Summary
Pyrenophora teres f. teres and P. teres f. maculata are significant pathogens that cause net blotch of barley. An increased number of loci involved in P. teres resistance or susceptibility responses of barley as well as interacting P. teres virulence effector loci have recently been identified through biparental and association mapping studies of both the pathogen and host. Characterization of the resistance/susceptibility loci in the host and the interacting effector loci in the pathogen will provide a path for targeted gene validation for better‐informed release of resistant barley cultivars. This review assembles concise consensus maps for all loci published for both the host and pathogen, providing a useful resource for the community to be used in pathogen characterization and barley breeding for resistance to both forms of P. teres.
Keywords: barley, effector, necrotrophic fungus, Pyrenophora teres f. maculata, Pyrenophora teres f. teres, resistance, susceptibility
Pyrenophora teres causes net blotch of barley. Here we review the latest progress on host resistance and pathogen virulence, providing a valuable resource for the barley net blotch research community.
Introduction
Pyrenophora teres is the causal agent of net blotch of barley and is found in two forms, P. teres f. teres and P. teres f. maculata, the causal agents of net form net blotch (NFNB) and spot form net blotch (SFNB), respectively (Smedegård‐Petersen, 1971). Net blotch is one of the most economically important diseases of barley, with typical yield losses ranging from 10% to 40% and possible complete crop failure in extreme cases (Galano et al., 2011; Jayasena et al., 2007; Jebbouj and El Yousfi, 2009; Khan, 1987; Moya et al., 2018; Piening and Kaufmann, 1969). Net blotch is present in all barley‐growing regions of the world (Shipton et al., 1973), including northern (Douiyssi et al., 1998), eastern (Zeleke, 2017) and southern Africa (Louw et al., 1995), Middle (Arabi et al., 2003) and Far‐East Asia (Sato and Takeda, 1997), Europe (Arabi et al., 1992), North (Tekauz, 1990) and South America (Moya et al., 2018), and Oceania (Cromey and Parkes, 2003; Williams et al., 1999). The potential economic loss to P. teres f. teres and P. teres f. maculata has been valued at approximately A$117 and A$192 million per annum, respectively, in Australia alone (Murray and Brennan, 2010).
Originally named Helminthosporium teres [Sacc.] in 1809, the fungus was renamed P. teres Drechs. (anamorph Drechsler teres [Sacc.] Shoem.) in 1930 (Shoemaker, 1959). P. teres was subsequently divided into the two forms P. teres f. teres and P. teres f. maculata by Smedegård‐Petersen (1971) based on the lesion types. P. teres f. teres develops necrotic lesions with distinct striations, developing the net‐like pattern for which it was named. P. teres f. maculata develops oval necrotic lesions with a chlorotic halo (Shipton et al., 1973; Smedegård‐Petersen, 1971). Arguably, these two forms could be classified as separate species based on their genetic isolation (Ellwood et al., 2012; Jalli, 2011; Rau et al., 2007; Syme et al., 2018) and the rarity of hybridization of the two forms in nature, despite coinhabiting the same fields (Akhavan et al., 2015; Campbell et al., 2002; Çelik Oğuz et al., 2018; Leišova et al., 2005; McLean et al., 2014; Poudel et al., 2019). Hybrids can be induced in the lab (Campbell and Crous, 2003), suggesting a natural barrier to fertility or a fitness penalty in nature (Syme et al., 2018). The two forms are reported to have diverged from each other 519 kya (±30) (Ellwood et al., 2012; Syme et al., 2018), and from one of their closest relatives Pyrenophora tritici‐repentis, the causal agent of tan spot on wheat 8.04 Mya (±138 kya) (Ellwood et al., 2012; Ohm et al., 2012).
One P. teres form is often dominant within a geographical area. P. teres f. maculata has become more prevalent in North Dakota (Liu and Friesen, 2010) and Idaho, USA (Marshall et al., 2015) and Victoria, Australia (McLean et al., 2010a), but this likely depends on the susceptibility of the predominant cultivars planted in each region. Multiple differential barley sets have been proposed to classify global pathotypes of P. teres f. teres (Afanasenko et al., 2009, 1995; Fowler et al., 2017) and P. teres f. maculata isolates in local populations (McLean et al., 2014, 2010b). The taxonomy, population diversity, symptoms, life cycle, infection cycle and control of, and preliminary insights into the P. teres–barley interaction were previously reviewed by Liu et al. (2011), and therefore these aspects will not be covered here. In this review, we have collated the recent literature that lays the foundation for the functional characterization of P. teres–barley interactions.
Genetics of barley reactions to P. teres f. teres
Resistance to P. teres f. teres was first shown to be quantitatively inherited by Geschele (1928). Schaller and Wiebe (1952) screened barley lines for resistance to P. teres, but it is unknown which form was used, concluding that lines, primarily of Manchurian origin, contained enough resistance for introgression into elite barley lines. The first resistance locus implicated in the P. teres–barley interaction was harboured by Tifang in a Tifang × Atlas cross, inherited as a single gene trait and effective against Californian P. teres isolates (Schaller, 1955). Mode and Schaller (1958) subsequently designated this locus Pt1 and identified two additional loci, designated Pt2 and Pt3 (Table 1). The lines Ming, Manchurian and Harbin harboured Pt2 as a single locus, whereas the lines Canadian Lake Shore and CI4922 carried Pt2 and Pt3 (Mode and Schaller, 1958). The Pt1 and Pt2 loci were shown to be closely linked with 2.57% recombination by using resistant by resistant crosses (Mode and Schaller, 1958). An additional locus, designated Pta, was discovered conferring resistance to the Australian isolate W.A.‐2 that was distinct from Pt1, Pt2 and Pt3 (Khan and Boyd, 1969a). Khan and Boyd (1969a) concluded that the genetic makeup of resistance in Harbin, Manchuria, Tifang and Ming was different depending on the isolate used based on the existing research at the time. Following the trisomic analysis of Bockelman et al. (1977) that failed to separate Pt1 and Pt2, and the revision of the barley gene nomenclature, the Pt1 and Pt2 loci were collapsed into the Rpt1 locus on the long arm of chromosome 3H with Rpt1.a and Rpt1.b alleles from Tifang/Harbin and CI9819, respectively (Table 1). Additional subsequent loci that were collapsed into the Rpt1 locus included Pta, QRpts3L and QNFNBSLR.Al/S‐3H from Igri (Graner et al., 1996), Arapiles (Raman et al., 2003) and Alexis (Lehmensiek et al., 2007), respectively (Table 1). The chromosome 3H resistance locus in Canadian Lake Shore mapped to the short arm of chromosome 3H (Dinglasan et al., 2019), indicating that the Canadian Lake Shore resistance is not Rpt1 (Fig. 1). Confusion over the Pt2 locus occurred when Rpt2.c reported by Bockelman et al. (1977) was assumed to be Pt2 reported by Mode and Schaller (1958). However, Rpt2.c in CI9819 is distinct from Pt2, which is allelic to other sources of resistance that map to the Rpt1 locus (Bockelman et al., 1977) (Table 1). Manninen et al. (2006) reported a major and a minor effect locus effective against different P. teres f. teres isolates on chromosomes 6H and 1H, respectively, in a Rolfi × CI9819 population, corroborating Rpt2.c on chromosome 1H (Table 1).
Table 1.
Summary of current designated barley genes defined in the Pyrenophora teres f. teres and P. teres f. maculata–barley pathosystem including the current and previous synonyms, known alleles, barley chromosome and form of P. teres effective against. Different loci are indicated by alternating grey scale.
| Designated locus (ref) * | Current synonym (ref) * | Previous synonyms (ref) * | Known alleles | Location | Form‡ |
|---|---|---|---|---|---|
|
Rpt1 (Bockelman et al., 1977) |
Pt1 (Mode and Schafer, 1958) Pt2 (Mode and Schafer, 1958) Pt1a/Rpt1a (Bockelman et al., 1977) Pt1b/Rpt1b (Bockelman et al., 1977) Pt,,a (Graner et al., 1996) QRpts3L (Raman et al., 2003) QNFNBSLR.Al/S‐3H (Lehmensiek et al., 2007) QNFNBSLR.W/Al‐3H (Lehmensiek et al., 2007) QNFNBAPR.Al/S‐3H (Lehmensiek et al., 2007) QNFNBAPR.W/Al‐3H (Lehmensiek et al., 2007) |
Rpt1.a Rpt1.b |
3H | P. teres f. teres | |
| Rpt2 (Bockelman et al., 1977) |
Pt2c/Rpt2c (Bockelman et al., 1977) |
Rpt2.c | 1H | P. teres f. teres | |
|
Rpt3 (Bockelman et al., 1977) |
Pt3 (Mode and Schaller, 1958) Pt.d/Pt3d/Rpt3d (Bockelman et al., 1977) QRpts2S (Raman et al., 2003 ) QRpts2Sa (Raman et al., 2003) QRptts2 (Grewal et al., 2008) QNFNBSLR.Al/S‐2H (Lehmensiek et al., 2007) QNFNBSLR.W/Al‐2H (Lehmensiek et al., 2007) QNFNBSLR.Ar/F‐2Ha (Lehmensiek et al., 2007) QNFNBAPR.Ar/F‐2H (Lehmensiek et al., 2007) QPt.2H‐1/2 (Vatter et al., 2017) |
Rpt3.d | 2H |
P. teres f. teres P. teres f. maculata |
|
|
QRpt7 (Grewal et al., 2008) QNFNBAPR.Al/S‐7Hb # (Lehmensiek et al., 2007) QNFNBAPR.W/Al‐7Hb # (Lehmensiek et al., 2007) QRpt7 (Tamang et al., 2015) QRptm7‐3/4/5/6/7/8 (Wang et al., 2015) NBP_QRPtt7‐2 (Wonneberger et al., 2017b) Qns‐7H.2 (Daba et al., 2019) QRptm‐7H‐119‐137 (Tamang et al., 2019 ) |
Rpt4.e | 7H |
P. teres f. teres P. teres f. maculata |
||
| Rpt5 (Manninen et al., 2006) | Spt1 (Richards et al., 2016) |
Pta (Khan and Boyd, 1969a) Pt,,d (Graner et al., 1996) QRpts6L (Raman et al., 2003) QRpt (Embiri et al., 2005) rpt.r/rpt.r (Abu Qamar et al., 2008) QRpt6 (Grewal et al., 2008, 2012) RptCiho2291/Nomini (O’Boyle et al., 2014) SPN1 (Liu et al., 2015) QRptm6‐2 (Wang et al., 2015) QPt.6H‐1/2 (Vatter et al., 2017) NBP_QRPtt6‐1 (Wonneberger et al., 2017b) QRptta‐6H‐49.79 (Amezrou et al., 2018) QRptt.6H‐54‐55 (Amezrou et al., 2018) Qns‐6H.5 (Daba et al., 2019) Qnfnb‐6H.1‐4 (Daba et al., 2019) Qsfnb‐6H (Daba et al., 2019) QRptm‐6H‐55‐64 (Tamang et al., 2019) |
Rpt5.f Spt1.k Spt1.r |
6H |
P. teres f. teres P. teres f. maculata |
| Rpt6 (Manninen et al., 2006) | ‐ | Rpt6.g | 5H |
P. teres f. teres P. teres f. maculata |
|
| Rpt7 (Franckowiak and Platz, 2013) |
Rpt‐4H‐5‐7 (Yun et al., 2005) QNFNBAPR.Al/S‐4Ha # (Lehmensiek et al., 2007) AL_QRptt4‐1 (Wonneberger et al., 2017a) QPt.4H‐3 # (Vatter et al., 2017) QRptm‐4H‐58‐64 (Tamang et al., 2019) |
Rpt7.h | 4HL |
P. teres f. teres P. teres f. maculata |
|
| Rpt8 (Franckowiak and Platz, 2013) |
QTL on 4H (Friesen et al., 2006) QRpts4 # (Grewal et al., 2008) † Qns‐4H.2 (Daba et al., 2019) |
Rpt8.j | 4HS | P. teres f. maculata |
First paper to use gene designation.
Not confirmed.
Multiple papers use this designation.
Bold indicates predominant form resistance is associated with.
Figure 1.
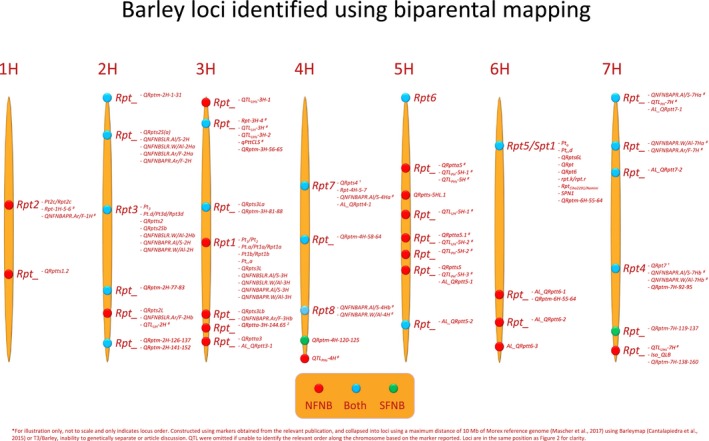
Consensus map of loci that are associated with net form net blotch (NFNB, red), spot form net blotch (SFNB, green) or both (blue) resistance/susceptibility across the barley genome identified using biparental mapping. Barley chromosome numbers are indicated at the top of each chromosome in sequential order. Designated loci are indicated by sequential numbering whereas previously undesignated loci are marked by an underscore (see also Table S3).
