Summary
Tomato yellow leaf curl virus (TYLCV), a begomovirus, causes large yield losses and breeding for resistance is an effective way to combat this viral disease. The resistance gene Ty‐1 codes for an RNA‐dependent RNA polymerase and has recently been shown to enhance transcriptional gene silencing of TYLCV. Whereas Ty‐1 was earlier shown to also confer resistance to a bipartite begomovirus, here it is shown that Ty‐1 is probably generic to all geminiviruses. A tomato Ty‐1 introgression line, but also stable transformants of susceptible tomato cv. Moneymaker and Nicotiana benthamiana (N. benthamiana) expressing the Ty‐1 gene, exhibited resistance to begomoviruses as well as to the distinct, leafhopper‐transmitted beet curly top virus, a curtovirus. Stable Ty‐1 transformants of N. benthamiana and tomato showed fewer symptoms and reduced viral titres on infection compared to wild‐type plants. TYLCV infections in wild‐type N. benthamiana plants in the additional presence of a betasatellite led to increased symptom severity and a consistent, slightly lowered virus titre relative to the high averaged levels seen in the absence of the betasatellite. On the contrary, in Ty‐1 transformed N. benthamiana viral titres increased in the presence of the betasatellite. The same was observed when these Ty‐1‐encoding plants were challenged with TYLCV and a potato virus X construct expressing the RNA interference suppressor protein βC1 encoded by the betasatellite. The resistance spectrum of Ty‐1 and the durability of the resistance are discussed in light of antiviral RNA interference and viral counter defence strategies.
Keywords: beet curly top virus, betasatellite, geminivirus, resistance, RNA interference, tomato yellow leaf curl virus, Ty‐1
Introduction
Tomato yellow leaf curl virus (TYLCV) is the representative of the Old World monopartite begomoviruses within the family Geminiviridae. The virus belongs to the most devastating plant viruses worldwide and causes major losses in economically important crops like tomato. The virus mainly occurs in the (sub)tropical and Mediterranean regions worldwide due to the distribution of its vector, the whitefly Bemisia tabaci (B. tabaci) (Moriones and Navas‐Castillo, 2000). TYLCV, and all monopartite begomoviruses, have a single, circular, single‐stranded DNA genome of c. 2.7 kb in size. In the field, monopartite begomoviruses are frequently observed with co‐replicating alpha‐ or betasatellites (Nawaz‐ul‐Rehman and Fauquet, 2009). Both satellites are approximately half the size of begomovirus genomes. While less is known about alphasatellites and their role in the pathogenesis of begomoviruses, co‐replication of betasatellites often leads to more severe disease symptoms (Zhou, 2013); therefore, betasatellites are regarded as pathogenicity determinants. Betasatellites code for one single protein, βC1, that is known to suppress the antiviral defence mechanism transcriptional gene silencing (TGS) (Yang et al., 2011; Zhou, 2013).
Because the vector B. tabaci is difficult to control, the most effective way to combat TYLCV infections is to breed for resistance. Six resistance genes are known, namely Ty‐1 to Ty‐6, and some of these are widely used for introgression breeding. Five of these genes are derived from wild tomato species: Ty‐1, Ty‐3, Ty‐4 and Ty‐6 are from Solanum chilense (S. chilense), Ty‐2 is from S. habrochaites, while ty‐5 was identified in an old commercial tomato cultivar Tyking (Hanson et al., 2006; Hutton and Scott, 2014; Hutton et al., 2012; Ji et al., 2007, 2009; Lapidot et al., 2015; Zamir et al., 1994). On viral challenge, plants containing any of these genes do not seem to completely abolish infection as reduced virus titres are still observed in comparison to susceptible cultivars, nor is a hypersensitive response seen (Castro et al., 2005; Maruthi et al., 2003; Picó et al., 1999, 2000). In recent years several of these genes have been cloned and characterized; only Ty‐4 and Ty‐6 are not. Ty‐2 encodes a nucleotide‐binding leucine‐rich repeat (NB‐LRR) protein, while the ty‐5 gene encodes an mRNA surveillance factor and contains a mutation that hampers viral protein translation (Lapidot et al., 2015; Yamaguchi et al., 2018). Ty‐1 and Ty‐3 are allelic and code for an RNA‐dependent RNA polymerase (RDR) from the γ‐class and distinct from the well‐established RDRs from the α‐class with recognized functions in the amplification of the antiviral RNA interference (RNAi) response (Verlaan et al., 2013). Until recently, no function was assigned to any of the RDRs from the γ‐class, but with the identification of Ty‐1 a new class of resistance genes was unveiled.
Besides a major role in gene regulation and chromosome dynamics, RNAi (also named RNA silencing) presents a major antiviral defence mechanism in plants (Shou‐Wei and Voinnet, 2007). The mechanism is induced by double‐stranded (ds) RNA molecules, which arise from viral replicative intermediates or secondary RNA folding structures in genomic or messenger RNA molecules. After their processing by Dicer‐like proteins (DCL) into small interfering (si) RNA molecules of 21–24 nucleotides (nt), one strand of the siRNA is uploaded into an RNA‐induced silencing complex (RISC), leading to its activation (Hammond, 2005). When this complex is loaded with a 21 nt siRNA strand and contains an Argonaute 1 (AGO1) core component, it is able to target RNA molecules with sequence complementarity to the siRNA strand, leading to their degradation or translational arrest (Mallory and Vaucheret, 2010). This process is generally referred to as post‐transcriptional gene silencing (PTGS). In contrast, AGO4 containing RISC is loaded with a 24 nt siRNA strand. This complex targets cytosine methyltransferases to complementary DNA molecules, causing cytosine methylation within the target sequence. This methylation leads to TGS of the DNA sequence (Mallory and Vaucheret, 2010).
The RNAi response in plants, in contrast to insects and animals, is amplified by RDRs (Donaire et al., 2008; Wang et al., 2010). During this process aberrant RNA molecules, resulting from the first cleavage by the slicer activity of AGO1, are recognized by RDRs and converted into more dsRNA molecules of the target sequence. Their processing by DCLs produces a second generation of siRNAs. Due to this amplification a strong (antiviral) RNAi response is mounted. In the Arabidopsis thaliana genome, six RDR genes have been annotated, encoding RDR1 to RDR6. From those, RDR1, RDR2 and RDR6 represent the α‐class, whereas RDR3, RDR4 and RDR5 fall into the γ‐class (Willmann et al., 2011). Silencing of RDR1 and RDR6 from the α‐class reduces the antiviral PTGS response and makes plants highly susceptible to RNA viruses, but the RNAi response against geminiviruses does not seem to be affected much by silencing of these RDRs (Aregger et al., 2012; Raja et al., 2008; Yu et al., 2003). This finding not only points to the involvement of other RDRs in the RNAi response against geminiviruses, but also supports the idea that the TGS pathway is the most important RNAi pathway against geminiviruses. In agreement with this hypothesis are findings showing that plants knocked out for proteins from the TGS pathway are hypersusceptible to geminivirus infection (Jackel et al., 2016; Raja et al., 2008). After the identification of Ty‐1 as an RDR γ‐class member, its involvement in the RNAi amplification was demonstrated by an increase of siRNA production and an elevated rate of cytosine methylation in viral DNA collected from TYLCV‐challenged Ty‐1 tomato plants (Butterbach et al., 2014).
While the Ty‐1 resistance gene is generally deployed to combat TYLCV, it has been shown to confer resistance to begomoviruses other than TYLCV (Barbieri et al., 2010; Butterbach et al., 2014; Pietersen and Smith, 2002; Prasanna et al., 2015; Shahid et al., 2013). However, whether the resistance is also effective against members of other genera, e.g. the Curtovirus genus, is still unknown. Furthermore, all previous studies on Ty‐1 so far have been conducted using introgression lines and therefore it still remains to be questioned whether the resistance is solely due to the expression of Ty‐1 or could involve other chromosomal introgression(s). Lastly, the earlier study by Butterbach et al. (2014) pointed towards an Achilles’ heel of Ty‐1 resistance, namely suppression of TGS by viral RNAi suppressor proteins. In that study the Ty‐1 resistance against TYLCV was compromised by a co‐infection with cucumber mosaic virus (CMV) (Butterbach et al., 2014). This effect was attributed to the CMV‐encoded RNAi suppressor 2b that was shown to inhibit AGO4 activity during TGS (González et al., 2010; Hamera et al., 2012). Since betasatellites encode suppressors of TGS, the compromising nature of betasatellites, by their encoded βC1 protein, on Ty‐1 resistance could be anticipated.
Here it is shown that Ty‐1 from a tomato introgression line, but also after stable transformation into susceptible tomato Moneymaker (MM) and Nicotiana benthamiana (N. benthamiana), confers resistance to a completely distinct leafhopper transmitted curtovirus, beet curly top virus (BCTV). Furthermore, TYLCV resistance in transgenic Ty‐1 plants is compromised not only by a co‐replicating betasatellite, but also by transient co‐expression of its encoded TGS suppressor protein only.
