Abstract
Protein folding within the endoplasmic reticulum (ER) exists in a delicate balance; perturbations of this balance can overload the folding capacity of the ER and disruptions of ER homoeostasis is implicated in numerous diseases. The unfolded protein response (UPR), a complex adaptive stress response, attempts to restore normal proteostasis, in part, through the up-regulation of various foldases and chaperone proteins including redox-active protein disulphide isomerases (PDIs). There are currently over 20 members of the PDI family each consisting of varying numbers of thioredoxin-like domains which, generally, assist in oxidative folding and disulphide bond rearrangement of peptides. While there is a large amount of redundancy in client proteins of the various PDIs, the size of the family would indicate more nuanced roles for the individual PDIs. However, the role of individual PDIs in disease pathogenesis remains uncertain. The following review briefly discusses recent findings of ER stress, the UPR and the role of individual PDIs in various respiratory disease states.
Keywords: disulphide bond, ER stress, PDI, pulmonary disease, UPR
The endoplasmic reticulum (ER) is a highly specialized organelle that plays numerous roles in the cell. It is the primary site of the synthesis of membrane-bound and secreted proteins, and as such maintains an oxidizing redox environment to facilitate the formation of disulphide bonds required for the stabilization of peptide structure (1). Additionally, numerous posttranslational modifications such as N-inked glycosylation occur solely within the ER (2). Approximately one-third of all proteins that traffic through the ER contain disulphide bonds and the ER contains a vast array of chaperones and foldases to assist in the folding of newly synthesized peptides (1, 3). Properly folded proteins are essential for the normal function of the cell, and potentially misfolded proteins are rapidly degraded by the ER-associated degradation (ERAD) (4). Under basal conditions, ∼30% of newly synthesized peptides are targeted for degradation (5).
Protein folding within the ER exists in a delicate balance, and the physiological states of increased protein synthesis can quickly overload the folding capacity of the ER leading to a buildup of unfolded or misfolded peptides in the ER lumen, termed ER stress. In an effort to combat this stress the cell activates the unfolded protein response (UPR), a highly conserved, multifaceted stress response aimed at restoring normal ER homoeostasis (6). Collectively the UPR attenuates normal protein synthesis, up-regulates ERAD machinery, increases the size of the ER and up-regulates various chaperones, including protein disulphide isomerases (PDIs) a large family of proteins that assists in the oxidative folding of nascent peptides (6). Failure of the UPR to restore normal ER homoeostasis leads to cell death through the activation of apoptotic pathways (6).
ER stress and activation of the UPR are common in the progression of numerous diseases including various cancers (7), neurodegenerative disorders (8) and viral infections (9). The individual aetiology of these disorders is as diverse as the cell’s response to each. And the exact intricacies of the molecular mechanisms underlying activation of the UPR and subsequent up-regulation of distinct PDIs remains poorly understood. A greater understanding of host pathways involved could potentially aid in the development of future treatments. The following review briefly discusses recent findings of ER stress, the UPR and the role of individual PDIs in various respiratory disease states.
The Unfolded Protein Response
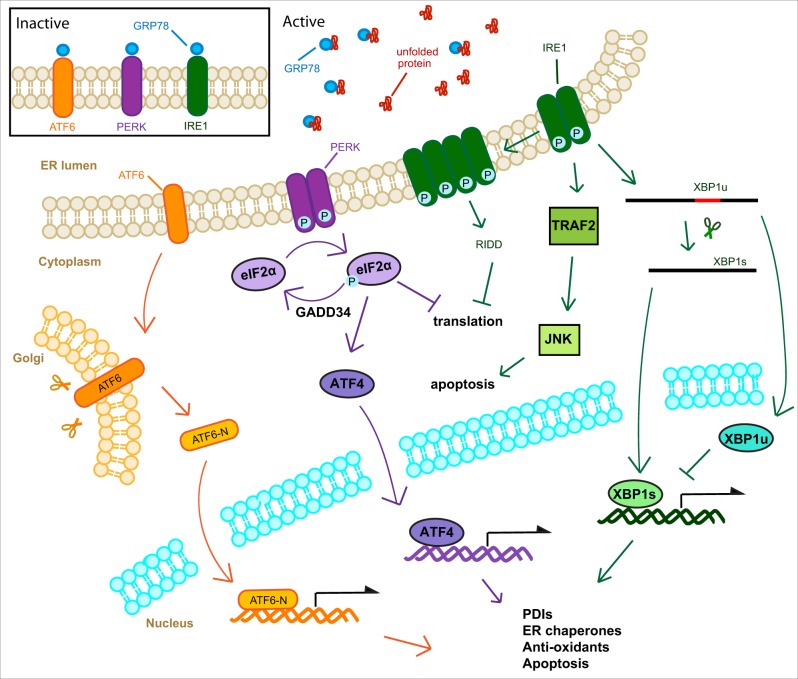
The UPR is a highly conserved collection of pathways responsible for monitoring the status of the ER. In mammals, the UPR consists of three pathways each controlled by a particular sensor, inositol-requiring protein 1 (IRE1), protein kinase RNA (PKR)-like ER kinase (PERK) and activating transcription factor-6 (ATF6) (Fig. 1). IRE1 exists in two isoforms α and β, this review focuses on solely IRE1 α, as the role the β isoform in activation of the UPR and induction of PDIs during pulmonary disease is less well characterized. These three pathways work in concert to decrease the protein load of the ER while simultaneously increasing its folding capacity. If the ER stress is too pronounced, or prolonged, the UPR directs the cell towards apoptosis (10–12).
Fig. 1.
Representation of canonical UPR signalling pathways. The UPR is activated by a buildup of unfolded protein within the ER lumen. GRP78 dissociates from the three ER stress sensors IRE1, ATF6 and PERK. Dimerization of IRE1 leads to autophosphorylation activating ribonuclease activity specific to XBP1 mRNA. This splicing generates XBP1s which is transported to the nuclease and induces the expression of UPR target genes. IRE1 phosphorylation also activates TRAF2 which directs the cell towards apoptosis through JNK signalling. IRE1 is also capable of associating into higher order structures which allow for non-specific degradation of ER associated mRNAs (RIDD). PERK dimerization leads to autophosphorylation activating kinase activity specific to eIF2a, halting protein translation. This loss of translation drives expression of ATF4 which acts as a transcription factor and induces the expression of UPR target genes. eIF2a is regenerated by GADD34. Upon dissociation of GRP78 from ATF6, ATF6 is transported to the Golgi where it is cleaved by cellular proteases to produce a transcription factor which induces the expression of UPR target genes.
Generally, the PERK pathway limits protein synthesis, the IRE1 pathway increases mRNA and protein degradation and the size of the ER, while the ATF6 pathway up-regulates chaperone proteins and protein degradation (10). Each of these transducers are integral membrane proteins residing in the ER membrane. Under normal conditions are held in inactive conformations by GRP78, an ER resident chaperone. Under conditions of ER stress unfolded or misfolded protein builds up within the ER lumen, GRP78 dissociates from the sensors owing to higher affinity to exposed hydrophobic residues on the unfolded proteins (11). While GRP78 is considered a master regulator of the UPR, the ER chaperone HSP47 has recently been shown as a selective regulator of the IRE1 arm of the UPR, displacing GRP78 and facilitating IRE1 oligomerization (13). Additionally, there is evidence that the individual transducers can bind unfolded protein directly (14).
Following GRP78 disassociation IRE1 dimerizes and undergoes trans-autophosphorylation of cytosolic kinase domains (11). Upon phosphorylation IRE1 displays endonucleolytic activity specifically targeting X-box binding protein 1 (XBP1) mRNA, this activity removes an intron and ultimately induces a frame shift by removing a stop codon. The spliced XBP1 (XBP1s) is translated and acts as a transcription factor driving the expression of ER chaperones, ERAD proteins and lipid synthesis (15). In mammals both spliced and un-spliced (XBP1u) are translated, interestingly while XBP1s acts a potent transcriptional activator of UPR effector genes, XBP1u acts as a repressor of the UPR (16) (Fig. 1).