The Rpt3.d locus located on chromosome 2H (Graner et al., 1996) is commonly used as a synonym for the Pt3/Pt.d locus identified by Mode and Schaller (1958) based on the Bockelman et al. (1977) reclassification in Tennessee Awnless D22‐5 (Franckowiak and Platz, 2013) (Table 1). The Rpt3.d locus also includes QRptts2 from TR251 (Grewal et al., 2012) (Table 1). Other reported loci identified on chromosome 2H that may be part of the Rpt3 locus include quantitative trait loci (QTLs) in Steptoe (Steffenson et al., 1996), Kaputar (Cakir et al., 2003), Franklin (Raman et al., 2003) and ND11213 (Emebiri et al., 2005) (Table S1).
The Pta locus was reclassified as Rpt5 on chromosome 6H with at least three alleles (Franckowiak and Platz, 2013; Khan and Boyd, 1969a; Manninen et al., 2006) (Table 1). Alleles of Rpt5 include Rpt5.f, Spt1.r (previously rpt5.k) and Spt1.k (previously rpt5.r) from CI5791/CI9819, Kombar and Rika, respectively (Abu Qamar et al., 2008; Koladia et al., 2017a; Manninen et al., 2000, 2006; Richards et al., 2016) (Table 1). Additional loci that may also be part of the Rpt5 locus and therefore contain additional alleles include QRpts6L, QRpt6, AL_QRptt6‐1, Rpt‐Nomini and Rpt‐CIho2291 in Halcyon (Raman et al., 2003), TR251 (Grewal et al., 2012), Lavrans (Wonneberger et al., 2017a), Nomini and CIho2291 (O’Boyle et al., 2014), respectively (Table 1), as well as undesignated QTLs from Steptoe (Steffenson et al., 1996), HOR 9088 (Richter et al., 1998), Kaputar, ND11213 (Cakir et al., 2003; Emebiri et al., 2005; Liu et al., 2015), Chevron (Ma et al., 2004), SM89010 (Friesen et al., 2006), M129 (St. Pierre et al., 2010), Baudin (Cakir et al., 2011), WPG8412, Pompadour, Stirling (Gupta et al., 2011), Falcon (Islamovic et al., 2017), H602 (Hisano et al., 2017) and UVC8 (Martin et al., 2018) (Table S1).
The Rpt5 locus has been reported by multiple studies and is considered to be a complex locus that is highly important in the P. teres f. teres–barley interaction (Abu Qamar et al., 2008; Adawy et al., 2013; Adhikari et al., 2019; Cakir et al., 2011, 2003; Dontsova et al., 2018; Emebiri et al., 2005; Friesen et al., 2006; Grewal et al., 2012, 2008; Gupta et al., 2011, 2010; Koladia et al., 2017a; Liu et al., 2015; Ma et al., 2004; Manninen et al., 2000, 2006; Martin et al., 2018; Molnar et al., 2000; Novakazi et al., 2019; O’Boyle et al., 2014; Raman et al., 2003; Rau et al., 2015; Richards et al., 2016; Richter et al., 1998; Rozanova et al., 2019; Spaner et al., 1998; St. Pierre et al., 2010; Steffenson et al., 1996; Wonneberger et al., 2017b, 2017a). Abu Qamar et al. (2008) found that Rika was resistant to P. teres f. teres isolate 15A and susceptible to isolate 6A, whereas for Kombar the reciprocal reactions occurred in that Kombar was resistant to 15A and susceptible to 6A. The two alleles rpt5.k. and rpt5.r were originally designated as recessive resistance due to a 1:3 resistant:susceptible segregation ratio and therefore the requirement of a homozygous state of alleles for resistance (Abu Qamar et al., 2008). The Rpt5 region that conferred susceptibility was delimited to a c. 5.9 cM region in a 118‐progeny doubled haploid (DH) population derived from Rika × Kombar (Abu Qamar et al., 2008). While attempting to map the susceptibility loci in Rika/Kombar, a single Rika × Kombar progeny line was resistant to both 15A and 6A, and a single progeny line was susceptible to both 15A and 6A. These putative recombinants led to the hypothesis that two closely linked genes, rpt.r and rpt.k were held in repulsion c. 1.8 cM apart and were responsible for susceptibility (Abu Qamar et al., 2008). Under a two‐gene scenario, rpt.r and rpt.k would each be responsible for initiating a susceptible response to an effector present in each parental isolate (Abu Qamar et al., 2008). Using the same Rika × Kombar DH population (Abu Qamar et al., 2008), 15 expressed sequence tag‐derived markers were added to the genetic map to refine the chromosome 6H region to a c. 3.3 cM genetic interval and the two suspected genes to a c. 1.6 cM genetic interval (Liu et al., 2010). Liu et al. (2010) concluded that the rpt.r/rpt.k region was most likely located close to the centromere on the long arm of chromosome 6H with high synteny to chromosomes 2 and 3 of rice and Brachypodium, respectively. The annotation list of the putative rpt5 region contained two leucine‐rich repeat (LRR) receptor‐like kinases, which are often associated with plant disease resistance (Liu et al., 2010).
In addition, the Rpt5 locus has also been designated Susceptibility to P. teres 1 (Spt1), indicating the dominant nature of susceptibility to P. teres f. teres in some barley lines (Richards et al., 2016) (Table 1). The standard nomenclature indicated Reaction to P. teres (Rpt), with capital R indicating dominant resistance and lower case r indicating recessive resistance (Franckowiak and Platz, 2013), and did not allow for the designation of dominant susceptibility. Therefore, both designations have been used for the Rpt5/Spt1 locus since the locus may provide dominant resistance in the case of CI5791 or dominant susceptibility in Rika/Kombar depending on the allele–effector combination (Abu Qamar et al., 2008; Manninen et al., 2006; Richards et al., 2016; Shjerve et al., 2014). Using immortal critical recombinants (ICRs) of the previously defined Rpt5/Spt1 locus, the region was fine mapped to a c. 0.24 cM interval, equating to a c. 466 kb genomic region of Brachypodium with 62 annotated genes (Richards et al., 2016). The genomic region of Brachypodium translated to c. 9.5 Mb of physical sequence in the barley genome containing 39 high‐confidence genes, leading to the hypothesis that the locus contained a single, two tightly linked or an ‘island’ of susceptibility genes that interact with effectors to varying specificities depending on the model (Richards et al., 2016). A total of six of the genes within the Rpt5/Spt1 locus coded for immunity receptor‐like proteins that could be responsible for a susceptible response (Richards et al., 2016). However, allelic analysis of candidate genes suggested that a divergent allelic series of a single gene rather than the two tightly linked genes or a ‘susceptibility island’ could be responsible for different effector specificities for the complex interactions with P. teres (Richards et al., 2016). In addition, the putative Rika × Kombar recombinants that were either resistant or susceptible to both 15A and 6A were later found to be suspect; therefore, this result no longer ruled out the hypothesis of one allelic gene at the Rpt5/Spt1 locus as concluded by Richards et al. (2016).
A separate approach lending support that Rpt5/Spt1 encoded a disease resistance gene that was targeted by a proteinaceous necrotrophic effector was shown by Liu et al. (2015) where the protein designated PttNE1 interacted with a gene at the same 6H (Rpt5/Spt1) locus and designated SPN1. In this study, the PttNE1–SPN1 interaction accounted for 31% of the disease variation (Liu et al., 2015). The isolates 15A, 0‐1, LDN07 Pt‐5, ND89‐19 and NB022 all exhibited a compatible reaction mapping to the SPN1 locus (Liu et al., 2015). In addition, the major QTL responsible for the isolate JPT9901 did not co‐localize with SPN1 but distally at a distinct locus, suggesting additional genes are harboured on chromosome 6H (Liu et al., 2015). This single Rpt5/Spt1 locus exhibits interactions that are reminiscent of the gene‐for‐gene model (Flor, 1971, 1955) and inverse gene‐for‐gene model (Friesen et al., 2007) in parallel. Therefore, the interaction between barley and P. teres can follow typical biotrophic models such as the gene‐for‐gene model (Flor, 1971, 1955) resulting in effector‐triggered immunity, as well as an inverse gene‐for‐gene model (Friesen et al., 2007) where necrotrophic effectors lead to necrotrophic effector‐triggered susceptibility (Liu et al., 2015; Richards et al., 2016).
The Rpt7 locus confers resistance to P. teres f. teres, located on the long arm of chromosome 4H in Halcyon (Raman et al., 2003; Read et al., 2003) (Table 1). Currently, it is unknown whether additional QTLs reported on chromosome 4H in Steptoe (Steffenson et al., 1996), Sloop (Lehmensiek et al., 2007), TR251 (Grewal et al., 2008), OUH602 (Yun et al., 2005), Arena/HOR9088 (Richter et al., 1998), PostxViresa/HOR9484 (König et al., 2014, 2013), Harrington/TR306 (Spaner et al., 1998) and Falcon (Islamovic et al., 2017) are at the same locus (Table S1). However, a second locus designated Rpt7 was identified by Tamang et al. (2015) conferring seedling resistance to two isolates of P. teres f. maculata, which will be covered later. As the Rpt7 locus on chromosome 4H was designated first, we have decided to keep to current nomenclature and the Rpt7 locus reported by Tamang et al. (2015) on 7H will become undesignated (Fig. 1).
A total of seven conventional genome‐wide association studies (GWAS) have been performed to investigate P. teres f. teres resistance in barley (Adhikari et al., 2019; Amezrou et al., 2018; Daba et al., 2019; Novakazi et al., 2019; Richards et al., 2017; Rozanova et al., 2019; Wonneberger et al., 2017b). Investigation of resistance to a set of diverse global isolates was performed on the Barley Core Collection (Muñoz‐Amatriaín et al., 2014; Richards et al., 2017), Nordic Barley Panel (Wonneberger et al., 2017b), ICARDA AM‐2014 Panel (Amezrou et al., 2018), Ethiopian, ICARDA and NDSU Barley Panel (Daba et al., 2019), Siberian Barley Panel (Rozanova et al., 2019), Ethiopian and Eritrean Barley Collection (Adhikari et al., 2019) and Vavilov Research Institute Collection (Novakazi et al., 2019). Between 7 and 31 unique genomic loci were identified in each GWAS (Fig. 2 and Table S1). Richards et al. (2017) revealed that the majority of the markers significantly associated with NFNB resistance localized to the centromeric region of chromosome 6H, further indicating the importance of this region.
Figure 2.
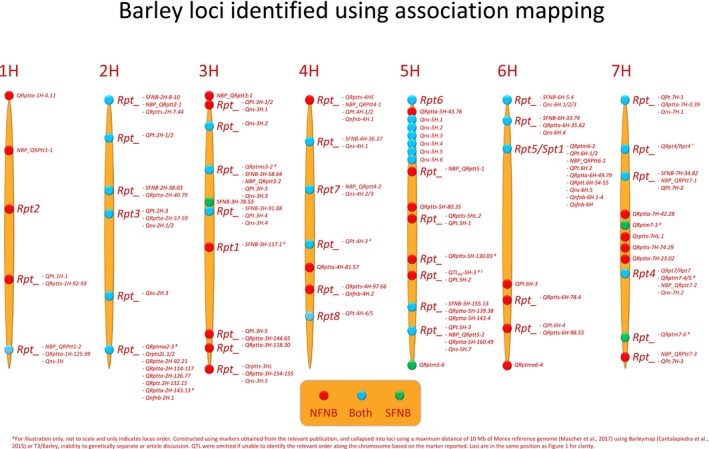
Consensus map of loci that are associated with net form net blotch (NFNB, red), spot form net blotch (SFNB, green) or both (blue) resistance/susceptibility across the barley genome identified using association mapping. Barley chromosome numbers are indicated at the top of each chromosome in sequential order. Designated loci are indicated by sequential numbering whereas previously undesignated loci are marked by an underscore (see also Table S3).
Vatter et al. (2017) performed a nested association mapping (NAM) study of barley resistance to P. teres f. teres. Vatter et al. (2017) used reaction type (Tekauz, 1985) and average ordinate to measure the infection of P. teres f. teres on the Halle Exotic Barley 25 (HEB‐25) population (Maurer et al., 2015), identifying 24 QTLs (Fig. 2). In both reaction type and average ordinate phenotyping, a QTL on chromosome 2H was responsible for the most phenotypic variation at 9.23% and 14.88%, respectively (Vatter et al., 2017), but this was not the same as the Rpt3 locus. The Rpt5/Spt1 locus was identified in the NAM approach but only contributed 0.08% and 0.39% of the phenotypic variation for reaction type and average ordinate phenotyping, respectively (Vatter et al., 2017). The low level of phenotypic variation explained by the Rpt5/Spt1 locus could be because few of the founding barley lines used in the HEB‐25 population contained alleles that provided resistance to P. teres f. teres.