Results
Broad‐spectrum resistance against geminiviruses in Ty‐1 introgression line
Ty‐1 has been shown to confer resistance not only to the monopartite TYLCV, but also to the bipartite begomovirus tomato severe rugose virus (ToSRV) (Butterbach et al., 2014). In light of its role in the amplification of RNAi, a mechanism that is antiviral and generic to all viruses, it was anticipated that Ty‐1 resistance would not be restricted to species from the TYLCV cluster. To test this hypothesis and determine the resistance spectrum of Ty‐1, a Ty‐1 breeding line was challenged with various TYLCV‐like viral species, i.e. TYLCV isolates from Spain and China (both belonging to the TYLCV Israel species), tomato yellow leaf curl Sardinia virus (TYLCSV), as well as a completely distinct geminivirus, namely the leafhopper‐transmitted BCTV, which is a representative of the Curtovirus genus. Plants were monitored for 7 weeks by scoring disease symptoms and viral titres were determined. Whereas the susceptible control MM showed severe disease symptoms after a challenge with all (curto‐ and begomo‐) viruses, the Ty‐1 breeding line remained (almost completely) symptomless on a challenge with the TYLCV Almeria isolate, TYLCV‐[CN:SH2], TYLCSV and BCTV (Fig. 1A). Subsequently, the virus titres were determined via quantitative PCR (qPCR) and the fold difference in titre between the susceptible MM plants and the Ty‐1 introgression line calculated using the ΔΔC t method (Livak and Schmittgen, 2001). While in MM plants titres were high for all four viruses (C t values between 14 and 16), their amounts were significantly reduced in Ty‐1 introgression lines (Fig. 1B). Ty‐1 resistance seemed most effective to TYLCSV and TYLCV‐[CN:SH2], for which (hardly) no virus was detected in Ty‐1 plants (C t values between 32 and 34). TYLCV and BCTV were still clearly detected in Ty‐1 plants (C t values between 18 and 20), but TYLCV titres dropped c. 18‐fold and BCTV titres c. 3‐fold in Ty‐1 plants compared to MM plants (Fig. 1B).
Figure 1.
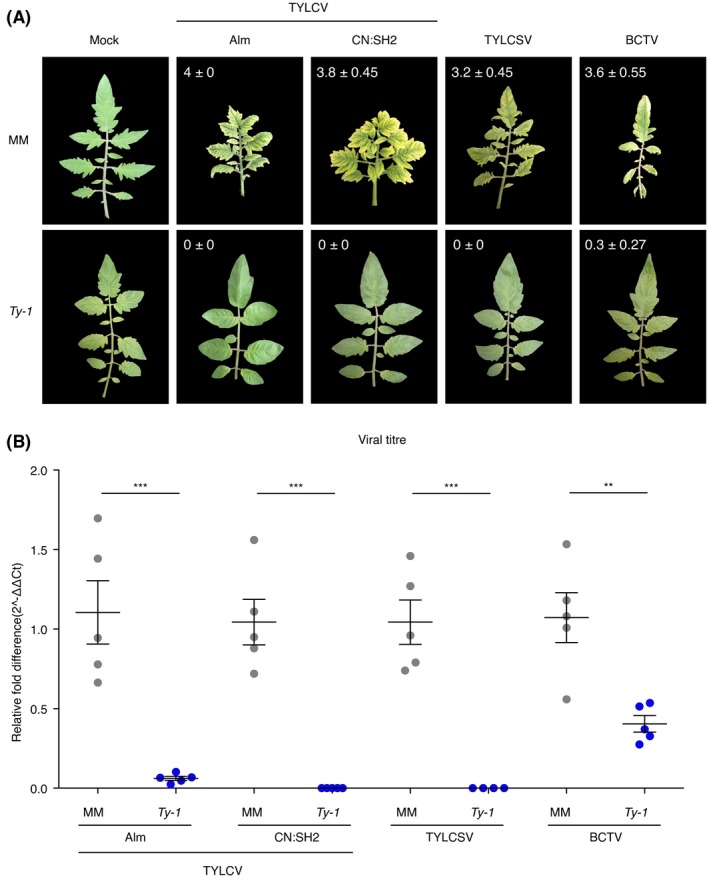
The Ty‐1 introgression line is resistant against a broad range of geminiviruses. (A) Symptoms upon infection with mock‐inoculation, TYLCV Almeria isolate (Alm), TYLCV‐[CN:SH2], TYLCSV and BCTV in susceptible control (Solanum lycopersicum 'Moneymaker', MM) and Ty‐1 introgression plants at 50 days post‐inoculation (dpi). In the left upper corners the symptom scores are depicted as mean of five biological replicates ± the standard deviation. (B) Virus titre quantification of TYLCV‐Alm, TYLCV‐[CN:SH2], TYLCSV and BCTV in MM and the Ty‐1 introgression line. Values were normalized relative to tomato EF1α and calibrated to the levels in MM plants (set to 1). Dots represent the relative virus titre of individual plants. Lines represent means and standard error of the mean of biological replicates. Asterisks indicate significant differences between Ty‐1 introgression line and MM according to one‐way analysis of variance (**P < 0.01, ***P < 0.001).
Transgenic expression of Ty‐1 in tomato confers broad‐spectrum resistance to geminiviruses
To rule out any other chromosomal introgression(s) being involved, and to ensure that the broad‐spectrum resistance was solely due to the expression of the Ty‐1 gene, susceptible tomato MM was transformed with a copy of the Ty‐1 gene for a functional complementation study. Agrobacterium‐mediated transformation of MM plants with a 35S promoter‐driven Ty‐1 construct resulted in nine primary transformants (T1). After selfing of the T1 plants, a T2 progeny was obtained that was challenged with TYLCV and monitored for phenotypic responses. In all the subsequent experiments, the Almeria isolate of TYLCV was used as representative of the Begomovirus genus, as was also done in previous Ty‐1 research (Butterbach et al., 2014; Verlaan et al., 2013). Several T2 families showed a clear segregation for TYLCV resistance (mild symptoms), correlated to the presence of the Ty‐1 transgene, as scored by a positive PCR amplification of the NPTII gene cassette and Ty‐1 insertion (data not shown). T2 plants showing resistance to TYLCV were used for selfing to produce a T3 generation. After another round of TYLCV challenges, two T3 families were selected from which all seedlings exhibited TYLCV resistance. These two families originated from two different primary transformants. A reverse transcription (RT)‐qPCR analysis on samples collected from these two families revealed a significant increase in the expression level of Ty‐1, which may vary between lines depending on integration site or number (Fig. 2A). The relative fold difference was calculated via the 2−ΔΔCt method, showing a 20‐ to 30‐fold increase in Ty‐1 expression levels in Ty‐1‐transformed lines relative to the ty‐1 allele (detected by the same primer set) from susceptible MM plants. Furthermore, a c. 10‐fold reduction was observed in TYLCV titres in the transgenic lines relative to the untransformed, susceptible MM plants (Fig. 2B). T3 individuals from these two lines that exhibited significantly higher Ty‐1 expression levels and reduced virus accumulation were selected and selfed to produce two homozygous T4 lines for further analysis (T4 lines 1 and 2).
Figure 2.