Tumour necrosis factor associated factor 2 (TRAF2) is known to interact with IRE1 under conditions of extended ER stress leading to activation of downstream inflammatory and apoptotic signalling (10).
IRE1 is also capable of forming higher order structures that utilize their endonucleolytic activity to degrade ER-localized mRNAs in a process called regulated IRE1 dependent degradation of mRNA (RIDD) (17).
Like IRE1, PERK also undergoes dimerization and transphosphorylation following release from GRP78. PERK then phosphorylates eukaryotic initiation factor 2a (eIF2a) attenuating cap-dependent protein translation. This decrease in global translation leads to the cap-independent translation of activating transcription factor 4 (ATF4). ATF4 leads to the expression of amino-acid transporters, genes important in protecting the cell against oxidative stress and XBP1 (10). ATF4 also leads to the expression of C/EBP homologous protein (CHOP), another transcription factor that drives the cell towards apoptosis (10).
Unlike both IRE1 and PERK, ATF6 can exist as either a monomer or an oligomer, stabilized by disulphide bonds, while still bound to GRP78. ATF6 contains a Golgi localization signal that is masked by GRP78, upon disassociation ATF6 is translocated to the Golgi body where it is consecutively cleaved by two proteases SP1 and SP2 (18). Interactions with PDIs ensure that only reduced monomeric ATF6 is moved to the Golgi. The proteases liberate the N-terminal cytosolic domain of ATF6 (ATF6-N). ATF6-N is a transcription factor that moves to the nucleus and induces expression of various UPR target genes. Chaperone proteins are the primary targets of ATF6-N, including GRP78, GRP94 and PDIs (10).
While it is useful to separate UPR signalling pathways into discrete units, there exists a large amount of crosstalk between them. Genes under the control of the UPR often contain ER stress response elements (ERSEs) in their promoter regions that are responsible for transcriptional induction. Both XBP1s and ATF6-N can bind to these elements, though ATF6-N binding requires additional transcription factors (19). Interestingly, XBP1s and ATF6-N can form heterodimers, which further complicates signalling (20). Moreover, all three UPR pathways often involve the same proteins (10). The PERK and ATF6 pathways lead to XBP1 expression, which is then processed by IRE1. And all three pathways converge on NF-κB activation, though each uses a distinct mechanism.
However, this does not mean the UPR exists in a binary state of either active or inactive. The individual pathways of the UPR can be activated independently of one another. For instance, numerous groups have shown differential pathway activation following influenza infection, suggesting distinct triggers for each signalling pathway (21, 22).
Furthermore, while UPR activation is classically thought to involve the accumulation of unfolded protein within the ER lumen, there are numerous studies demonstrating activation of the UPR in the absence of unfolded protein. Toll-like receptor (TLR)2 and 4 have been shown to activate IRE1 and subsequent XBP1 maturation in macrophages (23). Notably, this XBP1 activation did not induce expression of canonical ER stress genes but was required for the continued production of proinflammatory cytokines (23). Similarly, it has been reported dendritic cells constitutively activate the IRE1 arm of the UPR in the absence of ER stress, and this activation was required for homoeostasis of CD8α+ dendritic cells (24, 25). Additionally, mitochondrial reactive oxygen species have been shown to exacerbate TLR induced activation of the UPR (26). These findings are particularly interesting as the utilization of TLRs as an alternative activation pathway would suggest that pathogens themselves or pattern-associated molecular patterns (PAMPs) (27) and damage-associated molecular patterns (DAMPs) are directly capable of activating the UPR (28).
Protein Disulphide Isomerases
The UPR up-regulates a wide variety of chaperone proteins in an effort to restore normal proteostasis, among these are PDIs a large family of redox-active chaperones that play important roles in the formation, reduction and isomerization of disulphide bonds (29). Currently, there are over 20 members of the PDI family, each differentiated from one another by the number and organization of TRX domains (29, 30). Individual TRX domains are classified as catalytically active (a) or protein binding (b) by the presence or absence of a largely conserved CXXC sequence (Table I). The CXXC motif allows for the oxidoreductase activity of PDIs by alternating between an oxidized form, where both cysteines are linked through a disulphide bond, and a reduced form containing two free sulphydryl groups. This effectively transfers a disulphide to the client protein. While the individual catalytic sequences vary the overall mechanism remains the same, with the intervening residues modulating the pKa of the reactive residues. The N terminal Cys exists as a thiolate (-S−) anion due to its lower pKa, which mediates nucleophilic attack forming a mixed disulphide. The C terminal Cys is partially buried within the protein elevating its pKa relative to other thiols and preventing the reverse reaction from occurring. The pKa of this C terminal Cys is rapidly decreased by a conformational change in the protein itself, which brings the side chain of a highly conserved arginine in close proximity to the active site. This shift in pKa changes the C terminal Cys from a thiol to a thiolate anion, which acts to resolve the mixed disulphide through a subsequent nucleophilic attack. Additionally, salt bridges located beneath the active site also serve to modulate the pKa of the Cys residues. PDIs in the reduced dithiol state participate in isomerization reactions, shuffling disulphide bonds between Cys residues, whereas oxidized (-S-S-) PDIs introduce disulphide bonds into associated peptides. The introduction of a disulphide bond into the client protein leaves the oxidized PDI in the reduced state where it can be rapidly re-oxidized by an intricate network of enzymes (Fig. 2). The isomerization of disulphide bonds does not involve a net change in disulphides, so the enzyme remains in the reduced state following the reaction. The above process is extensively reviewed in the following references (31, 32).
Table I.
Characteristics of the human PDI gene family
| Protein name | Gene name | Accession number | Gene location | TRX domain organization | Catalytic sequence | ER locatlization | ER retention sequence | Molecular weight (Da) |
|---|---|---|---|---|---|---|---|---|
| PDIA1 | P4HB | P07237 | 17q25.3 | a-b-b′-a′ | CGHC, CGHC | ER lumen | Yes | 57,116 |
| PDIA2 | PDIA2 | Q13087 | 16p13.3 | a-b-b′-a′ | CGHC, CTHC | ER lumen | Yes | 58,206 |
| PDIA3 | PDIA3 | P30101 | 15q15.3 | a-b-b′-a′ | CGHC, CGHC | ER lumen | Yes | 56,782 |
| PDIA4 | PDIA4 | P13667 | 7q36.1 | a′-a-b-b′-a′ | CGHC, CGHC, CGHC | ER lumen | Yes | 72,932 |
| PDIA5 | PDIA5 | Q14554 | 3q21.1 | b-a′-a-a′ | CSMC, CGHC, CPHC | ER lumen | Yes | 59,594 |
| PDIA6 | PDIA6 | Q15084 | 2p25.1 | a′-a-b | CGHC, CGHC | ER lumen | Yes | 48,121 |
| PDIA7 | PDILT | Q8N807 | 16p12.1 | a-b-b′-a′ | SKQS, SKKC | ER lumen | Yes | 66,657 |
| PDIA8 | ERP27 | Q96DN0 | 12p12.3 | b-b′ | No catalytic site | ER lumen | Yes | 30,480 |
| PDIA9 | EPR29 | P30040 | 12q24.13 | b | No catalytic site | ER lumen | Yes | 28,993 |
| PDIA10 | ERP44 | Q9BS26 | 9q31.1 | a-b-b′ | CRFS | ER lumen | Yes | 46,971 |
| PDIA11 | TMX1 | Q9H3N1 | 14q22.1 | a | CPAC | Membrane bound | No | 31,791 |
| PDIA12 | TMX2 | Q9Y320 | 11q12.1 | a | SNDC | Membrane bound | Yes | 34,038 |
| PDIA13 | TMX3 | Q96JJ7 | 18q22.1 | a-b-b′ | CGHC | Membrane bound | Yes | 51,872 |
| PDIA14 | TMX4 | Q9H1E5 | 20p12.3 | a | CPSC | Membrane bound | Yes | 38,952 |
| PDIA15 | TXNDC5 | Q8NBS9 | 6p24.3 | a′-a-a′ | CGHC, CGHC, CGHC | ER lumen | Yes | 47,629 |
| PDIA16 | TXNDC12 | O95881 | 1p32.3 | a | CGAC | ER lumen | Yes | 19,206 |
| PDIA17 | AGR2 | O95994 | 7p21.1 | a | CPHS | ER lumen | Yes | 19,979 |
| PDIA18 | AGR3 | Q8TD06 | 7p21.1 | a | CQYS | ER lumen | Yes | 19,171 |
| PDIA19 | DNAJC10 | Q8IXB1 | 2q32.1 | a′-b-a′-a-a′ | CSHC, CPPC, CHPC, CGPC | ER lumen | Yes | 91,080 |
| PDIB1 | CASQ1 | P31415 | 1q23.2 | b-b-b′ | No catalytic site | ER lumen | No | 45,160 |
| PDIB2 | CASQ2 | O14958 | 1p13.1 | b-b-b′ | No catalytic site | ER lumen | No | 46,436 |
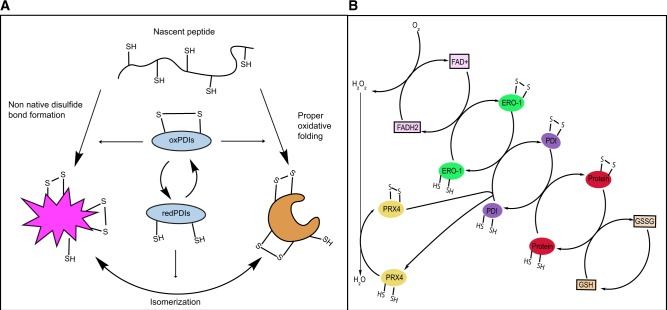
Fig. 2.
