquired symptomatic infections. Comparison of discriminative power associated with different genotyping markers in separate studies is complicated and often incomparable since heterozygosity values for a single marker can greatly vary between study sites [20, 72]. The choice of marker, genotyping technique and underlying genetic diversity of study groups affect outcomes of each study. Nonetheless, the heterozygosity displayed by six newly identified (0.542–0.922) and six previously reported (0.192–0.849) microsatellite markers for P. malariae [13, 14] could be used together to access higher polymorphisms with samples involving geographically large regions. Moreover, the combined set of markers would facilitate microsatellite genotype of P. malariae populations in similar ranks to those of P. vivax [67, 75] and P. falciparum [28, 76] which generally involves use of 10–13 sets of microsatellite markers. In particular, the newly identified marker Pm12_426 expressed HE value of 0.922 which was noticeably higher than previously reported highest HE of 0.811 [13]. The observed HE are promising, since the samples involved for validation were collected within same year and from geographically small region. Such features with high heterozygosity tend to make these markers suitable candidate for linkage mapping, which requires highly polymorphic markers [77]. The mean MOI for newly identified microsatellite markers (1.2 ± 0.1), was appreciable compared to previously published markers with mean MOI 1.12–1.32 [13]. Observed allelic diversity, high sensitivity and specificity (≥ 97%) for all six microsatellite markers suggests promising potency for population structure and epidemiological studies. All markers were unlikely to be in coding regions and were positioned within highly repetitive and AT-rich regions of the genome, which increases likelihood of these markers achieving higher heterozygosity in larger population size [78]. Likewise, the combined likelihood enhances the sensitivity of these markers by highly reducing identification of same genotype by chance, which enables utility of the markers in low transmission settings. The microsatellite genotyping method is relatively inexpensive compared to INDEL and SNP analysis [79, 80]. Moreover, the amplification and genotyping stages might be adopted to multiplex different loci, saving costs, time and facilitating large scale studies [23, 25].
VNTR-targeted INDEL analysis indicated promising ability of pmmsp1 gene polymorphisms to identify variants within the study population. The most frequently identified VNTRs with high heterozygosity were situated in pmmsp1 semi-conserved variable block 8, making it a potential genetic marker for P. malariae population study. The msp1 gene block 8 in previous study for P. falciparum and P. vivax had less than 45% interspecies identity 1 [40]. Presence of polymorphisms for msp1 block-2 have been reported for P. falciparum [81, 82], however, information on the P. malariae homologue is scarce. In present study, the pmmsp1 polymorphisms marker showed highly imbalanced distribution of its most common allele (40% for the A64 fragment) which might be explained by natural selection. The observed linkage disequilibrium for pmmsp1 polymorphisms is likely caused by SNPs grouped into haplotype blocks which often harbour limited number of distinct haplotypes [83]. Unlike microsatellites, the larger differences arising due to variation in copy numbers from VNTR in the amplified region is easily visualized by gel electrophoresis facilitating easy interpretation. However, template DNA required for INDEL analysis was more vulnerable to low DNA template quality which likely related to the larger amplicon size, and resulted in a smaller number of sample with successful amplification of the pmmsp1 gene. Nonetheless, the high allelic diversity and heterozygosity observed for well-sequenced samples indicated feasibility of this polymorphisms to be exploited for population genetic studies. The mean MOI for newly identified microsatellite markers were in range 1.10–1.20, similar to previously reported markers with mean MOI range of 1.12–1.32 [13]. The differences in observed genotypes and MOI by Pm12_426 and pmmsp1 markers for same population could indicate either greater transmission intensities or merely differences in the resolution of these molecular markers. Results from dendrograms and phylogenetic tree suggested similar outcomes while utilizing these markers to cluster population (Additional file 3: Fig. S1) and identify highly segregating variants.
Conclusions
In summary, the newly developed genotyping microsatellite markers and pmmsp1 gene polymorphisms may provide an important tool for studies in P. malariae. Practical applications include discrimination between disease recrudescence and reinfection in drug efficacy trials, studies on gene flow, parasite selection, population relatedness, signatures of selection and genetic diversity as a measure of transmission intensity and other genetic epidemiological questions. In addition to microsatellite typing, the VNTR-associated polymorphisms observed in semi-conserved block 8 of pmmsp1 gene are useful for assessing genetic diversity in P. malariae.