The most novel approach to mapping resistance QTLs in barley to P. teres f. teres used exome QTL‐seq (Hisano et al., 2017). For exome QTL‐seq, 10 highly resistant and 10 susceptible lines were pooled separately prior to exome capture library construction and sequencing (Hisano et al., 2017). Plotting the single nucleotide polymorphisms (SNPs) for QTL mapping identified two QTLs on chromosome 3H and one on chromosome 6H from barley line H602 (Hisano et al., 2017). Hisano et al. (2017) found that there were twice as many SNPs on chromosomes 3H and 6H within the resistant bulk sample compared to the number within the susceptible bulk sample, suggesting that these loci were undergoing diversifying selection resulting in resistant lines, potentially to avoid recognition of P. teres effectors that result in programmed cell death. The results of Hisano et al. (2017) are similar to those of Gupta et al. (2010), Koladia et al. (2017a) and Liu et al. (2015). The sheer number of reports of QTLs located at the centromere of chromosome 6H, now referred to as Rpt5/Spt1, warrants validation and characterization. Research to determine the alleles and validation of Rpt5/Spt1 in an array of barley lines is currently underway (Brueggeman et al., unpublished).
Genetics of barley reactions to P. teres f. maculata
Originally, the genetics conferring resistance to P. teres f. maculata contained a total of three major designated loci and therefore had been considered less complex in comparison to the P. teres f. teres–barley interaction. The continuous distribution of P. teres f. maculata infection responses indicate that quantitative loci in barley play a part in P. teres f. maculata resistance (Burlakoti et al., 2017) and occasionally previously identified resistance loci to P. teres f. teres have been found effective against P. teres f. maculata, although it is unconfirmed if the same genes are conferring resistance/susceptibility. All three of the major barley loci conferring resistance to P. teres f. maculata confer seedling resistance (Liu et al., 2011) and are designated Rpt4, Rpt6 and Rpt8, respectively (Table 1). The Rpt4 locus located on chromosome 7H was found in Galleon, CI9214, Keel, Tilga, Chebec (Williams et al., 2003, 1999), PI67381, PI84314 (Tamang et al., 2019) and possibly TR251 (Grewal et al., 2008). The Rpt6 locus was found on the short arm of chromosome 5H in CI9819 (Burlakoti et al., 2017; Manninen et al., 2006) and Rpt8 was found on chromosome 4H in Q21861 (Franckowiak and Platz, 2013; Friesen et al., 2006).
Breeding lines in Australia that used Rpt4 resistance were found to have lower levels of resistance to P. teres f. maculata than expected, indicating that additional genes were required for adult plant resistance (APR) (Williams et al., 2003). Although Rpt4 was found to contribute to APR in a Galleon × Huruna Nijo population, Rpt4 had a much smaller effect in the adult stage when compared to Rpt4 seedling resistance (Williams et al., 2003). At least three APR QTLs were mapped, one QTL on each of chromosomes 4H and 5H, and one to three QTLs on chromosome 7H (Williams et al., 2003) (Table S2). The barley lines Galleon, VB9104, CI9214, Kell and Chebec were found to have an APR locus on chromosome 7H, whereas Galleon and VB9104 also contained the APR loci on chromosomes 4H and 5H (Williams et al., 2003). The one to three APR loci on chromosome 7H mapped distal to Rpt4 (Williams et al., 2003), but based on the interval mapping approach used, all three loci are closely linked or overlap so it is difficult to conclude whether there are one, two or three APR loci in addition to Rpt4.
Four studies have used GWAS to investigate barley resistance to P. teres f. maculata (Burlakoti et al., 2017; Daba et al., 2019; Tamang et al., 2015; Wang et al., 2015). Evaluations of resistance to diverse global (Tamang et al., 2015), Australian (Wang et al., 2015) and single American isolates (Burlakoti et al., 2017; Daba et al., 2019) were performed on the Barley Core Collection (Muñoz‐Amatriaín et al., 2014; Neupane et al., 2015; Tamang et al., 2015), Northern Region Barley Breeding Program of Australia (Wang et al., 2015), Upper Midwest Breeding Programs (Burlakoti et al., 2017) and Ethiopian, ICARDA and NDSU Barley Panel (Daba et al., 2019), respectively. A total of 27 (Tamang et al., 2015), 29 (Wang et al., 2015), 11 (Burlakoti et al., 2017) and 1 (Daba et al., 2019) unique genomic loci were identified in each GWAS, respectively (Fig. 2 and Table S2).
The previously identified loci included QRpts4, QRpt6 corresponding to Rpt5/Spt1 (Grewal et al., 2008), Rpt4, Rpt6, Rpt7 (Williams et al., 2003) and QTLs on 4H (Friesen et al., 2006) corresponding to Rpt8 (Tamang et al., 2015). The Rpt7 locus reported by Tamang et al. (2015) on 7H was postulated to be the undesignated APR locus discovered by Williams et al. (2003), and therefore should remain an undesignated locus as the Rpt7 locus was previously designated on chromosome 4H (Table 1). In addition, the QRpt7 locus was previously reported by Grewal et al. (2008) and was established to be Rpt4 on chromosome 7H. QRpts4 (Grewal et al., 2008) and APR on chromosome 4H (Williams et al., 2003) may be the same locus as Rpt7 on chromosome 4H. The APR QTL located on chromosome 5H (Williams et al., 2003) is not Rpt6 and remains undesignated.
Four highly significant QTLs (QRptm7‐4, QRptm7‐6, QRptm7‐7 and QRptm7‐8) map to a 36 cM region that approximately encompasses the Rpt4 locus on chromosome 7H (Wang et al., 2015), similar to results reported by Williams et al. (2003). However, it is unknown if QTLs on chromosome 7H are multiple linked QTLs, multiple alleles of a single QTL or an artefact of population structure (Wang et al., 2015). Analysis of linkage decay of the QTLs located on chromosome 7H suggests that the four significant QTLs are independent of each other, but further verification is required (Wang et al., 2015).
Burlakoti et al. (2017) studied the effect of two‐ and six‐row barley, concluding that the proportion of six‐row barley (43%) resistant to P. teres f. maculata was higher than that of the two‐row barley (13%) tested. This difference is in contrast to Wang et al. (2015), who had comparable numbers of resistant lines (40–65%) in Australian two‐ and six‐row barley. The most significant QTL identified was novel, mapped to chromosome 2H and accounted for 4.3–6.6% of the phenotypic variation (Burlakoti et al., 2017). Daba et al. (2019) identified a single novel QTL on chromosome 6H, but this is attributed to most lines being susceptible to P. teres f. maculata. The association mapping studies of Tamang et al. (2015), Wang et al. (2015) and Burlakoti et al. (2017) indicated that the genetic interaction of P. teres f. maculata and barley was more complex than originally hypothesized. In addition, novel QTLs and therefore sources of resistance can be identified with the power of association mapping to evaluate more accessions in parallel.
Recently, studies to identify new sources of resistance to P. teres f. maculata have been undertaken by Çelik Oğuz et al. (2017) and Gyawali et al. (2018). Çelik Oğuz et al. (2017) found two six‐row landraces in Turkey that were resistant to all (three P. teres f. teres and three P. teres f. maculata) isolates tested that could be used in breeding programmes. Gyawali et al. (2018) screened 340 diverse ICARDA barley lines using one P. teres f. maculata isolate to identify 12 lines that were highly resistant and stable across environments to be used for resistance breeding. However, without mapping and subsequent allelic analysis it is unknown what resistance loci or alleles are present.
Effectors of P. teres
Effectors are molecules that interact with the host to manipulate the host response, and encompass secondary metabolites, peptides, small secreted proteins, enzymes and more recently small RNAs (Toruño et al., 2016). Effectors can be identified in multiple ways, including forward and reverse genetic approaches, including traditional genetic mapping, genomic mining and proteomics/metabolomic approaches. A genetic mapping approach to effector discovery is labour‐intensive and includes population development, genotyping and phenotyping but can generate highly useful candidate gene lists to functionally validate effectors that segregate within a given population. The use of genetic mapping is less effective in finding highly conserved effectors that do not segregate between parents of a population or those that become fixed within a natural population. Genomic mining or proteomic/metabolomic approaches can be used to identify these highly conserved effectors but result in large candidate lists of genes that need to be functionally validated.
For the successful colonization by necrotrophic fungi such as P. teres f. teres and P. teres f. maculata, necrosis and chlorosis of plant tissue is associated with a compatible reaction (reviewed in Liu et al., 2011). Chlorosis surrounding the necrotic lesion is often associated with diffusible phytotoxic secondary metabolites (Keon and Hargreaves, 1983; Sarpeleh et al., 2007). Four toxins, N‐(2‐amino‐2‐carboxyethyl)‐aspartic acid, anhydroaspergillomarasmine A, aspergillomarasmine A and aspergillomarasmine B, were isolated and their pathways subsequently identified (Bach et al., 1979; Friis et al., 1991; Sarpeleh et al., 2009; Smedegård‐Petersen, 1977) and used to screen for resistance to P. teres (Sharma, 1984; Weiergang et al., 2002). All four toxins identified were of the marasmine class that chelate iron ions and have been shown to be light‐ and temperature‐dependent, indicating a potential role targeting organelles such as the chloroplast (Sarpeleh et al., 2009). Secondary metabolites do not appear to be isolate‐specific and little host specificity exists apart from lines being either sensitive or insensitive (Sarpeleh et al., 2009, 2007).
Multiple studies have shown that different virulence profiles exist within P. teres populations around the globe (Akhavan et al., 2016; Arabi et al., 2003; Cromey and Parkes, 2003; Fowler et al., 2017; Gupta and Loughman, 2001; Harrabi and Kamel, 1990; Jalli and Robinson, 2000; Jonsson et al., 1997; Khan and Boyd, 1969b; Liu et al., 2012; McLean et al., 2010b; Robinson and Jalli, 1996; Sato and Takeda, 1997; Steffenson and Webster, 1992; Tekauz, 1990; Wu et al., 2003). Virulence variation among isolates on different barley lines appears to originate from their segregating effector repertoire, many of which are now believed to be proteinaceous in nature (Liu et al., 2015; Richards et al., 2016). Virulent P. teres f. teres and P. teres f. maculata isolates have increased production of proteases, possibly for faster establishment of the pathogen and increased sugar acquisition compared to less virulent isolates (Dikilitas et al., 2018). Two papers from the same group proposed that variations in virulence could be explained by fungal growth, ability to deliver effectors and differentially expressing effectors (Ismail et al., 2014a, 2014b). Ismail et al. (2014a) used six P. teres f. teres isolates and found that the four more virulent isolates had greater conidial germination and appressoria formation compared to the less virulent isolates yet extracted proteinaceous effectors from all isolates were able to induce necrosis. Three proteinaceous effectors were postulated in the virulent P. teres f. teres isolate 32/98 as PttXyn11A, PttCHFP1 and PttSP1 that showed homology to proteins involved in plant disease interactions (Ismail et al., 2014b). Using a combination of one‐ and two‐dimensional electrophoresis, Ismail and Able (2016) identified 63 proteins that were produced by 28 virulent isolates. Ismail and Able (2017) subsequently identified a transition between colonization and necrotrophy at approximately 48 h based on the in planta gene expression analysis of 222 proteins. As no functional validation was reported on these candidate genes/proteins, it is not yet known whether any of these are biologically relevant to the P. teres f. teres–barley interaction.
A proteinaceous necrotrophic effector, PttNE1, was isolated from intercellular wash fluid by Liu et al. (2015) using the susceptible barley line Hector inoculated with P. teres f. teres isolate 0‐1. PttNE1 exhibited direct association with disease and interacted with SPN1, and therefore Liu et al. (2015) concluded that host sensitivity resulted from necrotrophic effector‐triggered susceptibility, but the pathogen gene PttNE1 was not mapped in P. teres f. teres. The first mapped effector implicated in the P. teres f. teres–barley system, designated AvrHar, was identified by Weiland et al. (1999) in the P. teres f. teres 15A × 0‐1 population. The 15A allele of AvrHar was proposed to contribute low virulence on the barley line Harbin (Weiland et al., 1999) (Table 2). In an expanded population of 15A × 0‐1, the 15A allele of the AvrHar locus also conferred avirulence on Tifang and Canadian Lake Shore (Lai et al., 2007). In addition, two QTLs, designated AvrPra1 and AvrPra2, from 0‐1 were mapped and found to be functionally redundant to confer avirulence on Prato (Lai et al., 2007) (Table 2). The AvrHar and AvrPra2 loci mapped to the same locus on linkage group 7 (chromosome 5) (Lai et al., 2007; Wyatt et al., 2018), but whether the locus contains two tightly linked genes or alleles of the same gene is yet to be confirmed (Fig. 3). The AvrPra1 locus was mapped to linkage group 1 (Lai et al., 2007), currently designated chromosome 9 based on the 0‐1 assembly (Fig. 3, Wyatt et al., 2018). A separate avirulence gene, designated AvrHeartland, was identified in a 67‐progeny mapping population with the Canadian avirulent isolate WRS 1906 and the virulent isolate WRS 1607 (Beattie et al., 2007) (Table 2).
Table 2.