Functions of PDIs in oxidative folding. (A) Oxidized PDIs catalyse disulphide bond formation of nascent peptides in the ER. Leading to proper oxidative folding or non-native disulphide bond formation. Reduced PDIs facilitate isomerization of disulphide bonds. (B) PDIs are oxidized via interactions with ERO1. ERO1 uses FAD to transfer electrons to molecular oxygen generating hydrogen peroxide. PRX4 can also directly oxidize PDIs. PDIs transfer disulphides to client proteins. Glutathione contributes to disulphide bond reduction.
The re-oxidation of PDIs falls primarily to ERO1, which using FAD as a cofactor, transfers electrons from PDIs to molecular oxygen reducing it to hydrogen peroxide (33). Additional enzymes can oxidize PDIs: such as GPx7 and GPx8, two peroxidases that directly oxidize PDIs while reducing hydrogen peroxide (34). Glutathione is the primary redox buffer in the ER, and reduced glutathione is known to oxidize PDI in vitro (35). Interestingly oxidized glutathione has been shown to reduce PDIA3 in vivo, demonstrating its buffering role. The ability to reduce PDIA3 suggests glutathione possesses the ability to reduce other PDIs in vivo as well (36). While the non-catalytic b domains lack an active site, they nonetheless assist in the chaperone activity of PDIs by assisting in protein binding.
PDIs were originally characterized as ER resident proteins; most members of the family contain either a canonical KDEL sequence or a non-canonical retention sequence. Despite the near total presence of an ER retention sequence PDIs are commonly found throughout the cell, at the cell surface or even preferentially secreted from the cell (37). The dispersal throughout the cell despite the presence of a retention sequence may suggest unexplored roles for non-canonical retention sequences.
As one might expect, owing to the high degree of homology in the PDI family there exists a large amount of redundancy in terms of both functionality and client proteins. However, certain proteins appear to be clients of specific PDIs (38). PDIA3 has enhanced specificity towards glycoproteins owing to its association with both calreticulin and calnexin, two lectin-based chaperones within the ER lumen (39).
UPR and PDIs in asthma and pulmonary fibrosis
The UPR is initiated to manage the ER stress, but intense ER stress can result in apoptosis. Excessive ER stress and unhindered UPR can lead to apoptosis, proinflammatory signalling and epithelial-mesenchymal transition, features that have all been linked to lung fibrosis (40–43) and asthma (39, 44–46).
Although evidence is emerging, that downstream of UPR, PDIs are up-regulated in both asthma and pulmonary fibrosis, their function in the pathophysiology of lung diseases is not well understood. We have identified that various PDIs are up-regulated in allergic asthma (39, 45), and their increases correlated with the higher bronchodilator response or blood eosinophilic counts in allergic asthmatics (39, 45). Intriguingly our in-depth analysis of lung epithelial-specific knockouts of PDIA3 demonstrated that PDIA3 specifically regulate, eosinophilic and pro-fibrotic responses in lung epithelial cells by oxidizing cysteine sulphydryl (-SH) groups in eotaxin, periostin and epidermal growth factor (EGF) (45). Furthermore, we also demonstrated that PDIA3 facilitates -S-S- mediated oligomerization of pro-apoptotic BAK to induce intrinsic apoptosis in allergic airway disease models (45, 46). Ablation of Pdia3 specifically in lung epithelial cells attenuated, apoptotic, inflammatory and fibrotic responses in a model of allergic airway disease (45). These and other literature have led to the hypothesis that heterogeneous severe asthma could potentially be classified as an endotype of asthma (47).
Although, there is very little known about the impact of PDIs in pulmonary fibrosis recent literature has highlighted that PDIs potentially regulate disulphide bonds in many pro-apoptotic and pro-fibrotic proteins including collagen crosslinking enzyme lysyl oxidase like 2 (LOXL2) (45, 46, 48). Literature has also indicated that PDIA3 drives the trans-differentiation of murine alveolar epithelial cells and it is regulated by pro-fibrotic injury in mice (49). We have also identified that PDIA3 as a regulator of -S-S- bonds in death receptor CD95 (FAS) and inhibition or down-regulation of PDIA3 decreases -S-S- bonds in FAS, lung epithelial apoptosis and ultimately attenuation of pulmonary fibrosis in murine models of pulmonary fibrosis (46).
So far there are no proven therapeutics available to inhibit PDIs in the clinic, however, decades of research from various laboratories have identified many inhibitors that have shown in vivo and in vitro efficacy in inhibiting PDIs. Interestingly, rutinosides (plant flavonoids) that are known to inhibit PDIs are now being used in different clinical studies (50), also it is interesting to note that Dr Stockwell’s group have identified LOC14 as a specific inhibitor of PDIA1 and –A3 (51, 52). This literature suggests that UPR and subsequent induction of PDIs regulate pathology of various diseases and inhibiting PDIs may be a potential therapeutic approach that would benefit patients with chronic diseases.
UPR and PDIs in respiratory viral infection
Approximately 40 viruses are known to interact with the UPR, with many of these ultimately causing the induction of ER chaperone proteins (53). In this section, we highlight a few common respiratory viruses that display significant morbidity and mortality while also being known to cause exacerbations of lung diseases (54).
Influenza A virus (IAV) is known to activate different arms of the UPR depending on the model (21, 22, 55). Hassan et al. (21) demonstrated in isolated primary human tracheobronchial epithelial (HTBE) cells that IAV infection activated the IRE1 branch of the UPR but not the PERK or ATF6 branches. That same year Roberson et al. (22) showed in isolated primary mouse tracheal epithelial cells IAV infection strongly activated the ATF6 branch, but not PERK or IRE1. As indicated the models for these two studies are different which may account for the differences in UPR activation. However, one less explored difference between the two studies is the time post-viral infection in which the activation status of the UPR was explored. Hassan et al. examined UPR activation shortly after infection while Roberson et al. explored UPR activation at 24 and 48 h post-infection. This distinction may help explain the contradictory results as the two studies are examining the UPR at different points of the viral replication cycle where reproductive needs and thus utilization of host proteins may be quite different. A recent third study by Landeras-Bueno et al. (55) using chemical genomics showed IAV infection leads to the attenuation of the PERK branch of the UPR in A549 cells. Interestingly, in contrast to the previous studies, they showed no down-regulation of IRE1 or ATF6. Again, this may be due to differences in the time points examined.