Supplementary information
Additional file 1: Table S1. Genome-wide coverage and density of microsatellites in genome of five human malaria causing Plasmodium species. Table S2. Number and geographical origin of Plasmodium species and strains used in the present study. Table S3. Sensitivity and specificity estimates of the markers in method development.
Additional file 2. Schematic diagram of partial pmmsp1 gene amplification and VNTR alleles alignment. The diagram is representative of 14 alleles aligned to the partially sequenced pmmsp1 gene reference sequence (Accession no. FJ824669) using pmmsp1 marker described in Table 2. VNTR, variable number of tandem repeats; pmmsp1, Plasmodium malariae merozoite surface protein 1.
Additional file 3: Figure S1. Dendrogram and phylogenetic tree for P. malariae samples (A) Dendrogram was constructed for microsatellite markers with rows clustered using correlation distance and complete linkage. The columns were clustered using correlation distance and average linkage (N = 37). (B) Phylogenetic tree constructed ws constructed using neighbor joining method for pmmsp1 gene sequence INDEL polymorphisms (N = 27). Figure A and B are not equivalent representative of the population clustering as type of genetic markers and sample size used for analysis are different.
Acknowledgements
We would like to acknowledge the postdoctoral research sponsorship of Mahidol University for providing necessary funding for the research. We thank Miss. Kanokon Suwannasin, and Mr. Chanon Kunason for their help.
Abbreviations
- bp
base pair
- CDS
protein-coding-regions
- HE
heterozygosity
- Hd
genetic diversity
- INDEL
insertion deletion
- Kbp
kilo base pair
- Mbp
million base pair
- msp
merozoite surface protein
- ORFs
open reading frame
- RFU
relative fluorescent unit
- SNP
single nucleotide polymorphisms
- VNTR
variable number tandem repeat
Authors’ contributions
VBM and MI designed the study. VBM and SN preformed the experiments and data analysis and wrote first draft of the manuscript. WP, SN and MI assisted in part of data analysis. FS provided samples. MI, NJW and AMD assisted in logistic support and manuscript preparation. All authors read and approved the final manuscript.
Funding
The research was funded by postdoctoral research sponsorship of Mahidol University, Thailand, Thailand Science Research and Innovation (TSRI), Grant No. RTA6280006 and the Wellcome Trust of Great Britain, UK.
Availability of data and materials
The dataset generated during the current study are available from corresponding author on reasonable request.
Ethics approval and consent to participate
Ethical approval for the study was obtained from the ethical review board of the Faculty of Tropical Medicine, Mahidol University (TMEC 11-019).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Vivek Bhakta Mathema and Supatchara Nakeesathit contributed equally to this work
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12936-020-3122-2.
References
- 1.Lo E, Nguyen K, Nguyen J, Hemming-Schroeder E, Xu J, Etemesi H, et al. Plasmodium malariae prevalence and csp gene diversity, Kenya, 2014 and 2015. Emerg Infect Dis. 2017;23:601–610. doi: 10.3201/eid2304.161245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutledge GG, Marr I, Huang GKL, Auburn S, Marfurt J, Sanders M, et al. Genomic characterization of recrudescent Plasmodium malariae after treatment with artemether/lumefantrine. Emerg Infect Dis. 2017;23:1300–1307. doi: 10.3201/eid2308.161582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou M, Liu Q, Wongsrichanalai C, Suwonkerd W, Panart K, Prajakwong S, et al. High prevalence of Plasmodium malariae and Plasmodium ovale in malaria patients along the thai-myanmar border, as revealed by acridine orange staining and PCR-based diagnoses. Trop Med Int Health. 1998;3:304–312. doi: 10.1046/j.1365-3156.1998.00223.x. [DOI] [PubMed] [Google Scholar]