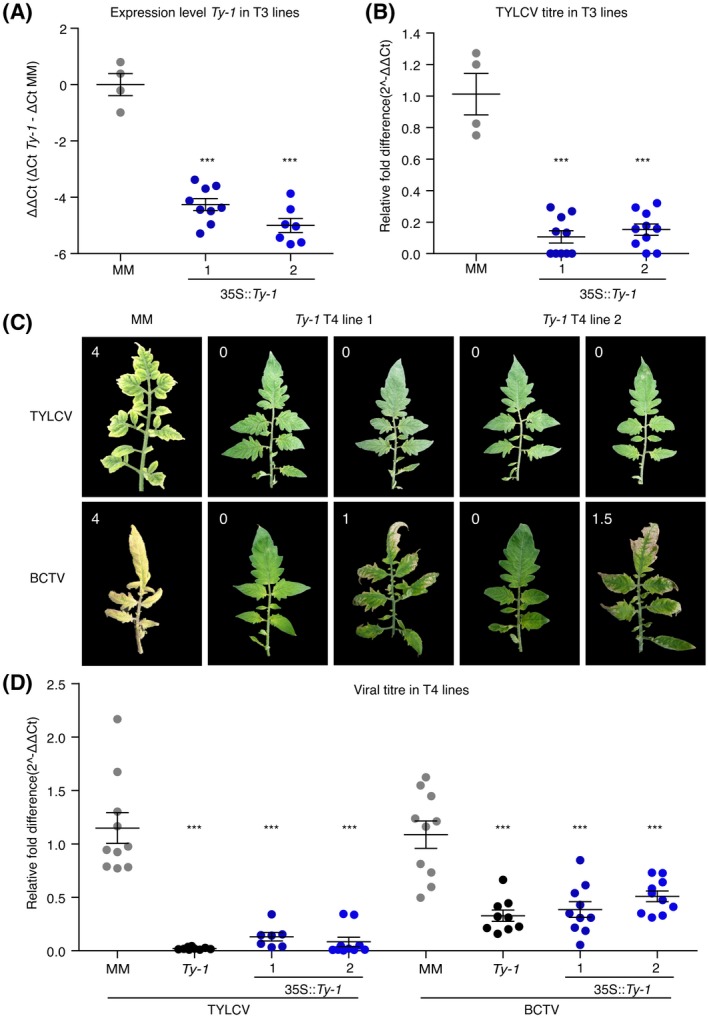
Transgenic tomato plants expressing Ty‐1 are resistant against both BCTV and TYLCV. For resistant control (Ty‐1 introgression line) and susceptible control (Solanum lycopersicum 'Moneymaker', MM), five plants were included; ten plants of each transgenic line were tested. In all graphs lines represent means and standard error of the mean of biological replicates, dots represent individual plants. (A) Transcript levels of Ty‐1 in T3 families. Values were normalized relative to EF1α. The ΔΔC t values (ΔC t Ty‐1 (C t Ty‐1 – C t EF1α) − ΔC t MM (C t Ty‐1 – C t EF1α)) are depicted, which represent Ty‐1 expression relative to the level of the susceptible ty‐1 allele in untransformed MM plants. Asterisks indicate significant differences between T3 and untransformed MM plants according to one‐way analysis of variance (***P < 0.001). (B) Virus titre quantification in T3 progeny of Ty‐1 transformants. Values were normalized relative to EF1α and calibrated to the levels in untransformed MM plants, which were set to 1. Asterisks indicate significant differences between T3 and untransformed MM plants according to one‐way analysis of variance (***P < 0.001). (C) Symptoms on infection with mock‐inoculation, TYLCV and BCTV in MM and in different individuals of two T4 families of Ty‐1‐transformed tomato plants at 50 days post‐inoculation (dpi). In the upper left corners, the disease symptom scores are depicted. (D) Virus titre quantification in plants from two T4 families (35S::Ty‐1), a Ty‐1 introgression line and untransformed MM. Values are calibrated to tomato EF1α and the fold difference was calculated compared to (susceptible) MM plants inoculated with TYLCV or BCTV (set to 1). Asterisks represent significant differences to the TYLCV‐ or BCTV‐challenged MM samples according to one‐way analysis of variance (***P < 0.001).
To complement and support the findings on the resistance spectrum of Ty‐1, batches of ten seedlings from the stable Ty‐1 transformants (T4 line 1 and 2) were challenged with TYLCV and BCTV, and monitored for their phenotypic responses. All seedlings from both lines showed resistance against TYLCV and displayed no disease symptoms (Fig. 2C). However, upon a challenge with BCTV, one out of ten plants from T4 line 1 and four out of ten from T4 line 2 ended up showing mild BCTV symptoms (Fig. 2C). Systemic leaves were collected at 50 days post‐inoculation (dpi) and viral titres of both TYLCV and BCTV were determined by qPCR in both lines and compared to the titres in MM and the Ty‐1 tomato introgression line. The results showed comparable levels of viral DNA accumulation for TYLCV and BCTV in both the Ty‐1 introgression line and Ty‐1 transgenic lines, and clearly reduced relative to the titres obtained from susceptible MM. However, the reduction in BCTV accumulation in both T4 lines was less profound than that of TYLCV (Fig. 2D). Plants from the two T4 lines that exhibited mild BCTV symptoms showed relatively higher BCTV titres compared to plants that remained symptomless.
Transgenic expression of Ty‐1 in N. benthamiana confers resistance to TYLCV and BCTV
To further substantiate the findings on broad‐spectrum resistance conferred by Ty‐1, and to test whether the Ty‐1 resistance gene also functions in other Solanaceae species, stable transformants of N. benthamiana plants were generated expressing Ty‐1 driven by a 35S promoter. In analogy to the transformation of tomato MM (previous section), stable transformants were selected based on a TYLCV resistant phenotype and selfed until T4. Expression levels of Ty‐1 were determined by RT‐qPCR in four transgenic lines selected from two independent transformations (Fig. 3A). The four transgenic lines showed a comparable high level of expression of Ty‐1. Whereas values for C t were in the range of c. 25 for all four transgenic lines, no signal was obtained from wild‐type (WT) N. benthamiana samples due to the absence of a ty‐1 homologue in WT N. benthamiana that could be amplified by the primers used. The C t values were normalized to the expression of the housekeeping gene Elongation Factor 1α (EF1α), and the ΔΔC t values were calculated to compare Ty‐1 transgenic plants with WT N. benthamiana, showing clear and high expression levels in Ty‐1 plants. On infection with TYLCV no clear symptoms were observed in all four lines (Fig. 3B), and when virus titres were determined a c. 100‐fold reduction was observed in the transgenic lines compared to WT N. benthamiana plants (Fig. 3C).
Figure 3.
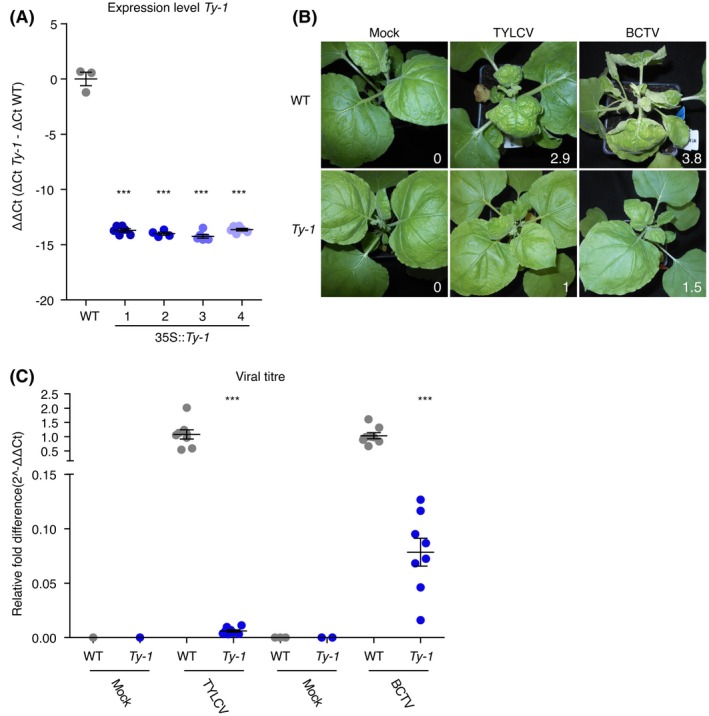
Ty‐1 transgenic Nicotiana benthamiana plants are resistant to TYLCV and BCTV. In all graphs, every point represents one plant and lines represent means and standard error of the mean of biological replicates. (A) Four independent transformant lines were generated and the expression level of Ty‐1 was measured. Values were normalized relative to EF1α. The ΔΔC t values (ΔC t Ty‐1 (C t Ty‐1 – C t EF1α) − ΔC t WT (Ct Ty‐1 – C t EF1α)) are depicted, which represent Ty‐1 expression relative to wild‐type (WT) plants. Asterisks indicate significant differences between WT plants and Ty‐1 transformations according to one‐way analysis of variance (***P < 0.001). (B) Symptoms upon infection with mock‐inoculation, TYLCV or BCTV in WT and Ty‐1 transgenic N. benthamiana plants. Pictures were taken at 18 days post‐inoculation (dpi) and the symptom scores are depicted in the lower right corners. (C) Virus titre quantification in WT and Ty‐1 transgenic N. benthamiana plants upon challenge with BCTV or TYLCV. 18 days post‐BCTV/TYLCV infection, young uninoculated leaves were harvested. Total DNA was isolated and viral titres were measured compared to the presence of genomic DNA with primers amplifying the gene for 25S rRNA. The relative fold difference is depicted, which is calculated via the 2−ΔΔCt method (WT infected plants set to 1). Note that the y‐axis is split to show the low titres. Asterisks indicate significant differences between WT plants and Ty‐1 transformants according to one‐way analysis of variance (***P < 0.001).
When the stable Ty‐1 N. benthamiana transformants were challenged with BCTV, only a slight curling and chlorosis (average symptom score 1.5) was observed at 18 dpi, whereas WT N. benthamiana revealed a strong curling and major chlorosis of systemic leaves (average symptom score 3.8) (Fig. 3B). BCTV titres showed a c. 15‐fold reduction relative to the titres obtained from susceptible N. benthamiana, which was smaller than the difference in TYLCV titres (between 70‐ and 180‐fold) (Fig. 3C). This smaller effect on BCTV compared to TYLCV was consistently observed during repeated experiments and resembled the observations in the Ty‐1 tomato introgression line and transgenic tomato.