The relationship between IAV and PDIs is more straightforward, in 2006, Solda et al. (38) clearly demonstrated PDIA3 was required for efficient folding of IAV HA in vitro. They also showed while other PDIs were able to act as surrogate chaperones for various proteins, efficient HA disulphide bond formation required PDIA3. Loss of IAV replication in cells treated with siRNA against PDIA3 supported this finding, though direct results on viral proteins were not examined (22). More recently expanded siRNA screens identified PDIA1 and four in addition to PDIA3 as having a role in IAV replication (56). The authors found substantial decreases in both IAV NP and M1 protein levels as well as significant decreases in viral transcript levels following siRNA treatment. Both M1 and NP lack disulphide bonds as part of their functional conformations (57), thus it is unclear whether the examined PDIs are interacting directly with the NP or M1.
Building on those results, we have shown PDIA3 plays an important role in IAV replication in vivo. Utilizing conditional PDIA3 knockout mice, we demonstrate PDIA3 deletion in lung epithelial cells significantly decreases levels of viral transcripts and proteins, as well as corresponding decreases in inflammatory cytokines and inflammatory and immune cells in the bronchoalveolar lavage fluid (BALF) (39). Moreover, we show ablation of Pdia3 in the lung epithelium diminishes IAV mediated methacholine induced AHR, providing a physiological readout illustrating the importance of PDIA3 in IAV replication (39).
Rhinovirus (RV) has recently been shown to cause impaired UPR activation in primary cystic fibrosis (CF) bronchial cells (58). However, this impairment of UPR activation does not appear to be cell intrinsic as activation with known chemical inducers of the UPR produces a robust UPR response as indicated by increased GRP78 and CHOP expression (58). Moreover, chemical activation of the UPR in RV infected primary cells significantly impeded viral replication and subsequent release of the virus. Suggesting that like IAV, RV is capable of directly activating the UPR. This is confirmed by a recent study from Song et al. (59) showing the human RV16 infection, specifically the non-structural protein 2B activates both PERK and ATF6 UPR pathways in H1-HeLa cells. Interestingly, this protein also simultaneously inactivates the IRE1 pathway by blocking phosphorylation and subsequent XBP1 splicing (59).
Like the above viruses, respiratory syncytial virus (RSV) infection activates the UPR. In both A549 cells and primary HTBE cells, UPR activation is characterized by increased GRP78 levels, and activation of both the IRE1 and ATF6 pathways, though no PERK activation was detected (60). However, a study involving a mouse model exploring RSV’s role in lung fibrosis found PERK activation 7 days post-infection along with elevated ATF6, GRP78 (61). XBP1 levels were not elevated and IRE1 activation was not explored. Intriguingly, while RSV activates the IRE1 branch of the UPR in both cell lines and isolated primary cells, IRE1 inhibits RSV replication (60). In both Ire1−/− mouse, embryonic fibroblasts and A549 cells treated with an IRE1 inhibitor viral transcript and protein levels were significantly higher than in RSV infected control cells (60). Though the exact mechanism for this is unclear, it may be related to IRE1 RIDD endonuclease activity.
Coronaviruses (CoV) are another virus family capable of causing respiratory exacerbation as well as significant illness on their own (62). SARS-CoV and MERS-CoV, in particular, have high pandemic potential and are associated with significant mortality (62). It is well-established CoV can induce the UPR in culture, this UPR activation has been linked to the spike (S) protein and for certain CoV, the exact amino acid domain responsible is known (63, 64). The S protein of SARS-CoV and CoV-HKU1, both beta-coronaviruses, induce the transcription of GRP78, GRP94 and CHOP through activation of the PERK pathway in vitro (63). Cells infected with MHV-A59, a model murine CoV in the same family as SARS and MERS, show activation of all three UPR branches, though ATF6 activation is limited later in infection (62). Relative activation of UPR branches varies between individual CoV as cells infected with SARS-CoV show limited XBP1 slicing and ATF6 cleavage (65, 66). Interestingly, an in vitro study using a selective PERK inhibitor found alleviating translational repression increased the levels of viral proteins but decreased viral titers (62). This was hypothesized to be due to increased translation of host anti-viral proteins.
There is currently little information on the relationship between RV, RSV and CoV and host PDIs. Thus, any potential interactions remain to be characterized, and any prospective anti-viral pharmacological influence need to be explored.
While we have limited ourselves to a discussion of respiratory viruses known to interact with the UPR, there are numerous other non-respiratory viruses that do so (53). Herpes Simplex Virus (67), Dengue Virus (68), Hepatitis B (69) and HIV (70) all interact with the UPR or PDIs despite all infecting drastically different cell types. These host-viral interactions may provide promising targets for the basis of the development of future antivirals.
UPR and PDIs in immune signalling
Both the UPR and PDIs play important roles in immune signalling and activation. Increased expression of proinflammatory cytokines during ER stress has been firmly established (71). And because it allows for ER expansion, increased protein production, and subsequent protein secretion, activation of the UPR is needed for the development and function of both secretory and immune cells (15, 72, 73). Additionally, GRP94 has been shown to be involved in the maturation of Toll-like receptors (74).
PDIA3 is a core component of the peptide loading complex, required for antigen presentation through MHC Class I (75). In addition, PDIs play important roles in cytokine folding and maturation. siRNA knockdown of PDIA3 in the lung epithelium has been shown to alter the oxidative folding of eotaxin and periostin in mice (45). PDIA1 is found at high levels in the secretory granules of eosinophils, suggesting a direct role in cytokine secretion (76).
UPR and PDIs in feedback regulation
There are an increasing number of papers exploring the redox regulation of the UPR sensors through thiol-disulphide exchange (77). In other words, modulation of the UPR itself through its downstream PDI effectors. A recent study determined PDIA1 and A3 are important in the regulation of PERK, utilizing lentiviral PDIA3 depletion in combination with small molecule-based PDI inhibition the authors concluded oxidized PDIA1 was an important activator of PERK, while PDIA3 was critical to regulating the oxidation state of PDIA1 (77).
One group found PDIA6 is important in modulating IRE1 signalling through interaction with Cys148 of the activated protein, facilitating its decay, thus acting as an attenuator of IRE1 signalling (78). Another study by the same group exploring the UPR and glucose-stimulated insulin secretion found PDIA6 regulates the RIDD activity of IRE1 (79). Utilizing shRNA against PDIA6, Eletto et al. (79) found that during UPR activation through chemical stressors, PDIA6 modulates the kinase activity of PERK as well as the XBP1 splicing ability of IRE1. However, upon activation due to glucose concentration, only RIDD activity of IRE1 is triggered, PDIA6 regulates this activity, and that this regulation was dependent of the enzymatic activity of PDIA6, rather than expression of PDIA6.
Another different study found PDIA5 is important for ATF6 activation and export during ER stress (80). Higa et al. (80) determined this activation was redox dependent and PDIA5 was involved in disulphide bond rearrangement of ATF6 which facilitated its transport to the Golgi. A very recent study utilizing trap mutants of various PDIs found PDIA16 modulates trafficking of ATF6 to the Golgi and assists in proteolytic cleavage (81). Another study using shRNA depletion of PDIs found PDIA4 regulates ATF6 activity, as increased GRP78 expression was detected specifically following PDIA4 deletion as opposed to other PDIs (82). Though, the exact nature of this control was not explored.
This avenue of research is promising as it demonstrates the possibility of controlling aberrant or exuberant UPR activation, or modulating specific arms of the UPR, through inhibition of individual PDIs.