Current Pyrenophora teres f. teres and P. teres f. maculata genes defined in the P. teres–barley pathosystem including the current and previous synonyms, known alleles, and which barley lines the allele is virulent or avirulent on along with phenotypic variation, the P. teres chromosome location of the effector and barley chromosome target
| Locus | Previous synonym | Alleles* | Virulent on† | Avirulent on‡ | R 2 (%)§ | Location‖ | Target |
|---|---|---|---|---|---|---|---|
| AvrHar | AvrHar (15A), AvrPra2 (0‐1) | Harbin, Tifang, Canadian Lake Shoreb, Pratoa | Harbin, Tifang, Canadian Lake Shorea, Pratob | – | Chr 5 | – | |
| AvrPra1 | AvrPra1 (0‐1) | Tifang, Canadian Lake Shore | Prato | – | Chr 9 | – | |
| AvrHeartland | AvrHeartland (WRS 1906) | Harrington | Heartland | – | Chr 1 | – | |
| VK1 | VK1 (15A) | Kombar | Rika | 0.26 | Chr 3 | Rpt5/Spt1 | |
| VK2 | VK2 (15A) | Kombar | Rika | 0.19 | Chr 2 | Rpt5/Spt1 | |
| VR1 | VR1 (6A) | Rika | Kombar | 0.35 | Chr 2 | Rpt5/Spt1 | |
| VR2 | VR2 (6A) | Rika | Kombar | 0.20 | Chr 10 | Rpt5/Spt1 | |
| PttTif1 | PttTif1 (FGOH04Ptt‐21) | Tifang, Manchurian, CI 4822 | Beecher | 0.45–0.67 | Chr 1 | – | |
| PttTif2 | PttTif3 (FGOH04Ptt‐21) | Tifang | Manchurian, CI 4822, Beecher, Pinnacle, Celebration, Hector, Stellar | 0.026 | Chr 8 | – | |
| PttBee1 | PttBee1 (FGOH04Ptt‐21) | Beecher | Tifang, Manchurian, CI 4822, Pinnacle, Celebration, Hector, Stellar | 0.56 | Chr 1 | – | |
| PttBee2 | PttBee2 (FGOH04Ptt‐21) | Beecher | Tifang, Manchurian, CI 4822, Pinnacle, Celebration, Hector, Stellar | 0.17 | Chr 5 | – | |
| PttPin1 | PttPin1 (BB25) | Pinnacle | Tifang, Manchurian, CI 4822, Beecher, Celebration, Hector, Stellar | 0.49 | Chr 3 | – | |
| PttPin2 | PttPin2 (FGOH04Ptt‐21) | Pinnacle | Tifang, Manchurian, CI 4822, Beecher, Celebration, Hector, Stellar | 0.11 | Chr 12 | – | |
| PttCel1 | PttCel1 (FGOH04Ptt‐21) | Celebration, Tifang | Manchurian, CI 4822, Beecher, Pinnacle, Hector, Stellar | 0.066–0.17 | Chr 8 | – | |
| PttCel2 | PttCel2 (FGOH04Ptt‐21) | Celebration | Tifang, Manchurian, CI 4822, Beecher, Pinnacle, Celebration, Hector, Stellar | 0.17 | Chr 9 | – | |
| PttHec1 | PttHec1 (FGOH04Ptt‐21) | Hector, Stellar | Tifang, Manchurian, CI 4822, Beecher, Pinnacle, Celebration | 0.11–0.18 | Chr 8 | – | |
| PtmSki1 |
vQTL1A vQTL1B vQTL1C |
PtmSki1 (FGOB10Ptm‐1) |
Skiff, TR326 81‐82/033 Welgevallen65‐31‐36 |
– – – |
0.21–0.23 0.21 0.34 |
Chr 1 Chr 1 Chr 1 |
– – – |
| PtmSki2 | vQTL2 | PtmSki2 (FGOB10Ptm‐1) | Skiff | Welgevallen65‐31‐36, 81‐82/033, TR326 | 0.22 | Chr 3 | – |
| PtmSki3 | vQTL3 | PtmSki3 (FGOB10Ptm‐1) | Skiff | Welgevallen65‐31‐36, 81‐82/033, TR326 | 0.20 | Chr 5 | – |
| PtmWel1 | vQTL4 | PtmWel1 (SG1) | Welgevallen65‐31‐36, 81‐82/033 | Skiff, TR326 | 0.30–0.37 | Chr 2 | – |
| PtmWel2 | vQTL5 | PtmWel2 (FGOB10Ptm‐1) | Welgevallen65‐31‐36, 81‐82/033, TR326 | Skiff | 0.26–0.34 | Chr 3 | – |
| PtmWel3 | vQTL6 | PtmWel3 (FGOB10Ptm‐1) | Welgevallen65‐31‐36 | Skiff, 81‐82/033, TR326 | 0.20 | Chr 4 | – |
Isolate contributing virulent allele.
Barley line the virulent isolate is known to be virulent on, additional lines may exist, a allele 1, b allele 2.
Barley line the virulent isolate is known to be avirulent on, additional lines may exist, a allele 1, b allele 2.
QTL effects containing ranges mean that the experiments were performed for multiple locations or multiple isolates and the effects for individual treatments fall into this range.
Inferred through current genome assembly and chromosome numbering (Wyatt et al., 2018).
No information is available for this entry.
Figure 3.
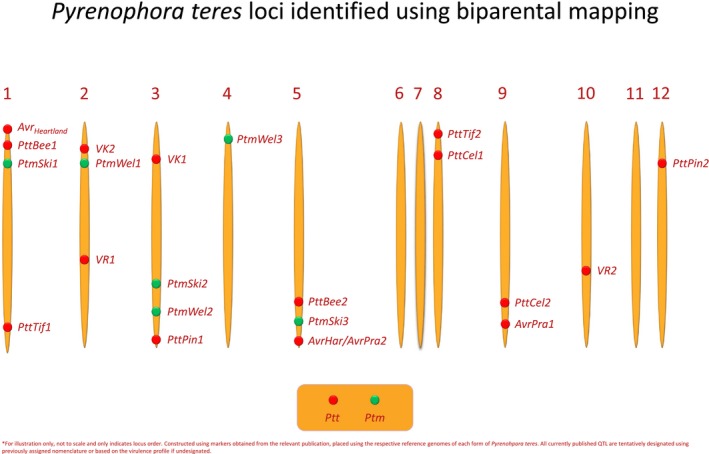
Map of Pyrenophora teres f. teres (Ptt, red) and P. teres f. maculata (Ptm, green) effectors across the P. teres genome identified using biparental mapping. Chromosome numbers are indicated at the top of each chromosome.
Afanasenko et al. (2007) proposed the presence of a suppressor of avirulence genes where virulence arose in 30 out of 84 progeny from an avirulent by virulent cross on the barley line Diamond. Based on an avirulent: virulent segregation ratio of 9:7, Afanasenko et al. (2007) suggested the presence of two avirulence genes and one suppressor gene. Based on phenotypic ratios alone, Afanasenko et al. (2007) proposed that up to four avirulence genes and up to two suppressor genes in various combinations were present in four P. teres f. teres crosses, but the corresponding loci were not mapped.
Shjerve et al. (2014) was the first study to begin using a restriction‐site associated‐digest–genotyping‐by‐sequencing (RAD‐GBS) approach detailed by Leboldus et al. (2015) that would allow the rapid construction of genetic maps by subsequent studies including Koladia et al. (2017b) and Carlsen et al. (2017) in P. teres f. teres and P. teres f. maculata, respectively. The RAD‐GBS approach facilitated the mapping of four QTL regions that have now been assigned chromosomal designations based on the recently updated 0‐1 reference genome assembly (Wyatt et al., 2018) including VK1 and VK2 (Virulence on Kombar 1 and 2) and VR1 and VR2 (Virulence on Rika 1 and 2). VK1 localized to chromosome 3, VK2 and VR1 to separate loci on chromosome 2 and VR2 on chromosome 10 (Fig. 3). Isolate 15A harboured the virulence alleles at VK1 and VK2 with each conferring virulence on Kombar but lacking virulence on Rika (Shjerve et al., 2014) (Table 2). The isolate 6A harboured virulence alleles at VR1 and VR2 and the reciprocal occurred, where VR1 and VR2 confer virulence on Rika but lacked virulence on Kombar (Shjerve et al., 2014) (Table 2). In addition, Shjerve et al. (2014) further characterized the progeny isolates of the 15A × 6A population to discover that VK1, VK2 and VR2 all targeted the same Spt1 host region on chromosome 6H. Isolates containing VK1 or VK2 alone were each virulent on Kombar containing Spt1.k, whereas the isolate that contained VR2 alone was avirulent (Shjerve et al., 2014). Conversely the isolate that contained VR2 alone was virulent on Rika that contained Spt1.r but isolates that contained VK1 and VK2 alone were avirulent (Shjerve et al., 2014).
Koladia et al. (2017b) identified nine unique QTLs in the P. teres f. teres cross between the Danish isolate BB25 and the North Dakotan isolate FGOH04Ptt‐21, of which only one of the virulence alleles was conferred by BB25 and eight were conferred by FGOH04Ptt‐21 (Table 2). A major QTL (PttTif1) with the virulent allele from FGOH04Ptt‐21 found on linkage group 10.1 (chromosome 1) contributed as much as 74% of the phenotypic variation on three lines (Manchurian, Tifang and CI4822) (Koladia et al., 2017b) (Fig. 3). The second major locus (PttBee1) conferring virulence from FGOH04Ptt‐21 was found on linkage group 1.1 (chromosome 1), contributing 56% of the phenotypic variation on the barley line Beecher (Koladia et al., 2017b) (Fig. 3). The BB25 isolate was only virulent on barley line Pinnacle with the BB25 virulence allele (PttPin1) located on linkage group 5.1 (chromosome 3), which explained 49% of the phenotypic variation (Koladia et al., 2017b) (Fig. 3). The virulence alleles of the remaining six loci all contributed less than 18% of the phenotypic variation and were distributed across multiple chromosomes (Koladia et al., 2017b). Currently, effector candidate genes underlying these major QTLs are being evaluated.
Carlsen et al. (2017) is currently the only study to map P. teres f. maculata virulence. The FGOB10Ptm‐1 × SG1 population was inoculated on commonly used SFNB differential barley lines Skiff, TR326, 81‐82/033 and PI392501 with a total of six loci identified across five of the 12 linkage groups (Carlsen et al., 2017) (Fig. 3 and Table 2). FGOB10Ptm‐1 and SG1 contributed the virulence allele at five and one QTL, respectively (Carlsen et al., 2017). The QTL with the virulence allele contributed by SG1 (vQTL4, PtmWel1) on linkage group 1.1 (chromosome 2) accounted for 30–37% of the phenotypic variation on barley lines 81‐82/033 and PI392501, whereas the largest effect QTL with the virulence allele contributed by FGOB10Ptm‐1 (vQTL5, PtmWel2) on linkage group 5.1 (chromosome 3) accounted for 26–34% of the phenotypic variation on barley lines 81‐82/033, TR326 and PI392501 (Carlsen et al., 2017) (Fig. 3 and Table 2). We have assigned tentative designations for this review until a robust naming nomenclature is established.
Advancement of genomic resources in barley and P. teres
The first assembled genome of barley was published by IBGSC (2012) using the American six‐row malting cultivar Morex. Again, Morex was used for a high‐quality reference assembly using chromosome conformation capture (Beier et al., 2017; Mascher et al., 2017), with subsequent analysis in the repetitive elements (Wicker et al., 2017). Additional lines that have been sequenced include the American two‐row spring feed barley line Bowman, the German two‐row spring malting barley line Barke, the Scottish two‐row winter malting barley line Igri (IBGSC, 2012), the Japanese two‐row spring malting barley line Haruna Nijo (IBGSC, 2012; Sato et al., 2016), and the Tibetan hulless cultivars Lasa Goumang (Zeng et al., 2015) and Zangqing320 (Dai et al., 2018; Nyima et al., 2018), but no published studies have compared these released barley genomes (Monat et al., 2018). Multiple studies have revealed genomic signatures of domestication using exome sequencing of 267 barley accessions (Russell et al., 2016), targeted resequencing of 433 barley accessions (Pankin et al., 2018) and whole‐genome sequencing of 172 barley accessions (Zeng et al., 2018). In the future we will be able to generate pan‐genomic resources that consolidate resistance loci to P. teres from different studies into their respective allelic series. Currently, large‐scale barley genome sequencing for pan‐genomic resources is hampered by the large amount of repetitive content in the barley genome (Monat et al., 2018). In this review, we consolidated mapped loci that have not currently been proven to be separate loci.
A wealth of P. teres genomic resources has been generated since the first genetic marker studies evaluating virulence/avirulence. The first P. teres genome was of the Canadian P. teres f. teres isolate 0‐1 using Illumina paired‐end reads (Ellwood et al., 2010). The size of 0‐1 was initially estimated at 41.95 Mb with an average sequence coverage of 20×, confirming the presence of at least nine chromosomes based on pulsed‐field gel electrophoresis and germ tube burst visualization (Ellwood et al., 2010). Wyatt et al. (2018) subsequently confirmed 12 chromosomes with a reference quality assembly of the same 0‐1 isolate using PacBio long‐read single‐molecule real‐time (SMRT) sequencing. The 0‐1 assembly produced a reported genome size of 46.5 Mb at an average coverage of 200× (Wyatt et al., 2018). Current genome assemblies include an additional eight P. teres f. teres isolates (W1‐1, NB29, NB85, NB73, 15A, 6A, FGOH04Ptt‐21 and BB25) and two P. teres f. maculata isolates (FGOB10Ptm‐1 and SG1) (Syme et al., 2018; Wyatt et al., 2019). Genome sizes currently are in the range of 39.27–41.28 Mb for P. teres f. maculata and 46.31–51.76 Mb in P. teres f. teres, concluding that P. teres f. maculata has a smaller genome due mostly to repetitive elements (Syme et al., 2018).