Co‐replication of a betasatellite compromises resistance by Ty‐1
Earlier, a co‐infection of CMV was shown to compromise Ty‐1 resistance (Butterbach et al., 2014), likely due to the abrogation of AGO4 activity by the CMV 2b RNAi suppressor protein. Under natural field conditions TYLCV is often observed with co‐replicating betasatellites, elements that encode suppressors of TGS (Yang et al., 2011). To test whether betasatellites also compromise Ty‐1 resistance, susceptible MM and Ty‐1 introgression tomato plants were co‐infected with TYLCV and an ageratum yellow vein virus associated betasatellite (AYVB) via agroinfiltration. In parallel, WT N. benthamiana plants were infected with either TYLCV alone or in combination with the betasatellite. At 21 dpi, no differences in viral symptoms were observed between TYLCV singly infected and TYLCV plus betasatellite co‐infected tomato plants (results not shown). However, PCR analysis of systemically infected tomato leaves for the presence of the betasatellite turned out negative (data not shown), indicating that the betasatellite did not co‐replicate with TYLCV in tomato. In contrast to tomato, at 19 dpi curling and yellowing of systemically infected leaves was more severe in N. benthamiana plants co‐infected with TYLCV and the betasatellite compared to plants only infected with TYLCV (Fig. 4A). The presence of the betasatellite in systemically infected leaves was confirmed by PCR (data not shown). For this reason, the compromising nature of betasatellites, and their encoded TGS suppressor protein, on Ty‐1 resistance was further analysed in stably transformed N. benthamiana Ty‐1 plants. To this end, Ty‐1 stable transformants, next to WT N. benthamiana plants, were challenged with TYLCV singly or in a mixed setting with the betasatellite. At 19 dpi WT plants again showed yellowing and curling of the top leaves in the presence of TYLCV, whereas the transgenic Ty‐1 N. benthamiana plants only exhibited very mild symptoms. In the additional presence of the betasatellite both WT and transgenic plants exhibited more severe yellowing and curling, although the symptoms in the transgenic plants infected with TYLCV and the betasatellite were less pronounced than in WT plants infected with TYLCV only (average symptom score WT + TYLCV of 2.7, Ty‐1 + TYLCV + betasatellite of 2, see Fig. 4A). When virus titres were determined by qPCR, a co‐infection with the betasatellite led to a c. 2‐fold increase of TYLCV titres in systemically infected leaves from Ty‐1 transgenic plants, while a reduction in virus titres was observed in WT (susceptible) plants (Fig. 4B). This difference was observed in all four transgenic lines and during two independent repetitions.
Figure 4.
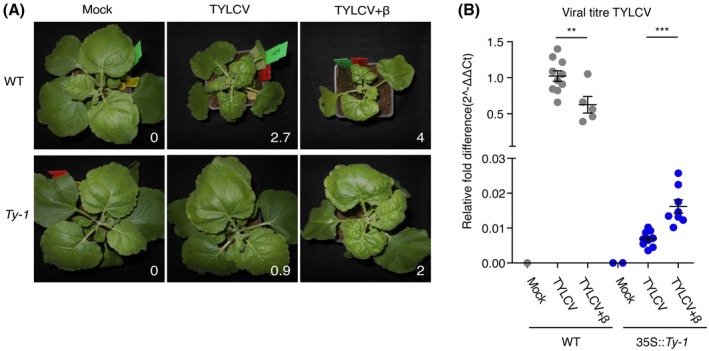
Breaking of resistance by co‐replication of AYVB in Ty‐1 transgenic Nicotiana benthamiana. (A) Pictures taken 19 days post‐inoculation (dpi) from wild‐type (WT) and 35S::Ty‐1 plants infected with TYLCV only or co‐infected with AYVB (denoted as β in the figure). The symptom score is indicated at the bottom right corner. (B) Systemically infected leaves of WT and 35S::Ty‐1 N. benthamiana plants were harvested 19 dpi and DNA was isolated. The viral titre was determined via quantitative PCR relative to the presence of genomic DNA (25S rRNA) and the relative fold difference was calculated via the 2−ΔΔCt method and normalized to the group WT infected with TYLCV (set to 1). Every point represents one plant. Lines represent means and standard error of the mean of biological replicates. Note that the y‐axis is split to show the low titres. Asterisks indicate significant differences between plants infected with TYLCV only or co‐infected with AYVB according to t‐test analysis (**P < 0.01; ***P < 0.001).
Co‐expression of βC1 is sufficient to compromise Ty‐1 resistance
To test whether the compromising effect of the betasatellite on Ty‐1 resistance against TYLCV was due to co‐replication of the betasatellite with TYLCV or to suppression of TGS by the betasatellite encoded protein βC1, experiments were repeated, but this time Ty‐1 stable transformants were challenged with TYLCV in the presence of a potato virus X (PVX) vector expressing the βC1 protein. Considering that so far βC1 protein TGS suppression activity has been reported only for tomato yellow leaf curl China virus associated betasatellite (TYLCCNB) and in the experiments here AYVB was used, TYLCCNB‐βC1 and a functionally deficient mutant were included as positive and negative control, respectively (Cui et al., 2005; Yang et al., 2011). Twenty‐five‐day‐old WT and Ty‐1 transgenic N. benthamiana plants were infected with TYLCV via agroinfiltration and at 10 dpi were challenged with PVX from which no protein, the βC1 protein of AYVB, the βC1 protein of TYLCCNB or the mutated TYLCCNB‐βC1 protein was expressed. The challenge with PVX was optimized at 10 days post‐TYLCV agroinfection due to PVX causing systemic infections much faster than TYLCV. Eight days post‐PVX infection plants were analysed for development of disease symptoms caused by TYLCV and/or PVX. Since PVX (empty vector) infections also caused chlorosis of the top leaves, these plants were more difficult to score for TYLCV symptoms. Whereas WT N. benthamiana plants infected with TYLCV showed symptoms as earlier observed (Fig. 5A), a mixed infection with PVX‐TYLCCNB‐βC1 or PVX‐AYVB‐βC1 intensified the curling and chlorosis (Fig. 5A) similarly to that during a co‐infection with the betasatellite (Fig. 4). During a co‐infection of TYLCV and the PVX‐TYLCCNB‐βC1 mutant (encoding a nonfunctional mutant βC1 TGS suppressor) only some necrosis and increased chlorosis was observed compared to TYLCV only. In the stably transformed Ty‐1 N. benthamiana plants, TYLCV only caused some slight chlorosis, but the additional presence of PVX‐AYVB‐βC1 or PVX‐TYLCCNB‐βC1, and not of PVX‐TYLCCNB‐βC1mutant, caused severe curling and chlorosis. To determine whether the increase in symptoms in the susceptible WT and Ty‐1 transgenic N. benthamiana plants also correlated with a higher TYLCV titre, samples were collected from systemically infected leaves and DNA purified for qPCR analysis. In WT N. benthamiana no significant change in virus titres was observed with TYLCV only versus a mixed infection with TYLCV and one of the PVX constructs (Fig. 5B). In the case of a mixed infection with PVX‐AYVB‐βC1 a trend of increased TYLCV titres was observed, which contrasted with the reduced titres seen in the presence of AYVB. In the Ty‐1 transgenic plants challenged with TYLCV, virus titres were again drastically reduced compared to WT plants, and these titres did not change with the addition of PVX‐empty or PVX‐TYLCCNB‐βC1 mutant. However, in the additional presence of PVX‐AYVB‐βC1 or PVX‐TYLCCNB‐βC1 TYLCV titres increased 2.7‐ and 2‐fold, respectively (Fig. 5B). These data indicate that the presence of a betasatellite compromises Ty‐1 resistance by means of its encoded TGS suppressor, the βC1 protein.
Figure 5.