- 4.Langford S, Douglas NM, Lampah DA, Simpson JA, Kenangalem E, Sugiarto P, et al. Plasmodium malariae infection associated with a high burden of anemia: a hospital-based surveillance study. PLoS Negl Trop Dis. 2015;9:e0004195. doi: 10.1371/journal.pntd.0004195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yman V, Wandell G, Mutemi DD, Miglar A, Asghar M, Hammar U, et al. Persistent transmission of Plasmodium malariae and Plasmodium ovale species in an area of declining Plasmodium falciparum transmission in Eastern Tanzania. PLoS Negl Trop Dis. 2019;13:e0007414. doi: 10.1371/journal.pntd.0007414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas NM, Lampah DA, Kenangalem E, Simpson JA, Poespoprodjo JR, Sugiarto P, et al. Major burden of severe anemia from non-falciparum malaria species in Southern Papua: a hospital-based surveillance study. PLoS Med. 2013;10:e1001575. doi: 10.1371/journal.pmed.1001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilles HM, Hendrickse RG. Nephrosis in Nigerian children. Role of Plasmodium malariae, and effect of antimalarial treatment. Br Med J. 1963;2:27–31. doi: 10.1136/bmj.2.5348.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nino CH, Cubides JR, Camargo-Ayala PA, Rodriguez-Celis CA, Quinones T, Cortes-Castillo MT, et al. Plasmodium malariae in the Colombian Amazon region: you don’t diagnose what you don’t suspect. Malar J. 2016;15:576. doi: 10.1186/s12936-016-1629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schindler T, Robaina T, Sax J, Bieri JR, Mpina M, Gondwe L, et al. Molecular monitoring of the diversity of human pathogenic malaria species in blood donations on Bioko island, Equatorial Guinea. Malar J. 2019;18:9. doi: 10.1186/s12936-019-2639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roman DNR, Rosalie NNA, Kumar A, Luther KMM, Singh V, Albert MS. Asymptomatic Plasmodium malariae infections in children from suburban areas of Yaounde, Cameroon. Parasitol Int. 2018;67:29–33. doi: 10.1016/j.parint.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Camargo M, Soto-De Leon SC, Del Rio-Ospina L, Paez AC, Gonzalez Z, Gonzalez E, et al. Micro-epidemiology of mixed-species malaria infections in a rural population living in the Colombian Amazon region. Sci Rep. 2018;8:5543. doi: 10.1038/s41598-018-23801-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roucher C, Rogier C, Sokhna C, Tall A, Trape JF. A 20-year longitudinal study of Plasmodium ovale and Plasmodium malariae prevalence and morbidity in a west African population. PLoS ONE. 2014;9:e87169. doi: 10.1371/journal.pone.0087169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruce MC, Macheso A, Galinski MR, Barnwell JW. Characterization and application of multiple genetic markers for Plasmodium malariae. Parasitology. 2007;134:637–650. doi: 10.1017/S0031182006001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruce MC, Macheso A, McConnachie A, Molyneux ME. Comparative population structure of Plasmodium malariae and Plasmodium falciparum under different transmission settings in Malawi. Malar J. 2011;10:38. doi: 10.1186/1475-2875-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camargo-Ayala PA, Cubides JR, Nino CH, Camargo M, Rodriguez-Celis CA, Quinones T, et al. High Plasmodium malariae prevalence in an endemic area of the Colombian Amazon region. PLoS ONE. 2016;11:e0159968. doi: 10.1371/journal.pone.0159968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayner JC. Plasmodium malariae malaria: From monkey to man? EBioMedicine. 2015;2:1023–1024. doi: 10.1016/j.ebiom.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutledge GG, Bohme U, Sanders M, Reid AJ, Cotton JA, Maiga-Ascofare O, et al. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature. 