Abstract
Heterotrimeric guanine nucleotide–binding proteins (G proteins), which are composed of α, β, and γ subunits, are versatile, guanine nucleotide–dependent, molecular on-off switches. In animals and fungi, the exchange of GDP for GTP on Gα controls G protein activation and is crucial for normal cellular responses to diverse extracellular signals. The model plant Arabidopsis thaliana has a single canonical Gα subunit, AtGPA1. We found that, in planta, the constitutively active, GTP-bound AtGPA1(Q222L) mutant and the nucleotide-free AtGPA1(S52C) mutant interacted with Gβγ1 and Gβγ2 dimers with similar affinities, suggesting that G protein heterotrimer formation occurred independently of nucleotide exchange. In contrast, AtGPA1(Q222L) had a greater affinity than that of AtGPA1(S52C) for Gβγ3, suggesting that the GTP-bound conformation of AtGPA1(Q222L) is distinct and tightly associated with Gβγ3. Functional analysis of transgenic lines expressing either AtGPA1(S52C) or AtGPA1(Q222L) in the gpa1-null mutant background revealed various mutant phenotypes that were complemented by either AtGPA1(S52C) or AtGPA1(Q222L). We conclude that, in addition to the canonical GDP-GTP exchange–dependent mechanism, plant G proteins can function independently of nucleotide exchange.
INTRODUCTION
The heterotrimeric guanine nucleotide–binding protein (G protein) complex consists of a guanine nucleotide–bound Gα subunit and a Gβγ dimer tethered to the cytoplasmic side of the plasma membrane where it relays extracellular signals from cell surface receptors to initiate various signaling pathways. In animals, G protein–coupled receptors (GPCRs) catalyze the exchange of guanosine diphosphate (GDP) for guanosine triphosphate (GTP) on the Gα subunit, leading to a specific conformational change in Gα that reduces its affinity for Gβγ while increasing its affinity for downstream effectors (1). These effectors, such as adenylyl cyclases, cyclic guanosine 3′,5′-monophosphate (cGMP) phosphodiesterase, phospholipase Cβ isoforms, and Rho guanine nucleotide exchange factors (GEFs), specifically recognize the GTP-bound conformation of Gα (2, 3). Because of its intrinsic guanosine triphosphatase (GTPase) activity, the Gα subunit eventually hydrolyzes GTP to GDP and returns to Gβγ to reform the heterotrimer. Thus, in this classical model, GDP or GTP bound to Gα defines the inactive or active mode of signaling, respectively. GTP binding and hydrolysis by Gα require five structurally conserved loops termed the G1 to G5 boxes (4, 5). The G1 to G3 boxes coordinate the α-, β-, and γ-phosphate moieties of guanine nucleotides and the Mg2+ ion, whereas the G4 and G5 boxes are important for interaction with the guanine ring. In humans, mutations and posttranslational modifications that affect the GDP-GTP cycle lead to severe clinical abnormalities (6-9).
In plants, heterotrimeric G proteins are found only in multicellular species ranging from Charophyte green algae (Chara braunii) (10) to land plants (11-13), and they play direct and indirect roles in developmental and stress-related responses (14-16). The Arabidopsis Gα subunit (AtGPA1) has a highly similar crystal structure to that of animal Gαi (17) and achieves GTP- and GDP-bound conformations that are toggled with the assistance of GTPase-activating proteins, such as regulator of G protein signaling 1 (RGS1) and phospholipase Dα1 (18-20). Unlike animal Gα subunits, AtGPA1 binds and hydrolyzes GTP with kobs and kcat (single turnover) values of ~14 and 0.12 min−1, respectively, which means that GTP hydrolysis, not GDP release, is the rate-limiting step of the G protein cycle under the reported conditions (19). Representatives of vascular plant Gα subunits also show rapid GDP release and GTP binding in vitro without the help of GEFs (13), indicating that the fast nucleotide exchange is an evolutionarily conserved trait of plant Gα.
GTP binding induces a conformational change in Gα that typically decreases its affinity for Gβγ and promotes heterotrimer disassembly (21). Therefore, the disassembly is indicative of heterotrimer activation, whereas an intact heterotrimer is considered to be inactive. However, some heterotrimer combinations do not disassemble upon GTP binding but rather undergo a structural rearrangement (22-24). In rice, Gα (RGA1) bound to nonhydrolyzable GTPγS exists as a free form, as does the RGA1(Q223L) mutant, which is incapable of GTP hydrolysis (25). Similarly in maize, the Gα Compact plant 2, CT2(Q223L) variant, abolishes its interaction with the Gβγ dimer (16). Both examples support the dissociation-upon-activation model. In contrast, the Arabidopsis constitutively GTP-bound AtGPA1(Q222L) variant can interact with Gβγ, suggesting that the heterotrimer does not invariably disassemble upon activation (26). Arabidopsis G protein subunits are found in large protein complexes (~700 kDa), and treatment with GTPγS, which supposedly activates the heterotrimer, promotes only partial dissociation of AtGPA1 from the complex (27). These observations point to a complex relationship between GTP binding and dissociation of the heterotrimeric G proteins in plants, providing an argument against the use of dissociation as an indication of activation.
In addition, on the basis of functional analyses of constitutively GTP-bound Gα subunits in rice, maize, and Arabidopsis, it is unclear whether GTP binding always indicates activation of G protein–mediated signaling (16, 25, 28, 29). In rice, expression of RGA1(Q223L) driven by the native RGA1 promoter in a null RGA1 mutant (d1) background complements the d1 dwarf phenotype only to wild-type (WT) height instead of producing taller plants as might be expected from a constitutively active RGA1 (28). In maize, CT2(Q223L) only partially complements ct2 phenotypes, such as dwarfism, shortened leaves, and enlarged shoot apical meristem, whereas WT CT2 fully rescues all of the mutant phenotypes (16), leading to the conclusion that CT2(Q223L) functions as a weak, rather than a constitutively active, variant (16). These observations are inconsistent with the assumption that GTP binding by the Gα subunit activates G protein–mediated signaling. However, although expressing RGA1(Q223L) in a rice d1 background complements plant height only to that observed in the WT plant, the grains of plants expressing the mutant Gα are longer than the those of plants expressing WT Gα (28). This hypercomplementation of the d1 short seed phenotype is consistent with constitutive activation of the signaling pathway that mediates this trait (28). In Arabidopsis, ectopic expression of AtGPA1(Q222L) results in longer etiolated hypocotyls and primary roots than those of plants expressing WT AtGPA1 (18, 30), as well as increased stomatal density in the hypocotyl epidermis and in the abaxial epidermis of cotyledons (31), which is also consistent with the assumption of constitutive signaling. Nonetheless, interpretation of these results is complicated by the fact that these studies in Arabidopsis used a strong constitutive promoter overexpressing AtGPA1(Q222L) in a WT background, which could result in enhanced or aberrant traits.
Although the constitutively GTP-bound Gα has been widely studied, there are no reports on inactive Gα variants in plants. In mammalian systems, the G1 box (P-loop) makes contacts with the guanine nucleotide at the α and β phosphates (5). The serine/threonine residue at the final position of the G1 box consensus sequence G-X-X-X-X-G-K-(S/T) is particularly important for the β-phosphate contact and coordination of Mg2+. Mutations in that position virtually abolish GTP binding and prevent the variant Gα from achieving a stable active conformation (7, 32, 33). These mutations exert a dominant-negative effect and are well characterized in multiple human Gα subunits (Gs, Gi, Go and Gt), as well as in small, monomeric Ras G proteins (34).
Here, we studied two Arabidopsis Gα mutant variants: a presumably inactive AtGPA1(S52C) variant with abolished guanine nucleotide binding and a presumed constitutively active AtGPA1(Q222L) variant, which is incapable of GTP hydrolysis. We showed that AtGPA1(S52C) did not bind to GTP or GDP and that AtGPA1(Q222L) was GTP bound. The interactions of these mutant proteins with Gβγ dimers were then examined using yeast three-hybrid (Y3H) and in planta assays. Comparative analysis of transgenic plants expressing the mutated and native AtGPA1 subunits in a gpa1-null mutant background revealed a number of phenotypes that were complemented by AtGPA1(S52C). Our results provide evidence that nucleotide exchange is not essential for several functions of AtGPA1.