Due to the diversity of the natural P. teres f. teres population, evident from the mapped effectors mentioned earlier, a pan‐genomic sequencing approach of four P. teres f. teres isolates 15A, 6A, FGOH04Ptt‐21 and BB25 was used to evaluate the genomic architecture and gene content in comparison to 0‐1 (Wyatt et al., 2019). Each P. teres f. teres isolate was predicted to contain approximately 200 effectors using EffectorP (Wyatt et al., 2019), but EffectorP prediction is known to underestimate effectors due to stringent criteria. The majority of published virulence QTLs have been identified in subtelomeric regions where chromosome rearrangements and repetitive elements are found at higher frequency, allowing rapid development of polymorphisms (Wyatt et al., 2019). Wyatt et al. (2019) showed that 14 of 15 currently published P. teres f. teres QTLs span accessory genomic compartments with ten of 14 accessory genomic compartment QTLs specifically localizing to subtelomeric regions. This observation highlights the importance of the subtelomeric regions as drivers of virulence for P. teres f. teres. Additionally, a whole chromosome fusion was found between chromosomes 1 and 2 of isolate BB25, which is predicted to be a recent event and not inherited through ancestry and lost in other lineages (Wyatt et al., 2019). Chromosome fusions have been recently identified in the closely related species P. tritici‐repentis in reference isolate M4 and may be a common feature in the Pyrenophora genus (Moolhuijzen et al., 2018).
Conclusion and Future Direction
Liu et al. (2011) stated that the following characteristics facilitate the use of P. teres as a candidate model organism: the worldwide economic importance of the disease, the ability to produce biparental mapping populations and clone genes from the pathogen, the amenability of the pathogen to transformation for gene validation, relatively abundant genomic resources in both the host and pathogen, and the large numbers of host mapping populations to decipher the interactions and diversity of both the host and pathogen. These points were in their infancy and have been built on since Liu et al. (2011). As noted, P. teres has become an increasing problem worldwide, leading to the expansion of pathogen and host mapping studies that have identified loci important for pathogen virulence and host resistance/susceptibility using biparental and association mapping studies. In summary, over 340 and 140 QTLs have been published in relation to barley responses to P. teres f. teres and P. teres f. maculata isolates, respectively, of which eight loci are sequentially designated from Rpt1 to Rpt8 on chromosomes 3H, 1H, 2H, 7H, 6H, 5H, 4H and 4H, respectively (Table 1). Despite fine‐mapping of loci such as Rpt5/Spt1 (Richards et al., 2016), delimiting loci within the barley pan‐genome remains difficult due to the multitude of studies that report large overlapping genomic regions (Figs 1 and 2). Additionally, a large proportion of identified QTLs are collapsed into 39 consensus loci, resulting in a total of 47 loci (Figs 1 and 2). In contrast, a total of 22–23 loci have been published in P. teres that follow a tentative nomenclature of (a)virulence on a specific barley line (Table 2). An exponential growth of genomic resources for P. teres has occurred since the first genome assembly reported by Ellwood et al. (2010), with reference quality genome assemblies and analysis (Syme et al., 2018; Wyatt et al., 2019). Adoption of the P. teres–barley pathosystem as an agriculturally significant model has allowed rapid advancement in the abundance of genomic sequence, biparental and natural populations, and marker trait associations/QTLs to validate and characterize genes in both host and pathogen. Despite some overlap in functional resistance/susceptibility loci to P. teres f. teres and P. teres f. maculata in barley, each form of P. teres has a novel repertoire of effectors at their disposal. Therefore, breeding for resistance in barley to P. teres f. teres and P. teres f. maculata should proceed separately based on the most effective loci to the respective local P. teres populations.
Supporting information
Table S1 Studies mapping barley resistance/susceptibility genes to Pyrenophora teres f. teres. Different populations are indicated by alternating grey scale with the parent contributing the resistance allele in bold. Plant stage indicates whether the resistance is effective at the seedling or adult stage. If a single genotype isolate is used, this is indicated by the name and country of origin, whereas natural infection is indicated by specific location. Barley chromosome, phenotypic variation and designation of each locus are displayed, if available, from the corresponding reference. The inferred locus designation is reported based on markers obtained from the relevant publication and collapsed into loci using a maximum distance of 10 Mb of the Morex reference genome (Mascher et al., 2017) using BarleyMap (Cantalapiedra et al., 2015) or T3/Barley (Fig. 1).
Table S2 Studies mapping barley resistance/susceptibility genes to Pyrenophora teres f. maculata. Different populations are indicated by alternating grey scale with the parent contributing the resistance allele in bold. Plant stage indicates whether the resistance is effective at the seedling or adult stage. If a single genotype isolate is used this is indicated by the name and country of origin, whereas natural infection is indicated by specific location. Barley chromosome, phenotypic variation and designation of each locus are displayed if available from the corresponding reference. The inferred locus designation is reported based on markers obtained from the relevant publication and collapsed into loci using a maximum distance of 10 Mb of the Morex reference genome (Mascher et al., 2017) using BarleyMap (Cantalapiedra et al., 2015) or T3/Barley (Fig. 1).
Table S3 Genomic positions of markers reported in biparental or association mapping studies that could be anchored to the Morex barley reference genome (Mascher et al., 2017), including the locus designation (from publication), the corresponding marker, the assigned chromosome and chromosome position, and the reference.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.
References
- Abu Qamar, M. , Liu, Z.H. , Faris, J.D. , Chao, S. , Edwards, M.C. , Lai, Z. , Franckowiak, J.D. and Friesen, T.L. (2008) A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theor. Appl. Genet. 117, 1261. [DOI] [PubMed] [Google Scholar]
- Adawy, S.S. , Diab, A.A. , Sayed, A.‐H.I. , Shafik, D.I. , El‐Morsy, S.I. and Saker, M.M. (2013) Constuction of genetic linkage map and QTL analysis of net blotch resistance in barley. Int. J. Adv. Biotechnol. Res. 4, 348–363. [Google Scholar]
- Adhikari, A. , Steffenson, B.J. , Smith, M.J. and Dill‐Macky, R. (2019) Genome‐wide association mapping of seedling net form net blotch resistance in an Ethiopian and Eritrean barley collection. Crop Sci. 59, 1625–1638. [Google Scholar]
- Afanasenko, O.S. , Hartleb, H. , Guseva, N.N. , Minarikova, V. and Janosheva, M. (1995) A set of differentials to characterize populations of Pyrenophora teres Drechs. for international use. J. Phytopathol. 143, 501–507. [Google Scholar]
- Afanasenko, O. , Mironenko, N. , Filatova, O. , Kopahnke, D. , Krämer, I. and Ordon, F. (2007) Genetics of host–pathogen interactions in the Pyrenophora teres f. teres (net form)–barley (Hordeum vulgare) pathosystem. Eur. J. Plant Pathol. 117, 267–280. [Google Scholar]
- Afanasenko, O.S. , Jalli, M. , Pinnschmidt, H.O. , Filatova, O. and Platz, G.J. (2009) Development of an international standard set of barley differential genotypes for Pyrenophora teres f. teres . Plant Pathol. 58, 665–676. [Google Scholar]
- Akhavan, A. , Turkington, T.K. , Kebede, B. , Tekauz, A. , Kutcher, H.R. , Kirkham, C. , Xi, K. , Kumar, K. , Tucker, J.R. and Strelkov, S.E. (2015) Prevalence of mating type idiomorphs in Pyrenophora teres f. teres and P. teres f. maculata populations from the Canadian prairies. Can. J. Plant Pathol. 37, 52–60. [Google Scholar]
- Akhavan, A. , Turkington, T.K. , Askarian, H. , Tekauz, A. , Xi, K. , Tucker, J.R. , Kutcher, H.R. and Strelkov, S.E. (2016) Virulence of Pyrenophora teres populations in western Canada. Can. J. Plant Pathol. 38, 183–196. [Google Scholar]
- Amezrou, R. , Verma, R.P.S. , Chao, S. , Brueggeman, R.S. , Belqadi, L. , Arbaoui, M. , Rehman, S. and Gyawali, S. (2018) Genome‐wide association studies of net form of net blotch resistance at seedling and adult plant stages in spring barley collection. Mol. Breed. 38, 58. [Google Scholar]
- Arabi, M.I. , Barrault, G. , Sarrafi, A. and Albertini, L. (1992) Variation in the resistance of barley cultivars and in the pathogenicity of Drechslera teres f. sp. maculata and D. teres f. sp. teres isolates from France. Plant Pathol. 41, 180–186. [Google Scholar]
- Arabi, M.I.E. , Al‐Safadi, B. and Charbaji, T. (2003) Pathogenic variation among isolates of Pyrenophora teres, the causal agent of barley net blotch. J. Phytopathol. 151, 376–382. [Google Scholar]
- Bach, E. , Christensen, S. , Dalgaard, L. , Larsen, P.O. , Olsen, C.E. and Smedegård‐Petersen, V. (1979) Structures, properties and relationship to the aspergillomarasmines of toxins produced by Pyrenophora teres . Physiol. Plant Pathol. 14, 41–46. [Google Scholar]
- Beattie, A.D. , Scoles, G.J. and Rossnagel, B.G. (2007) Identification of molecular markers linked to a Pyrenophora teres avirulence gene. Phytopathology, 97, 842–849. [DOI] [PubMed] [Google Scholar]
- Beier, S. , Himmelbach, A. , Colmsee, C. , Zhang, X.‐Q. , Barrero, R.A. , Zhang, Q. , Li, L. , Bayer, M. , Bolser, D. , Taudien, S. , Groth, M. , Felder, M. , Hastie, A. , Šimková, H. , Staňková, H. , Vrána, J. , Chan, S. , Muñoz‐Amatriaín, M. , Ounit, R. , Wanamaker, S. , Schmutzer, T. , Aliyeva‐Schnorr, L. , Grasso, S. , Tanskanen, J. , Sampath, D. , Heavens, D. , Cao, S. , Chapman, B. , Dai, F. , Han, Y. , Li, H. , Li, X. , Lin, C. , McCooke, J.K. , Tan, C. , Wang, S. , Yin, S. , Zhou, G. , Poland, J.A. , Bellgard, M.I. , Houben, A. , Doležel, J. , Ayling, S. , Lonardi, S. , Langridge, P. , Muehlbauer, G.J. , Kersey, P. , Clark, M.D. , Caccamo, M. , Schulman, A.H. , Platzer, M. , Close, T.J. , Hansson, M. , Zhang, G. , Braumann, I. , Li, C. , Waugh, R. , Scholz, U. , Stein, N. and Mascher, M. (2017) Construction of a map‐based reference genome sequence for barley, Hordeum vulgare L. Sci. Data, 4, 170044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockelman, H.E. , Sharp, E.L. and Eslick, R.F. (1977) Trisomic analysis of genes for resistance to scald and net blotch in several barley cultivars. Can. J. Bot. 55, 2142–2148. [Google Scholar]
- Burlakoti, R.R. , Gyawali, S. , Chao, S. , Smith, K.P. , Horsley, R.D. , Cooper, B. , Muehlbauer, G.J. and Neate, S.M. (2017) Genome‐wide association study of spot form of net blotch resistance in the Upper Midwest barley breeding programs. Phytopathology, 107, 100–108. [DOI] [PubMed] [Google Scholar]
- Cakir, M. , Gupta, S. , Platz, G.J. , Ablett, G.A. , Loughman, R. , Emebiri, L.C. , Poulsen, D. , Li, C.D. , Lance, R.C.M. , Galwey, N.W. , Jones, M.G.K. and Appels, R. (2003) Mapping and validation of the genes for resistance to Pyrenophora teres f. teres in barley (Hordeum vulgare L.). Aust. J. Agric. Res. 54, 1369–1377. [Google Scholar]
- Cakir, M. , Gupta, S. , Li, C. , Hayden, M. , Mather, D.E. , Ablett, G.A. , Platz, G.J. , Broughton, S. , Chalmers, K.J. , Loughman, R. , Jones, M.G.K. and Lance, R.C.M. (2011) Genetic mapping and QTL analysis of disease resistance traits in the barley population Baudin × AC Metcalfe. Crop Pasture Sci. 62, 152–161. [Google Scholar]