2017;542:101–104. doi: 10.1038/nature21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maguire JD, Sumawinata IW, Masbar S, Laksana B, Prodjodipuro P, Susanti I, et al. Chloroquine-resistant Pasmodium malariae in South Sumatra, Indonesia. Lancet. 2002;360:58–60. doi: 10.1016/S0140-6736(02)09336-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Nyunt MH, Yun SG, Lu F, Cheng Y, Han JH, et al. Variable number of tandem repeats of 9 Plasmodium vivax genes among Southeast Asian isolates. Acta Trop. 2017;170:161–168. doi: 10.1016/j.actatropica.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, Mayxay M, et al. Contrasting genetic structure in Plasmodium vivax populations from asia and South America. Int J Parasitol. 2007;37:1013–1022. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/S0031182099004552. [DOI] [PubMed] [Google Scholar]
- 22.Jarne P, Lagoda PJ. Microsatellites, from molecules to populations and back. Trends Ecol Evol. 1996;11:424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- 23.Vieira ML, Santini L, Diniz AL, Munhoz Cde F. Microsatellite markers: what they mean and why they are so useful. Genet Mol Biol. 2016;39:312–328. doi: 10.1590/1678-4685-GMB-2016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 25.Guichoux E, Lagache L, Wagner S, Chaumeil P, Leger P, Lepais O, et al. Current trends in microsatellite genotyping. Mol Ecol Resour. 2011;11:591–611. doi: 10.1111/j.1755-0998.2011.03014.x. [DOI] [PubMed] [Google Scholar]
- 26.Sutton PL, Luo Z, Divis PCS, Friedrich VK, Conway DJ, Singh B, et al. Characterizing the genetic diversity of the monkey malaria parasite Plasmodium cynomolgi. Infect Genet Evol. 2016;40:243–252. doi: 10.1016/j.meegid.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Figan CE, Sa JM, Mu J, Melendez-Muniz VA, Liu CH, Wellems TE. A set of microsatellite markers to differentiate Plasmodium falciparum progeny of four genetic crosses. Malar J. 2018;17:60. doi: 10.1186/s12936-018-2210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 29.Greenhouse B, Myrick A, Dokomajilar C, Woo JM, Carlson EJ, Rosenthal PJ, et al. Validation of microsatellite markers for use in genotyping polyclonal Plasmodium falciparum infections. Am J Trop Med Hyg. 2006;75:836–842. doi: 10.4269/ajtmh.2006.75.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menegon M, Bardaji A, Martinez-Espinosa F, Botto-Menezes C, Ome-Kaius M, Mueller I, et al. Microsatellite genotyping of Plasmodium vivax isolates from pregnant women in four malaria endemic countries. PLoS ONE. 2016;11:e0152447. doi: 10.1371/journal.pone.0152447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez JC, McNamara DT, Bockarie MJ, Baird JK, Carlton JM, Zimmerman PA. Identification of a polymorphic Plasmodium vivax microsatellite marker. Am J Trop Med Hyg. 2003;69:377–379. doi: 10.4269/ajtmh.2003.69.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soontarawirat I, Andolina C, Paul R, Day NPJ, Nosten F, Woodrow CJ, et al. Plasmodium vivax genetic diversity and heterozygosity in blood samples and resulting oocysts at the Thai–Myanmar border. Malar J. 2017;16:355. doi: 10.1186/s12936-017-2002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miles A, Iqbal Z, Vauterin P, Pearson R, Campino S, Theron M, et al. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 2016;26:1288–1299. doi: 10.1101/gr.203711.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Escalante AA, Grebert HM, Chaiyaroj SC, Riggione F, Biswas S, Nahlen BL, et al. Polymorphism in the gene encoding the pfs48/45 antigen of Plasmodium falciparum. Xi. Asembo bay cohort project. Mol Biochem Parasitol. 2002;119:17–22. doi: 10.1016/S0166-6851(01)00386-3. [DOI] [PubMed] [Google Scholar]