RESULTS
Substitution of Ser52 abolishes guanine nucleotide binding, whereas that of Gln222 abolishes GTP hydrolysis in AtGPA1
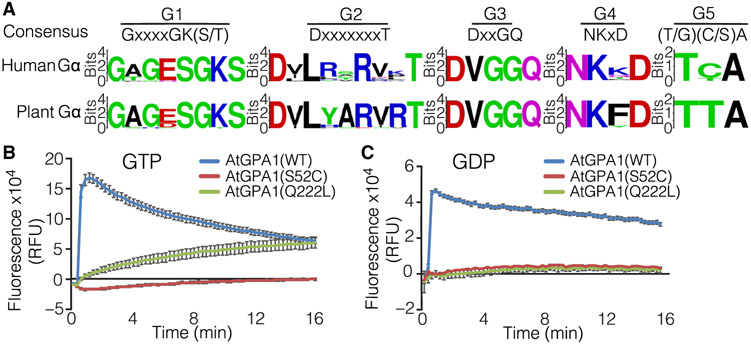
GTP binding and hydrolysis by Gα are mediated by five loop-like structures that are formed by highly conserved sequences termed G1 to G5 boxes (5). Sequence analysis of canonical Gα subunits from 24 plant species representing all classes of land plants and of 16 human Gα subunits (fig. S1) revealed a high degree of conservation in all five G boxes (Fig. 1A). Sequence conservation suggests the functional importance of GTP binding and is consistent with multiple studies associating amino acid substitutions in several of these boxes with health disorders in humans (7, 34). Substitutions of serine or threonine in the G1 box (P-loop) or glutamine in the G3 box result in reduced GTP affinity or GTPase activity of Gα, respectively (34-36). We produced the corresponding mutations, AtGPA1(S52C) and AtGPA1(Q222L), and assayed their ability to bind GTP in vitro. Incubation of purified recombinant AtGPA1(S52C) with fluorescent BODIPY-GTP did not result in an increase in fluorescence above basal, whereas fluorescence was increased in the presence of either WT AtGPA1 or AtGPA1(Q222L) (Fig. 1B and fig. S2A). WT AtGPA1 exhibited a rapid increase in fluorescence, indicating BODIPY-GTP binding, which was followed by a slow decrease, implying GTP hydrolysis (Fig. 1B). In the case of AtGPA1(Q222L), fluorescence did not decrease, which is consistent with its inability to hydrolyze GTP (Fig. 1B). The observed differences in GTP binding dynamics are consistent with those from previous reports for AtGPA1 (37) and maize Gα (CT2) (16). To determine whether AtGPA1(S52C) and AtGPA1(Q222L) could bind to GDP, we performed binding experiments with fluorescent BODIPY-GDP. GDP binding to either AtGPA1(S52C) or AtGPA1(Q222L) was negligible compared to the amount of GDP that bound to WT AtGPA1 (Fig. 1C and fig. S2B). These data suggest that AtGPA1(S52C) likely exists as a guanine nucleotide–free form, whereas AtGPA1(Q222L) is likely in a constitutively GTP-bound state. The impaired GTP-binding capacity of AtGPA1(S52C) compared to that of WT AtGPA1 was also observed in experiments with radiolabeled GTPγS (fig. S3). These data showed that AtGPA1(S52C) did not bind GTP at the concentrations that usually occur under physiological conditions; however, at increased concentrations, a limited amount of radiolabeled GTPγS might be bound to the protein.
Fig. 1. Sequence conservation of G boxes in Gα subunits and in vitro GDP and GTP binding of AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L).
(A) Consensus sequences for the conserved five polypeptide loops (G1 to G5 boxes) of Gα proteins from 24 plant species and 16 human Gα subunits. (B and C) Real-time fluorescence assays of AtGPA1(WT), AtGPA1(S52C), and AtGPA1(Q222L) upon addition of (B) GTP or (C) GDP conjugated to the BODIPY-FL fluorophore. Guanine nucleotide binding was assessed by the increase in relative fluorescence units (RFUs). Data are means ± SEM from three independent experiments.
AtGPA1(S52C) and AtGPA1(Q222L) interact with Gβγ dimers in planta
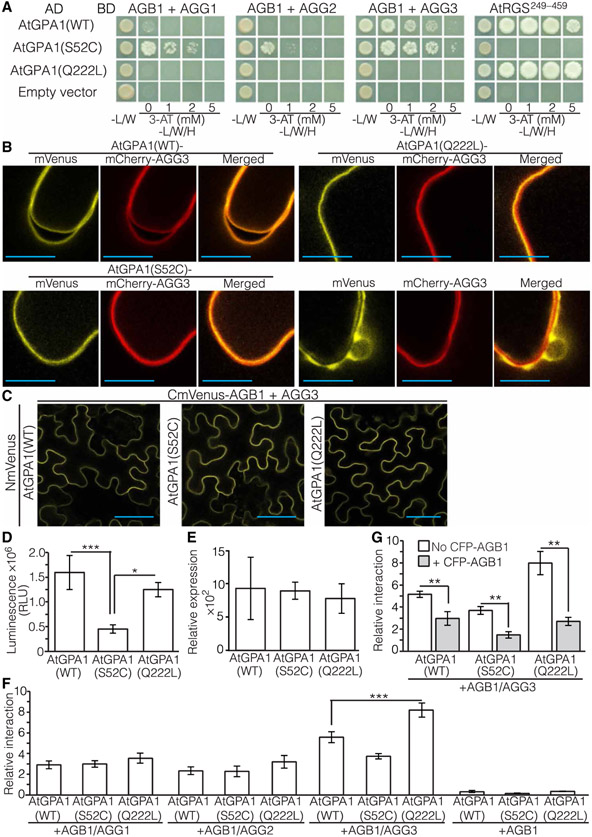
Heterotrimeric G proteins toggle between active and inactive states to control the signaling flow. In animal systems, these states are determined by the Gα subunit conformation, with GTP-bound Gα being active and GDP-bound Gα being inactive. For animal G proteins, in vivo GTP binding and subsequent activation can occur both with and without heterotrimer disassembly (38). Studies in plants showed inconsistent results, with GTP-bound Gα being partially or completely dissociated from the Gβγ dimer (25, 27) or bound to it (26). Such apparent contradictions could be explained by differences between the protein-protein interaction assays that were used, or they could reflect the distinct characteristics of plant heterotrimeric G proteins. To gain further insights into the dynamics of these subunit interactions, we tested the ability of WT AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L) to interact with Arabidopsis Gβ (AGB1) coexpressed with Gγ subunits (AGGs). Y3H assays showed that AtGPA1(S52C) had stronger interactions with the three different AGB1-AGG dimers compared to those of WT AtGPA1, as demonstrated by yeast growth on higher concentrations of the histidine synthesis competitive inhibitor 3-amino-1,2,4-triazole (3-AT) (Fig. 2A), whereas AtGPA1(Q222L) did not interact with any of the three dimers (Fig. 2A). To confirm that the lack of interaction was not due to the result of a lack of AtGPA1(Q222L) expression in yeast, we showed an interaction between AtGPA1(Q222L) and the cytoplasmic domain of AtRGS1, including the RGS box, an interaction that was not exhibited by AtGPA1(S52C) (Fig. 2A). Therefore, these data suggest that similar to animal and fungal Gα subunits, AtGPA1 can adopt distinct conformational states in a nucleotide-dependent manner.
Fig. 2. Gβγ-binding properties of AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L).