- Campbell, G.F. and Crous, P.W. (2003) Genetic stability of net × spot hybrid progeny of the barley pathogen Pyrenophora teres . Australas. Plant Pathol. 32, 283–287. [Google Scholar]
- Campbell, G.F. , Lucas, J.A. and Crous, P.W. (2002) Evidence of recombination between net‐ and spot‐type populations of Pyrenophora teres as determined by RAPD analysis. Mycol. Res. 106, 602–608. [Google Scholar]
- Cantalapiedra, C.P. , Boudiar, R. , Casas, A.M. , Igartua, E. and Contreras‐Moreira, B. (2015) BARLEYMAP: physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol. Breed. 35, 13. [Google Scholar]
- Carlsen, S.A. , Neupane, A. , Wyatt, N.A. , Richards, J.K. , Faris, J.D. , Xu, S.S. , Brueggeman, R.S. and Friesen, T.L. (2017) Characterizing the Pyrenophora teres f. maculata–barley interaction using pathogen genetics. G3 Genes|Genomes|Genetics, 7, 2615–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çelik Oğuz, A. , Karakaya, A. , Ergün, N. and Sayim, İ. (2017) Turkish barley landraces resistant to net and spot forms of Pyrenophora teres . Phytopathol. Mediterr. 56, 217–223. [Google Scholar]
- Çelik Oğuz, A. , Ölmez, F. and Karakaya, A. (2018) Mating type idiomorphs of Pyrenophora teres in Turkey. Zemdirbyste‐Agriculture, 105, 271–278. [Google Scholar]
- Cromey, M.G. and Parkes, R.A. (2003) Pathogenic variation in Drechslera teres in New Zealand. New Zeal. Plant Prot. 56, 251–256. [Google Scholar]
- Daba, S.D. , Horsley, R. , Brueggeman, R. , Chao, S. and Mohammadi, M. (2019) Genome‐wide association studies and candidate gene identification for leaf scald and net blotch in barley (Hordeum vulgare L.). Plant Dis. 103, 880–889. [DOI] [PubMed] [Google Scholar]
- Dai, F. , Wang, X. , Zhang, X.‐Q. , Chen, Z. , Nevo, E. , Jin, G. , Wu, D. , Li, C. and Zhang, G. (2018) Assembly and analysis of a qingke reference genome demonstrate its close genetic relation to modern cultivated barley. Plant Biotechnol. J. 16, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikilitas, M. , Çelík Oğuz, A. and Karakaya, A. (2018) Extracellular protease activity and glucose production in isolates of net blotch pathogens differing in virulence. Zemdirbyste‐Agriculture, 105, 89–94. [Google Scholar]
- Dinglasan, E. , Hickey, L. , Ziems, L. , Fowler, R. , Anisimova, A. , Baranova, O. , Lashina, N. and Afanasenko, O. (2019) Genetic characterization of resistance to Pyrenophora teres f. teres in the international barley differential Canadian lake shore. Front. Plant Sci. 10, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontsova, A.A. , Alabushev, A.V. , Lebedeva, M.V. and Potokina, E.K. (2018) Analysis of polymorphism of microsatellite markers linked to a long‐term net form of net blotch resistance gene in winter barley varieties in the south of Russia. Ind. J. Genet. 78, 317–323. [Google Scholar]
- Douiyssi, A. , Rasmusson, D.C. and Roelfs, A.P. (1998) Responses of barley cultivars and lines to isolates of Pyrenophora teres . Plant Dis. 82, 316–321. [DOI] [PubMed] [Google Scholar]
- Ellwood, S.R. , Liu, Z. , Syme, R.A. , Lai, Z. , Hane, J.K. , Keiper, F. , Moffat, C.S. , Oliver, R.P. and Friesen, T.L. (2010) A first genome assembly of the barley fungal pathogen Pyrenophora teres f. teres . Genome Biol. 11, R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood, S.R. , Syme, R.A. , Moffat, C.S. and Oliver, R.P. (2012) Evolution of three Pyrenophora cereal pathogens: recent divergence, speciation and evolution of non‐coding DNA. Fungal Genet. Biol. 49, 825–829. [DOI] [PubMed] [Google Scholar]
- Emebiri, L.C. , Platz, C.G. and Moody, D.B. (2005) Disease resistance genes in a doubled haploid population of two‐rowed barley segregating for malting quality attributes. Aust. J. Agric. Res. 56, 49–56. [Google Scholar]
- Flor, H.H. (1955) Host‐parasite interaction in flax rust ‐ its genetics and other implications. Phytopathology, 45, 680–685. [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Fowler, R.A. , Platz, G.J. , Bell, K.L. , Fletcher, S.E.H. , Franckowiak, J.D. and Hickey, L.T. (2017) Pathogenic variation of Pyrenophora teres f. teres in Australia. Australas. Plant Pathol. 46, 115–128. [Google Scholar]
- Franckowiak, J.D. and Platz, G.J. (2013) International database for barley genes and barley genetic stocks. Barley Genet. Newsl. 43, 48–223. [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Lai, Z. and Steffenson, B.J. (2006) Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a doubled‐haploid barley population. Genome, 49, 855–859. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Meinhardt, S.W. and Faris, J.D. (2007) The Stagonospora nodorum‐wheat pathosystem involves multiple proteinaceous host‐selective toxins and corresponding host sensitivity genes that interact in an inverse gene‐for‐gene manner. Plant J. 51, 681–692. [DOI] [PubMed] [Google Scholar]
- Friis, P. , Olsen, C.E. and Møller, B.L. (1991) Toxin production in Pyrenophora teres, the ascomycete causing the net‐spot blotch disease of barley (Hordeum vulgare L.). J. Biol. Chem. 266, 13329–13335. [PubMed] [Google Scholar]
- Galano, T. , Bultosa, G. and Fininsa, C. (2011) Malt quality of 4 barley (Hordeum vulgare L.) grain varieties grown under low severity of net blotch at Holetta, west Shewa. Ethiopia. African J. Biotechnol. 10, 797–806. [Google Scholar]
- Geschele, E.E. (1928) The response of barley to parasitic fungi Helminthosporium teres Sacc. Bull. Appl. Bot. Genet. Plant Breed. 19, 371–384 (in Rev. Appl. Mycol. 8, 165). [Google Scholar]
- Graner, A. , Foroughi‐Wehr, B. and Tekauz, A. (1996) RFLP mapping of a gene in barley conferring resistance to net blotch (Pyrenophora teres). Euphytica, 91, 229–234. [Google Scholar]
- Grewal, T.S. , Rossnagel, B.G. , Pozniak, C.J. and Scoles, G.J. (2008) Mapping quantitative trait loci associated with barley net blotch resistance. Theor. Appl. Genet. 116, 529–539. [DOI] [PubMed] [Google Scholar]
- Grewal, T.S. , Rossnagel, B.G. and Scoles, G.J. (2012) Mapping quantitative trait loci associated with spot blotch and net blotch resistance in a doubled‐haploid barley population. Mol. Breed. 30, 267–279. [Google Scholar]
- Gupta, S. and Loughman, R. (2001) Current virulence of Pyrenophora teres on barley in Western Australia. Plant Dis. 85, 960–966. [DOI] [PubMed] [Google Scholar]
- Gupta, S. , Li, C.D. , Loughman, R. , Cakir, M. , Platz, G. , Westcott, S. , Bradley, J. , Broughton, S. and Lance, R. (2010) Quantitative trait loci and epistatic interactions in barley conferring resistance to net type net blotch (Pyrenophora teres f. teres) isolates. Plant Breed. 129, 362–368. [Google Scholar]
- Gupta, S. , Li, C. , Loughman, R. , Cakir, M. , Westcott, S. and Lance, R. (2011) Identifying genetic complexity of 6H locus in barley conferring resistance to Pyrenophora teres f. teres . Plant Breed. 130, 423–429. [Google Scholar]
- Gyawali, S. , Amezrou, R. , Verma, R.P.S. , Brueggeman, R. , Rehman, S. , Belqadi, L. , Arbaoui, M. , Tamang, P. and Singh, M. (2018) Seedling and adult stage resistance to spot form of net blotch (SFNB) in spring barley and stability of adult stage resistance to SFNB in Morocco. Eur. J. Plant Pathol. 153, 475–487. [Google Scholar]
- Harrabi, M. and Kamel, A. (1990) Virulence spectrum to barley in some isolates of Pyrenophora teres from the Mediterranean region. Plant Dis. 74, 230–232. [Google Scholar]
- Hisano, H. , Sakamoto, K. , Takagi, H. , Terauchi, R. and Sato, K. (2017) Exome QTL‐seq maps monogenic locus and QTLs in barley. BMC Genom. 18, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBGSC (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature, 491, 711–716. [DOI] [PubMed] [Google Scholar]
- Islamovic, E. , Bregitzer, P. and Friesen, T.L. (2017) Barley 4H QTL confers NFNB resistance to a global set of P. teres f. teres isolates. Mol. Breed. 37, 29. [Google Scholar]
- Ismail, I.A. and Able, A.J. (2016) Secretome analysis of virulent Pyrenophora teres f. teres isolates. Protemics, 16, 2625–2636. [DOI] [PubMed] [Google Scholar]
- Ismail, I.A. and Able, A.J. (2017) Gene expression profiling of virulence‐associated proteins in planta during net blotch disease of barley. Physiol. Mol. Plant Pathol. 98, 69–79. [Google Scholar]
- Ismail, I.A. , Godfrey, D. and Able, A.J. (2014a) Fungal growth, proteinaceous toxins and virulence of Pyrenophora teres f. teres on barley. Australas. Plant Pathol. 43, 535–546. [Google Scholar]
- Ismail, I.A. , Godfrey, D. and Able, A.J. (2014b) Proteomic analysis reveals the potential involvement of xylanase from Pyrenophora teres f. teres in net form net blotch disease of barley. Australas. Plant Pathol. 43, 715–726. [Google Scholar]
- Jalli, M. (2011) Sexual reproduction and soil tillage effects on virulence of Pyrenophora teres in Finland. Ann. Appl. Biol. 158, 95–105. [Google Scholar]
- Jalli, M. and Robinson, J. (2000) Stable resistance in barley to Pyrenophora teres f. teres isolates from the Nordic‐Baltic region after increase on standard host genotypes. Euphytica, 113, 71–77. [Google Scholar]
- Jayasena, K.W. , Van Burgel, A. , Tanaka, K. , Majewski, J. and Loughman, R. (2007) Yield reduction in barley in relation to spot‐type net blotch. Australas. Plant Pathol. 36, 429–433. [Google Scholar]
- Jebbouj, R. and El Yousfi, B. (2009) Barley yield losses due to defoliation of upper three leaves either healthy or infected at boot stage by Pyrenophora teres f. teres . Eur. J. Plant Pathol. 125, 303–315. [Google Scholar]
- Jonsson, R. , Bryngelsson, T. and Gustafsson, M. (1997) Virulence studies of Swedish net blotch isolates (Drechslera teres) and identification of resistant barley lines. Euphytica, 94, 209–218. [Google Scholar]
- Keon, J.P.R. and Hargreaves, J.A. (1983) A cytological study of the net blotch disease of barley caused by Pyrenophora teres . Physiol. Plant Pathol. 22, 321–329. [Google Scholar]
- Khan, T.N. (1987) Relationship between net blotch (Drechslera teres) and losses in grain yield of barley in Western Australia. Aust. J. Agric. Res. 38, 671–769. [Google Scholar]
- Khan, T.N. and Boyd, W.J.R. (1969a) Inheritance of resistance to net blotch in barley. II. Genes conditioning resistance against race W.A.‐2. Can. J. Genet. Cytol. 11, 592–597. [Google Scholar]
- Khan, T.N. and Boyd, W.J.R. (1969b) Physiologic specialization in Drechslera teres . Aust. J. Biol. Sci. 22, 1229–1236. [Google Scholar]
- Koladia, V.M. , Faris, J.D. , Richards, J.K. , Brueggeman, R.S. , Chao, S. and Friesen, T.L. (2017a) Genetic analysis of net form net blotch resistance in barley lines CIho 5791 and Tifang against a global collection of P. teres f. teres isolates. Theor. Appl. Genet. 130, 163–173. [DOI] [PubMed] [Google Scholar]
- Koladia, V.M. , Richards, J.K. , Wyatt, N.A. , Faris, J.D. , Brueggeman, R.S. and Friesen, T.L. (2017b) Genetic analysis of virulence in the Pyrenophora teres f. teres population BB25×FGOH04Ptt‐21. Fungal Genet. Biol. 107, 12–19. [DOI] [PubMed] [Google Scholar]
- König, J. , Perovic, D. , Kopahnke, D. and Ordon, F. (2013) Development of an efficient method for assessing resistance to the net type of net blotch (Pyrenophora teres f. teres) in winter barley and mapping of quantitative trait loci for resistance. Mol. Breed. 32, 641–650. [Google Scholar]
- König, J. , Perovic, D. , Kopahnke, D. and Ordon, F. (2014) Mapping seedling resistance to net form of net blotch (Pyrenophora teres f. teres) in barley using detached leaf assay. Plant Breed. 133, 356–365. [Google Scholar]
- Lai, Z. , Faris, J.D. , Weiland, J.J. , Steffenson, B.J. and Friesen, T.L. (2007) Genetic mapping of Pyrenophora teres f. teres genes conferring avirulence on barley. Fungal Genet. Biol. 44, 323–329. [DOI] [PubMed] [Google Scholar]