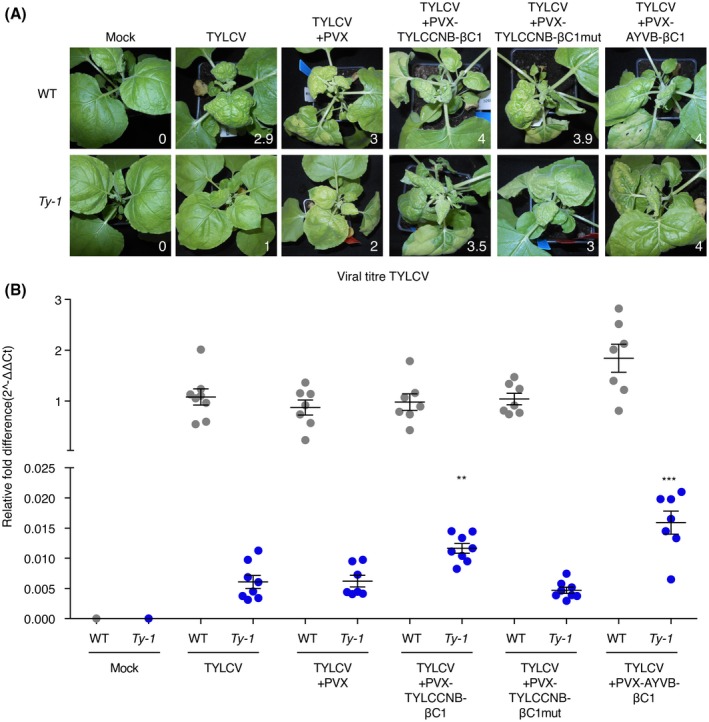
βC1 protein expressed via a PVX vector compromises resistance against TYLCV in Ty‐1 transgenic Nicotiana benthamiana. (A) Wild‐type (WT) and Ty‐1 transgenic N. benthamiana plants were infected with TYLCV and at 10 days post‐inoculation (dpi) were challenged with PVX expressing TYLCCNB‐βC1, TYLCCNB‐βC1 mutant, AYVB‐ βC1 or empty PVX vector. Eight days post‐PVX infection, pictures were taken and plants were scored for disease symptoms, indicated in the bottom right corner. (B) From the plants described under 5a, samples of systemically infected leaves were collected, DNA was isolated and the titre of TYLCV was determined via quantitative PCR. The viral titre was normalized to the presence of genomic DNA (25S rRNA) and the fold difference was calculated according to the 2−ΔΔCt method relative to WT plant infected with TYLCV only (set to 1). Every point represents one plant. Lines represent means and standard error of the mean of biological replicates. Note that the y‐axis is split to show the low titres. Asterisks indicate significant differences according to one‐way analysis of variance. In the analysis WT TYLCV‐only infected plants were compared with the different WT co‐infected groups and the same was done for the Ty‐1 transformants (**P < 0.01; ***P < 0.001).
Discussion
Earlier we showed that Ty‐1 confers resistance against the monopartite TYLCV (Israel strain, Almeria isolate) and the bipartite begomovirus ToSRV. We also showed that Ty‐1 resistance involves an amplification of the antiviral RNAi response, leading to increased levels of viral siRNA production and concomitant cytosine methylation within the viral DNA genome (Butterbach et al., 2014). Here it is shown that, besides TYLCV, Ty‐1 also confers resistance to TYLCSV, another species from the TYLCV cluster of begomoviruses, and to the completely distinct leafhopper‐transmitted curtovirus BCTV. Furthermore, stable transformants of susceptible tomato MM and N. benthamiana expressing a Ty‐1 transgene exhibited the same resistance spectrum as tomato Ty‐1 introgression lines, excluding the involvement of other resistance loci in this broad‐spectrum resistance. In the presence of a betasatellite, but also on trans‐complementation of the βC1 protein expressed from a PVX virus vector, Ty‐1 resistance against TYLCV was compromised. Altogether, these results demonstrate that Ty‐1 provides resistance to a wide range of geminiviruses, which is not restricted to begomoviruses only, and is compromised by suppressors of RNAi that interfere at TGS.
Whereas this study shows for the first time that Ty‐1 also hampers the replication of a curtovirus, the findings on the effectiveness of Ty‐1 against a group of TYLCV‐like viruses are supported and in agreement with the outcome of other studies. Others have reported on the efficacy of Ty‐1 against the monopartite TYLCV‐Mld (Shahid et al., 2013), TYLCSV (Barbieri et al., 2010), tomato curly stunt virus (ToCSV), tomato leaf curl Bangalore virus (ToLCBV), honeysuckle yellow vein mosaic virus (HYVMV) and tobacco leaf curl Japan virus (TbLCJV) (Pietersen and Smith, 2002; Prasanna et al., 2015; Shahid et al., 2013) and to multiple bipartite begomoviruses including tomato mottle virus (ToMoV), tomato leaf curl New Delhi virus (ToLCNDV) and tomato leaf curl Palampur virus (ToLCPalV) (Prasanna et al., 2015). In our earlier study, Ty‐1 was shown to confer resistance to the bipartite begomovirus ToSRV as well (Butterbach et al., 2014).
Ty‐1 does not confer resistance to RNA viruses, as recently shown for tomato spotted wilt virus (TSWV) and CMV (Butterbach et al., 2014). Also in the study presented here, the symptoms caused by a PVX infection were not different between WT and stably transformed Ty‐1 N. benthamiana. However, RNA viruses may compromise Ty‐1 resistance against geminiviruses, as shown earlier during a co‐infection of TYLCV and CMV. The observed increase in TYLCV titres in the latter case was explained to be due to inhibition of the TGS response by the CMV 2b RNAi suppressor protein (Butterbach et al., 2014; González et al., 2010; Hamera et al., 2012). This idea is strengthened by the current study, showing a compromising effect on Ty‐1 resistance by AYVB, and also solely by the AYVB βC1 and TYLCCNB βC1 proteins expressed from a PVX virus vector. TYLCCNB βC1 protein has been demonstrated to suppress TGS via inhibition of the enzyme S‐adenosylhomocysteine hydrolase (SAHH), an enzyme that is required for production of the methyl donor used in the TGS pathway (Yang et al., 2011). CMV 2b in contrast has been shown to hamper TGS via inhibition of AGO4 (González et al., 2010; Hamera et al., 2012). Despite the fact that these viral suppressors act differently, the ability to inhibit TGS at (any) different step(s) of the pathway apparently is sufficient to compromise Ty‐1‐mediated resistance.
Recently, Conflon et al. (2018) reported on the compromising effect of a co‐replicating betasatellite on Ty‐1 resistance. Ty‐1 tomato lines co‐infected with different TYLCV strains and a cotton leaf curl Gezira betasatellite revealed an increase in disease symptoms, but interestingly titres only increased in the mixed infection case with TYLCV‐Il, while those of TYLCV‐Mld were decreased on addition of the betasatellite. How to explain these effects caused by one and the same betasatellite remains to be further investigated, although it is likely that this has to do with the virus strains and not the betasatellite. Considering that the efficacy of Ty‐1 is compromised by trans‐complementing TGS suppressors, the suppressors encoded by the geminiviral DNA genome of TYLCV‐Il and TYLCV‐Mld will also affect Ty‐1 resistance. If their suppressors of RNAi differ in strength, Ty‐1‐mediated TGS will be suppressed to varying degrees, leading to different levels of reduction in virus titres. Although speculative, whether this also explains the observed difference in efficacy of Ty‐1 resistance towards TYLCV and BCTV remains to be analysed. To this end, initial attempts to detect TGS suppression by BCTV and TYLCV in 16C‐TGS transgenic plants (as shown by Buchmann et al., 2009) failed and remain to be repeated.
Recently several articles have been published on Ty‐1 resistance‐breaking strains of TYLCV in cultivations of Ty‐1‐bearing tomato. Samples of Ty‐1 plants showing TYLCV‐like symptoms collected in Morocco, Italy and Spain revealed the presence of viruses derived from a recombination event between TYLCV and TYLCSV in which a noncoding region between the origin of replication and the start of the coding region of V2 were exchanged (Belabess et al., 2015; Granier et al., 2019; Panno et al., 2018; Torre et al., 2019). In Morocco, this recombinant replaced both parental strains, but also under laboratory conditions this recombinant was positively selected in Ty‐1‐bearing plants (Belabess et al., 2015, 2016). So far, the mechanism behind the improved fitness of this resistance‐breaking strain is unknown, but the region of recombination covers the promoter region of the coat protein. The resistance breaking, therefore, might be due to (a combination of) changes in replication efficiency or viral gene expression, or the region of recombination might be less prone to TGS.
In conclusion, Ty‐1 is a unique resistance gene that confers resistance to a broad spectrum of geminiviruses, as demonstrated for begomoviruses and a curtovirus. Although Ty‐1 is likely to confer resistance to all geminiviruses, suppression of RNAi by co‐replicating betasatellites, or other RNA viruses, but also by suppressors encoded from the geminiviral DNA genome itself, seems to present the Achilles’ heel of the resistance mechanism. Whether Ty‐1 resistance can also be compromised by an infection involving a bacterial or fungal/oomycete pathogen, some of which are shown to either cause hypomethylation or suppress (the amplification of) RNAi (Hou et al., 2019; Navarro et al., 2008; Pavet et al., 2006), remains an interesting question. The effect of co‐infections on Ty‐1 resistance is important in light of disease management strategies, and simultaneously underlines the importance for monitoring the presence of other pathogens that encode suppressors of RNAi and interfere at the level of TGS during cultivation of Ty‐1‐bearing tomato plants.