(A) Interaction between Arabidopsis Gα variants and Gβγ dimers in Y3H assays. AtGPA1(WT), AtGPA1(S52C), and AtGPA1(Q222L) fused to the GAL4 activation domain (AD) were coexpressed with AGB1 fused to the GAL4-binding domain (BD) and AGG1, AGG2, AGG3, or the C-terminal domain of AtRGS1 (RGS1249–459). Images are representative of three independent experiments. (B) Subcellular localization of mVenus-tagged AtGPA1 variants coexpressed with the plasma membrane protein AGG3 tagged with mCherry in N. benthamiana leaves. The localization of untagged mVenus coexpressed with mCherry-AGG3 is included for reference. Images are representative of three independent experiments. Scale bars, 10 μm. (C) BiFC assays for the interaction between AtGPA1 variants containing the N-terminal mVenus fragment (NmVenus) and AGB1 fused to the C-terminal mVenus fragment (CmVenus) in the presence of AGG3 in N. benthamiana leaves. Images are representative of five independent experiments. Scale bars, 50 μm. (D) Relative amounts of firefly luciferase–tagged AtGPA1(WT), AtGPA1(S52C), and AtGPA1(Q222L) by transient expression in N. benthamiana leaves evaluated as RLUs. Data are means ± SEM from nine independent replicate experiments. (E) qRT-PCR analysis of the expression of AtGPA1 variants fused to firefly luciferase and transiently expressed in N. benthamiana leaves. Data are means ± SEM from three independent replicate experiments. (F) Relative interactions between AtGPA1 variants double-tagged with a Cluc and an 11–amino acid peptide tag (HiBiT) and AGB1 tagged with an Nluc in N. benthamiana leaves. mCherry-tagged AGG1, AGG2, or AGG3 was coexpressed (+AGB1/AGG) or absent (+AGB1). Data are means ± SEM from 10 (+AGB1/AGG1 and +AGB1/AGG2), 12 (+AGB1/AGG3), or 4 (+AGB1) independent replicate experiments. (G) CFP-AGB1 coexpression effects on the interactions between AtGPA1 variants tagged with Cluc and HiBiT and AGB1 tagged with Nluc coexpressed with mCherry-AGG3 (+AGB1/AGG3) in N. benthamiana leaves. Data are means ± SEM from at least four independent replicate experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by one-way analysis of variance (ANOVA) with the Tukey’s multiple comparisons method [for (D) to (F)] and by unpaired two-tailed Student’s t test comparing between with and without CFP-AGB1 for each AtGPA1 variant interaction (G).
AtGPA1 forms a complex with the AGB1-AGG1 dimer at the plasma membrane (26). To test whether nucleotide-binding impairment or constitutive GTP binding affected the plasma membrane localization of AtGPA1, we inserted mVenus into the αB-αC loop of the helical domain of AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L) and analyzed their subcellular localization in Nicotiana benthamiana leaves at high resolution without ectopic coexpression of AGB1. All of the AtGPA1 variants displayed a pattern that was consistent with plasma membrane localization (Fig. 2B). Next, we assayed the AtGPA1-AGB1-AGG3 interaction in planta by bimolecular fluorescence complementation (BiFC). The N-terminal fragment of the yellow fluorescent protein mVenus was introduced into the αB-αC loop of the helical domain in AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L), whereas its C-terminal fragment was fused to AGB1. These AtGPA1 and AGB1 fusion proteins were transiently coexpressed together with AGG3 tagged with the red fluorescent protein mCherry in the leaves of N. benthamiana. BiFC analysis showed interactions between AGB1 and AtGPA1, AtGPA1(S52C), or AtGPA1(Q222L) in the presence of AGG3 (Fig. 2C), whereas no fluorescence was observed in the absence of AGG3 (fig. S4).
An interaction between AtGPA1(Q222L) and AGB1-AGG1 in planta was reported previously (26) but is in contrast with the results from our Y3H experiments (Fig. 2A). This discrepancy prompted us to perform additional protein-protein interaction studies in planta using the quantitative split firefly luciferase complementation (SFLC) assay. SFLC assays are used to quantify interactions because of an almost linear relationship between the light intensity emitted upon luciferin oxidation and the amount of luciferase protein (39). It is nevertheless important to ascertain the amounts of the individual fusion proteins because these ultimately affect the amount of reconstituted luciferase. To estimate the amount of each AtGPA1 variant produced in transient expression assays, we inserted firefly luciferase in the αB-αC loop of the helical domains of AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L) and expressed these fusion proteins in N. benthamiana leaves. The relative luminescence unit (RLU) values that we obtained from the AtGPA1(S52C) samples were consistently less than those obtained from the WT AtGPA1 and AtGPA1(Q222L) samples (Fig. 2D).
Our quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis revealed that the mRNA abundances of all three transiently expressed AtGPA1 variants were similar (Fig. 2E), which suggested that the AtGPA1(S52C) protein was less stable than WT AtGPA1 and AtGPA1(Q222L), perhaps because of its nucleotide-free conformation (7). To address the issue of unequal protein amount, we designed a different approach that enabled us to evaluate the amount of AtGPA1 interacting with AGB1, as well as the total amount of AtGPA1 in the same sample for normalization purposes. The C-terminal fragment of firefly luciferase (CLuc) (40, 41) was fused to the HiBiT sequence (42) and introduced into the αB-αC loop of the helical domain of AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L), whereas the N-terminal fragment of firefly luciferase (NLuc) was fused to AGB1. The constructs were coexpressed together with each of the three mCherry-tagged AGG constructs in N. benthamiana leaves. To evaluate the strength of each interaction, luciferin was added to the wells of 96-well plates containing leaf samples and light emission was then measured. Then, Nano-Glo LgBiT protein and reagent (Promega) were then added to the wells, and the second light emission was measured. The intensity of the luciferase light emission indicated the relative amount of AtGPA1 bound to AGB1-AGG, whereas the second reading evaluated the light emitted by the HiBiT-LgBiT complex, which was proportional to the total amount of AtGPA1. To compare the interaction stringency among different subunits, we normalized the luciferase emission values (first reading) using the HiBiT-LgBiT emission intensity (second reading). We found that both the nucleotide-free and GTP-bound conformations of AtGPA1 interacted with all three AGB1-AGG dimers (Fig. 2F), confirming our BiFC results and previously reported data (26). When no AGG subunit was coexpressed, a low level of light emission was observed, indicative of an interaction between AtGPA1 and AGB1 (Fig. 2F) perhaps caused by native N. benthamiana Gγ subunits targeting a small portion of NLuc-AGB1 to the plasma membrane where an interaction with AtGPA1 could take place.
To ensure that the luciferase complementation was not due to surface concentration effects, we performed a competition assay by assessing the effect of coexpressing cyan fluorescent protein (CFP)–fused AGB1 on the interaction between AtGPA1 and AGB1 fused to the luciferase fragments. In the case of a specific interaction, CFP-AGB1 should compete with NLuc-AGB1, thereby reducing the light intensity produced by reconstituted luciferase. We found that coexpression of CFP-AGB1 decreased the light intensity values for all three AtGPA1 variants (Fig. 2G). Therefore, the SFLC between the AtGPA1 variants and AGB1 was specific and not due to surface concentration. We observed that values of luciferase complementation were higher when AtGPA1(Q222L) and AGB1-AGG3 were coexpressed compared to that of either AtGPA1(WT) or AtGPA1(S52C) and AGB1/AGG3 (Fig. 2F), suggesting a greater binding affinity of AtGPA1(Q222L) for AGB1-AGG3. The luciferase complementation was similar for the three AtGPA1 variants when the AGB1-AGG1 and AGB1-AGG2 dimers were used (Fig. 2F). Our findings on the interaction between AtGPA1 and AGB1-AGG3 were unexpected and may be specific for the plant system, because AGG3 is a divergent, plant-specific form of Gγ with a transmembrane topology (43, 44). In addition, plant G proteins are found in large, 400- or 700-kDa protein complexes (25, 27), which suggests the presence of additional proteins in the native system. Proteins such as RGS1 could affect the interaction between AtGPA1 and AGB1-AGG. To test this hypothesis, we fused the N-terminal fragment of luciferase to the C terminus of AtRGS1 and quantified its interaction with the three AtGPA1 variants. The interaction of AtGPA1(Q222L) with AtRGS1 was stronger than those of the other AtGPA1 variants (fig. S5). We also demonstrated that AtRGS1 interacted with three distinct AGB1-AGG dimers (fig. S6), supporting the possibility that AtRGS1 mediates interactions between AtGPA1 and AGB1-AGG dimers. Together, these observations suggest that AtGPA1 exhibits distinct nucleotide-dependent conformations, although GTP binding provides a different outcome for its interaction with AGB1-AGG3 depending on the cellular environment of the assay (Fig. 2, A and F).