Small molecule inhibition of PDIs
Given the scope of PDIs, it is not surprising there is plentiful investigation into modulating their activity. Numerous chemicals have been shown to inhibit PDI activity, from antibodies (83) and hormones (84) to antibiotics (85). In the past few years, there has been an influx of inhibitors identified from small-molecule screen libraries with increasing specificity towards PDIs (86–91). Most of these molecules are identified as potential chemotherapeutic agents and have been characterized against PDIA1, the prototypical member of the PDI family (92). However, given the high degree of homology between PDI family members some inhibitory activity against other PDIs is often predicted. A full description of currently available small molecule PDI inhibitors can be found in Table II, the following section will highlight a few key inhibitors.
Table II.
Characteristics of various PDI inhibitors
| Compound name | Mode of action | Clinical trial a | References |
|---|---|---|---|
| Bacitracin | Competitive inhibitor binds to free thiols in substrate binding region. Cell impermeable | Yes (57) | 85 |
| 16F16 | Irreversibly binds to cysteine residues in active site. Cell permeable | No | 52 |
| LOC14 | Allosteric inhibitor. Binds adjacent to active site, forces protein to maintain oxidized conformation. Reversible. Cell permeable | No | 39, 51 |
| PACMA31 | Irreversibly binds to cysteine residues in active site. Cell permeable | No | 86 |
| CCF642 | Allosteric inhibitor. Irreversibly binds to conserved lysine directly adjacent to the active site. | No | 7 |
| Cell permeable | |||
| P1 | Irreversibly binds to cysteine residues in active site. Cell permeable | No | 87 |
| E64FC26 | Pan-PDI inhibitor mechanism unknown. cell permeable | No | 93 |
| KSC-34 | PDIA1 inhibitor selective for C53 in a domain active site. Cell permeable | No | 92 |
| ML359 | PDIA1 inhibitor mechanism unknown. Reversible. Cell permeable | No | 88 |
| RB-11-ca | PDIA1 inhibitor selective for C53 in a domain active site. Cell permeable | No | 89 |
| Juniferdin | PDIA1 inhibitor mechanism unknown. Cell permeable | No | 88 |
| Eupatorin | Flavonoid compound binds to tryptophan residues near the active site of PDIA3 | No | 94, 95 |
| Eupatorin-5-methyl ether | Flavonoid compound binds to tryptophan residues near the active site of PDIA3. | No | 94, 95 |
| Quercetin-3-rutinoside | PDIA1 inhibitor binds to b' domain. Reversible. Cell impermeable | No | 50 |
| T8 | Allosteric inhibitor binds near active site. Reversible. Cell permeable | No | 90 |
| RL90 | Anti-PDIA1 antibody | No | 83 |
| 17β-estradiol | Binds to bb' domain | Yes (2,054) | 84 |
| 35G8 | Believed to bind with cysteine residues in active site | No | 91 |
Clinicaltrial.gov.
16F16 was the first of this new wave of inhibitors, it acts by covalently binding to reactive Cys residues in the active site of PDIs (52). Hoffstrom et al. (52) utilized it to explore mechanisms linking PDIs to apoptotic cell death. E64FC26 is a novel PDI inhibitor-based off of an indene moiety that acts as a pan inhibitor of numerous PDIs though the inhibitory mechanism in unclear (93). The authors found E64FC26 improved survival in a mouse model of multiple myeloma, with little adverse effects. KSC-34 is a particularly interesting inhibitor, it displays enhanced specificity towards PDIA1, which is not unusual, but KSC-34 is selective towards the N-terminal CGHC active site of the protein (92). This means KSC-34 can be used to explore specific functionalities and protein interactions of each individual active site. Additionally, the specificity of this inhibitor is encouraging in that is suggests the possibility of identifying novel inhibitors with the same level of specificity targeted towards other PDIs.
LOC14 is unique, it is a reversible PDI inhibitor that binds adjacent to the CGHC active site and locks the enzyme in an oxidized conformation, though the exact residues remain to be elucidated (51). LOC14 has been found to be neuroprotective in both cell culture and animal models and displays high stability (51). We have recently demonstrated LOC14 is also capable of inhibiting PDIA3 as well as PDIA1, and that treatment with LOC14 significantly decreased Influenza replication and the maturation of viral proteins (39). Another group has demonstrated the redox modulating effects of flavonoid compounds on PDIA3 (94). Rather than directly interacting with the active site these compounds bind to residues on the protein binding b and b’ domain absent on other PDIs (94, 95). These flavonoids are interesting as they may provide a structural basis for the design of specific PDIA3 inhibitors.
The above compounds represent an expansive range of mechanisms and targets within the PDI family, from E64FC26 acting as a pan PDI inhibitor (93) to KSC-34 acting towards a specific active site on PDIA1 (92). PDIs are being found to play increasingly important roles in a wide variety of conditions and cellular activities, though current techniques to elucidate their exact role remain limited. Conventional RNA interference techniques take time to ensure sufficient knockdown of a target gene and targeting multiple genes simultaneously can pose additional problems. Employing targeted inhibitors allows for rapid decreases in protein activity and could theoretically be titrated to achieve a desired activity level. We have already demonstrated the efficacy of these inhibitors by utilizing them to determine the role of PDIs in oxidative folding of influenza proteins (39), while Cole et al. (92) explored the specific role of the PDIA1 A site on protein folding and secretion. Current compounds are able to target some of the PDI family, though inhibitory action against all PDIs has yet to be explored, and highly specific inhibitors towards PDIs other than PDIA1 remain to be developed. Nonetheless, the development of increasingly specific inhibitors specifically targeted towards individual PDIs would provide an invaluable tool to determine the distinct role of unique PDIs during disease pathogenesis.
Conclusion
The UPR and PDIs play critical roles in numerous cellular processes. Much remains unknown regarding their role in normal responses and their impact on the development of disease. Investigation into the branches of the UPR and their downstream effectors remains challenging. However, small molecule PDI inhibitors provide an exciting opportunity to tease apart the molecular mechanisms of ER stress and provide potential platforms for the development of future therapeutics.
Acknowledgements
We thank members of the Anathy laboratory for their inputs and critical comments on this review.
Funding
This work was supported by NIH R01 awards; HL122383 and HL141364 to V.A. N.C. was supported by NIH T32 fellowship HL076122.
Conflict of Interest
V.A. hold patents: U.S. Patent No. 8,679,811, ‘Treatments Involving Glutaredoxins and Similar Agents’ and U.S. Patent, 9,907,828, ‘Treatments of oxidative stress conditions’. V.A. have received consulting fees and research funds (contracts) from Celdara Medical LLC, N.H. for his contributions with the commercialization of glutaredoxin for the treatment of pulmonary fibrosis.