- Leboldus, J.M. , Kinzer, K. , Richards, J. , Ya, Z. , Yan, C. , Friesen, T.L. and Brueggeman, R. (2015) Genotype‐by‐sequencing of the plant‐pathogenic fungi Pyrenophora teres and Sphaerulina musiva utilizing ion torrent sequence technology. Mol. Plant Pathol. 16, 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmensiek, A. , Platz, G.J. , Mace, E. , Poulsen, D. and Sutherland, M.W. (2007) Mapping of adult plant resistance to net form of net blotch in three Australian barley populations. Aust. J. Agric. Res. 58, 1191–1197. [Google Scholar]
- Leišova, L. , Minariˇḱova, V. , Kučera, L. and Ovesná, J. (2005) Genetic diversity of Pyrenophora teres isolates as detected by AFLP analysis. J. Phytopathol. 153, 569–578. [Google Scholar]
- Liu, Z.H. and Friesen, T.L. (2010) Identification of Pyrenophora teres f. maculata, causal agent of spot type net blotch of barley in North Dakota. Plant Dis. 94, 480. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Faris, J.D. , Edwards, M.C. and Friesen, T.L. (2010) Development of expressed sequence tag (EST)‐based markers for genomic analysis of a barley 6H region harboring multiple net form net blotch resistance genes. Plant Genome, 3, 41–52. [Google Scholar]
- Liu, Z. , Ellwood, S.R. , Oliver, R.P. and Friesen, T.L. (2011) Pyrenophora teres: profile of an increasingly damaging barley pathogen. Mol. Plant Pathol. 12, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.H. , Zhong, S. , Stasko, A.K. , Edwards, M.C. and Friesen, T.L. (2012) Virulence profile and genetic structure of a North Dakota population of Pyrenophora teres f. teres, the causal agent of net form net blotch of Barley. Phytopathology, 102, 539–546. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Holmes, D.J. , Faris, J.D. , Chao, S. , Brueggeman, R.S. , Edwards, M.C. and Friesen, T.L. (2015) Necrotrophic effector‐triggered susceptibility (NETS) underlies the barley–Pyrenophora teres f. teres interaction specific to chromosome 6H. Mol. Plant Pathol. 16, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw, J.P.J. , Victor, D. , Crous, P.W. , Holz, G. and Janse, B.J.H. (1995) Characterization of Pyrenophora isolates associated with spot and net type lesions on barley in South Africa. J. Phytopathol. 143, 129–134. [Google Scholar]
- Ma, Z.Q. , Lapitan, N.L.V. and Steffenson, B. (2004) QTL mapping of net blotch resistance genes in a doubled‐haploid population of six‐rowed barley. Euphytica, 137, 291–296. [Google Scholar]
- Manninen, O. , Kalendar, R. , Robinson, J. and Schulman, A.H. (2000) Application of BARE‐1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol. Gen. Genet. 264, 325–334. [DOI] [PubMed] [Google Scholar]
- Manninen, O.M. , Jalli, M. , Kalendar, R. , Schulman, A. , Afanasenko, O. and Robinson, J. (2006) Mapping of major spot‐type and net‐type net‐blotch resistance genes in the Ethiopian barley line CI 9819. Genome, 49, 1564–1571. [DOI] [PubMed] [Google Scholar]
- Marshall, J.M. , Kinzer, K. and Brueggeman, R.S. (2015) First report of Pyrenophora teres f. maculata the cause of spot form net blotch of barley in Idaho. Plant Dis. 99, 1860. [Google Scholar]
- Martin, A. , Platz, G.J. , de Klerk, D. , Fowler, R.A. , Smit, F. , Potgieter, F.G. and Prins, R. (2018) Identification and mapping of net form of net blotch resistance in South African barley. Mol. Breed. 38, 53. [Google Scholar]
- Mascher, M. , Gundlach, H. , Himmelbach, A. , Beier, S. , Twardziok, S.O. , Wicker, T. , Radchuk, V. , Dockter, C. , Hedley, P.E. , Russell, J. , Bayer, M. , Ramsay, L. , Liu, H. , Haberer, G. , Zhang, X.‐Q. , Zhang, Q. , Barrero, R.A. , Li, L. , Taudien, S. , Groth, M. , Felder, M. , Hastie, A. , Šimková, H. , Staňková, H. , Vrána, J. , Chan, S. , Muñoz‐Amatriaín, M. , Ounit, R. , Wanamaker, S. , Bolser, D. , Colmsee, C. , Schmutzer, T. , Aliyeva‐Schnorr, L. , Grasso, S. , Tanskanen, J. , Chailyan, A. , Sampath, D. , Heavens, D. , Clissold, L. , Cao, S. , Chapman, B. , Dai, F. , Han, Y. , Li, H. , Li, X. , Lin, C. , McCooke, J.K. , Tan, C. , Wang, P. , Wang, S. , Yin, S. , Zhou, G. , Poland, J.A. , Bellgard, M.I. , Borisjuk, L. , Houben, A. , Doležel, J. , Ayling, S. , Lonardi, S. , Kersey, P. , Langridge, P. , Muehlbauer, G.J. , Clark, M.D. , Caccamo, M. , Schulman, A.H. , Mayer, K.F.X. , Platzer, M. , Close, T.J. , Scholz, U. , Hansson, M. , Zhang, G. , Braumann, I. , Spannagl, M. , Li, C. , Waugh, R. and Stein, N. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature, 544, 427–433. [DOI] [PubMed] [Google Scholar]
- Maurer, A. , Draba, V. , Jiang, Y. , Schnaithmann, F. , Sharma, R. , Schumann, E. , Kilian, B. , Reif, J.C. and Pillen, K. (2015) Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genom. 16, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, M.S. , Howlett, B.J. and Hollaway, G.J. (2010a) Spot form of net blotch, caused by Pyrenophora teres f. maculata, is the most prevalent foliar disease of barley in Victoria, Australia. Australas. Plant Pathol. 39, 46–49. [Google Scholar]
- McLean, M.S. , Keiper, F.J. and Hollaway, G.J. (2010b) Genetic and pathogenic diversity in Pyrenophora teres f. maculata in barley crops of Victoria, Australia. Australas. Plant Pathol. 39, 319–325. [Google Scholar]
- McLean, M.S. , Martin, A. , Gupta, S. , Sutherland, M.W. , Hollaway, G.J. and Platz, G.J. (2014) Validation of a new spot form of net blotch differential set and evidence for hybridisation between the spot and net forms of net blotch in Australia. Australas. Plant Pathol. 43, 223–233. [Google Scholar]
- Mode, C.J. and Schaller, C.W. (1958) Two additional factors for host resistance to net blotch in barley. Agron. J. 50, 15–18. [Google Scholar]
- Molnar, S.J. , James, L.E. and Kasha, K.J. (2000) Inheritance and RAPD tagging of multiple genes for resistance to net blotch in barley. Genome, 43, 224–231. [PubMed] [Google Scholar]
- Monat, C. , Schreiber, M. , Stein, N. and Mascher, M. (2018) Prospects of pan‐genomics in barley. Theor. Appl. Genet. 132, 785–796. [DOI] [PubMed] [Google Scholar]
- Moolhuijzen, P.M. , See, P.T. , Oliver, R.P. and Moffat, C.S. (2018) Genomic distribution of a novel Pyrenophora tritici‐repentis ToxA insertion element. PLoS ONE, 13, e0206586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya, P.A. , Girotti, J.R. , Toledo, A.V. and Sisterna, M.N. (2018) Antifungal activity of Trichoderma VOCs against Pyrenophora teres, the causal agent of barley net blotch. J. Plant Prot. Res. 58, 45–53. [Google Scholar]
- Muñoz‐Amatriaín, M. , Cuesta‐Marcos, A. , Endelman, J.B. , Comadran, J. , Bonman, J.M. , Bockelman, H.E. , Chao, S. , Russell, J. , Waugh, R. , Hayes, P.M. and Muehlbauer, G.J. (2014) The USDA barley core collection: genetic diversity, population structure, and potential for genome‐wide association studies. PLoS ONE, 9, e94688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, G.M. and Brennan, J.P. (2010) Estimating disease losses to the Australian barley industry. Australas. Plant Pathol. 39, 85–96. [Google Scholar]
- Neupane, A. , Tamang, P. , Brueggeman, R.S. and Friesen, T.L. (2015) Evaluation of a barley core collection for spot form net blotch reaction reveals distinct genotype‐specific pathogen virulence and host susceptibility. Phytopathology, 105, 509–517. [DOI] [PubMed] [Google Scholar]
- Novakazi, F. , Afanasenko, O. , Anisimova, A. , Platz, G.J. , Snowdon, R. , Kovaleva, O. , Zubkovich, A. and Ordon, F. (2019) Genetic analysis of a worldwide barley collection for resistance to net form of net blotch disease (Pyrenophora teres f. teres). Theor. Appl. Genet. 132, 2633–2650. [DOI] [PubMed] [Google Scholar]
- Nyima, T. , Zeng, X. , Li, X. , Bai, L. , Wang, Y. , Xu, T. , Yuan, H. , Xu, Q. , Zha, S. , Wei, Z. , Qimei, W. , Basang, Y. , Dunzhu, J. and Yu, M. (2018) An improved high‐quality genome assembly and annotation of Qingke, Tibetan Hulless Barley. bioRxiv, 409136, 10.1101/409136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle, P.D. , Brooks, W.S. , Barnett, M.D. , Berger, G.L. , Steffenson, B.J. , Stromberg, E.L. , Maroof, M.A.S. , Liu, S.Y. and Griffey, C.A. (2014) Mapping net blotch resistance in ‘Nomini’ and CIho 2291 barley. Crop Sci. 54, 2596–2602. [Google Scholar]
- Ohm, R.A. , Feau, N. , Henrissat, B. , Schoch, C.L. , Horwitz, B.A. , Barry, K.W. , Condon, B.J. , Copeland, A.C. , Dhillon, B. , Glaser, F. , Hesse, C.N. , Kosti, I. , LaButti, K. , Lindquist, E.A. , Lucas, S. , Salamov, A.A. , Bradshaw, R.E. , Ciuffetti, L. , Hamelin, R.C. , Kema, G.H.J. , Lawrence, C. , Scott, J.A. , Spatafora, J.W. , Turgeon, B.G. , de Wit, P.J.G.M. , Zhong, S. , Goodwin, S.B. and Grigoriev, I.V. (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 8, e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankin, A. , Janine, A. , Christian, B. and Maria, K. (2018) Targeted resequencing reveals genomic signatures of barley domestication. New Phytol. 218, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piening, L. and Kaufmann, M.L. (1969) Comparison of the effectors of net blotch and leaf removal on yield in barley. Can. J. Plant Sci. 49, 731–735. [Google Scholar]
- Poudel, B. , McLean, M.S. , Platz, G.J. , McIlroy, J.A. , Sutherland, M.W. and Martin, A. (2019) Investigating hybridisation between the forms of Pyrenophora teres based on Australian barley field experiments and cultural collections. Eur. J. Plant Pathol. 153, 465–473. [Google Scholar]
- Raman, H. , Platz, G.J. , Chalmers, K.J. , Raman, R. , Read, B.J. , Barr, A.R. and Moody, D.B. (2003) Mapping of genomic regions associated with net form of net blotch resistance in barley. Aust. J. Agric. Res. 54, 1359–1367. [Google Scholar]
- Rau, D. , Attene, G. , Brown, A.H.D. , Nanni, L. , Maier, F.J. , Balmas, V. , Saba, E. , Schäfer, W. and Papa, R. (2007) Phylogeny and evolution of mating‐type genes from Pyrenophora teres, the causal agent of barley “net blotch” disease. Curr. Genet. 51, 377–392. [DOI] [PubMed] [Google Scholar]
- Rau, D. , Rodriguez, M. , Leonarda Murgia, M. , Balmas, V. , Bitocchi, E. , Bellucci, E. , Nanni, L. , Attene, G. and Papa, R. (2015) Co‐evolution in a landrace meta‐population: two closely related pathogens interacting with the same host can lead to different adaptive outcomes. Sci. Rep. 5, 12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, B.J. , Raman, H. , McMichael, G. , Chalmers, K.J. , Ablett, G.A. , Platz, G.J. , Raman, R. , Genger, R.K. , Boyd, W.J.R. , Li, C.D. , Grime, C.R. , Park, R.F. , Wallwork, H. , Prangnell, R. and Lance, R.C.M. (2003) Mapping and QTL analysis of the barley population Sloop × Halcyon. Aust. J. Agric. Res. 54, 1145–1153. [Google Scholar]
- Richards, J. , Chao, S. , Friesen, T. and Brueggeman, R. (2016) Fine mapping of the barley chromosome 6H net form net blotch susceptibility locus. G3 Genes|Genomes|Genetics, 6, 1809–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, J.K. , Friesen, T.L. and Brueggeman, R.S. (2017) Association mapping utilizing diverse barley lines reveals net form net blotch seedling resistance/susceptibility loci. Theor. Appl. Genet. 130, 915–927. [DOI] [PubMed] [Google Scholar]
- Richter, K. , Schondelmaier, J. and Jung, C. (1998) Mapping of quantitative trait loci affecting Drechslera teres resistance in barley with molecular markers. Theor. Appl. Genet. 97, 1225–1234. [Google Scholar]
- Robinson, J. and Jalli, M. (1996) Diversity among Finnish net blotch isolates and resistance in barley. Euphytica, 92, 81–87. [Google Scholar]