The GTP binding–impaired AtGPA1(S52C) variant complements some, but not all, gpa1-null mutant phenotypes
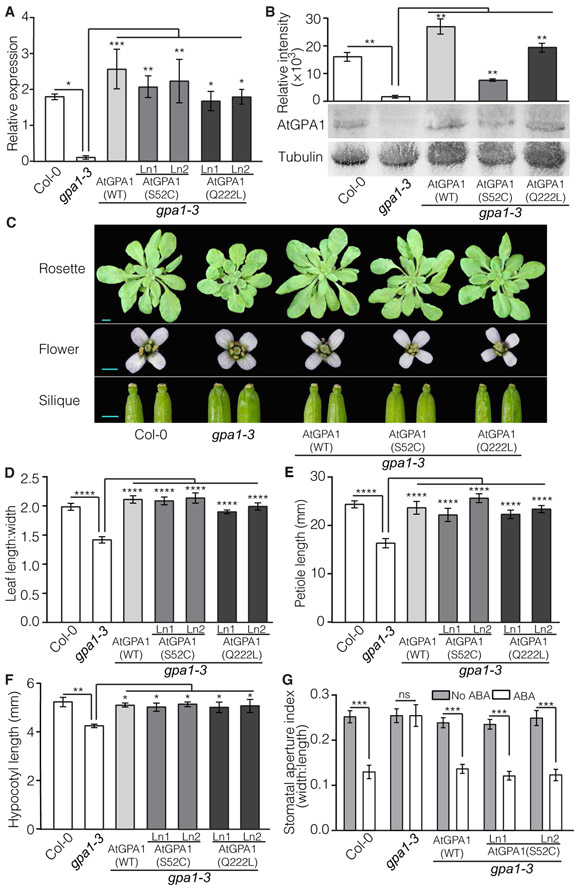
Our earlier results indicated that AtGPA1(S52C) is deficient in binding to both GTP and GDP (Fig. 1, B and C) and therefore cannot adopt the canonical active conformation. Knockout of AtGPA1 results in various phenotypes (14, 29). We queried whether plants expressing AtGPA1(S52C) were functionally equivalent to AtGPA1 knockout plants, assuming that if AtGPA1(S52C) is inactive, it should not complement the mutant phenotypes. We transformed the null gpa1-3 mutant with AtGPA1(WT), AtGPA1(S52C), or AtGPA1(Q222L) under the control of the native AtGPA1 promoter. Multiple T3 homozygous lines were obtained for each construct, and transgene expression was evaluated by qRT-PCR analysis. Lines with similar expression to that of the native AtGPA1 transcripts in WT (Col-0) plants were selected for further analyses (Fig. 3A). Western blotting analyses confirmed the presence of AtGPA1(WT), AtGPA1(S52C), and AtGPA1(Q222L) proteins in the transgenic lines, although the amount of AtGPA1(S52C) was less than that of the other two variants (Fig. 3B), consistent with the results obtained in the transient expression studies in N. benthamiana leaves (Fig. 2D).
Fig. 3. Complementation of gpa1-3 mutant phenotypes by AtGPA1(S52C) and AtGPA1(Q222L).
(A) qRT-PCR analysis of the expression of AtGPA1 variants relative to SAND (AT2G28390) in 14-day-old Arabidopsis transgenic lines (Ln) in the gpa1-3 background. Data are means ± SEM from five experiments. (B) Western blotting analysis of AtGPA1 variants was performed on 6-day-old seedlings of Col-0, gpa1-3, and gpa1-3–expressing GPA1(WT), AtGPA1(S52C) Ln1, and AtGPA1(Q222L) Ln1. The bar graph represents the intensity of bands corresponding to the AtGPA1 variants relative to those representing α-tubulin. Data are means ± SEM from three experiments. **P < 0.01 by unpaired two-tailed Student’s t test in comparison with gpa1-3. Blots are representative of three experiments. (C) Representative rosette, flower, and silique morphologies in Col-0, gpa1-3, and gpa1-3–expressing AtGPA1(WT), AtGPA1(S52C) Ln1, and AtGPA1(Q222L) Ln1. Scale bars, 1 mm. (D) Ratios of rosette leaf length:width of 5-week-old plants. Data are means ± SEM of 14 to 18 plants per genotype. (E) Petiole lengths of 5-week-old plants. Data are means ± SEM of 12 to 15 plants per genotype. (F) Hypocotyl lengths of 2-day-old etiolated seedlings. Data are means ± SEM from three experiments with at least 10 to 24 seedlings per genotype per experiment. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by one-way ANOVA with Tukey’s multiple comparisons test [for (A) and (D) to (F)]. (G) Stomatal opening index evaluated by the width:length ratio of stomatal apertures after exposure to 50 μM ABA for 3 hours in the presence of white light (120 μmol/m2 s1). Data are means ± SEM from three plants with 26 to 49 stomatal apertures measured per sample. ***P < 0.005 by unpaired two-tailed Student’s t test for comparison between ABA-treated and untreated plants per genotype. ns, not significant.
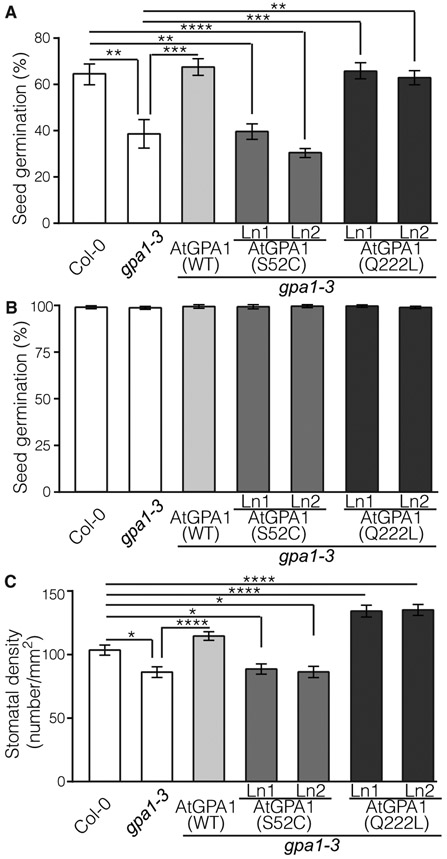
The AtGPA1 knockout mutant exhibits characteristic morphological abnormalities in above-ground organs, including smaller rosettes and flowers, rounder leaves with shorter petioles, altered silique morphology with a distinct flattened tip, and shorter dark-grown hypocotyls (14). AtGPA1(S52C) rescued all of these morphological alterations in the gpa1-3 mutant, producing plants with a WT appearance (Fig. 3C). Quantitative measures of leaf length:width ratio, petiole length, and hypocotyl length confirmed the complementation by AtGPA1(S52C) and showed that AtGPA1(WT) and AtGPA1(Q222L) also complemented the gpa1-3–null mutant phenotypes to a similar extent as AtGPA1(S52C) (Fig. 3, D to F), suggesting that nucleotide exchange on AtGPA1 is not necessary for these functions. AtGPA1 also mediates responses to abscisic acid (ABA), with gpa1-3 mutants displaying hyposensitivity to ABA-triggered inhibition of stomatal opening (45) and hypersensitivity to ABA-induced inhibition of seed germination (46). Stomatal opening sensitivity to ABA was restored by AtGPA1(S52C) and AtGPA1(WT) (Fig. 3G). In contrast, AtGPA1(S52C) failed to rescue the hypersensitivity to ABA inhibition of seed germination displayed by gpa1-3 mutants, whereas AtGPA1(WT) and AtGPA1(Q222L) complemented this phenotype (Fig. 4A). Germination rates without ABA supplementation were similar in all genotypes (Fig. 4B). It is tempting to suggest that this phenotype requires nucleotide exchange and hence nucleotide exchange–dependent activation of AtGPA1, although we cannot discard the possibility that the reduced amount of AtGPA1(S52C) in the transgenic lines could be responsible for the complementation failure. AtGPA1 is a regulator of transpiration efficiency in Arabidopsis, with gpa1 mutants displaying reduced stomatal density (31, 47). We found that AtGPA1(S52C) failed to rescue the reduced stomatal density phenotype, whereas AtGPA1(WT) fully restored it and AtGPA1(Q222L) caused a statistically significant increase in stomatal density compared to that in Col-0 (Fig. 4C). The combined results whereby AtGPA1(Q222L) outperformed AtGPA1(WT) but AtGPA1(S52C) failed to complement the mutation support a nucleotide exchange–dependent mechanism controlling stomatal production. Together, these results suggest that AtGPA1, and hence plant G proteins, can function using nucleotide exchange–dependent and nucleotide exchange–independent mechanisms.
Fig. 4. AtGPA1(S52C) does not complement some gpa1-3 mutant phenotypes.