Experimental Procedures
Plant material and virus sources
Throughout this study S. lycopersicum and N. benthamiana plants were maintained under greenhouse conditions at 23 °C during the day and at 21 °C at night (16 h light/8 h dark regime) and at a relative humidity of 60%. Tomato cv. Moneymaker (MM) was used as susceptible control and a Ty‐1 introgression line was derived from S. chilense LA1969 (Verlaan et al., 2013). Plants were infected with geminiviruses by means of agroinoculation of infectious clones. Agroinfectious clones used in this study were from two isolates of TYLCV, namely the Israel strain isolated from Almeria, Spain, as described by Morilla et al. (2005) (GenBank AJ489258.1), and the Israel strain isolated from Shanghai, China, named TYLCV‐[CN:SH2] [GenBank AM282874, (Zhang et al., 2009)], an isolate of TYLCSV [GenBank X61153.1 (Noris et al., 1998)] and an isolate of BCTV California Logan [GenBank M24597.2 (Stanley et al., 1986)]. Co‐infections were performed with an agroinfectious clone of AYVB [GenBank AJ252072.1 (Saunders et al., 2000)].
Transformation of tomato and N. benthamiana plants with Ty‐1
To generate a construct for transformation purposes, the Ty‐1 full‐length coding sequence was amplified from complementary DNA (cDNA) of Ty‐1 introgression lines with forward primer Ty‐1‐CDS‐F and reverse primer Ty‐1‐CDS‐R (for primer sequences see Table S1) (Verlaan et al., 2013). PCR products were cloned into the pENTR/D‐TOPO vector (Invitrogen, Carlsbad, CA, USA) and verified by sequence analysis prior to recombination into pK7WG2 via an LR reaction (Invitrogen). The resulting plasmid was transferred to Agrobacterium tumefaciens strain AGL1 by electroporation. Transformation of cotyledons from MM and N. benthamiana was carried out according to the method described by Appiano et al. (2015). Plants regenerated from kanamycin‐resistant primary transformants (T1) were selected and self‐pollinated to produce T2 families. From each segregating T2 family, transgenic lines were selected by PCR amplification of the NPTII gene cassette [using primers NPT3 and NPT4 (Heilersig et al., 2006), amplifying a 722‐bp fragment] and a partial Ty‐1 gene sequence (using primers CaMV 35S promoter primer 35S‐F and gene insert‐specific primer Ty‐1‐R, amplifying a 822‐bp fragment).
PVX constructs
PVX infectious clones encoding the betasatellite C1 protein gene were constructed as follows. The open reading frame of AYVB C1 was amplified from the infectious clone (GenBank AJ252072.1) using primers AYVB‐C1‐F and AYVB‐C1‐R (Table S1). Sequences for C1 and a C1 mutant (Cui et al., 2005) of the TYLCCNB were ordered as a gene block from Integrated DNA Technologies (IDT, Leuven, Belgium) and were based on GenBank accession AJ421621.1. The gene blocks were cloned into pJet1.2 according to the manufacturers’ protocol (Thermo Fisher, Waltham, MA, USA), and their sequences verified. Next, the TYLCCNB C1 gene was amplified with primers TYLCCNB‐C1‐F and TYLCCNB‐C1‐R containing a 5’ ClaI or SalI restriction site. Amplicons were purified and subsequently digested with SalI and ClaI prior to cloning into the SalI‐ and ClaI‐digested PVX vector pGR107 (Jones et al., 1999). Positive clones were selected and verified by sequence analysis. The Agrobacterium strain GV3101, containing the helper plasmid pSoup, was transformed with the constructs PVX‐AYVB‐βC1, PVX‐TYLCCNB‐βC1, PVX‐TYLCCNB‐βC1mut or with the empty pGR107 plasmid for subsequent agroinfiltration of plants.
Agrobacterium transient transformation assay
Agrobacterium transient transformation assays were performed following a slightly modified protocol of Bucher et al. (2003). In brief, A. tumefaciens was grown overnight at 28 °C in 3 mL LB3 (10 g/L tryptone, 5 g/L yeast, 4 g/L NaCl, 1 g/L KCl, 3 g/L MgSO4.2H2O) medium containing proper antibiotic selection pressure. From this culture, 600 μL was transferred and incubated overnight in 3 mL induction medium [10.5 g/L K2HPO4, 4.5 g/L KH2PO4, 1 g/L(NH4)2SO4, 0.5 g/L sodium citrate.2H2O, 1 mM MgSO4.7H2O, 0.2% (w/v) glucose, 0.5% (v/v) glycerol, 50 μM acetosyringone, 10 mM 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.6]. The next day bacteria were pelleted by centrifugation (15 min, 2670 g) and resuspended in MS MES buffer [Murashige and Skoog medium (Duchefa Biochemie, Haarlem, Netherlands) supplemented with 150 μM acetosyringone, 10 mM MES and 87 mM sucrose] at an OD600 of 0.5 per construct. One hour after plants were watered in excess, the first two true leaves were fully infiltrated with this mixture by pressure inoculation with a needleless syringe on the abaxial side of the leaf.
Disease assessment
Plant responses were scored several times during the whole disease assay on viral symptom development. The time span of the disease assay varied per experiment and lasted up to 50 days. Depending on the lines and experiments, five to ten plants were challenged. Each plant was rated using a 0 to 4 disease severity index (DSI) described by Friedmann et al. (1998), where 0 indicates no viral disease symptoms and 4 means severe symptoms. Intermediate scores (0.5, 1.5, 2.5 and 3.5) were incorporated for more precise scoring.
Nucleic acid purification
Systemically infected leaves were harvested and snap frozen in liquid nitrogen. The samples were ground either by mortar and pestle or by using precellys (Bertin Instruments, Montigny‐le‐Bretonneux, France) for 10 s at 5000 rpm. To determine geminivirus titres, DNA was isolated following the cetyltrimethyl ammonium bromide (CTAB) method of Doyle and Doyle (1987) with slight modifications as described by Fulton et al. (1995). In brief, ground plant material was incubated for 1 h at 65 °C with CTAB buffer (0.1 M Tris, 0.7 M NaCl, 0.01 M EDTA, 2% CTAB). After chloroform/isoamyl alcohol extraction and spinning of the samples, the upper aqueous phase was transferred to a new tube and the DNA precipitated from the suspension by adding isopropanol in a 1:1 ratio. DNA was pelleted by centrifugation for 10 min at 9000 g, dried and dissolved in Milli‐Q water. For Ty‐1 expression analysis, RNA from N. benthamiana plants was extracted using TRIzol according to the manufacturers’ protocol (Invitrogen) and RNA from tomato plants by using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. DNA and RNA concentrations were measured with a NanoDrop ND‐1000 device.
Detection of the betasatellite
DNA of AYVB was amplified using AYVB‐specific primers AYVB‐F1, AYVB‐F2 and AYVB‐R (Table S1). The combination of AYVB‐F1 and AYVB‐R resulted in a product of 823 bp, whereas AYVB‐F2 and AYVB‐R resulted in a product of 457 bp. The PCR was performed with GoTaq polymerase according to the manufacturers’ protocol (Promega, Madison, WI, USA), using the following cycling conditions: 5 min at 95 °C; 40 cycles of 30 s at 95 °C, 30 s at 55 °C, 60 s at 72 °C; 7 min at 72 °C. Amplified PCR products were visualized in a 1% agarose gel using ethidium bromide.
Virus titration
Relative virus titres of the TYLCV Almeria isolate, TYLCSV, TYLCV‐[CN:SH2] and BCTV were determined by qPCR using 25S rRNA (N. benthamiana) or EF1α (S. lycopersicum, Solyc06g005060) as internal control. To measure samples of N. benthamiana, the reaction mixture contained 1× Sybr Select (Applied Biosystems, Foster City, CA, USA), 375 nM forward primer, 375 nM reverse primer and 10 ng genomic DNA, while for S. lycopersicum samples the iQ SYBR Green supermix (Bio‐Rad, Hercules, CA, USA) was used. For PCR amplification the following primers were used (Table S1): for TYLCV Almeria isolate primers TYLCV‐Alm‐qPCR‐F and TYLCV‐Alm‐qPCR‐R (Powell et al., 2012), for TYLCSV primers TYLCSV‐qPCR‐F and TYLCSV‐qPCR‐R, for TYLCV‐[CN:SH2] primers TYLCV‐CN‐qPCR‐F and TYLCV‐CN‐qPCR‐R, for BCTV either primers BCTV‐qPCR‐F1 and BCTV‐qPCR‐R1 or BCTV‐qPCR‐F2 and BCTV‐qPCR‐R2. During qPCR 25S rRNA was amplified using primers 25S‐qPCR‐F and 25S‐qPCR‐R, and EF1α using primers SlEF1a‐qPCR‐F and SlEF1a‐qPCR‐R.
The qPCR was performed in a Bio‐Rad CFX384 using the following cycling conditions: 2 min at 95 °C, 40 cycles of 15 s at 95 °C and 1 min at 60 °C, followed by a melting curve with 0.5 °C steps from 60 to 95 °C to determine PCR specificity. Relative viral titres were calculated using the ΔΔC t method (Livak and Schmittgen, 2001). Values were normalized relative to the internal control 25S rRNA or EF1α, and calibrated to levels of the control plants, which were set as 1.