- 35.Nateghpour M, Haghi AM, Naderi B, Sepehrizadeh Z. Genetic polymorphism of Plasmodium vivax duffy binding protein in malarious areas in Southeastern of Iran. J Parasit Dis. 2017;41:1132–1138. doi: 10.1007/s12639-017-0946-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zakeri S, Barjesteh H, Djadid ND. Merozoite surface protein-3alpha is a reliable marker for population genetic analysis of Plasmodium vivax. Malar J. 2006;5:53. doi: 10.1186/1475-2875-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le HG, Kang JM, Moe M, Jun H, Thai TL, Lee J, et al. Genetic polymorphism and natural selection of circumsporozoite surface protein in Plasmodium falciparum field isolates from Myanmar. Malar J. 2018;17:361. doi: 10.1186/s12936-018-2513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, et al. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/S0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 39.Saralamba N, Mayxay M, Newton PN, Smithuis F, Nosten F, Archasuksan L, et al. Genetic polymorphisms in the circumsporozoite protein of Plasmodium malariae show a geographical bias. Malar J. 2018;17:269. doi: 10.1186/s12936-018-2413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.del Portillo HA, Longacre S, Khouri E, David PH. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci USA. 1991;88:4030–4034. doi: 10.1073/pnas.88.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice BL, Acosta MM, Pacheco MA, Carlton JM, Barnwell JW, Escalante AA. The origin and diversification of the merozoite surface protein 3 (msp3) multi-gene family in Plasmodium vivax and related parasites. Mol Phylogenet Evol. 2014;78:172–184. doi: 10.1016/j.ympev.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beeson JG, Drew DR, Boyle MJ, Feng G, Fowkes FJ, Richards JS. Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol Rev. 2016;40:343–372. doi: 10.1093/femsre/fuw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Zhao Z, Zhang X, Li X, Zhu M, Li P, et al. Naturally acquired antibody responses to Plasmodium vivax and Plasmodium falciparum merozoite surface protein 1 (msp1) c-terminal 19 kDa domains in an area of unstable malaria transmission in Southeast Asia. PLoS ONE. 2016;11:e0151900. doi: 10.1371/journal.pone.0151900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ockenhouse CF, Angov E, Kester KE, Diggs C, Soisson L, Cummings JF, et al. Phase i safety and immunogenicity trial of fmp1/as02a, a Plasmodium falciparum msp-1 asexual blood stage vaccine. Vaccine. 2006;24:3009–3017. doi: 10.1016/j.vaccine.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 45.Pattaradilokrat S, Sawaswong V, Simpalipan P, Kaewthamasorn M, Siripoon N, Harnyuttanakorn P. Genetic diversity of the merozoite surface protein-3 gene in Plasmodium falciparum populations in Thailand. Malar J. 2016;15:517. doi: 10.1186/s12936-016-1566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cochrane AH, Collins WE, Nussenzweig RS. Monoclonal antibody identifies circumsporozoite protein of Plasmodium malariae and detects a common epitope on Plasmodium brasilianum sporozoites. Infect Immun. 1984;45:592–595. doi: 10.1128/IAI.45.3.592-595.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Zhou RM, Zhang YL, Wang DQ, Li SH, Yang CY, et al. Analysis of polymorphisms in the circumsporozoite protein gene of Plasmodium vivax isolates from Henan province, China. Malar J. 2018;17:103. doi: 10.1186/s12936-018-2237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rayner JC, Huber CS, Feldman D, Ingravallo P, Galinski MR, Barnwell JW. Plasmodium vivax merozoite surface protein PvMSP-3β is radically polymorphic through mutation and large insertions and deletions. Infect Genet Evol. 2004;4:309–319. doi: 10.1016/j.meegid.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Zilversmit MM, Volkman SK, DePristo MA, Wirth DF, Awadalla P, Hartl DL. Low-complexity regions in Plasmodium falciparum: missing links in the evolution of an extreme genome. Mol Biol Evol. 2010;27:2198–2209. doi: 10.1093/molbev/msq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ord R, Polley S, Tami A, Sutherland CJ. High sequence diversity and evidence of balancing selection in the pvmsp3alpha gene of Plasmodium vivax in the Venezuelan Amazon. Mol Biochem Parasitol. 