(A and B) Seed germination rate (%) of Col-0, gpa1-3, and Arabidopsis transgenic lines (Ln) expressing AtGPA1(WT), AtGPA1(S52C), and AtGPA1(Q222L) in the gpa1-3 background after exposure to 5 μM ABA for 48 hours (A) or after 48 hours without treatment (B). Data are means ± SEM from three independent replicates (plates) with 100 to 200 seeds per genotype used for each replicate. (C) Number of stomata per square millimeter on the rosettes of 5-week-old plants of the indicated genotypes. Data are means ± SEM from at least seven plants. *P < 0.05, **P < 0.005, ***P < 0.001, and ****P < 0.0001 by one-way ANOVA with Tukey’s multiple comparisons test.
DISCUSSION
The finding that the nucleotide-free AtGPA1(S52C) variant and the constitutively GTP-bound AtGPA1(Q222L) variant rescued gpa1 phenotypes (Fig. 3, C to F) leads us to the conclusion that AtGPA1 has nucleotide exchange–independent functions. Nevertheless, not all of the studied phenotypes showed nucleotide-independent behavior (Fig. 4C), indicating the coexistence of nucleotide-dependent and nucleotide-independent functions for AtGPA1. Although nucleotide exchange–dependent G protein signaling exists in plants, it seems that the plant G protein cycle does not simply mirror the animal model. The absence of prototypical GPCRs with GEF activity, the high rate of GDP release, and the low rate of GTP hydrolysis shown by AtGPA1 argue against the existence of the canonical guanine nucleotide exchange activation mechanism for plant G proteins (13, 19, 48). Several models for plant-specific, noncanonical signaling mechanisms involving guanine nucleotide exchange have been proposed (26, 48-50).
Guanine nucleotide–independent activation of Gα (51, 52) and heterotrimer disassembly (38) is known in animal systems, although the activation of GDP-bound Gα is disputed (53). In plants, inactive AtGPA1 within its heterotrimeric state attenuates cell division in primary roots (54). There is a growing body of evidence suggesting the involvement of G proteins in plant responses that are initiated by receptors that do not resemble canonical GPCRs (55), and the hypothesis that plant G proteins could be activated by phosphorylation has been proposed (56). A number of single-transmembrane receptor-like kinases (RLKs) and receptor-like proteins interact directly with plant G protein subunits (50, 57-63), and G protein subunits are phosphorylated in vivo (60, 61, 64, 65). The RLK brassinosteroid-insensitive 1–associated receptor kinase 1 (BAK1) phosphorylates AtGPA1 at a conserved Tyr166 residue (64). A phosphomimetic substitution at this position stabilizes the interaction between AtRGS1 and AtGPA1 in its GDP-bound conformation, providing a putative phosphorylation-dependent mechanism that circumvents the need for GDP-GTP exchange. It is also possible to contemplate alternative scenarios in which the rescue of the mutant phenotypes could be due to an indirect signaling role for AtGPA1. Scaffolding proteins are common in signaling pathways, providing spatial proximity to multiple signaling components (66). Plant Gα subunits could have functions as scaffolds for other proteins without the need for cycling between active and inactive conformations. In this scenario, knockout mutant phenotypes could be rescued by any conformation of Gα subunit provided that it provides a binding surface for the proteins involved.
G proteins are constituents of multiprotein complexes at the plasma membrane (25, 27). We hypothesize that these complexes are diverse and that the composition of each complex determines the nature of the activation mechanism, signaling pathway, and ultimately the function of the G protein heterotrimer. For example, complexes that recruit AtRGS1 presumably favor nucleotide exchange–dependent activation of AtGPA1 to mediate processes such as stomatal development (Fig. 4C) and root cell division (54). In contrast, AtGPA1 activity is likely modulated by phosphorylation when the heterotrimer directly couples to RLKs, whereas in other complexes, AtGPA1 could function without a specific activation step. The existence of complexes with different modes of action explains some apparent inconsistencies in the published literature. Briefly, in rice, Gα subunits are part of 400-kDa complexes, treatment with GTPγS releases almost 100% of Gα from these complexes, and Gα(Q223L) exists as a free form (25). In contrast, only 30% of Arabidopsis total Gα is in multiprotein complexes of 700 kDa, treatment with GTPγS releases only a third of the Gα subunits from these complexes (27), and Gα(Q222L) is bound to Gβγ (Fig. 2, C and F) (26). The first and most obvious difference between rice and Arabidopsis is the physical size of the Gα-containing complexes. We argue that G protein–containing complexes in rice and Arabidopsis are different in nature. In rice leaves, the predominant Gγ subunit is RGG2 (67), which is a type B Gγ that is characterized by lack of the isoprenylation motif that is essential for animal Gγ subunits (12). In contrast, Arabidopsis is deficient in type B Gγ (12), and the predominant complexes in leaves likely contain type A Gγ subunits, namely, AGG1 or AGG2. Gγ subunits determine the interaction specificity between Gβγ and AtGPA1 (Fig. 2, A and F) and provide functional selectivity for heterotrimeric G proteins (43, 68). Therefore, the expected inherent differences between the G protein complexes present in rice and Arabidopsis leaves could explain the differences observed upon treatment with GTPγS. Our hypothesis predicts at least four modes of action for different multiprotein complexes: nucleotide exchange–dependent activation with and without dissociation, as well as nucleotide exchange–independent activation with and without dissociation. The report that treatment with GTPγS released about 30% of AtGPA1 from multiprotein complexes in Arabidopsis leaves (27) is consistent with models in which (i) heterotrimer disassembly occurs in only a subset of GTPγS-bound complexes and (ii) AtGPA1 is not always activated by GTPγS binding. Our results showed that GTP-bound AtGPA1(Q222L) did not interact with AGB1-AGG dimers in the Y3H system (Fig. 2A); however, these interactions did occur in planta (Fig. 2, C and F) (26). We posit that complexes with ectopically expressed plant G protein subunits would naturally display different dynamics in planta and yeast. Specifically, the disassembly of the G protein heterotrimer would be determined by the presence or absence of accessory proteins. Thus, depending on the experimental context, such as cell type and species, the analyzed complexes would differ, leading to distinct dynamics in plant heterotrimers. The existence of the different multiprotein complexes also supports the multiple roles reported for plant G proteins, which may explain how a small repertoire of G protein subunits provides specific signaling to a multitude of processes.
In conclusion, we showed that plant canonical Gα subunits can be functional without the requirement for guanine nucleotide exchange. This departure from the textbook paradigm of G protein activation changes our understanding of G proteins, at least in plants. On the practical side, given that plant G proteins are major determinants of essential agronomic traits, such as yield and nitrogen use efficiency in cereals (69-72), establishing their activation mechanisms can provide new avenues to improve agricultural productivity and sustainability.
MATERIALS AND METHODS
Sequence analysis
The amino acid sequences of Gα subunits from 24 diverse plant species representing all major taxa of green plants [Viridiplantae (taxid:33090)] were obtained by homology searches using Basic Local Alignment Search Tool (BLAST) on the database Nucleotide collection (nr/nt) and Expressed Sequence Tags (EST) with AtGPA1 as a query sequence. Sixteen human Gα subunits representing the four classes of animal Gα subunits were obtained from the National Center for Biotechnology Information (NCBI) protein database. The sequences were aligned using the multiple sequence alignment tool Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/).