Ty‐1 gene expression analysis
Prior to gene expression analysis 1 µg of purified total RNA of N. benthamiana samples was treated with TURBO DNase (Invitrogen) following the manufacturer’s instructions. First‐strand cDNA was synthesized using random hexamers (Roche, Basel, Switzerland) and M‐MLV reverse transcriptase according to the manufacturers’ protocol (Promega, Madison, WI, USA). The RT‐qPCR was performed in a total volume of 10 µL, containing 1 µL of 5× diluted cDNA, 1× Sybr Select (Applied Biosystems), 375 nM forward primer and 375 nM reverse primer. Total RNA of S. lycopersicum was treated with DNase I, Amplification Grade (Invitrogen) following the manufacturer’s instructions. cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio‐Rad) and RT‐qPCR performed with the iQ SYBR Green supermix (Bio‐Rad). To determine Ty‐1 expression levels, either Ty‐1‐qPCR‐F1 and Ty‐1‐qPCR‐R1 or Ty‐1‐qPCR‐F2 and Ty‐1‐qPCR‐R2 primers were used. The gene expression was measured relative to the housekeeping gene EF1α by using the primers nbEF1α‐qPCR‐F and nbEF1α‐qPCR‐R in the case of N. benthamiana samples or SlEF1a‐qPCR‐F and SlEF1a‐qPCR‐R in the case of S. lycopersicum samples (Table S1). RT‐qPCR was performed in a Bio‐Rad CFX384 using the same conditions as for the virus titration. Relative gene expression was calculated using the formula 2^−(ΔC t Ty‐1 (C t Ty‐1 – C t EF1α) − ΔC t MM (C t Ty‐1 – C t EF1α)) (Livak and Schmittgen, 2001).
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Table S1 List of sequences of primers used in this research.
Acknowledgements
The authors would like to thank Prof. Eduardo Rodríguez Bejarano (Universidad de Málaga, Malaga, Spain) for providing the infectious TYLCV (Almeria isolate) clone, Dr Emanuela Noris (Institute of Plant Virology, Torino, Italy) for providing the infectious clone of TYLCSV and Dr Keith Saunders (John Innes Centre, Norwich, UK) for providing the infectious clone of BCTV and AYVB. The authors would like to thank Annelies E.H.M. Loonen for making the primary tomato and N. benthamiana transformants. Monique van Oers (Wageningen University) is acknowledged for critically reading the manuscript prior to submission. This research was financially supported by NWO‐CNPq within the Joint Research Project Biobased Economy (729.004.011), Plant Breeding of Wageningen University & Research and a vegetable breeding company.
References
- Appiano, M. , Pavan, S. , Catalano, D. , Zheng, Z. , Bracuto, V. , Lotti, C. , Visser, R.G.F. , Ricciardi, L. and Bai, Y. (2015) Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1 . Transgenic Res. 24, 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aregger, M. , Borah, B.K. , Seguin, J. , Rajeswaran, R. , Gubaeva, E.G. , Zvereva, A.S. , Windels, D. , Vazquez, F. , Blevins, T. , Farinelli, L. and Pooggin, M.M. (2012) Primary and secondary siRNAs in geminivirus‐induced gene silencing. PLoS Pathog. 8, e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri, M. , Acciarri, N. , Sabatini, E. , Sardo, L. , Accotto, G.P. and Pecchioni, N. (2010) Introgression of resistance to two Mediterranean virus species causing tomato yellow leaf curl into a valuable traditional tomato variety. J. Plant Pathol. 92, 485–493. [Google Scholar]
- Belabess, Z. , Dallot, S. , El‐Montaser, S. , Granier, M. , Majde, M. , Tahiri, A. , Blenzar, A. , Urbino, C. and Peterschmitt, M. (2015) Monitoring the dynamics of emergence of a non‐canonical recombinant of tomato yellow leaf curl virus and displacement of its parental viruses in tomato. Virology, 486, 291–306. [DOI] [PubMed] [Google Scholar]
- Belabess, Z. , Peterschmitt, M. , Granier, M. , Tahiri, A. , Blenzar, A. and Urbino, C. (2016) The non‐canonical tomato yellow leaf curl virus recombinant that displaced its parental viruses in southern Morocco exhibits a high selective advantage in experimental conditions. J. Gen. Virol. 97, 3433–3445. [DOI] [PubMed] [Google Scholar]
- Bucher, E. , Sijen, T. , Haan, P.De , Goldbach, R. and Prins, M. (2003) Negative‐strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J. Virol. 77, 1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann, R.C. , Asad, S. , Wolf, J.N. , Mohannath, G. and Bisaro, D.M. (2009) Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome‐wide reductions in cytosine methylation. J. Virol. 83, 5005–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterbach, P. , Verlaan, M.G. , Dullemans, A. , Lohuis, D. , Visser, R.G.F. , Bai, Y. and Kormelink, R. (2014) Tomato yellow leaf curl virus resistance by Ty‐1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. USA. 111, 12942–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, A.P.De , Díez, M.J. and Nuez, F. (2005) Evaluation of breeding tomato lines partially resistant to tomato yellow leaf curl Sardinia virus and tomato yellow leaf curl virus derived from Lycopersicon chilense . Can. J. Plant Pathol. 27, 268–275. [Google Scholar]
- Conflon, D. , Granier, M. , Tiendrébéogo, F. , Gentit, P. , Peterschmitt, M. and Urbino, C. (2018) Accumulation and transmission of alphasatellite, betasatellite and tomato yellow leaf curl virus in susceptible and Ty‐1‐resistant tomato plants. Virus Res. 253, 124–134. [DOI] [PubMed] [Google Scholar]
- Cui, X. , Li, G. , Wang, D. , Hu, D. and Zhou, X. (2005) A begomovirus DNAβ‐encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 79, 10764–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire, L. , Barajas, D. , Martínez‐García, B. , Martínez‐Priego, L. , Pagán, I. and Llave, C. (2008) Structural and genetic requirements for the biogenesis of tobacco rattle virus‐derived small interfering RNAs. J. Virol. 82, 5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J.J. and Doyle, J.L. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Friedmann, M. , Lapidot, M. , Cohen, S. and Pilowsky, M. (1998) A novel source of resistance to tomato yellow leaf curl virus exhibiting a symptomless reaction to viral infection. J. Am. Soc. Hortic. Sci. 123, 1004–1007. [Google Scholar]
- Fulton, T.M. , Chunwongse, J. and Tanksley, S.D. (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Report. 13, 207–209. [Google Scholar]
- González, I. , Martínez, L. , Rakitina, D.V. , Lewsey, M.G. , Atencio, F.A. , Llave, C. , Kalinina, N.O. , Carr, J.P. , Palukaitis, P. and Canto, T. (2010) Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol. Plant–Microbe Interact. 23, 294–303. [DOI] [PubMed] [Google Scholar]
- Granier, M. , Tomassoli, L. , Manglli, A. , Nannini, M. , Peterschmitt, M. and Urbino, C. (2019) First report of TYLCV‐IS141, a tomato yellow leaf curl virus recombinant infecting tomato plants carrying the Ty‐1 resistance gene in Sardinia (Italy). Plant Dis. 103, 1437. [Google Scholar]
- Hamera, S. , Song, X. , Su, L. , Chen, X. and Fang, R. (2012) Cucumber mosaic virus suppressor 2b binds to AGO4‐related small RNAs and impairs AGO4 activities. Plant J. 69, 104–115. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M. (2005) Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 579, 5822–5829. [DOI] [PubMed] [Google Scholar]
- Hanson, P. , Green, S.K. and Kuo, G. (2006) Ty‐2, a gene on chromosome 11 conditioning geminivirus resistance in tomato. Tomato Genet. Coop. Rep. 56, 17–18. [Google Scholar]
- Heilersig, H.J.B. , Loonen, A. , Bergervoet, M. , Wolters, A.M.A. and Visser, R.G.F. (2006) Post‐transcriptional gene silencing of GBSSI in potato: effects of size and sequence of the inverted repeats. Plant Mol. Biol. 60, 647–662. [DOI] [PubMed] [Google Scholar]
- Hou, Y. , Zhai, Y. , Feng, L. , Karimi, H.Z. , Rutter, B.D. , Zeng, L. , Choi, D.S. , Zhang, B. , Gu, W. , Chen, X. , Ye, W. , Innes, R.W. , Zhai, J. and Ma, W. (2019) A Phytophthora effector suppresses trans‐kingdom RNAi to promote disease susceptibility. Cell Host Microbe, 25, 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton, S.F. and Scott, J.W. (2014) Ty‐6, a major begomovirus resistance gene located on chromosome 10. Rep. Tomato Genet. Coop. 64, 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton, S.F. , Scott, J.W. and Schuster, D.J. (2012) Recessive resistance to tomato yellow leaf curl virus from the tomato cultivar Tyking is located in the same region as Ty‐5 on chromosome 4. HortScience, 47, 324–327. [Google Scholar]