Glossary
Abbreviations
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor-6
- CF
cystic fibrosis
- CHOP
C/EBP homologous protein
- CoV
coronaviruses
- eIF2a
eukaryotic initiation factor 2a
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERSEs
ER stress response elements
- HTBE
human tracheobronchial epithelial
- IAV
influenza A virus
- IRE1
inositol-requiring protein 1
- PDIs
protein disulphide isomerases
- PERK
(PKR)-like ER kinase
- PKR
protein kinase RNA
- RIDD
regulated IRE1 dependent degradation of mRNA
- RSV
respiratory syncytial virus
- RV
rhinovirus
- TRAF2
tumour necrosis factor associated factor 2
- UPR
unfolded protein response
- XBP1
X-box binding protein 1
References
- 1. Gaut J.R., Hendershot L.M. (1993) The modification and assembly of proteins in the endoplasmic reticulum. Curr. Opin. Cell. Biol. 5, 589–595 [DOI] [PubMed] [Google Scholar]
- 2. Aebi M. (2013) N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 [DOI] [PubMed] [Google Scholar]
- 3. Fewell S.W., Travers K.J., Weissman J.S., Brodsky J.L. (2001) The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 35, 149–191 [DOI] [PubMed] [Google Scholar]
- 4. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) ERAD: the long road to destruction. Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 5. Schubert U., Antón L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 [DOI] [PubMed] [Google Scholar]
- 6. Nakada E.M., Bhakta N.R., Korwin-Mihavics B.R., Kumar A., Chamberlain N., Bruno S.R., Chapman D.G., Hoffman S.M., Daphtary N., Aliyeva M., Irvin C.G., Dixon A.E., Woodruff P.G., Amin S., Poynter M.E., Desai D.H., Anathy V. (2019) Conjugated bile acids attenuate allergen-induced airway inflammation and hyperresposiveness by inhibiting UPR transducers. JCI Insight 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vatolin S., Phillips J.G., Jha B.K., Govindgari S., Hu J., Grabowski D., Parker Y., Lindner D.J., Zhong F., Distelhorst C.W., Smith M.R., Cotta C., Xu Y., Chilakala S., Kuang R.R., Tall S., Reu F.J. (2016) Novel protein disulfide isomerase inhibitor with anticancer activity in multiple myeloma. Cancer Res. 76, 3340–3350 [DOI] [PubMed] [Google Scholar]
- 8. Zhou X., Li G., Kaplan A., Gaschler M.M., Zhang X., Hou Z., Jiang M., Zott R., Cremers S., Stockwell B.R., Duan W. (2018) Small molecule modulator of protein disulfide isomerase attenuates mutant huntingtin toxicity and inhibits endoplasmic reticulum stress in a mouse model of Huntington's disease. Hum. Mol. Genet. 27, 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith J.A. (2018) Regulation of cytokine production by the unfolded protein response; implications for infection and autoimmunity. Front. Immunol. 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 11. Schroder M., Kaufman R.J. (2005) ER stress and the unfolded protein response. Mutat. Res. 569, 29–63 [DOI] [PubMed] [Google Scholar]
- 12. Minakshi R., Rahman S., Jan A.T., Archana A., Kim J. (2017) Implications of aging and the endoplasmic reticulum unfolded protein response on the molecular modality of breast cancer. Exp. Mol. Med. 49, e389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sepulveda D., Rojas-Rivera D., Rodríguez D.A., Groenendyk J., Köhler A., Lebeaupin C., Ito S., Urra H., Carreras-Sureda A., Hazari Y., Vasseur-Cognet M., Ali M.M.U., Chevet E., Campos G., Godoy P., Vaisar T., Bailly-Maitre B., Nagata K., Michalak M., Sierralta J., Hetz C. (2018) Interactome screening identifies the ER luminal chaperone Hsp47 as a regulator of the unfolded protein response transducer IRE1alpha. Mol. Cell 69, 238–252.e237 [DOI] [PubMed] [Google Scholar]
- 14. Wang P., Li J., Tao J., Sha B. (2018) The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization. J. Biol. Chem. 293, 4110–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bettigole S.E., Glimcher L.H. (2015) Endoplasmic reticulum stress in immunity. Annu. Rev. Immunol. 33, 107–138 [DOI] [PubMed] [Google Scholar]
- 16. Yoshida H., Oku M., Suzuki M., Mori K. (2006) pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172, 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maurel M., Chevet E., Tavernier J., Gerlo S. (2014) Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 39, 245–254 [DOI] [PubMed] [Google Scholar]
- 18. Gardner B.M., Pincus D., Gotthardt K., Gallagher C.M., Walter P. (2013) Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 5, a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thuerauf D.J., Morrison L.E., Hoover H., Glembotski C.C. (2002) Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J. Biol. Chem. 277, 20734–20739 [DOI] [PubMed] [Google Scholar]
- 20. Shoulders M.D., Ryno L.M., Genereux J.C., Moresco J.J., Tu P.G., Wu C., Yates J.R. 3rd, Su A.I., Kelly J.W., Wiseman R.L. (2013) Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3, 1279–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hassan I.H., Zhang M.S., Powers L.S., Shao J.Q., Baltrusaitis J., Rutkowski D.T., Legge K., Monick M.M. (2012) Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J. Biol. Chem. 287, 4679–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roberson E.C., Tully J.E., Guala A.S., Reiss J.N., Godburn K.E., Pociask D.A., Alcorn J.F., Riches D.W., Dienz O., Janssen-Heininger Y.M., Anathy V. (2012) Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-beta release in lung epithelial cells. Am. J. Respir. Cell Mol. Biol. 46, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martinon F., Chen X., Lee A.H., Glimcher L.H. (2010) TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 11, 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osorio F., Tavernier S.J., Hoffmann E., Saeys Y., Martens L., Vetters J., Delrue I., De Rycke R., Parthoens E., Pouliot P., Iwawaki T., Janssens S., Lambrecht B.N. (2014) The unfolded-protein-response sensor IRE-1α regulates the function of CD8α+ dendritic cells. Nat. Immunol. 15, 248. [DOI] [PubMed] [Google Scholar]
- 25. Tavernier S.J., Osorio F., Vandersarren L., Vetters J., Vanlangenakker N., Van Isterdael G., Vergote K., De Rycke R., Parthoens E., van de Laar L., Iwawaki T., Del Valle J.R., Hu C.C., Lambrecht B.N., Janssens S. (2017) Regulated IRE1-dependent mRNA decay sets the threshold for dendritic cell survival. Nat. Cell Biol. 19, 698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mogilenko D.A., Haas J.T., L’Homme L., Fleury S., Quemener S., Levavasseur M., Becquart C., Wartelle J., Bogomolova A., Pineau L., Molendi-Coste O., Lancel S., Dehondt H., Gheeraert C., Melchior A., Dewas C., Nikitin A., Pic S., Rabhi N., Annicotte J.-S., Oyadomari S., Velasco-Hernandez T., Cammenga J., Foretz M., Viollet B., Vukovic M., Villacreces A., Kranc K., Carmeliet P., Marot G., Boulter A., Tavernier S., Berod L., Longhi M.P., Paget C., Janssens S., Staumont-Sallé D., Aksoy E., Staels B., Dombrowicz D. (2019) Metabolic and innate immune cues merge into a specific inflammatory response via the UPR. Cell 177, 1201–1216.e1219 [DOI] [PubMed] [Google Scholar]
- 27. Medzhitov R. (2001) Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1, 135–145 [DOI] [PubMed] [Google Scholar]
- 28. Roh J.S., Sohn D.H. (2018) Damage-associated molecular patterns in inflammatory diseases. Immune Netw. 18, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Appenzeller-Herzog C., Ellgaard L. (2008) The human PDI family: versatility packed into a single fold. Biochim. Biophys. Acta 1783, 535–548 [DOI] [PubMed] [Google Scholar]
- 30. Galligan J.J., Petersen D.R. (2012) The human protein disulfide isomerase gene family. Hum. Genomics 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oka O.B.V., Yeoh H.Y., Bulleid N.J. (2015) Thiol-disulfide exchange between the PDI family of oxidoreductases negates the requirement for an oxidase or reductase for each enzyme. Biochem. J. 469, 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hatahet F., Ruddock L.W. (2009) Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 11, 2807–2850 [DOI] [PubMed] [Google Scholar]
- 33. Zito E. (2015) ERO1: a protein disulfide oxidase and H2O2 producer. Free Radic. Biol. Med. 83, 299–304 [DOI] [PubMed] [Google Scholar]