Purification of AtGPA1 protein and in vitro guanine nucleotide–binding assays
The complementary DNA (cDNA) fragments encoding AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L) were subcloned into the pDEST17 vector. The proteins were expressed in ArcticExpress cells upon induction with 80 or 250 μM isopropyl-β-d-thiogalactopyranoside (IPTG) at 14°C. The cells were harvested by centrifugation and lysed in extraction buffer [50 mM tris-HCl (pH 8.0), 100 mM NaCl, 2 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF), leupeptin (2 μg/ml), 10 μM GDP, lysozyme (250 μg/ml), and 0.2% C12E10]. The bacterial lysate was centrifuged at 100,000g for 1 hour and then incubated with nickel-NTA agarose. The proteins were eluted from the resin with elution buffer [20 mM tris-HCl (pH 8.0), 250 mM NaCl, 5 mM 2-mercaptoethanol, 1 mM PMSF, and 10% glycerol] with 100 or 250 mM imidazole. The eluted proteins were dialyzed in buffer [25 mM tris-HCl (pH 7.6), 50 mM NaCl, 2 mM MgCl2, 1 mM EDTA, 5 mM 2-mercaptoethanol, 1 mM PMSF, and 10 μM GDP] and frozen at −80°C. The amount of GTPγS bound to the recombinant proteins was assessed using radiolabeled [35S]GTPγS (PerkinElmer Inc.). Recombinant AtGPA1 proteins at 0.5 μM were mixed with a series of concentrations of [35S]GTPγS (1.25, 2.5, 5, 10, 20, 40, and 80 μM) in 60 μl of reaction buffer [50 mM tris-HCl (pH 7.4), 1 mM EDTA, 1 mM dithiothreitol (DTT), and 5 mM MgCl2]. The lowest concentration of [35S]GTPγS used (1.25 μM) was still 2.5-fold greater than the concentration of GPA1 proteins (500 nM = 0.5 μM). After incubation at room temperature for 2.5 hours, 50 μl of the reaction mixture was transferred into 450 μl of ice-cold wash buffer [20 mM tris-HCl (pH 7.4), 100 mM NaCl, and 25 mM MgCl2] with 0.1 mM GTP and then applied to a nitrocellulose membrane filter. The filter was washed three times with ice-cold wash buffer. The amount of [35S]GTPγS on the membrane was measured by scintillation counting. For BODIPY-FL-GDP- and BODIPY-FL-GTP-binding assays, the proteins were purified as described earlier, with the modifications that protein expression was induced with 500 μM IPTG, that GDP was excluded from the extraction buffer, that the lysates were centrifuged at 30,000g for 15 min, and that buffer exchange was performed into 25 mM tris-HCl (pH 7.7), 50 mM NaCl, and 5% glycerol using a centrifugal filter with a 10,000 cutoff. Assays were run at 25°C with a Berthold TriStar2 LB 942 plate reader with 485-nm excitation and 535-nm emission filters, a 15-s cycle time, a 0.1-s counting time per measurement, and 60% lamp intensity. Proteins were diluted to 340 nM in 100 μl of dialysis buffer [25 mM tris-HCl (pH 7.7), 50 mM NaCl, and 5% glycerol] supplemented with 10 mM MgCl2. Binding was initiated with the addition of 100 μl of 100 nM BODIPY-FL-GDP or BODIPY-FL-GTP in 20 mM tris-HCl (pH 8.0) at cycle 2 of 61. Raw data were normalized by fluorescence values obtained for bovine serum albumin, which was used as a negative control for nucleotide binding.
Protein-protein interaction assays and subcellular localization
Y3H assays were conducted in a semiquantitative manner using a 3-AT curve, as previously described (73). To perform in planta subcellular localization and protein interaction analyses, Agrobacterium tumefaciens GV3101 was transformed with designated plasmids. The bacteria were grown for 16 to 24 hours at 30°C and harvested by centrifugation. They were resuspended in infiltration buffer (10 mM MgCl2 and 100 μM acetosyringone), diluted to give an OD600 (optical density at 600 nm) = 0.2, and infiltrated into 4-week-old N. benthamiana leaves using a syringe without a needle. All tests contained plasmids with the gene silencing suppressor p19. For subcellular localization studies, mVenus was inserted in the αB-to-αC loop (between Leu131 and Thr132) of AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L) and subcloned into pCambia1305.1. mCherry was fused to AGG3 and coexpressed as a plasma membrane marker. For BiFC assays, mVenus was split into N- and C-terminal fragments at Lys210 (74). The N-terminal mVenus fragment, NmVenus, was inserted into the αB-to-αCloop (between Leu131 and Thr132) of AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L), and the resultant constructs were subcloned into pCambia1305.1. The sequence encoding CmVenus was fused to that encoding AGB1, and the resultant construct was subcloned in pCambia1305.1. These constructs were introduced into leaves of 3- to 4-week-old N. benthamiana plants together with a plasmid encoding mCherry-tagged AGG3. At 48 hours after infiltration, leaf samples were observed under a Zeiss LSM 700 confocal microscope. Photographs were taken with a 510/550-nm (yellow fluorescent protein) excitation/emission filter and/or with a 580/630-nm (mCherry) excitation/emission filter. The SFLC assay used two plasmids, pLuciN and pLuciC, which were generated for this study with two fragments of firefly luciferase reported previously (41). The cDNA of AGB1 was subcloned into pLuciN. Two cDNA fragments each of AtGPA1, AtGPA1(S52C), and AtGPA1(Q222L) splitting the proteins in the αB-to-αC loop at position Leu131 were sequentially subcloned into pLuciC, resulting in the insertion of the firefly luciferase C-terminal fragment into the αB-to-αC loop of the AtGPA1 subunits. A sequence encoding an 11–amino acid tag, HiBiT (42), was subcloned next to the sequence encoding the firefly luciferase C-terminal fragment in each AtGPA1 construct. Infiltrations included mCherry-tagged AGG1, AGG2, or AGG3. Seventy-two hours after infiltration, leaf discs of 6 mm in diameter were excised and each disc was placed in a well of a 96-well plate. Luciferin (40 μl; Promega, catalog number E1601) diluted to 0.2 mM in sterile water was added into the wells, and luminescence was measured using a GloMax Reader (Promega) for 3 hours at 5 s per well. The maximum luminescence value for each disc was selected (first reading). Then, the luciferin solution was removed and 40 μl of Nano-Glo HiBiT Lytic Reagent (Promega, catalog number N3030), which was prepared according to the manufacturer’s instructions, was added. The luminescence was measured using a GloMax Reader for 1 hour at 5 s per well. The maximum luminescence value for each disc was selected (second reading). Every combination of A. tumefaciens carrying constructs of interest was infiltrated into 4 to 12 leaves as independent replicate experiments. To calculate the relative interaction, we divided the first reading values by the values of the second reading and multiplied by 100. For each independent replicate experiment, at least three leaf discs were measured and the resultant values per disc were used to calculate a mean value of every independent replicate experiment.
Plant materials and growth conditions
The Arabidopsis transfer DNA insertion null mutant used in this study was gpa1-3, which was previously described (75). To produce transgenic lines expressing AtGPA1 variants, the AtGPA1 promoter sequence 1438 base pairs upstream of the coding region was PCR-amplified from WT (Col-0) genomic DNA. The AtGPA1 coding sequence was amplified from Col-0 cDNA, and the AtGPA1(Q222L) coding sequence was amplified from a plasmid provided by H. Okamoto (University of Southampton, United Kingdom) using primers 5′-TCTGCAGTAGAAGTCGACATCATACT-3′ and 5′-TCCATGGTCATAAAAGGCCAGCCTCCAGT-3′. The AtGPA1(S52C) coding sequence was generated by site-directed mutagenesis PCR using two pairs of primers: 5′-TCTGCAGTAGAAGTCGACATCATACT-3′ with 5′-TTGTACATTTTCCAGATTCCCCAGCA-3′ and 5′-TGCTGGGGAATCTGGAAAATGTACAA-3′ with 5′-TCCATGGTCATAAAAGGCCAGCCTCCAGT-3′. The promoter, coding sequence, and the nopaline synthase terminator were introduced into the binary vector pART27. Stable transgenic Arabidopsis lines were produced by the floral dip inoculation method, as described previously (76). T3 and T4 homozygous lines were used for analyses. Unless otherwise specified, plants were grown under a short-day photoperiod (8 hours/16 hours) at 23°C/19°C.
Gene expression analysis by qRT-PCR
RNA was extracted from 2-week-old seedlings using a Maxwell RSC Plant RNA kit (Promega). Total RNA (1 μg per sample) was processed to produce cDNA using iScript Reverse Transcription Supermix (Bio-Rad). PCRs using SYBR Green RT-PCR Reagent (Roche, catalog number 06402712001) were set up using the following primers: 5′-ATCAGCGAGTACGACCAAAC-3′ and 5′-GTTTCAGGACCCAGTCGAAT-3′ (AtGPA1) and 5′-GTTGGGTCACACCAGATTTTG-3′ and 5′-GCTCCTTGCAAGAACACTTCA-3′ (MON1). MON1 (monensin sensitivity 1) (AT2G28390) was used as a reference gene (77).