- Jackel, J.N. , Storer, J.M. , Coursey, T. and Bisaro, D.M. (2016) Arabidopsis RNA polymerases IV and V are required to establish H3K9 methylation, but not cytosine methylation, on geminivirus chromatin. J. Virol. 90, 7529–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y. , Schuster, D.J. and Scott, J.W. (2007) Ty‐3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty‐1 on chromosome 6 of tomato. Mol. Breed. 20, 271–284. [Google Scholar]
- Ji, Y. , Scott, J.W. and Schuster, D.J. (2009) Molecular mapping of Ty‐4, a new tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. J. Am. Soc. Hortic. Sci. 134, 281–288. [Google Scholar]
- Jones, L. , Hamilton, A.J. , Voinnet, O. , Thomas, C.L. , Maule, A.J. and Baulcombe, D.C. (1999) RNA–DNA interactions and DNA methylation in post‐transcriptional gene silencing. Plant Cell, 11, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot, M. , Karniel, U. , Gelbart, D. , Fogel, D. , Evenor, D. , Kutsher, Y. , Makhbash, Z. , Nahon, S. , Shlomo, H. , Chen, L. , Reuveni, M. and Levin, I. (2015) A novel route controlling begomovirus resistance by the messenger RNA surveillance factor Pelota. PLoS Genet. 11, e1005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the −2 ΔΔCt method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mallory, A. and Vaucheret, H. (2010) Form, function, and regulation of ARGONAUTE proteins. Plant Cell, 22, 3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthi, M.N. , Czosnek, H. , Vidavski, F. , Tarba, S.Y. , Milo, J. , Leviatov, S. , Venkatesh, H.M. , Padmaja, A.S. , Kulkarni, R.S. and Muniyappa, V. (2003) Comparison of resistance to tomato leaf curl virus (India) and tomato yellow leaf curl virus (Israel) among Lycopersicon wild species, breeding lines and hybrids. Eur. J. Plant Pathol. 109, 1–11. [Google Scholar]
- Morilla, G. , Janssen, D. , García‐Andrés, S. , Moriones, E. , Cuadrado, I.M. and Bejarano, E.R. (2005) Pepper (Capsicum annuum) is a dead‐end host for tomato yellow leaf curl virus. Phytopathology, 95, 1089–1097. [DOI] [PubMed] [Google Scholar]
- Moriones, E. and Navas‐Castillo, J. (2000) Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 71, 123–134. [DOI] [PubMed] [Google Scholar]
- Navarro, L. , Jay, F. , Nomura, K. , He, S.Y. and Voinnet, O. (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science, 321, 964–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz‐ul‐Rehman, M.S. and Fauquet, C.M. (2009) Evolution of geminiviruses and their satellites. FEBS Lett. 583, 1825–1832. [DOI] [PubMed] [Google Scholar]
- Noris, E. , Vaira, A.M. , Caciagli, P. , Masenga, V. , Gronenborn, B. and Accotto, G.P. (1998) Amino acids in the capsid protein of tomato yellow leaf curl virus that are crucial for systemic infection, particle formation, and insect transmission. J. Virol. 72, 10050–10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panno, S. , Caruso, A.G. and Davino, S. (2018) The nucleotide sequence of a recombinant tomato yellow leaf curl virus strain frequently detected in Sicily isolated from tomato plants carrying the Ty‐1 resistance gene. Arch. Virol. 163, 795–797. [DOI] [PubMed] [Google Scholar]
- Pavet, V. , Quintero, C. , Cecchini, N.M. , Rosa, A.L. and Alvarez, M.E. (2006) Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae . Mol. Plant–Microbe Interact. 19, 577–587. [DOI] [PubMed] [Google Scholar]
- Picó, B. , Ferriol, M. , Díez, M.J. and Nuez, F. (1999) Developing tomato breeding lines resistant to tomato yellow leaf curl virus. Plant Breed. 118, 537–542. [DOI] [PubMed] [Google Scholar]
- Picó, B. , Sifres, A. , Elia, M. , Diez, M.J. and Nuez, F. (2000) Searching for new resistance sources to tomato yellow leaf curl virus within a highly variable wild Lycopersicon genetic pool. Acta Physiol. Plant. 22, 344–350. [Google Scholar]
- Pietersen, G. and Smith, M.F. (2002) Tomato yellow leaf curl virus resistant tomatoes show resistance to tomato curly stunt virus. Plant Dis. 86, 528–534. [DOI] [PubMed] [Google Scholar]
- Powell, M.E. , Cuthbertson, A.G.S. , Bell, H.A. , Boonham, N. , Morris, J. and Northing, P. (2012) First record of the Q biotype of the sweetpotato whitefly, Bemisia tabaci, intercepted in the UK. Eur. J. Plant Pathol. 133, 797–801. [Google Scholar]
- Prasanna, H.C. , Sinha, D.P. , Rai, G.K. , Krishna, R. , Kashyap, S.P. , Singh, N.K. , Singh, M. and Malathi, V.G. (2015) Pyramiding Ty‐2 and Ty‐3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathol. 64, 256–264. [Google Scholar]
- Raja, P. , Sanville, B.C. , Buchmann, R.C. and Bisaro, D.M. (2008) Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82, 8997–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, K. , Bedford, I.D. , Briddon, R.W. , Markham, P.G. , Wong, S.M. and Stanley, J. (2000) A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA. 97, 6890–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid, M.S. , Ito, T. , Kimbara, J. , Onozato, A. , Natsuaki, K.T. and Ikegami, M. (2013) Evaluation of tomato hybrids carrying Ty‐1 and Ty‐2 loci to Japanese monopartite begomovirus species. J. Phytopathol. 161, 205–209. [Google Scholar]
- Shou‐Wei, D. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley, J. , Markham, P.G. , Callis, R.J. and Pinner, M.S. (1986) The nucleotide sequence of an infectious clone of the geminivirus beet curly top virus. EMBO J. 5, 1761–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre, C. , Agüero, J. and Aranda, M.A. (2019) First evidence of tomato yellow leaf curl virus‐Israel IS76 recombinant isolates associated with severe yellow leaf curl epidemics in resistant tomatoes in Spain. Plant Dis. 103, 780–781. [Google Scholar]
- Verlaan, M.G. , Hutton, S.F. , Ibrahem, R.M. , Kormelink, R. , Visser, R.G.F. , Scott, J.W. , Edwards, J.D. and Bai, Y. (2013) The tomato yellow leaf curl virus resistance genes Ty‐1 and Ty‐3 are allelic and code for DFDGD‐class RNA‐dependent RNA polymerases. PLoS Genet. 9, e1003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.‐B. , Wu, Q. , Ito, T. , Cillo, F. , Li, W.‐X. , Chen, X. , Yu, J.‐L. and Ding, S.‐W. (2010) RNAi‐mediated viral immunity requires amplification of virus‐derived siRNAs in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA. 107, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann, M.R. , Endres, M.W. , Cook, R.T. and Gregory, B.D. (2011) The functions of RNA‐dependent RNA polymerases in Arabidopsis. Arab. B. 9, e0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, H. , Ohnishi, J. , Saito, A. , Ohyama, A. , Nunome, T. , Miyatake, K. and Fukuoka, H. (2018) An NB‐LRR gene, TYNBS1, is responsible for resistance mediated by the Ty‐2 begomovirus resistance locus of tomato. Theor. Appl. Genet. 131, 1345–1362. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Xie, Y. , Raja, P. , Li, S. , Wolf, J.N. , Shen, Q. , Bisaro, D.M. and Zhou, X. (2011) Suppression of methylation‐mediated transcriptional gene silencing by βC1–sahh protein interaction during geminivirus‐betasatellite infection. PLoS Pathog. 7, e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D. , Fan, B. , MacFarlane, S.A. and Chen, Z. (2003) Analysis of the involvement of an inducible Arabidopsis RNA‐dependent RNA polymerase in antiviral defense. Mol. Plant–Microbe Interact. 16, 206–216. [DOI] [PubMed] [Google Scholar]
- Zamir, D. , Ekstein‐Michelson, I. , Zakay, Y. , Navot, N. , Zeidan, M. , Sarfatti, M. , Eshed, Y. , Harel, E. , Pleban, T. , Van‐Oss, H. and Kedar, N. (1994) Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY‐1 . Theor. Appl. Genet. 88, 141–146. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Gong, H. and Zhou, X. (2009) Molecular characterization and pathogenicity of tomato yellow leaf curl virus in China. Virus Genes, 39, 249–255. [DOI] [PubMed] [Google Scholar]
- Zhou, X. (2013) Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 51, 357–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 List of sequences of primers used in this research.