- 34. Wang L., Zhang L., Niu Y., Sitia R., Wang C.-C. (2013) Glutathione peroxidase 7 utilizes hydrogen peroxide generated by Ero1α to promote oxidative protein folding. Antioxid. Redox Signal. 20, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bass R., Ruddock L.W., Klappa P., Freedman R.B. (2004) A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J. Biol. Chem. 279, 5257–5262 [DOI] [PubMed] [Google Scholar]
- 36. Ali Khan H., Mutus B. (2014) Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front. Chem. 2, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bartels A.K., Gottert S., Desel C., Schafer M., Krossa S., Scheidig A.J., Grotzinger J., Lorenzen I. (2019) KDEL receptor 1 contributes to cell surface association of protein disulfide isomerases. Cell Physiol. Biochem. 52, 850–868 [DOI] [PubMed] [Google Scholar]
- 38. Solda T., Garbi N., Hammerling G.J., Molinari M. (2006) Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J. Biol. Chem. 281, 6219–6226 [DOI] [PubMed] [Google Scholar]
- 39. Chamberlain N., Korwin-Mihavics B.R., Nakada E.M., Bruno S.R., Heppner D.E., Chapman D.G., Hoffman S.M., van der Vliet A., Suratt B.T., Dienz O., Alcorn J.F., Anathy V. (2019) Lung epithelial protein disulfide isomerase A3 (PDIA3) plays an important role in influenza infection, inflammation, and airway mechanics. Redox Biol. 22, 101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burman A., Tanjore H., Blackwell T.S. (2018) Endoplasmic reticulum stress in pulmonary fibrosis. Matrix Biol. 68-69, 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kropski J.A., Blackwell T.S. (2018) Endoplasmic reticulum stress in the pathogenesis of fibrotic disease. J. Clin. Invest. 128, 64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tanjore H., Cheng D.S., Degryse A.L., Zoz D.F., Abdolrasulnia R., Lawson W.E., Blackwell T.S. (2015) Alveolar epithelial cells undergo epithelial-to-mesenchymal transition in response to endoplasmic reticulum stress. J. Biol. Chem. 290, 3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lawson W.E., Cheng D.S., Degryse A.L., Tanjore H., Polosukhin V.V., Xu X.C., Newcomb D.C., Jones B.R., Roldan J., Lane K.B., Morrisey E.E., Beers M.F., Yull F.E., Blackwell T.S. (2011) Endoplasmic reticulum stress enhances fibrotic remodeling in the lungs. Proc. Natl. Acad. Sci. U S A 108, 10562–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bhakta N.R., Christenson S.A., Nerella S., Solberg O.D., Nguyen C.P., Choy D.F., Jung K.L., Garudadri S., Bonser L.R., Pollack J.L., Zlock L.T., Erle D.J., Langelier C., Derisi J.L., Arron J.R., Fahy J.V., Woodruff P.G. (2018) IFN-stimulated gene expression, type 2 inflammation, and endoplasmic reticulum stress in asthma. Am. J. Respir. Crit. Care Med. 197, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoffman S.M., Chapman D.G., Lahue K.G., Cahoon J.M., Rattu G.K., Daphtary N., Aliyeva M., Fortner K.A., Erzurum S.C., Comhair S.A.A., Woodruff P.G., Bhakta N., Dixon A.E., Irvin C.G., Janssen-Heininger Y.M.W., Poynter M.E., Anathy V. (2016) Protein disulfide isomerase-endoplasmic reticulum resident protein 57 regulates allergen-induced airways inflammation, fibrosis, and hyperresponsiveness. J. Allergy Clin. Immunol. 137, 822–832.e827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anathy V., Roberson E., Cunniff B., Nolin J.D., Hoffman S., Spiess P., Guala A.S., Lahue K.G., Goldman D., Flemer S., van der Vliet A., Heintz N.H., Budd R.C., Tew K.D., Janssen-Heininger Y.M. (2012) Oxidative processing of latent Fas in the endoplasmic reticulum controls the strength of apoptosis. Mol. Cell Biol. 32, 3464–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jeong J.S., Kim S.R., Cho S.H., Lee Y.C. (2019) A novel insight on endotyping heterogeneous severe asthma based on endoplasmic reticulum stress: beyond the “type 2/non-type 2 dichotomy”. Int. J. Mol. Sci. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jessop C.E., Watkins R.H., Simmons J.J., Tasab M., Bulleid N.J. (2009) Protein disulphide isomerase family members show distinct substrate specificity: p 5 is targeted to BiP client proteins. J. Cell Sci. 122, 4287–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mutze K., Vierkotten S., Milosevic J., Eickelberg O., Konigshoff M. (2015) Enolase 1 (ENO1) and protein disulfide-isomerase associated 3 (PDIA3) regulate Wnt/beta-catenin-driven trans-differentiation of murine alveolar epithelial cells. Dis. Model Mech. 8, 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jasuja R., Passam F.H., Kennedy D.R., Kim S.H., van Hessem L., Lin L., Bowley S.R., Joshi S.S., Dilks J.R., Furie B., Furie B.C., Flaumenhaft R. (2012) Protein disulfide isomerase inhibitors constitute a new class of antithrombotic agents. J. Clin. Invest. 122, 2104–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaplan A., Gaschler M.M., Dunn D.E., Colligan R., Brown L.M., Palmer A.G. 3rd, Lo D.C., Stockwell B.R. (2015) Small molecule-induced oxidation of protein disulfide isomerase is neuroprotective. Proc. Natl. Acad. Sci. U S A 112, E2245–E2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hoffstrom B.G., Kaplan A., Letso R., Schmid R.S., Turmel G.J., Lo D.C., Stockwell B.R. (2010) Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat. Chem. Biol. 6, 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li S., Kong L., Yu X. (2015) The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit. Rev. Microbiol. 41, 150–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Busse W.W., Lemanske R.F. Jr,, Gern J.E. (2010) Role of viral respiratory infections in asthma and asthma exacerbations. Lancet 376, 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Landeras-Bueno S., Fernández Y., Falcón A., Oliveros J.C., Ortín J. (2016) Chemical genomics identifies the PERK-mediated unfolded protein stress response as a cellular target for influenza virus inhibition. MBio 7, e00085–00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim Y., Chang K.O. (2018) Protein disulfide isomerases as potential therapeutic targets for influenza A and B viruses. Virus Res. 247, 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Selimova L.M., Zaides V.M., Zhdanov V.M. (1982) Disulfide bonding in influenza virus proteins as revealed by polyacrylamide gel electrophoresis. J. Virol. 44, 450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schogler A., Caliaro O., Brugger M., Oliveira Esteves B.I., Nita I., Gazdhar A., Geiser T., Alves M.P. (2019) Modulation of the unfolded protein response pathway as an antiviral approach in airway epithelial cells. Antiviral Res. 162, 44–50 [DOI] [PubMed] [Google Scholar]
- 59. Song J., Chi M., Luo X., Song Q., Xia D., Shi B., Han J. (2019) Non-structural protein 2B of human rhinovirus 16 activates both PERK and ATF6 rather than IRE1 to trigger ER stress. Viruses 11, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hassan I., Gaines K.S., Hottel W.J., Wishy R.M., Miller S.E., Powers L.S., Rutkowski D.T., Monick M.M. (2014) Inositol-requiring enzyme 1 inhibits respiratory syncytial virus replication. J. Biol. Chem. 289, 7537–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang L., Cheng W., Zhang Z. (2017) Respiratory syncytial virus infection accelerates lung fibrosis through the unfolded protein response in a bleomycin-induced pulmonary fibrosis animal model. Mol. Med. Rep. 16, 310–316 [DOI] [PubMed] [Google Scholar]
- 62. Irigoyen N., Franaszek K., Dinan A.M., Moore N.A., Siddell S.G., Brierley I., Firth A.E. (2018) Activation of the unfolded protein response and inhibition of translation initiation during coronavirus infection. bioRxiv, 292979
- 63. Siu K.L., Chan C.P., Kok K.H., Woo P.C., Jin D.Y. (2014) Comparative analysis of the activation of unfolded protein response by spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus HKU1. Cell Biosci. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jin D.Y., Woo P.C. (2016) Modulation of cell signalling by human coronavirus HKU1 S and M proteins. Hong Kong Med. J. 22, 22–24 [PubMed] [Google Scholar]