Western blotting
Seedlings were grown on half-strength Murashige and Skoog (MS) medium containing 1% sucrose and 1% agar (pH 6.0) under 12-hour/12-hour light/dark conditions at 23°C for 6 days and then were ground in liquid nitrogen. The samples were incubated in extraction buffer [50 mM tris-HCl (pH 7.5), 50 mM NaCl, 5 mM EGTA, 2 mM DTT, 1% Triton X-100, 1% SDS, and protease inhibitor cocktail (Sigma-Aldrich, catalog number P9599)] for 2 hours at 4°C, which was followed by centrifugation at 17,000g for 20 min at 4°C. Total protein extracts were resolved by 12% SDS–polyacrylamide gel electrophoresis, transferred onto Hybond-C extra nitrocellulose (GE Healthcare Lifescience), and incubated with a primary antibody against AtGPA1 (Agrisera, catalog number AS122370) at a 1:2000 dilution or with an antibody against tubulin α chain (Agrisera, catalog number AS10680) at a 1:1000 dilution, which was followed by incubation with alkaline phosphatase–conjugated anti-rabbit secondary antibody. Signals were visualized by chromogenic substrate [bromochloroindolyl phosphate–nitro blue tetrazolium (BCIP-NBT)] according to the manufacturer’s protocol (Thermo Fisher Scientific, catalog number WB7105). Band intensities for each antibody were quantified with ImageJ software, and relative intensities compared to those of bands corresponding to α-tubulin were plotted.
Hypocotyl elongation
Surface-sterilized seeds were sown onto petri dishes with half-strength MS medium, 1% sucrose, and 1% agar (pH 6.0) and stratified at 4°C for 5 days. Germination was induced by 16-hour light treatment at 22°C, and then the plates were covered with foil and held vertically to facilitate hypocotyl elongation. Photographs were taken after 48 hours. Hypocotyl lengths were measured from captured images using ImageJ software.
ABA inhibition of seed germination
Plants of the designated genotypes were grown side by side to minimize environmental variabilities in a short-day photoperiod (8 hours/16 hours) at 23°C/19°C for 4 weeks, which was followed by a long-day photoperiod (16 hours/8 hours) at 23°C for flowering induction. The sets of dry-harvested seeds were surface-sterilized and sown onto plates with half-strength MS medium, 1% sucrose, and 1% agar (pH 6.0) with or without 5 μM ABA. Seeds were stratified at 4°C for 4 days and then were transferred to long-day light conditions at 22°C. The germination rate was scored at 48 hours after light induction.
Stomatal analyses
The eighth or ninth leaves of 5-week-old plants were used to prepare abaxial epidermal strips, which were stained with an aqueous solution of toluidine blue (2 mg/ml). Stomatal density was scored as the number of stomata per 1 mm2. ABA inhibition of stomatal opening assays was performed following a previously described protocol (78). The stained epidermal samples were photographed with a Zeiss Axio Scope.A1 with a 20× objective lens. The lengths and widths of the stomata were measured with ImageJ software, and the stomatal aperture index (the width:length ratio) was calculated.
Supplementary Material
Fig. S1. Conserved motifs in Gα subunits.
Fig. S2. In vitro BODIPY-GTP– and BODIPY-GDP–binding studies.
Fig. S3. Analysis of GTPγS binding to AtGPA1 and AtGPA1(S52C).
Fig. S4. Negative controls for BiFC assays.
Fig. S5. Assessment of interactions between AtRGS1 and AtGPA1 variants in planta.
Fig. S6. Assessment of the interaction between AtRGS1 and AGB1 in planta and of the effects of AGG subunits.
Acknowledgments:
We thank D. Greenway (University of Queensland) for providing an independent confirmation that appropriate statistical tests were used to analyze the data in this study. We thank A. M. Jones (University of North Carolina) for discussing our original data and hypothesis. We thank H. Okamoto (University of Southampton, United Kingdom) for providing the AtGPA1(Q222L) clone.
Funding: The research was supported by the University of Queensland PhD scholarship (to N.M.) and by the National Institute of General Medical Sciences of the NIH under award number R01GM126079 (to S.M.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Oldham WM, Hamm HE, Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol 9, 60–71 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Aittaleb M, Boguth CA, Tesmer JJG, Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol. Pharmacol 77, 111–125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodmann E-L, Wolters V, Bünemann M, Dynamics of G protein effector interactions and their impact on timing and sensitivity of G protein-mediated signal transduction. Eur. J. Cell Biol 94, 415–419 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Bourne HR, Sanders DA, McCormick F, The GTPase superfamily: Conserved structure and molecular mechanism. Nature 349, 117–127 (1991). [DOI] [PubMed] [Google Scholar]
- 5.Sprang SR, G protein mechanisms: Insights from structural analysis. Annu. Rev. Biochem 66, 639–678 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Iiri T, Herzmark P, Nakamoto JM, van Dop C, Bourne HR, Rapid GDP release from Gsα in patients with gain and loss of endocrine function. Nature 371, 164–168 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Marivin A, Leyme A, Parag-Sharma K, DiGiacomo V, Cheung AY, Nguyen LT, Dominguez I, Garcia-Marcos M, Dominant-negative Gα subunits are a mechanism of dysregulated heterotrimeric G protein signaling in human disease. Sci. Signal 9, ra37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milligan G, Kostenis E, Heterotrimeric G-proteins: A short history. Br. J. Pharmacol 147, S46–S55 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Hayre M, Vázquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Gutkind JS, The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer 13, 412–424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackenberg D, Sakayama H, Nishiyama T, Pandey S, Characterization of the heterotrimeric G-protein complex and its regulator from the green alga Chara braunii expands the evolutionary breadth of plant G-protein signaling. Plant Physiol. 163, 1510–1517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anantharaman V, Abhiman S, de Souza RF, Aravind L, Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene 475, 63–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trusov Y, Chakravorty D, Botella JR, Diversity of heterotrimeric G-protein γ subunits in plants. BMC. Res. Notes 5, 608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM, G protein activation without a GEF in the plant kingdom. PLOS Genet. 8, e1002756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urano D, Chen J-G, Botella JR, Jones AM, Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 3, 120186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urano D, Maruta N, Trusov Y, Stoian R, Wu Q, Liang Y, Jaiswal DK, Thung L, Jackson D, Botella JR, Jones AM, Saltational evolution of the heterotrimeric G protein signaling mechanisms in the plant kingdom. Sci. Signal 9, ra93 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Wu Q, Regan M, Furukawa H, Jackson D, Role of heterotrimeric Gα proteins in maize development and enhancement of agronomic traits. PLOS Genet. 14, e1007374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones JC, Duffy JW, Machius M, Temple BR, Dohlman HG, Jones AM, The crystal structure of a self-activating G protein α subunit reveals its distinct mechanism of signal initiation. Sci. Signal 4, ra8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP, A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301, 1728–1731 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen J-G, Siderovski DP, Jones AM, Willard FS, GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc. Natl. Acad. Sci. U.S.A 104, 17317–17322 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhury SR, Pandey S, The role of PLDα1 in providing specificity to signal-response coupling by heterotrimeric G-protein components in Arabidopsis. Plant J. 86, 50–61 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Gilman AG, G proteins: Transducers of receptor-generated signals. Annu. Rev. Biochem 56, 615–649 (1987). [DOI] [PubMed] [Google Scholar]
- 22.Bünemann M, Frank M, Lohse MJ, Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc. Natl. Acad. Sci. U.S.A 100, 16077–16082 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Digby GJ, Lober RM, Sethi PR, Lambert NA, Some G protein heterotrimers physically dissociate in living cells. Proc. Natl. Acad. Sci. U.S.A 103, 17789–17794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein S, Reuveni H, Levitzki A, Signal transduction by a nondissociable heterotrimeric yeast G protein. Proc. Natl. Acad. Sci. U.S.A 97, 3219–3223 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato C, Mizutani T, Tamaki H, Kumagai H, Kamiya T, Hirobe A, Fujisawa Y, Kato H, Iwasaki Y, Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J. 38, 320–331 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Adjobo-Hermans MJ, Goedhart J, Gadella TW Jr., Plant G protein heterotrimers require dual lipidation motifs of Gα and Gγ and do not dissociate upon activation. J. Cell Sci 119, 5087–5097 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Assmann SM, Fedoroff NV, Characterization of the Arabidopsis heterotrimeric G protein. J. Biol. Chem 283, 13913–13922 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Oki K, Fujisawa Y, Kato H, Iwasaki Y, Study of the constitutively active form of the α subunit of rice heterotrimeric G proteins. Plant Cell Physiol. 46, 381–386 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Urano D, Miura K, Wu Q, Iwasaki Y, Jackson D, Jones AM, Plant morphology of heterotrimeric G protein mutants. Plant Cell Physiol. 57, 437–445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto H, Matsui M, Deng XW, Overexpression of the heterotrimeric G-protein α-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13, 1639–1652 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Hu G, Cheng Y, Huang J, Heterotrimeric G protein α and β subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Dev. Biol 324, 68–75 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Natochin M, Barren B, Artemyev NO, Dominant negative mutants of transducin-α that block activated receptor. Biochemistry 45, 6488–6494 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slepak VZ, Quick MW, Aragay AM, Davidson N, Lester HA, Simon MI, Random mutagenesis of G protein α subunit Goα. Mutations altering nucleotide binding. J. Biol. Chem 268, 21889–21894 (1993). [PubMed] [Google Scholar]