2005;144:86–93. doi: 10.1016/j.molbiopara.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Ndong Ngomo JM, M’Bondoukwe NP, Yavo W, Bongho Mavoungou LC, Bouyou-Akotet MK, Mawili-Mboumba DP. Spatial and temporal distribution of pfmsp1 and pfmsp2 alleles and genetic profile change of Plasmodium falciparum populations in Gabon. Acta Trop. 2018;178:27–33. doi: 10.1016/j.actatropica.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 52.Brito CF, Ferreira MU. Molecular markers and genetic diversity of Plasmodium vivax. Mem Inst Oswaldo Cruz. 2011;106(Suppl 1):12–26. doi: 10.1590/S0074-02762011000900003. [DOI] [PubMed] [Google Scholar]
- 53.Kadekoppala M, Holder AA. Merozoite surface proteins of the malaria parasite: the msp1 complex and the msp7 family. Int J Parasitol. 2010;40:1155–1161. doi: 10.1016/j.ijpara.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Babiker HA, Lines J, Hill WG, Walliker D. Population structure of Plasmodium falciparum in villages with different malaria endemicity in East Africa. Am J Trop Med Hyg. 1997;56:141–147. doi: 10.4269/ajtmh.1997.56.141. [DOI] [PubMed] [Google Scholar]
- 55.Some AF, Bazie T, Zongo I, Yerbanga RS, Nikiema F, Neya C, et al. Plasmodium falciparum msp1 and msp2 genetic diversity and allele frequencies in parasites isolated from symptomatic malaria patients in Bobo-Dioulasso, Burkina Faso. Parasit Vectors. 2018;11:323. doi: 10.1186/s13071-018-2895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guimaraes LO, Wunderlich G, Alves JM, Bueno MG, Rohe F, Catao-Dias JL, et al. Merozoite surface protein-1 genetic diversity in Plasmodium malariae and Plasmodium brasilianum from Brazil. BMC Infect Dis. 2015;15:529. doi: 10.1186/s12879-015-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Araujo MS, Messias MR, Figueiro MR, Gil LH, Probst CM, Vidal NM, et al. Natural Plasmodium infection in monkeys in the state of Rondonia (Brazilian Western Amazon) Malar J. 2013;12:180. doi: 10.1186/1475-2875-12-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fandeur T, Volney B, Peneau C, de Thoisy B. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology. 2000;120:11–21. doi: 10.1017/S0031182099005168. [DOI] [PubMed] [Google Scholar]
- 59.Smithuis F, Kyaw MK, Phe O, Win T, Aung PP, Oo AP, et al. Effectiveness of five artemisinin combination regimens with or without primaquine in uncomplicated falciparum malaria: an open-label randomised trial. Lancet Infect Dis. 2010;10:673–681. doi: 10.1016/S1473-3099(10)70187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srisutham S, Saralamba N, Malleret B, Renia L, Dondorp AM, Imwong M. Four human Plasmodium species quantification using droplet digital PCR. PLoS ONE. 2017;12:e0175771. doi: 10.1371/journal.pone.0175771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snounou G, Singh B. Nested pcr analysis of Plasmodium parasites. Methods Mol Med. 2002;72:189–203. doi: 10.1385/1-59259-271-6:189. [DOI] [PubMed] [Google Scholar]
- 62.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-B. [DOI] [PubMed] [Google Scholar]
- 63.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. Stard 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin Chem. 2015;61:1446–1452. doi: 10.1373/clinchem.2015.246280. [DOI] [PubMed] [Google Scholar]
- 64.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. Plasmodb: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simbaqueba J, Sanchez P, Sanchez E, Nunez Zarantes VM, Chacon MI, Barrero LS, et al. Development and characterization of microsatellite markers for the Cape Gooseberry Physalis peruviana. PLoS ONE. 2011;6:e26719. doi: 10.1371/journal.pone.0026719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayer C. Phobos—a tandem repeat search tool for complete genomes. 2010:http://www.ruhr-uni-bochum.de/spezzoo/cm. Accessed 25 Apr 2018.