- Rozanova, I.V. , Lashina, N.M. , Mustafin, Z.S. , Gorobets, S.A. , Efimov, V.M. , Afanasenko, O.S. and Khlestkina, E.K. (2019) SNPs associated with barley resistance to isolates of Pyrenophora teres f. teres . BMC Genom. 20, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, J. , Mascher, M. , Dawson, I.K. , Kyriakidis, S. , Calixto, C. , Freund, F. , Bayer, M. , Milne, I. , Marshall‐Griffiths, T. , Heinen, S. , Hofstad, A. , Sharma, R. , Himmelbach, A. , Knauft, M. , van Zonneveld, M. , Brown, J.W.S. , Schmid, K. , Kilian, B. , Muehlbauer, G.J. , Stein, N. and Waugh, R. (2016) Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 48, 1024. [DOI] [PubMed] [Google Scholar]
- Sarpeleh, A. , Wallwork, H. , Catcheside, D.E.A. , Tate, M.E. and Able, A.J. (2007) Proteinaceous metabolites from Pyrenophora teres contribute to symptom development of barley net blotch. Phytopathology, 97, 907–915. [DOI] [PubMed] [Google Scholar]
- Sarpeleh, A. , Tate, M.E. , Wallwork, H. , Catcheside, D. and Able, A.J. (2009) Characterisation of low molecular weight phytotoxins isolated from Pyrenophora teres . Physiol. Mol. Plant Pathol. 73, 154–162. [Google Scholar]
- Sato, K. and Takeda, K. (1997) Net blotch resistance in wild species of Hordeum . Euphytica, 95, 179–185. [Google Scholar]
- Sato, K. , Tanaka, T. , Shigenobu, S. , Motoi, Y. , Wu, J. and Itoh, T. (2016) Improvement of barley genome annotations by deciphering the Haruna Nijo genome. DNA Res. 23, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, C.W. (1955) Inheritance of resistance to net blotch of barley. Phytopathology, 45, 174–176. [Google Scholar]
- Schaller, C.W. and Wiebe, G.A. (1952) Sources of resistance to net blotch of barley. Agron. J. 334–336. [Google Scholar]
- Sharma, H.S.S. (1984) Assessment of the reaction of some spring barley cultivars to Pyrenophora teres using whole plants, detached leaves and toxin bioassay. Plant Pathol. 33, 371–376. [Google Scholar]
- Shipton, W.A. , Khan, T.N. and Boyd, W.J.R. (1973) Net blotch of barley. Rev. Plant Pathol. 52, 269–290. [Google Scholar]
- Shjerve, R.A. , Faris, J.D. , Brueggeman, R.S. , Yan, C. , Zhu, Y. , Koladia, V. and Friesen, T.L. (2014) Evaluation of a Pyrenophora teres f. teres mapping population reveals multiple independent interactions with a region of barley chromosome 6H. Fungal Genet. Biol. 70, 104–112. [DOI] [PubMed] [Google Scholar]
- Shoemaker, R.A. (1959) Nomenclature of Drechslera and Bipolaris, grass parasites segregated from “Helminthosporium”. Can. J. Bot. 37, 879–887. [Google Scholar]
- Smedegård‐Petersen, V. (1971) Pyrenophora teres f. maculata f. nov. and Pyrenophora teres f. teres on barley in Denmark. Kgl. Vet. Landbohojsk. Arsskr. 124–144. [Google Scholar]
- Smedegård‐Petersen, V. (1977) Isolation of two toxins produced by Pyrenophora teres and their significance in disease development of net‐spot blotch of barley. Physiol. Plant Pathol. 10, 203–211. [Google Scholar]
- Spaner, D. , Shugar, L.P. , Choo, T.M. , Falak, I. , Briggs, K.G. , Legge, W.G. , Falk, D.E. , Ullrich, S.E. , Tinker, N.A. , Steffenson, B.J. and Mather, D.E. (1998) Mapping of disease resistance loci in barley on the basis of visual assessment of naturally occurring symptoms. Crop Sci. 38, 843–850. [Google Scholar]
- St. Pierre, S. , Gustus, C. , Steffenson, B. , Dill‐Macky, R. and Smith, K.P. (2010) Mapping net form net blotch and Septoria speckled leaf blotch resistance loci in barley. Phytopathology, 100, 80–84. [DOI] [PubMed] [Google Scholar]
- Steffenson, B.J. and Webster, R.K. (1992) Pathotype diversity of Pyrenophora teres f. teres on barley. Phytopathology, 82, 170–177. [DOI] [PubMed] [Google Scholar]
- Steffenson, B.J. , Hayes, P.M. and Kleinhofs, A. (1996) Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor. Appl. Genet. 92, 552–558. [DOI] [PubMed] [Google Scholar]
- Syme, R.A. , Martin, A. , Wyatt, N.A. , Lawrence, J.A. , Muria‐Gonzalez, M.J. , Friesen, T.L. and Ellwood, S.R. (2018) Transposable element genomic fissuring in Pyrenophora teres is associated with genome expansion and dynamics of host–pathogen genetic interactions. Front. Genet. 9, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang, P. , Neupane, A. , Mamidi, S. , Friesen, T. and Brueggeman, R. (2015) Association mapping of seedling resistance to spot form net blotch in a worldwide collection of barley. Phytopathology, 105, 500–508. [DOI] [PubMed] [Google Scholar]
- Tamang, P. , Richards, J.K. , Alhashal, A. , Sharma Poudel, R. , Horsley, R.D. , Friesen, T.L. and Brueggeman, R.S. (2019) Mapping of barley susceptibility/resistance QTL against spot form net blotch caused by Pyrenophora teres f. maculata using RIL populations. Theor. Appl. Genet. 132, 1953–1963. [DOI] [PubMed] [Google Scholar]
- Tekauz, A. (1985) A numerical scale to classify reactions of barley to Pyrenophora teres . Can. J. Plant Pathol. 7, 181–183. [Google Scholar]
- Tekauz, A. (1990) Characterization and distribution of pathogenic variation in Pyrenophora teres f. teres and P. teres f. maculata from western Canada. Can. J. Plant Pathol. 12, 141–148. [Google Scholar]
- Toruño, T.Y. , Stergiopoulos, I. and Coaker, G. (2016) Plant‐pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatter, T. , Maurer, A. , Kopahnke, D. , Perovic, D. , Ordon, F. and Pillen, K. (2017) A nested association mapping population identifies multiple small effect QTL conferring resistance against net blotch (Pyrenophora teres f. teres) in wild barley. PLoS ONE, 12, e0186803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Mace, E.S. , Platz, G.J. , Hunt, C.H. , Hickey, L.T. , Franckowiak, J.D. and Jordan, D.R. (2015) Spot form of net blotch resistance in barley is under complex genetic control. Theor. Appl. Genet. 128, 489–499. [DOI] [PubMed] [Google Scholar]
- Weiergang, I. , Lyngs Jørgensen, H.J. , Møller, I.M. , Friis, P. and Smedegaard‐petersen, V. (2002) Correlation between sensitivity of barley to Pyrenophora teres toxins and susceptibility to the fungus. Physiol. Mol. Plant Pathol. 60, 121–129. [Google Scholar]
- Weiland, J.J. , Steffenson, B.J. , Cartwright, R.D. and Webster, R.K. (1999) Identification of molecular genetic markers in Pyrenophora teres f. teres associated with low virulence on ‘Harbin’ barley. Phytopathology, 89, 176–181. [DOI] [PubMed] [Google Scholar]
- Wicker, T. , Schulman, A.H. , Tanskanen, J. , Spannagl, M. , Twardziok, S. , Mascher, M. , Springer, N.M. , Li, Q. , Waugh, R. , Li, C. , Zhang, G. , Stein, N. , Mayer, K.F.X. and Gundlach, H. (2017) The repetitive landscape of the 5100 Mbp barley genome. Mob. DNA, 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, K.J. , Lichon, A. , Gianquitto, P. , Kretschmer, J.M. , Karakousis, A. , Manning, S. , Langridge, P. and Wallwork, H. (1999) Identification and mapping of a gene conferring resistance to the spot form of net blotch (Pyrenophora teres f. maculata) in barley. Theor. Appl. Genet. 99, 323–327. [Google Scholar]
- Williams, K.J. , Platz, G.J. , Barr, A.R. , Cheong, J. , Willsmore, K. , Cakir, M. and Wallwork, H. (2003) A comparison of the genetics of seedling and adult plant resistance to the spot form of net blotch (Pyrenophora teres f. maculata). Aust. J. Agric. Res. 54, 1387–1394. [Google Scholar]
- Wonneberger, R. , Ficke, A. and Lillemo, M. (2017a) Mapping of quantitative trait loci associated with resistance to net form net blotch (Pyrenophora teres f. teres) in a doubled haploid Norwegian barley population. PLoS ONE, 12, e0175773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonneberger, R. , Ficke, A. and Lillemo, M. (2017b) Identification of quantitative trait loci associated with resistance to net form net blotch in a collection of Nordic barley germplasm. Theor. Appl. Genet. 130, 2025–2043. [DOI] [PubMed] [Google Scholar]
- Wu, H.‐L. , Steffenson, B.J. , Zhong, S. , Li, Y. and Oleson, A.E. (2003) Genetic variation for virulence and RFLP markers in Pyrenophora teres . Can. J. Plant Pathol. 25, 82–90. [Google Scholar]
- Wyatt, N.A. , Richards, J.K. , Brueggeman, R.S. and Friesen, T.L. (2018) Reference assembly and annotation of the Pyrenophora teres f. teres isolate 0–1. G3 Genes|Genomes|Genetics, 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt, N.A. , Richards, J.K. , Brueggeman, R.S. and Friesen, T.L. (2019) A comparative genomic analysis of the barley pathogen Pyrenophora teres f. teres identifies sub‐telomeric regions as drivers of virulence. Mol. Plant‐Microbe Interact. 10.1094/MPMI-05-19-0128-R. [DOI] [PubMed] [Google Scholar]
- Yun, S.J. , Gyenis, L. , Hayes, P.M. , Matus, I. , Smith, K.P. , Steffenson, B.J. and Muehlbauer, G.J.M. (2005) Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci. 45, 2563–2572. [Google Scholar]
- Zeleke, T. (2017) Evaluation of host reaction and yield performance of malt barley cultivars to net blotch, Pyrenophora teres in Bale Highlands. Ethiopia. J. Plant Sci. 5, 43–47. [Google Scholar]
- Zeng, X. , Long, H. , Wang, Z. , Zhao, S. , Tang, Y. , Huang, Z. , Wang, Y. , Xu, Q. , Mao, L. , Deng, G. , Yao, X. , Li, X. , Bai, L. , Yuan, H. , Pan, Z. , Liu, R. , Chen, X. , WangMu, Q. , Chen, M. , Yu, L. , Liang, J. , DunZhu, D. , Zheng, Y. , Yu, S. , LuoBu, Z. , Guang, X. , Li, J. , Deng, C. , Hu, W. , Chen, C. , TaBa, X. , Gao, L. , Lv, X. , Abu, Y.Ben , Fang, X. , Nevo, E. , Yu, M. , Wang, J. and Tashi, N. (2015) The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc. Natl Acad. Sci. USA 112, 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X. , Guo, Y. , Xu, Q. , Mascher, M. , Guo, G. , Li, Shuaicheng , Mao, L. , Liu, Q. , Xia, Z. , Zhou, J. , Yuan, H. , Tai, S. , Wang, Y. , Wei, Z. , Song, L. , Zha, S. , Li, Shiming , Tang, Y. , Bai, L. , Zhuang, Z. , He, W. , Zhao, S. , Fang, X. , Gao, Q. , Yin, Y. , Wang, J. , Yang, H. , Zhang, J. , Henry, R.J. , Stein, N. and Tashi, N. (2018) Origin and evolution of qingke barley in Tibet. Nat. Commun. 9, 5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Studies mapping barley resistance/susceptibility genes to Pyrenophora teres f. teres. Different populations are indicated by alternating grey scale with the parent contributing the resistance allele in bold. Plant stage indicates whether the resistance is effective at the seedling or adult stage. If a single genotype isolate is used, this is indicated by the name and country of origin, whereas natural infection is indicated by specific location. Barley chromosome, phenotypic variation and designation of each locus are displayed, if available, from the corresponding reference. The inferred locus designation is reported based on markers obtained from the relevant publication and collapsed into loci using a maximum distance of 10 Mb of the Morex reference genome (Mascher et al., 2017) using BarleyMap (Cantalapiedra et al., 2015) or T3/Barley (Fig. 1).
Table S2 Studies mapping barley resistance/susceptibility genes to Pyrenophora teres f. maculata. Different populations are indicated by alternating grey scale with the parent contributing the resistance allele in bold. Plant stage indicates whether the resistance is effective at the seedling or adult stage. If a single genotype isolate is used this is indicated by the name and country of origin, whereas natural infection is indicated by specific location. Barley chromosome, phenotypic variation and designation of each locus are displayed if available from the corresponding reference. The inferred locus designation is reported based on markers obtained from the relevant publication and collapsed into loci using a maximum distance of 10 Mb of the Morex reference genome (Mascher et al., 2017) using BarleyMap (Cantalapiedra et al., 2015) or T3/Barley (Fig. 1).
Table S3 Genomic positions of markers reported in biparental or association mapping studies that could be anchored to the Morex barley reference genome (Mascher et al., 2017), including the locus designation (from publication), the corresponding marker, the assigned chromosome and chromosome position, and the reference.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