- 34.Barren B, Artemyev NO, Mechanisms of dominant negative G-protein α subunits. J. Neurosci. Res 85, 3505–3514 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Kleuss C, Raw AS, Lee E, Sprang SR, Gilman AG, Mechanism of GTP hydrolysis by G-protein α subunits. Proc. Natl. Acad. Sci. U.S.A 91, 9828–9831 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masters SB, Miller RT, Chi M-H, Chang F-H, Beiderman B, Lopez NG, Bourne HR, Mutations in the GTP-binding site of GSα alter stimulation of adenylyl cyclase. J. Biol. Chem 264, 15467–15474 (1989). [PubMed] [Google Scholar]
- 37.Pandey S, Nelson DC, Assmann SM, Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136, 136–148 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Lambert NA, Dissociation of heterotrimeric G proteins in cells. Sci. Signal 1, re5 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Paulmurugan R, Umezawa Y, Gambhir SS, Noninvasive imaging of protein–protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. U.S.A 99, 15608–15613 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J-F, Bush J, Xiong Y, Li L, McCormack M, Large-scale protein-protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PLOS ONE 6, e27364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulmurugan R, Gambhir SS, Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein–protein interactions. Anal. Chem 79, 2346–2353 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, Wood MG, Eggers CT, Encell LP, Wood KV, NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol 11, 400–408 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR, An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 67, 840–851 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Wolfenstetter S, Chakravorty D, Kula R, Urano D, Trusov Y, Sheahan MB, McCurdy DW, Assmann SM, Jones AM, Botella JR, Evidence for an unusual transmembrane configuration of AGG3, a class C Gγ subunit of Arabidopsis. Plant J. 81, 388–398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra G, Zhang W, Deng F, Zhao J, Wang X, A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312, 264–266 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Pandey S, Chen J-G, Jones AM, Assmann SM, G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 141, 243–256 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Nilson SE, Assmann SM, The α-subunit of the Arabidopsis heterotrimeric G protein, GPA1, is a regulator of transpiration efficiency. Plant Physiol. 152, 2067–2077 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, Taylor JP, Jones AM, Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat. Cell Biol 14, 1079–1088 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urano D, Jones AM, Heterotrimeric G protein–xcoupled signaling in plants. Annu. Rev. Plant Biol 65, 365–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choudhury SR, Pandey S, Phosphorylation-dependent regulation of G-protein cycle during nodule formation in soybean. Plant Cell 27, 3260–3276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh M, Peterson YK, Lanier SM, Smrcka AV, Receptor- and nucleotide exchange–independent mechanisms for promoting G protein subunit dissociation. J. Biol. Chem 278, 34747–34750 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Uğur Ö, Öner SS, Molinari P, Ambrosio C, Sayar K, Onaran HO, Guanine nucleotide exchange-independent activation of Gs protein by β2-adrenoceptor. Mol. Pharmacol 68, 720–728 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Wieland T, Michel MC, Can a GDP-liganded G-protein be active? Mol. Pharmacol 68, 559–562 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Chen J-G, Gao Y, Jones AM, Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol. 141, 887–897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakravorty D, Assmann SM, G protein subunit phosphorylation as a regulatory mechanism in heterotrimeric G protein signaling in mammals, yeast, and plants. Biochem. J 475, 3331–3357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trusov Y, Botella JR, Plant G-proteins come of age: Breaking the bond with animal models. Front. Chem 4, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aranda-Sicilia MN, Trusov Y, Maruta N, Chakravorty D, Zhang Y, Botella JR, Heterotrimeric G proteins interact with defense-related receptor-like kinases in Arabidopsis. J. Plant Physiol 188, 44–48 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Bommert P, Je BI, Goldshmidt A, Jackson D, The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502, 555–558 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Ishida T, Tabata R, Yamada M, Aida M, Mitsumasu K, Fujiwara M, Yamaguchi K, Shigenobu S, Higuchi M, Tsuji H, Shimamoto K, Hasebe M, Fukuda H, Sawa S, Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep. 15, 1202–1209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, Zhang Y, Zhou J-M, Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5, e13568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng Y, Chen L, Li S, Zhang Y, Xu R, Liu Z, Liu W, Kong J, Huang X, Wang Y, Cheng B, Zheng L, Li Y, BRI1 and BAK1 interact with G proteins and regulate sugar-responsive growth and development in Arabidopsis. Nat. Commun 9, 1522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu Y, Chakravorty D, Assmann SM, The G protein β-subunit, AGB1, interacts with FERONIA in RALF1-regulated stomatal movement. Plant Physiol. 176, 2426–2440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tunc-Ozdemir M, Jones AM, Ligand-induced dynamics of heterotrimeric G protein-coupled receptor-like kinase complexes. PLOS ONE 12, e0171854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li B, Tunc-Ozdemir M, Urano D, Jia H, Werth EG, Mowrey DD, Hicks LM, Dokholyan NV, Torres MP, Jones AM, Tyrosine phosphorylation switching of a G protein. J. Biol. Chem 293, 4752–4766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tunc-Ozdemir M, Urano D, Jaiswal DK, Clouse SD, Jones AM, Direct modulation of heterotrimeric G protein-coupled signaling by a receptor kinase complex. J. Biol. Chem 291, 13918–13925 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langeberg LK, Scott JD, Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol 16, 232–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miao J, Yang Z, Zhang D, Wang Y, Xu M, Zhou L, Wang J, Wu S, Yao Y, Du X, Gu F, Gong Z, Gu M, Liang G, Zhou Y, Mutation of RGG2, which encodes a type B heterotrimeric G protein γ subunit, increases grain size and yield production in rice. Plant Biotechnol. J 17, 650–664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen J-G, Jones AM, Botella JR, Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell 19, 1235–1250 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Botella JR, Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 17, 563–568 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q, GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet 112, 1164–1171 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X, Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet 41, 494–497 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Sun H, Qian Q, Wu K, Luo J, Wang S, Zhang C, Ma Y, Liu Q, Huang X, Yuan Q, Han R, Zhao M, Dong G, Guo L, Zhu X, Gou Z, Wang W, Wu Y, Lin H, Fu X, Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet 46, 652–656 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Chakravorty D, Gookin TE, Milner MJ, Yu Y, Assmann SM, Extra-large G proteins expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. Plant Physiol. 169, 512–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gookin TE, Assmann SM, Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J. 80, 553–567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones AM, Ecker JR, Chen J-G, A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol. 131, 1623–1627 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clough SJ, Bent AF, Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 77.Lilly ST, Drummond RS, Pearson MN, MacDiarmid RM, Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana. Mol. Plant Microbe Interact 24, 294–304 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Zhu M, Jeon BW, Geng S, Yu Y, Balmant K, Chen S, Assmann SM, Preparation of epidermal peels and guard cell protoplasts for cellular, electrophysiological, and -omics assays of guard cell function. Methods Mol. Biol 1363, 89–121 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Conserved motifs in Gα subunits.
Fig. S2. In vitro BODIPY-GTP– and BODIPY-GDP–binding studies.
Fig. S3. Analysis of GTPγS binding to AtGPA1 and AtGPA1(S52C).
Fig. S4. Negative controls for BiFC assays.
Fig. S5. Assessment of interactions between AtRGS1 and AtGPA1 variants in planta.
Fig. S6. Assessment of the interaction between AtRGS1 and AGB1 in planta and of the effects of AGG subunits.