- 67.Gunawardena S, Karunaweera ND, Ferreira MU, Phone-Kyaw M, Pollack RJ, Alifrangis M, et al. Geographic structure of Plasmodium vivax: microsatellite analysis of parasite populations from Sri Lanka, Myanmar, and Ethiopia. Am J Trop Med Hyg. 2010;82:235–242. doi: 10.4269/ajtmh.2010.09-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haubold B, Hudson RR. Lian 3.0: Detecting linkage disequilibrium in multilocus data. Linkage analysis. Bioinformatics. 2000;16:847–848. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- 69.Metsalu T, Vilo J. Clustvis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar S, Stecher G, Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koepfli C, Mueller I, Marfurt J, Goroti M, Sie A, Oa O, et al. Evaluation of Plasmodium vivax genotyping markers for molecular monitoring in clinical trials. J Infect Dis. 2009;199:1074–1080. doi: 10.1086/597303. [DOI] [PubMed] [Google Scholar]
- 73.Okonechnikov K, Golosova O, Fursov M, team U Unipro ugene: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 74.Trimarsanto H, Benavente ED, Noviyanti R, Utami RA, Trianty L, Pava Z, et al. Vivaxgen: an open access platform for comparative analysis of short tandem repeat genotyping data in Plasmodium vivax populations. PLoS Negl Trop Dis. 2017;11:e0005465. doi: 10.1371/journal.pntd.0005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orjuela-Sanchez P, Sa JM, Brandi MC, Rodrigues PT, Bastos MS, Amaratunga C, et al. Higher microsatellite diversity in Plasmodium vivax than in sympatric Plasmodium falciparum populations in Pursat, Western Cambodia. Exp Parasitol. 2013;134:318–326. doi: 10.1016/j.exppara.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abukari Z, Okonu R, Nyarko SB, Lo AC, Dieng CC, Salifu SP, et al. The diversity, multiplicity of infection and population structure of P. falciparum parasites circulating in asymptomatic carriers living in high and low malaria transmission settings of Ghana. Genes (Basel) 2019 doi: 10.3390/genes10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ball AD, Stapley J, Dawson DA, Birkhead TR, Burke T, Slate J. A comparison of SNPs and microsatellites as linkage mapping markers: lessons from the zebra finch (Taeniopygia guttata) BMC Genomics. 2010;11:218. doi: 10.1186/1471-2164-11-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hamilton WL, Claessens A, Otto TD, Kekre M, Fairhurst RM, Rayner JC, et al. Extreme mutation bias and high at content in Plasmodium falciparum. Nucleic Acids Res. 2017;45:1889–1901. doi: 10.1093/nar/gkw1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schindel DE, Miller SE. DNA barcoding a useful tool for taxonomists. Nature. 2005;435:17. doi: 10.1038/435017b. [DOI] [PubMed] [Google Scholar]
- 80.Castagnone-Sereno P, Danchin EG, Deleury E, Guillemaud T, Malausa T, Abad P. Genome-wide survey and analysis of microsatellites in nematodes, with a focus on the plant-parasitic species Meloidogyne incognita. BMC Genomics. 2010;11:598. doi: 10.1186/1471-2164-11-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller LH, Roberts T, Shahabuddin M, McCutchan TF. Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1) Mol Biochem Parasitol. 1993;59:1–14. doi: 10.1016/0166-6851(93)90002-F. [DOI] [PubMed] [Google Scholar]
- 82.Tanabe K, Mackay M, Goman M, Scaife JG. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol. 1987;195:273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 83.Takeuchi F, Yanai K, Morii T, Ishinaga Y, Taniguchi-Yanai K, Nagano S, et al. Linkage disequilibrium grouping of single nucleotide polymorphisms (SNPs) reflecting haplotype phylogeny for efficient selection of tag SNPs. Genetics. 2005;170:291–304. doi: 10.1534/genetics.104.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Genome-wide coverage and density of microsatellites in genome of five human malaria causing Plasmodium species. Table S2. Number and geographical origin of Plasmodium species and strains used in the present study. Table S3. Sensitivity and specificity estimates of the markers in method development.
Additional file 2. Schematic diagram of partial pmmsp1 gene amplification and VNTR alleles alignment. The diagram is representative of 14 alleles aligned to the partially sequenced pmmsp1 gene reference sequence (Accession no. FJ824669) using pmmsp1 marker described in Table 2. VNTR, variable number of tandem repeats; pmmsp1, Plasmodium malariae merozoite surface protein 1.
Additional file 3: Figure S1. Dendrogram and phylogenetic tree for P. malariae samples (A) Dendrogram was constructed for microsatellite markers with rows clustered using correlation distance and complete linkage. The columns were clustered using correlation distance and average linkage (N = 37). (B) Phylogenetic tree constructed ws constructed using neighbor joining method for pmmsp1 gene sequence INDEL polymorphisms (N = 27). Figure A and B are not equivalent representative of the population clustering as type of genetic markers and sample size used for analysis are different.
Data Availability Statement
The dataset generated during the current study are available from corresponding author on reasonable request.