- 65. Fung T.S., Liao Y., Liu D.X. (2014) The endoplasmic reticulum stress sensor IRE1alpha protects cells from apoptosis induced by the coronavirus infectious bronchitis virus. J Virol. 88, 12752–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. DeDiego M.L., Nieto-Torres J.L., Jiménez-Guardeño J.M., Regla-Nava J.A., Álvarez E., Oliveros J.C., Zhao J., Fett C., Perlman S., Enjuanes L. (2011) Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 7, e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang P., Su C., Jiang Z., Zheng C. (2017) Herpes simplex virus 1 UL41 protein suppresses the IRE1/XBP1 signal pathway of the unfolded protein response via its RNase activity. J. Virol. 91, e02056–02016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Perera N., Miller J.L., Zitzmann N. (2017) The role of the unfolded protein response in dengue virus pathogenesis. Cell Microbiol. 19 [DOI] [PubMed] [Google Scholar]
- 69. Li Y., Xia Y., Cheng X., Kleiner D.E., Hewitt S.M., Sproch J., Li T., Zhuang H., Liang T.J. (2019) Hepatitis B surface antigen activates unfolded protein response in forming ground glass hepatocytes of chronic hepatitis B. Viruses 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cerutti N., Killick M., Jugnarain V., Papathanasopoulos M., Capovilla A. (2014) Disulfide reduction in CD4 domain 1 or 2 is essential for interaction with HIV glycoprotein 120 (gp120), which impairs thioredoxin-driven CD4 dimerization. J. Biol. Chem. 289, 10455–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wheeler M.C., Rizzi M., Sasik R., Almanza G., Hardiman G., Zanetti M. (2008) KDEL-retained antigen in B lymphocytes induces a proinflammatory response: a possible role for endoplasmic reticulum stress in adaptive T cell immunity. J. Immunol. 181, 256–264 [DOI] [PubMed] [Google Scholar]
- 72. Lee A.H., Chu G.C., Iwakoshi N.N., Glimcher L.H. (2005) XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24, 4368–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bettigole S.E., Lis R., Adoro S., Lee A.H., Spencer L.A., Weller P.F., Glimcher L.H. (2015) The transcription factor XBP1 is selectively required for eosinophil differentiation. Nat. Immunol. 16, 829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu S., Hong F., Gewirth D., Guo B., Liu B., Li Z. (2012) The molecular chaperone gp96/GRP94 interacts with Toll-like receptors and integrins via its C-terminal hydrophobic domain. J. Biol. Chem. 287, 6735–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stepensky D., Bangia N., Cresswell P. (2007) Aggregate formation by ERp57-deficient MHC class I peptide-loading complexes. Traffic 8, 1530–1542 [DOI] [PubMed] [Google Scholar]
- 76. Dias F.F., Amaral K.B., Carmo L.A., Shamri R., Dvorak A.M., Weller P.F., Melo R.C. (2014) Human eosinophil leukocytes express protein disulfide isomerase in secretory granules and vesicles: ultrastructural studies. J. Histochem. Cytochem. 62, 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kranz P., Neumann F., Wolf A., Classen F., Pompsch M., Ocklenburg T., Baumann J., Janke K., Baumann M., Goepelt K., Riffkin H., Metzen E., Brockmeier U. (2017) PDI is an essential redox-sensitive activator of PERK during the unfolded protein response (UPR). Cell Death Dis. 8, e2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Eletto D., Eletto D., Dersh D., Gidalevitz T., Argon Y. (2014) Protein disulfide isomerase A6 controls the decay of IRE1α signaling via disulfide-dependent association. Mol. Cell 53, 562–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Eletto D., Eletto D., Boyle S., Argon Y. (2016) PDIA6 regulates insulin secretion by selectively inhibiting the RIDD activity of IRE1. FASEB J. 30, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Higa A., Taouji S., Lhomond S., Jensen D., Fernandez-Zapico M.E., Simpson J.C., Pasquet J.-M., Schekman R., Chevet E. (2014) Endoplasmic reticulum stress-activated transcription factor ATF6α requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol. Cell. Biol. 34, 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Oka O.B., Lith M., Rudolf J., Tungkum W., Pringle M.A., Bulleid N.J. (2019) ERp18 regulates activation of ATF6alpha during unfolded protein response. EMBO J. 38, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Paxman R., Plate L., Blackwood E.A., Glembotski C., Powers E.T., Wiseman R.L., Kelly J.W. (2018) Pharmacologic ATF6 activating compounds are metabolically activated to selectively modify endoplasmic reticulum proteins. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kaetzel C.S., Rao C.K., Lamm M.E. (1987) Protein disulphide-isomerase from human placenta and rat liver. Purification and immunological characterization with monoclonal antibodies. Biochem J. 241, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tsibris J.C., Hunt L.T., Ballejo G., Barker W.C., Toney L.J., Spellacy W.N. (1989) Selective inhibition of protein disulfide isomerase by estrogens. J. Biol. Chem. 264, 13967–13970 [PubMed] [Google Scholar]
- 85. Dickerhof N., Kleffmann T., Jack R., McCormick S. (2011) Bacitracin inhibits the reductive activity of protein disulfide isomerase by disulfide bond formation with free cysteines in the substrate-binding domain. FEBS J. 278, 2034–2043 [DOI] [PubMed] [Google Scholar]
- 86. Xu S., Butkevich A.N., Yamada R., Zhou Y., Debnath B., Duncan R., Zandi E., Petasis N.A., Neamati N. (2012) Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc. Natl. Acad. Sci. U S A 109, 16348–16353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ge J., Zhang C.J., Li L., Chong L.M., Wu X., Hao P., Sze S.K., Yao S.Q. (2013) Small molecule probe suitable for in situ profiling and inhibition of protein disulfide isomerase. ACS Chem. Biol. 8, 2577–2585 [DOI] [PubMed] [Google Scholar]
- 88. Khodier C., VerPlank L., Nag P.P., Pu J., Wurst J., Pilyugina T., Dockendorff C., Galinski C.N., Scalise A.A., Passam F., van Hessem L., Dilks J., Kennedy D.R., Flaumenhaft R., Palmer M.A.J., Dandapani S., Munoz B., Schrieber S.L. (2010) Identification of ML359 as a small molecule inhibitor of protein disulfide isomerase in Probe Reports from the NIH Molecular Libraries Program. National Center for Biotechnology Information (US), Bethesda, MD: [PubMed] [Google Scholar]
- 89. Banerjee R., Pace N.J., Brown D.R., Weerapana E. (2013) 1,3,5-Triazine as a modular scaffold for covalent inhibitors with streamlined target identification. J. Am. Chem. Soc. 135, 2497–2500 [DOI] [PubMed] [Google Scholar]
- 90. Eirich J., Braig S., Schyschka L., Servatius P., Hoffmann J., Hecht S., Fulda S., Zahler S., Antes I., Kazmaier U., Sieber S.A., Vollmar A.M. (2014) A small molecule inhibits protein disulfide isomerase and triggers the chemosensitization of cancer cells. Angew. Chem. Int. Ed. 53, 12960–12965 [DOI] [PubMed] [Google Scholar]
- 91. Kyani A., Tamura S., Yang S., Shergalis A., Samanta S., Kuang Y., Ljungman M., Neamati N. (2018) Discovery and mechanistic elucidation of a class of protein disulfide isomerase inhibitors for the treatment of glioblastoma. ChemMedChem 13, 164–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cole K.S., Grandjean J.M.D., Chen K., Witt C.H., O’Day J., Shoulders M.D., Wiseman R.L., Weerapana E. (2018) Characterization of an A-site selective protein disulfide isomerase A1 inhibitor. Biochemistry 57, 2035–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Robinson R.M., Reyes L., Duncan R.M., Bian H., Reitz A.B., Manevich Y., McClure J.J., Champion M.M., Chou C.J., Sharik M.E., Chesi M., Bergsagel P.L., Dolloff N.G. (2019) Inhibitors of the protein disulfide isomerase family for the treatment of multiple myeloma. Leukemia 33, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Giamogante F., Marrocco I., Romaniello D., Eufemi M., Chichiarelli S., Altieri F. (2016) Comparative analysis of the interaction between different flavonoids and PDIA3. Oxid. Med. Cell. Longev. 2016, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Giamogante F., Marrocco I., Cervoni L., Eufemi M., Chichiarelli S., Altieri F. (2018) Punicalagin, an active pomegranate component, is a new inhibitor of PDIA3 reductase activity. Biochimie 147, 122–129 [DOI] [PubMed] [Google Scholar]